Abstract
Identifying key cellular events that facilitate stem cell function and tissue organization is crucial for understanding the process of regeneration. Planarians are powerful model system to study regeneration and stem cell (neoblast) function. Here, using planaria, we show that the initial events of regeneration, such as epithelialization and epidermal organization are critically regulated by a novel cytoplasmic poly A-binding protein, SMED-PABPC2. Knockdown of smed-pabpc2 leads to defects in epidermal lineage specification, disorganization of epidermis and ECM, and deregulated wound healing, resulting in the selective failure of neoblast proliferation near the wound region. Polysome profiling suggests that epidermal lineage transcripts, including zfp-1, are translationally regulated by SMED-PABPC2. Together, our results uncover a novel role for SMED-PABPC2 in the maintenance of epidermal and ECM integrity, critical for wound healing and subsequent processes for regeneration.
KEY WORDS: Planaria, Epidermis, Poly (A)-binding proteins, Neoblast, Regeneration
Summary: PABPC2 plays a crucial role in the maintenance of epidermal and extracellular matrix integrity, which is essential for neoblast function and regeneration in planarians.
INTRODUCTION
Planarians have emerged as a tractable model system with which to study regeneration and stem cell biology. Their remarkable regenerative prowess comes from a population of adult somatic stem cells, called neoblasts, present throughout the mesenchyme of the animal (Baguñà, 2012; Wolff, 1962). Upon amputation or injury, neoblasts proliferate in two characteristic mitotic peaks in order to meet the demands of the regenerative process (Wenemoser and Reddien, 2010). The first mitotic peak is a body-wide response to injury and occurs within 12 h post-amputation (hpa) and declines after 24 hpa. The second mitotic peak is localized near the wound region and is observed at 48 hpa. Neoblast progeny initially form an undifferentiated, unpigmented tissue, called blastema, at the site of amputation, which is devoid of neoblast cells (Aboobaker, 2011; Rink, 2013). The blastema grows and differentiates further to form missing tissues and organs. Recent studies have also highlighted the role of non-neoblasts cells in instructing neoblasts for planarian regeneration. For example, position control genes (PCGs) that provide positional cues to neoblasts are expressed in the muscle cells (Witchley et al., 2013).
Recent efforts have uncovered broad roles for different post-transcriptional processes in stem cell/neoblast maintenance and differentiation (Guo et al., 2006; Palakodeti et al., 2008; Rouhana et al., 2010; Sasidharan et al., 2013; Solana et al., 2013; Lakshmanan et al., 2016; Wang et al., 2010). Here, we describe the role of cytoplasmic poly (A)-binding protein (PABPC) in neoblast function. PABPCs are RNA-binding proteins conserved across eukaryotes, and their role in translational regulation has been extensively studied (Gorgoni et al., 2011; Goss and Kleiman, 2013; Wang et al., 2010). However, PABPC also regulates mRNA turnover, nonsense-mediated decay and miRNA-mediated repression (Gorgoni et al., 2011; Goss and Kleiman, 2013).
Metazoans express multiple genes encoding PABPC: five in humans, three in Xenopus, two each in C. elegans and mouse, and one in yeast (Mangus et al., 2003). These genes are spatiotemporally regulated and have varied functions (Goss and Kleiman, 2013). In the planarian Dugesia japonica, two genes encoding cytoplasmic poly (A)-binding proteins, dj-pabpc1 and dj-pabpc2 have been reported. dj-pabpc1 expression is ubiquitous and its knockdown leads to severe regeneration defects. Knockdown animals fail to form the blastema and lyse within 2 days post amputation (dpa) (Rouhana et al., 2010). However, the mechanism by which it regulates regeneration is not known. In Schmidtea mediterranea, smed-pabpc1 is expressed in the germline tissue, and its knockdown leads to a block in meiotic progression, suggesting a role for SMED-PABPC1 in spermatogenesis (Wang et al., 2010). Our study identified an additional cytoplasmic poly (A)-binding protein, smed-pabpc2 (referred to as pabpc2) from the Schmidtea mediterranea transcriptome (Resch et al., 2012), which is a homologue of dj-pabpc1.
pabpc2 is expressed in majority of the cell types, apart from the pharynx and terminally differentiated epidermal cells (NB.22.1e+ and laminB+). Knockdown of pabpc2 leads to drastic regeneration and homeostatic defects in Schmidtea mediterranea. As PABPC in eukaryotes is a known translation initiator, we performed polysome profiling followed by transcriptome sequencing from control and knockdown animals to identify the targets of PABPC2. Strikingly, a specific set of transcripts that are essential for epidermal lineage determination, including zfp-1, were translationally repressed in knockdown animals. Extensive molecular characterization of pabpc2 knockdown phenotypes revealed defects in the epidermal turnover that led to defective organization of the epidermal tissue along with the loss of extracellular matrix (ECM) integrity. Further analysis showed that the epidermal defects in knockdown animals led to sustained injury response, which subsequently resulted in the failure of activation of the second mitotic peak that is essential for blastema formation. Taken together, our data points to a crucial role for PABPC2 in the maintenance of epidermal and ECM integrity, which in turn is essential for neoblast function and regeneration in planarians.
RESULTS
pabpc2 showed enriched expression in epidermal lineage, gut and neoblasts
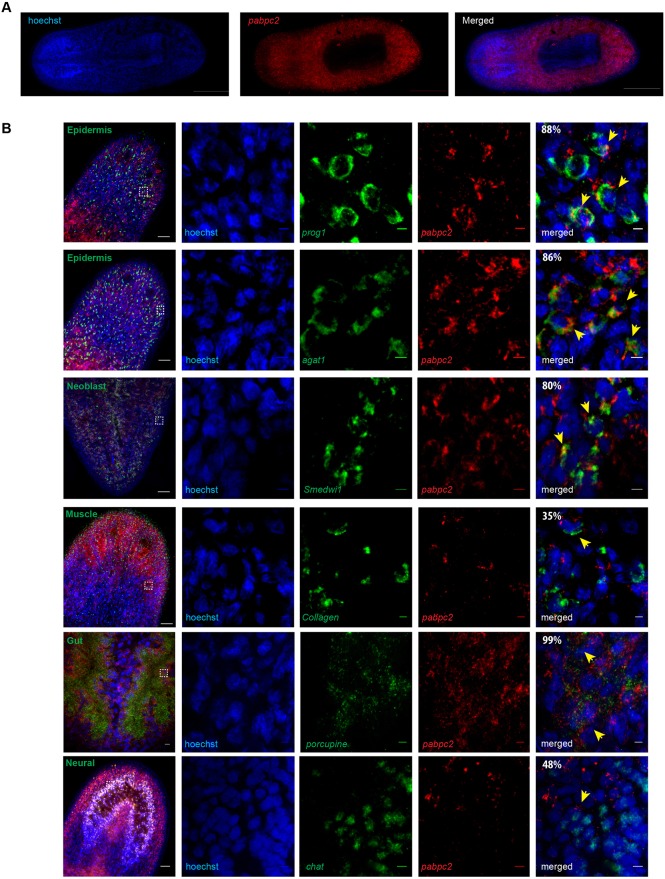
Smed-PABPC2 protein has well-conserved RRM domains and shows 44% identity to mammalian cytoplasmic poly (A)-binding protein 1 (Fig. S1A,B). Fluorescent in situ hybridization to study the expression pattern revealed that pabpc2 is mostly expressed in all the cell types of planarians, except in the pharynx (Fig. 1A). Fluorescent in situ hybridization on pabpc2 knockdown animals showed the absence of pabpc2 staining, suggesting that the pabpc2 probe is specific (Fig. S1C). We then investigated the expression of pabpc2 in different cell types from the available single cell sequencing (SCS) data (https://radiant.wi.mit.edu/app/), which identified transcripts enriched in different cell types (Wurtzel et al., 2015). In general, pabpc2 expression is observed in all the cell types, but is enriched in the epidermal lineage, gut and neoblasts (Fig. S1D). The expression of pabpc2 in different cell lineages was also confirmed by colocalization studies using lineage and tissue-specific markers for epidermal lineage (prog1, agat1), brain (chat), gut (porcupine), muscle (collagen), and neoblast (smedwi1). Co-expression studies revealed that 88%, 86%, 80% and 99% of pabpc2+ cells colocalized with prog1, agat1, smedwi1 and porcupine, respectively (Fig. 1B). However, 35% and 48% of the pabpc2+ cells colocalized with collagen and chat, respectively, confirming the single-cell sequencing data, which showed enriched expression of pabpc2 in the epidermal lineage, gut and neoblasts. Interestingly, pabpc2 did not colocalize with terminally differentiated epidermal cells (laminB+ and Nb.22.1e+) (Fig. S1E). Next, we investigated its functional role in planarian regeneration and homeostasis.
Fig. 1.
