Abstract
IMPORTANCE
Guidelines for cancer genetic testing based on family history may miss clinically actionable genetic changes with established implications for cancer screening or prevention.
OBJECTIVE
To determine the proportion and potential clinical implications of inherited variants detected using simultaneous sequencing of the tumor and normal tissue (“tumor-normal sequencing”) compared with genetic test results based on current guidelines.
DESIGN, SETTING, AND PARTICIPANTS
From January 2014 until May 2016 at Memorial Sloan Kettering Cancer Center, 10 336 patients consented to tumor DNA sequencing. Since May 2015, 1040 of these patients with advanced cancer were referred by their oncologists for germline analysis of 76 cancer predisposition genes. Patients with clinically actionable inherited mutations whose genetic test results would not have been predicted by published decision rules were identified. Follow-up for potential clinical implications of mutation detection was through May 2017.
EXPOSURE
Tumor and germline sequencing compared with the predicted yield of targeted germline sequencing based on clinical guidelines.
MAIN OUTCOMES AND MEASURES
Proportion of clinically actionable germline mutations detected by universal tumor-normal sequencing that would not have been detected by guideline-directed testing.
RESULTS
Of 1040 patients, the median age was 58 years (interquartile range, 50.5–66 years), 65.3% were male, and 81.3% had stage IV disease at the time of genomic analysis, with prostate, renal, pancreatic, breast, and colon cancer as the most common diagnoses. Of the 1040 patients, 182 (17.5%; 95%CI, 15.3%–19.9%) had clinically actionable mutations conferring cancer susceptibility, including 149 with moderate- to high-penetrance mutations; 101 patients tested (9.7%; 95%CI, 8.1%–11.7%) would not have had these mutations detected using clinical guidelines, including 65 with moderate- to high-penetrance mutations. Frequency of inherited mutations was related to case mix, stage, and founder mutations. Germline findings led to discussion or initiation of change to targeted therapy in 38 patients tested (3.7%) and predictive testing in the families of 13 individuals (1.3%), including 6 for whom genetic evaluation would not have been initiated by guideline-based testing.
CONCLUSIONS AND RELEVANCE
In this referral population with selected advanced cancers, universal sequencing of a broad panel of cancer-related genes in paired germline and tumor DNA samples was associated with increased detection of individuals with potentially clinically significant heritable mutations over the predicted yield of targeted germline testing based on current clinical guidelines. Knowledge of these additional mutations can help guide therapeutic and preventive interventions, but whether all of these interventions would improve outcomes for patients with cancer or their family members requires further study.
TRIAL REGISTRATION
clinicaltrials.gov Identifier: NCT01775072
In addition to guiding therapy, simultaneous DNA sequence analysis of tumor-normal pairs (“tumor-normal sequencing”) reveals inherited cancer predisposition mutations in 3% to 12.6% of pediatric and adult patients with cancer.1–7 It remains unknown, however, how many inherited mutations would be detected by multigene tumor-normal analysis compared with a traditional family history–based approach to genetic counseling and testing, and what the clinical implications of these findings would be. Selection for genetic testing is traditionally based on pathologic features of the tumor, age at diagnosis, family history of cancer, and other factors represented in clinical practice guidelines.8–10 Studies have not determined whether inherited mutations found by tumor-normal sequencing would have been detected by traditional approaches to selection for genetic testing.1–5
This study presents the results of analyses of inherited (“germline”) DNA performed in a prospective analysis of patients with advanced cancer tested by a targeted tumor-normal sequencing panel as previously described.11,12 The goals of the study were to determine the incremental proportion of clinically actionable mutations detected by concurrent germline analysis in patients with cancer undergoing universal tumor profiling compared with selective germline testing based on existing practice guidelines and to assess the association of identified mutations with therapeutic management and targeted cancer prevention in family members.
Methods
Patient Cohort
The cohort comprised patients with advanced cancer at Memorial Sloan Kettering Cancer Center undergoing tumor and normal DNA sequencing using MSK-IMPACT (Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets), a 410-gene panel.1,11,12 From May 2015, in the context of an institutional review board–approved protocol, patients with selected tumor types were prospectively offered secondary germline analysis after consenting to tumor genetic analysis. Germline analysis included 76 genes on the MSK-IMPACT panel associated with hereditary cancer predisposition, including all cancer-predisposing genes identified in the American College of Medical Genetics and Genomics guidelines.13–15 Nine mutation carriers were included in a prior study,1 and 23 and 51 mutation carriers were included in recent series of prostate cancer,16,17 although clinical annotation including family history and treatment information was not available for those articles.
Ascertainment, Consent, Sequencing, Variant Calling, and Results Reporting
Through their physicians, patients were offered participation in a research study, using a video consent aid explaining risks and benefits of testing for inherited mutations (germline testing). Eligibility was restricted to those who also consented to tumor sequencing, with emphasis on patients with advanced (stage III-IV) disease; however, physicians had discretion for patient referral. Genetic testing reports were issued to the medical record, and individuals with pathogenic or likely pathogenic variants (henceforth referred to as “pathogenic variants”) were invited for genetic counseling where at-risk family members were identified (eAppendix 1 in Supplement 1). Tumor DNA and nontumor DNA were sequenced and variants were reported as described previously11 (eAppendix 2 in the Supplement 1). Variants were interpreted based on American College of Medical Genetics and Genomics criteria18 by a clinical molecular geneticist or molecular pathologist; variants of unknown significance (eTable in Supplement 2) were not included in clinical reports. Loss of heterozygosity (LOH), defined as loss of the normal allele in the tumor at the locus of the inherited mutation, was assessed by the FACETS algorithm as published previously19; hypermutated status was defined as 20 or more mutations; and microsatellite instability high (MSI-H) was defined as more than 10% of loci on the MSK-IMPACT panel demonstrating microsatellite instability.20
In this study, not all pathogenic mutations (associated with disease causation) were considered clinically actionable. Clinical actionability of pathogenic variants was defined by evidence for their utility in cancer prevention (eAppendix 3 in Supplement 1) and/or potential utility as therapeutic targets. For this study, all germline mutations of established low, moderate, or high risk (penetrance) were considered clinically actionable8–10,16,21,22 (eTable 1 in Supplement 1). Mutations were classified as having high penetrance (relative risk >4), moderate penetrance (relative risk 2–4), or low penetrance (relative risk <2) as well as being recessive or of uncertain clinical actionability based on known disease-associated risks and current modeling.8–10,16,21
Comparison of Conventional Family History–Based and Agnostic Testing for Cancer Predisposition Syndromes
Detailed clinical annotation, including self-reported religion and race, was abstracted for each patient record. Religion and race information was collected to analyze genetic effects of population stratification. Ashkenazi ancestry was determined by self-report of religious preference, via a list of choices, at the time of registration or by specific report of Eastern European Jewish background of relatives at the time of genetic counseling. Race was self-reported by fixed categories at the time of registration. Three-generation family history information was assessed at the time of results communication or from records at the time of genetic testing. Published guidelines and syndrome-specific genetic testing algorithms8–10,23–25 were used to determine which genetic tests would be indicated based on tumor histologic features, bilaterality, multiple metachronous cancers, age at onset of cancer, family history of cancer (including age at which relatives were affected), and self-reported Ashkenazi ancestry. Where indicated, multigene panels (eTable 2 in Supplement 1) were considered standard of care and were applied using decision rules based on published guidelines (eAppendix 4 in Supplement 1). A pathogenic variant in secondary analysis was considered incremental if it would not have been detected by testing that would have been ordered based on application of these decision rules, with additional decision rules for cases harboring 2 variants (eAppendix 5 in Supplement 1).
