Summary
The GLYcan Data Exchange (GLYDE) standard has been developed for the representation of the chemical structures of monosaccharides, glycans and glycoconjugates using a connection table formalism formatted in XML. This format allows structures, including those that do not exist in any database, to be unambiguously represented and shared by diverse computational tools. GLYDE implements a partonomy model based on human language along with rules that provide consistent structural representations, including a robust namespace for specifying monosaccharides. This approach facilitates the reuse of data processing software at the level of granularity that is most appropriate for extraction of the desired information. GLYDE-II has already been used as a key element of several glycoinformatics tools. The philosophical and technical underpinnings of GLYDE-II and recent implementation of its enhanced features are described.
Keywords: Bioinformatics, Glycan, Structure, Representation, XML
Introduction
Glycobiology, broadly defined as the study of the structure, biosynthesis and biological functions of glycans and glycoconjugates, is an emerging field of research that has found increasing applications in diverse technologies ranging from medicine to biofuels (Varki and Sharon, 2009; Walt et al., 2012). Glycomics, which focuses on the structures and abundances of specific glycans in various biological samples, has been enabled by developments in molecular analysis that make it possible to detect, identify and quantify glycans as free molecules or as components of glycoconjugates (Cummings and Pierce, 2014). However, the development of glycomics has lagged behind genomics and proteomics in large part due to analytical challenges that stem from the structural complexity of glycans. Glycan structures cannot be inferred directly from genomic data, as the structure of each glycan is the result of an often complex metabolic pathway whose individual steps are catalyzed by sundry glycosyl-transferases and other glycan modifying enzymes (Varki and Sharon, 2009).
Progress in glycomics has also been slowed by lack of robust informatics tools to archive, retrieve, analyze, mine and transfer the large amounts of multifaceted data that is generated in the course of this research (York et al., 2014). The development of databases and ontologies that contain knowledge regarding glycan structures and their relationships to biological and physical phenomena is especially challenging. Nevertheless, several isolated databases that archive information about carbohydrate structure, biosynthesis and function have been developed. The integration of this diverse information is a major bioinformatics challenge that must be addressed if glycobiology is to reach its potential to address critical issues in biomedicine, biofuels and other domains.
The existing carbohydrate databases implement diverse structural representation protocols. One reason for the development of different glycan sequence formats is the diversity of molecular building blocks (monosaccharides) and the frequent existence of branches. A second reason is that few of the sequence formats used in the various databases have been published. Unfortunately, this lack of accessibility has led to the proliferation of formats rather than to the establishment of a single standard. More than a dozen sequence formats have been developed, and more than one can be used in a single database or software application. These sequence formats include linearized representations of the branched sequences, such as LINUCS (Bohne-Lang et al., 2001), the BCSDB format (Egorova and Toukach, 2014) and LinearCode® (Banin et al., 2002), as well as connection table representations, such as GlycoCT (Herget et al., 2008), WURCS (Tanaka et al., 2014) and KCF (Hashimoto et al., 2006) and XML representations, such as CabosML (Kikuchi et al., 2005). The development of GlycomeDB (Ranzinger et al., 2011), a meta-database for glycan structures, has been a major step towards the integration of this structural data. More recently, the GlyTouCan glycan structure repository (Aoki-Kinoshita et al., 2016) has been implemented to provide a robust and stable semantic basis for describing glycan structure. Nevertheless, communication between different data acquisition, storage and processing systems still requires well-defined methods for transferring structural information that may not be included in an existing database. The GLYcan Data Exchange format (GLYDE) has been developed as a standard XML (Extensible Markup Language) based format to address these concerns. GLYDE has been widely accepted by the glycoinformatics community (Packer et al., 2008) and is a frequently used format for exchanging glycan structure information (Eavenson et al., 2015). This paper describes the philosophical and technical background for GLYDE and the recently implemented enhancements of this standard.
Results
The GLYDE XML format is defined via two schema specifications: an XSD (XML Schema Definition) schema and a complementary but more flexible DTD (Document Type Definition) schema. These define a specification framework, which we call PARCHMENT (PARtonomy of CHeMical ENTities), which allows the structure of biological molecules (including complex glycans) to be completely and unambiguously specified at several levels of granularity. Notably, this provides a unified format for the concise and complete structural representation of each molecule in the vast, naturally occurring combinatorial set of glycoconjugates that arises by the attachment of a large number of distinct glycans to a large number of distinct non-carbohydrate moieties.
