Abstract
Children with neurodevelopmental disorders benefit most from early interventions and treatments. The development and validation of brain-based biomarkers to aid in objective diagnosis can facilitate this important clinical aim. The objective of this review is to provide an overview of current progress in the use of neuroimaging to identify brain-based biomarkers for autism spectrum disorder (ASD) and attention-deficit/hyperactivity disorder (ADHD), two prevalent neurodevelopmental disorders. We summarize empirical work that has laid the foundation for using neuroimaging to objectively quantify brain structure and function in ways that are beginning to be used in biomarker development, noting limitations of the data currently available. The most successful machine learning methods that have been developed and applied to date are discussed. Overall, there is increasing evidence that specific features (for example, functional connectivity, gray matter volume) of brain regions comprising the salience and default mode networks can be used to discriminate ASD from typical development. Brain regions contributing to successful discrimination of ADHD from typical development appear to be more widespread, however there is initial evidence that features derived from frontal and cerebellar regions are most informative for classification. The identification of brain-based biomarkers for ASD and ADHD could potentially assist in objective diagnosis, monitoring of treatment response and prediction of outcomes for children with these neurodevelopmental disorders. At present, however, the field has yet to identify reliable and reproducible biomarkers for these disorders, and must address issues related to clinical heterogeneity, methodological standardization and cross-site validation before further progress can be achieved.
Introduction
In clinical research, the term ‘biomarker’ or ‘biological marker’ refers to a broad category of medical signs, or objective indications of medical state, that can be measured accurately and reproducibly and may influence and predict the incidence and outcome of disease.1 Increasingly, clinical neuroscience has shifted from a focus on identifying neural correlates of psychiatric conditions to using metrics derived from brain imaging to predict diagnostic category, disease progression, or response to intervention. Over the past several years, researchers have begun to integrate sophisticated machine learning approaches into studies of brain structure and function, with the result that several candidate ‘brain-based biomarkers’ associated with specific disorders have been proposed. The purpose of this review is to summarize recent progress in identifying brain-based biomarkers for autism spectrum disorder (ASD) and attention-deficit/hyperactivity disorder (ADHD), and highlight roadblocks that must be overcome before further progress can be made. Of note, the term ‘biomarker’ has in some cases been loosely applied in studies that do not strictly meet the definition of the term. In the interest of surveying all potentially relevant contributions to the literature, we review studies that have used supervised learning approaches to classify individuals into clinical categories based on neuroimaging data. As highlighted in other recent reviews of brain-based biomarkers in translational neuroimaging,2, 3 we note the significant challenges that lie ahead, including the development of classifiers that generalize across studies, sites and heterogeneous clinical populations.
ASD and ADHD are common disorders affecting youth, and share a high degree of comorbidity.4 Given their high prevalence (ASD affects an estimated 1.5% of children,5 and ADHD affects an estimated 11% of children and adolescents aged 4–17[ref. 6]) and the cost and consequences of delays in treatment,7 it is imperative to diagnose and predict outcomes for children with a high degree of accuracy as early as is feasible.8 Both ASD and ADHD are currently diagnosed on the basis of parent interview and clinical observation,9, 10, 11 with considerable and sometimes troubling differences emerging between sites. In the case of ASD, significant site differences have been reported in best-estimate clinical diagnoses such that even across sites with well-documented fidelity using standardized diagnostic instruments, clinical distinctions were not reliable.12 The identification of objective, reliable biomarkers is thus a critical yet elusive goal for neuroimaging researchers investigating these neurodevelopmental disorders.
What should a biomarker predict?
In order for a biomarker to be developed, it must be demonstrated that the marker is present before the onset of symptoms, and that it is specific to the disorder.13 We proceed with the caveat that very few of the neuroimaging studies reviewed here would fit this strict definition. Few have used prior-onset features to predict whether a child will later develop ASD or ADHD (but see ref. 14), and most published classification studies only distinguished patients from controls rather than distinguishing among different patient groups. Further, in all classification studies to date, group labels (for example, patient vs control) were determined by clinical diagnosis and reflect a binary designation that may not reflect the heterogeneity and dimensionality inherent to complex neurodevelopmental disorders. Although diagnosis following the Diagnostic and Statistical Manual of Mental Disorders (DSM-5)11 has been the norm in child psychiatry, it is worth noting that there has been a recent shift towards considering dimensions of behavior, consistent with the Research Domain Criteria (RDoC) approach.15 Although there has been some push back from the clinical community regarding the utility of adopting an RDoC framework,16 there is increasing recognition of the fact that dimensions of behavior can cut across traditional diagnostic categories. A few existing studies have focused on dimensions of symptoms using approaches such as support vector regression (see ‘Overview of classifiers’) that allow one to examine behaviors in a continuous rather than categorical manner. Here, we aim to review commonly used classifiers in neuroimaging, summarize current findings relevant to the development of brain-based biomarkers for ASD and ADHD, and discuss open challenges.
Overview of classifiers in neuroimaging
Neuroimaging research in clinical populations has recently begun testing the potential of the attributes of neuroimaging data sets to classify participants into a clinical disorder group and a typical control group. These machine-assisted techniques are often referred to as classifiers. Classifiers fall under the broad branch of machine learning, where computers use algorithms to learn which patterns account for the differences between two or more groups. This may involve establishing a rule whereby a given observation can be classified or sorted into an existing group. These rules can be thought of as features that cause the classifier to distinguish one group from another. By analyzing which features are driving the decisions of the classifier, the patterns of differences between groups can be inferred. Thus, pattern classification analysis of neuroimaging data is important in testing the diagnostic utility of neuroimaging-based markers of psychiatric disorders.
In classifying clinical populations, the classifier searches for differences in neural patterns between a healthy control group and a patient group. These patterns can be activation or lack of activation of a specific brain area, connectivity of a specific brain network, volumetric differences across brain areas, or other metrics derived from neuroimaging data. In other words, classifiers use individual features (such as brain activity, brain morphology or white matter orientation) to predict group membership of a given participant. By examining which features are most important to the classification algorithm, researchers can understand the specific patterns that account for the largest variance between clinical populations and controls. A limitation of this approach, however, is that the classifiers are multivariate combinations of different features, which can make interpretation of specific anatomical contributions difficult. When carefully constructed, biomarkers have the potential to provide insight into the neurological mechanisms of the disorder and targets for future treatment.
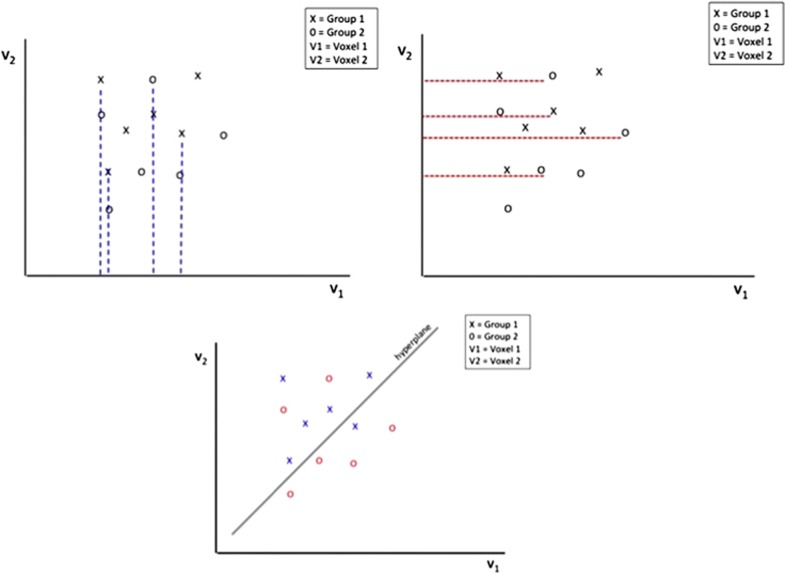
Several of the most commonly used classifiers in neuroimaging include: support vector machine (SVM), logistic regression, decision tree and random forest (RF). SVM is a supervised classification method that uses a subset of data for training, which can then be applied to new data. SVM locates the hyperplane (a high-dimensional plane) that optimally separates data into two groups (for example, patients versus controls) based on features of the data (Figure 1). In the case of neuroimaging data, each point in feature space corresponds to an individual subject. Individuals thus become points in a high-dimensional space, and a hyperplane can then be used for discrimination purposes.17 SVMs have been applied successfully in many brain state and disease state classification problems using functional magnetic resonance imaging (fMRI).18, 19 Logistic regression uses maximum likelihood to estimate the logistic function (an S-shaped curve); this models the probability that an observation belongs to a particular category.20 The decision tree method21 is a supervised learning strategy which builds a classifier from a set of training samples with a list of features (or attributes) and a class label. RF is a popular machine learning algorithm22 that is an ensemble learning method that randomly samples the data with replacement (bootstrapping) to construct many decision trees. Each decision tree is constructed, or grown, using a random subset of the input features. The majority vote of the trees makes up the ‘forest’ or the prediction. Although other types of classifiers can in principle be used in neuroimaging research, those reviewed here have been the most widely used to date.