Identification and characterization of SMED-PABPC2. (A) Fluorescent in situ hybridization to study the expression pattern of smed-pabpc2. Scale bars: 500 μm. (B) Double fluorescent in situ hybridization showing co-expression of pabpc2 with prog1, agat1, smedwi1, collagen, porcupine and chat. The images in the first column were taken at 20× magnification. Scale bars: 50 μm. The white boxes indicate the area magnified in the columns to the right. The percentage of colocalization is shown in the last panel. Probes are indicated; yellow arrows indicate co-labeled cells. Scale bars: 5 μm. n=6. See also Fig. S1.
pabpc2 is essential for planarian regeneration and homeostasis
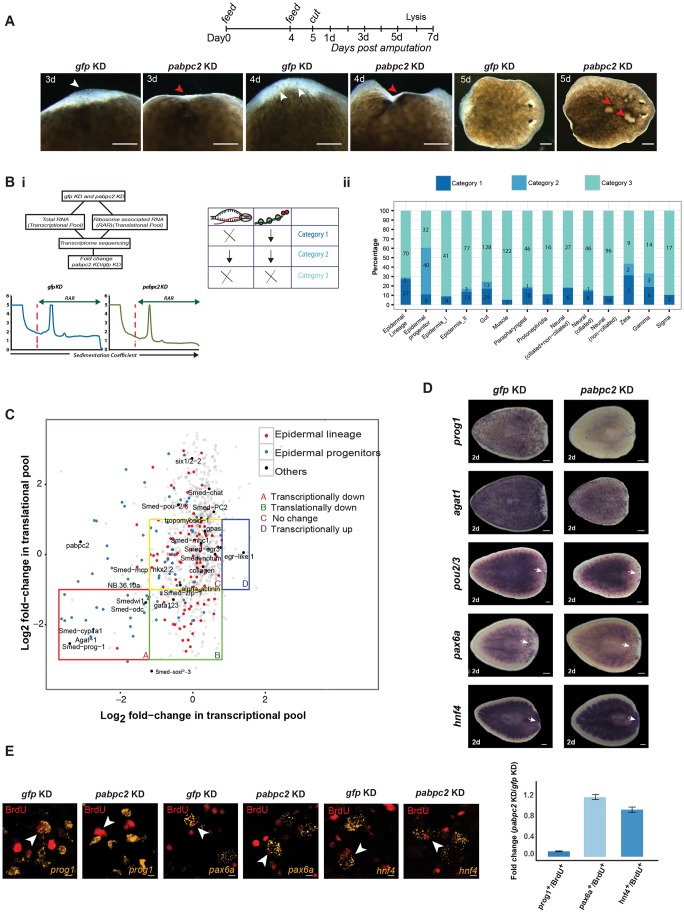
Bacteria expressing pabpc2 dsRNA were fed to the animals for knockdown of the gene. Green fluorescent protein (gfp) dsRNA was used as a negative control. Knockdown of pabpc2 in uncut animals resulted in the formation of lesions within 5-6 days post-2nd feed and the animals underwent lysis by day 11 (Fig. S2A). In regenerating animals, pabpc2 knockdown resulted in a smaller blastema that regressed by 3-4 dpa and the animals subsequently lysed by 5-7 dpa (Fig. 2A). The control animals (gfp dsRNA-treated animals) showed no defects even after 3 weeks post-feeding. In summary, these results clearly show that pabpc2 is crucial for homeostasis and regeneration in planarians.
Fig. 2.
PABPC2 regulates epidermal lineage transcripts. (A) Timeline showing RNAi feed schedule. Images were taken at 3, 4 and 5 dpa after dsRNA treatment. White arrowheads show normal blastema in control animals. Red arrowheads highlight the defective blastema and the lesions in knockdown animals (100/100). Scale bars: 200 μm. (Bi) The method used to identify the transcripts associated with the ribosome and cellular transcripts in the gfp and pabpc2 knockdown animals. ×, no change; ↓, decrease. (Bii) Stack bar depicting number and percentage of transcripts that belong to different categories across various cell types. Fold change was calculated by taking the ratio of the normalized number of transcripts between pabpc2 and gfp knockdown animals. (C) Scatter plot of fold-change values (PABPC/GFP) showing the distribution of transcripts between the transcriptional and translational pool. Transcripts from different categories are marked as four quadrants A, B, C and D. Transcripts that belong to epidermal lineage and epidermal progenitors are highlighted in red and blue, respectively. Some well-known markers belonging to different cell types and wound-healing genes are labeled in the scatter plot. (D) Whole-mount in situ hybridization using different progenitor markers such as prog1, agat1, pou2/3, hnf4 and pax6a at 2 dpa in gfp and pabpc2 knockdown animals. Epidermal progenitors (prog1 and agat1) showed a significant reduction in the expression upon pabpc2 knockdown, unlike other progenitors (hnf4, pax6a and pou2/3). White arrows indicate staining in the blastema. Scale bars: 200 μm (n=10). (E) Confocal images showing BrdU and progenitor (prog1, hnf4 and pax6a)-positive cells in gfp and pabpc2 knockdown animals. BrdU was injected post-2nd feed and animals were fixed 2 days post-BrdU injections. Equal numbers of BrdU cells were counted in gfp and pabpc2 knockdown animals and the numbers of colocalized cells were counted for each progenitor. The histogram depicts the fold change in colocalized cells in gfp and pabpc2 knockdown animals. Error bars were calculated from biological replicates. Yellow arrows indicate co-labeled cells. Scale bars: 5 μm. n=6. See also Fig. S2.
pabpc2 regulates transcripts essential for epidermal lineage during planarian regeneration
PABPCs enhance translation initiation by interacting with the translation initiation factor eIF4G and form a ‘closed loop’ mRNA conformation that is essential for the ribosome assembly (Tarun and Sachs, 1996). To identify the transcripts that were translationally repressed upon pabpc2 knockdown during regeneration, ribosome-associated RNA (RAR) was analyzed at 24 hpa in control and pabpc2 knockdown animals. mRNA from the ribosomal complexes (monosomes and polysomes) was purified, and deep sequencing was performed to identify the transcripts depleted from the translational pool in pabpc2 knockdown animals. We also performed deep sequencing of cellular mRNA from the control and knockdown animals at 24 hpa to identify the transcripts that were transcribed but not translated (Fig. 2Bi).
The reads were mapped to the transcriptome database (see Materials and Methods) to identify the transcripts that were translationally repressed in pabpc2 knockdown animals. A total of 4412 transcripts with an adjusted P-value of less than 0.05 and a minimum of 10 reads mapping to each of the transcripts after normalization were considered for further analysis (Table S1). Recent efforts based on the single-cell transcriptome analysis assigned 4787 transcripts to 13 different cell types from planaria (Wurtzel et al., 2015). From our transcriptome analysis, we identified 991 of these 4787 transcripts with coverage for each cell type varying from ∼6 to 44% (Fig. S2B).
The transcripts were classified into three broad categories based on a change of at least twofold calculated between the pabpc2 knockdown and mock-treated animals. The categories include: (1) transcripts that were translationally downregulated; (2) transcripts that were transcriptionally downregulated; and (3) transcripts that were either unaffected or upregulated both transcriptionally and translationally. We observed that 462 out of 991 (46.7%) transcripts fell into the 3rd category, representing all cell types (Fig. 2Bii). This suggests that PABPC2 is not a global regulator of translation in planaria but regulates translation of specific transcripts.
In pabpc2 knockdown animals, category 1 transcripts (actively transcribed but not translated) were found in all the cell types to varying extents (Fig. 2Bii). The majority of these transcripts belonged to epidermal lineage (27%) followed by gut (17%). In spite of the low coverage (10%) of epidermal lineage transcripts from our transcriptome pool (Fig. S2B), the majority of these transcripts belonged to category 1, suggesting that PABPC2 could potentially be an active regulator of epidermal lineage transcripts. As further proof, we also found that among different classes of the neoblast population, the majority of transcripts in the category 1 belonged to the zeta class (essential for epidermal lineage) and gamma class (essential for gut lineage). Some of the epidermal lineage transcripts depleted from the translational pool include gata123, soxP-3 and zfp-1, which are essential for the epidermal lineage formation (Fig. 2C) (van Wolfswinkel et al., 2014). Next, we probed the nature of the translationally repressed transcripts belonging to other cell types, such as neural, gut, muscle and protonephridia. Interestingly, none of the well-characterized transcripts essential for neural (chat, PC2, gpas), gut (nkx 2.2), muscle (tropomyosin, smed-mhc-1) and protonephridia (pou2/3, rootletin, six1/2-2) specification was affected (Fig. 2C) (Forsthoefel et al., 2012; Scimone et al., 2014; Witchley et al., 2013).
We also investigated the transcripts belonging to category 2, which were transcriptionally downregulated. Strikingly, majority of the category 2 transcripts belonged to either epidermal progenitors (49.4%) or gut cells (7.6%) (Fig. 2Bii). Some of the well-characterized epidermal progenitor markers such as prog-1, agat-1, odc and cyp1a1 were transcriptionally downregulated (Fig. 2C).
The RAR data suggest that in pabpc2 knockdown animals, the zfp-1 translation was defective but the transcript levels remain unchanged, whereas the prog1 and agat1 transcript levels were significantly reduced (Fig. 2C). We confirmed these results by real-time PCR on control and knockdown animals at 24 hpa and we found no change in overall zfp-1 transcript level (Fig. S2C). However, the levels of prog1 and agat1 were dramatically reduced (Fig. S2D). Here, we assume that the decrease in levels of transcripts such as zfp-1 from the translation pool subsequently leads to the failure of the formation of epidermal progenitors in the knockdown animals. Interestingly, the transcripts from other cell types that were translationally downregulated did not result in overt phenotypes.