The GLYDE standard also includes a set of rules, naming conventions for the parts, and enumeration of chemical entities that are acceptable parts at various levels of granularity. These implementation rules are absolutely required for representational consistency and disambiguation, as purely syntactic enforcement of these rules (e.g. solely by the GLYDE schema) is not possible.
History
The GLYDE format for the representation of glycan structure was developed to take advantage of the hierarchical syntax and extensibility of XML. The first version of GLYDE (Sahoo et al., 2005) used a hierarchical XML tree to intuitively mirror tree-like structures of branched glycan structures. However, this approach was found to be insufficient to represent all of the structural variation observed in glycans and glycoconjugates.
GLYDE-II was developed to account for this structural variation by using XML to implement a connection table approach. In this context, GLYDE-II provides a consistent namespace for monosaccharides and the ability to represent repeating structural features. Notably, the GLYDE-II syntax enforces a partonomic model in which structures are specified as collections of parts (Fig. 1), which can themselves be collections of smaller parts, facilitating the representation of highly complex molecules such as glycoconjugates. GLYDE-II specifies molecular topology by defining explicit connections (i.e. links) between the parts of the molecule. Version 1.2 of GLYDE introduces two important modifications: (1) classification of molecular parts using semantics that are more consistent with the way biochemists view biopolymer structure and processing and (2) optional specification of molecular geometry using a general approach that is consistent over all granularity levels and for all different classes of molecular parts. These modifications make it possible for the glycoscientist to take advantage of the GLYDE partonomy model to create and interpret representations of glycoconjugate structure that are consistent with the way biochemists think about these complex molecules and their interactions with other molecules.
Figure 1.
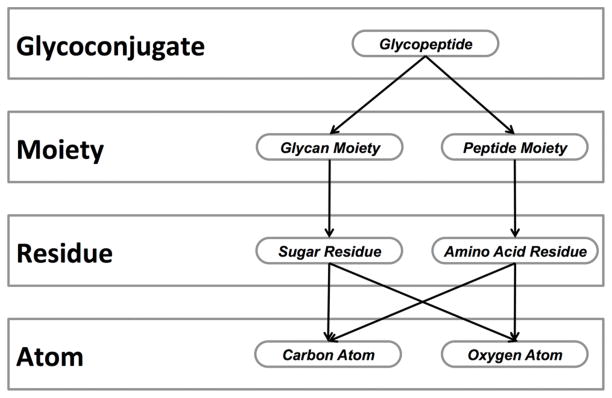
Some of the partonomy relationships implemented by GLYDE-II. Each object (rounded rectangles) is classified (as an atom, residue, etc.) according to its complexity. Arrows emanating from an object point to its parts. Each object can have many parts, only a few of which are shown. The structure of each part is defined by reference to an archetypal object. For example, the structure of each of the distinct carbon atom objects in a sugar residue is specified by reference to the archetypal carbon atom.
Partonomy and archetyping
One aspect that distinguishes the different glycan structure representation formats that have been developed is the way different parts of these molecules are grouped and specified. This stems from the fact that glycans are often found as components of more complex structures commonly referred to as glycoconjugates, which include glycoproteins, glycolipids and chemically modified glycans, such as oligosaccharides that have a fluorescent tag attached to the reducing end. Furthermore, the complex, branched nature of glycans has led to semantic differences in the way glycans and non-glycan moieties are described. However, GLYDE is based on semantics that are common to diverse types of biopolymers, providing an integrated way to represent the different parts of a glycoconjugate structure. This approach allows the representation and processing of structures that are specified at different levels of granularity (e.g. to identify specific components or calculate molecular masses) to be performed using a common set of algorithms. To make this practical, structural granularity must be rigorously defined using the concept of partonomy (Casati and Varzi, 1999), in which each object is described as a collection of parts (Fig. 1).