Figure 1.
Illustration of support vector machine (SVM). SVM is one machine learning approach that is often used in classification studies. If there is a population of subjects (x=autism spectrum disorders, o=typically developing) with voxel values (v1 and v2, for example), then evaluation of one voxel at a time would not differentiate the two groups because there is a substantial amount of overlap between the two groups on each dimension (as shown by the dashed red and blue lines). A univariate analysis evaluating data one voxel at a time (e.g., either v1 alone v2) would not be able to detect group differences in such a scenario. However, if v1 and v2 are considered together, a plane separating the two groups can be constructed, thereby identifying a neighborhood where the two groups differ in spatial patterns of the anatomical or functional measures of interest.
There are many metrics that measure the performance of classifiers, but the most commonly used metric is accuracy. Accuracy assesses the percentage of participants that were correctly classified as being in the clinical or control group. In addition to accuracy, sensitivity and specificity are commonly reported measures of classifier performance. Sensitivity measures the proportion of clinical cases that are correctly identified, and in the binary clinical-control classifier, sensitive indicates the accuracy for the clinical group. Specificity measures the proportion of controls that are correctly identified, and for binary classifiers, indicates accuracy for the control group.
Although machine learning algorithms have opened up a new avenue to neuroimaging research by providing classification of disease states, methodological rigor needs to be ensured for accurate interpretation and application of findings. The goal of machine learning methods is to fit an algorithm to a set of training data in a way that the algorithm can subsequently be applied to new data. Fitting an algorithm to training data can be a relatively simple process, whereas creating a classifier that can generalize beyond the training data set is quite challenging. One of the commonly seen effects involves the classifier exhibiting high accuracy on the training data set but plummeting to chance-level accuracy when presented with new data. This problem is referred to as overfitting, and it often indicates that the algorithm has been fit to random noise of the data rather than its truly classifying features.23 Overfitting occurs when the classifier exhibits good performance on the training data but has poor generalizability to new data. Under-fitting, on the other hand, occurs when the classifier exhibits poor performance on both the training data and the testing data. Under-fitting indicates poor overall fit.
To ensure that a classifier can perform accurately on new data, it is important to reserve data specifically for testing. For example, a classifier could be trained on half of the data and then tested on the other half. However, it is important to maximize the information available for building the classifier, and it can be detrimental to reserve too much data for testing. To deal with this dilemma, a procedure referred to as cross-validation, the standard approach to measure predictive power of a data set, is used. In cross-validation, the available data are split into a training set, used to train the model, and a testing set, unseen by the model during training and used to compute a prediction error.24 One form of cross-validation, known as the ‘leave-one-out’ method, involves leaving one example out of the data set, training the classifier on the remaining items, and then testing one example. This is repeated for each example in the set, and the accuracy is computed from the performance on each of the examples. As this procedure can be computationally expensive with large data sets, ‘k-fold cross-validation’ can be used instead, which involves dividing the data into k-parts (for example, 5, 10) that are each held out for testing.20
In addition, a desirable quality of a machine learning algorithm is high stability. A classifier is deemed to have higher stability if modifying the training data does not largely change the resulting classification algorithm. For example, if removing a single example from the training set results in large perturbations in the resulting classification algorithm, it reflects the relative instability of the classifier. Stability is an important characteristic of a classifier that needs to be ensured in studies of classification analyses, as unstable predictions may result in poor reproducibility of findings.
Brain-based biomarkers of ASD
ASD is now thought to affect 1 in 68 children,25 making early diagnosis an urgent public health concern. The gold standard for clinical assessment includes administration of the Autism Diagnostic Observation Schedule 2 (ADOS-2)26 and the Autism Diagnostic Interview-Revised (ADI-R),10 which assess behavior by semi-structured play-based interviews and parent interviews, respectively. No objective, biological markers exist for diagnosing ASD. Although biomarkers can in principle also be derived from task-based fMRI features, these are less likely to be applied in a clinical setting where task difficulty may preclude some children from participation. Thus, we limit the current discussion to reviewing progress in the development of structural and intrinsic functional connectivity MRI-based biomarkers (Table 1). Although structural MRI and resting-state fMRI data are easier to collect from participants than task fMRI data, it should be noted that one major limitation of the existing literature is that nearly all studies focus on those with ASD without intellectual disability (ID) due to the requirement to stay still for extended periods of time and comply with verbal instructions in the scanner environment. Most studies consequently only recruit high-functioning individuals with ASD.
Table 1. Neuroimaging-based classification studies of ASD.
| Study | Number of subjects ASD(F)/TD (F) | Age range (years) | IQ range (FIQ) | Classification method | Features | Validation scheme | ASD:control performance accuracy, sensitivity, specificity | Subgroup performance accuracy |
|---|---|---|---|---|---|---|---|---|
| Akshoomoff et al.,76 | 52 (0)/15 (0) | 1.7–5.2 | NR | Discrimi NRnt function a NRlysis | GM and WM volumes | NR | NR, 95.8, 92.3 | HFA vs TD & LFA: 68 LFA vs TD & HFA : 85 |
| Neeley et al.,77 | 33 (0)/24 (0) | ASD mean 13.94 TD mean 13.29 | ASD mean 97.14 TD mean 100.22 (PIQ) | Classification and regression tree, LR | GM and WM temporal lobe volume | Leave-one-out cross-validation | TD vs ASD: NR, 85, 83 RD vs ASD: 73, NR, NR | |
| Singh et al.,78 | 16 (NR)/11 (NR) | NR | NR | Linear programming boosting | Cortical thickness | 10 iterations of 9-fold CV, | ~90, NR, NR | |
| Ecker et al.,79 | 20 (0)/20 (0) | 20–68 | 76–141 | SVM (linear kernel) | Cortical thickness | Leave-two-out cross-validation | 90, 90, 80 | |
| Ecker et al.,80 | 22 (0)/22 (0) | 18–42 | ASD mean 104, TD mean 111 | SVM (linear kernel) | GM and WM | Leave-two-out cross-validation, training set | 81, 88, 86 | |
| Jiao81 | 22 (3)/16 (3) | 6–15 | ASD mean 102, TD mean 107.5 | SVM, MLP, FT, LMT | Regional cortical thickness | 10-fold cross-validation, holdout set | 87, 95, 75 | |
| Anderson et al.,27 | 40 (0)/40 (0) | 8–42 | 63–140 (VIQ) | Leave-one-out classifier | Resting-state functional connectivity, ROI | Leave-one-out cross-validation, independent validation set | 79, 83, 75 | |
| Uddin et al.,28 | 24 (2)/24 (2) | 8–18 | ASD mean 100.9, TD mean 104.6 | SVM (with radial basis function kernel) | GM in default mode network regions | 10-fold cross-validation, holdout set | 90, NR, NR | |
| Calderoni et al.,82 | 38 (38)/19 (19) | 2–8 | 40–114 | SVM (linear kernel) | Voxel-wise GM maps | Leave-pair-out CV, holdout group. | AUC=80 | |
| Ingalhalikar et al.,83 | 96 (NR)/42 (NR) | NR | NR | Ensemble classifiers, LDA | Functional features from MEG and structural connectivity features from DTI | 5-fold CV, holdout set. | 88.4, NR, NR | |
| Desphande et al.,84 | 15 (NR)/15 (NR) | 16–34 | 80–140 | RCE-SVM | Causal connectivity weights during theory of mind task, assessment scores, fractional anisotropy | 10-fold cross-validation, holdout set. | 95.9, 96.9, 94.8 | |
| Nielsen et al.,29 | 447 (52)/517 (91) | 6–64 | ASD mean 105, TD mean 112 (VIQ) | Leave-one-out classifier | Resting-state functional connectivity, ROI | Leave-one-out CV using binning system | 60, 62, 58 | |