To validate the RAR data, which showed the potential role of PABPC2 in epidermal lineage formation, we performed whole-mount in situ hybridization on gfp and pabpc2 knockdown animals with several known progenitors for epidermis (prog1, agat1), gut (hnf4), neurons (pax6a) and protonephridia (pou2/3) at 2 dpa. In accordance with RAR and transcriptome data, we observed a dramatic decrease only in the epidermal progenitors in the knockdown animals (Fig. 2D). To further confirm the crucial role of PABPC2 in the differentiation of neoblasts into epidermal progenitors, we also performed BrdU labeling on uncut control and knockdown animals. We administered BrdU at 24 h post-2nd feeding and fixed the animals after 2 days. Around a 90% decrease in the formation of new epidermal progenitor cells was observed in pabpc2 knockdown animals compared with control. However, no defect was observed in the formation of progenitors of gut and brain (Fig. 2E). Taken together, our results demonstrate the role of PABPC2 in the generation of epidermal lineage, crucial for both epidermal regeneration and turnover.
pabpc2 knockdown animals have disorganized epidermal tissue and extracellular matrix
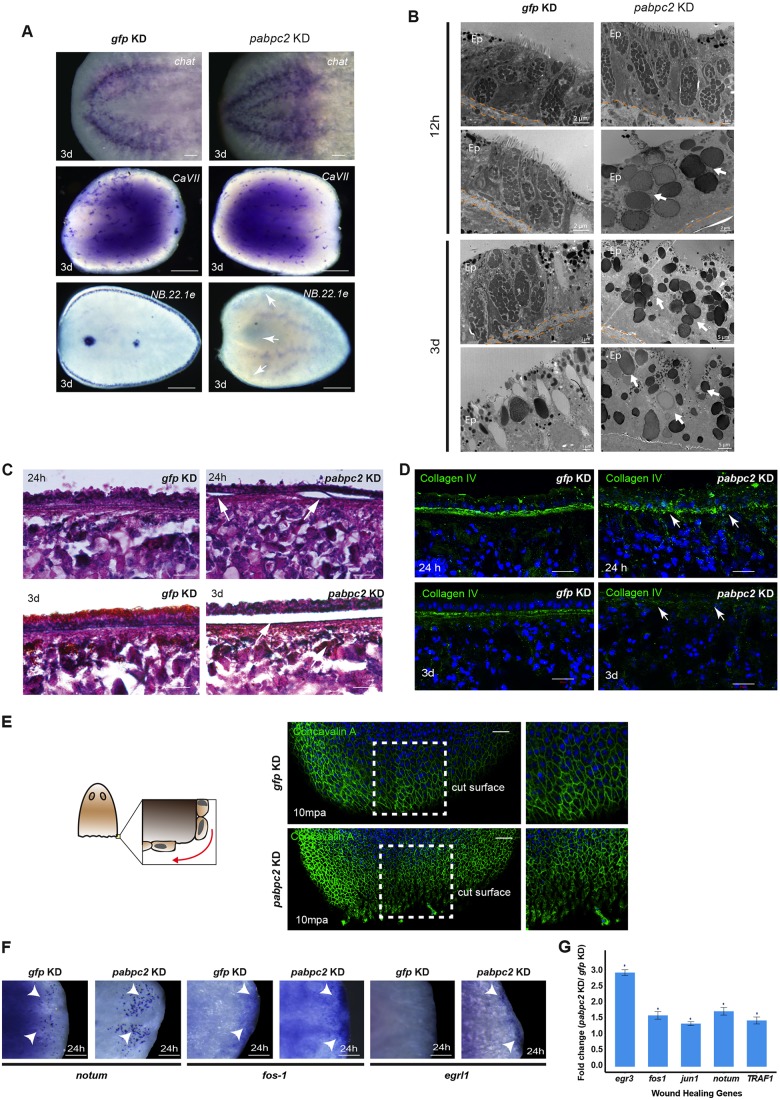
Our results based on the expression pattern and molecular function of PABPC2 categorically highlight its role in epidermal maintenance and/or turnover. To further confirm this, we investigated the effect of pabpc2 knockdown on the epidermal tissue. Whole-mount in situ hybridization on control and pabpc2 knockdown animals at 3 dpa for the epidermal marker NB.22.1e, which marks marginal adhesive glands and ventral mouth opening (Reddien et al., 2007; Tazaki et al., 2002), revealed a complete absence of its expression, suggesting a drastic defect in the epidermal tissue. In contrast, other tissue-specific markers, such as cavII, which marks the tubules of the protonephridia, and chat, which marks the bilobed structure and the ventral nerve cord of CNS (Rink et al., 2011; van Wolfswinkel et al., 2014; Wagner et al., 2011), showed normal expression patterns in both control and knockdown animals (Fig. 3A). Furthermore, we also validated the disorganization of epidermal cells in knockdown animals by immunostaining with anti-rootletin antibody, which marks the ciliated epidermal cells. In control animals, immunostaining revealed a concentric organization of rootlets in the ciliated epidermal cells. However, in the knockdown animals the organization of the rootlets was disrupted, revealing the abnormal organization of the epidermal cells (Fig. S3A).
Fig. 3.
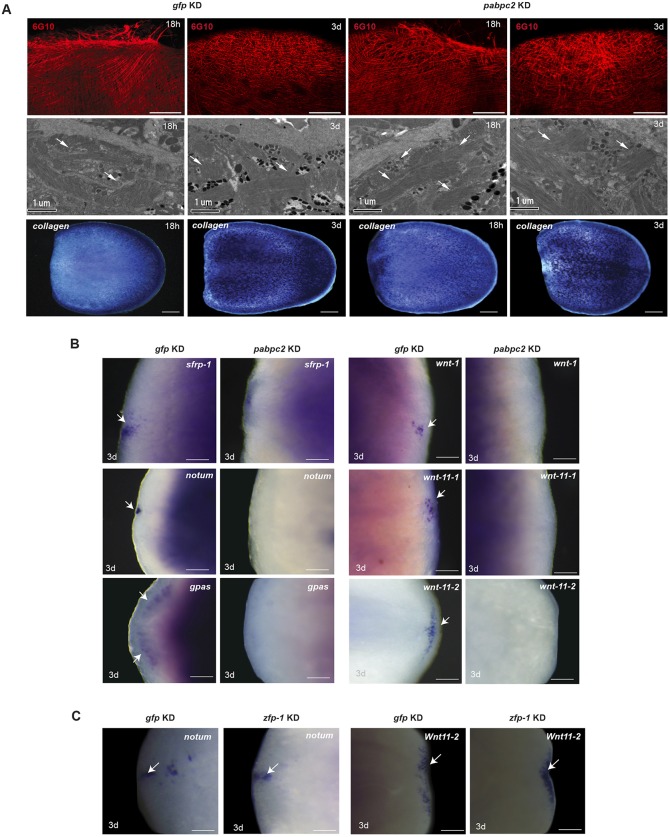
SMED-PABPC2 is essential for maintaining epidermal and ECM integrity and epithelialization. (A) Whole-mount in situ hybridization showing expression of the differentiated tissue markers chat, cavII and NB.22.1e in gfp and pabpc2 knockdown animals. Scale bars: 500 μm. White arrows indicate loss of NB.22.1e expression in pabpc2 knockdown animals. n=10. (B) EM images showing organization of the epidermal tissue on the non-regenerating side at 12 hpa and 3 dpa in gfp and pabpc2 knockdown animals. Arrows show rhabdite-like cells in knockdown animals. Ep, epidermis. n=5. (C) Histological sections showing the organization of epidermis after pabpc2 knockdown in regenerating animals. Sagittal sections made from the regenerating animals at 24 h and 3 days post-amputation were stained with Hematoxylin and Eosin. Arrows show peeling of the epidermis in pabpc2 knockdown animals in regions away from amputation. Scale bars: 20 µm. n=9. (D) Maximum intensity projections of z-stacks of gfp and pabpc2 knockdown sagittal sections stained with collagen IV antibody at 24 h and 3 days post amputation. Arrows showing disorganization of ECM in pabpc2 knockdown animals. Scale bars: 20 µm. n=10. (E) Schematic showing stretching of epithelial cells near the wound region. Confocal images of gfp and pabpc2 knockdown animals stained with concavalin A-FITC showing the organization of dorsal epidermal tissue near the amputated region at 10 min post-amputation (mpa). The image is tiled. Scale bars: 100 µm. n=5. (F) Whole-mount in situ hybridization showing upregulation of early wound-healing genes such as notum, fos-1 and egr like 1 (egrl1) near the blastema at 24 hpa in pabpc2 knockdown animals. Scale bars: 50 µm. n=10. Arrowheads show the expression of transcripts in the blastema region. (G) Quantification of level of expression of wound-healing genes by qRT-PCR. Fold-change of wound-healing gene levels in pabpc2 knockdown animals at 24 hpa. The error bars are drawn from biological triplicates and indicate s.e.m. *P<0.05. See also Fig. S3.
Epidermal tissue is primarily made up of two cell types: epithelial cells and gland cells. Interspersed between the epithelial cells are the gland cells that contain rod-shaped secretory granules called rhabdites. Studies have shown that rhabdite-forming cells contribute to the successive renewal of epidermal cells (Hori, 1979; Skaer, 1965). Rhabdites are formed in the mesenchyme and migrate to the epidermal tissue through ducts of the gland cells. Once in the epidermis, they are rapidly released out through the exterior opening of the gland cells to form the mucous layer (Martin, 1978). Electron microscopy (EM) studies to investigate the organization of epidermal tissue in pabpc2 knockdown revealed disorganization of epidermal tissue as early as 12 hpa in knockdown animals (Fig. 3B). The knockdown animals lacked epithelial cells and had rhabdite-like bodies throughout the epidermis, which showed a dramatic increase by 3 dpa (Fig. 3B). Similar results were also observed in the uncut knockdown animals (Fig. S3B).
