Abstract
The amygdala brain region has been implicated in the pathophysiology of schizophrenia through emotion processing. However, transcriptome messages in the amygdala of schizophrenia patients have not been well studied. We used RNA sequencing to investigate gene-expression profiling in the amygdala tissues, and identified 569 upregulated and 192 downregulated genes from 22 schizophrenia patients and 24 non-psychiatric controls. Gene functional enrichment analysis demonstrated that the downregulated genes were enriched in pathways such as 'synaptic transmission' and 'behavior', whereas the upregulated genes were significantly over-represented in gene ontology pathways such as 'immune response' and 'blood vessel development'. Co-expression-based gene network analysis identified seven modules including four modules significantly associated with 'synaptic transmission', 'blood vessel development' or 'immune responses'. Taken together, our study provides novel insights into the molecular mechanism of schizophrenia, suggesting that precision-tailored therapeutic approaches aimed at normalizing the expression/function of specific gene networks could be a promising option in schizophrenia.
Introduction
Schizophrenia is a brain disorder, characterized by abnormal social behavior and failure to differentiate real from unreal. Schizophrenia typically presents with symptoms of unclear or confused thinking, lack of reality recognition, auditory hallucinations, reduced social engagement and attenuated emotional expression.
The amygdala region has an important role in the processing of emotions and has been implicated in the pathophysiology of schizophrenia.1, 2, 3, 4 As perturbations of emotional responsiveness present a major clinical feature of schizophrenia, dysfunction within the amygdala region of the brain may contribute to the behavioral phenotypes of patients diagnosed with schizophrenia. For example, functional neuroimaging studies have demonstrated altered amygdala activity as an underlying factor of emotional dysfunction in schizophrenia.5, 6, 7 Reduced amygdala volumes have also been demonstrated among patients with early-course schizophrenia.8
To explore the molecular mechanism underlying schizophrenia, a number of transcriptome studies using microarray or RNA sequencing (RNA-seq) techniques have been conducted, using tissues from different brain regions, including the superior temporal gyrus,9 dorsolateral prefrontal cortex,10, 11, 12, 13 cerebellar cortex,14 hippocampus15 and the anterior cingulate cortex.16 Most of the transcriptome studies have pointed to the abnormal activation of the immune system in the brain of schizophrenia patients.10, 11, 13, 15, 17, 18 Gene markers such as SERPINA3, IFITM1, IFITM2 and IFITM3 have been consistently replicated in multiple studies.9, 10, 11, 13, 15, 18 However, gene-expression alterations in the amygdala of schizophrenia patients have not been thoroughly studied. Although a previous transcriptome study of amygdala using microarray chips suggested that genes from the cytomatrix-active zone may be upregulated,19 our understanding of the overall gene dysregulation in the amygdala of schizophrenia patients is still unclear.
In this study, we investigated gene-expression profiling in the amygdala tissues from 22 patients with schizophrenia and 24 non-psychiatric controls, using RNA-seq. We uncovered several dysregulated genes and pathways that are associated with the pathophysiology of schizophrenia. We further performed weighted gene correlation network analysis (WGCNA) to construct a gene co-expression network and identify functional modules of highly correlated genes.20 Our study provides convergent support for the established role of immune system in schizophrenia, as well as novel insight into the specific molecular mechanisms involving synaptic transmission in schizophrenia, with potential implications for the development of novel treatment for patients diagnosed with schizophrenia.
Materials and methods
Post-mortem brain samples
Amygdala of post-mortem brain was obtained from the Lieber Brain bank (http://www.libd.org) with consent. The Research Ethics Board of CHOP approved the study, and written informed consent was obtained from all subjects. Cases in this study included nine, seven, five and one patients with diagnosis of undifferentiated, disorganized, paranoid schizophrenia and schizoaffective disorder, respectively. Control subjects were individuals without psychiatric diagnoses. The post-mortem interval, age and gender of cases and controls were well matched (Supplementary Table S1). Non-parametric tests showed that the age and post-mortem interval are not associated with schizophrenia case/control phenotypes (P-value=0.17 and 0.09, respectively), and χ2-test showed that gender is not associated with case/control groups either (P-value=0.71). The sample description and RNA-seq data are available at https://www.ncbi.nlm.nih.gov/bioproject/379666.
Library preparation and mRNA-seq
RNA-seq libraries were constructed using Illumina TruSeq RNA sample Prep Kit (Illumina, San Diego, CA, USA) following the manufacturer's instruction. The poly-A-containing mRNA molecules were purified from 300 to 500 ng DNAse-treated total RNA using oligo (dT) beads. Following the purification steps, the mRNA was fragmented into small pieces using divalent cations under elevated temperature (94 °C) for 2 min. Under these conditions, fragment lengths range from 130 to 290 bp with a median length of 185 bp. Reverse transcriptase and random primers were used to generate the first-strand complementary DNA from the cleaved RNA fragments. The second strand DNA was synthesized using DNA Polymerase I and RNaseH. These complementary DNA fragments then went through an end-repair process using T4 DNA polymerase, T4 PNK and Klenow DNA polymerase, and the addition of a single ‘A’ base using Klenow exo (3' to 5' exo minus), followed by ligation of the illumine PE adapters using T4 DNA Ligase. An index was inserted into illumina adapters so that multiple samples can be sequenced in a single lane. These products were then purified and enriched with polymerase chain reaction to create the final complementary DNA library for high-throughput DNA sequencing using Highseq2000 (Illumina). The concentration of RNA-seq libraries was measured by Qubit (Invitrogen, Waltham, MA, USA) and quantified with quantitative polymerase chain reaction. The quality of RNA-seq library was measured by LabChipGX (Caliper, MA, USA) using HT DNA 1 K/12 K/Hi-sensitivity LabChip. The libraries are multiplexed and loaded on a flowcell for cluster generation on cBot (Illumina). During sequencing, the Illumina Real Time Analysis module was used to perform image analysis and base calling, and the BCL Converter (CASAVA v1.8.2, Illumina) were followed to generate FASTQ files, which contain the sequence reads. Sequencing depth was over 80 million (2 × 100-bp 40 million paired-end) mappable sequencing reads.