Adjustment for Founder Mutations and DNA Repair Genes
To adjust for effects of founder mutations, a set of Ashkenazi and European founder mutations in BRCA1/2, APC, MSH2, MSH6, CHEK2, or MUTYH were included in the overall analysis but also identified so as to allow subset analysis (eBox in Supplement 1).
Mutations in ATM, BAP1, BARD1, BRCA1/2, BRIP1, CHEK2, FAM175A, MLH1, MSH2, MSH6, NBN, PALB2, PMS2, and RAD51D recently associated with advanced prostate cancer16 were coded for separate analysis of patients with advanced prostate cancer, and to assess patients potentially amenable to targeted therapies.
Statistical Analysis
Variant frequencies in cases were compared with allele frequencies in noncancer controls from public databases and stratified by European and Ashkenazi subsets.26–30 Allele frequencies were compared by Fisher exact 2-sided binomial test in R version 3.3 software (R Foundation) using RStudio version 0.99.903 (RStudio). To estimate findings resulting from a different case mix, tumor type–specific rates of pathogenic variants were multiplied by cancer rates in the general population31 as well as rates of discordant variants observed in a separate ascertainment1 (eAppendix 6 in Supplement 1). Where proportions are presented, 95% confidence intervals were derived. Clinical variables were compared with regard to genetically defined subsets, mutational load as measured by number of somatic mutations, and time from diagnosis to tumor-normal analysis for metastatic disease, using 2-sample t test for independent samples, with 2-tailed P values significant at P < .05. Rates of incremental findings were compared between subsets by Fisher exact test, and proportions of germline findings were compared by stage of disease using χ2 test.
Results
Cohort Characteristics
As part of an institutional review board–approved protocol, from January 1, 2014, until May 31, 2016, 10 336 patients consented to genetic analysis of their tumors. The distribution of cancer diagnoses, sex, age, and stage of disease in the 1040 patients who consented to secondary germline testing from May 1, 2015, through May 31, 2016, is shown in Table 1. Of the 1040 patients, 65.3% were male and 34.7% female. The median age of those who consented to genetic analysis of their tumors was 58 years (interquartile range [IQR], 47–67 years), and the median age of those who consented to secondary germline testing was 58 years (IQR, 50.5–66 years). The median age was similar for all tumor subsets except prostate cancer, for which patients consenting to germline analysis were slightly younger than those who underwent tumor variant calling only (median age, 61 [IQR, 55–67] vs 63 [IQR, 56–68] years, respectively; P = .04). Of the 1040 patients, the proportions with stage 0, I, II, III, and IV disease at the time of genomic analysis were 0.3%, 3.3%, 7.9%, 7.2%, and 81.3%, respectively. Self-identification of Jewish ancestry was more common in those consenting to secondary germline analysis than in those undergoing tumor variant calling only (26.9% vs 18.1%, respectively).
Table 1.
Characteristics of the Study Cohort
| Characteristic | No. (%) | ||
|---|---|---|---|
| Total (N = 1040) | Results of 76-Gene Germline Sequencing | ||
| Tested Negative (n = 835) | Tested Positive (n = 205) | ||
| Sex | |||
| Male | 679 (65.3) | 548 (65.6) | 131 (63.9) |
| Female | 361 (34.7) | 287 (34.4) | 74 (36.1) |
| Age at diagnosis, median (IQR), y | 58 (50.5–66) | 58 (51–67) | 58 (49–65) |
| Tumor typea | |||
| Adrenocortical | 7 (0.7) | 6 (0.7) | 1 (0.5) |
| Ampullary | 3 (0.3) | 2 (0.2) | 1 (0.5) |
| Biliary | 27 (2.6) | 21 (2.5) | 6 (2.9) |
| Bladder | 16 (1.5) | 7 (0.8) | 9 (4.4) |
| Breast | 101 (9.7) | 84 (10.0) | 17 (8.3) |
| Colorectal | 65 (6.2) | 57 (6.8) | 8 (3.9) |
| Endometrial | 25 (2.4) | 21 (2.5) | 4 (1.9) |
| Esophagogastric | 34 (3.3) | 28 (3.4) | 6 (2.9) |
| Melanoma | 3 (0.3) | 2 (0.2) | 1 (0.5) |
| Mesothelioma | 4 (0.4) | 3 (0.4) | 1 (0.5) |
| Paraganglioma | 2 (0.2) | 1 (0.1) | 1 (0.5) |
| Non–small cell lung | 2 (0.2) | 0 | 2 (1.0) |
| Ovarian | 19 (1.8) | 13 (1.6) | 6 (2.9) |
| Pancreatic | 176 (16.9) | 132 (15.8) | 44 (21.5) |
| Prostate | 362 (34.8) | 291 (34.9) | 71 (34.6) |
| Renal cell | 140 (13.5) | 117 (14.0) | 23 (11.2) |
| Small-bowel | 5 (0.5) | 3 (0.4) | 2 (1.0) |
| Unknown primary | 10 (1.0) | 8 (1.0) | 2 (1.0) |
| Other | 39 (3.7) | 39 (4.7) | 0 |
| Stagea,b | |||
| 0 | 3 (0.3) | 3 (0.4) | 0 |
| I | 34 (3.3) | 31 (3.7) | 3 (1.5) |
| II | 82 (7.9) | 74 (8.8) | 8 (3.9) |
| III | 75 (7.2) | 70 (8.4) | 5 (2.4) |
| IV | 846 (81.3) | 657 (78.7) | 189 (92.2) |
Abbreviation: IQR, interquartile range.
The number of patients with each diagnosis and stage are listed according to mutation status and corresponding to organ sites listed under tumor types.
Stage was determined at the time of genomic testing.
Variants Detected
Of the 1040 patients undergoing secondary germline analysis, 205 patients (19.7%) harbored pathogenic variants conferring cancer predisposition. Of the 205 patients with pathogenic variants, 182 carried clinically actionable mutations of high (n = 97), moderate (n = 52), or low (n = 33) penetrance, 8 were carriers of variants associated with recessive syndromes, and 15 carried variants in genes of unproven clinical actionability. Table 2 shows the distribution of 220 variants in these 205 patients, listed by penetrance class and by tumor type. For tumor types with more than 10 cases tested, the incidence of patients with inherited pathogenic variants ranged from 56.3% (9 of 16 patients) for bladder cancers (including urothelial carcinomas) to 25.0% (44 of 176 patients) for pancreatic cancer, 19.6% (71 of 362 patients) for prostate cancer, 16.4% (23 of 140 patients) for renal cancer, and 9.2% (8 of 65 patients) for colon cancer. Each patient had an average of 1.8 variants of uncertain significance in the 76 genes tested, with 833 of 1040 patients (80%) having at least 1 variant of uncertain significance (eTable in Supplement 2). Of the 1040 patients, 15 had more than 1 variant (eTable in Supplement 3).