The defect in the epidermal tissue in the knockdown animals is potentially due to the failure in the generation of the epidermal progenitors. This was supported by the whole-mount in situ hybridization carried out in pabpc2 knockdown animals as early as 6 hpa, which showed decrease in the epidermal progenitors (Fig. S2D). We further tested the sensitivity of epidermal progenitor formation to the PABPC2 levels by feeding low doses of pabpc2 dsRNA to the animals (one dose of dsRNA feed). The animals fed with low dose of dsRNA showed significant decrease in the prog1+ and agat1+ cells (Fig. S3C), suggesting that PABPC2 is crucial for the epidermal progenitor formation. In planarians, the epidermal layer is attached to the sub-epidermal tissue via an extracellular matrix (ECM) (Hori, 1979; Hori, 1991). As pabpc2 knockdown disrupts organization of the epidermal cells, we also investigated the attachment of epidermal tissue to the ECM in the knockdown animals. Sagittal sections stained with Hematoxylin and Eosin revealed severe blistering and detachment of the epidermis from the underlying ECM in knockdown animals as early as 24 hpa, which dramatically increased by 3 dpa (Fig. 3C). We next investigated whether the epidermal detachment could be due to the disorganization of the ECM using collagen as a marker. Collagen is the major component of the ECM and is secreted by the epidermal cells in planaria (Hori, 1980). Mammalian collagen IV antibody, which crossreacts with the planarian collagen in the ECM, was used to study the ECM organization. The control animals showed an intact, closely packed collagen staining between epidermal and sub-epidermal layers. However, pabpc2 knockdown animals showed diffused staining by 24 hpa, which was spread across epidermal and sub-epidermal layer by 3 dpa (Fig. 3D). These results clearly demonstrate that PABPC2 is essential for the maintenance of epidermis and ECM integrity.
pabpc2 knockdown animals showed sustained wound response during regeneration and homeostasis
Next, we investigated the role of PABPC2 in wound healing because of its requirement for the maintenance of the epidermal tissue. In planarians, it has been reported that within 10-15 min post-amputation (mpa) epidermal cells near the injury site close the wound as a result of muscle contraction (Chandebois, 1980). This is followed by passive stretching of the pre-existing epithelial cells near the wound region to form a thin film of cells (Chandebois, 1980; Morita et al., 1969). However, the importance of the epidermis in wound closure and its impact on the stem cell function has not been elucidated. The epidermal organization near the cut surface in pabpc2 knockdown animals was studied by staining the animals with the lectin concavalin A, which marks the boundaries of the epidermal cells (Zayas et al., 2010). In the control treated animals, the epidermal cells near the wound region were elongated and subsequently lead to the closure of the wound (Fig. 3E, Fig. S3D). Interestingly in pabpc2 knockdown animals, pre-existing epithelial cells near the wound region failed to stretch and showed a rounded appearance as early as 10 mpa (minutes post-amputation) (Fig. 3E). This defect in the knockdown animals persisted at a later time point, 24 hpa (Fig. S3D), suggesting a defect in the wound closure.
We reasoned that the knockdown animals might sense the epithelialization defect as a sustained injury leading to the deregulation of some of the early wound-response genes. Whole-mount in situ hybridization analysis of pabpc2 knockdown animals indeed showed an upregulation of early wound-response genes such as notum and fos-1 at 24 hpa (Fig. 3F) compared with the control animals. In addition, egr like 1 (egrl1), an early wound-response gene, the expression of which is normally seen until 1 hpa (Wenemoser et al., 2012), showed sustained expression even at 24 hpa in pabpc2 knockdown animals (Fig. 3F). qRT-PCR analysis on some of the early wound healing further corroborated with whole-mount in situ hybridization (Fig. 3G). Interestingly, pabpc2 knockdown animals that were not injured or amputated also showed expression of wound healing genes such as jun-1 and fos-1, which was not observed in the control uncut animals (Fig. S3E). Furthermore, the transcriptome and qRT-PCR analysis also validated the upregulation of early wound-response genes such as egrl1, notum, TRAF1 and egr3 in the pabpc2 knockdown animals (Fig. S3F,G). Taken together, these results suggest a crucial role for PABPC2 in the organization of epidermal tissue near the site of injury that is essential for the wound response.
pabpc2 knockdown leads to a defect in neoblast proliferation near the wound region
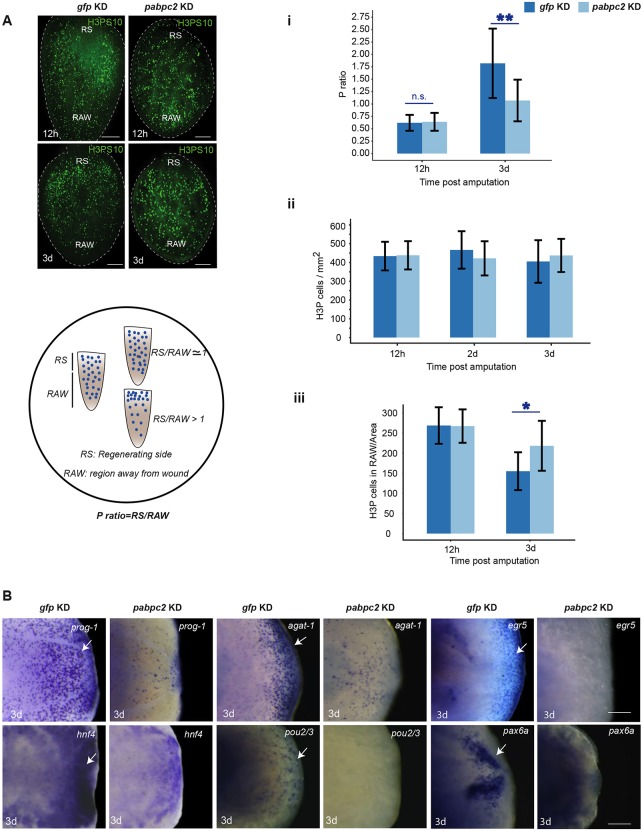
Neoblast proliferation in Schmidtea mediterranea occurs in two distinct phases during regeneration: first, body-wide proliferation at 4-12 hpa; and second, specific proliferation near the amputated region, which occurs at 2-3 dpa (Wenemoser and Reddien, 2010). The second mitotic phase is crucial for blastema formation. As pabpc2 knockdown animals showed regression of the blastema, we investigated the status of the two mitotic peaks during regeneration in the control and pabpc2 knockdown animals. The changes in the two mitotic peaks can be measured by calculating the ratio of the number of proliferating cells (H3PS10 stained) near the wound region compared with away from the wound region per unit area (P-ratio). A P-ratio close to one indicates the first mitotic peak, which is a body-wide response because of the even distribution of proliferating neoblasts across the regenerating animal. A P-ratio greater than one indicates second mitotic peak, because of the increased number of neoblasts present near the wound region (Fig. 4A). We measured the P-ratios at 12 hpa (first mitotic peak) and 3 dpa (second mitotic peak) in pabpc2 and gfp knockdown animals. The P-ratio was close to one at 12 hpa in both gfp and pabpc2 knockdown animals, suggesting normal activation of the first mitotic peak (Fig. 4Ai). Interestingly, the P-ratio remained close to one at 3 dpa in pabpc2 knockdown animals, whereas the P-ratio in the control animals was greater than one, indicating that PABPC2 is crucial for the second mitotic peak in the regenerating animals (Fig. 4Ai). However, there was no significant reduction in the overall number of the H3P+ cells, suggesting that pabpc2 knockdown has no effect on the overall proliferation of neoblasts upon amputation (Fig. 4Aii). We also performed whole-mount in situ hybridization with a neoblast marker, smedwi-1, at 3 dpa and found no observable changes in the expression of smedwi-1 (Fig. S4A). We predicted that the reduction in the number of H3P+ cells near the wound region would be compensated for by the increase in the number of dividing cells away from the wound region. This prediction was tested by counting the number of H3P+ cells in the region away from the wound site. We observed a 1.4-fold (P≤0.001) increase in the number of mitotic cells away from the wound region in pabpc2 knockdown animals at 3 dpa (Fig. 4Aiii). This suggests that, although the total number of neoblasts in pabpc2 knockdown animals is similar to control animals, the defect in mitosis near the wound region is compensated for by an increase in mitosis away from the wound region in knockdown animals. Taken together, the data presented clearly demonstrate that the knockdown of pabpc2 leads to the defect in the localized proliferation of neoblasts near the wound region, resulting in the regression of the blastema.
Fig. 4.
Effect of smed-pabpc2 knockdown on neoblast proliferation and blastema formation. (A) Max intensity projections of confocal images showing H3PS10+ cells in regenerating animals in gfp and pabpc2 knockdown animals. Scale bars: 100 µm. n=21. Schematic showing the procedure of calculating the P-ratio. Animals were divided in the ratio of 1:2 from the cut side and considered as regenerating side (RS) and region away from wound (RAW), respectively. Mitotic cells were calculated in both the regions and normalized to per unit animal area. P ratio=RS/RAW. (i) The P ratio at 12 hpa and 3 dpa in gfp and pabpc2 knockdown animals. pabpc2 knockdown animals showed a P ratio close to 1 even at 3 dpa, unlike control animals. The difference between the P ratios in control and knockdown animals at 3 dpa was significant (**P<0.0001). n.s., non-significant. (ii) The total number of mitotic cells in gfp and smed-pabpc2 knockdown animals at 12 hpa, 2 dpa and 3 dpa. (iii) Cell numbers in the region away from the wound at 12 hpa and 3 dpa. A significant increase was observed in cell number in RAW in pabpc2 knockdown animals at 3 dpa (*P<0.05) (n=21). (B) Whole-mount in situ hybridization showing expression of progenitor markers, prog-1, agat-1, egr5, hnf4, pax6A and pou2/3 in pabpc2 knockdown and gfp knockdown animals at 3 dpa. White arrows indicate the staining observed in the blastema. Scale bars: 50 μm. n=10. See also Fig. S4.
Our previous result showed that the knockdown of pabpc2 in the uncut animals led to the expression of early wound-response genes, suggesting a sustained injury throughout the animals. The sustained early wound response has been implicated in the prolonged global proliferation in the regeneration animals (Lin and Pearson, 2017). We predicted that the rate of neoblast proliferation would increase in the uncut animals after pabpc2 knockdown due to the sustained expression of early wound-response genes. We indeed found 1.5-fold increase in the number of H3P+ cells in the knockdown animals compared with the control animals (Fig. S4B). The sustained expression of the early wound-response genes in the regenerating animals could possibly explain the prolonged global proliferation and a failure in localized proliferation.
pabpc2 knockdown animals show subsequent loss of progenitors in the blastema
In regenerating planarians, the bimodal pattern of neoblast proliferation was shown to be crucial for blastema formation (Wenemoser and Reddien, 2010). The regenerating blastema is typically characterized by the presence of neoblast progenitors (Rink, 2013). As we observed a failure in the localized proliferation of neoblasts leading to a defective blastema formation, we investigated the presence of several known progenitors near the wound region at 3 dpa in gfp and pabpc2 knockdown animals. Whole-mount in situ hybridization for epidermal progenitors (prog1, agat1 and egr5) (Tu et al., 2015), and progenitors of other tissues, such as gut (hnf4), protonephridia (pou2/3) and brain (pax6A) (Scimone et al., 2011, 2014; Wagner et al., 2011) showed a drastic reduction in their expression at 3 dpa near the blastema (Fig. 4B). Together, these results confirm that pabpc2 knockdown animals, which showed failure in the localized proliferation, had a defective blastema.
pabpc2 knockdown animals showed no gross defects in sub-epidermal muscle layer but failed to express PCGs at 3 dpa
The pronounced defect in ECM and epidermis in pabpc2 knockdown animals led us to investigate the effect of pabpc2 knockdown on sub-epidermal cells. Planarian sub-epidermal layer is made up of circular, longitudinal and diagonal fibers that form the muscle net (Cebrià et al., 1997). We used 6G10 antibody that marks muscle fibers to study the organization of sub-epidermal muscle cells (Ross et al., 2015). Immunostaining of the control and pabpc2 knockdown animals showed no gross defect in the muscle organization (Fig. 5A). This result was further corroborated by the EM studies, which showed the normal appearance of muscles in the sub-epidermal layers (Fig. 5A). Whole-mount in situ hybridization to study the expression pattern of collagen, which is a marker for the muscle cells, also showed no significant changes at 18 hpa and 3 dpa in control and pabpc2 knockdown animals (Fig. 5A). We also investigated the expression of collagen in uncut pabpc2 knockdown animals and found no observable change (Fig. S5A). Thus, our results suggest that the knockdown of pabpc2 did not have a profound effect on the organization of the sub-epidermal muscle layer in planaria.