Sequencing data analysis
Genomic Short-read Nucleotide Alignment Program (version 2015-12-31.v9) was used to map the reads to the reference human genome (hg19). Common single-nucleotide polymorphisms (SNPs) recorded in dbSNP137 were taken into account to improve the alignment accuracy.21 The output SAM files were converted to BAM files, sorted by index. Duplication reads were removed using the SAMtools pipeline,22 and only unique, paired reads were used in the analysis. NCBI reference sequences (RefSeq) were used for gene annotation.23 Both fragments per kilobase of transcript per million mapped reads (FPKM)-based and count-based algorithms were used to identify the differentially expressed genes between cases and controls. The FPKM-based algorithm was implemented in the Cuffdiff package of Cufflinks2.2.1 (ref. 24) and two count-based algorithms were implemented in R packages edgeR25 and DEseq2.26 Default parameters of Cuffdiff, edgeR and DEseq2 were used for the gene differential expression analysis.
Weighted gene co-expression network analysis
Co-expressed gene modules were detected by WGCNA. First, the normalized FPKM values of transcripts were calculated by the cuffnorm package of cufflinks.24 A matrix of correlations between all pairs of differentially expressed genes was generated by the FPKM values, and further converted into an adjacency matrix with a power function, so that the connection strength between two genes xi and xj was defined as aij=|cor(xi, xj)|β. The parameter was determined by the criterion that the resulting adjacency matrix approximately fits a scale-free topological feature according to a model-fitting index proposed previously.27 The model-fitting index of a perfect scale-free network is 1. The value β of was 15, which is the minimum value required to make the model-fitting index above 0.8.27
The adjacency matrix was further transformed into a topological overlap matrix, which captures not only the direct interaction between two genes but also their indirect interactions through all the other genes in the network. A similarity measure was defined: TOMij=(Σu aiuauj+aij)/(min(ki,kj)+1−aij), where ki=Σu aiu was the node connectivity.28, 29 1−TOMij was used as a distance matrix in the hierarchical clustering of the transcript units for module detection.28
The module eigengene is the first principal component of the matrix of expression values of a given module, which was adopted to characterize the gene-expression profile of the module.
Gene functional enrichment analysis
Gene functional enrichment analysis was applied on the differentially expressed genes and gene modules identified by WGCNA. DAVID (http://david.abcc.ncifcrf.gov/) was used to test enrichment in gene sets with gene ontology (GO) terms compared with the background list of all genes.30 Benjamini–Hochberg procedure was performed for multiple testing.
Results
Identification of differentially expressed genes
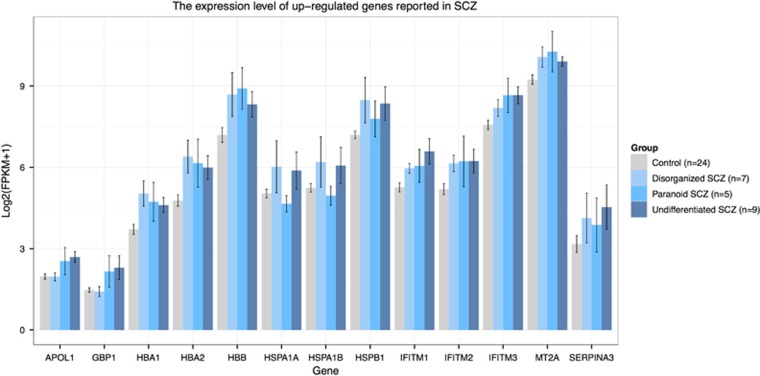
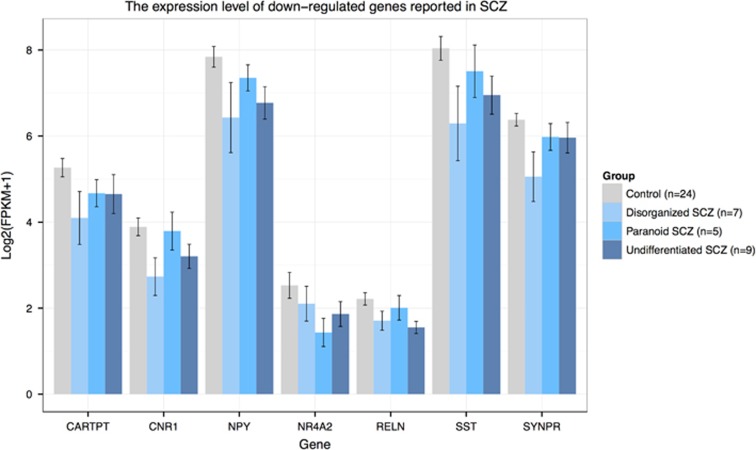
To search for differentially expressed genes, we employed multiple state-of-the-art statistical RNA-seq analysis workflows based on FPKM values and read counts,31, 32 built from the most mature and widely used computational tools, including Cuffdiff,24 edgeR25 and DEseq2.26 Cuffidiff, edgeR and DEseq2 identified 88, 99 and 104 differentially expressed genes, respectively, which have a P-value<0.05 after multiple testing corrections (Supplementary Figure S1 and Supplementary Table S2). Among those, 25 genes were identified by all three methods, including HBA1, HBA2, HBB and IFITM1, all of which have been reported to be differentially expressed in previous transcriptome studies in schizophrenia (Figures 1 and 2 and Supplementary Table S3).15, 17, 33 Several other and more highly differentially expressed genes showed significant P-values by one or two methods—all supported by previous studies. These include GBP1, HSPA1A, HSPA1B, HSPB1, IFITM2, IFITM3, NPY, SST and SERPINA3 (Figures 1 and 2 and Supplementary Table S3).9, 11, 12, 13, 15, 19 In addition to the differentially expressed genes that met rigid statistical thresholds, multiple genes demonstrated nominal significance, several of which are involved in biological pathways of interest to the pathophysiology of schizophrenia. Further analysis of genes with a P-value<0.05, notwithstanding multiple testing corrections, included 761 genes shown to be of nominal significance by all three methods, and involved in downstream pathway and gene network functions of relevance to schizophrenia (Supplementary Figures S1 and 2 and Supplementary Table S4).