Along with partonomy, object archetyping is another core feature of GLYDE. For example, the glycan moiety of a glycoconjugate (e.g. glycopeptide) is defined by reference to a glycan molecule (e.g. the glycan released from a glycopeptide by the enzyme PNGase-F). Such use of a small archetype molecule to describe a part of a larger molecule is consistent with conventional chemical language. For example, a glucose residue in a glycan is customarily denoted by reference to the molecule glucose (a monosaccharide). These semantics are based on concepts that are both historical (e.g. the characterization of small biomolecular building blocks such as purines, sugars and amino acids by Fischer Kunz, 2002) and biochemical (e.g. the transformation of such small molecules into so-called “residues” when they are incorporated into biopolymers). This approach makes the structural representations much more concise by allowing detailed structural information (and/or references to external sources) to be specified one time for the archetype and inferred thereafter when the archetype is referenced. Archetyping atoms makes the structural representation somewhat more concise. However, archetyping larger structures (like sugar residues or glycan moieties) makes the structural representation substantially more concise. This is an important consideration when using a format such as XML that is verbose by nature.
Implementation of the GLYDE-II partonomy model
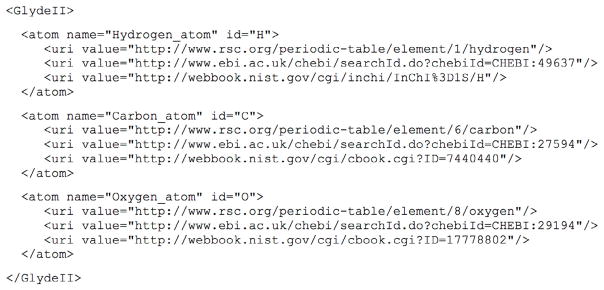
A key function of GLYDE is to provide a basis for the operational classification of molecular objects, using a vocabulary indicated by italics in the text below. At the most fundamental level, GLYDE specifies structures as collections of atoms. Fig. 2 illustrates a minimal GLYDE-II representation of three atom objects. By definition, an atom object is not connected to another atom object by a molecular bond. In some cases, an unattached atom can be specified as a part of a GLYDE structure, but more frequently an atom is used as an archetype for a bound atom, which is, by definition, connected to at least one other bound atom by a covalent bond in the context of a molecule, which is a collection of molecular parts that are connected by covalent bonds. However, a molecule is defined such that it is not connected to any other molecule by covalent bonds.