Table 2.
Distribution of 220 Pathogenic or Likely Pathogenic Variants in 205 Patients With Advanced Cancer Consenting to Tumor-Normal Genomic Analysis
| Gene Mutated in Tumor Type | Patients With Advanced Cancer With Pathogenic Variants | Total Pathogenic Variants per Gene, No. | |
|---|---|---|---|
| Total per Gene, No. | By Tumor Type (No. of Pathogenic Variants Detected by Gene and Tumor Type) | ||
| High Penetrance | |||
| BAP1 | 3 | Mesothelioma (1), renal cell (2) | 3 |
| BRCA1 | 14 | Bladder (1), breast (2), esophagogastric (1), ovarian (2), pancreatic (6), prostate (2) | 14 |
| BRCA2 | 45 | Ampullary (1), biliary (3), breast (1), esophagogastric (1), melanoma (1), ovarian (3), pancreatic (11), prostate (24) | 45 |
| CDH1 | 2 | Esophagogastric (2) | 2 |
| CDKN2A | 3 | Pancreatic (3) | 3 |
| FH | 3 | Renal cell (3) | 3 |
| FLCN | 1 | Prostate (1) | 1 |
| MEN1 | 1 | Non–small cell lung (1) | 1 |
| MLH1 | 1 | Bladder (1) | 1 |
| MSH2 | 7 | Bladder (4), colorectal (2), prostate (1) | 7 |
| MSH6 | 4 | Bladder (2), prostate (1), renal cell (1) | 4 |
| PALB2 | 5 | Biliary (1), ovarian (1), pancreatic (1), prostate (1), renal cell (1) | 5 |
| PMS2 | 2 | Prostate (1), small-bowel (1) | 2 |
| SDHA | 2 | Breast (1), renal cell (1) | 2 |
| SDHB | 2 | Paraganglioma (1), renal cell (1) | 2 |
| VHL | 2 | Renal cell (2) | 2 |
| Moderate Penetrance | |||
| ATM | 15 | Breast (1), colorectal (1), esophagogastric (1), pancreatic (5), prostate (7) | 15 |
| BRIP1 | 4 | Breast (1), colorectal (1), prostate (2) | 4 |
| CHEK2 | 28a | Breast (9), colorectal (1), endometrial (2), pancreatic (7), prostate (8), renal cell (5) | 32 |
| MITF | 2 | Prostate (2) | 2 |
| NBN | 2 | Prostate (2) | 2 |
| RAD51D | 1 | Non–small cell lung (1) | 1 |
| Low Penetrance | |||
| APC | 19a | Biliary (1), bladder (1), breast (1), colorectal (3), ovarian (1), pancreatic (7), prostate (7), renal cell (1), small-bowel (1), unknown primary (1) | 24 |
| MUTYH | 14a | Adrenocortical (1), biliary (1), breast (1), colorectal (1), pancreatic (3), prostate (5), renal cell (3), unknown primary (1) | 16 |
| Recessive Alleles | |||
| FHb | 3a | Pancreatic (1), prostate (4) | 5 |
| RECQL4 | 5a | Breast (1), colorectal (1), endometrial (1), esophagogastric (1), prostate (1), renal cell (1) | 6 |
| Uncertain Clinical Actionability | |||
| BARD1 | 3 | Pancreatic (1), prostate (1), renal cell (1) | 3 |
| CHEK2c | 11 | Bladder (1), breast (1), endometrial (1), pancreatic (2), prostate (5), renal cell (1) | 11 |
| FAM175A | 0a | Pancreatic (1) | 1 |
| RAD50 | 1 | Pancreatic (1) | 1 |
| Total | |||
| Pathogenic variants by tumor type | 205 | Adrenocortical (1), ampullary (1), biliary (6), bladder (10), breast (19), colorectal (10), endometrial (4), esophagogastric (6), melanoma (1), mesothelioma (1), non–small cell lung (2), ovarian (7), pancreatic (49), paraganglioma (1), prostate (75), renal cell (23), small-bowel (2), unknown primary (2) | 220 |
For the determination of the total number of patients with advanced cancer with pathogenic variants per gene, patients with 2 pathogenic variants were classified according to the higher-penetrance variant (eAppendix 5 in Supplement 1), resulting in different values compared with the total number of pathogenic variants by gene.
Refers to the FH c. 1431_1433dupAAA (p. Lys477dup) mutation considered as a recessive allele.
Refers to the CHEK2 c.470T>C (p.Ile157Thr) mutation classified as a variant of uncertain clinical actionability.
Pathogenic variants of genes involved in DNA repair pathways16 were observed in 49 of 362 patients (13.5%) with prostate cancer, all of whom had advanced disease, but also in 87 of 678 patients (12.8%) with cancers other than prostate cancer. Of the 220 mutations detected in 205 patients, 83 mutations (37.7%; in 79 patients) were known founder mutations (including Ashkenazi founder mutations, and European founder mutations in MUTYH and the CHEK2 c.1100delC). Cases with founder mutations represented 75.8% of low-penetrance, 40.4% of moderate-penetrance, and 34% of high-penetrance pathogenic alleles (eTable 3 in Supplement 1).
Comparisons With Phenotype-Directed Approaches
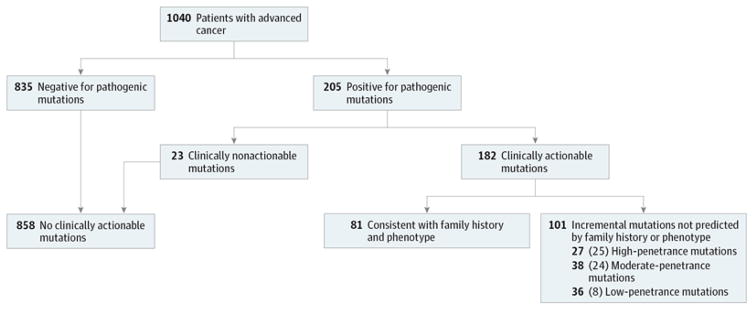
A total of 101 cases had incremental clinically actionable findings that would not have been detected by phenotype-directed testing using current clinical criteria, representing 9.7%(95%CI, 8.1%–11.7%) of the 1040 cases overall and 55.5% (95% CI, 48.2%–62.5%) of the 182 patients with clinically actionable mutations (Figure). Had the case mix conformed to population cancer incidence rates,31 the 55.5% proportion of incremental findings would have been 49.1%. Of the 101 patients with incremental clinically actionable findings, 27, 38, and 36 patients carried high-, moderate-, or low-penetrance mutations, respectively (eTable 4 in Supplement 1). Thus, of the 101 incremental findings, 65 were moderate- or high-penetrance mutations. There was no difference in the proportion of patients with incremental findings in the 85.2% of probands with family history assessed by genetic counselors or the 14.8% assessed from physician records.