Fig. 5.
smed-pabpc2 does not affect sub-epidermal muscle cells but affects PCGs expression. (A) Confocal images showing the organization of muscle cells stained with anti-6G10 antibody near the blastema after 18 h and 3 dpa in gfp and pabpc2 knockdown animals. Images were taken using LSM700 confocal microscope. Scale bars: 100 µm. n=6. EM images and whole-mount in situ hybridization with collagen showing muscle organization in gfp and pabpc2 knockdown animals at 18 hpa and 3 dpa. Arrows indicate sub-epidermal muscle cells. Scale bars: 200 µm. n=5. (B) Whole-mount in situ hybridization showing expression of anterior (sfrp-1, notum and gpas) and posterior (wnt-1, wnt11-1 and wnt11-2) PCGs in control and pabpc2 knockdown animals in the blastema at 3 dpa. Arrows mark the expression of PCGs in the blastema. Scale bars: 50 µm. (C) Whole-mount in situ hybridization showing expression of notum and wnt11-2 in zfp-1 knockdown animals at 3 dpa in the blastema region. Unlike pabpc2 knockdown animals, zfp-1 knockdown animals expressed PCGs in the blastema. Scale bars: 50 µm. n=8. Arrows indicate PCG expression in the blastema.
In planarians, the sub-epidermal muscle layer expresses most of the PCGs that provide positional cues essential for neoblast differentiation (Witchley et al., 2013). We investigated the expression of the PCGs such as notum, sfrp and gpas, which specify anteriority, and wnt1, wnt11-1 and wnt11-2, which specify posteriority (Gurley et al., 2010; Mii and Taira, 2009; Petersen and Reddien, 2011). Whole-mount in situ hybridization on pabpc2 knockdown animals at 3 dpa showed a complete absence of PCG expression both in the anterior and posterior regenerating tissue (blastema) (Fig. 5B). The reduction in the expression of PCGs could be either due to the disorganization of epidermal tissue or to their translational downregulation resulting from pabpc2 knockdown. We tested both these possibilities by knockdown of zfp-1 and by transcriptome sequencing of RAR. The zfp-1 knockdown animals, which have perturbed epidermal organization, did not show any loss of PCG expression (wnt11-1 and notum) at 3 dpa (Fig. 5C), suggesting that the disruption of epidermal organization per se does not affect PCG expression. We also investigated the translational status of the PCGs in pabpc2 knockdown animals. The transcriptome sequencing of RAR revealed more than twofold upregulation in the association of the PCG transcripts (wnt11-5, sfrp3, evi/WIs and slit1) with the translational pool in the knockdown animals at 24 hpa. This suggests that the PABPC2 is not required for the translation of PCG transcripts (Fig. S5B). Although we observed upregulation of notum at 24 hpa in the knockdown animals, its expression was subsequently downregulated by 3 dpa (Fig. 3F). It is very well known that some of the PCGs encoding members of Wnt and TGFβ signaling pathways are also wound-response genes that are expressed after 6 hpa (Wenemoser et al., 2012). As pabpc2 knockdown animals have sustained expression of early wound-response genes, the absence of PCG expression seen at 3 dpa could be an effect of deregulated wound response.
In pabpc2 knockdown animals, we also observed disorganization of the ECM, which was not observed in the zfp-1 knockdown animals (Fig. S5C). Here, we speculate that the loss of PCGs expression in the sub-epidermal tissue could be due to the disorganization of the ECM. This was supported by the studies in vertebrate models, which showed that the ECM interaction with the tissue types was crucial for the regulation of gene expression (Boudreau et al., 1995; Spencer et al., 2007). However the crosstalk between ECM and sub-epidermis that is crucial for the regulation of PCG expression in planaria needs to be further investigated.
DISCUSSION
Regeneration is a complex process mediated by stem cell function and tissue organization. Studies from hydra, amphibian and planaria have shown that non-stem cells express various tissue patterning genes essential for regeneration (Lengfeld et al., 2009; McCusker and Gardiner, 2014; Takahashi and Fujisawa, 2009; Witchley et al., 2013). However, the potential role of non-stem cells during regeneration remains largely unexplored. In this study, we show the pivotal role of epidermal integrity and the crucial role played by the RNA-binding protein PABPC2 in epithelialization and epidermal turnover during planarian regeneration and homeostasis.
smed-pabpc2 is expressed in multiple cell types, except pharynx and terminally differentiated epidermal cells (NB.22.1e+ and laminB+). Strikingly, pabpc2 knockdown animals showed perturbation mainly in epidermal tissue despite its absence in the epidermal cells. This defect in the epidermal organization could be either due to the defect in the formation of the epidermal progenitors and/or maintenance of the epidermal integrity. Whole-mount in situ hybridization for epidermal progenitors on animals fed with a low dose of pabpc2 dsRNA (1st feed) showed substantial reduction in the progenitor population. However, the other progenitors remained seemingly unaffected, even after 48 h post-amputation, suggesting that epidermal progenitors are exquisitely sensitive to pabpc2 knockdown compared with the other progenitors. This result was also supported by the BrdU-labeling experiments, which showed failure in differentiation of neoblasts to epidermal progenitors. Furthermore, we also observed no detectable expression of pabpc2 in epidermal cells, which suggests that PABPC2 might not be directly involved in the maintenance of epidermal tissue. Taken together, our results indicate that the defects observed in the epidermal tissue are most likely due to the defect in the epithelial lineage specification. However, it is difficult to rule out the direct role of PABPC2 in epidermal organization exclusively based on the lack of expression of pabpc2 in NB.22.1e+ and laminB+ cells. Currently, the lack of markers to study various epidermal cell types limits our understanding regarding the expression of pabpc2 in those cell types.
Epidermis is well characterized for its protective function. Upon amputation/injury, epidermal cells provide the first response by covering the wound within a few hours (Chandebois, 1980; Morita and Best, 1974). It has been shown that the epithelial cells near the site of injury come into direct contact to the underlying sub-epidermal muscle (Schurmann and Peter, 1998). In pabpc2 knockdown animals, we observed the rounded appearance of the epithelial cells near the wound region, suggesting a defect in the attachment of the epithelial cells to the underlying sub-epidermal muscle. In addition, we also observed blistering of the epidermal tissue and the disorganization of the ECM in the regions away from the wound in the pabpc2 knockdown animals. RNA-binding proteins such as PABPs have been implicated in formation of integrin-rich focal adhesion complexes that are essential for epidermis-ECM interactions (Babic et al., 2009; de Hoog et al., 2004; Lee et al., 2009). Depletion of these RNA-binding proteins alters cell morphology and the ability of the cell to spread post-adhesion (Chicurel et al., 1998; de Hoog et al., 2004; Katz et al., 2012). Thus, we speculate that the knockdown of pabpc2 may result in the disruption of cell matrix adhesion, resulting in a defect in epithelialization. However, it is not clear whether the blistering defects observed are either a consequence of a defect in the epidermal turnover or a direct effect of pabpc2 knockdown on the epidermal organization.
Furthermore, in pabpc2 knockdown animals, we also observed prolonged expression of the early wound-response genes (jun-1, fos-1, egrl1 and notum). In the regenerating animals, these genes are upregulated within 30 mpa to 3 hpa, and their expression is subsequently downregulated by 12 hpa (Wenemoser et al., 2012; Wurtzel et al., 2015). We believe the epidermal defect in the knockdown animals was perceived as a sustained injury leading to the upregulation and prolonged expression of the early wound-response genes. This was also supported by the enriched expression of wound-response genes jun-1 and fos-1 in pabpc2 knockdown uncut animals. However, it is also possible that PABPC2 directly regulates expression of these wound-healing genes, which is currently difficult to address due to the lack of antibody.
Localized proliferation of neoblasts occurs only in the amputated animals near the site of injury (Wenemoser and Reddien, 2010). However, the factors essential for the switch from global to localized proliferation remains unknown. Lin and Pearson (2017) have shown that the prolonged expression of the early wound-response genes in yorkie knockdown animals affected the bimodal proliferation of neoblast. Yorkie is a transcriptional co-activator in the Hippo kinase cascade, the knockdown of which led to prolonged global proliferation of neoblasts and delayed second phase of proliferation, resulting in the defective blastema formation (Lin and Pearson, 2017). Similarly, knockdown of follistatin, an early wound-response gene, showed failure in second phase of neoblast proliferation and blastema formation (Gaviño et al., 2013; Roberts-Galbraith and Newmark, 2015). Together, these studies suggest a strong correlation between the regulation of wound response and proliferation of neoblasts crucial for blastema formation. In pabpc2 knockdown animals, the sustained global neoblast proliferation and failure in localized proliferation could be an outcome of prolonged early wound response. The other possibility could be the downregulation of the intrinsic factors in the neoblasts upon pabpc2 knockdown, resulting in the failure of localized proliferation.
