Abstract
Branching morphogenesis creates arborized epithelial networks. In the mammalian kidney, an epithelial progenitor pool at ureteric branch tips (UBTs) creates the urine-transporting collecting system. Using region-specific mouse reporter strains, we performed an RNA-seq screen, identifying tip- and stalk-enriched gene sets in the developing collecting duct system. Detailed in situ hybridization studies of tip-enriched predictions identified UBT-enriched gene sets conserved between the mouse and human kidney. Comparative spatial analysis of their UBT niche expression highlighted distinct patterns of gene expression revealing novel molecular heterogeneity within the UBT progenitor population. To identify kidney-specific and shared programs of branching morphogenesis, comparative expression studies on the developing mouse lung were combined with in silico analysis of the developing mouse salivary gland. These studies highlight a shared gene set with multi-organ tip enrichment and a gene set specific to UBTs. This comprehensive analysis extends our current understanding of the ureteric branch tip niche.
KEY WORDS: Branching morphogenesis, Ureteric bud, Tip progenitor, RNA-seq
Summary: RNA-seq and in situ studies of progenitor pools driving the morphogenesis of mammalian kidney epithelial networks highlight gene sets linked to kidney and cross-organ branching programs.
INTRODUCTION
Epithelial branching morphogenesis shapes the development of a wide variety of organ systems in invertebrate and vertebrate embryos (Ochoa-Espinosa and Affolter, 2012; Iber and Menshykau, 2013). The airways of the Drosophila tracheal system and mammalian lung, and the urine-transporting network of the collecting duct system of the mammalian kidney, are particularly well-studied examples (Affolter and Caussinus, 2008; Costantini, 2012; Little and McMahon, 2012). In each, branching growth generates a complex tubular network for fluid transport laying down an organic-specific pattern of cellular diversity with a characteristic system-specific morphology. Branching within each system is stereotypical to varying degrees (Metzger et al., 2008; Short et al., 2014). In the salivary gland, a distinct process of epithelial clefting generates branch tips (Patel and Hoffman, 2014). Despite differences in the cellular responses and the interplay with adjacent cell populations, common regulatory themes have been identified in multi-organ control of branching growth, most notably fibroblast growth factor (FGF) pathway signaling (Trueb et al., 2013; Affolter and Caussinus, 2008).
In the mammalian kidney, active development initiates with the swelling and outgrowth of a subset of cells from the nephric duct, the ureteric bud (UB), into a pre-specified population of metanephric mesenchyme (MM) (McMahon, 2016). The cells of the UB generate the highly branched ureteric epithelium of the collecting duct network to which MM-derived nephrons attach to form the complete tubular network of the kidney's filtration and transport system. Outgrowth of the UB, and subsequent branching morphogenesis of the ureteric epithelium, involves a complex regulatory interplay among the collecting duct progenitors within the ureteric branch tips (UBTs) and the adjacent mesenchymal progenitors for nephron and interstitial cell lineages within the MM (Little and McMahon, 2012). UBTs lay down a complexity of epithelial cell types over the course of development through the differentiation of cells emerging from the UBT progenitor pool (Costantini, 2012). Together, the interactions within the UBT niche coordinate growth and differentiation of distinct cell lineages to generate a functional organ of appropriate size, shape and cellular diversity.
In the mammalian lung, mesenchymal-epithelial interactions drive an initially invariant pattern of branching growth at lung branch tips (LBTs) controlling airway growth, differentiation and morphogenesis (Herriges and Morrisey, 2014; Rock and Hogan, 2011; Metzger et al., 2008). Unlike the mammalian kidney where UBT-derived Wnt signals trigger the transformation of mesenchymal nephron progenitors to epithelial nephrons responsible for much of the mass and functional role of the kidney, the lung mesenchyme remains mesenchymal and shows a more limited differentiation (McCulley et al., 2015). Thus, each organ shows common developmental themes, such as the mesenchyme-mediated control of epithelial growth and branching, and the maintenance and controlled commitment of UBT- and LBT-localized progenitors, that might be underpinned by shared regulatory processes. However, the different morphology of UBT- and LBT-derived epithelial cells, the distinct cell types generated by these populations, and the different properties and responses of their adjacent mesenchyme populations predict organ-specific mechanisms within each tip population. Active branch outgrowth is not the only mechanism of branching morphogenesis. In the salivary gland, salivary branch tips (SBTs) arise as epithelial clefts that subdivide the epithelium into end buds and ducts (Patel and Hoffman, 2014).
UBT, LBT and SBT development is stimulated by mesenchyme-derived signals that enhance and maintain high proliferative activity within tip progenitor populations. In the kidney, MM production of glial-derived neurotrophic growth factor (GDNF) initiates RET/GFRA1 receptor pathway signaling in the ureteric bud (Costantini and Shakya, 2006). RET encodes a receptor tyrosine kinase and a variety of studies have shown a larger number of receptor tyrosine kinases, including FGFR, MET, EGFR and VEGFR2 (KDR), contribute to varying degrees at different stages of ureteric epithelial branching (Walker et al., 2016; Tufro et al., 2007; Ishibe et al., 2009). In the lung and salivary gland, mesenchyme-derived FGF signaling, predominantly mesenchyme-derived FGF10 acting through FGFR2, plays the central role in branching morphogenesis (Min et al., 1998; Sekine et al., 1999; Arman et al., 1999; De Moerlooze et al., 2000; Ohuchi et al., 2000). GDNF and FGF pathways converge on ETV- and SOX-regulated transcriptional pathways in both UBT and LBT progenitors though organ-specific differences in the use of ETV and SOX family members are observed (Reginensi et al., 2011; Lu et al., 2009; Herriges et al., 2015; Zhu et al., 2012). UBTs and LBTs also differ in the signals they produce: for example, Wnt11 in UBTs and sonic hedgehog in LBTs; both signals target the underlying mesenchyme in organ-specific reciprocal signaling networks between epithelium and mesenchyme to regulate branching processes (Majumdar et al., 2003; Pepicelli et al., 1998).
Importantly, genetic analyses in mouse and human have highlighted conserved roles within each organ for specific regulatory actions. In particular, congenital anomalies of the kidney and urinary tract (CAKUT) comprise a relatively common syndrome (20-30% of all congenital malformations) (Brown et al., 1987; Queisser-Luft et al., 2002). CAKUT is associated with mutations in RET, BMP4 and ROBO2, and a number of other genes that are known to alter branching growth of the ureteric network in the mouse kidney (Nicolaou et al., 2015).
To gain a more comprehensive understanding of epithelial branch tip regulation, we have extended previous studies using micro-dissection and microarray techniques to identify tip-specific regulatory factors in the developing mammalian kidney (Schwab et al., 2003; Stuart et al., 2003; Schmidt-Ott et al., 2005). The screen performed here identified a large number of previously unknown UBT-enriched genes, as well as a large gene set enriched in maturing regions of the non-branching ureteric epithelial network. Temporal and spatial analysis of their expression in the mammalian kidney confirmed UBT enrichment and defined spatial subdomains within UBTs and conservation of UBT expression between mouse and human. Comparative studies with the lung and salivary gland highlight gene sets that are UBT specific and those enriched within epithelial tips of multiple organ systems. The stratified gene sets identified here provide a rich resource for mechanistic investigation of branching programs within the developing kidney and across other mammalian organs. Furthermore, the gene sets highlight novel candidates to explore in the context of human congenital anomalies and diseases. Mechanistic analysis of the novel gene sets is likely to shed new light on how branch tip programs regulate the size, cellular diversity and distinct morphologies of mammalian organ systems.
RESULTS
RNA-sequencing analysis identifies UBT-enriched and stalk-enriched genes
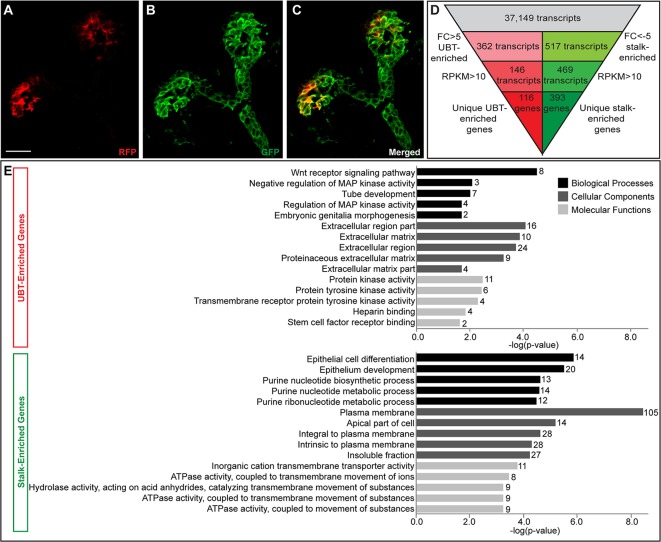
To identify genes that are differentially expressed in tip and stalk regions of the developing ureteric epithelium of the mouse kidney, we utilized two fluorescently labeled transgenic mouse strains: Wnt11-RFP (Harding et al., 2011) and Hoxb7-GFP (Srinivas et al., 1999). Wnt11, a target of RET/GDNF signaling in kidney development (Pepicelli et al., 1997; Majumdar et al., 2003), is expressed within the tip population whereas Hoxb7 is expressed throughout the entire UB epithelium (Fig. 1A-C). Kidneys from a single litter were pooled together for each biological replicate from embryonic day (E) 16.5 day Wnt11-RFP; Hoxb7-GFP embryos and dissociated to single cells; tip (RFP+, GFP+) and stalk (RFP−, GFP+) enriched cell fractions were separated by fluorescence-activated cell sorting (FACS), and RNA sequencing (RNA-seq) was performed on three biological replicates of each cell population. Primary analysis of RNA-seq data mapped 37,149 transcripts. To identify more specifically RNAs strongly enriched in each population, we focused on only those genes with a reads per kilobases per million (RPKM) value greater than ten in at least one sample, and at least a 5-fold expression differential between the UBT and stalk fractions (Fig. 1D).