Figure. Clinical Actionability, Concordance With Family History and Phenotype, Penetrance, and Founder Mutations in 1040 Patients Undergoing Sequencing of Germline and Tumor DNA.
Representation of the 1040 cases that carried clinically actionable pathogenic or presumed pathogenic variants, comprising 182 cases broken into subsets of no incremental cases (in which mutations would have been detected using genetic testing guidelines based on phenotype and family history) and incremental cases (in which mutations would not have been detected using guideline-based approaches). The incremental cases are categorized by high-, moderate-, or low-penetrance mutations. Numbers in parentheses indicate the numbers of patients in that category without Ashkenazi Jewish or European founder mutations.
Had all individuals been screened for population-specific founder mutations in addition to guideline-directed testing, the proportion of incremental clinically actionable findings would have declined to 57 of 182 patients (31.3%; 95%CI, 25.0%–38.4%). Had patients with prostate cancer also been screened with a panel of DNA repair genes, the resulting proportion of findings considered incremental would be 35 of 182 patients (19.2%; 95%CI, 14.2%–25.6%) or 3.4%(95%CI, 2.4%–4.7%) of the 1040 cases overall (eTable 4 in Supplement 1). The highest proportion of actionable findings that would not have been detected based on clinical guidelines were observed in biliary, prostate, colorectal, renal, and pancreatic tumors (eTable 4 in Supplement 1). Table 3 shows all cases with incremental actionable findings, excluding known founder mutations, which would not have been detected using guideline directed approaches for high-penetrance mutations (BRCA2, CDKN2A, PALB2, VHL, SDHA, MSH2, MSH6, and BAP1), moderate-penetrance mutations (CHEK2, ATM, MITF, BRIP1, and RAD51D), and low-penetrance mutations (APC, MUTYH). Twenty-two percent of germline BRCA1/2 mutations (13 of 59 mutations) and 42.8% of mismatch repair gene mutations (6 of 14 mutations), including founder mutations, were seen in patients who would not have been referred for testing using existing guidelines.
Table 3.
Cases With Germline Actionable Incremental Nonfounder Variants Not Predicted by Phenotype-Based Guidelines, Arranged According to Penetrance and Including Tumor Status of the Gene With Inherited Mutationa
| Case No. | Tumor Typeb | Gene With Inherited Mutation | Tumor Status of Gene With Inherited Mutationc |
|---|---|---|---|
| High Penetrance (n = 26 Cases) | |||
| R56 | Biliary | PALB2 | WT |
| R155d | Biliary | BRCA2 | WT |
| R80 | Bladder | MSH6 | Second mutation (h+, m−) |
| R102 | Bladder | MSH2 | WT (h+, m+) |
| R116 | Bladder | MSH6 | LOH (h+, m−) |
| APC | WT | ||
| R197 | Breast | SDHA | LOH |
| R26d | Pancreatic | BRCA2 | LOH |
| R101 | Pancreatic | CDKN2A | WT |
| R106 | Pancreatic | BRCA2 | WT |
| R190 | Pancreatic | BRCA2 | ND |
| R50 | Prostate | BRCA1e | ND |
| R53 | Prostate | BRCA2 | LOH |
| R79d | Prostate | BRCA2e | LOH |
| R111 | Prostate | MSH2e | LOH (h+, m+) |
| FHe | WT | ||
| R132 | Prostate | PALB2e | ND |
| R135f | Prostate | MSH6e | WT (h−, m−) |
| R151d | Prostate | BRCA2e | Second mutation |
| R166d | Prostate | PMS2e | LOH (h+, m+) |
| R182 | Prostate | BRCA2e | WT |
| R192 | Prostate | BRCA2e | WT |
| R205 | Prostate | BRCA2e | Second mutation |
| R6 | Renal cell | PALB2 | WT |
| R117 | Renal cell | VHL | LOH |
| R180 | Renal cell | SDHA | CN-LOH |
| R134d | Mesothelioma | BAP1 | LOH |
| R164d | Ampullary | BRCA2 | LOH |
| Moderate Penetrance (n = 24 Cases) | |||
| R136 | Colorectal | ATM | WT |
| APCg | WT | ||
| R69 | Colorectal | BRIP1 | WT |
| R201 | Esophagogastric | ATM | ND |
| R38 | Pancreatic | ATM | LOH |
| R173 | Pancreatic | CHEK2 | WT |
| R188 | Pancreatic | ATM | Second mutation |
| R87 | Prostate | MITF | WT |
| R118 | Prostate | MITF | WT |
| R4 | Prostate | CHEK2e | WT |
| R17d | Prostate | NBNe | WT |
| R18 | Prostate | CHEK2e | WT |
| R31 | Prostate | BRIP1e | LOH |
| R55 | Prostate | BRIP1e | LOH |
| R62 | Prostate | ATMe | WT |
| R66 | Prostate | NBNe | LOH |
| R71 | Prostate | ATMe | Second mutation |
| R108d | Prostate | ATMe | ND |
| R120 | Prostate | ATMe | ND |
| R150d | Prostate | ATMe | ND |
| R195 | Prostate | ATMe | LOH |
| R147d | Renal cell | CHEK2 | ND |
| R189 | Renal cell | CHEK2 | WT |
| R129d | Non–small cell lung | RAD51D | ND |
| R185d | Endometrial | CHEK2 | ND |
| Low Penetrance (n = 8 Cases) | |||
| R20d | Colorectal | MUTYH | ND |
| R91 | Pancreatic | MUTYH | WT |
| R59 | Prostate | MUTYH | LOH |
| R127 | Prostate | MUTYH | ND |
| R133 | Prostate | MUTYH | WT |
| R128 | Renal cell | MUTYH | LOH |
| R82 | Unknown primary | MUTYH | WT |
| R149 | Adrenocortical | MUTYH | LOH |
Abbreviations: CN, copy neutral; LOH, loss of heterozygosity; ND, not determined; WT, wild type.
The breakdown of cases by penetrance type (26 high-penetrance, 24 moderate-penetrance, and 8 low-penetrance cases) corresponds to those listed at the bottom of the Figure in parentheses with the exception of the single case noted in footnote f.
Represents the sample for which tumor profiling had been performed.
The following indicate the tumor status: LOH, status of the second allele in the tumor of the gene mutated in the germline; WT, that the second allele was not mutated; second mutation, the second allele was mutated in the tumor in the same gene demonstrating a germline variant; CN-LOH, LOH in the tumor not accompanied by changes of copy number in the chromosomal region around the mutation; ND, cases in which zygosity was not determined; h+, hypermutated tumor profile (≥20 mutations); h−, not hypermutated tumor profile (<20 mutations); m+, microsatellite instability high (MSI sensor positive; ≥10% of loci on the sequencing panel); and m−, microsatellite instability negative (MSI sensor negative; <10% of loci on the sequencing panel).