Studies have shown that the localized proliferation is essential for blastema formation (Gaviño et al., 2013; Lin and Pearson, 2017; Roberts-Galbraith and Newmark, 2015). In pabpc2 knockdown animals, failure in the neoblast proliferation near the wound region explains the defect in blastema formation, which was verified by the absence of progenitors in the blastema at 3 dpa. Our result suggests that the defect in the blastema could be a consequence of defect in epithelial-basement membrane attachments, which have been suggested to have an instructive role in planarian blastema formation (Reddien and Sanchez Alvarado, 2004).
The best characterized function of poly (A)-tail-bound PABPC is enhancing translation initiation by interacting with translation initiation factors bound at the 5′-end of the mRNA. Although considered as a ‘global’ effector of translation, the individual mRNAs that are translationally regulated vary among different PABPCs. For example, in Xenopus, the knockdown of the three different PABPCs showed distinct cellular and developmental phenotypes (Smith et al., 2014). Studies in C. elegans have also shown that out of the two PABP isoforms, PAB-1, but not PAB-2, is essential for fertility (Smith et al., 2014). This suggests that each PABPC has a specific set of targets that it regulates, although the factors that impart specificity are not well understood. RAR analysis in control and pabpc2 knockdown animals revealed that PABPC2 regulates the translation of specific set of targets, such as zfp-1, gata123 and odc-1, that are essential for epidermal lineage formation. Thus, these results highlight the pivotal role of PABPC2 in regulating the translation of the transcripts that are essential for the formation of epidermal progenitors in planarians, and this is crucial for the maintenance of epidermal organization.
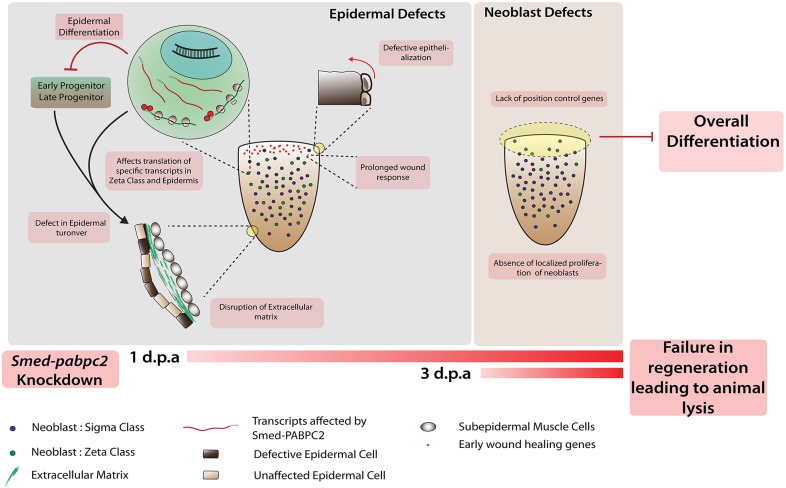
The facts that the animals lyse within one week post-amputation and that PABPC is a multi-functional protein make it difficult to delineate the primary and secondary consequences. Given that epidermal defects manifest themselves much earlier than other tissues, it is most likely the primary consequence of pabpc2 knockdown. Thus, we speculate that the epithelial phenotype might be the cause of all other defects, including the defect in localized proliferation of the neoblast (Fig. 6).
Fig. 6.
Model showing the crucial role of smed-pabpc2 in epidermal integrity and neoblast function. smed-pabpc2 knockdown animals show failure of epidermal organization due to failure of epidermal turnover. The epidermal defects leads to several other defects, such as loss of ECM integrity, defective wound closure and prolonged wound response. These potentially lead to a neoblast proliferation defect near the wound region and absence of PCGs, which subsequently affects overall differentiation and planarian regeneration.
The cellular events that facilitate regeneration in planaria are comparable with the events crucial to tissue regeneration in higher metazoans. For example, the events essential for wound healing in planaria, such as epithelialization of the wound surface, are also observed during the cutaneous wound healing in vertebrates (Arwert et al., 2012; Jamora, 2014). Similarly, mobilization of the neoblast stem cells to the site of injury, which is crucial for planarian regeneration, has also been shown to be essential in metazoans (Arwert et al., 2012; Jamora, 2014). However, the possible influence of epidermal and ECM organization during regeneration has not been comprehensively investigated in higher metazoans. In the current study, we uncover a novel role for PABPC2 in epidermal turnover and organization, which is essential for planarian regeneration.
MATERIALS AND METHODS
Planarian culture
Animals used in this study belonged to the sexual strain of the species Schmidtea mediterranea. They were maintained at 20°C in planarian media (2 mM NaCl, 0.1 mM KCl, 0.1 mM MgSO4, 0.12 mM NaHCO3 in distilled water), and fed beef liver paste twice a week. Animals were starved one week prior to any experiments.
RNAi experiments
RNAi was carried out using the feeding protocol described previously (Reddien et al., 2005). Post-feeding, animals were cut into three fragments: head, trunk and tail.
Whole-mount in situ hybridization and double fluorescence in situ hybridization
Digoxigenin- or fluorescein-labeled RNA probes were synthesized using an in vitro transcription kit (Roche). Whole-mount in situ hybridization and double fluorescence in situ hybridization were carried out as described previously (Pearson et al., 2009; King and Newmark, 2013).
Whole-mount immunostaining
Animals were treated in 2% HCl for 5 min on ice. They were then incubated in Carnoy's fixative for 2.5 to 3 h. One rinse in absolute methanol and overnight bleaching in 6% hydrogen peroxide followed fixation. The animals were then rehydrated in graded methanol PBS washes, and incubated in blocking solution (10% horse serum in PBSTx) for 6 h. Primary antibody (anti phospho-histone H3 ser10, Abcam 06-570; 6G10, DSHB; and rootletin antibody, a kind gift from Dr Jochen Rink, Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany) incubation was carried out overnight at room temperature at dilutions of 1:250 (rootletin antibody) or 1:100 (H3PS10 antibody). After four washes (2 h each) at room temperature with blocking solution, animals were incubated in secondary antibody solution (Alexa Fluor donkey anti mouse-488 and Alexa Fluor donkey anti rabbit-488, Molecular Probes) overnight at room temperature. For concavalin A-FITC staining, animals were incubated for 4 h at room temperature after blocking in Carbo-free blocking solution (Vector Labs) at a dilution of 1:2500. For 6G10 immunostaining, bleached animals were blocked in 10% horse serum in PBSTx blocking solution and were incubated with anti 6G10 antibody (diluted 1:5) overnight at 4°C.
Image acquisition and quantitation
Images were acquired using the LSM 780 laser scanning confocal microscope. Quantitation of H3P-positive cells was carried out using the multiple pointer tool on ImageJ. RS/RAW quantitation was carried out by counting H3P-positive cells on both the regenerating as well as non-regenerating ends of the animal. The animals were divided in the ratio of 1:2 for RS and RAW, respectively, and the number of cells was counted in respective regions and was normalized to the body area. Colocalization of the different progenitor and tissue population agat1, prog1, collagen, smedwi1, chat and porcupine with pabpc2 was quantified using MATLAB (Mathworks). (See supplementary Materials and Methods for details.)
Embedding and cryosectioning
Regenerating fragments were treated with 2% HCl and fixed in Carnoy's fixative. The dehydrated animals were given two washes for 30 min in 100% xylene. Xylene was replaced by melted paraffin wax for 1 h at 70°C and the animals were embedded in paraffin wax in commercially available molds (Tissue-Tek, 4566). Animals were subsequently sectioned sagittally at 16 µm using a Cryostat Leica 1850.
Immunostaining on sections
Slides containing planarian sections were fixed with 4% formaldehyde for 20 min. They were washed thrice in PBS and subsequently blocked for 1 h in 10% horse serum in PBSTx (PBS+0.3% Triton-X). Overnight incubation with primary antibody was performed at 4°C (rabbit anti-mouse collagen IV, Abcam, ab-756p; 1:200). Secondary antibody was incubated for 45 min (Alexa-Fluor anti-rabbit 488, Molecular Probes). Sections were then washed and stained with DAPI.
Transmission electron microscopy
Samples were prepared as described previously (Brubacher et al., 2014). N-block staining using 2% urnayl acetate was carried out for 1 h prior to dehydration with graded ethanol series. Ultrathin sections of 60 nm were cut using an RMC cryo ultra-microtome equipped with a diamond knife; sections were then placed on 200 mesh copper grids. Stained sections were observed with a TEM (Tecnai G2 Spirit Bio-TWIN) operating at 100 kV. The electron micrographs were digitized using Adobe Photoshop by adjusting the contrast and the brightness balance.
Polysome profiling
Animals were soaked in cycloheximide (CHX) for 2 h at 10 µg/ml concentration in planaria media. They were than macerated in cold CMF (Ca2+/Mg2+-free media) with CHX followed by collagenase treatment for 30 min to dissociate them into cells. The cells were than pelleted and washed three times with CMF+CHX and resuspended in 500 µl of lysis buffer (6 M urea, 2 M thiourea, 50 mM DTT, 5% glycerol, 1× PIC, RNAse inhibitor and CHX). Cells were incubated in lysis buffer for 30 min at 4°C and then vigorously pipetted up and down for complete lysis of cells. Planarian lysate was loaded onto a sucrose gradient (15%-45%) and centrifuged at 276,960 g for 2 h. The sucrose gradient was then loaded on to an ISCO Teledyne Gradient fractionator. Sixty percent sucrose was pumped from below to push the sucrose gradient to the UV chamber. Constant absorbance was measured and the fractions were collected.
qRT-PCR analysis
Total RNA from samples of five or six animals was extracted with Trizol reagent (Invitrogen) and cDNAs were synthesized with SuperScriptII Reverse Transcriptase (Invitrogen). qRT-PCR experiments were then performed using the Absolute qRT-PCR SYBR Green Master Mix (Thermo Scientific). Experiments were performed on three biological replicates per time point. Each biological replicate was technically replicated three times in each reaction. smed-actin was used for normalization. Students t-test was used to test for statistical significance.