Figure 1.
Bar plot showing expression levels of upregulated genes reported in previous schizophrenia transcriptome studies.
Figure 2.
Bar plot showing expression levels of downregulated genes reported in previous schizophrenia transcriptome studies.
Upregulation of gene pathways related to blood vessel development and immune response
Of the 761 (75%) genes analyzed, 569 were overexpressed in the amygdala of individuals with schizophrenia. Functional enrichment analysis of the 569 upregulated genes revealed that genes involved in 'blood vessel development' (GO: 0001568, Pcorrected=1.50 × 10−7), 'immune response' (GO: 0006955, Pcorrected=1.04 × 10−4), 'major histocompatibility complex (MHC) class I receptor activity' (GO: 0032393, Pcorrected=4.99 × 10−3) and 'inflammatory response' (GO: 0006954, Pcorrected=6.83 × 10−3) were significantly over-represented in cases compared with controls (Table 1). As the immune system activation has been repeatedly observed in many transcriptome studies of schizophrenia, we investigated whether the previously reported genes associated with immune systems were also dysregulated in amygdala. We found the expression of SERPINA3, IFITM1, IFITM2 and IFITM3, HSPA1A, HSPA1B, HSPB1, GBP1, MT2A and APOL1 to be upregulated in the amygdala of schizophrenia patients. Besides SERPINA3, two members of the serine proteinase inhibitor superfamily SERPINE1 and SERPING1 were also overexpressed. The paralogous genes of GBP1, MT2A and APOL1, including GBP2, MT1G, MT1E, MT1F, MT1L and APOLD1, were also upregulated in the schizophrenia cases (Supplementary Table S4).
Table 1. Functional enrichment of differentially expressed genes.
| Expression | Term | Fold enrichment | P-value | BenjaminiQ-value |
|---|---|---|---|---|
| Upregulation | GO:0001568~blood vessel development | 4.06 | 6.06E−11 | 1.50E−07 |
| GO:0001944~vasculature development | 3.96 | 1.13E−10 | 1.40E−07 | |
| GO:0001525~angiogenesis | 4.62 | 1.03E−08 | 8.46E−06 | |
| GO:0048514~blood vessel morphogenesis | 3.83 | 1.75E−08 | 1.08E−05 | |
| GO:0009615~response to virus | 5.14 | 6.40E−08 | 3.17E−05 | |
| GO:0005576~extracellular region | 1.66 | 1.08E−07 | 3.56E−05 | |
| GO:0030198~extracellular matrix organization | 5.08 | 1.90E−07 | 7.84E−05 | |
| GO:0042127~regulation of cell proliferation | 2.13 | 2.20E−07 | 7.78E−05 | |
| GO:0044421~extracellular region part | 2.04 | 2.29E−07 | 3.77E−05 | |
| GO:0006955~immune response | 2.21 | 3.36E−07 | 1.04E−04 | |
| GO:0009611~response to wounding | 2.29 | 2.95E−06 | 8.10E−04 | |
| GO:0022610~biological adhesion | 2.09 | 2.98E−06 | 7.37E−04 | |
| GO:0051270~regulation of cell motion | 3.38 | 3.95E−06 | 8.88E−04 | |
| GO:0007155~cell adhesion | 2.04 | 6.72E−06 | 1.28E−03 | |
| GO:0032393~MHC class I receptor activity | 13.23 | 7.98E−06 | 4.99E−03 | |
| GO:0006952~defense response | 2.02 | 3.79E−05 | 5.50E−03 | |
| GO:0006954~inflammatory response | 2.49 | 4.98E−05 | 6.83E−03 | |
| GO:0019882~antigen processing and presentation | 4.50 | 6.58E−05 | 8.10E−03 | |
| Downregulation | GO:0007268~synaptic transmission | 5.94 | 2.33E−08 | 2.59E−05 |
| GO:0007610~behavior | 4.44 | 9.56E−08 | 5.30E−05 | |
| GO:0019226~transmission of nerve impulse | 5.05 | 2.15E−07 | 7.94E−05 | |
| GO:0006928~cell motion | 3.72 | 1.15E−05 | 2.54E−03 | |
| GO:0030534~adult behavior | 9.68 | 1.75E−05 | 3.23E−03 | |
| GO:0007218~neuropeptide signaling pathway | 8.95 | 2.92E−05 | 4.62E−03 | |
| GO:0005509~calcium ion binding | 2.62 | 4.63E−05 | 7.89E−03 | |
| GO:0051969~regulation of transmission of nerve impulse | 6.37 | 8.08E−05 | 9.91E−03 | |
| GO:0007409~axonogenesis | 5.39 | 9.55E−05 | 1.05E−02 | |
| GO:0031644~regulation of neurological system process | 6.12 | 1.07E−04 | 1.07E−02 | |
| GO:0050877~neurological system process | 2.24 | 1.68E−04 | 1.54E−02 | |
| GO:0048667~cell morphogenesis involved in neuron differentiation | 4.98 | 1.75E−04 | 1.48E−02 | |
| GO:0048812~neuron projection morphogenesis | 4.89 | 2.01E−04 | 1.58E−02 | |
| GO:0046873~metal ion transmembrane transporter activity | 3.83 | 2.81E−04 | 3.15E−02 | |
| GO:0005261~cation channel activity | 4.19 | 2.82E−04 | 2.39E−02 | |
| GO:0005216~ion channel activity | 3.53 | 2.96E−04 | 2.01E−02 | |
| GO:0050804~regulation of synaptic transmission | 6.12 | 3.21E−04 | 2.34E−02 | |
| GO:0007186~G-protein-coupled receptor protein signaling pathway | 2.22 | 3.61E−04 | 2.47E−02 |
Abbreviations: GO, gene ontology; MHC, major histocompatibility complex.