Patients had additional cancer diagnoses.
Cases with DNA repair genes associated with advanced prostate cancer in the article by Pritchard et al.16
The MSH6 variant in this case is a founder variant (see eTable in Supplement 3) and was not counted toward the total of high-penetrance cases with nonfounder variants, but it is included here to illustrate one of the incremental cases with hypermutation status and MSI sensor data.
Refers to a second nonincremental variant.
Clinical Implications of Results Transmitted
At a median time to transmission of results to patients after testing of 1 month (range, 0–16 months), as of May 2017, germline results have been communicated to 193 of 205 patients (94.1%) with pathogenic variants detected and 175 of 182 patients (96.2%) with clinically actionable findings. For 29 patients (probands) found to have actionable mutations, genetic testing had been offered to relatives; for 13 of these families, the index patient’s genetic findings would not have been discovered through guideline-based genetic evaluation. In half (15 of 29) of the families tested, the mutation has been detected in at least 1 relative; for 20 relatives with mutations, increased surveillance or risk-reducing surgical procedures have been recommended, with oophorectomy documented in a daughter of a proband with advanced prostate cancer. Of the 182 probands with actionable findings, 132 had mutations in DNA repair genes, resulting in discussion or initiation of a change to US Food and Drug Administration–approved or off-label use of targeted therapy in 38 patients. Of these 38 patients, 11 received treatment with poly adenosine diphosphate–ribose polymerase (PARP) inhibitors and/or platinum-based chemotherapy as of May 2017, and such treatments were discussed or in planning in the remaining 27 patients (Table 4 and eTable 5 in Supplement 1). In an additional patient in his mid-60s with advanced prostate cancer with more than 20 somatic mutations and a PMS2 germline mutation, immunotherapy was planned but not yet administered. Of the subset of 101 cases with actionable findings not predicted by phenotype, 59 involved DNA repair genes. This resulted in discussion or initiation of targeted therapy in 17 patients, with targeted treatment being administered in 6 of these 17 individuals (Table 4 and eTable 5 in Supplement 1).
Table 4.
Cases for Which Germline Genetic Information Affected Discussion or Implementation of Therapeutic Strategies
| Outcome | Cases, No. | |
|---|---|---|
| With Clinically Actionable Mutations (n = 182) | With Clinically Actionable Mutations Incremental to Phenotype-Based Assessment (n = 101) | |
| Discussion or initiation of targeted therapy | 38 | 17 |
| Targeted therapy administered | 11a | 6a |
| PARP inhibitors | 9b | 4c |
| Platinum-based therapy | 4d | 2e |
Abbreviation: PARP, poly adenosine diphosphate–ribose polymerase.
Not including 1 case of hypermutated prostate cancer with a PMS2 variant with planning of immunotherapy treatment in progress. Patients may have received more than 1 targeted therapy.
Includes 3 cases of prostate cancer with ATM mutations, 2 cases of prostate cancer with BRCA2 mutations, and 1 case of biliary cancer and 3 cases of pancreatic cancer with BRCA1/2 mutations.
Includes 1 case of biliary cancer with BRCA2 mutation, 1 case of pancreatic cancer with BRCA2 mutation, and 2 cases of prostate cancer with ATM mutations.
Includes 3 cases of prostate cancer with ATM and BRCA2 mutations and 1 case of pancreatic cancer with BRCA1 mutation (2 of these cases also received PARP inhibitors).
Includes 1 case of prostate cancer with ATM mutation and 1 case of prostate cancer with BRCA2 mutation.
Comparison of Incremental Findings by Stage of Disease
Among 194 stage 0 to III cases, 16 (8.3%; 95%CI, 5.1%–13.0%) had likely pathogenic germline variants, compared with 189 of 846 patients (22.3%; 95%CI, 19.7%–25.3%) with stage IV disease (P < .001). Among 166 clinically actionable pathogenic variants in stage IV cases, 93 (56.0%; 95% CI, 48.4%–63.4%) were incremental, compared with 8 of 16 incremental actionable variants (50.0%; 95% CI, 28.0%–72.0%) among patients with stage 0 to III disease (P = .64). This analysis yielded similar results by all tumor types except colon cancer, in which Lynch mutations were more common in patients with earlier stage disease (eTable 6 in Supplement 1). The median number of tumor (somatic) mutations was 4 (IQR, 2–6) in 205 cases with pathogenic variants and 3 (IQR, 2–5) in 835 cases without pathogenic variants (P = .31), with a median of 3 mutations observed in both stage 0 through III disease (IQR, 1–5) and stage IV disease (IQR, 2–6) (P = .07). The median time from diagnosis to tumor-normal testing for metastatic disease was 2 years for both the 181 patients with pathogenic variants (IQR, 1–6 years) and the 648 patients without such variants (IQR, 1–5 years) (P = .91).
Analysis of Cohorts of Ashkenazi and Non-Ashkenazi Background
Of 205 patients with pathogenic variants, 192 self-reported ancestry (68 Ashkenazi, 124 non-Ashkenazi). Had guideline directed approaches been augmented with testing for population-specific founder mutations, 5 of 68 patients (7.4%; 95% CI, 3.2%–16.1%) of Ashkenazi ancestry with clinically actionable variants would have had incremental findings not predicted by phenotype or ancestry, compared with 46 of 124 patients (37.1%;95%CI, 29.1%–45.9%) who were not of Ashkenazi ancestry (P < .001) (eTable 7 in Supplement 1).
Comparisons With Public Databases
Among patients of non-Ashkenazi ancestry, a set of CHEK2 mutations, predicted by family history in only a single patient, was over represented in prostate and pancreatic cancers compared with controls from the Exome Aggregation Consortium.26,27 MUTYH mutations were enriched in non-Ashkenazi patients with prostate cancer and not predicted by family history. Among patients of Ashkenazi ancestry, there was enrichment for CHEK2 mutations in patients with pancreatic cancer, compared with a public database of 2177 controls of Ashkenazi ancestry30 (eTable 8 in Supplement 1).
Tumor-Germline Correlations
In the 205 patients with a pathogenic variant, 93 of 170 tumors (54.7%) showed LOH or a pathogenic somatic second mutation in the same gene as the pathogenic variant. Of 180 evaluable pathogenic variants in 170 tumors, 82 variants showed LOH in the tumors and 13 tumors demonstrated a loss-of-function mutation or a previously reported deleterious missense variant.32,33 Concurrent somatic LOH or a second mutation at the same locus accompanied the germline mutation in 37 of 48 evaluable tumors (77.1%) in patients with BRCA1/2 germline mutations, 9 of 12 patients (75.0%) with mismatch repair gene variants, and 13 of 36 patients (36.1%) with germline CHEK2 variants, including 5 of 10 patients (50.0%) with the founder CHEK2 c.1100delC variant. Of the 103 pathogenic BRCA mutations identified in the tumors of the 1040 patients in this study, only 59 were germline in origin, whereas 44 were detected in the tumor.