| Sato et al.,85 | 82 (0)/84 (0) | 18–42 | ASD mean 110, TD mean 114 | Support vector regression with radial basis function | Inter-regional cortical thickness correlation | Leave-one-out CV | r=0.362 (correlation between predicted and observed ADOS) | |
| Uddin et al.,30 | 20 (4)/20 (4) | 7–12 | 78–148 | LR | Resting-state functional connectivity, network components | Independent validation set. | 83, 67, 100 | |
| Plitt et al.,86 | 59 (0)/59 (0) | ASD mean 18.3, TD mean 17.66 | ASD mean 115.76, TD mean 111.02 | RF, KNN, linear SVM, rbf-SVM, L1LR, L2LR, ENLR, GNB), LDA | Resting-state functional connectivity, ROI | Leave-one-out CV, replication samples | 76.67, 75, 78.33 | |
| Segovia et al.,87 | 52 (17)/40 (20) | 12–18 | 73–146 | Searchlight approach and SVM | Gray matter | 10-fold CV | TD vs ASD: 77.2 TD vs ASD siblings: 82.5 | |
| Wee et al.,31 | 59 (13), 58 (13) | 4–22 | NR | SVM | Regional and Inter-regional cortical and subcortical features | Two-fold CV, holdout set | 96.27, 95.5, 97 | |
| Zhou et al.,32 | 127 (24.1)/153 (24.8) | ASD mean 13.5, TD mean 14.5 | ASD mean 104.3, TD mean 111.7 | 67 classifiers tested | 22 structural and functional features | CV using different folds (2–10) or percentage splits (10–90), holdout set. | 68, ~100, ~100 | |
| Chen et al.,33 | 126 (18), 126 (31) | 6–35 | 37–155 (PIQ) | SVM, PSO, RFE, RF | Resting-state functional connectivity, ROI | Bootstrapping out-of-bag, independent validation set | 91, 89, 83 | |
| Ghiassian et al.49 | 430 (11)/45 8(17) | ASD mean 17.3, TD mean 17.1 | ASD mean 106, TD mean 111 | fMRI HOG-feature-based patient classification (MHPC) learning algorithm | Histogram of oriented gradients features from functional and structural MRI | 5-fold CV, holdout set | 65, 71.3, 58.3 | |
| Gori et al.,88 | 21 (0)/20 (0) | 2–6 | 32–123 (NVIQ) | SVM | Volume, thickness, surface area, voxel values of GM | Leave-pair-out CV, holdout set. | AUC of (80±7), 75, 70 | |
| Iidaka89 | 312 (39)/328 (61) | 6–19 | 41–148 | PNN | Resting-state functional connectivity, ROI | Leave-one-out CV, V-fold cross-validation | 90, 92, 87 | |
| Libero et al.,90 | 19 (4)/18 (4) | 19–40 | ASD mean 115.4, TD mean 117.1 | Decision-tree classification | Structural MRI, DTI, and 1H-MRS | Leave-one-out CV | 91.9, NR, NR | |
| Lombardo et al.,91 | 60 (NR)/24 (NR), also included 19 language-delayed | 1–4 | NR | Partial least squares LDA | fMRI speech-related activation and clinical intake measures | 5-fold CV | ASD poor language vs ASD good language : 80, 88, 75 | |
| Subbaraju et al.,92 | 508 (59)/546 (95) | NR | NR | Extended metacognitive radial basis function neural classifier | GM | Holdout set | 83.24, NR, NR | |
| Abraham et al.,93 | 871 subjects (144 F) | 6–64 | NR | Linear SVM | Resting-state functional connectivity, ROI | 10-fold CV, holdout set | 66.8, 61, 72.3 | |
| Yahata et al.,45 | 74(16)/107)34), Validation 54(2)/52(2) | 18–40 | ASD mean 110.0, TD mean 113.3 | L1-SCCA, sparse LR | Resting-state functional connectivity, ROI | Leave-one-out CV, holdout set, independent validation set | Original cohort: 85.0, 80.0, 89.0, Validation cohort: 75.0, NR, NR | |
| Haar et al.,34 | 539 (0)/573 (0) | 6–65 | ASD mean 106, TD-matched | LDA, QDA | Regional volume, surface area and cortical thickness | 10-fold CV, leave-two-out CV, randomization analysis | <60, NR, NR | |
| Katuwal et al.,35 | 361 (0)/373 (0) | 6–64 | ASD mean 105.2, TD mean 11.8 | RF, GBM | Morphometric features from structural MRI | 10-fold CV, holdout set | 60, NR, NR | |
| Wang et al.,94 | 54 (7)/57 (17) | <15 | ASD mean 107.9, TD mean 114.4 | SVM | GM and WM volumes | 10-fold CV, holdout set | 75.4, 74.63, 75.96 |
Abbreviations: ASD, autism spectrum disorder; CV, cross-validation; F, female; FIQ, full-scale intelligence quotient; fMRI, functional magnetic resonance imaging; FT, functional trees; GM, gray matter; GNB, Gaussian Naive Bayes; HFA, high-functioning autism; KNN, k-nearest neighbor; L1LR, L1-regularized logistic regression; L1-SCCA, L1-norm regularized sparse canonical correlation analysis ENLR, elastic-net-regularized logistic regression; L2LR, L2-regularized logistic regression; LDA, linear discriminant analysis; LFA, low functioning autism; LMT, logistic model trees; LR, logistic regression; MLP, multilayer perceptrons; NR, information not reported; NVIQ, nonverbal IQ; PIQ, performance IQ; PNN, probabilistic neural network; PSO, particle swarm optimization; QDA, quadratic discriminant analysis; rbf-SVM, Gaussian kernel support vector machines; RD, Reading Disorder; RF, random forest; RFE, recursive feature elimination, gradient boosting method; rs-fMRI, resting-state fMRI; SVM, support vector machines; TD, typical development; VIQ, verbal IQ; WM, white matter.
Structural MRI
In one of the first detailed analyses of potential brain biomarkers in adults with ASD, Ecker et al.36 examined five morphological parameters including volumetric (cortical thickness and surface area) and geometric (average convexity or concavity, mean curvature and metric distortion of cortex) features derived from structural MRI data. Using SVM and a multiparameter classification approach, they found that all five parameters together produced up to 85% accuracy, 90% sensitivity and 80% specificity for discriminating individuals with ASD from neurotypical (NT) controls. The authors further demonstrated that the classifier built to discriminate ASD from NT individuals was clinically specific in that it somewhat successfully categorized individuals with ADHD as non-autistic. These results represent an important first step towards identifying structural biomarkers in ASD but are limited in their clinical utility in that only adults (aged 20–68) were examined. As ASD is a disorder with early life onset and variable developmental trajectory, studies of younger individuals are critical for disentangling disorder-specific neural signatures.
In a study of children and adolescents with autism, gray matter volume was used as a structural feature for classification. Using SVM searchlight classification, this study found that gray matter within default mode network regions (posterior cingulate cortex and medial temporal lobes: 92%, medial prefrontal cortex: 88%) could be used to discriminate between clinical and control groups with high accuracy.28 The earlier study of adults with ASD also reported morphometric abnormalities in the posterior cingulate that contributed to classification, suggesting that this cortical midline region may be a locus of dysfunction throughout the lifespan.
A more recent study of children used a unique approach, multi-kernal SVM, combining regional (cortical thickness, gray matter volume) and interregional (morphological change patterns between pairs of ROIs) features to achieve 96% classification accuracy.31 In this study, the neuroanatomical features contributing the most to classification were subcortical structures including putamen and accumbens, unlike the studies in older individuals.
Although the studies by Uddin and Wee converge in finding that white matter volume was a poor feature for classification of ASD, others have had considerably greater success using features derived from white matter. Lange et al.37 report high sensitivity (94%), specificity (90%) and accuracy (92%) using diffusion tensor imaging (DTI)-derived metrics in a discovery cohort and a small replication sample. This region-of-interest-based study specifically examined white matter microstructure in the superior temporal gyrus and temporal stem.
A recent study utilizing 590 6–35-year-old participants from the Autism Brain Imaging Data Exchange (ABIDE38) data set, however, was more pessimistic. Using linear and non-linear discriminant analysis to perform multivariate classification based on anatomical measures (gray matter volume, cortical thickness and cortical surface area) the authors achieved only 56 and 60% accuracy based on subcortical volumes and cortical thickness measures, respectively. The authors take these poor decoding accuracies to indicate that anatomical differences offer very limited diagnostic value in ASD.34
Another study also using the ABIDE data set examined morphometric features from structural MRIs of 361 individuals with ASD and 373 controls. Using a RF classifier, the authors demonstrate that only modest classification could be achieved using brain structural properties alone, but that sub-grouping individuals by verbal IQ, autism severity, and age significantly improved classification accuracy.35 This suggests that to achieve the highest classification accuracies in ASD, multiple different structural features may need to be combined with behavioral indices.