Suppression of gene pathways related to synaptic transmission and behavior
Of the 761 (25%) analyzed genes, 192 were downregulated in the amygdala of schizophrenia patients in comparison with controls. Functional enrichment analysis demonstrated that the 192 genes were significantly enriched in biological pathways including 'synaptic transmission' (GO: 0007268, Pcorrected=2.59 × 10−5), 'behavior' (GO: 0007610, Pcorrected=5.30 × 10−5), 'neuropeptide signaling pathway' (GO: 0007218, Pcorrected=4.62 × 10−3) and 'calcium ion-binding' (GO: 0005509, Pcorrected=7.89 × 10−3;Table 1). Notably, several GABAergic genes, which exhibited reduced expression in the prefrontal cortex of subjects with schizophrenia, were uncovered in our study, including NPY, SST and RELN.34, 35, 36, 37 Moreover, expression levels of genes involved in dopaminergic transmission, such as CNR1, HTR2A, HTR2C, NR4A2, NTSR1, NXPH3 and SLC17A8, were also reduced (Supplementary Table S4).38, 39, 40, 41, 42, 43
Comparison with previous RNA-seq studies in schizophrenia
We compared our results with two RNA-seq studies conducted on the prefrontal cortex and hippocampus of individuals with schizophrenia, respectively.10, 15 Fillman et al. identified 399 upregulated and 201 downregulated genes. Of those, 34 upregulated genes significantly overlapped with the upregulated genes in our study (P=2.82 × 10−7). Hwang et al. detected 123 upregulated and 21 downregulated genes. Among them, 50 upregulated genes significantly overlapped with the upregulated genes in our study (P=3.40 × 10−42). No downregulated genes overlapped between our study and any of the two previous studies. Gene functional enrichment analysis indicated that the upregulated genes identified by Fillman et al. were significantly enriched in 'inflammatory response' (GO: 0006954, Pcorrected=0.012) and 'blood vessel development' (GO: 0001568, Pcorrected=0.039), and nominally enriched in 'immune response' (GO: 0006955, P=6.13 × 10−4, Pcorrected=0.071; Supplementary Table S5). The upregulated genes identified by Hwang et al. were also nominally enriched in 'immune response' (GO: 0006955, P=1.06 × 10−4, Pcorrected=0.095; Supplementary Table S6). The enriched pathways of downregulated genes in our study were not significant in any of the two previous studies.
Detection of functional modules in co-expression-based gene networks
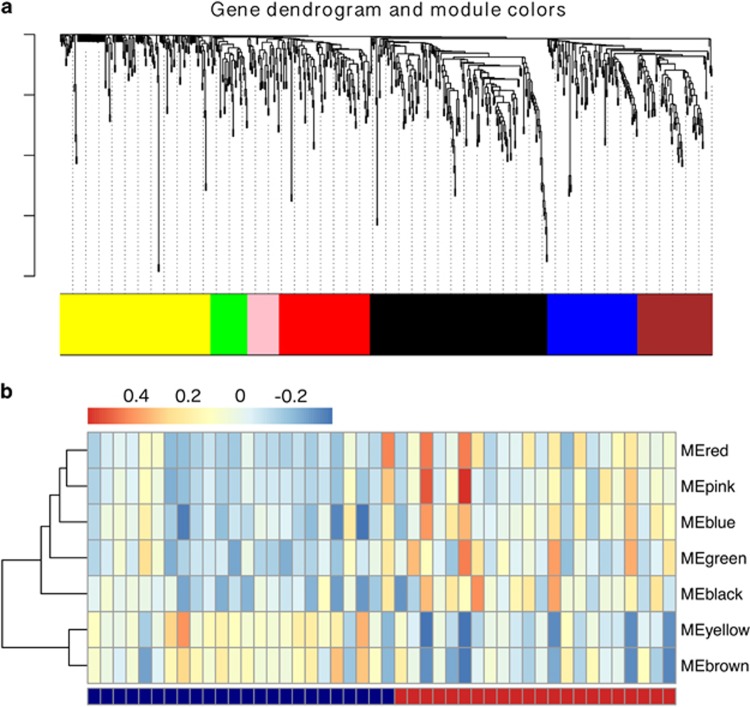
We further analyzed our data with WGCNA, which has been widely applied to study co-expression-based gene networks, both construction and module detection. Seven co-expression modules were identified including four modules that were significantly enriched in genes with specific pathway roles (see Figure 3 and Table 2: 'Module yellow', 176 genes, 'synaptic transmission' and 'calcium ion binding' 'Module green', 43 genes, 'blood vessel development' 'Module pink', 37 genes, 'immune response' 'Module red', 106 genes, 'immune response' and 'blood vessel development'). For each module, the eigengene was calculated to represent the overall expression profile of the genes in a certain module. The expression level of 'Module yellow' was low in schizophrenia cases, whereas expression levels of 'Module pink', 'Module red' and 'Module green' were high in cases, suggesting a high consistency between the pathway and co-expression-based gene network analysis.