A hypermutated tumor profile (defined as >20 somatic mutations) or MSI-H (defined as >10% of loci by MSIsensor20) was observed in 51 of 1040 patients (4.9%). Germline pathogenic variants in mismatch repair genes were identified in 12 of these 51 patients (23.5%). Two additional patients with germline MSH6 truncating mutations had tumors that were not hypermutated or MSI-H by MSI-sensor (1 with renal cell cancer and 1 with prostate cancer). Among the hypermutated or MSI-H cases, 14 of 51 (27.5%) fulfilled clinical criteria for Lynch syndrome (revised Bethesda guidelines and/or Amsterdam II criteria); 6 of 12 patients (50.0%) with an MSI-H tumor and an identified germline pathogenic variant in a mismatch repair gene met these criteria. Of the 14 cases with germline mismatch repair mutations, 6 were incrementally detected by germline analysis in the absence of a family history diagnostic of Lynch syndrome, including 3 patients with bladder cancer and 3 with prostate cancer. Of these 6 cases, 3 demonstrated LOH or a second somatic mutation (Table 3).
Discussion
This study identified clinically actionable inherited mutations in 17.5% of patients with advanced cancer, compared with 3% to 12.6% in prior series.1–7 Of the 1040 patients, 101 (9.7%) would not have been detected using clinical guidelines, which represented 55.5% of the total of 182 patients with actionable findings in the series. Germline findings led to discussion or initiation of change to targeted therapy in 38 (3.7%) of the 1040 patients tested and predictive testing in the families of 13 individuals (1.3%), including 6 for whom genetic evaluation would not have been initiated by guideline-based testing.
The prevalence of germline variants in a clinical setting will be affected by stage, case mix, ethnic ancestry of the population, and methods of variant classification. There was a significantly greater overall prevalence of germline mutations observed in patients with metastatic disease, although the proportion of germline findings that were incremental to predictions based on family history (approximately 50%) was similar in metastatic and nonmetastatic disease. This interesting association may reflect more aggressive biological features (eg, via greater mutational load of deleterious variants) or improved chance of survival with metastases in those with germline mutations. However, in the cohort studied here, there was no association of inherited mutations with increased tumor mutational load or difference in time from diagnosis to metastatic disease; further molecular profiling and prospective studies of treatment response may provide an explanation for the increased prevalence of germline mutations in metastatic disease. In this series, there was an abundance of late-stage prostate, pancreatic, renal, and breast cancers in which germline variants were more frequent, probably accounting for the higher observed prevalence of pathogenic variants found here compared with prior studies. While these findings reflect the experience at a referral cancer center, had the case mix more closely resembled population cancer incidence rates, the proportion of incremental findings would have been 49.1%. Had the entire cohort been screened for ancestry-specific (Ashkenazi and northern European) mutations, the proportion of incremental actionable findings would have been 31.3%, approximating the rate in a hypothetical cohort with no population diversity. However, if family history assessment by clinicians is less complete than the 3-generation information used in this analysis, the proportion of apparent incremental findings may be higher in practice. Thus, guideline-based testing will fail to detect a third to half (31.3%–55.5%) of genetic findings found by tumor-normal sequencing, taking into account case mix, disease stage, and ethnic ancestry.
The complementary role of tumor and germline sequencing was exemplified by colon and breast cancer testing. Tumor-normal testing was able to diagnose Lynch syndrome in patients who would not otherwise have been tested. The sensitivity of tumor-derived hypermutation or MSI-H status for detection of Lynch syndrome was 85.7%, with germline sequencing resulting in the diagnosis of Lynch syndrome in an additional 2 patients, 1 of whom would not have been diagnosed by family history. Half of the 12 cases with pathogenic variants in mismatch repair genes in the setting of a hyper-mutated or MSI-H tumor would not have met guideline-based criteria for tumor-directed immunohistochemical analysis. For breast cancer cases, had tumor-only testing been performed, approximately 40% of patients with BRCA1/2 variants detected with tumor sequencing would have been referred for a germline confirmation test that would have been negative; conversely “subtraction” of germline from tumor DNA sequence would have obscured 59 germline BRCA1/2 cases. These observations support the rationale for combined tumor-normal sequencing.3,34
Although epigenetic mechanisms of loss of the second allelemay also exist, concurrent somatic alterations in the same gene or LOH in matched tumors support the pathogenic role of many of the germline variants observed in nonsyndromic settings. For example, germline CHEK2 mutations were observed in 6 patients with renal cancer; the second allele was mutated in 3 of 5 evaluable tumors. Among novel nonsyndromic associations seen here, mutations in CHEK2 were enriched in prostate and pancreas cancers, MUTYH heterozygous mutations were enriched in prostate cancer, and mutations in MSH6, BARD1, PALB2, MITF, and SDHB, were observed absent the typical family history associated with these genes. Other unanticipated findings reported here will require further functional and genetic epidemiological genomic exploration, including the observation of the recurrent mutation FH c.1431_1433dupAAA (p.Lys477dup) and RECQL4 loss-of-function variants.35–37
To our knowledge, this study marks the first large-scale effort to return germline findings in the context of tumor-normal sequencing to patients. Such an approach in patients with advanced cancer was found to uncover potentially actionable germline variants that would not be detected using existing guidelines for genetic risk assessment. Less resource intensive strategies than tumor-normal sequencing could be applied but would result in lower sensitivity. For example, testing all patients for a dozen DNA repair genes and several APC and MUTYH founder mutations, combined with standard phenotypic assessment, would have detected 92.3% of patients with clinically actionable germline variants.
This study has several limitations. Among these is the lack of sufficient follow-up to assess the effect of the genetic information on patient or family outcomes, including potential harms due to false-positive results of screening. In addition, there was physician discretion for referral for tumor sequencing, potentially favoring enrollment of those who may have been eligible for targeted therapies. The usual care comparator in this study was synthetic and not the actual yield of testing a population according to guidelines. Also, interpretation of detailed family history information was by expert reviewers using reproducible but complex algorithms. In addition, the study had unique demographic characteristics and case mix. These factors will limit generalizability of findings to a community practice environment, where germline testing panels are also being introduced.38
Conclusions
In this referral population with selected advanced cancers, universal sequencing of a broad panel of cancer-related genes in paired germline and tumor DNA samples was associated with increased detection of individuals with potentially clinically significant heritable mutations over the predicted yield of targeted germline testing based on current clinical guidelines. Knowledge of these additional mutations can help guide therapeutic and preventive interventions, but whether all of these interventions would improve outcomes for patients with cancer or their family members requires further study.
Supplementary Material
Key Points.
Question
How many additional individuals with inherited cancer-predisposing mutations might be detected by DNA sequencing of tumor and normal tissue in patients with advanced cancer compared with restricting genetic testing to clinical guideline–directed testing?
Findings
In this case series of 1040 patients with advanced cancer, 101 of 182 patients with clinically actionable inherited mutations detected by tumor-normal sequencing would not have been detected by guideline-directed testing based on family history, age, and tumor type.