Resting-state functional MRI
Resting-state fMRI (rs-fMRI) has been increasingly used over the past decade to study the development of functional brain circuits, and to better understand the large-scale organization of the typically and atypically developing brain.39 Resting-state fMRI entails collecting functional imaging data from participants as they lay in the MRI scanner, typically fixating gaze on a cross-hair or with their eyes closed, and refraining from engaging in any specific cognitive task.40 Some of the advantages of using rs-fMRI in pediatric and clinical populations are that functional brain organization can be examined independent of task performance, imaging data can be acquired from otherwise difficult-to-scan populations41 and a full data set can be collected in as little as 5 min.42
In one of the first studies to use rs-fMRI to attempt to discriminate autism from typical development, Anderson and colleagues used pairwise functional connectivity measures from regions of interest (ROIs) across the entire brain to demonstrate that classification accuracy of 79% could be obtained using data from participants aged 8–42. For individuals under the age of 20, the classifier performed at 89% accuracy. The most informative connections contributing to successful classification were in areas of the default mode network, anterior insula, fusiform gyrus and superior parietal lobule.27 This work emphasizes the potential for classifiers to be more accurate and most informative when applied to subsamples within more restricted age ranges.
Another study examining a younger cohort (age 7–12) using a logistic regression classifier also found that the salience network, including the anterior insular cortices, contained information that could be used to discriminate children with ASD from TD children with 78% accuracy.30 This study used independent component analysis (ICA) maps as features for classification. Data collected at another institution could be classified with 80% accuracy based on the classifier built by the authors. This type of cross-site validation of classifiers is essential for clinical utility, but has proven to be difficult. Nielsen et al.29 utilized pairwise connectivity data from 964 subjects collected at 16 different sites to obtain 60% classification accuracy. As in the earlier study from Anderson et al., connections involving the DMN, parahippocampal and fusiform gyri, and insula contained the most information necessary for accurate classification. This study demonstrates the challenges of building classifiers that can perform accurately across multi-site data sets. Another recent study utilizing data from ABIDE found that the RF approach produced a classification accuracy of 91% based on a functional connectivity matrix of 220 ROIs across the brain. In this study, informative features were found to be located in somatosensory, default mode network, and visual and subcortical regions.33 The most recent study using resting-state fMRI data from the ABIDE database computed whole-brain connectomes (functional connectivity matrices between brain regions of interest) from several atlases to achieve classification accuracy (with support vector classification approaches) of 67%.43 Connections within the default mode network and parieto-insular connections contributed most to prediction of diagnostic category. Finally, a recent study conducted on a large Japanese cohort achieved 85% accuracy and found that discriminating features included functional connectivity between regions of the cingulo-opercular network.44 This rs-fMRI classification study used a unique combination of two machine learning algorithms, L1-regularized sparse canonical correlation analysis (L1-SCCA) and sparse logistic regression, and had fair generalizability across samples, achieving 75% accuracy in an independent validation cohort.45 Taken together, the emerging picture from rs-fMRI studies is that discriminating patterns of connectivity in ASD may reside in DMN and salience/cingulo-opercular network regions.
Multimodal MRI
Owing to the sparse nature of the ASD biomarker literature there is very little information on the use of multimodal MRI to classify ASD. However, there seems to be potential in combining structural and resting-state MRI features to discriminate between ASD and TD groups. A study investigating a wide range of classifiers reports that a random tree classifier using combined cortical thickness and functional connectivity measures resulted in improved classification and prediction accuracy compared to classification using the single imaging features.32 This study was comprehensive in its inclusion of classifiers and features, and the findings suggest an integrative model could be fruitful. However, a more systematic approach to evaluation of classification algorithms is necessary for further progress.
Brain-based biomarkers of ADHD
Compared with the ASD biomarker literature, the biomarker literature for ADHD has a greater number of studies (Table 2). This is in part due to the aggregation of a large (N=973), multi-site, publicly available data set called ADHD-200 (http://fcon_1000.projects.nitrc.org/indi/adhd200/)46 with which an orchestrated global machine learning competition was implemented. The ADHD-200 Global Competition provided a platform for the development of diagnostic classification tools for ADHD using structural and functional MRI data. The best classifier of the 21 competitors in terms of specificity, or the ability to accurately classify TD individuals, was from Eloyan et al.,47 who reported 61% accuracy, 94% specificity and 21% sensitivity to predict diagnosis (TD, ADHD-Inattentive or ADHD-Combined). Surprisingly, when only using phenotypic measures such as site of data collection, sex, age, handedness and IQ, another study48 achieved higher classification accuracy than any imaging-based classifiers (62.52%), whereas a more recent study demonstrated that combining phenotypic and functional imaging data achieved the better accuracy (65%) than phenotypic data alone (59.6%).49 As this competition, researchers have continued to test novel classifiers to improve upon these initial results. We highlight the most successful classifiers below and the features that led to successful prediction of ADHD diagnosis, focusing on binary (TD vs ADHD) classification.
Table 2. Neuroimaging-based classification studies of ADHD.
| Study | Number of subjects ADHD (F)/TD (F) | Age range (years) | IQ range (FIQ) | Classification method | Features | Validation scheme | ADHD:control performancea accuracy, sensitivity, specificity (%)a | Subgroup performancea accuracy (%) | Multiclass performancea ADHD-C vs ADHD-I vs TD |
|---|---|---|---|---|---|---|---|---|---|
| Zhu et al.,50 | 9 (0)/11 (0) | 11–16.5 | >80 | PCA-based Fisher discriminative analysis | ReHo | Leave on out cross-validation, Holdout set | 85, 78, 91b | ||
| Bohland et al.,51 | 272 (53)/482 (227) | 7–21 | NR | Linear SVM classifier | Local anatomical attributes, resting-state measures | 2-fold cross-validation, holdout set | 76, NR, NRc | ||
| Brown et al.,48 | ADHD-C 141 (24), ADHD-I 98 (26)/429 (204) | ADHD-C mean 11.4, ADHD-I mean 12.1, Control mean 12.4 | ADHD-C mean 107.5, ADHD-I mean 104.2, TD mean 114.3 | Logistic classifier, linear SVM, quadratic SVM, cubic SVM, and RBF SVM | rs-fMRI, phenotypic data | 10-fold cross-validation, holdout set | 75, NR, NR | 69 | |
| Colby et al.,52 | 285 (NR)/491 (NR) | NR | NR | SVM RFE, RBF SVM | Surface, volume, functional connectivity, graph metrics | 10-fold cross-validation, holdout set | 55, 33, 80 | ||
| Dai et al.,53 | 222 (48)/402 (194) | ADHD-C mean 11.32, ADHD-I mean 12.02, TD mean 12.47 | NR | SVM-RBF, multi-kernel learning | Cortical thickness, volume, ReHo, functional connectivity | Nested cross-validation, holdout set, | 65.9, 22.5, 89.8 | 56.9 | |
| Eloyan et al.,47 | ADHD-I 125 (NR), ADHD-H 84 (NR)/363 (NR) | 7–26 | NR | Singular value decompositions, CUR decompositions, random forest, gradient boosting, bagging, SVMs | rs-fMRI, structural MRI | Holdout set | 78, 21, 94d | ADHD-I vs ADHD-C: 80 | |
| Igual et al.,95 | 39 (4)/39 (12) | ADHD mean 10.8, TD mean 11.7 | NR | SVM | Structural features of caudate nucleus segmentation | 5-fold cross-validation, holdout set | 94.04, 96.2, 91.2 | ||
| Sato et al.,96 | 383 (87)/546 (260) | ADHD mean 11.6, TD mean 12.3 | NR | AdaBoostM1, Bagging, LogitBoost, SVM, logistic regression | ReHo, ALFF, resting-state networks | leave-one-out and K-fold cross-validation | 57.9, NR, NR | 67 | |
| Sidhu et al.,97 | 141 (NR) ADHD-C, 98 (NR) ADHD-I/429 (NR) | NR | NR | SVM | Resting-state connectivity and phenotypic data | 10-fold cross-validation, holdout set | 76, NR, NR | 68.6 | |
| Lim et al.,54 | 29 (0)/29 (0) | 10–17 | 81–125 | Linear binary Gaussian process classification | Gray matter volume | Leave-one-out cross-validation, holdout set | 79.3, 75.9, 82.8 e | ADHD vs ASD: 85.2 | 68.2 |
| Peng et al.,55 | 55 (NR)/55 (NR) | 9–14 | >80 | Extreme learning machine, linear SVM, SVM-RBF | Cortical thickness and volume measures | Leave-one-out cross-validation, permutation tests. | 90.2, NR, NR | ||
| Wang et al.,98 | 23 (5)/23 (5) | ADHD mean 35.1, TD mean 43 | NR | Linear support vector classifier | ReHo, resting-state | Leave-out cross-validation feature selection loop, holdout set | 80, 87, 74 | ||