Figure 3.
(a) The co-expression modules identified by weighted gene correlation network analysis (WGCNA). (b) The Eigengene heatmap of co-expression modules. Bottom legend: cases and controls are colored in red and blue, respectively. Top legend: Eigengene values of co-expression modules.
Table 2. Functional enrichment of genes in co-expression modules.
| Module | Term | Fold enrichment | P-value | BenjaminiQ-value |
|---|---|---|---|---|
| Yellow | GO:0005576~extracellular region | 2.56 | 3.59E−11 | 6.79E−09 |
| GO:0044421~extracellular region part | 2.99 | 2.08E−07 | 1.97E−05 | |
| GO:0007268~synaptic transmission | 5.01 | 1.63E−06 | 2.21E−03 | |
| GO:0010033~response to organic substance | 3.17 | 2.51E−06 | 1.70E−03 | |
| GO:0005509~calcium ion binding | 2.85 | 7.29E−06 | 2.29E−03 | |
| GO:0019226~transmission of nerve impulse | 4.26 | 1.05E−05 | 4.72E−03 | |
| GO:0005615~extracellular space | 2.89 | 4.95E−05 | 3.12E−03 | |
| GO:0007155~cell adhesion | 2.84 | 6.54E−05 | 2.19E−02 | |
| GO:0022610~biological adhesion | 2.84 | 6.67E−05 | 1.79E−02 | |
| GO:0016477~cell migration | 4.32 | 9.98E−05 | 2.23E−02 | |
| Green | GO:0001568~blood vessel development | 15.78 | 6.40E−09 | 3.65E−06 |
| GO:0001944~vasculature development | 15.40 | 7.90E−09 | 2.25E−06 | |
| GO:0048514~blood vessel morphogenesis | 14.65 | 7.65E−07 | 1.45E−04 | |
| GO:0001525~angiogenesis | 15.67 | 3.16E−05 | 4.49E−03 | |
| GO:0005886~plasma membrane | 2.01 | 3.00E−04 | 3.13E−02 | |
| Pink | GO:0006955~immune response | 6.76 | 6.38E−06 | 1.87E−03 |
| Red | GO:0006955~immune response | 3.84 | 2.92E−06 | 4.13E−03 |
| GO:0032403~protein complex binding | 8.08 | 3.46E−06 | 8.55E−04 | |
| GO:0002684~positive regulation of immune system process | 6.80 | 4.34E−06 | 3.07E−03 | |
| GO:0031349~positive regulation of defense response | 14.10 | 9.30E−06 | 4.38E−03 | |
| GO:0001775~cell activation | 5.64 | 2.23E−05 | 7.88E−03 | |
| GO:0006952~defense response | 3.59 | 5.81E−05 | 1.64E−02 | |
| GO:0030198~extracellular matrix organization | 9.90 | 7.01E−05 | 1.64E−02 | |
| GO:0005886~plasma membrane | 1.67 | 1.51E−04 | 2.96E−02 | |
| GO:0048584~positive regulation of response to stimulus | 5.61 | 1.86E−04 | 3.70E−02 | |
| GO:0045321~leukocyte activation | 5.47 | 2.21E−04 | 3.84E−02 | |
| GO:0007155~cell adhesion | 3.15 | 2.28E−04 | 3.53E−02 | |
| GO:0022610~biological adhesion | 3.15 | 2.31E−04 | 3.23E−02 | |
| GO:0001568~blood vessel development | 5.40 | 2.40E−04 | 3.05E−02 | |
| GO:0005911~cell−cell junction | 6.33 | 2.48E−04 | 2.43E−02 | |
| GO:0045089~positive regulation of innate immune response | 15.98 | 2.52E−04 | 2.93E−02 | |
| GO:0033256~I-kappaB/NF-kappaB complex | 112.78 | 2.54E−04 | 1.67E−02 | |
| GO:0001944~vasculature development | 5.27 | 2.83E−04 | 3.04E−02 | |
| GO:0046649~lymphocyte activation | 5.91 | 3.82E−04 | 3.80E−02 | |
| GO:0045088~regulation of innate immune response | 13.62 | 4.69E−04 | 4.33E−02 | |
| GO:0001525~angiogenesis | 6.95 | 4.81E−04 | 4.17E−02 | |
| GO:0046637~regulation of alpha−beta T-cell differentiation | 23.53 | 6.08E−04 | 4.95E−02 | |
| GO:0030098~lymphocyte differentiation | 8.57 | 6.43E−04 | 4.94E−02 |
Abbreviations: GO, gene ontology; NF-kappaB, nuclear factor-κappaB.
A few highly connected nodes, which can hold the whole network together, represent the hubs of the network. Similarly, genes with a high number of interactions are considered to have an important role in organizing the biological network. Unlike protein–protein interaction networks, WGCNA defines a whole network connectivity measure (kTotal) for each gene according to its Pearson correlation coefficient with all of the other genes, and an intramodular connectivity measure (kWithin) when only considering the connection strength of each individual gene with all the other genes within the same module.
We further investigated whether the intramodular hub genes of a co-expression module were associated with the enriched pathways of the respective modules. Interestingly, two intramodular hubs of 'Module yellow' with the highest values of connectivity were GABA-related genes NPY and SST, and the top three connected intramodule hubs in 'Module green' were ESAM, CDH5 and MMRN2, all of which are associated with angiogenesis or vascular diseases. The top intramodule hubs in 'Module pink' and 'Module red' were also strongly linked to the immune system (Supplementary Table S7).