Figure 2.
GLYDE representation of a collection of three atom objects (hydrogen, carbon, oxygen). External sources of information about the structure of each atom are indicated by uri tags.
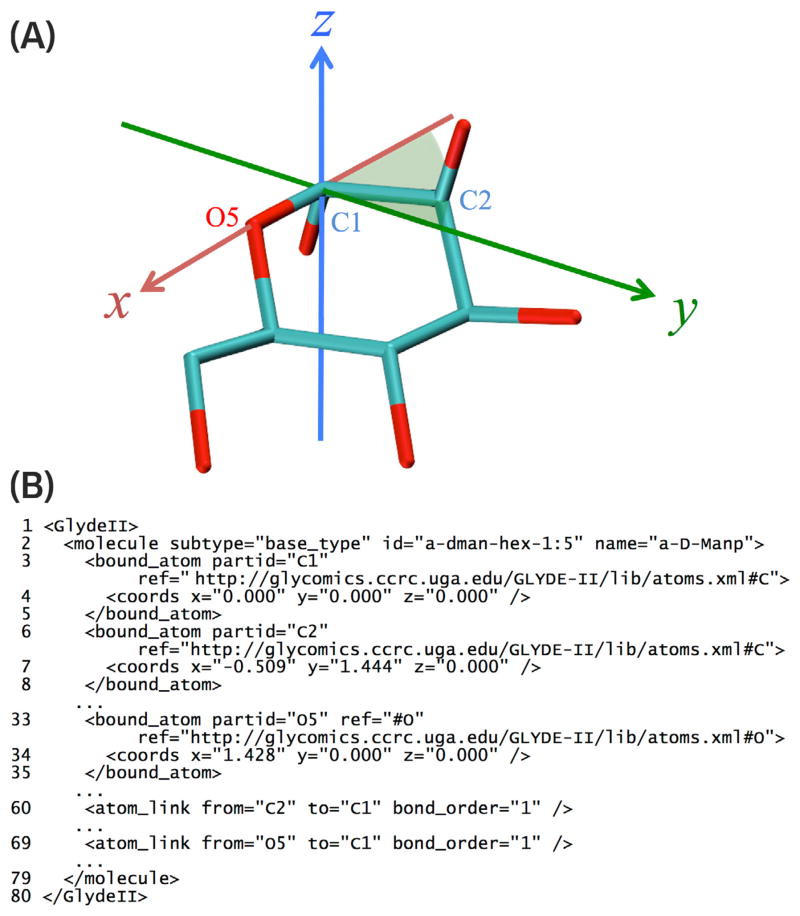
In general, the GLYDE specification of a molecule consists of a list of its parts and the links between those parts. A molecule is most directly represented in GLYDE as a collection of bound atom objects and the atom link objects that connect them, as illustrated for a monosaccharide molecule (α-D-Manp) in Fig. 3. Within such a molecule representation, the structure of each bound atom is specified by its ref attribute, which indicates the id attribute of the atom object (Fig. 2) that serves as the archetype for the bound atom. A molecule can also be specified as a collection of parts that are more complex than atoms. For example, a molecule can be composed of residue objects (Fig. 4), whose structures are themselves specified by reference to molecule objects (e.g. monosaccharides), which in turn are specified as collections of bound atom and atom link objects (Fig. 3). Connections between residue objects are specified by residue link objects, which encompass atom link objects that specify the atoms involved in the covalent bonds connecting the residue objects (Fig. 4).
Figure 3.
Stick model (A) and abbreviated GLYDE-II representation (B) of an α-D-Manp molecule. (Several lines of XML code are omitted for brevity; the remaining lines are numbered.) The structure of each bound atom is specified by its ref attribute (lines 3, 6 and 33), which points to a GLYDE-II representation of the atom (Fig. 2) serving as the archetype for the bound atom. The molecular topology is fully specified by atom link objects (e.g. lines 60 and 69), which connect bound atom objects. The molecular configuration (stereochemistry) is specified explicitly by listing the coordinates of each bound atom. Alternatively, stereochemistry of the molecule can be inferred from its id (line 2), which by rule corresponds to its representation using a format based on GlycoCT. The conventional orientation and position of the Cartesian axes in the atomistic GLYDE-II representation is defined by the alignment of three key bound atom objects: the anomeric carbon (C1 in this case) is at the origin, the ring oxygen (O5 in this case) is on the x-axis, and the highest-numbered carbon (C2 in this case) that is directly linked to the anomeric carbon is in the first or second quadrant of the x,y-plain (where y > 0).