Fig. 1.
Identification of UBT- and stalk-enriched genes from RNA-sequencing data set. (A-C) Immunofluorescence of a Wnt11RFP;Hoxb7GFP E15.5 ureteric bud used to acquire UBT and stalk cell populations for RNA sequencing. (A) RFP-positive cells restricted to the UBTs. (B) GFP-positive cells labeling the entire ureteric epithelium. (C) Merged image displaying the UBT cells (RFP+, GFP+) and stalk cells (GFP+). (D) Filtering strategy to narrow down to a stringent list of UBT- and stalk-enriched genes for further analysis. (E) Gene ontology (GO) term analysis of UBT-enriched and stalk-enriched gene lists (number of genes for each term are listed at the end of the bar). Scale bar: 20 μm.
This analysis generated a stringent list of 116 unique tip-enriched and 393 unique stalk-enriched genes (Tables S1 and S2, Fig. S1). This tip-enriched gene list includes many genes previously characterized for UBT activity, including Ret, Gfra1, Wnt11, Etv4, Etv5 and Sox8 (for a review, see O'Brien and McMahon, 2014). Gene ontology (GO) analysis of the UBT-enriched set recovered terms associated with signaling pathways and structural aspects known to be important in UBTs, and more generally in branching morphogenesis (Fig. 1E). For instance, the top GO terms for biological processes are associated with well-established signaling pathways in the embryonic kidney, such as Wnt and MAP kinase (MAPK) pathways. Moreover, the GO terms for cellular components are linked to the extracellular matrix. During branching morphogenesis, matrix remodeling and degradation is likely to facilitate branching events (Kim and Nelson, 2012). For molecular functions, key actions such as heparin binding and tyrosine kinase activity relate to FGF signaling and downstream signal transduction pathways.
As expected, given the FACS approach and high levels of expression in UBTs, the most highly enriched UBT gene was Wnt11 (Table S1). Furthermore, the list includes many well-characterized genes encoding receptor complexes for GDNF and transcriptional mediators of UBT signaling pathways that are known to be UBT enriched and have been shown to play central roles in UBT regulation, including Ret, Gfra1, Etv4 and Etv5 (Pachnis et al., 1993; Schuchardt et al., 1994; Cacalano et al., 1998; Enomoto et al., 1998; Keefe et al., 2013; Lu et al., 2009). Sox8 co-regulates UBT development together with a UBT-enriched partner Sox9 but Sox9 does not appear in this high stringency list (Table S1). However, Sox9 expression was enriched in the UBT fraction below the 5-fold threshold cutoff [fold change (FC)=4.15, RPKM=23.92]. This result indicates that among the set of genes with <5-fold enrichment there are likely to be additional UBT-enriched genes of biological significance to UBT-directed developmental processes.
The most highly stalk-enriched gene sets includes a number of genes associated with differentiation of mature ureteric epithelial cell types, including Aqp2 (encoding a principal cell-specific water channel protein) (Kwon et al., 2013); Foxq1 (encoding a transcriptional determinant of pH-regulating intercalated cells) (Blomqvist et al., 2004); several uroplakin (Upk) genes, which encode components forming an impermeable epithelial barrier within the urinary tract that extends from the medullary collecting duct to the bladder (Wu et al., 2009); and signal-encoding genes such as Wnt7b and Shh that drive morphogenesis of the deep medullary ureteric epithelium and development of smooth muscle associated with this epithelium, respectively (Yu et al., 2002, 2009; Table S2). In summary, UBT and stalk epithelial populations reflect distinct cell types with expected differences in gene expression and predicted biological action giving confidence that the differential expression screen achieved our primary goal of generating robust datasets for regional gene activity in the developing ureteric system.
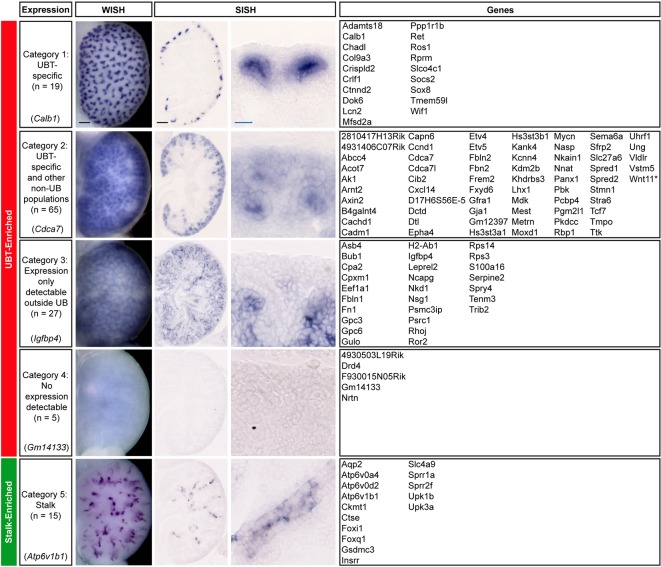
Categorization of the UBT-enriched gene set by in situ hybridization
To compare expression of all the UBT-enriched genes from the stringent dataset with a subset of the most stalk-enriched gene set (FC≤−50), whole-mount in situ hybridization (WISH) was performed on E15.5 urogenital systems (UGSs) using Wnt11 (UBT specific, high expression) and Shh (stalk specific, low expression) as positive controls for sensitivity, probe penetration and regional expression. We chose E15.5 for follow up in situ analysis because E15.5 samples enable more effective complete penetration in WISH procedures; this stage has been the standard for whole-mount screens in large scale projects within the Genito-Urinary Molecular Anatomy Project (GUDMAP; www.GUDMAP.org) (McMahon et al., 2008; Yu et al., 2012). Furthermore, the expectation is that branching programs are similar at E15.5 and E16.5; to our knowledge no differences in tip gene expression have been described between any active stage of branching tip outgrowth in the mammalian kidney. We also compared our data with data in GUDMAP and Eurexpress (a transcriptome atlas database for mouse embryo; www.eurexpress.org) to evaluate and corroborate our new datasets (McMahon et al., 2008; Yu et al., 2012; Harding et al., 2011; Diez-Roux et al., 2011).
Predicted UBT-enriched genes were stratified into four kidney expression categories: (1) specifically expressed in UBTs, (2) UBT-enriched expression and additional non-ureteric epithelial population(s) expression, (3) expression only detectable outside of the ureteric epithelium, and (4) no expression detectable (Fig. 2). To ensure that none of the genes in categories 3 and 4 scored as negative from a mismatch between the E16.5 screen and the E15.5 in situ analysis, all genes in these categories (32 total) were secondarily screened for UBT expression at E16.5; as expected, none showed detectable UBT expression at E16.5 (data not shown). Furthermore, online (www.gudmap.org and www.eurexpress.org) section in situ hybridization (SISH) and microarray profiling of many of these genes verifies the absence of tip expression. These genes likely reflect true false positives from the primary screen or potentially low levels of expression below the threshold for robust detection.
Fig. 2.
Whole-mount and section in situ hybridization expression pattern analysis of UBT-enriched and stalk-enriched genes identified from the RNA-seq dataset on Swiss Webster E15.5 kidneys. Expression patterns shown are example genes for each category as indicated on the left. Genes listed on the right have the expression pattern described in the categories on the left. Leprel2, also known as P3h3. *In addition to ureteric tip-restricted expression in the ureteric epithelium, Wnt11 is activated in interstitial mesenchyme cells, coincident with medullary remodeling from E15.5 (Yu et al., 2009). Scale bars: 100 μm (black); 20 μm (blue).
Eighty-four of the predicted UBT-enriched genes (72%) showed UBT-specific expression patterns: 19 in category 1 expression and 65 in category 2 expression. Genes in these categories were assessed further by SISH on E15.5 UGSs to establish a more detailed view of which specific cell types show expression (Fig. 2). The SISH results confirmed the general WISH annotation but also provide more insights into other expression domains outside of the ureteric epithelium, which include the cap mesenchyme, interstitial cells, and segments of the developing nephron, and give more detailed spatial information on the domain of expression within the UBT.
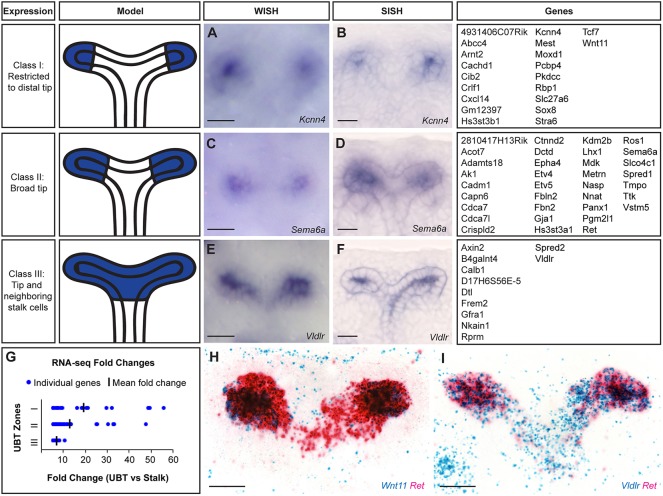
A close examination of UBT expression identified three distinct categories of genes based on their expression domain within the UB: Class I (20 genes) with expression restricted to the distal tip such as Kcnn4, which encodes a calcium-activated potassium channel protein (Ishii et al., 1997; Joiner et al., 1997); Class II (34 genes) with broader tip expression such as Sema6a, which encodes a transmembrane protein linked to axon guidance (Leighton et al., 2001; Suto et al., 2007; Matsuoka et al., 2011; Belle et al., 2016); and Class III (11 genes), for which gene expression extends beyond the UBT into the near adjacent stalk epithelium, such as Vldlr, which encodes a reelin-binding lipoprotein receptor (Bock and May, 2016; Fig. 3A-F). Class I genes, with the most distal expression, have the highest mean fold change (Fig. 3G). The mean fold change decreases as the expression zones widen in Class II and III. Thus, there is a relationship between the spatial distribution of gene activity within the UBTs and UBT enrichment in the datasets. To confirm the differing zones of UBT expression observed, the RNAscope Duplex Detection kit was utilized to show the expression of two genes on a single tissue section. Wnt11 (Class I) is restricted to the distal tip whereas Ret (Class II) has a broader expression pattern (Fig. 3H). The expression of Vldlr (Class III) extends further than Ret into the neighboring stalk cells (Fig. 3I). Collectively, these data provide strong evidence for molecular heterogeneity among cells within the branch tips.