Discussion
Although multiple gene-expression studies have been performed on a variety of post-mortem brain tissues using both microarray and RNA-seq technologies, little attention has been devoted to examine transcriptome changes in the amygdala region of schizophrenia patients. To the best of our knowledge, no previous transcriptome study has specifically focused on subtypes of schizophrenia. Given the high degree of clinical heterogeneity of schizophrenia and the relatively small sample sizes of previous transcriptome studies (<50 samples), transcriptome changes in schizophrenia could be better understood by focusing on subgroups of patients with similar clinical manifestations.
In this study, we replicated many differentially expressed genes previously reported in schizophrenia, such as IFITM1, IFITM2, IFITM3, NPY, SST and SERPINA3. Elevated gene expression of the immune-related genes, including IFITM1, IFITM2, IFITM3 and SERPINA3, has been repeatedly observed in the prefrontal cortex and hippocampus of patients with schizophrenia.9, 10, 11, 13, 15 Previous transcriptome studies have also revealed that genes involved in activation of the immune system are differentially expressed in schizophrenia, suggesting that they may contribute to the disease pathogenesis.10, 11, 13, 15, 17, 18 Here, we confirmed the previously observed overexpression of 'immune response' and 'inflammatory response' GO pathways in the amygdala region, using gene enrichment analysis. Our enrichment analysis also supported that upregulation of 'MHC class I receptor activity' results in keeping with the highly significant genome-wide association study association of the MHC region with schizophrenia. Although enrichment analysis of schizophrenia subtypes only showed upregulation of the 'immune response' pathway in undifferentiated and paranoid schizophrenia, we found that most differentially expressed genes involved in 'immune response', 'inflammatory response' and 'MHC class I receptor activity' pathways, including IFITM1, IFITM2, IFITM3 and SERPINA3, are overexpressed in all three subtypes (Figure 1 and Supplementary Figures S3 and 4), suggesting that the activation of the immune system may be a common denominator in diverse subtypes of schizophrenia.
We additionally identified upregulated gene expression in the 'blood vessel development' pathway in schizophrenia. Further analyses of schizophrenia subtypes indicated that most differentially expressed genes involved in 'blood vessel development' are overexpressed in all three subtypes (Supplementary Figure S5). Enrichment analysis of schizophrenia subtypes also suggested elevated gene expression of the 'blood vessel development' pathway in all three subtypes. Moreover, we replicated the result by re-analyzing the differentially expressed genes from an independent RNA-seq study of the prefrontal cortex.10 The observation of altered gene expressions in immune system and angiogenesis lends supports to the vascular-inflammatory theory of schizophrenia. This theory hypothesized that inflammation involving the microvascular system leads to dysregulation of cerebral blood flow and damage to the blood–brain barrier, predisposing to the development of schizophrenia.44
Our study also confirmed that there is suppression of GABAgeric markers (that is, downregulation in NPY, SST and RELN expression) and showed that gene networks involving 'synaptic transmission', 'behavior', 'neuropeptide signaling pathway' and 'calcium ion-binding' pathways were also significantly downregulated in schizophrenia. Further analyses of schizophrenia subtypes indicated that most differentially expressed genes involved in 'synaptic transmission' and 'behavior' are downregulated in all three subtypes (Supplementary Figure S6). In contrast to the altered expression of immune-related gene pathways, the dysregulation of synaptic transmission or behavior pathway has not been previously reported in transcriptome-wide studies of schizophrenia.9, 10, 11, 13, 14, 15, 17 We also studied the differentially expressed gene sets of two published RNA-seq studies on the prefrontal cortex and hippocampus, respectively, in individuals with schizophrenia obeserving upregulation of immune pathways. Although speculative, the deficit of synaptic and behavioral genes in the amygdala may be more likely to associate with the behavioral phenotypes in schizophrenia, whereas the activation of the immune system may be associated with the more common phenotypes in schizophrenia.
We further investigated the differentially expressed genes among three subtypes of schizophrenia (Supplementary Note). Interestingly, our results showed that several genes previously implicated in schizophrenia are oppositely regulated in subtypes of schizophrenia. It is difficult to identify unified gene-expression profiles when analyzing a group of patients with diverse symptoms in schizophrenia. For example, genes involved in the neuregulin 1–ErbB4 signaling pathway, including DOCK7, EGFR, PTPRZ1 and the key regulator ERBB4,45, 46, 47 were downregulated in undifferentiated and disorganized schizophrenia, while they were upregulated in paranoid schizophrenia (Supplementary Figure S7). Several genes that encode subunits of the NMDA receptor (GRIN2D), GABA receptor (GABRA2 and GABRB1), GABA transporter (SLC6A1 and SLC6A11) and glutamate transporter (SLC1A3) also displayed the same expression pattern. NTRK2, a susceptibility gene of paranoid schizophrenia,48 was not only overexpressed in paranoid schizophrenia, but also suppressed in undifferentiated and disorganized schizophrenia. In addition, the expression of GRM3 was significantly reduced in undifferentiated schizophrenia, while it was increased in disorganized and paranoid schizophrenia (Supplementary Figure S7). Elevated expression of CNTN2 and EFHD1 was specifically found in disorganized schizophrenia (Supplementary Figure S7). In summary, the above gene-expression profiles appear to be associated with particular clinical phenotypes or subsets of schizophrenia patients, and could potentially serve as biomarkers to identify therapeutic targets for particular subtypes of schizophrenia.
