Abstract
Nucleotide excision repair is a major DNA repair mechanism in all cellular organisms. In this repair system, the DNA damage is removed by concerted dual incisions bracketing the damage and at a precise distance from the damage. Here, we review the basic mechanisms of excision repair in Escherichia coli and humans and the recent genome-wide mapping of DNA damage and repair in these organisms at single-nucleotide resolution.
Keywords: damage recognition, kinetic proofreading, damage mapping, repair mapping, carcinogenesis, chemotherapy
Introduction
In a retrospective on the 21st anniversary of the historical paper describing the double helical structure of DNA, Francis Crick wrote that “We totally missed the possible role of enzymes in repair although, due to Claud Rupert's early very elegant work on photoreactivation (italics added), I later came to realize that DNA is so precious that probably many distinct repair mechanisms would exist.” (1). In fact, following the landmark paper by Rupert describing photolyase (2), which mediates photoreactivation, many other enzymes and enzymatic mechanisms that repair DNA were discovered, and there is a consensus that without DNA repair life would not exist (3–5). Indeed, DNA is very reactive and hence continuously altered by physical (light and heat) and chemical (ranging from water to polyaromatic hydrocarbons) agents.
DNA repair is the elimination of chemically or physically damaged (altered) or mismatched nucleotides or correction of the abnormal DNA structures. Based on the types of damages processed and the mechanistic features of the repair reactions, five types of DNA repair mechanisms (pathways) have been defined (6) as follows: direct repair; base excision repair; nucleotide excision repair; double-strand break/cross-link repair; and mismatch repair. In direct repair, the chemical bond(s) that constitute damage are broken. In base excision repair, the glycosidic bond linking the damaged base to the phosphodiester backbone is broken; the resulting apurinic/apyrimidinic (AP) site deoxyribose moiety is removed, and the missing nucleotide is replaced by DNA polymerase and ligated. In nucleotide excision repair, the damaged base(s) are removed by concerted dual incision of phosphodiester bonds bracketing the lesion at relatively precise distances from the damage to generate oligonucleotides of 12–13 nt2 (prokaryotes) or 26–27 nt (eukaryotes), and the resulting gap is filled and ligated. In double-strand break/cross-link repair, the phosphodiester bonds in both strands of the duplex are broken by physical or chemical agents, genome-remodeling enzymes, or by enzymatic means during repair of interstrand cross-links caused by both intrinsic metabolites and external DNA-damaging agents. The breaks are repaired by either direct end joining or homologous recombination. In mismatch repair, mismatched bases resulting from replication errors, recombination, or base deamination are corrected by exonucleolytic removal of the mismatch followed by gap filling and ligation.
The basic biochemical mechanisms of all these pathways have been worked out in considerable detail. However, how these repair events are modulated within the genomic/chromatin context remains to be investigated. Some of the parameters that affect damage formation and repair have been identified using methods that map damage and repair at high resolution in short genomic segments or at low resolution and genome-wide (7–10). Recently, we developed methods to map genome-wide and at single-nucleotide resolution sites of damage by bulky lesions that are substrates for excision repair and sites where these bulky lesions are repaired (11, 12). In the following, we will present a brief overview of Escherichia coli and human excision repair mechanisms followed by a description of these recently developed damage and repair mapping methods. Finally, we present mapping results that address some fundamental questions pertaining to the contribution of various repair/replication/transcription enzymes to E. coli excision repair and the effect of chromatin states on damage and repair in humans.