| Anderson et al.,50 | 276 (55)/472 (227) | 7.1–21.8 | NR | Non-negative matrix factorization | Default mode structure and graph theory metrics | Leave-one-out cross-validation | 66.8, 76.2 50.6, | ||
| Ghiassian et al.,49 | 279 (21)/490 (47) | ADHD mean 11.6, TD mean12.2 | ADHD mean 107, TD mean 114 | (f)MRI HOG-feature-based patient classification (MHPC) learning algorithm | Personal characteristics and structural brain features | 5-fold cross-validation, holdout set | 69.6, 79.8, 57.1 | ||
| Johnston et al.,56 | 34 (0)/34(0) | 8.5–18.4 | ADHD mean 99.8, TD mean 103.7 | Non-linear SVM | Structural MRI, gray and white matter volume | Leave-one-out cross-validation, holdout set. | 93, 100, 85 | ||
| Deshpande et al,57 | 173 (NR) ADHD-I, 260 (NR) ADHD-C/744 (NR) | NR | NR | Fully connected cascade (FCC) artificial neural network (ANN) architectur | Functional brain connectivity | Leave-one-out cross-validation, holdout set. | TD : ADHD-I: 90, NR, NR TD : ADHD-C: 90, NR, NR | ADHD-I : ADHD-C: 95 | |
| Qureshi et al.,58 | 53 (9) ADHD-I, 53 (9) ADHD-C, 53 (9) TD | 7–14 | NR | Hierarchical extreme learning machine classifier | Cortical thickness | 70/30 partitioning and 10-fold cross-validation. | TD: ADHD-I: 85.3, NR, NR TD: ADHD-C: 79.4, NR, NR | ADHD-I: ADHD-C: 80.3 | 60.8 |
| Xiao et al.,99 | 32 (NR)/15 (NR) | NR | NR | Mutual information-based feature ranking and Lasso-based feature selection | Cortical thickness features | leave-one-out cross-validation, holdout set | 81, 81, 80 | ||
| Qureshi et al.,59 | 53 (9) ADHD-I, 53 (9) ADHD-C, 53 (9) TD | 7–14 | NR | Extreme learning machine classifier | Structural features | Permutation test | TD:ADHD-I: 89.3, 92.9, 85.7 TD:ADHD-C: 92.9, 85.7, 100 | ADHD-I: ADHD-C: 85.7 | 76.2 |
Abbreviations: ADHD, attention-deficit/hyperactivity disorder; ENLR, elastic-net-regularized logistic regression; F, female; FIQ, Full-scale intelligence quotient; fMRI, functional magnetic resonance imaging; FT, functional trees; GM, gray matter; GNB, Gaussian naive Bayes; gradient boosting method; KNN, k-nearest neighbor; L1LR, L1-regularized logistic regression; L2LR, L2-regularized logistic regression; LDA, linear discriminant analysis; LMT, logistic model trees; LR, logistic regression; MLP, multilayer perceptrons; NR, information not reported; NVIQ, nonverbal IQ; PIQ, performance IQ; PNN, probabilistic neural network; PSO, particle swarm optimization; QDA, quadratic discriminant analysis; RBF-radial basis function; RF, random forest; RFE, recursive feature elimination; rs-fMRI, resting-state fMRI; SVM, support vector machines; TD, typical development; VIQ, verbal IQ; WM, white matter.
Refers to classifier with greatest accuracy.
Generalization rate reported instead of accuracy.
Note table reports best accuracy however authors use AUC as preferred method of evaluation.
Accuracy defined as ‘percent total points’. Note distinction from standard definition of overall percentage of correct classifications. 4.
Control group included both ASD and TD subjects.
Structural MRI
Structural MRI shows promise as a potential biomarker for ADHD. Several research groups have achieved impressive classification accuracies using predictors such as gray matter volume and surface area. One of the best-performing classifiers for ADHD used white matter alone to achieve 93% accuracy, with a sensitivity of 100% and specificity of 85%.56 The researchers reported reduced white matter in the central pons, which was predictive of ADHD diagnosis. Similarly, Peng et al.55 reported high classification accuracy (90.18%) using an extreme learning machine (ELM) algorithm with multiple cortical features. Discriminative brain regions included inferior frontal, temporal, occipital and insular cortices. Another study that used whole-brain gray matter volume to classify boys with ADHD and TD boys reported 79.3% accuracy, with 75.9% sensitivity and 82.8% specificity.54 Similar to the Peng et al.55 study, Lim and colleagues reported that the ventrolateral frontal cortex and insula were discriminative. In addition, this group also reported that limbic regions such as hippocampus, amygdala, hypothalamus and ventral striatum were predictive of ADHD status.54
The structural studies mentioned above were single site studies, possibly contributing to their high performance. Of note, one multi-site study using a variety of structural measures obtained high accuracies for binary classification (TD vs ADHD-I: 85.29%, TD vs ADHD-C: 79.40%), demonstrating that even multi-site studies have achieved good performance using structural measures.58
Resting-state functional MRI
On one of the largest data sets used to date in ADHD classification (N=1177)57 achieved 90% accuracy using measures of whole-brain functional connectivity and an artificial neural network algorithm. Of note, these researchers assessed accuracy of binary classification for TD/ADHD-I and TD/ADHD-C separately, which may have contributed to their success. Interestingly, the researchers regressed out the effects of age, IQ, handedness, sex and site from each feature set prior to classification, indicating that imaging-specific features predicted diagnostic status unique from phenotypic characteristics. They found that OFC-cortical and cortico-cerebellar functional connectivity was most discriminative. Similarly, using local and long-distance measures of functional connectivity, Cheng et al.60 found that frontal and cerebellar regions were most discriminative in classifying ADHD and TD children. Zhu et al.61 used a measure of local connectivity, regional homogeneity, to discriminate between ADHD and controls, and found that the most discriminative brain regions included the PFC, ACC, and cerebellum. Overall, functional connectivity of frontal and cerebellar brain regions appear to be good candidates for future use as features in discriminating individuals with ADHD from controls.
Multimodal MRI
Few studies have begun to combine structural and rsMRI data to classify individuals with ADHD from TD individuals. Of those that have, classifier performance tends to be poorer overall than for studies utilizing structural or functional data alone.50, 51, 52, 53, 59 One exception to this pattern was a study by Qureshi et al.,59 which used structural and functional data as predictors for an impressively high multi-class classification (TD, ADHD-I, ADHD-C; one vs all: 76.19%).59 Their success may have been due to employing rigorous feature selection in addition to testing more than one classifier, which demonstrated that the ELM algorithm outperformed the more traditional SVM.
Comorbidity of ASD and ADHD and cross-diagnostic classification
One particularly problematic issue in developing specific biomarkers is the presence of comorbidity across disorders. Rates of comorbid ADHD symptoms in children with ASD range from 37–85%,62 with ADHD the second-most common comorbidity in ASD.63 Conversely, rates of comorbid ASD in children with ADHD are lower, at about 22%.64, 65 Thus, it will be necessary to parse heterogeneity within these disorders prior to attempting to identify biomarkers.
Importantly, the Lim et al.54 study is the only study to date to test an ADHD-specific classifier by discriminating adolescents with ADHD from those with ASD. When classifying these two disorders, they report even higher accuracy than when discriminating ADHD from TD (accuracy 85.2 vs 79.3%). To our knowledge, this study is also the first to attempt to discriminate adolescents with ADHD, ASD, and healthy controls, achieving a balanced accuracy of 68.2%. One other study also considered both ASD and ADHD, but tested each disorder against healthy controls separately, missing an opportunity to employ cross-diagnostic classification.49 A recent study applied machine learning to scores derived from the Social Responsiveness Scale to differentiate between ASD and ADHD,66 but brain-based features were not evaluated.
Limitations of current approaches
Machine-assisted classification of neuroimaging data has provided a new direction to ASD and ADHD research that has important implications for diagnosis and treatment. First, the identification of reliable biomarkers can help provide mechanistic explanations of etiology and behavioral symptomatology. Second, in the long run, using such markers could better assist behavior-based diagnosis. This is especially important for complex or borderline cases, where misdiagnosis is not rare. Third, biomarker screenings could be used on infants and young children to assess their risk of developing a disorder, which is helpful in identifying children at high risk before they show symptoms.67 Thus, children at high risk could receive early, targeted treatment and intervention that would positively impact outcomes of disorders. Infant sibling designs aimed at studying high-risk children are among the most promising avenues for future research. For example, a recent prospective neuroimaging study found that hyperexpansion of the cortical surface between 6 and 12 months of age precedes brain volume overgrowth observed between 12 and 24 months in high-risk infants diagnosed with ASD at 24 months.14 This type of longitudinal work will be necessary to develop true biomarkers for any neurodevelopmental disorder.