Fig. 3.
Classification of UBT expression from whole-mount and section in situ hybridization within E15.5 kidneys highlighting three distinct zones. (A,B) Kcnn4 is restricted to the very distal ends of the UBT. (C,D) Sema6a is expressed widely throughout the tip domain. (E,F) Vldlr is expressed throughout the UBT domain and weakly into the near adjacent stalk regions. Schematics illustrating the expression domains for each class are shown on the left. Expression patterns shown in A-F are example genes for each class as indicated. Genes categorized within each class are listed on the right. (G) Distribution of RNA-seq fold changes for each zone. (H) Expression pattern of Wnt11 (Class I) and Ret (Class II). (I) Expression pattern of Ret (Class II) and Vldlr (Class III). Scale bar: 20 μm.
Comparative expression analysis of UBT-specific genes in the developing mouse and human kidney
In the human kidney, UB outgrowth and branching initiates at gestation week 5 and is thought to continue until around week 14-15 when UBTs continue to extend cortically, undergoing growth and differentiation, but in the absence of repeated branching (Osathanondh and Potter, 1963). We used SISH to examine the expression of a selected group of UBT-restricted genes in 15-16 week human fetal kidney samples (Fig. 4A-R). Of these, WNT11, ETV4, ETV5, HS3ST3A1, HS3ST3B1, CRLF1 and SOX8 showed UBT-restricted expression at UBTs in the human kidney (Fig. 4A-P). In contrast, KDM2B, which encodes a lysine demethylase linked to Wnt pathway regulation, was the only gene we observed from this subset that was not detected in the human UBTs at this time, though strong conserved expression was observed within early forming nephron structures (Fig. 4Q,R).
Fig. 4.
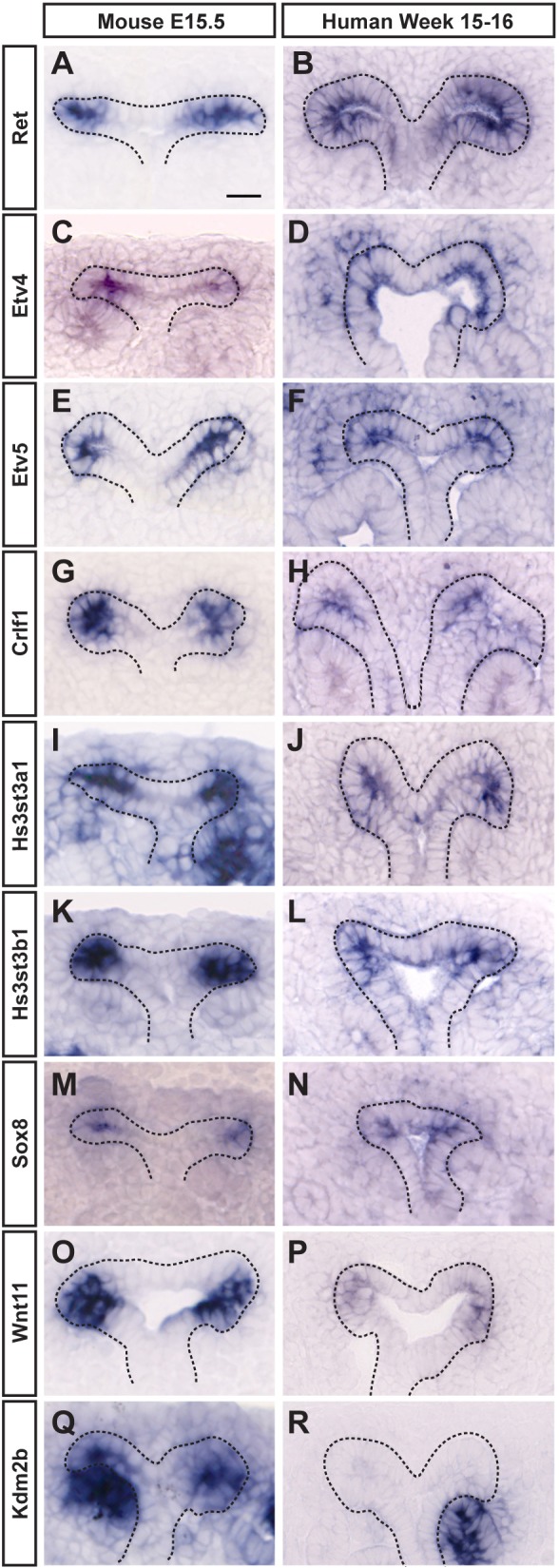
SISH comparison of UBT expression between mouse (E15.5) and human (week 15-16) kidneys. (A-N) Ret, Etv4, Etv5, Crlf1, Hs3st3a1, Hs3st3b1 and Sox8 show strong UBT expression in both species. (O,P) Wnt11 has strong UBT expression in the mouse and much weaker UBT expression in the human kidney. (Q,R) Kdm2b shows UBT expression only in the mouse kidney. Note that expression in the distal portion of the renal vesicle is conserved between the species. Scale bar: 20 μm.
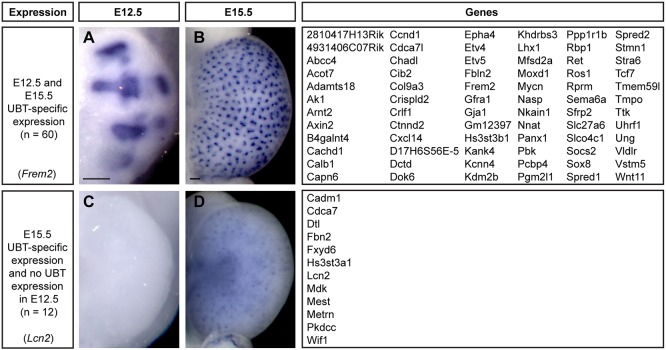
Construction of a spatiotemporal gene expression map of tip-specific genes
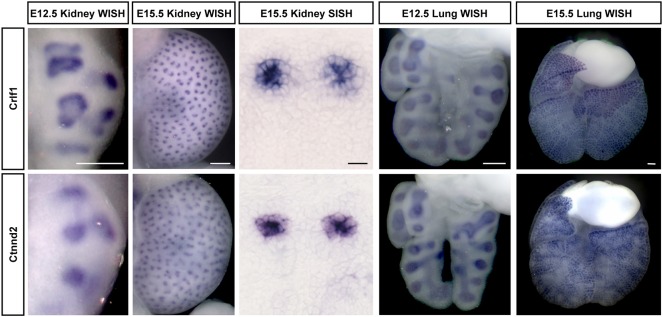
To obtain a broader view of the expression of UBT-specific genes at different stages of the branching process, we examined the expression of category 1 and 2 genes at E12.5 by WISH, shortly after the initiation of UB branching, when a maximum of six branching generations have taken place (Short et al., 2014). Sixty genes (86%) had similar expression at both embryonic stages (Fig. 5): these included genes known to be associated with receptor tyrosine kinase (RTK) (Ret, Gfra1, Etv4, Etv5, Sox8, Spred1, Spred2) and Wnt signaling (Axin2, Ccnd1, Ctnnd2, Sfrp2, Tcf7, Wnt11). For the subset of genes not detected at E12.5, this could reflect dynamic temporal expression such that the gene is off early, or below the limits of detection, at this earlier time point.
Fig. 5.
WISH UBT expression in Swiss Webster kidneys at E12.5 and E15.5. (A,B) Frem2 has UBT-specific expression in the kidney at both E12.5 and E15.5. (C,D) Lcn2 only shows UBT expression in the E15.5 kidney. Scale bars: 100 μm.
Cross-organ analysis of expression of the UBT-enriched gene set categories 1 and 2 in the embryonic lung and salivary gland
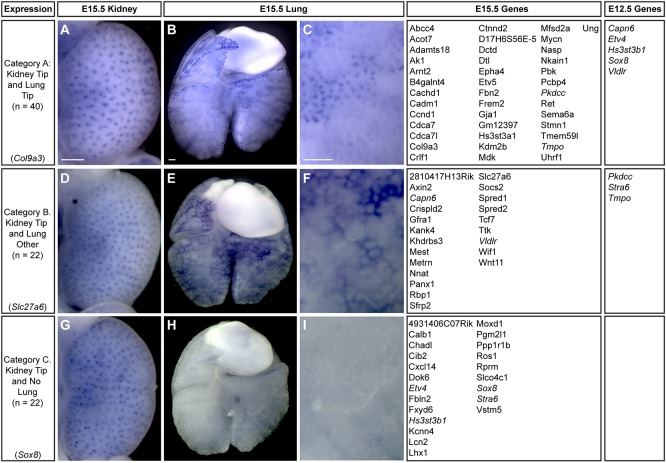
To determine which UBT-specific genes in the kidney may have broader roles in growth and branching in other organs, we extended WISH expression studies for this set to E12.5 and E15.5 lungs. Comparative analysis highlighted three distinct categories of expression: (A) genes with branch tip-specific expression in both the kidney and lung, (B) genes with branch tip-specific expression in the kidney and expression in a non-tip population in lung, and (C) genes with branch tip-specific expression in the kidney and no detectable expression in the lung (Fig. 6). Forty genes (48%) of the E15.5 UBT-enriched gene set were expressed also within LBTs, 22 genes (26%) showed expression outside of the LBTs, and 22 genes (26%) were not detectably expressed within lung samples. Category A genes are candidates for playing conserved, generic roles in epithelial branch-tip regulation across organ systems, whereas those genes with kidney-restricted expression are candidates for kidney-specific actions that could link to different branching events driving the distinct pattern of kidney morphogenesis, or distinct tip-niche interactions that distinguish the developing kidney from the lung. Interestingly, a subset of the UBT-enriched genes was detected in other cell type(s) within the lung close to the LBT epithelium (category B). Seventy-six (90%) genes had a similar expression pattern between E12.5 and E15.5 lungs, demonstrating that the developmental process is largely consistent over time. Fig. 7 shows a comparative summary of kidney and lung expression of a selected set of genes at each developmental stage.