Molecular mechanisms of nucleotide excision repair
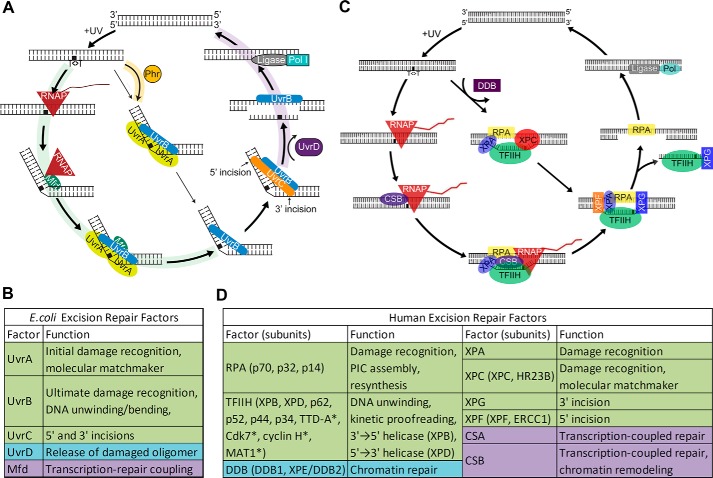
Nucleotide excision repair (excision repair) has been characterized in considerable detail in prokaryotes and eukaryotes (5, 13–18), but it remains under-explored in Archaea. The basic steps of nucleotide excision repair are as follows: (a) damage recognition; (b) dual incisions bracketing the lesion to form a 12–13-nt oligomer in prokaryotes (19) and a 26–27-nt oligomer in eukaryotes (20–23); (c) release of the excised oligomer; (d) repair synthesis to fill the gap; and (e) ligation. Damage recognition and excision are carried out by three proteins in E. coli and six repair factors encompassing 15–16 proteins in humans (Fig. 1). In addition, there are accessory proteins facilitating release of the excised oligomer in prokaryotes, damage access in eukaryotes, and transcription-associated repair both in prokaryotes and eukaryotes. It should be noted that the excision repair proteins proper, in contrast to proteins of all other DNA repair pathways, are not evolutionarily conserved from prokaryotes to eukaryotes (5).
Figure 1.
Molecular mechanisms of nucleotide excision repair. A, E. coli excision repair. In addition to the three core dual incision proteins, photolyase (Phr) aids in recognition of CPDs both in vivo and in vitro and accelerates the rate of CPD repair. Mfd translocase couples transcription to repair, and UvrD helicase releases the excised oligomer, freeing UvrB and UvrC for catalytic turnover (58). B, E. coli excision repair factors. C, human excision repair. In addition to the six core excision repair factors, the DDB heterodimer stimulates repair of CPDs in vivo but not in vitro by a poorly defined mechanism. In TCR, stalled RNAPII with the aid of CSA and CSB acts as the damage sensor and accelerates the rate of repair of the transcribed strand. D, human excision repair factors. * indicates TFIIH subunits not essential for excision repair. B and D, green, core repair factors; purple, transcription-repair coupling factors; blue, other repair proteins involved in excision repair.
Damage recognition
Damage recognition is the rate-limiting step in excision repair because for many lesions processed by nucleotide excision repair, the difference between damaged and normal bases is often minor (15). Thus, damage recognition proteins associate with damaged and much more abundant undamaged DNA at comparable levels. Specificity in excision repair is achieved by multistep damage recognition mechanisms, including molecular matchmaking (24, 25) and kinetic proofreading, and by employing molecular proxies.
Molecular matchmaker
A molecular matchmaker is a protein that uses the energy released from ATP hydrolysis to bring two compatible but otherwise solitary macromolecules together, and it promotes their association for productive engagement and then dissociates from the complex (15).
Kinetic proofreading
Kinetic proofreading is a mechanism that achieves high specificity, beyond the level that can be achieved by the free energy difference between correct and incorrect interactions (equilibrium discrimination), by the presence of unidirectional energy-utilizing irreversible intermediate steps, and at each step the reaction can be aborted to the original reactants (26, 27). Kinetic proofreading differs from other multistep kinetic schemes of the Michaelis-Menten type in which every step, except the ultimate one, is reversible to the preceding one. As an illustration of the power of kinetic proofreading in conferring specificity, a 10-fold difference between the off rates for specific and nonspecific complexes (e.g. 0.1 and 1.0 s−1) can provide an ∼104-fold difference in the effector reaction by interposing five steps between the binding and catalysis steps. The end result is that kinetic proofreading by utilizing energy from ATP hydrolysis and other sources achieves biologically acceptable specificity at a physiologically relevant rate. It is an essential specificity determinant in replication (28), recombination (29), transcription (30), splicing (31), translation (32–37), T cell receptor signaling (38, 39), microtubule and actin filament formation (40), and anaphase-promoting complex-mediated cell cycle control (41) and excision repair (15, 42–44).