Despite the promise and potential of neuroimaging-based markers, inconsistency in the current classification literature suggests that more empirical work must be undertaken. Several factors including participant age, type of classifier used, and sample size contribute to these inconsistencies. In the case of sample size, it is difficult to compare two studies where one tested 1000 participants (for example) and achieved a classification accuracy of 60% and another tested 40 participants and achieved a classification accuracy of 80%. Furthermore, as most studies used different classification algorithms and included different neuroimaging features, it is not possible at present to directly compare results from various studies. A recent review highlights these issues, pointing out that almost all studies that have reported high classification accuracies had sample sizes smaller than 100.3
It is important to note that biomarkers must meet several criteria before being used clinically. First, a biomarker should be present before symptoms begin to serve predictive value. All studies reviewed here were conducted in school-age children and older individuals. Sampling younger children is necessary to examine the predictive power of early-identified brain features. Furthermore, it is quite possible that features identified in the existing literature might reflect either causal or compensatory differences in brain function and structure, and that these features may not be the same as those that are most predictive at early ages. Studies should ideally prospectively screen young children and longitudinally track the occurrence of symptoms. It has already been shown in studies of high-risk infants that developmental trajectories of white matter are most predictive of later ASD diagnosis.68 This suggests that critical information necessary for accurate classification may be overlooked in cross-sectional investigations.
Second, a biomarker must be defined independently of diagnostic symptoms; otherwise, the classifier is validated using the same features that created it. In other words, the relationship between the biomarker and neuropathology must be clear.69 So far, neuroimaging studies have found an array of potential markers and there is not yet convergence on proposed mechanisms. Third, the biomarker must be specific to the disorder rather than a hallmark of general pathology. For instance, abnormal DMN functional connectivity has been associated with both ASD and schizophrenia.70, 71 There are also overlapping biomarkers in children with autism and their unaffected siblings.72, 73 To achieve specificity, the classifier must be tested on large samples that include a variety of disorders. Neuroimaging databases, such ABIDE and the UCLA Multimodal Connectivity Database, can help provide these large samples to test classifiers on. A classification model should be precisely defined and validated across research-sites and populations. Further, a biomarker should have high diagnostic performance in classification, as measured by sensitivity and specificity.69 One recent study in ASD has demonstrated that classification via behavioral measures (in this case scores on the Social Responsiveness Scale) outperformed classification based on analysis of rs-fMRI data.74 However, brain-based biomarkers are at a clear disadvantage compared with behavioral measures, as behavioral measures were designed based on diagnostic criteria. Once diagnoses are more biologically grounded rather than relying solely on observation and interview, it is the hope that early and more objective diagnoses can be achieved.
Other limitations of current approaches include the practice of recruitment of equal case and control samples. In almost all of the studies reviewed here, equal numbers of individuals with ASD or ADHD were included. However, if one wants to apply a classifier to the general population where the prevalence of the disorder ranges from 1.5 to 11%, this can lead to ascertainment bias. For example, with a near ideal classifier of 95% sensitivity and specificity, in 1000 individuals we would accurately identify about 14 of the 15 children with ASD (true positive), but we would inaccurately identify 49 of the remaining 986 as having ASD (false positive) if we assume no other disorders are present. In this example, only 14/63 (22%) of positive identifications would be true cases of ASD.
Biomarkers will not replace clinical assessments, which characterize the extent of specific deficits, but could potentially change treatment goals and methods.67 There are financial barriers to this development, however. MRI is an expensive tool that is unlikely to be used regularly outside of dense urban areas or major hospitals. In ‘Future directions’, we highlight alternative neuroimaging approaches that may prove more feasible in the clinical setting.
In the future, a biomarker could potentially be necessary to receive treatment or insurance coverage, but this will surely create issues if a subset of patients has symptoms but no identifiable biomarker. Finally, families should be given accurate information on what the presence of a biomarker means for their child. It is important not to develop a deterministic view, as it is likely that a biomarker only signifies an increased risk of developing the disorder. This will influence the choices that parents and physicians make regarding treatment.
Future directions: increasing sample heterogeneity
All of the reviewed studies recruited individuals without ID who could successfully complete MRI scans. Of course, this biases the studies to high-functioning individuals, and it is not clear how well the results generalize to the broader population of children with ASD and ADHD. Even within high-functioning individuals, MRI success rates can vary.41 There are a few approaches that in principle could be applied to a greater range of children at varying levels of intellectual function. Sedation and natural sleep75 have been used and can facilitate the collection of neuroimaging data from otherwise inaccessible children. In addition, neuroimaging approaches such as electroencephalography, magnetoencephalography or functional near-infrared spectroscopy (fNIRS), which do not place as stringent requirements on participants to remain motionless, are potentially useful tools with which to derive brain features for classification. Though these methods have limited spatial resolution, making it difficult to assess the contributions of specific brain regions, they can in principle be utilized to a greater extent in future work in biomarker development.
At present, the field has not yet reached the point where we can use brain-based biomarkers to diagnose individuals with a specific disorder. However, the studies reviewed here are instrumental in identifying key dysfunctional brain regions and circuits, moving us closer to understanding the biological basis of these prevalent neurodevelopmental disorders.
Acknowledgments
This work was supported by awards K01MH092288 and R01MH107549 from the National Institute of Mental Health, a Slifka/Ritvo Innovation in Autism Research Award, and a NARSAD Young Investigator Grant to L.Q.U.
Footnotes
The authors declare no conflict of interest.
References
- Strimbu K, Tavel JA. What are biomarkers? Curr Opin HIV AIDS 2010; 5: 463–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo CW, Chang LJ, Lindquist MA, Wager TD. Building better biomarkers: brain models in translational neuroimaging. Nat Neurosci 2017; 20: 365–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbabshirani MR, Plis S, Sui J, Calhoun VD. Single subject prediction of brain disorders in neuroimaging: promises and pitfalls. NeuroImage. 2017; 145(Pt B): 137–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson JL, Rieske RD, Williams LW. The relationship between autism spectrum disorders and attention-deficit/hyperactivity disorder: an overview. Res Dev Disabil 2013; 34: 2475–2484. [DOI] [PubMed] [Google Scholar]
- Christensen DL, Baio J, Van Naarden Braun K, Bilder D, Charles J, Constantino JN et al. Prevalence and characteristics of autism spectrum disorder among children aged 8 years—autism and developmental disabilities monitoring network, 11 sites, United States, 2012. MMWR Surveill Summ 2016; 65: 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser SN, Danielson ML, Bitsko RH, Holbrook JR, Kogan MD, Ghandour RM et al. Trends in the parent-report of health care provider-diagnosed and medicated attention-deficit/hyperactivity disorder: United States, 2003-2011. J Am Acad Child Adoles Psychiatry 2014; 53: 34–46 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlin C, Falkmer M, Parsons R, Albrecht MA, Falkmer T. The cost of autism spectrum disorders. PLoS ONE 2014; 9: e106552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klin A, Klaiman C, Jones W. Reducing age of autism diagnosis: developmental social neuroscience meets public health challenge. Rev Neurol 2015; 60(Suppl 1): S3–11. [PMC free article] [PubMed] [Google Scholar]
- Lord C, Risi S, Lambrecht L, Cook EH Jr, Leventhal BL, DiLavore PC et al. The autism diagnostic observation schedule-generic: a standard measure of social and communication deficits associated with the spectrum of autism. J Autism Dev Disord 2000; 30: 205–223. [PubMed] [Google Scholar]
- Lord C, Rutter M, Le Couteur A. Autism diagnostic interview-revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord 1994; 24: 659–685. [DOI] [PubMed] [Google Scholar]
- American Psychiatric AssociationDiagnostic and Statistical Manual of Mental Disorders. 5th edn. American Psychiatric Publishing: Arlington, VA, 2013. [Google Scholar]