We subsequently applied the unbiased WGCNA method on the differentially expressed genes to identify the key modules of highly co-expressed genes. Consistent with the results proposed above, four of seven detected modules were associated with immune response, vascular development or synaptic transmission, indicating that dysregulated genes that are involved in the same pathway are highly co-expressed. Further investigation of the intramodular hubs also confirmed that the genes that function as key hubs in a module have an important role in the enriched pathways of that module, such as NPY, SST in synaptic transmission, and ESAM, CDH5 and MMRN2 in blood vessel development, suggesting that the intramodular hub genes could function as potential biomarkers or be lead target sites for future therapeutic interventions. For example, modulators of NPY or SST signaling and drugs that increase the activity of the GABAergic pathway have been reviewed as attractive therapeutic opportunities for schizophrenia.49, 50, 51
In conclusion, we performed the first transcriptome study of amygdala in schizophrenia patients, using RNA-seq, and observed significant associations between specific transcriptome changes in amygdala and schizophrenia subtypes. Those include, but are not limited to, upregulation of genes that regulate activation of immune responses and angiogenesis pathways, and suppression of genes involved in synaptic transmission. Taken together, these gene networks may co-contribute in a significant way to the pathophysiology of schizophrenia. We conclude that our study provides new insights into the genomic underpinnings of schizophrenia and may allow for novel precision therapeutic development strategies targeting schizophrenia to be implemented in future studies.
Acknowledgments
We thank Joel Kleinman, Daniel R Weinberger and Thomas M Hyde for their assistance in recruiting and sequencing subjects for participation in the study. This study is funded by an Institutional Development Award to the Center for Applied Genomics from The Children's Hospital of Philadelphia; Adele and Daniel Kubert donation; MH096891-03S1 (NIMH); RC2 MH089924-02 (NIMH); R01 MH097284-03 (NIMH); U01-HG008684 (NIH).
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
The authors declare no conflict of interest.
Supplementary Material
References
- Pankow A, Friedel E, Sterzer P, Seiferth N, Walter H, Heinz A et al. Altered amygdala activation in schizophrenia patients during emotion processing. Schizophr Res 2013; 150: 101–106. [DOI] [PubMed] [Google Scholar]
- Phelps EA, LeDoux JE. Contributions of the amygdala to emotion processing: from animal models to human behavior. Neuron 2005; 48: 175–187. [DOI] [PubMed] [Google Scholar]
- Rahm C, Liberg B, Reckless G, Ousdal O, Melle I, Andreassen OA et al. Negative symptoms in schizophrenia show association with amygdala volumes and neural activation during affective processing. Acta Neuropsychiatr 2015; 27: 213–220. [DOI] [PubMed] [Google Scholar]
- Salgado-Pineda P, Fakra E, Delaveau P, Hariri AR, Blin O. Differential patterns of initial and sustained responses in amygdala and cortical regions to emotional stimuli in schizophrenia patients and healthy participants. J Psychiatr Neurosci 2010; 35: 41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkquist OA, Olsen EK, Nelson BD, Herbener ES. Altered amygdala-prefrontal connectivity during emotion perception in schizophrenia. Schizophr Res 2016; 175: 35–41. [DOI] [PubMed] [Google Scholar]
- Williams LM, Das P, Harris AW, Liddell BB, Brammer MJ, Olivieri G et al. Dysregulation of arousal and amygdala-prefrontal systems in paranoid schizophrenia. Am J Psychiatry 2004; 161: 480–489. [DOI] [PubMed] [Google Scholar]
- Anticevic A, Repovs G, Barch DM. Emotion effects on attention, amygdala activation, and functional connectivity in schizophrenia. Schizophr Bull 2012; 38: 967–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich AM, Cho YT, Tang Y, Savic A, Krystal JH, Wang F et al. Amygdala volume is reduced in early course schizophrenia. Psychiatry Res 2016; 250: 50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JQ, Wang X, Beveridge NJ, Tooney PA, Scott RJ, Carr VJ et al. Transcriptome sequencing revealed significant alteration of cortical promoter usage and splicing in schizophrenia. PLoS ONE 2012; 7: e36351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillman SG, Cloonan N, Catts VS, Miller LC, Wong J, McCrossin T et al. Increased inflammatory markers identified in the dorsolateral prefrontal cortex of individuals with schizophrenia. Mol Psychiatry 2013; 18: 206–214. [DOI] [PubMed] [Google Scholar]
- Arion D, Unger T, Lewis DA, Levitt P, Mirnics K. Molecular evidence for increased expression of genes related to immune and chaperone function in the prefrontal cortex in schizophrenia. Biol Psychiatry 2007; 62: 711–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KH, Elashoff M, Higgs BW, Song J, Kim S, Sabunciyan S et al. Putative psychosis genes in the prefrontal cortex: combined analysis of gene expression microarrays. BMC Psychiatry 2008; 8: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saetre P, Emilsson L, Axelsson E, Kreuger J, Lindholm E, Jazin E. Inflammation-related genes up-regulated in schizophrenia brains. BMC Psychiatry 2007; 7: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudge J, Miller NA, Khrebtukova I, Lindquist IE, May GD, Huntley JJ et al. Genomic convergence analysis of schizophrenia: mRNA sequencing reveals altered synaptic vesicular transport in post-mortem cerebellum. PLoS ONE 2008; 3: e3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang Y, Kim J, Shin JY, Kim JI, Seo JS, Webster MJ et al. Gene expression profiling by mRNA sequencing reveals increased expression of immune/inflammation-related genes in the hippocampus of individuals with schizophrenia. Transl Psychiatry 2013; 3: e321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Chen X, Jin L, Xiong M. Canonical correlation analysis for RNA-seq co-expression networks. Nucleic Acids Res 2013; 41: e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Sun J, Chen J, Wang L, Li A, Helm M et al. RNA-Seq analysis implicates dysregulation of the immune