In E. coli excision repair, first-order damage recognition is accomplished in an ATP-dependent manner by UvrA2 in the UvrA2UvrB1 complex. Within this complex UvrA2 functions as the molecular matchmaker, mediating the formation of a UvrB-DNA complex using the energy of ATP hydrolysis. Both UvrA and UvrB have ATPase activities, and in addition UvrB has translocase function. The two proteins thus perform the kinetic proofreading steps leading to formation of a very stable UvrB-DNA complex at the damage site concomitant with the dissociation of UvrA from the complex (molecular matchmaker) to enable UvrC binding and dual incision (24, 25). First-order damage recognition in humans is accomplished by RPA, XPA, and XPC along with the TFIIH factor composed of 6–10 subunits, two of which, XPB and XPD, are helicases (13, 14, 45). Within this complex, XPC is the molecular matchmaker that plays a key role in damage recognition and, together with the kinetic proofreading function of the XPB and XPD helicases, helps create a stable complex (preincision complex 1, PIC1) in which the DNA is unwound by about 25 bp around the damage site (14, 15). Then, XPG enters the complex as XPC comes off (PIC2), followed by entry of XPF-ERCC1 (PIC3). Within PIC1, -2, and -3, ATP hydrolysis by XPB and XPD is used for kinetic proofreading to ensure specificity (46, 47). Following the final proofreading step, the dual incisions take place.
Recognition by proxy and transcription-coupled repair (TCR)
In vivo, the rate of damage recognition and therefore repair both in E. coli and in humans is affected by multiple factors in addition to the excision repair proteins proper. These include DNA binding of transcription factors and other DNA-binding proteins in both E. coli and humans and compaction in chromatin, nucleosomes, and post-translational histone modifications in humans (48, 49). Some DNA-binding proteins such as photolyase in E. coli stimulate excision repair by facilitating the assembly of the excision repair proteins. However, nucleosomes and transcription factor binding in general interfere with damage recognition. In contrast to these factors that play a limited role in excision repair or exert nonspecific effects on repair, transcription has a unique and specific effect (50, 51); in E. coli RNA polymerase and in humans RNA polymerase II stimulate the repair of the transcribed strand. It has been found that both of these polymerases, upon encountering a lesion in the transcribed strand, arrest at the damage site forming a ternary RNA pol-RNA-DNA complex with a half-life of ∼20 h (52, 53). Thus, RNA pol has been referred to as “the most specific damage recognition protein” (54). In fact, this complex on its own inhibits repair in E. coli and may do so in humans, although reconstituted transcription-repair reactions have shown that RNAPII stalled at a lesion in the template does not inhibit its repair and may in fact stimulate repair of transcription-blocking lesions by removing the inhibitory effect of histones on excision repair (53, 55). Importantly, RNA pol stalled at a template strand lesion is specifically recognized by proteins at the interface of transcription and repair: Mfd translocase in E. coli (56, 57) and CSB translocase in humans (58). These proteins help recruit the repair proteins and in doing so accelerate the rate of damage recognition and hence rate of repair. This is most evident for lesions that are poorly recognized by excision repair proteins proper, such as cyclobutane pyrimidine dimers (CPD) and platinum-d(GpG) diadducts. TCR has only a modest effect on lesions that are efficiently recognized by the core repair machinery, such as (6–4) photoproducts (11) and BPDE-guanine (benzo[α]pyrene diol epoxide-guanine) adducts (59).
Dual incisions
In E. coli, the dual incisions are made by UvrC within the UvrB1-UvrC1-DNA complex; the 3′-incision is made by the GIY-YIG nuclease motif in the N-terminal domain of the protein (60, 61) three to four nucleotides 3′ to the damage (19), and the 5′-incision is made by the Endo-V nuclease motif in the C-terminal domain of the protein (62) seven nucleotides 5′ to the damage (19). The dual incisions are concerted but asynchronous (62). In humans, within PIC3 XPG makes the 3′-incision by its Flap endonuclease domain five to six nucleotides 3′ to the damage, and XPF makes the 5′-incision 19–22 nucleotides 5′ to the damage by its SMX family structure-specific endonuclease active site (13, 14, 21).