Figure 4.
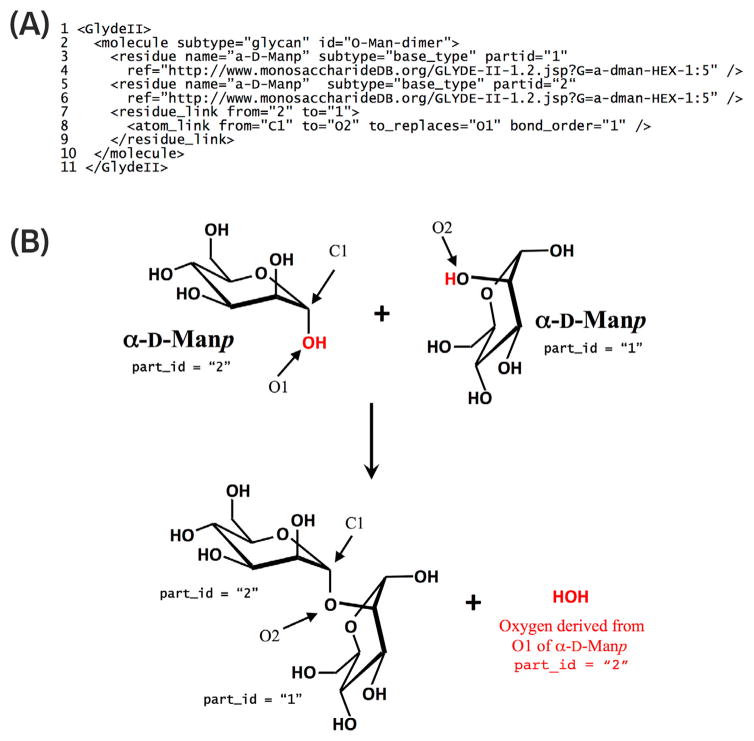
(A) GLYDE-II representation of the disaccharide molecule that is used as an archetype for the disaccharide moiety shown in Fig. 5. The properties of each of the residue objects in this molecule are specified by reference to the archetypal α-D-Manp molecule shown in Fig. 3. The residue link (line 7) formally indicates a directed bond from one residue to another. This residue link, in turn, encompasses an atom link (line 8) from C1 of one of the α-D-Manp residue objects (part id = “2”) to O2 of the other α-D-Manp residue object (part id = “1”). These directional semantics allow unambiguous interpretation of the attributes (e.g., from = “C1” and to = “O2”) in the atom link. That is, the bond extends from “C1” of α-D-Manp residue (part id = “1”) to “O2” of α-D-Manp residue (part id = “2”). This code also infers that, in the context of this atom link, “to” is a synonym for “O2 of α-D-Manp residue #1” and “from” is a synonym for “C1 of α-D-Manp residue #2”. By rule, an atom in one residue can replace an atom in the other residue when a residue link is formed (see Panel B). The atom link attribute to replaces = “O1” can thus be unambiguously interpreted as “O2 of α-D-Manp residue #1 replaces O1 of α-D-Manp residue #2”. (B) Formation of the glycosidic bond connecting the two α-D-Manp residue objects in the disaccharide encoded by the text in (A). The GLYDE convention dictates that the geometry of each archetypal monosaccharide molecule is retained when it is transformed into a residue. During this transformation, the glycosidic oxygen of one residue (e.g. O1 of the α-D-Manp residue #2) is released as a water molecule and replaced by an atom in the other residue (e.g. O2 of α-D-Manp residue #1).
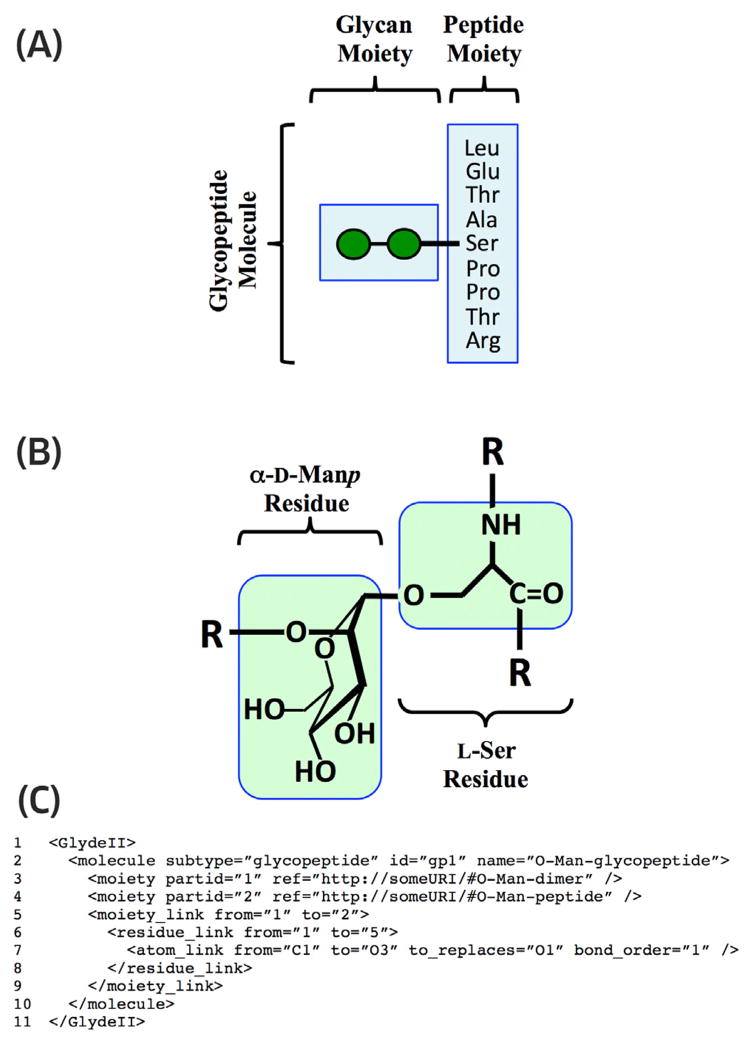
The structures of complex molecule objects such as glycoconjugates are specified using the same hierarchical approach (Fig. 5), in which larger structures are constructed as collections of smaller structures. Connections between the larger structures embody connections between the smaller structures that they contain. For example, moiety link objects embody residue link objects.