- Lord C, Petkova E, Hus V, Gan W, Lu F, Martin DM et al. A multisite study of the clinical diagnosis of different autism spectrum disorders. Arch Gen Psychiatry 2012; 69: 306–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerys BE, Pennington BF. How do we establish a biological marker for a behaviorally defined disorder? Autism as a test case. Autism Res 2011; 4: 239–241. [DOI] [PubMed] [Google Scholar]
- Hazlett HC, Gu H, Munsell BC, Kim SH, Styner M, Wolff JJ et al. Early brain development in infants at high risk for autism spectrum disorder. Nature 2017; 542: 348–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insel TR. The NIMH Research Domain Criteria (RDoC) Project: precision medicine for psychiatry. Am J Psychiatry 2014; 171: 395–397. [DOI] [PubMed] [Google Scholar]
- Lilienfeld SO, Treadway MT. Clashing diagnostic approaches: DSM-ICD versus RDoC. Ann Rev Clin Psychol 2016; 12: 435–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burges CJC. A tutorial on support vector machines for pattern recognition. Data Min Knowledge Discov 1998; 2: 121–167. [Google Scholar]
- Craddock RC, Holtzheimer PE 3rd, Hu XP, Mayberg HS. Disease state prediction from resting state functional connectivity. Magn Reson Med 2009; 62: 1619–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaConte SM. Decoding fMRI brain states in real-time. NeuroImage 2011; 56: 440–454. [DOI] [PubMed] [Google Scholar]
- Pereira F, Mitchell T, Botvinick M. Machine learning classifiers and fMRI: a tutorial overview. NeuroImage. 2009; 45(1 Suppl): S199–S209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan J. Induction of decision trees. Mach Learn 1986; 1: 81–106. [Google Scholar]
- Breiman L. Random forests. Mach Learn 2001; 45: 4–32. [Google Scholar]
- Domingos P. A few useful things to know about machine learning. Commun ACM 2012; 55: 78. [Google Scholar]
- Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. Springer: New York, NY, USA, 2009. [Google Scholar]
- Developmental Disabilities Monitoring Network Surveillance Year Principal Investigators. Prevalence of autism spectrum disorder among children aged 8 years—autism and developmental disabilities monitoring network, 11 sites, United States, 2010. MMWR Surveill Summ 2014; 63(Suppl 2): 1–21. [PubMed] [Google Scholar]
- Lord C, Rutter M, DiLavore P, Risi S, Gotham K, Bishop S. Autism Diagnostic Observation Schedule–2nd Edition (ADOS-2). Western Psychological Corporation: Los Angeles, CA, USA, 2012. [Google Scholar]
- Anderson JS, Nielsen JA, Froehlich AL, DuBray MB, Druzgal TJ, Cariello AN et al. Functional connectivity magnetic resonance imaging classification of autism. Brain 2011; 134(Pt 12): 3742–3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin LQ, Menon V, Young CB, Ryali S, Chen T, Khouzam A et al. Multivariate searchlight classification of structural magnetic resonance imaging in children and adolescents with autism. Biol Psychiatry 2011; 70: 833–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen JA, Zielinski BA, Fletcher PT, Alexander AL, Lange N, Bigler ED et al. Multisite functional connectivity MRI classification of autism: ABIDE results. Front Hum Neurosci 2013; 7: 599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin LQ, Supekar K, Lynch CJ, Khouzam A, Phillips J, Feinstein C et al. Salience network-based classification and prediction of symptom severity in children with autism. JAMA Psychiatry 2013; 70: 869–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee CY, Wang L, Shi F, Yap PT, Shen D. Diagnosis of autism spectrum disorders using regional and interregional morphological features. Hum Brain Map 2014; 35: 3414–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Yu F, Duong T. Multiparametric MRI characterization and prediction in autism spectrum disorder using graph theory and machine learning. PLoS ONE 2014; 9: e90405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CP, Keown CL, Jahedi A, Nair A, Pflieger ME, Bailey BA et al. Diagnostic classification of intrinsic functional connectivity highlights somatosensory, default mode, and visual regions in autism. NeuroImage Clin 2015; 8: 238–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haar S, Berman S, Behrmann M, Dinstein I. Anatomical abnormalities in autism? Cereb Cortex 2016; 26: 1440–1452. [DOI] [PubMed] [Google Scholar]
- Katuwal GJ, Baum SA, Cahill ND, Michael AM. Divide and conquer: sub-grouping of ASD improves ASD detection based on brain morphometry. PLoS ONE 2016; 11: e0153331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker C, Marquand A, Mourao-Miranda J, Johnston P, Daly EM, Brammer MJ et al. Describing the brain in autism in five dimensions—magnetic resonance imaging-assisted diagnosis of autism spectrum disorder using a multiparameter classification approach. J Neurosci 2010; 30: 10612–10623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange N, Dubray MB, Lee JE, Froimowitz MP, Froehlich A, Adluru N et al. Atypical diffusion tensor hemispheric asymmetry in autism. Autism Res 2010; 3: 350–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino A, Yan CG, Li Q, Denio E, Castellanos FX, Alaerts K et al. The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol Psychiatry 2014; 19: 659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin LQ, Supekar K, Menon V. Typical and atypical development of functional human brain networks: insights from resting-state FMRI. Front Syst Neurosci 2010; 4: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med 1995; 34: 537–541. [DOI] [PubMed] [Google Scholar]
- Yerys BE, Jankowski KF, Shook D, Rosenberger LR, Barnes KA, Berl MM et al. The fMRI success rate of children and adolescents: typical development, epilepsy, attention deficit/hyperactivity disorder, and autism spectrum disorders. Hum Brain Map 2009; 30: 3426–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk KR, Hedden T, Venkataraman A, Evans KC, Lazar SW, Buckner RL. Intrinsic functional connectivity as a tool for human connectomics: theory, properties, and optimization. J Neurophysiol 2009; 103: 297–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham A, Milham MP, Di Martino A, Craddock RC, Samaras D, Thirion B et al. Deriving reproducible biomarkers from multi-site resting-state data: an autism-based example. NeuroImage 2017; 147: 736–745. [DOI] [PubMed] [Google Scholar]
- Dosenbach NU, Fair DA, Cohen AL, Schlaggar BL, Petersen SE. A dual-networks architecture of top-down control. Trends Cogn Sci 2008; 12: 99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahata N, Morimoto J, Hashimoto R, Lisi G, Shibata K, Kawakubo Y et al. A small number of abnormal brain connections predicts adult autism spectrum disorder. Nat Commun 2016; 7: 11254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ADHD-200 Consortium. The ADHD-200 Consortium: a model to advance the translational potential of neuroimaging in clinical neuroscience. Front Syst Neurosci 2012; 6: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eloyan A, Muschelli J, Nebel MB, Liu H, Han F, Zhao T et al. Automated diagnoses of attention deficit hyperactive disorder using magnetic resonance imaging. Front Syst Neurosci 2012; 6: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MR, Sidhu GS, Greiner R, Asgarian N, Bastani M, Silverstone PH et al. ADHD-200 Global Competition: diagnosing ADHD using personal characteristic data can outperform resting state fMRI measurements. Front Syst Neurosci 2012; 6: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiassian S, Greiner R, Jin P, Brown MR. Using functional or structural magnetic resonance images and personal characteristic data to identify ADHD and autism. PLoS ONE 2016; 11: e0166934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson A, Douglas PK, Kerr WT, Haynes VS, Yuille AL, Xie J et al. Non-negative matrix factorization of multimodal MRI, fMRI and phenotypic data reveals differential changes in default mode subnetworks in ADHD. NeuroImage 2014; 102(Pt 1): 207–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohland JW, Saperstein S, Pereira F, Rapin J, Grady L. Network, anatomical, and non-imaging measures for the prediction of ADHD diagnosis in individual subjects. Front Syst Neurosci 2012; 6: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby JB, Rudie JD, Brown JA, Douglas PK, Cohen MS, Shehzad Z. Insights into multimodal imaging classification of ADHD. Front Syst Neurosci 2012; 6: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai D, Wang J, Hua J, He H. Classification of ADHD children through multimodal magnetic resonance imaging. Front Syst Neurosci 2012; 6: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L, Marquand A, Cubillo AA, Smith AB, Chantiluke K, Simmons A et al. Disorder-specific predictive classification of adolescents with attention deficit hyperactivity disorder (ADHD) relative to autism using structural magnetic resonance imaging. PLoS ONE 2013; 8: e63660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Lin P, Zhang T, Wang J. Extreme learning machine-based classification of ADHD using brain structural MRI data. PLoS ONE 2013; 8: e79476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston BA, Mwangi B, Matthews K, Coghill D, Konrad K, Steele JD. Brainstem abnormalities in attention deficit hyperactivity disorder support high accuracy individual diagnostic classification. Hum Brain Map 2014; 35: 5179–5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande G, Wang P, Rangaprakash D, Wilamowski B. Fully connected cascade artificial neural network architecture for attention deficit hyperactivity disorder classification from functional magnetic resonance imaging data. IEEE Trans Cybernet 2015; 45: 2668–2679. [DOI] [PubMed] [Google Scholar]