Following primers were used for qRT-PCR: smed-jun1, 5′-TCCAGTAACCAGCCACAACT-3′ and 5′-AAAGCGCGTTGTTTTCTTGT-3′; smed-fos1, 5′-CCGGTAACTGCAACTAAGCC-3′ and 5′-ACTGAAATTGGCGTCGTTCA-3′; smed-traf1, 5′-AATAGTGTGGCCGTTTCGAC-3′ and 5′-GGCTGACCTGCTCCTACATT-3′; smed-egr3, 5′-TCGTCGGGATGAATTGAAAAGA-3′ and 5′-ATGTCGCAACCTTTCGTCTG-3′; smed-notum, 5′-AAACCGGCAAGTCTCCATGT-3′ and 5′-TGGGAAAGCGGTGAACATGT-3′; smed-zfp-1, 5′-AGCCAAAATAGTCCAGTACCCA-3′ and 5′-TGGTGTTGATTTTCGCTTCTGT-3′.
mRNA purification and transcriptome sequencing
Total RNA was purified using TRIzol reagent from the desired polysome fraction and also from the whole animal, and sequenced using Illumina TrueSeq RNA Samole preparation kit v2 on HiSeq. Details of RNA-seq analysis can be found in the supplementary Materials and Methods.
BrdU labeling and immunofluorescence
10 mg/ml BrdU solution was prepared in 1× planaria media containing 10% DMSO, which was injected in planarians. BrdU staining was carried out as described previously (Vasquez-Doorman and Petersen, 2014) with few modifications. After fluorescence in situ hybridization development as described previously (King and Newmark, 2013), POD was inactivated with 2N HCl for 45 min, followed by antibody labeling of BrdU. Rat anti-BrdU (Abcam) was used at 1:1000 dilution.
Acknowledgements
We thank Dr Ramkumar Sambasivan, Dr Aziz Aboobaker, Dr Jochen Rink and Dr Colin Jamora for reviewing the manuscript, Nishan Shettigar for confocal imaging and all the lab members for manuscript comments. High-throughput sequencing was performed at the Next-Generation Sequencing facility at Genotypic, Bangalore. Confocal imaging was carried out at the CIFF facility, NCBS, Bangalore.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: D.B., A.G., S.R., D.P.; Methodology: D.B., J.G., R.P., A.G., D.P.; Software: V.L., D.P.; Validation: D.B., J.K.; Formal analysis: D.B., V.L., S.D., A.G., S.R., D.P.; Investigation: D.B., J.K., K.N., S.K., V.S.; Resources: D.P.; Data curation: V.L.; Writing - original draft: D.B., D.P.; Writing - review & editing: D.B., J.K., S.K., S.R.; Visualization: J.K., D.P.; Supervision: A.G., S.R., D.P.; Project administration: D.B., D.P.; Funding acquisition: D.P.
Funding
This work was generously supported by the Wellcome Trust/DBT India Alliance (500160/Z/09/Z). D.P. is supported by funding from a Wellcome Trust/DBT India Alliance Intermediate Fellowship. D.B. and J.K. are supported by a Council for Scientific and Industrial Research fellowship. Deposited in PMC for immediate release.
Data availability
Data have been deposited in the Sequence Read Archive (SRA, https://www.ncbi.nlm.nih.gov/sra/) under SRP078776.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.152942.supplemental
References
- Aboobaker A. A. (2011). Planarian stem cells: a simple paradigm for regeneration. Trends Cell Biol. 21, 304-311. 10.1016/j.tcb.2011.01.005 [DOI] [PubMed] [Google Scholar]
- Arwert E. N., Hoste E. and Watt F. M. (2012). Epithelial stem cells, wound healing and cancer. Nature reviews. Cancer 12, 170-180. 10.1038/nrc3217 [DOI] [PubMed] [Google Scholar]
- Babic I., Sharma S. and Black D. L. (2009). A role for polypyrimidine tract binding protein in the establishment of focal adhesions. Mol. Cell. Biol. 29, 5564-5577. 10.1128/MCB.00590-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baguñà J. (2012). The planarian neoblast: the rambling history of its origin and some current black boxes. Int. J. Dev. Biol. 56, 19-37. 10.1387/ijdb.113463jb [DOI] [PubMed] [Google Scholar]
- Boudreau N., Myers C. and Bissell M. J. (1995). From laminin to lamin: regulation of tissue-specific gene expression by the ECM. Trends Cell Biol. 5, 1-4. 10.1016/S0962-8924(00)88924-2 [DOI] [PubMed] [Google Scholar]
- Brubacher J. L., Vieira A. P. and Newmark P. A. (2014). Preparation of the planarian Schmidtea mediterranea for high-resolution histology and transmission electron microscopy. Nat. Protoc. 9, 661-673. 10.1038/nprot.2014.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebrià F., Vispo M., Newmark P., Bueno D. and Romero R. (1997). Myocyte differentiation and body wall muscle regeneration in the planarian Girardia tigrina. Dev. Genes Evol. 207, 306-316. 10.1007/s004270050118 [DOI] [PubMed] [Google Scholar]
- Chandebois R. (1980). The dynamics of wound closure and its role in programming of planarian regeneration II- Distalization. Dev. Growth Differ. 22, 693-704. 10.1111/j.1440-169X.1980.00693.x [DOI] [PubMed] [Google Scholar]
- Chicurel M. E., Singer R. H., Meyer C. J. and Ingber D. E. (1998). Integrin binding and mechanical tension induce movement of mRNA and ribosomes to focal adhesions. Nature 392, 730-733. 10.1038/33719 [DOI] [PubMed] [Google Scholar]
- de Hoog C. L., Foster L. J. and Mann M. (2004). RNA and RNA binding proteins participate in early stages of cell spreading through spreading initiation centers. Cell 117, 649-662. 10.1016/S0092-8674(04)00456-8 [DOI] [PubMed] [Google Scholar]
- Forsthoefel D. J., James N. P., Escobar D. J., Stary J. M., Vieira A. P., Waters F. A. and Newmark P. A. (2012). An RNAi screen reveals intestinal regulators of branching morphogenesis, differentiation, and stem cell proliferation in planarians. Dev. Cell 23, 691-704. 10.1016/j.devcel.2012.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaviño M. A., Wenemoser D., Wang I. E. and Reddien P. W. (2013). Tissue absence initiates regeneration through follistatin-mediated inhibition of activin signaling. Elife 2, e00247 10.7554/eLife.00247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoni B., Richardson W. A., Burgess H. M., Anderson R. C., Wilkie G. S., Gautier P., Martins J. P. S., Brook M., Sheets M. D. and Gray N. K. (2011). Poly(A)-binding proteins are functionally distinct and have essential roles during vertebrate development. Proc. Natl. Acad. Sci. USA 108, 7844-7849. 10.1073/pnas.1017664108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss D. J. and Kleiman F. E. (2013). Poly(A) binding proteins: are they all created equal? Wiley interdisciplinary reviews. RNA 4, 167-179. 10.1002/wrna.1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T., Peters A. H. and Newmark P. A. (2006). A Bruno-like gene is required for stem cell maintenance in planarians. Dev. Cell 11, 159-169. 10.1016/j.devcel.2006.06.004 [DOI] [PubMed] [Google Scholar]
- Gurley K. A., Elliott S. A., Simakov O., Schmidt H. A., Holstein T. W. and Sanchez Alvarado A. (2010). Expression of secreted Wnt pathway components reveals unexpected complexity of the planarian amputation response. Dev. Biol. 347, 24-39. 10.1016/j.ydbio.2010.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori I. (1979). Regeneration of the epidermis and basement membrane of the planarian Dugesia japonica after total-body X irradiation. Radiat. Res. 77, 521-533. 10.2307/3575163 [DOI] [PubMed] [Google Scholar]
- Hori I. (1980). Localization of newly synthesized precursors of basal lamina in the regenerating planarian as revealed by autoradiography. Tissue Cell 12, 513-521. 10.1016/0040-8166(80)90040-3 [DOI] [PubMed] [Google Scholar]
- Hori I. (1991). Differentiation of epidermal cells in the regenerating planarian Dugesia japonica. Hydrobiologia 227, 19-24. 10.1007/BF00027576 [DOI] [Google Scholar]
- Jamora C. (2014). Mechanisms of wound repair. In Stem Cells: From Basic Research to Therapy. Tissue Homeostasis and Regeneration During Adulthood, Applications Legislation and Ethics (ed. Calegari, F. and Waskow, C.) Vol. 2, pp. 67-103. [Google Scholar]
- Katz Z. B., Wells A. L., Park H. Y., Wu B., Shenoy S. M. and Singer R. H. (2012). beta-Actin mRNA compartmentalization enhances focal adhesion stability and directs cell migration. Genes Dev. 26, 1885-1890. 10.1101/gad.190413.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King R. S. and Newmark P. A. (2013). In situ hybridization protocol for enhanced detection of gene expression in the planarian Schmidtea mediterranea. BMC Dev. Biol. 13, 8 10.1186/1471-213X-13-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmanan V., Bansal D., Kulkarni J., Poduval D., Krishna S., Sasidharan V., Anand P., Seshasayee A. and Palakodeti D. (2016). Genome-wide analysis of polyadenylation events in schmidtea mediterranea. G3 (Bethesda) 6, 3035-3048. 10.1534/g3.116.031120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H., Rangarajan E. S., Yogesha S. D. and Izard T. (2009). Raver1 interactions with vinculin and RNA suggest a feed-forward pathway in directing mRNA to focal adhesions. Structure 17, 833-842. 10.1016/j.str.2009.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengfeld T., Watanabe H., Simakov O., Lindgens D., Gee L., Law L., Schmidt H. A., Ozbek S., Bode H. and Holstein T. W. (2009). Multiple Wnts are involved in Hydra organizer formation and regeneration. Dev. Biol. 330, 186-199. 10.1016/j.ydbio.2009.02.004 [DOI] [PubMed] [Google Scholar]
- Lin A. Y. T. and Pearson B. J. (2017). Yorkie is required to restrict the injury responses in planarians. PLoS Genet. 13, e1006874 10.1371/journal.pgen.1006874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus D. A., Evans M. C. and Jacobson A. (2003). Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 4, 223 10.1186/gb-2003-4-7-223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G. G. (1978). A new function of rhabdites: mucus production for ciliary gliding. Zoomorphologie 91, 235-248. 10.1007/BF00999813 [DOI] [Google Scholar]