Figure 5.
The structure of a glycopeptide molecule illustrated as a hierarchical collection of GLYDE parts (bound atom, residue and moiety) connected by links. Residues in the glycan moiety are represented using the CFG graphical format for glycan structure. (A) The glycan moiety and peptide moiety are connected by a moiety link, which embodies a residue link connecting one of the α-D-Manp residue objects (green circle) in the gly-can moiety to the Ser residue in the peptide moiety. (B) The two residues that connect the moieties are shown in atomic detail. The residue link connecting these residues embodies an atom link connecting C1 of the α-D-Manp residue to O3 of the Ser residue. (C) GLYDE-II representation of the glycopeptide. The moiety link indicates that the “O-Man-dimer” (Fig. 4) is covalently attached to the “O-Man-peptide”, whose structure is specified in “http://someURI”. The enclosed residue link indicates that the α-D-Manp residue (partid = “1” in the GLYDE-II specification of the “O-Man-dimer”) is linked to the L-Ser residue (partid = “5” in the GLYDE-II specification of the “O-Man-peptide”). The further enclosed atom link indicates that the bound atom C1 (partid = “C1” in the referenced GLYDE-II specification of α-D-Manp residue objects in the “O-Man-dimer” moiety) is covalently attached to the bound atom O3 (par-tid = “O3” in the referenced GLYDE-II specification of the L-Ser residue objects in the “O-Man-peptide”).
Molecular geometry in GLYDE-II
Several different systematic nomenclatures (R/S, α/β, D/L, etc.) have been developed to specify molecular stereochemistry. However, assigning the stereochemistry of an asymmetric atom (or generating an explicit geometric interpretation of such an assignment) using a systematic nomenclature can be computationally challenging. GLYDE addresses this issue by allowing the stereochemistry of a molecule to be specified using two different methods (1) explicitly defining the three-dimensional geometry of each its parts using Cartesian coordinates; (2) parsing the ref attribute of each residue component of a molecule. Method (1) provides a unified way to specify the configurational geometry of all the atoms in a complex biopolymer without having to parse a systematic stereochemical nomenclature (or combination of different nomenclatures). Method (2) is much more efficient in contexts where an atomistic representation is not required. In the case of a monosaccharide residue, the value of the ref attribute is a string corresponding to its GlycoCT-based representation, which was developed as part of the EUROCarbDB initiative. This allows, for example, α-linked glucose residues to be distinguished from β-linked glucose residues in a trivial manner without any need to generate and compare atomistic representations, which are rarely required for logical inference. Thus, GLYDE fully supports both atomistic and abstract representations of molecular geometry, and notably, provides a logical and consistent framework for both representations at different levels of granularity. The conventions defined in GLYDE facilitate the implementation of algorithms to interconvert the GLYDE representation with fully atomistic representations (see Supporting information).
Classification of parts
Judicious selection of the appropriate granularity in specifying or parsing a GLYDE representation will facilitate the development of algorithms to identify correlations between the structure of a biomolecule and its biological function or physical properties. Therefore, parts at each level of granularity are distinguished using chemical and biosynthetic criteria. For example, several different moiety types are defined, including glycan moiety, peptide moiety and lipid moiety. A glycan moiety is composed primarily of monosaccharide residues (typically connected by glycosidic bonds) while a peptide moiety is composed primarily of amino acid residues (typically connected by amide bonds). Although these semantics are not required to completely define the chemical structure of a biopolymer, they are useful for identifying chemical and computational contexts in order to facilitate the use of the structural information for data processing and knowledge discovery.