Fig. 6.
Expression pattern analysis of tip-enriched genes in Swiss Webster E15.5 kidneys and lungs. (A-C) Col9a3 shows tip expression in both kidneys and lungs at E15.5. (D-F) Slc27a6 is UBT specific in the kidney and is expressed outside of the tip in the lung. (G-I) Sox8 is expressed within the kidney UBT and is absent from the lung. Expression patterns shown are example genes for each category as indicated on the left. Italicized genes have a different tip expression pattern at E12.5 compared with E15.5, which are categorized on the right under ‘E12.5 Genes’. Scale bars: 200 μm.
Fig. 7.
Examples of kidney and lung expression of two UBT-specific genes characterized by in situ hybridization at E12.5 and E15.5. Scale bars: 200 μm (white); 20 μm (black).
The mammalian salivary gland is another branched epithelial organ system that shares some regulatory mechanisms with the lung and kidney (Varner and Nelson, 2014). However, a major distinction is that SBTs arise by a process of clefting, where invaginations of the epithelial basement membrane establish new branching events. This highly dynamic process requires extracellular matrix (ECM) remodeling, including fibronectin assembly and rearrangements in cell-cell and cell-matrix adhesions (Patel and Hoffman, 2014). We investigated the expression of the UBT-enriched genes from categories 1 and 2 within the salivary gland utilizing two online resources: Eurexpress and the Salivary Gland Molecular Anatomy Project (http://sgmap.nidcr.nih.gov) (Diez-Roux et al., 2011; Musselmann et al., 2011) (Table S3). Fifty-five percent (46 genes) of the UBT-enriched genes were also enriched within the developing SBTs, and 31% (26 genes) showed conserved epithelial branch tip-enriched expression in the kidney, lung and salivary gland highlighting potential core branch program regulatory factors (Table S3). Notably, 25 of the 84 (30%) UBT-enriched genes were only detected in UBTs, which could indicate kidney-specific roles for this cohort.
DISCUSSION
Prior microarray analysis studies examining gene expression in subpopulations of the embryonic kidney offer informative insights into the regulatory networks involved in kidney development (Schwab et al., 2003; Stuart et al., 2003; Schmidt-Ott et al., 2005). Our UBT-enriched list expands these previous studies using RNA-seq technology. This gene set includes well-verified UBT-specific genes, such as Wnt11, Etv4, Etv5, Gfra1, Ret and Sox8/9. Although the RNA-sequencing screen identified established UBT-enriched genes, the majority of the genes identified here have not been documented previously in kidney development and/or branching morphogenesis. The expanded insight into gene expression linked to UBT cells reflects both the improved sensitivity of the RNA-seq-based approach and the absence of inherent limitations using defined probe sets on microarrays.
UBT-enriched list identifies novel genes with distinct expression domains in epithelial branch tips
Whole-mount in situ hybridization on E15.5 mouse kidneys revealed that 84/116 (72%) of the primary UBT-enriched gene set from RNA-seq analysis display tip-restricted expression patterns within the ureteric epithelium. A number of these genes are associated with signaling pathways important in branching morphogenesis, notably RTK, Wnt and retinoic acid pathways (Table S4). Several genes have been implicated in the regulation of the cell cycle and cell proliferation; for example, Ccnd1, Mycn and Cdca7 (Ma et al., 2015; Eilers and Eisenman, 2008; Gill et al., 2013). This observation is consistent with the enhanced rates of cell proliferation seen in UBTs, which is likely to be important for growth and morphogenesis of the ureteric epithelium (Michael and Davies, 2004). The ECM is also of special interest given the requirement of branch tips to move through the extracellular space; several genes in the UBT-enriched gene set are either components of the matrix (Col9a3, Flbn2, Frem2) (Brachvogel et al., 2013; Olijnyk et al., 2014; Timmer et al., 2005) or predicted to remodel matrix components (Adamts18, Cadm1) (Kelwick et al., 2015; Moiseeva et al., 2014) (Table S4).
A subset of UBT-specific genes has been suggested to play a role in kidney development (Table S5). Several of these genes are well-established in kidney development, such as Wnt11, Ret, Gfra1, Etv4 and Etv5. The remaining genes have been linked through individual patient case studies and/or sparse research (Arnt2, Crispld2, Crlf1, Epha4, Mdk, Mycn, Ros1, Sfrp2) (Table S5); their presence in the UBT during kidney development is interesting. Lastly, most genes with UBT-enriched expression have not been implicated in kidney development previously. These novel genes expand our understanding of the molecular features of kidney development identifying new targets for mechanistic follow-up studies.
Distinct zones of gene expression within the UBT are highlighted by distinct subsets of UBT-enriched genes. Class I and II genes demarcate the UBT population discretely and therefore are potentially part of the ‘tip identity’. By contrast, Class III genes extend beyond the UBT into neighboring stalk cells. These differences could reflect distinct regulatory actions in the dynamic events of branching morphogenesis, or in the maintenance and commitment of progenitor cells within the UBT niche. For example, a subset of tip cells lose ‘tip identity’ generating stalk cells in a process that is poorly understood (Shakya et al., 2005; Riccio et al., 2016). Interestingly, Spred2 and Axin2, both Class III genes and negative regulators of MAPK and Wnt signaling, respectively (Wakioka et al., 2001; Yamamoto et al., 1998), show persistent expression in branch tips that could link to an ongoing suppression of UBT signaling promoting branch tip progenitors.
On the other hand, Class I gene expression, exemplified by Wnt11, Tcf7 and Sox8, is tightly restricted to the most distal ends of the UBT. Notably, these genes are all positive activators of branching morphogenesis (Pepicelli et al., 1997; van de Wetering et al., 1997; Reginensi et al., 2011). The broader epithelial expression of negative regulators and tight, tip-most expression domain for positive regulators of branch tip-associated signaling activity could ensure appropriate spatial regulation of these signaling pathways.
Though the temporal analysis was limited, the finding that 86% of genes showed UBT-enriched expression at both E12.5 and E15.5 kidneys argues against a dramatic change in UBT regulatory programs during early stages of mammalian kidney development. Branching growth of the UBT continues until approximately the day of birth (E19.5) in the mouse (Cebrián et al., 2004; Short et al., 2014). However, the UBT niche persists for another 2-3 days of postnatal development, a critical period as over half the nephron precursors (the renal vesicles) form at this time.
Kidney-specific and general branching morphogenesis pathways and components
Comparing the gene expression pattern of UBT-enriched genes between kidneys and lungs has illuminated potential kidney-specific regulators as well as general regulators of branching morphogenesis. Of the 84 genes that display UBT-specific expression in kidneys, 40 have LBT-specific expression. Furthermore, of these genes with tip expression in both kidney and lung, 67% also show expression in the embryonic mouse salivary gland epithelial tip (according to publicly available online SISH and/or microarray data; www.eurexpress.com; sgmap.nidcr.nih.gov) (Table S3) though the salivary gland forms branches through a distinct process of epithelial cleft formation. This dataset is likely to highlight core branching components. Of note, many of these shared tip-specific genes are known to be associated with ECM properties (Adamts18, Cadm1, Col9a3, Hs3st3a1), and cell cycle and proliferation (Ccnd1, Cdca7, Cdca7l, Mdk, Nasp, Uhrf1) highlighting the importance of specific matrix components to branching and enhanced growth (Michael and Davies, 2004).
A few of these genes have been connected to lung and/or salivary gland branching morphogenesis. For example, Etv5 has been previously implicated downstream of FGF signaling in the regulation of lung branching (Liu et al., 2003), and Vldlr, Hs3st3a1 and Hs3st3b1 have been shown to increase FGFR2b-mediated signaling and proliferation during salivary gland branching morphogenesis (Rebustini et al., 2012; Patel et al., 2014). Furthermore, conditional ablation of Ctnnd2 in the salivary gland resulted in decreased levels of E-cadherin (cadherin 1), and defects in cell-cell adhesion and resulting salivary gland morphology (Davis and Reynolds, 2006).
Interestingly, 25 (30%) UBT-enriched genes appear to be absent from LBTs and SBTs (Table S3). These genes include Gfra1, Sox8 and Wnt11, which have been characterized for their roles in branching morphogenesis in the kidney (Cacalano et al., 1998; Enomoto et al., 1998; Reginensi et al., 2011; Pepicelli et al., 1997). However, no functional analysis has been reported for most of this gene set. As RET/GDNF signaling is a central, kidney-specific pathway stimulating UBT branching and progenitor maintenance, differences in the RTK signaling profile are expected and these are reflected by the kidney-specific expression of Dok6, Gfra1, Sox8, Wnt11, and probably several other genes.
Surprisingly, several genes associated with calcium-related biology appear to be kidney specific. These include Cib2, encoding a calcium- and integrin-binding protein that is associated with photoreceptor and hair cell maintenance (Riazuddin et al., 2012), and Capn6, which belongs to the calpain gene family and encodes a calcium-dependent cysteine protease linked to microtubule stabilization (Tonami et al., 2007).