Excision/release of the excised oligomer
In E. coli, following the dual incision the UvrB-C-excised oligomer complex remains bound to the duplex (25, 63). The complex is displaced by the UvrD helicase (25, 63, 64), and the excised oligomer is degraded by exonucleases. The released UvrB and UvrC proteins enter new rounds of repair. In the absence of UvrD, the number of excised oligomers is virtually stoichiometric with the number of UvrC molecules in the cell, because of the three excision repair proteins UvrC is the least abundant (17, 19). In humans, the dual incision takes place within the context of an excision repair bubble (∼25 nt) around the damage site (24, 46), and in contrast to E. coli, the excised oligomer is released from the duplex without the need for an additional helicase (46). However, the excised oligomer is released in a tight complex with TFIIH-XPG and dissociates from these proteins with a half-life of ∼3 h in vitro (65) and ∼10 min in vivo (66), and then it is rapidly degraded by nucleases.
Repair synthesis
In E. coli, the excision gap is filled by DNA polymerase I (63). The repair patch matches the size of the excision gap (67); hence, there is no strand displacement during gap filling. In the absence of DNA pol I, the gap can be filled by DNA pol II and DNA pol III (13). In humans, the excision gap is filled by DNA pol δ/ϵ in proliferating cells (13, 14). In non-proliferating cells, other DNA polymerases such as pol κ/λ may fill the gap (68). In any event, as in E. coli, in humans the size of the repair patch matches the size of the excision gap (∼26–27 nt) indicating that no nick translation occurs (69). However, in a small fraction of cases the excision gap is enlarged to a size of >50 nt by Exo I, and the ssDNA is covered by RPA to constitute a signal for ATR checkpoint kinase (70).
Ligation
In E. coli, the nick at the 3′-end of the repair patch is ligated by E. coli ligase (63, 67). In humans, DNA ligase I is the primary ligase for sealing the 3′-nick (13, 14), although the XRCC1-ligase3 complex may carry out ligation in certain cell types and physiological states (71).
Methods for genome-wide maps of excision repair
Recently, methods were developed for mapping UV, cisplatin, and BPDE-induced DNA damages and their repair genome-wide in humans and in E. coli (11, 12, 59, 64, 72–74). These methods will be described in general outlines and then their applications to the E. coli and human genomes will be presented.
Damage-sequencing (Damage-seq)
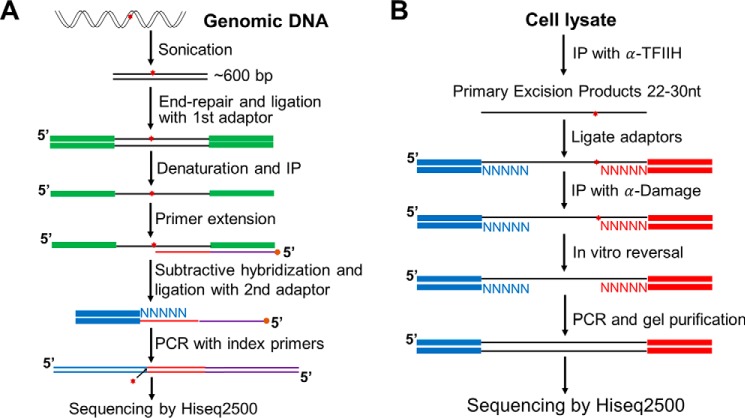
This method is based on the fact that high-fidelity DNA polymerases arrest at bulky damage sites. Fig. 2A shows a scheme of the damage-seq method (12, 73). After treating cells with a DNA-damaging agent, genomic DNA is isolated and fragmented. Next, the fragments are end-repaired and ligated to the first adaptor. Then, the DNA is denatured and immunoprecipitated with anti-damage antibodies. Next, a biotinylated primer is annealed to the adaptor and extended by a high-fidelity DNA polymerase, which is blocked by DNA damage. The extended primers are purified by streptavidin-coated beads. The extension products from undamaged strands contain sequence complementary to the 5′-sequence of the first adaptor and thus can be removed by subtractive hybridization with an oligomer bearing the 5′-sequence of the first adaptor. The second adaptor is then ligated, followed by PCR with index primers. Because of the immunoprecipitation and subtractive hybridization steps, nonspecific products from the undamaged strand are minor, can be identified by the existence of the 5′-sequence of the first adaptor, and thus discarded at the data analysis step.