system in schizophrenia. BMC Genomics 2012; 13(Suppl 8): S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Mirnics K. Immune system disturbances in schizophrenia. Biol Psychiatry 2014; 75: 316–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidenhofer J, Bowden NA, Scott RJ, Tooney PA. Altered gene expression in the amygdala in schizophrenia: up-regulation of genes located in the cytomatrix active zone. Mol Cell Neurosci 2006; 31: 243–250. [DOI] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008; 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 2010; 26: 873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009; 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 2007; 35(Database issue): D61–D65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 2013; 31: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010; 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 2005; 4, Article17. [DOI] [PubMed] [Google Scholar]
- Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabasi AL. Hierarchical organization of modularity in metabolic networks. Science 2002; 297: 1551–1555. [DOI] [PubMed] [Google Scholar]
- Chang X, Xu T, Li Y, Wang K. Dynamic modular architecture of protein-protein interaction networks beyond the dichotomy of 'date' and 'party' hubs. Sci Rep 2013; 3: 1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012; 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc 2013; 8: 1765–1786. [DOI] [PubMed] [Google Scholar]
- Glatt SJ, Everall IP, Kremen WS, Corbeil J, Sasik R, Khanlou N et al. Comparative gene expression analysis of blood and brain provides concurrent validation of SELENBP1 up-regulation in schizophrenia. Proc Natl Acad Sci USA 2005; 102: 15533–15538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti A, Auta J, Davis JM, Di-Giorgi-Gerevini V, Dwivedi Y, Grayson DR et al. Decrease in reelin and glutamic acid decarboxylase67 (GAD67) expression in schizophrenia and bipolar disorder: a postmortem brain study. Arch Gen Psychiatry 2000; 57: 1061–1069. [DOI] [PubMed] [Google Scholar]
- Fatemi SH, Stary JM, Earle JA, Araghi-Niknam M, Eagan E. GABAergic dysfunction in schizophrenia and mood disorders as reflected by decreased levels of glutamic acid decarboxylase 65 and 67 kDa and Reelin proteins in cerebellum. Schizophr Res 2005; 72: 109–122. [DOI] [PubMed] [Google Scholar]
- Hashimoto T, Arion D, Unger T, Maldonado-Aviles JG, Morris HM, Volk DW et al. Alterations in GABA-related transcriptome in the dorsolateral prefrontal cortex of subjects with schizophrenia. Mol Psychiatry 2008; 13: 147–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellios N, Huang HS, Baker SP, Galdzicka M, Ginns E, Akbarian S. Molecular determinants of dysregulated GABAergic gene expression in the prefrontal cortex of subjects with schizophrenia. Biol Psychiatry 2009; 65: 1006–1014. [DOI] [PubMed] [Google Scholar]
- Franklin JM, Carrasco GA. Cannabinoid-induced enhanced interaction and protein levels of serotonin 5-HT(2A) and dopamine D(2) receptors in rat prefrontal cortex. J Psychopharmacol 2012; 26: 1333–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolozzi A, Diaz-Mataix L, Scorza MC, Celada P, Artigas F. The activation of 5-HT receptors in prefrontal cortex enhances dopaminergic activity. J Neurochem 2005; 95: 1597–1607. [DOI] [PubMed] [Google Scholar]
- Xing G, Zhang L, Russell S, Post R. Reduction of dopamine-related transcription factors Nurr1 and NGFI-B in the prefrontal cortex in schizophrenia and bipolar disorders. Schizophr Res 2006; 84: 36–56. [DOI] [PubMed] [Google Scholar]
- Sakae DY, Marti F, Lecca S, Vorspan F, Martin-Garcia E, Morel LJ et al. The absence of VGLUT3 predisposes to cocaine abuse by increasing dopamine and glutamate signaling in the nucleus accumbens. Mol Psychiatry 2015; 20: 1448–1459. [DOI] [PubMed] [Google Scholar]
- Nishimura K, Murayama S, Takahashi J. Identification of neurexophilin 3 as a novel supportive factor for survival of induced pluripotent stem cell-derived dopaminergic progenitors. Stem Cell Transl Med 2015; 4: 932–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Chan NG, Bannon MJ, Orozco-Barrios CE, Escobedo L, Zamudio S, De la Cruz F et al. Neurotensin-polyplex-mediated brain-derived neurotrophic factor gene delivery into nigral dopamine neurons prevents nigrostriatal degeneration in a rat model of early Parkinson's disease. J Biomed Sci 2015; 22: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson DR, Gottesman II. Theories of schizophrenia: a genetic-inflammatory-vascular synthesis. BMC Med Genet 2005; 6: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai Y, Janas JA, Wang CL, Van Aelst L. Regulation of chandelier cell cartridge and bouton development via DOCK7-mediated ErbB4 activation. Cell Rep 2014; 6: 254–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakura Y, Nawa H. ErbB1-4-dependent EGF/neuregulin signals and their cross talk in the central nervous system: pathological implications in schizophrenia and Parkinson's disease. Front Cell Neurosci 2013; 7: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxbaum JD, Georgieva L, Young JJ, Plescia C, Kajiwara Y, Jiang Y et al. Molecular dissection of NRG1-ERBB4 signaling implicates PTPRZ1 as a potential schizophrenia susceptibility gene. Mol Psychiatry 2008; 13: 162–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Su Y, Zhang C, Xing M, Ding W, Liao L et al. The interaction of BDNF and NTRK2 gene increases the susceptibility of paranoid schizophrenia. PLoS ONE 2013; 8: e74264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves J, Martins J, Baptista S, Ambrosio AF, Silva AP. Effects of drugs of abuse on the central neuropeptide Y system. Addict Biol 2016; 21: 755–765. [DOI] [PubMed] [Google Scholar]
- Brothers SP, Wahlestedt C. Therapeutic potential of neuropeptide Y (NPY) receptor ligands. EMBO Mol Med 2010; 2: 429–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantazopoulos H, Wiseman JT, Markota M, Ehrenfeld L, Berretta S. Decreased numbers of somatostatin-expressing neurons in the amygdala of subjects with bipolar disorder or schizophrenia: relationship to circadian rhythms. Biol Psychiatry 2017; 81: 536–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.