Comparison of mouse and human kidney biology
The majority of developmental kidney studies have used the mouse as the standard model system for human kidney development. However, there are important differences that may reflect different regulatory actions in mouse and human UBTs (Osathanondh and Potter, 1963; Saxen and Sariola, 1987). Whereas the mouse kidney is unilobular, the human kidney is a fusion of multiple independently developing lobes. The differences here may indicate variability in the earliest branching events. Furthermore, much of the cortical expansion of the human kidney beyond week 15 (the human kidney initiates UB outgrowth at around 5 weeks and continues active nephrogenesis until 36 weeks; Osathanondh and Potter, 1963), occurs without further branching.
Despite these marked differences, our initial studies revealed conservation in the expression of a mouse UB-enriched gene set in the human kidney at 15-16 weeks. Thus, human and mouse kidneys likely utilize conserved regulatory mechanisms within and outside of UBTs. Interestingly, Kdm2b, which encodes a lysine demethylase linked to regulation of the Wnt signaling pathway, was not detected in human UBTs (Lu et al., 2015; Boulard et al., 2016). Given our rudimentary knowledge of the actual connection between discrete spatial signaling mediated by WNTs, GDNF, FGFs and retinoic acid, and the distinct cellular outcomes of branching and proliferative cell growth in the mouse kidney, subtle modulation in signaling responses through regulatory factors such as Kdm2b could play a role in the differential dynamics of mouse and human UBTs.
Future exploration of novel UBT genes
Our study has uncovered a large number of genes of potential interest in kidney-specific or organ-wide programs of epithelial branching growth in the mammalian embryo. For example, Adamts18 is highly enriched in the UBT population (FC=47.47, RPKM=15.25), and also shows elevated expression in LBTs and the epithelial tips in the developing salivary gland. Given that Adamts18 encodes a protease in a family that targets ECM components (Kelwick et al., 2015), it is an attractive candidate for a conserved role across different organs to facilitate matrix remodeling in branching growth.
Hs3st3a1 and Hs3st3b1 encode closely related heparan sulfate 3-O-sulfotransferases; the former is enriched in the tips of the kidney, lung and salivary gland whereas the latter is only enriched in SBTs and UBTs. These genes encode enzymes that catalyze the addition of a sulfate group to heparan sulfates. Given the requirement for heparan sulfate as a co-receptor for FGF receptor activation, and the shared requirement for FGF signaling in branching growth across multiple organs, these genes are predicted to play a conserved role in branching morphogenesis. Indeed, initial evidence points to a role for these enzymes in salivary gland development (Patel et al., 2014).
The next phase of analysis requires a detailed systematic functional dissection of the gene sets identified here in the context of mammalian organogenesis. Although much progress has been made in mutational studies in the mouse, this remains a considerable challenge in studying in vivo development, especially where conditional mutagenesis is required to reveal organ-specific roles. Kidney and lung organoid models could facilitate mutational studies (Takasato et al., 2015; Dye et al., 2015) if it can be shown that these organoid models recapitulate normal epithelial morphogenesis. Connecting these gene sets with genome-wide association studies suggesting gene-disease associations within specific organs systems when taken together with insights from the analyses here may help in prioritizing specific targets of interest for functional analysis.
MATERIALS AND METHODS
RNA sequencing
UBT cells, and their committed epithelial stalk descendants, were isolated from E16.5 kidneys carrying Wnt11-RFP [Harding et al., 2011; B6;D-Tg(Wnt11-TagRFP/cre/ERT2)28Amc/J] and Hoxb7-GFP [Srinivas et al., 1999; 129S.Cg-Tg(Hoxb7-EGFP)33Cos/J] transgenes by FACS. All mouse handling, husbandry and procedures were performed in compliance with the guidelines set forth by the Institutional Animal Care and Use Committees (IACUC) at the University of Southern California. In the ureteric epithelium at this time, Wnt11 is expressed specifically in UBTs, whereas Hoxb7 is present throughout the entire ureteric epithelium [Wnt11 is also expressed within the interstitial mesenchyme cells starting from E15.5 (Yu et al., 2009)]. RNA was isolated from UBT-enriched (RFP+, GFP+) and stalk-enriched (RFP−, GFP+) fractions from three biological replicates, and mRNA samples subjected to Next Generation RNA sequencing: 150 bases, paired-end reads were obtained on a HiSeq 2000. Raw sequencing reads were trimmed with the Quality Score method. TopHat2 was utilized to map these trimmed reads to the mouse genome (mm10 annotation). The Partek E/M method was applied using Partek suite bioinformatics tool to quantify the aligned reads to genes/transcripts with Ensembl 72 annotation. Upper quartile normalization was used to normalize the read counts per gene/transcript in all samples (Bullard et al., 2010). A minimum expression filter was used to discard any transcripts if total raw reads across all samples was less than ten. This data was analyzed for differential expression using the PartekGene Specific Analysis method. The resulting RNA-seq data set comprised 37,149 mapped transcripts. We narrowed down these transcripts to focus on those genes that most likely modulate kidney tip morphogenesis by applying a reasonably stringent cutoff with a minimum RPKM value of ten in at least one of the two cell populations and a 5-fold or greater difference in tip or stalk expression to identify robust tip- and stalk-enriched gene sets. The RNA-seq data are available in Gene Expression Omnibus (accession number GSE93267).
Riboprobe generation
Total RNA was extracted from Swiss Webster E15.5 kidneys or human fetal kidney tissues (de-identified human tissue was donated anonymously from elective terminations with informed consent and internal review board approval obtained for the experimental use of the tissue samples) using the RNeasy micro kit (Qiagen). RNA integrity was confirmed using a NanoDrop 2000c (Thermo Scientific) and the SuperScript VILO cDNA synthesis kit (Invitrogen) was used to generate cDNA from the total RNA. Primers for each gene of interest in the RNA expression screen were designed using Primer3, adding a T7 promoter sequence to the 5′ end of the reverse primer. A cDNA probe template was amplified using these primers, the appropriate PCR product confirmed by gel electrophoresis, and digoxigenin-labeled RNA synthesized was from each DNA template using T7 RNA polymerase. Digoxigenin-labeled RNA probes were purified on Micro Bio-Spin columns (Bio-Rad) and the transcript integrity confirmed by gel electrophoresis. Probes were diluted to a concentration of 10 ng/µl in pre-hybridization buffer (50% formamide, 5× SSC, 1% SDS, 50 μg/ml heparin, 50 μg/ml yeast tRNA) and stored long-term at −80°C.
In situ hybridization (ISH)
In situ hybridization was performed using our previously reported procedure (Yu et al., 2012). Briefly, for WISH, urogenital systems and lungs of E12.5 and E15.5 Swiss Webster mice were harvested and fixed overnight in 4% paraformaldehyde (PFA), then dehydrated through a methanol series and stored in methanol at −20°C before use. Samples were rehydrated and bleached with 6% hydrogen peroxide, incubated in proteinase K (10 μg/ml), fixed in 4% PFA, and then pre-hybridized in hybridization buffer for several hours at 70°C before hybridization with each RNA probe. Samples were transferred to a BioLane HTI machine for formamide washes, antibody incubation, and MBST [100 mM maleic acid, 150 mM NaCl, 0.1% Tween-20 (pH 7.5)] washes. To reveal in situ hybridization, samples were incubated with BM Purple for up to 48 h, post-fixed in 4% PFA, then transferred to 80% glycerol for imaging on an AxioZoom.V16 stereozoom microscope (Zeiss).
For SISH, mouse and human fetal tissues (anonymously donated from elective terminations and obtained in accordance with institutional guidelines) were harvested and briefly fixed in 4% PFA. Tissues were placed in 30% sucrose overnight at 4°C, embedded in OCT, and sectioned at 12 μm on a Zeiss Microm HM550 cryostat. Slides were fixed in 4% PFA overnight, treated with proteinase K, followed by a brief 4% PFA fixation. To decrease background staining, tissues were acetylated in an acetylation solution (1 M triethanolamine, 0.65% HCl and 0.375% acetic anhydride) and then dehydrated in 95% ethanol. Probes were applied to the slides and incubated overnight at 70°C, then washed with 50% formamide, TNE [10 mM Tris (pH 7.5), 500 mM NaCl, 1 mM EDTA], 2× SSC, 0.2× SSC and MBST [100 mM maleic acid, 150 mM NaCl, 0.1× Tween-20 (pH 7.5)] solutions. Slides were incubated with blocking solution [2% blocking reagent (Roche), 20% heat-inactivated sheep serum] for 1 h, then overnight at 4°C in antibody solution. Samples were stained with BM Purple for up to 14 days, fixed in 4% PFA, and mounted with Glycergel (Dako). SISH samples were imaged on an AxioScan.Z1 (Zeiss).
To compare expression of two genes on the same section, we utilized an RNAscope 2.5 Duplex Detection Kit (Advanced Cell Diagnostics). Following routine dissection, embedding and sectioning as for other SISH experiments, tissues were serially incubated in hydrogen peroxide and protease in the kit according to manufacturer's instructions, then hybridized with RNAscope's probes at 40°C in the HybEZ oven (Advanced Cell Diagnostics). Slides were briefly stained as appropriate for each probe labeling following manufacturer's recommendations, and then mounted with VectaMount (Vector Laboratories). Tissues were imaged on an AxioImager.Z1 (Zeiss).
Salivary branch tip (SBT) expression online resources analyses
Two publicly available online resources were utilized to analyze SBT expression. Eurexpress (www.eurexpress.org) provides E14.5 whole embryo SISH data. Salivary Gland Molecular Anatomy Project (http://sgmap.nidcr.nih.gov) showcases microarray data from a range of developmental stages from microdissected regions of the submandibular gland (Diez-Roux et al., 2011; Musselmann et al., 2011). E13.0 epithelial bud to duct expression ratio was calculated, and ratios above 1.15 were considered SBT-enriched.