Figure 2.
Damage-seq and XR-seq methods for high-resolution genome-wide mapping of DNA damage and repair. A, damage-seq. The key step is the arrest of a high-fidelity DNA polymerase at the nucleotide 3′ to the damaged base (12). B, XR-seq. The key step is the capture of the excised oligomer by TFIIH co-immunoprecipitation followed by damage-specific immunoprecipitation (IP) (11).
Excision repair-sequencing (XR-seq)
In this method, the 12–13-nt-long excision products in E. coli and the ∼26–27-nt-long excision products in humans generated by excision repair are isolated (66, 75, 76), processed, sequenced, and aligned to the respective genome to generate repair maps. Fig. 2B shows a scheme of the method. In E. coli, the excised oligomer is released from the duplex free of a protein (45). Thus, to isolate the excised oligomer, cells are gently lysed, and oligonucleotide-sized fragments are separated from genomic DNA for further processing. In contrast, in humans the excised oligomer is released in a tight complex with TFIIH-XPG (45, 65, 66) from which it is released with a half-life of 10 min and degraded to smaller oligomers. Hence, the “primary” excision products of human excision repair are obtained by co-immunoprecipitation with TFIIH or XPG antibodies. The oligomers are then deproteinized and ligated to 5′- and 3′-adaptors. Next, the ligation products are immunoprecipitated with damage-specific antibodies followed by reversing the damage by either photolyases (CPD and (6-4)PP) or by NaCN (cisplatin and oxaliplatin adducts). Instead of reversal, the lesions can also be bypassed by translesion synthesis DNA polymerase (77) to generate a damage-free strand (59). Finally, the oligonucleotides are amplified, sequenced, and aligned to the genome.
Repair maps of E. coli and human genomes
Excision repair map of the E. coli genome at single-nucleotide resolution
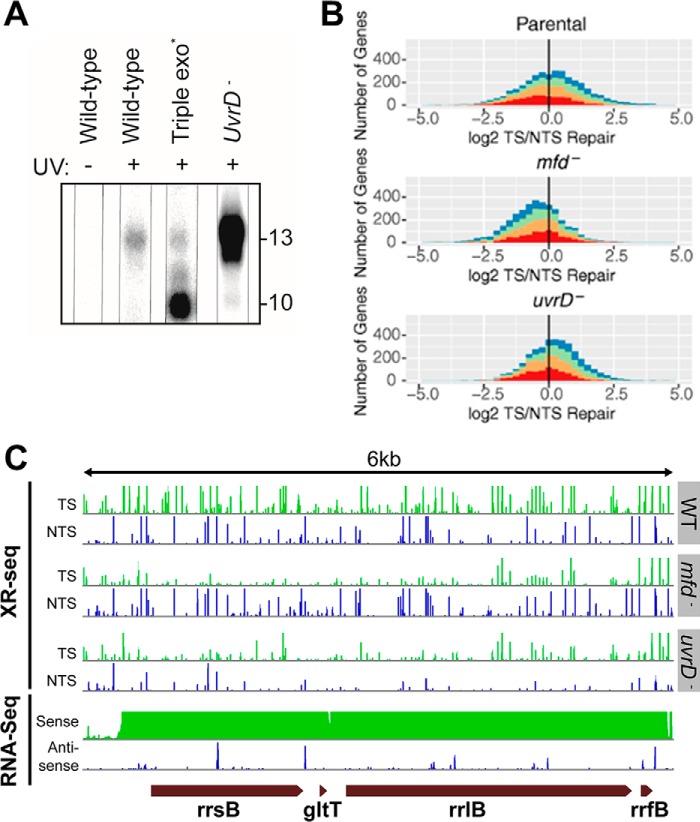
The XR-seq method originally developed for mammalian cells (11) was subsequently used to analyze excision repair in E. coli (64) to address some fundamental question regarding the molecular mechanism of excision repair and the role of various candidate genes in excision repair. This study provided some valuable insights into the processing of the excised oligomer and brought clarity to the roles of various proteins implicated in E. coli transcription-coupled repair (64).