GLYDE-II rules
The GLYDE syntax, described above, is not in itself sufficient to ensure that structural representations of glycoconjugates will be consistent. Additional rules are required to standardize parameters such as the part id attribute of a bound atom and the id of an archetype molecule (Supporting information). As mentioned above, the id attribute of a monosaccharide molecule corresponds to the GlycoCT-based representation of that molecule. This provides a convenient way to compare monosaccharide residue compositions or stereochemistry without generating a fully atomistic representation of the glycoconjugate. This is possible since the GlycoCT namespace for monosaccharides is machine-readable, providing unique, unambiguous names that encode the salient chemical properties of these molecules. A large collection of GlycoCT representations are maintained and curated by MonosaccharideDB (http://www.monosaccharidedb.org/). By rule, the ref attribute of a GLYDE monosaccharide residue is a uniform resource identifier (URI) consisting of the following three parts: (1) the uniform resource locator (URL) of MonosaccharideDB (“http://www.monosaccharideDB.org/”); (2) a series of characters corresponding to an http GET request (“GLYDE-II-1.2.jsp?G=“) (3) the GlycoCT string representing the molecule (e.g. “b-dglc-HEX-1:5”).
Applications
The GLYDE-II format is currently supported in the widely used MS annotation software GlycoWorkbench as an option to export and import glycan structures. It is also being used as the standard format for common web service interfaces that are being developed to enable exchange of carbohydrate structural information among databases and software applications. These include the GlycO ontology at the CCRC, the database of the Consortium for Functional Glycomics (CFG), the Kyoto Encyclopedia of Genes and Genomes (KEGG) via the RINGS portal at Soka University (http://rings.t.soka.ac.jp/), UniCarb-DB, EUROCarbDB, the meta-database of carbohydrate structure GlycomeDB and the recently initiated GlyTouCan glycan structure registry. Support and utilization of GLYDE-II is a core feature of our Qrator software, which provides an intuitive interface that facilitates the human curation of glycan structures (Eavenson et al., 2015). We continue to develop software that fully supports the GLYDE-II format.
Supplementary Material
Acknowledgments
This work is supported by the NIH/NIGMS-funded National Center for Glycomics and Glycoproteomics (8P41GM103490).
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.pisc.2016.05.013.
References
- Aoki-Kinoshita K, Agravat S, Aoki NP, Arpinar S, Cummings RD, Fujita A, Fujita N, Hart GM, Haslam SM, Kawasaki T, Matsubara M, Moreman K, Okuda W, Pierce S, Ranzinger M, Shikanai R, Solovieva T, Suzuki E, Tsuchiya Y, Yamada S, York I, Zaia WS, Narimatsu J. GlyTouCan 1.0—the international glycan structure repository. Nucleic Acids Res. 2016;44:D1237–D1242. doi: 10.1093/nar/gkv1041. http://dx.doi.org/10.1093/nar/gkv1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banin E, Neuberger Y, Altshuler Y, Halevi A, Inbar O, Dotan N, Dukler A. A novel LinearCode nomenclature for complex carbohydrates. Trends Glycosci Glycotechnol. 2002;14:127–137. http://dx.doi.org/10.4052/tigg.14.127. [Google Scholar]
- Bohne-Lang A, Lang E, Förster T, von der Lieth CW. LINUCS: linear notation for unique description of carbohydrate sequences. Carbohydr Res. 2001;336:1–11. doi: 10.1016/s0008-6215(01)00230-0. http://dx.doi.org/10.1016/s0008-6215(01)00230-0. [DOI] [PubMed] [Google Scholar]
- Casati R, Varzi AC. Parts and Places: The Structures of Spatial Representations. MIT Press; 1999. [Google Scholar]