Acknowledgements
We thank Yibu Chen and Meng Li for their help with the RNA sequencing, and Gohar Saribekyan for sectioning the human tissue.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: J.-D.B., A.P.M.; Methodology: E.A.R., J.-D.B., A.P.M.; Validation: E.A.R.; Formal analysis: E.A.R., J.-D.B.; Investigation: E.A.R., J.-D.B.; Writing - original draft: E.A.R.; Writing - review & editing: E.A.R., J.-D.B., A.P.M.; Visualization: E.A.R.; Supervision: A.P.M.; Project administration: A.P.M.; Funding acquisition: E.A.R., A.P.M.
Funding
Work in A.P.M.'s laboratory was funded by a grant from the National Institutes of Health [DK054364]. E.R. was supported by a graduate student fellowship from the National Institutes of Health [5T32HD060549] and by the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health [F31DK107216]. Deposited in PMC for release after 12 months.
Data availability
The RNA-seq data are available at Gene Expression Omnibus under accession number GSE93267. All kidney in situ data will be made available through GUDMAP (www.gudmap.org) and all lung in situ data will be made available through LUNG-MAP (www.lung-map.org).
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.149112.supplemental
References
- Affolter M. and Caussinus E. (2008). Tracheal branching morphogenesis in Drosophila: new insights into cell behaviour and organ architecture. Development 135, 2055-2064. 10.1242/dev.014498 [DOI] [PubMed] [Google Scholar]
- Arman E., Haffner-Krausz R., Gorivodsky M. and Lonai P. (1999). Fgfr2 is required for limb outgrowth and lung-branching morphogenesis. Proc. Natl. Acad. Sci. USA 96, 11895-11899. 10.1073/pnas.96.21.11895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belle M., Parray A., Belle M., Chédotal A. and Nguyen-Ba-Charvet K. T. (2016). PlexinA2 and Sema6A are required for retinal progenitor cell migration. Dev. Growth Differ. 58, 492-502. 10.1111/dgd.12298 [DOI] [PubMed] [Google Scholar]
- Blomqvist S. R., Vidarsson H., Fitzgerald S., Johansson B. R., Ollerstam A., Brown R., Persson A. E. G., Bergström G. G. and Enerbäck S. (2004). Distal renal tubular acidosis in mice that lack the forkhead transcription factor Foxi1. J. Clin. Invest. 113, 1560-1570. 10.1172/JCI20665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock H. H. and May P. (2016). Canonical and non-canonical reelin signaling. Front. Cell Neurosci. 10, 166 10.3389/fncel.2016.00166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulard M., Edwards J. R. and Bestor T. H. (2016). Abnormal X chromosome inactivation and sex-specific gene dysregulation after ablation of FBXL10. Epigenetics Chromatin 9, 22 10.1186/s13072-016-0069-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachvogel B., Zaucke F., Dave K., Norris E. L., Stermann J., Dayakli M., Koch M., Gorman J. J., Bateman J. F. and Wilson R. (2013). Comparative proteomic analysis of normal and collagen IX null mouse cartilage reveals altered extracellular matrix composition and novel components of the collagen IX interactome. J. Biol. Chem. 288, 13481-13492. 10.1074/jbc.M112.444810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown T., Mandell J. and Lebowitz R. L. (1987). Neonatal hydronephrosis in the era of sonography. AJR Am. J. Roentgenol. 148, 959-963. 10.2214/ajr.148.5.959 [DOI] [PubMed] [Google Scholar]
- Bullard J. H., Purdom E., Hansen K. D. and Dudoit S. (2010). Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinform. 11, 94 10.1186/1471-2105-11-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacalano G., Farinas I., Wang L.-C., Hagler K., Forgie A., Moore M., Armanini M., Phillips H., Ryan A. M., Reichardt L. F. et al. (1998). GFRalpha1 is an essential receptor component for GDNF in the developing nervous system and kidney. Neuron 21, 53-62. 10.1016/S0896-6273(00)80514-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebrián C., Borodo K., Charles N. and Herzlinger D. A. (2004). Morphometric index of the developing murine kidney. Dev. Dyn. 231, 601-608. 10.1002/dvdy.20143 [DOI] [PubMed] [Google Scholar]
- Costantini F. (2012). Genetic controls and cellular behaviors in branching morphogenesis of the renal collecting system. Wiley Interdiscip. Rev. Dev. Biol. 1, 693-713. 10.1002/wdev.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini F. and Shakya R. (2006). GDNF/Ret signaling and the development of the kidney. BioEssays. 28, 117-127. 10.1002/bies.20357 [DOI] [PubMed] [Google Scholar]
- Davis M. A. and Reynolds A. B. (2006). Blocked acinar development, E-cadherin reduction, and intraepithelial neoplasia upon ablation of p120-catenin in the mouse salivary gland. Dev. Cell 10, 21-31. 10.1016/j.devcel.2005.12.004 [DOI] [PubMed] [Google Scholar]
- De Moerlooze L., Spencer-Dene B., Revest J. M., Hajihosseini M., Rosewell I. and Dickson C. (2000). An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development 127, 483-492. [DOI] [PubMed] [Google Scholar]
- Diez-Roux G., Banfi S., Sultan M., Geffers L., Anand S., Rozado D., Magen A., Canidio E., Pagani M., Peluso I. et al. (2011). A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 9, e1000582 10.1371/journal.pbio.1000582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye B. R., Hill D. R., Ferguson M. A., Tsai Y. H., Nagy M. S., Dyal R., Wells J. M., Mayhew C. N., Nattiv R., Klein O. D. et al. (2015). In vitro generation of human pluripotent stem cell derived lung organoids. Elife, doi: 10.7554/eLife.05098 10.7554/eLife.05098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M. and Eisenman R. N. (2008). Myc's broad reach. Genes Dev. 22, 2755-2766. 10.1101/gad.1712408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto H., Araki T., Jackman A., Heuckeroth R. O., Snider W. D., Johnson E. M. Jr. and Milbrandt J. (1998). GFR alpha1-deficient mice have deficits in the enteric nervous system and kidneys. Neuron 21, 317-324. 10.1016/S0896-6273(00)80541-3 [DOI] [PubMed] [Google Scholar]
- Gill R. M., Gabor T. V., Couzens A. L. and Scheid M. P. (2013). The MYC-associated protein CDCA7 is phosphorylated by AKT to regulate MYC-dependent apoptosis and transformation. Mol. Cell. Biol. 33, 498-513. 10.1128/MCB.00276-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding S. D., Armit C., Armstrong J., Brennan J., Cheng Y., Haggarty B., Houghton D., Lloyd-MacGilp S., Pi X., Roochun Y.. et al. (2011). The GUDMAP database--an online resource for genitourinary research. Development 138, 2845-2853. 10.1242/dev.063594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herriges M. and Morrisey E. E. (2014). Lung development: orchestrating the generation and regeneration of a complex organ. Development 141, 502-513. 10.1242/dev.098186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herriges J. C., Verheyden J. M., Zhang Z., Sui P., Zhang Y., Anderson M. J., Swing D. A., Lewandoski M. and Sun X. (2015). FGF-regulated ETV transcription factors control FGF-SHH feedback loop in lung branching. Dev. Cell 35, 322-332. 10.1016/j.devcel.2015.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iber D. and Menshykau D. (2013). The control of branching morphogenesis Open Biol. 3, 130088 10.1098/rsob.130088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibe S., Karihaloo A., Ma H., Zhang J., Marlier A., Mitobe M., Togawa A., Schmitt R., Czyczk J., Kashgarian M. et al. (2009). Met and the epidermal growth factor receptor act cooperatively to regulate final nephron number and maintain collecting duct morphology. Development 136, 337-345. 10.1242/dev.024463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T. M., Silvia C., Hirschberg B., Bond C. T., Adelman J. P. and Maylie J. (1997). A human intermediate conductance calcium-activated potassium channel. Proc. Natl. Acad. Sci. USA 94, 11651-11656. 10.1073/pnas.94.21.11651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joiner W. J., Wang L.-Y., Tang M. D. and Kaczmarek L. K. (1997). hSK4, a member of a novel subfamily of calcium-activated potassium channels. Proc. Natl. Acad. Sci. USA 94, 11013-11018. 10.1073/pnas.94.20.11013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keefe Davis T., Hoshi M. and Jain S. (2013). Stage specific requirement of Gfralpha1 in the ureteric epithelium during kidney development. Mech. Dev. 130, 506-518. 10.1016/j.mod.2013.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelwick R., Desanlis I., Wheeler G. N. and Edwards D. R. (2015). The ADAMTS (A Disintegrin and Metalloproteinase with Thrombospondin motifs) family. Genome Biol. 16, 113 10.1186/s13059-015-0676-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. Y. and Nelson C. M. (2012). Extracellular matrix and cytoskeletal dynamics during branching morphogenesis. Organogenesis 8, 56-64. 10.4161/org.19813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon T.-H., Frøkiær J. and Nielsen S. (2013). Regulation of aquaporin-2 in the kidney: a molecular mechanism of body-water homeostasis. Kidney Res. Clin. Pract. 32, 96-102. 10.1016/j.krcp.2013.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leighton P. A., Mitchell K. J., Goodrich L. V., Lu X., Pinson K., Scherz P., Skarnes W. C. and Tessier-Lavigne M. (2001). Defining brain wiring patterns and mechanisms through gene trapping in mice. Nature 410, 174-179. 10.1038/35065539 [DOI] [PubMed] [Google Scholar]