- McCusker C. D. and Gardiner D. M. (2014). Understanding positional cues in salamander limb regeneration: implications for optimizing cell-based regenerative therapies. Dis. Models Mech. 7, 593-599. 10.1242/dmm.013359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mii Y. and Taira M. (2009). Secreted Frizzled-related proteins enhance the diffusion of Wnt ligands and expand their signalling range. Development 136, 4083-4088. 10.1242/dev.032524 [DOI] [PubMed] [Google Scholar]
- Morita M. and Best J. B. (1974). Electron microscopic studies of planarian regeneration. II. Changes in epidermis during regeneration. J. Exp. Zoolog. 187, 345-373. 10.1002/jez.1401870305 [DOI] [PubMed] [Google Scholar]
- Morita M., Best J. B. and Noel J. (1969). Electron microscopic studies of planarian regeneration. I. Fine structure of neoblasts in Dugesia dorotocephala. J. Ultrastruct. Res. 27, 7-23. 10.1016/S0022-5320(69)90017-3 [DOI] [PubMed] [Google Scholar]
- Palakodeti D., Smielewska M., Lu Y.-C., Yeo G. W. and Graveley B. R. (2008). The PIWI proteins SMEDWI-2 and SMEDWI-3 are required for stem cell function and piRNA expression in planarians. RNA 14, 1174-1186. 10.1261/rna.1085008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson B. J., Eisenhoffer G. T., Gurley K. A., Rink J. C., Miller D. E., Sanchez, and Alvarado A. (2009). Formaldehyde-based whole-mount in situ hybridization method for planarians. Dev. Dyn. 238, 443-450. 10.1002/dvdy.21849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen C. P. and Reddien P. W. (2011). Polarized notum activation at wounds inhibits Wnt function to promote planarian head regeneration. Science 332, 852-855. 10.1126/science.1202143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien P. W. and Sanchez Alvarado A. (2004). Fundamentals of planarian regeneration. Annu. Rev. Cell Dev. Biol. 20, 725-757. 10.1146/annurev.cellbio.20.010403.095114 [DOI] [PubMed] [Google Scholar]
- Reddien P. W., Bermange A. L., Murfitt K. J., Jennings J. R. and Sánchez Alvarado A. (2005). Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev. Cell 8, 635-649. 10.1016/j.devcel.2005.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien P. W., Bermange A. L., Kicza A. M. and Sanchez Alvarado A. (2007). BMP signaling regulates the dorsal planarian midline and is needed for asymmetric regeneration. Development 134, 4043-4051. 10.1242/dev.007138 [DOI] [PubMed] [Google Scholar]
- Resch A. M., Palakodeti D., Lu Y.-C., Horowitz M. and Graveley B. R. (2012). Transcriptome analysis reveals strain-specific and conserved stemness genes in Schmidtea mediterranea. PloS ONE 7, e34447 10.1371/journal.pone.0034447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink J. C. (2013). Stem cell systems and regeneration in planaria. Dev. Genes Evol. 223, 67-84. 10.1007/s00427-012-0426-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink J. C., Vu H. T.-K. and Sanchez Alvarado A. (2011). The maintenance and regeneration of the planarian excretory system are regulated by EGFR signaling. Development 138, 3769-3780. 10.1242/dev.066852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts-Galbraith R. H. and Newmark P. A. (2015). On the organ trail: insights into organ regeneration in the planarian. Curr. Opin. Genet. Dev. 32, 37-46. 10.1016/j.gde.2015.01.009 [DOI] [PubMed] [Google Scholar]
- Ross K. G., Omuro K. C., Taylor M. R., Munday R. K., Hubert A., King R. S. and Zayas R. M. (2015). Novel monoclonal antibodies to study tissue regeneration in planarians. BMC Dev. Biol. 15, 2 10.1186/s12861-014-0050-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhana L., Shibata N., Nishimura O. and Agata K. (2010). Different requirements for conserved post-transcriptional regulators in planarian regeneration and stem cell maintenance. Dev. Biol. 341, 429-443. 10.1016/j.ydbio.2010.02.037 [DOI] [PubMed] [Google Scholar]
- Sasidharan V., Lu Y. C., Bansal D., Dasari P., Poduval D., Seshasayee A., Resch A. M., Graveley B. R. and Palakodeti D. (2013). Identification of neoblast- and regeneration-specific miRNAs in the planarian Schmidtea mediterranea. RNA 19, 1394-1404. 10.1261/rna.038653.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurmann W. and Peter R. (1998). Inhibition of regeneration in the planarian Dugesia polychroa (Schmidt) by treatment with magnesium chloride: a morphological study of wound closure. Hydrobiologia 383, 111-116. [Google Scholar]
- Scimone M. L., Srivastava M., Bell G. W. and Reddien P. W. (2011). A regulatory program for excretory system regeneration in planarians. Development 138, 4387-4398. 10.1242/dev.068098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone M. L., Kravarik K. M., Lapan S. W. and Reddien P. W. (2014). Neoblast specialization in regeneration of the planarian Schmidtea mediterranea. Stem Cell Rep. 3, 339-352. 10.1016/j.stemcr.2014.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaer R. J. (1965). The origin and continuous replacement of epidermal cells in the planarian Polycelis tenuis (Iijima). J. Embryol exp Morph 13, 129-139. [PubMed] [Google Scholar]
- Smith R. W. P., Blee T. K. P. and Gray N. K. (2014). Poly(A)-binding proteins are required for diverse biological processes in metazoans. Biochem. Soc. Trans. 42, 1229-1237. 10.1042/BST20140111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solana J., Gamberi C., Mihaylova Y., Grosswendt S., Chen C., Lasko P., Rajewsky N. and Aboobaker A. A. (2013). The CCR4-NOT complex mediates deadenylation and degradation of stem cell mRNAs and promotes planarian stem cell differentiation. PLoS Genet. 9, e1004003 10.1371/journal.pgen.1004003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer V. A., Xu R. and Bissell M. J. (2007). Extracellular matrix, nuclear and chromatin structure, and gene expression in normal tissues and malignant tumors: a work in progress. Adv. Cancer Res. 97, 275-294. 10.1016/S0065-230X(06)97012-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T. and Fujisawa T. (2009). Important roles for epithelial cell peptides in hydra development. BioEssays 31, 610-619. 10.1002/bies.200800163 [DOI] [PubMed] [Google Scholar]
- Tarun S. Z. Jr. and Sachs A. B. (1996). Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 15, 7168-7177. [PMC free article] [PubMed] [Google Scholar]
- Tazaki A., Kato K., Orii H., Agata K. and Watanabe K. (2002). The body margin of the planarian Dugesia japonica: characterization by the expression of an intermediate filament gene. Dev. Genes Evol. 212, 365-373. 10.1007/s00427-002-0253-0 [DOI] [PubMed] [Google Scholar]
- Tu K. C., Cheng L. C., T K Vu H, Lange J. J., McKinney S. A., Seidel C. W. and Sanchez Alvarado A. (2015). Egr-5 is a post-mitotic regulator of planarian epidermal differentiation. Elife 4, e10501 10.7554/eLife.10501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wolfswinkel J. C., Wagner D. E. and Reddien P. W. (2014). Single-cell analysis reveals functionally distinct classes within the planarian stem cell compartment. Cell Stem Cell 15, 326-339. 10.1016/j.stem.2014.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez-Doorman C. and Petersen C. P. (2014). zic-1 Expression in Planarian neoblasts after injury controls anterior pole regeneration. PLoS Genet. 10, e1004452 10.1371/journal.pgen.1004452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D. E., Wang I. E. and Reddien P. W. (2011). Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science 332, 811-816. 10.1126/science.1203983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Stary J. M., Wilhelm J. E. and Newmark P. A. (2010). A functional genomic screen in planarians identifies novel regulators of germ cell development. Genes Dev. 24, 2081-2092. 10.1101/gad.1951010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenemoser D. and Reddien P. W. (2010). Planarian regeneration involves distinct stem cell responses to wounds and tissue absence. Dev. Biol. 344, 979-991. 10.1016/j.ydbio.2010.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenemoser D., Lapan S. W., Wilkinson A. W., Bell G. W. and Reddien P. W. (2012). A molecular wound response program associated with regeneration initiation in planarians. Genes Dev. 26, 988-1002. 10.1101/gad.187377.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witchley J. N., Mayer M., Wagner D. E., Owen J. H. and Reddien P. W. (2013). Muscle cells provide instructions for planarian regeneration. Cell Rep. 4, 633-641. 10.1016/j.celrep.2013.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff E. (1962). Recent Researches on the Regeneration of Planaria. New York: Ronald Press. [Google Scholar]
- Wurtzel O., Cote L. E., Poirier A., Satija R., Regev A. and Reddien P. W. (2015). A Generic and Cell-Type-Specific Wound Response Precedes Regeneration in Planarians. Dev. Cell 35, 632-645. 10.1016/j.devcel.2015.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayas R. M., Cebrià F., Guo T., Feng J. and Newmark P. A. (2010). The use of lectins as markers for differentiated secretory cells in planarians. Dev. Dyn. 239, 2888-2897. 10.1002/dvdy.22427 [DOI] [PMC free article] [PubMed] [Google Scholar]