- Qureshi MN, Min B, Jo HJ, Lee B. Multiclass classification for the differential diagnosis on the ADHD subtypes using recursive feature elimination and hierarchical extreme learning machine: structural MRI study. PLoS ONE 2016; 11: e0160697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi MNI, Oh J, Min B, Jo HJ, Lee B. Multi-modal, multi-measure, and multi-class discrimination of ADHD with hierarchical feature extraction and extreme learning machine using structural and functional brain MRI. Front Hum Neurosci 2017; 11: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W, Ji X, Zhang J, Feng J. Individual classification of ADHD patients by integrating multiscale neuroimaging markers and advanced pattern recognition techniques. Front Syst Neurosci 2012; 6: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu CZ, Zang YF, Cao QJ, Yan CG, He Y, Jiang TZ et al. Fisher discriminative analysis of resting-state brain function for attention-deficit/hyperactivity disorder. NeuroImage 2008; 40: 110–120. [DOI] [PubMed] [Google Scholar]
- Leitner Y. The co-occurrence of autism and attention deficit hyperactivity disorder in children - what do we know? Front Hum Neurosci 2014; 8: 268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonoff E, Pickles A, Charman T, Chandler S, Loucas T, Baird G. Psychiatric disorders in children with autism spectrum disorders: prevalence, comorbidity, and associated factors in a population-derived sample. J Am Acad Child Adolesc Psychiatry 2008; 47: 921–929. [DOI] [PubMed] [Google Scholar]
- van der Meer JM, Oerlemans AM, van Steijn DJ, Lappenschaar MG, de Sonneville LM, Buitelaar JK et al. Are autism spectrum disorder and attention-deficit/hyperactivity disorder different manifestations of one overarching disorder? Cognitive and symptom evidence from a clinical and population-based sample. J Am Acad Child Adolesc Psychiatry 2012; 51: 1160–72 e3. [DOI] [PubMed] [Google Scholar]
- Ronald A, Simonoff E, Kuntsi J, Asherson P, Plomin R. Evidence for overlapping genetic influences on autistic and ADHD behaviours in a community twin sample. J Child Psychol Psychiatry 2008; 49: 535–542. [DOI] [PubMed] [Google Scholar]
- Duda M, Haber N, Daniels J, Wall DP. Crowdsourced validation of a machine-learning classification system for autism and ADHD. Transl Psychiatry 2017; 7: e1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh P, Elsabbagh M, Bolton P, Singh I. In search of biomarkers for autism: scientific, social and ethical challenges. Nat Rev Neurosci 2011; 12: 603–612. [DOI] [PubMed] [Google Scholar]
- Wolff JJ, Gu H, Gerig G, Elison JT, Styner M, Gouttard S et al. Differences in white matter fiber tract development present from 6 to 24 months in infants with autism. Am J Psychiatry 2012; 169: 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo CW, Wager TD. Neuroimaging-based biomarker discovery and validation. Pain 2015; 156: 1379–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdaugh DL, Shinkareva SV, Deshpande HR, Wang J, Pennick MR, Kana RK. Differential deactivation during mentalizing and classification of autism based on default mode network connectivity. PLoS ONE 2012; 7: e50064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity AG, Pearlson GD, McKiernan K, Lloyd D, Kiehl KA, Calhoun VD. Aberrant "default mode" functional connectivity in schizophrenia. Am J Psychiatry 2007; 164: 450–457. [DOI] [PubMed] [Google Scholar]
- Spencer MD, Holt RJ, Chura LR, Suckling J, Calder AJ, Bullmore ET et al. A novel functional brain imaging endophenotype of autism: the neural response to facial expression of emotion. Transl Psychiatry 2011; 1: e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser MD, Hudac CM, Shultz S, Lee SM, Cheung C, Berken AM et al. Neural signatures of autism. Proc Natl Acad Sci USA 2010; 107: 21223–21228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plitt M, Barnes KA, Martin A. Functional connectivity classification of autism identifies highly predictive brain features but falls short of biomarker standards. NeuroImage Clin 2015; 7: 359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce K. Early functional brain development in autism and the promise of sleep fMRI. Brain Res 2011; 1380: 162–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akshoomoff N, Lord C, Lincoln AJ, Courchesne RY, Carper RA, Townsend J et al. Outcome classification of preschool children with autism spectrum disorders using MRI brain measures. J Am Acad Child Adolesc Psychiatry 2004; 43: 349–357. [DOI] [PubMed] [Google Scholar]
- Neeley ES, Bigler ED, Krasny L, Ozonoff S, McMahon W, Lainhart JE. Quantitative temporal lobe differences: autism distinguished from controls using classification and regression tree analysis. Brain Dev 2007; 29: 389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V, Mukherjee L, Chung MK. Cortical surface thickness as a classifier: boosting for autism classification. Med Image Comput Comput Assist Interv 2008; 11(Pt 1): 999–1007. [DOI] [PubMed] [Google Scholar]
- Ecker C, Marquand A, Mourao-Miranda J, Johnston P, Daly EM, Brammer MJ et al. Describing the brain in autism in five dimensions—magnetic resonance imaging-assisted diagnosis of autism spectrum disorder using a multiparameter classification approach. J Neurosci 2010. a; 30: 10612–10623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker C, Rocha-Rego V, Johnston P, Mourao-Miranda J, Marquand A, Daly EM et al. Investigating the predictive value of whole-brain structural MR scans in autism: a pattern classification approach. Neuroimage 2010. b; 49: 44–56. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Chen R, Ke X, Chu K, Lu Z, Herskovits EH. Predictive models of autism spectrum disorder based on brain regional cortical thickness. Neuroimage 2010; 50: 589–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderoni S, Retico A, Biagi L, Tancredi R, Muratori F, Tosetti M. Female children with autism spectrum disorder: an insight from mass-univariate and pattern classification analyses. Neuroimage 2012; 59: 1013–1022. [DOI] [PubMed] [Google Scholar]
- Ingalhalikar M1, Parker WA, Bloy L, Roberts TP, Verma R. Using multiparametric data with missing features for learning patterns of pathology. Med Image Comput Comput Assist Interv 2012; 15(Pt 3): 468–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande G, Libero LE, Sreenivasan KR, Deshpande HD, Kana RK. Identification of neural connectivity signatures of autism using machine learning. Front Hum Neurosci 2013; 7: 670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato JR, Hoexter MQ, Oliveira PP Jr, Brammer MJ, AIMS MRC. Consortium, Murphy D, Ecker C. Inter-regional cortical thickness correlations are associated with autistic symptoms: a machine-learning approach. J Psychiatr Res 2013; 47: 453–459. [DOI] [PubMed] [Google Scholar]
- Plitt M, Barnes KA, Martin A. Functional connectivity classification of autism identifies highly predictive brain features but falls short of biomarker standards. Neuroimage Clin 2014; 7: 359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segovia F, Holt R, Spencer M, Górriz JM, Ramírez J, Puntonet CG et al. Identifying endophenotypes of autism: a multivariate approach. Front Comput Neurosci 2014; 8: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gori I, Giuliano A, Muratori F, Saviozzi I, Oliva P, Tancredi R et al. Gray matter alterations in young children with autism spectrum disorders: comparing morphometry at the voxel and regional level. J Neuroimaging 2015; 25: 866–874. [DOI] [PubMed] [Google Scholar]
- Iidaka T. Resting state functional magnetic resonance imaging and neural network classified autism and control. Cortex 2015; 63: 55–67. [DOI] [PubMed] [Google Scholar]
- Libero LE, DeRamus TP, Lahti AC, Deshpande G, Kana RK. Multimodal neuroimaging based classification of autism spectrum disorder using anatomical, neurochemical, and white matter correlates. Cortex 2015; 66: 46–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo MV, Pierce K, Eyler LT, Carter Barnes C, Ahrens-Barbeau C, Solso S et al. Different functional neural substrates for good and poor language outcome in autism. Neuron 2015; 86: 567–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaraju V, Sundaram S, Narasimham S, Suresh MB. Accurate detection of autism spectrum disorder from structural MRI using extended metacognitive radial basis function network. Exp Syst Appl 2015; 42: 8775–8790. [Google Scholar]
- Abraham A, Milham MP, Di Martino A, Craddock RC, Samaras D, Thirion B et al. Deriving reproducible biomarkers from multi-site resting-state data: an autism-based example. Neuroimage 2017; 147: 736–745. [DOI] [PubMed] [Google Scholar]
- Wang L, Wee CY, Tang X, Yap PT, Shen D. Multi-task feature selection via supervised canonical graph matching for diagnosis of autism spectrum disorder. Brain Imaging Behav 2016; 10: 33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igual L, Soliva JC, Escalera S, Gimeno R, Vilarroya O, Radeva P. Automatic brain caudate nuclei segmentation and classification in diagnostic of attention-deficit/hyperactivity disorder. Comput Med Imaging Graph 2012; 36: 591–600. [DOI] [PubMed] [Google Scholar]
- Sato JR, Hoexter MQ, Fujita A, Rohde LA. Evaluation of pattern recognition and feature extraction methods in ADHD prediction. Front Syst Neurosci 2012; 6: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu GS, Asgarian N, Greiner R, Brown MR. Kernel principal component analysis for dimensionality reduction in fMRI-based diagnosis of ADHD. Front Syst Neurosci 2012; 6: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Jiao Y, Tang T, Wang H, Lu Z. Altered regional homogeneity patterns in adults with attention-deficit hyperactivity disorder. Eur J Radiol 2013; 82: 1552–1557. [DOI] [PubMed] [Google Scholar]
- Xiao C, Bledsoe J, Wang S, Chaovalitwongse WA, Mehta S, Semrud-Clikeman M et al. An integrated feature ranking and selection framework for ADHD characterization. Brain Inform 2016; 3: 145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]