- Little M. H. and McMahon A. P. (2012). Mammalian kidney development: principles, progress, and projections. Cold Spring Harb. Perspect. Biol. 4, a008300 10.1101/cshperspect.a008300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. Z., Wada J., Kumar A., Carone F. A., Takahashi M. and Kanwar Y. S. (1996). Comparative role of phosphotyrosine kinase domains of c-ros and c-ret protooncogenes in metanephric development with respect to growth factors and matrix morphogens. Dev. Biol. 178, 133-148. 10.1006/dbio.1996.0204 [DOI] [PubMed] [Google Scholar]
- Liu Y., Jiang H., Crawford H. C. and Hogan B. L. (2003). Role for ETS domain transcription factors Pea3/Erm in mouse lung development. Dev. Biol. 261, 10-24. 10.1016/S0012-1606(03)00359-2 [DOI] [PubMed] [Google Scholar]
- Lu B. C., Cebrian C., Chi X., Kuure S., Kuo R., Bates C. M., Arber S., Hassell J., MacNeil L., Hoshi M. et al. (2009). Etv4 and Etv5 are required downstream of GDNF and Ret for kidney branching morphogenesis. Nat. Genet. 41, 1295-1302. 10.1038/ng.476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Gao Y., Zhang Z., Cao Q., Zhang X., Zou J. and Cao Y. (2015). Kdm2a/b lysine demethylases regulate canonical wnt signaling by modulating the stability of nuclear beta-catenin. Dev. Cell 33, 660-674. 10.1016/j.devcel.2015.04.006 [DOI] [PubMed] [Google Scholar]
- Ma J., Cui B., Ding X., Wei J. and Cui L. (2015). Over-expression of cyclin D1 promotes NSCs Proliferation and induces the differentiation into astrocytes via Jak-STAT3 pathways. Neurochem. Res. 40, 1681-1690. 10.1007/s11064-015-1635-9 [DOI] [PubMed] [Google Scholar]
- Majumdar A., Vainio S., Kispert A., McMahon J. and McMahon A. P. (2003). Wnt11 and Ret/Gdnf pathways cooperate in regulating ureteric branching during metanephric kidney development. Development 130, 3175-3185. 10.1242/dev.00520 [DOI] [PubMed] [Google Scholar]
- Matsuoka R. L., Nguyen-Ba-Charvet K. T., Parray A., Badea T. C., Chédotal A. and Kolodkin A. L. (2011). Transmembrane semaphorin signalling controls laminar stratification in the mammalian retina. Nature 470, 259-263. 10.1038/nature09675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulley D., Wienhold M. and Sun X. (2015). The pulmonary mesenchyme directs lung development. Curr. Opin. Genet. Dev. 32, 98-105. 10.1016/j.gde.2015.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon A. P. (2016). Development of the mammalian kidney. Curr. Top. Dev. Biol. 117, 31-64. 10.1016/bs.ctdb.2015.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon A. P., Aronow B. J., Davidson D. R., Davies J. A., Gaido K. W., Grimmond S., Lessard J. L., Little M. H., Potter S. S., Wilder E. L. et al. (2008). GUDMAP: the genitourinary developmental molecular anatomy project. J. Am. Soc. Nephrol. 19, 667-671. 10.1681/ASN.2007101078 [DOI] [PubMed] [Google Scholar]
- Metzger R. J., Klein O. D., Martin G. R. and Krasnow M. A. (2008). The branching programme of mouse lung development. Nature 453, 745-750. 10.1038/nature07005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael L. and Davies J. A. (2004). Pattern and regulation of cell proliferation during murine ureteric bud development. J. Anat. 204, 241-255. 10.1111/j.0021-8782.2004.00285.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min H., Danilenko D. M., Scully S. A., Bolon B., Ring B. D., Tarpley J. E., DeRose M. and Simonet W. S. (1998). Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes Dev. 12, 3156-3161. 10.1101/gad.12.20.3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moiseeva E. P., Straatman K. R., Leyland M. L. and Bradding P. (2014). CADM1 controls actin cytoskeleton assembly and regulates extracellular matrix adhesion in human mast cells. PLoS ONE 9, e85980 10.1371/journal.pone.0085980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselmann K., Green J. A., Sone K., Hsu J. C., Bothwell I. R., Johnson S. A., Harunaga J. S., Wei Z. and Yamada K. M. (2011). Salivary gland gene expression atlas identifies a new regulator of branching morphogenesis. J. Dent. Res. 90, 1078-1084. 10.1177/0022034511413131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaou N., Renkema K. Y., Bongers E. M., Giles R. H. and Knoers N. V. (2015). Genetic, environmental, and epigenetic factors involved in CAKUT. Nat. Rev. Nephrol. 11, 720-731. 10.1038/nrneph.2015.140 [DOI] [PubMed] [Google Scholar]
- O'Brien L. L. and McMahon A. P. (2014). Induction and patterning of the metanephric nephron. Semin. Cell Dev. Biol. 36, 31-38. 10.1016/j.semcdb.2014.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa-Espinosa A. and Affolter M. (2012). Branching morphogenesis: from cells to organs and back. Cold Spring Harb. Perspect. Biol. 4, a008243 10.1101/cshperspect.a008243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohuchi H., Hori Y., Yamasaki M., Harada H., Sekine K., Kato S. and Itoh N. (2000). FGF10 acts as a major ligand for FGF receptor 2 IIIb in mouse multi-organ development. Biochem. Biophys. Res. Commun. 277, 643-649. 10.1006/bbrc.2000.3721 [DOI] [PubMed] [Google Scholar]
- Olijnyk D., Ibrahim A. M., Ferrier R. K., Tsuda T., Chu M.-L., Gusterson B. A., Stein T. and Morris J. S. (2014). Fibulin-2 is involved in early extracellular matrix development of the outgrowing mouse mammary epithelium. Cell. Mol. Life Sci. 71, 3811-3828. 10.1007/s00018-014-1577-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osathanondh V. and Potter E. L. (1963). Development of human kidney as shown by microdissection. Iii. Formation and interrelationship of collecting tubules and nephrons . Arch. Pathol. 76, 290-302. [PubMed] [Google Scholar]
- Pachnis V., Mankoo B. and Costantini F. (1993). Expression of the c-ret proto-oncogene during mouse embryogenesis. Development. 119, 1005-1017. [DOI] [PubMed] [Google Scholar]
- Patel V. N. and Hoffman M. P. (2014). Salivary gland development: a template for regeneration. Semin. Cell Dev. Biol. 25-26, 52-60. 10.1016/j.semcdb.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel V. N., Lombaert I. M. A., Cowherd S. N., Shworak N. W., Xu Y., Liu J. and Hoffman M. P. (2014). Hs3st3-modified heparan sulfate controls KIT+ progenitor expansion by regulating 3-O-sulfotransferases. Dev. Cell 29, 662-673. 10.1016/j.devcel.2014.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepicelli C. V., Kispert A., Rowitch D. H. and McMahon A. P. (1997). GDNF induces branching and increased cell proliferation in the ureter of the mouse. Dev. Biol. 192, 193-198. 10.1006/dbio.1997.8745 [DOI] [PubMed] [Google Scholar]
- Pepicelli C. V., Lewis P. M. and McMahon A. P. (1998). Sonic hedgehog regulates branching morphogenesis in the mammalian lung. Curr. Biol. 8, 1083-1086. 10.1016/S0960-9822(98)70446-4 [DOI] [PubMed] [Google Scholar]
- Qiu L., Hyink D. P., Gans W. H., Amsler K., Wilson P. D. and Burrow C. R. (2004). Midkine promotes selective expansion of the nephrogenic mesenchyme during kidney organogenesis. Organogenesis. 1, 14-21. 10.4161/org.1.1.979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queißer-Luft A., Stolz G., Wiesel A., Schlaefer K. and Spranger J. (2002). Malformations in newborn: results based on 30,940 infants and fetuses from the Mainz congenital birth defect monitoring system (1990-1998). Arch. Gynecol. Obstet. 266, 163-167. 10.1007/s00404-001-0265-4 [DOI] [PubMed] [Google Scholar]
- Quinlan J., Kaplan F., Sweezey N. and Goodyer P. (2007). LGL1, a novel branching morphogen in developing kidney, is induced by retinoic acid. Am. J. Physiol. Renal Physiol. 293, F987- F993. 10.1152/ajprenal.00098.2007 [DOI] [PubMed] [Google Scholar]
- Rebustini I. T., Hayashi T., Reynolds A. D., Dillard M. L., Carpenter E. M. and Hoffman M. P. (2012). miR-200c regulates FGFR-dependent epithelial proliferation via Vldlr during submandibular gland branching morphogenesis. Development 139, 191-202. 10.1242/dev.070151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reginensi A., Clarkson M., Neirijnck Y., Lu B., Ohyama T., Groves A. K., Sock E., Wegner M., Costantini F., Chaboissier M.-C. et al. (2011). SOX9 controls epithelial branching by activating RET effector genes during kidney development. Hum. Mol. Genet. 20, 1143-1153. 10.1093/hmg/ddq558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riazuddin S., Belyantseva I. A., Giese A. P. J., Lee K., Indzhykulian A. A., Nandamuri S. P., Yousaf R., Sinha G. P., Lee S., Terrell D. et al. (2012). Alterations of the CIB2 calcium- and integrin-binding protein cause Usher syndrome type 1J and nonsyndromic deafness DFNB48. Nat. Genet. 44, 1265-1271. 10.1038/ng.2426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccio P., Cebrian C., Zong H., Hippenmeyer S. and Costantini F. (2016). Ret and Etv4 promote directed movements of progenitor cells during renal branching morphogenesis. PLoS Biol. 14, e1002382 10.1371/journal.pbio.1002382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock J. R. and Hogan B. L. M. (2011). Epithelial progenitor cells in lung development, maintenance, repair, and disease. Annu. Rev. Cell Dev. Biol. 27, 493-512. 10.1146/annurev-cellbio-100109-104040 [DOI] [PubMed] [Google Scholar]