- Cummings RD, Pierce JM. The challenge and promise of glycomics. Chem Biol. 2014;21:1–15. doi: 10.1016/j.chembiol.2013.12.010. http://dx.doi.org/10.1016/j.chembiol.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eavenson M, Kochut KJ, Miller JA, Ranzinger R, Tiemeyer M, Aoki K, York WS. Qrator: a web-based curation tool for glycan structures. Glycobiology. 2015;25:66–73. doi: 10.1093/glycob/cwu090. http://dx.doi.org/10.1093/glycob/cwu090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egorova KS, Toukach PV. Expansion of coverage of carbohydrate structure database (CSDB) Carbohydr Res. 2014;389:112–114. doi: 10.1016/j.carres.2013.10.009. http://dx.doi.org/10.1016/j.carres.2013.10.009. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Goto S, Kawano S, Aoki-Kinoshita KF, Ueda N, Hamajima M, Kawasaki T, Kanehisa M. KEGG as a glycome informatics resource. Glycobiology. 2006;16:63R–70R. doi: 10.1093/glycob/cwj010. http://dx.doi.org/10.1093/glycob/cwj010. [DOI] [PubMed] [Google Scholar]
- Herget S, Ranzinger R, Maass K, von der Lieth CW. GlycoCT—a unifying sequence format for carbohydrates. Carbohydr Res. 2008;343:2162–2171. doi: 10.1016/j.carres.2008.03.011. http://dx.doi.org/10.1016/j.carres.2008.03.011. [DOI] [PubMed] [Google Scholar]
- Kikuchi N, Kameyama A, Nakaya S, Ito H, Sato T, Shikanai T, Takahashi Y, Narimatsu H. The carbohydrate sequence markup language (CabosML): an XML description of carbohydrate structures. Bioinformatics. 2005;21:1717–1718. doi: 10.1093/bioinformatics/bti152. http://dx.doi.org/10.1093/bioinformatics/bti152. [DOI] [PubMed] [Google Scholar]
- Kunz H. Emil Fischer—unequalled classicist, master of organic chemistry research, and inspired trailblazer of biological chemistry. Angew Chem Int Ed. 2002;41:4439–4451. doi: 10.1002/1521-3773(20021202)41:23<4439::AID-ANIE4439>3.0.CO;2-6. http://dx.doi.org/10.1002/1521-3773(20021202)41:23<4439::aid-anie4439>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Packer NH, von der Lieth CW, Aoki-Kinoshita KF, Lebrilla CB, Paulson JC, Raman R, Rudd P, Sasisekharan R, Taniguchi N, York WS. Frontiers in glycomics: bioinformatics and biomarkers in disease. Proteomics; An NIH white paper prepared from discussions by the focus groups at a workshop on the NIH campus; Bethesda MD. September 11–13, 2006; 2008. pp. 8–20. [DOI] [PubMed] [Google Scholar]
- Ranzinger R, Herget S, von der Lieth CW, Frank M. GlycomeDB—a unified database for carbohydrate structures. Nucleic Acids Res. 2011;39:D373–D376. doi: 10.1093/nar/gkq1014. http://dx.doi.org/10.1093/nar/gkq1014 (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo SS, Thomas C, Sheth A, Henson C, York WS. GLYDE—an expressive XML standard for the representation of glycan structure. Carbohydr Res. 2005;340:2802–2807. doi: 10.1016/j.carres.2005.09.019. http://dx.doi.org/10.1016/j.carres.2005.09.019. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Aoki-Kinoshita KF, Kotera M, Sawaki H, Tsuchiya S, Fujita N, Shikanai T, Kato M, Kawano S, Yamada I, Narimatsu H. WURCS: the Web3 unique representation of carbohydrate structures. J Chem Inf Model. 2014;54:1558–1566. doi: 10.1021/ci400571e. http://dx.doi.org/10.1021/ci400571e. [DOI] [PubMed] [Google Scholar]
- Varki A, Sharon N. Historical background and overview. In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of Glycobiology. 2. Cold Spring Harbor Laboratory Press; Cold Spring Harbor (NY): 2009. [PubMed] [Google Scholar]
- Walt DA, Aoki-Kinoshita AF, Bendiak B, Bertozzi CR, Boons G-J, Darvill A, Hart G, Kiessling LL, Lowe J, Moon RJ, Paulson JC, Sasisekharan R, Varki AP, Wong C-H, Bowman K, Friedman D, Siddiqui S, Yancey R. Transforming Glycoscience: A Roadmap for the Future. The National Academies Press; 2012. [PubMed] [Google Scholar]
- York WS, Agravat S, Aoki-Kinoshita KF, McBride R, Campbell MP, Costello CE, Dell A, Feizi T, Haslam SM, Karlsson N, Khoo KH, Kolarich D, Liu Y, Novotny M, Packer NH, Paulson JC, Rapp E, Ranzinger R, Rudd PM, Smith DF, Struwe WB, Tiemeyer M, Wells L, Zaia J, Kettner C. MIRAGE—the minimum information required for a glycomics experiment. Glycobiology. 2014;24:402–406. doi: 10.1093/glycob/cwu018. http://dx.doi.org/10.1093/glycob/cwu018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.