Meaning
In selected populations of patients with advanced cancer, universal sequencing of germline and tumor DNA for a broad panel of cancer-related genes may detect more potentially clinically significant heritable mutations than a targeted approach based on current clinical guidelines. It is not known if this will result in improved outcomes.
Acknowledgments
Funding/Support: This study was supported by the Robert and Kate Niehaus Center for Inherited Cancer Genomics at Memorial Sloan Kettering Cancer Center, the Andrew Sabin Family Foundation, the Sharon Levine Corzine Research Fund from Memorial Sloan Kettering Cancer Center, the Breast Cancer Research Foundation, Marie-Josée and Henry R. Kravis, the J. Randall and Kathleen L. MacDonald Renal Cancer Research Fund, the David M. Rubenstein Center for Pancreatic Cancer Research, and Cancer Center Support Grant P30 CA008748-50 from the National Cancer Institute.
Role of the Funder/Sponsor: The sponsors had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Additional Information: “MSK-IMPACT” is trademarked as a description, but there is no patent on the test or reagents. Memorial Sloan Kettering investigators have published the gene list and genomic intervals and also permit other laboratories to purchase the IMPACT probes directly from commercial laboratories, with no financial benefit to Memorial Sloan Kettering or any researcher at Memorial Sloan Kettering.
Author Contributions: Dr Offit had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. Drs Mandelker and Zhang and Ms Kemel are co–first authors. Drs Robson and Offit are co–senior authors.
Concept and design: Mandelker, Kemel, Stadler, Walsh, Lipkin, Cambria, Galle, Arcila, Leach, Scher, Baselga, Klimstra, Solit, Hyman, Berger, Robson, Offit.
Acquisition, analysis, or interpretation of data: Mandelker, Zhang, Kemel, Stadler, Joseph, Zehir, Pradhan, Arnold, Walsh, Li, Balakrishnan, Syed, Prasad, Nafa, Carlo, Cadoo, Sheehan, Fleischut, Salo-Mullen, Trottier, Lincoln, Mukherjee, Ravichandran, Abida, Arcila, Benayed, Shah, Yu, Bajorin, Coleman, Lowery, Garcia-Aguilar, Kantoff, Sawyers, Dickler, Saltz, Motzer, O’Reilly, Scher, Baselga, Solit, Hyman, Berger, Ladanyi, Robson, Offit.
Drafting of the manuscript: Mandelker, Kemel, Stadler, Pradhan, Walsh, Syed, Carlo, Lincoln, Cambria, Galle, Shah, Yu, Scher, Solit, Robson, Offit.
Critical revision of the manuscript for important intellectual content: Mandelker, Zhang, Kemel, Stadler, Joseph, Zehir, Pradhan, Arnold, Walsh, Li, Balakrishnan, Prasad, Nafa, Carlo, Cadoo, Sheehan, Fleischut, Salo-Mullen, Trottier, Lipkin, Mukherjee, Ravichandran, Abida, Arcila, Benayed, Bajorin, Coleman, Leach, Lowery, Garcia-Aguilar, Kantoff, Sawyers, Dickler, Saltz, Motzer, O’Reilly, Scher, Baselga, Klimstra, Solit, Hyman, Berger, Ladanyi, Robson, Offit.
Statistical analysis: Kemel, Joseph, Zehir, Pradhan, Syed, Mukherjee, Ravichandran, Offit.
Obtained funding: Coleman, Sawyers, Motzer, Solit, Offit.
Administrative, technical, or material support: Zhang, Kemel, Pradhan, Arnold, Walsh, Li, Balakrishnan, Syed, Prasad, Nafa, Sheehan, Fleischut, Salo-Mullen, Trottier, Lincoln, Cambria, Galle, Abida, Arcila, Benayed, Shah, Bajorin, Coleman, Leach, Lowery, Garcia-Aguilar, Motzer, O’Reilly, Scher, Klimstra, Solit, Hyman, Berger, Ladanyi, Offit.
Supervision: Mandelker, Kemel, Lipkin, Kantoff, Motzer, Scher, Baselga, Klimstra, Solit, Berger, Ladanyi, Offit.
Conflict of Interest Disclosures: All authors have completed and submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Ms Kemel reported prior employment at Bioreference Laboratories. Dr Lowery reported serving on advisory boards for Agios Pharmaceuticals and Celgene. Dr Sawyers reporting serving on the board of directors for Novartis. Dr Dickler reported serving on advisory boards for Genentech/Roche, Novartis, Pfizer, Puma Biotech, AstraZeneca, and TapImmune. Dr Saltz reported receiving a grant from Taiho Pharmaceutical. Dr Scher reported serving as a consultant to Astellas, BIND Therapeutics, Clovis Oncology, Ferring Pharmaceuticals, Janssen Research and Development, Medivation, Merck, Roche, Sanofi Aventis, Takeda Millennium, and WIRB-Copernicus Group; receiving support to the Memorial Sloan Kettering Cancer Center from Medivation, Janssen, Illumina, and Innocrin Pharmaceuticals; and serving on the board of directors for Asterias Biotherapeutics. Dr Solit reported serving as a consultant to Pfizer and Loxo Oncology. Dr Hyman reported serving as a consultant to Chugai, CytomX, Boehringer Ingelheim, and Atara Biotherapeutics; and receiving grants from AstraZeneca, Puma Biotechnology, and Loxo Oncology. Dr Ladanyi reported receiving personal fees from Boehringer Ingelheim and AstraZeneca through the National Comprehensive Cancer Network; and a receiving a grant from Loxo Oncology. No other disclosures were reported.
Additional Contributions: The following persons, all affiliated with Memorial Sloan Kettering Cancer Center, made substantial contributions to the work reported in this article through data collection, program support, or editing assistance: Jacob Musinsky, BA (data collection), Carolyn Stewart, BA (data collection, editing assistance), Christina Tran, BS (data collection), Melissa Salerno, MEd (data collection), Farzeen Aslam, MHA (program support), and Ederlinda Paraiso, MPA (program support); they received no compensation other than as employees of Memorial Sloan Kettering Cancer Center. We thank the Exome Aggregation Consortium and the groups that provided exome variant data for comparison. A full list of contributing groups can be found at http://exac.broadinstitute.org/about. The data shown here are also in part based on data generated by The Cancer Genome Atlas (TCGA) Research Network (http://cancergenome.nih.gov/).