- Sallstrom J., Peuckert C., Gao X., Larsson E., Nilsson A., Jensen B. L., Onozato M. L., Persson A. E. G., Kullander K. and Carlstrom M. (2013). Impaired EphA4 signaling leads to congenital hydronephrosis, renal injury, and hypertension. Am. J. Physiol. Renal Physiol. 305, F71-F79. 10.1152/ajprenal.00694.2012 [DOI] [PubMed] [Google Scholar]
- Saxen L. and Sariola H. (1987). Early organogenesis of the kidney. Pediatr. Nephrol. 1, 385-392. 10.1007/BF00849241 [DOI] [PubMed] [Google Scholar]
- Schmidt-Ott K. M., Yang J., Chen X., Wang H., Paragas N., Mori K., Li J. Y., Lu B., Costantini F., Schiffer M. et al. (2005). Novel regulators of kidney development from the tips of the ureteric bud. J. Am. Soc. Nephrol. 16, 1993-2002. 10.1681/ASN.2004121127 [DOI] [PubMed] [Google Scholar]
- Schuchardt A., D'Agati V., Larsson-Blomberg L., Costantini F. and Pachnis V. (1994). Defects in the kidney and enteric nervous system of mice lacking the tyrosine kinase receptor Ret. Nature 367, 380-383. 10.1038/367380a0 [DOI] [PubMed] [Google Scholar]
- Schwab K., Patterson L. T., Aronow B. J., Luckas R., Liang H.-C. and Potter S. S. (2003). A catalogue of gene expression in the developing kidney. Kidney Int. 64, 1588-1604. 10.1046/j.1523-1755.2003.00276.x [DOI] [PubMed] [Google Scholar]
- Sekine K., Ohuchi H., Fujiwara M., Yamasaki M., Yoshizawa T., Sato T., Yagishita N., Matsui D., Koga Y., Itoh N. et al. (1999). Fgf10 is essential for limb and lung formation. Nat. Genet. 21, 138-141. 10.1038/5096 [DOI] [PubMed] [Google Scholar]
- Skinner M. A., Safford S. D., Reeves J. G., Jackson M. E. and Freemerman A. J. (2008). Renal aplasia in humans is associated with RET mutations. Am. J. Hum. Genet. 82, 344-351. 10.1016/j.ajhg.2007.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakya R., Watanabe T. and Costantini F. (2005). The role of GDNF/Ret signaling in ureteric bud cell fate and branching morphogenesis. Dev. Cell 8, 65-74. 10.1016/j.devcel.2004.11.008 [DOI] [PubMed] [Google Scholar]
- Short K. M., Combes A. N., Lefevre J., Ju A. L., Georgas K. M., Lamberton T., Cairncross O., Rumballe B. A., McMahon A. P., Hamilton N. A. et al. (2014). Global quantification of tissue dynamics in the developing mouse kidney. Dev. Cell 29, 188-202. 10.1016/j.devcel.2014.02.017 [DOI] [PubMed] [Google Scholar]
- Srinivas S., Goldberg M. R., Watanabe T., D'Agati V., al-Awqati Q. and Costantini F. (1999). Expression of green fluorescent protein in the ureteric bud of transgenic mice: a new tool for the analysis of ureteric bud morphogenesis. Dev. Genet. 24, 241-251. [DOI] [PubMed] [Google Scholar]
- Stuart R. O., Bush K. T. and Nigam S. K. (2003). Changes in gene expression patterns in the ureteric bud and metanephric mesenchyme in models of kidney development. Kidney Int. 64, 1997-2008. 10.1046/j.1523-1755.2003.00383.x [DOI] [PubMed] [Google Scholar]
- Suto F., Tsubio M., Kamiya H., Mizuno H., Kiyama Y., Komai S., Shimizu M., Sanbo M., Yagi T., Hiromi Y. et al. (2007). Interactions between plexin-A2, plexin-A4, and semaphorin 6A control lamina-restricted projection of hippocampal mossy fibers. Neuron 53, 535-547. 10.1016/j.neuron.2007.01.028 [DOI] [PubMed] [Google Scholar]
- Takasato M., Er P. X., Chiu H. S., Maier B., Baillie G. J., Ferguson C., Parton R. G., Wolvetang E. J., Roost M. S., Chuva de Sousa Lopes S. M. et al. (2015). Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526, 564-568. 10.1038/nature15695 [DOI] [PubMed] [Google Scholar]
- Timmer J. R., Mak T. W., Manova K., Anderson K. V. and Niswander L. (2005). Tissue morphogenesis and vascular stability require the Frem2 protein, product of the mouse myelencephalic blebs gene. Proc. Natl. Acad. Sci. USA 102, 11746-11750. 10.1073/pnas.0505404102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonami K., Kurihara Y., Aburatani H., Uchijima Y., Asano T. and Kurihara H. (2007). Calpain 6 is involved in microtubule stabilization and cytoskeletal organization. Mol. Cell. Biol. 27, 2548-2561. 10.1128/MCB.00992-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueb B., Amann R. and Gerber S. D. (2013). Role of FGFRL1 and other FGF signaling proteins in early kidney development. Cell. Mol. Life Sci. 70, 2505-2518. 10.1007/s00018-012-1189-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufro A., Teichman J., Banu N. and Villegas G. (2007). Crosstalk between VEGF-A/VEGFR2 and GDNF/RET signaling pathways. Biochem. Biophys. Res. Commun. 358, 410-416. 10.1016/j.bbrc.2007.04.146 [DOI] [PubMed] [Google Scholar]
- van de Wetering M., Cavallo R., Dooijes D., van Beest M., van Es J., Loureiro J., Ypma A., Hursh D., Jones T., Bejsovec A. et al. (1997). Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell 88, 789-799. 10.1016/S0092-8674(00)81925-X [DOI] [PubMed] [Google Scholar]
- Varner V. D. and Nelson C. M. (2014). Cellular and physical mechanisms of branching morphogenesis. Development 141, 2750-2559 10.1242/dev.104794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilar J., Lalou C., Duong Van Huyen J. P., Charrin S., Hardouin S., Raulais D., Merlet-Benichou C. and Lelievre-Pegorier M. (2002). Midkine is involved in kidney development and in its regulation by retinoids. J. Am. Soc. Nephrol. 13, 668-676. [DOI] [PubMed] [Google Scholar]
- Wakioka T., Sasaki A., Kato R., Shouda T., Matsumoto A., Miyoshi K., Tsuneoka M., Komiya S., Baron R. and Yoshimura A. (2001). Spred is a Sprouty-related suppressor of Ras signalling. Nature 412, 647-651. 10.1038/35088082 [DOI] [PubMed] [Google Scholar]
- Walker K. A., Sims-Lucas S. and Bates C. M. (2016). Fibroblast growth factor receptor signaling in kidney and lower urinary tract development. Pediatr. Nephrol. 31, 885-895. 10.1007/s00467-015-3151-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb E. A., AlMutair A., Kelberman D., Bacchelli C., Chanudet E., Lescai F., Andoniadou C. L., Banyan A., Alsawaid A., Alrifai M. T. et al. (2013). ARNT2 mutation causes hypopituitarism, post-natal microcephaly, visual and renal anomalies. Brain 136, 3096-3105. 10.1093/brain/awt218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss A.-C., Airik R., Bohnenpoll T., Greulich F., Foik A., Trowe M.-O., Rudat C., Costantini F., Adams R. H. and Kispert A. (2014). Nephric duct insertion requires EphA4/EphA7 signaling from the pericloacal mesenchyme. Development 141, 3420-3430. 10.1242/dev.113928 [DOI] [PubMed] [Google Scholar]
- Williams R. D., Chagtai T., Alcaide-German M., Apps J., Wegert J., Popov S., Vujanic G., van Tinteren H., van den Heuvel-Eibrink M. M., Kool M. et al. (2015). Multiple mechanisms of MYCN dysregulation in Wilms tumour. Oncotarget 6, 7232-7243. 10.18632/oncotarget.3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X.-R., Kong X.-P., Pellicer A., Kreibich G. and Sun T.-T. (2009). Uroplakins in urothelial biology, function, and disease. Kidney Int. 75, 1153-1165. 10.1038/ki.2009.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H., Kishida S., Uochi T., Ikeda S., Koyama S., Asashima M. and Kikuchi A. (1998). Axil, a member of the Axin family, interacts with both glycogen synthase kinase 3beta and beta-catenin and inhibits axis formation of Xenopus embryos. Mol. Cell. Biol. 18, 2867-2875. 10.1128/MCB.18.5.2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshino K., Rubin J. S., Higinbotham K. G., Uren A., Anest V., Plisov S. Y. and Perantoni A. O. (2001). Secreted Frizzled-related proteins can regulate metanephric development. Mech. Dev. 102, 45-55. 10.1016/S0925-4773(01)00282-9 [DOI] [PubMed] [Google Scholar]
- Yu J., Carroll T. J. and McMahon A. P. (2002). Sonic hedgehog regulates proliferation and differentiation of mesenchymal cells in the mouse metanephric kidney. Development 129, 5301-5312. [DOI] [PubMed] [Google Scholar]
- Yu J., Carroll T. J., Rajagopal J., Kobayashi A., Ren Q. and McMahon A. P. (2009). A Wnt7b-dependent pathway regulates the orientation of epithelial cell division and establishes the cortico-medullary axis of the mammalian kidney. Development 136, 161-171. 10.1242/dev.022087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Valerius M. T., Duah M., Staser K., Hansard J. K., Guo J. J., McMahon J., Vaughan J., Faria D., Georgas K. et al. (2012). Identification of molecular compartments and genetic circuitry in the developing mammalian kidney. Development 139, 1863-1873. 10.1242/dev.074005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Li Y., Jun Wei J. W. and Liu X. (2012). The role of Sox genes in lung morphogenesis and cancer. Int. J. Mol. Sci. 13, 15767-15783. 10.3390/ijms131215767 [DOI] [PMC free article] [PubMed] [Google Scholar]