References
- 1.Schrader KA, Cheng DT, Joseph V, et al. Germline variants in targeted tumor sequencing using matched normal DNA. JAMA Oncol. 2016;2(1):104–111. doi: 10.1001/jamaoncol.2015.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seifert BA, O’Daniel JM, Amin K, et al. Germline analysis from tumor-germline sequencing dyads to identify clinically actionable secondary findings. Clin Cancer Res. 2016;22(16):4087–4094. doi: 10.1158/1078-0432.CCR-16-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones S, Anagnostou V, Lytle K, et al. Personalized genomic analyses for cancer mutation discovery and interpretation. Sci Transl Med. 2015;7(283):283ra53. doi: 10.1126/scitranslmed.aaa7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parsons DW, Roy A, Yang Y, et al. Diagnostic yield of clinical tumor and germline whole-exome sequencing for children with solid tumors. JAMA Oncol. 2016;2(5):616–624. doi: 10.1001/jamaoncol.2015.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mody RJ, Wu Y-M, Lonigro RJ, et al. Integrative clinical sequencing in the management of refractory or relapsed cancer in youth. JAMA. 2015;314(9):913–925. doi: 10.1001/jama.2015.10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meric-Bernstam F, Brusco L, Daniels M, et al. Incidental germline variants in 1000 advanced cancers on a prospective somatic genomic profiling protocol. Ann Oncol. 2016;27(5):795–800. doi: 10.1093/annonc/mdw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J, Walsh MF, Wu G, et al. Germline mutations in predisposition genes in pediatric cancer. N Engl J Med. 2015;373(24):2336–2346. doi: 10.1056/NEJMoa1508054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hampel H, Bennett RL, Buchanan A, Pearlman R, Wiesner GL Guideline Development Group, American College of Medical Genetics and Genomics Professional Practice and Guidelines Committee and National Society of Genetic Counselors Practice Guidelines Committee. A practice guideline from the American College of Medical Genetics and Genomics and the National Society of Genetic Counselors: referral indications for cancer predisposition assessment. Genet Med. 2015;17(1):70–87. doi: 10.1038/gim.2014.147. [DOI] [PubMed] [Google Scholar]
- 9.National Comprehensive Cancer Network. [Accessed September 23, 2016];Genetic/Familial High-Risk Assessment: Breast and Ovarian, Version 1.2017. 2016 Sep 19; https://www.nccn.org/professionals/physician_gls/pdf/genetics_screening.pdf.
- 10.National Comprehensive Cancer Network. [Accessed September 23, 2016];Genetic/Familial High-Risk Assessment: Colorectal, Version 2.2016. 2016 Sep 26; https://www.nccn.org/professionals/physician_gls/pdf/genetics_colon.pdf.
- 11.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17(3):251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23(6):703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rahman N. Realizing the promise of cancer predisposition genes. Nature. 2014;505(7483):302–308. doi: 10.1038/nature12981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalia SS, Adelman K, Bale SJ, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med. 2017;19(2):249–255. doi: 10.1038/gim.2016.190. [DOI] [PubMed] [Google Scholar]
- 15.Cheng DT, Prasad M, Chekaluk Y, et al. Comprehensive detection of germline variants by MSK-IMPACT, a clinical diagnostic platform for solid tumor molecular oncology and concurrent cancer predisposition testing. BMC Med Genomics. 2017;10(1):33. doi: 10.1186/s12920-017-0271-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pritchard CC, Mateo J, Walsh MF, et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med. 2016;375(5):443–453. doi: 10.1056/NEJMoa1603144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abida W, Armenia J, Gopalan A, et al. Prospective genomic profiling of prostate cancer across disease states reveals germline and somatic alterations that may affect clinical decision making. JCO Precis Oncol. 2017;(1):1–16. doi: 10.1200/PO.17.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richards S, Aziz N, Bale S, et al. ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 2016;44(16):e131. doi: 10.1093/nar/gkw520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niu B, Ye K, Zhang Q, et al. MSI sensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics. 2014;30(7):1015–1016. doi: 10.1093/bioinformatics/btt755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.UpToDate. [Accessed October 19, 2016];Search. https://www.uptodate.com/contents/search.
- 22.American Society of Clinical Oncology. [Accessed October 19, 2016];Cancer Genetics Program. http://university.asco.org/cancer-genetics-program.
- 23.Menko FH, van Steensel MAM, Giraud S, et al. European BHD Consortium. Birt-Hogg-Dubé syndrome: diagnosis and management. Lancet Oncol. 2009;10(12):1199–1206. doi: 10.1016/S1470-2045(09)70188-3. [DOI] [PubMed] [Google Scholar]
- 24.Pilarski R, Cebulla CM, Massengill JB, et al. Expanding the clinical phenotype of hereditary BAP1 cancer predisposition syndrome, reporting three new cases. Genes Chromosomes Cancer. 2014;53(2):177–182. doi: 10.1002/gcc.22129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thakker RV, Newey PJ, Walls GV, et al. Endocrine Society. Clinical practice guidelines for multiple endocrine neoplasia type 1 (MEN1) J Clin Endocrinol Metab. 2012;97(9):2990–3011. doi: 10.1210/jc.2012-1230. [DOI] [PubMed] [Google Scholar]
- 26.Exome Aggregation Consortium. [Accessed August 24, 2016];ExAC Browser Beta. http://exac.broadinstitute.org/about.
- 27.Lek M, Karczewski KJ, Minikel EV, et al. Exome Aggregation Consortium. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.International Genome Sample Resource. [Accessed August 24, 2016];IGSR and the 1000 Genomes Project. http://www.internationalgenome.org/
- 29.National Center for Biotechnology Information. [Accessed August 24, 2016];dbSNP Short Genetic Variations. https://www.ncbi.nlm.nih.gov/projects/SNP/
- 30.IBD Exomes Portal. [Accessed October 5, 2016];IBD Exomes browser. https://ibd.broadinstitute.org/
- 31.American Cancer Society. [Accessed December 19, 2016];Cancer facts and figures. 2016 https://www.cancer.org/acs/groups/content/@research/documents/document/acspc-047079.pdf.
- 32.Forbes SA, Beare D, Gunasekaran P, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43(database issue):D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wellcome Trust Sanger Institute Genome Research Ltd. [Accessed August 24, 2016];COSMIC: Catalogue of Somatic Mutations in Cancer. http://cancer.sanger.ac.uk/cosmic.
- 34.Bombard Y, Robson M, Offit K. Revealing the incidentalome when targeting the tumor genome. JAMA. 2013;310(8):795–796. doi: 10.1001/jama.2013.276573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang LL, Gannavarapu A, Kozinetz CA, et al. Association between osteosarcoma and deleterious mutations in the RECQL4 gene in Rothmund-Thomson syndrome. J Natl Cancer Inst. 2003;95(9):669–674. doi: 10.1093/jnci/95.9.669. [DOI] [PubMed] [Google Scholar]
- 36.Ng AJM, Walia MK, Smeets MF, et al. The DNA helicase RECQL4 is required for normal osteoblast expansion and osteosarcoma formation. PLoS Genet. 2015;11(4):e1005160. doi: 10.1371/journal.pgen.1005160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang LL, Plon SE. Rothmund-Thomson syndrome. In: Pagon RA, Adam MP, Ardinger HH, et al., editors. GeneReviews. Seattle, WA: University of Washington; 1993. [Google Scholar]
- 38.Offit K. The future of clinical cancer genomics. Semin Oncol. 2016;43(5):615–622. doi: 10.1053/j.seminoncol.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.