Production and fate of the excised oligomer
When we attempted to isolate the excised 12–13-mer from UV-irradiated E. coli cells, we found the following: 1) in wild-type cells the excised oligomer was rapidly degraded from 12 to 13 nts to 10-mers and smaller species not detectable by the 3′-radiolabeling method used; 2) in cells with mutations in the three major E. coli exonucleases (Exo I, Exo VII, and RecJ), the oligomer was digested processively from the 3′-end generating a 10-nt-long oligomer as the major product consisting of, in addition to CPD, 7 nt 5′ and 1 nt 3′ to the damage where the 3′- to 5′-exonucleases stop; 3) the most striking finding, however, was the status of the excised oligomer in uvrD− mutant; in this background the excised oligomer was much more abundant and was almost exclusively in the form of the 12–13-nt-long primary excision product (Fig. 3A). This finding confirmed the role of UvrD in excision repair that was deduced from in vitro experiments with purified proteins (25, 63); following the dual incision by UvrC, the UvrB-UvrC-“excised oligonucleotide” complex remains bound to the duplex; UvrD (helicase II) acting as a 3′- to 5′-helicase releases UvrC and the excised oligonucleotide and in concert with DNA pol I displaces UvrB from the excision gap, which is filled by DNA pol I and ligated. Thus, it appears that UvrB-UvrC protect the excised oligomer from nucleases, explaining the high yield of primary 12–13-nt excision product in the uvrD− mutant.
Figure 3.
XR-seq analysis of E. coli genome. A, effect of genetic background on recovery of the excised oligomer. In WT cells, 12–13-mers are captured at low yield. In triple exonuclease mutant deficient in major ssDNA exonucleases, the excised oligomer is degraded to a 10-mer by a 3′- to 5′-exonuclease that stops at a nucleotide 3′ to the dimer. Most strikingly, in the uvrD− mutant the excised oligomer is much more abundant than in other strains and almost exclusively 12–13-nt in length, consistent with the idea that the “excised” oligomer is not released from the UvrB-UvrC-DNA complex in the absence of UvrD and hence is protected from ssDNA nucleases. B, frequency distribution of log2-transformed TS/NTS repair in all annotated genes. E. coli genes in three indicated strains are colored by sense strand transcription levels going from the lowest quartile RNA-seq count colored in red, to orange, to green, and to the highest transcription quartile in blue. The log2 (TS/NTS) means for each strain are 1.16 for WT, 0.68 for mfd−, and 1.17 for uvrD− mutant. The vertical black line represents the border where TS repair level is equal to NTS repair. C, screen shots of rRNA operon (including gltT RNA gene) transcription (RNA-seq) and excision repair maps (XR-seq) for E. coli WT, and mfd−, and uvrD− mutants. Note the reversal of the TS/NTS repair ratio in mfd− versus WT and the enhancement of the TS/NTS ratio in uvrD− mutant even though overall repair is reduced in this strain (58).
Excision repair map of E. coli and mechanism of transcription-repair coupling
Excision repair maps of WT, mfd− (78, 79), and uvrD− strains were generated by XR-seq to assess the roles of Mfd translocase and UvrD helicase in transcription-coupled repair. The maps revealed a rather complex genome-wide repair pattern vis à vis annotated genes because of the apparently widespread functional and spurious antisense transcription throughout most of the E. coli genome (80). This study confirmed the TCR model based on in vitro data (56, 79, 81), including single-molecule experiments (82), and generated a repair map to complement the curated transcription map of E. coli. Importantly, this study revealed unexpected features of E. coli repairome. First of all, it showed that preferential repair of the transcribed strands (TS) is a predominant feature of excision repair and that in genes with no confounding features, TCR is absolutely dependent on transcription-repair coupling factor (TRCF) encoded by the mfd gene (Fig. 3B) (78, 81). In the absence of TRCF, the preferential repair of TS is abolished. Moreover, in the absence of TRCF, the NTS (non-template strand) becomes the strand of preferential repair. This is because in the absence of TRCF, RNAP stops at damage site and interferes with the repair of the TS (52), whereas the NTS is repaired more efficiently in the mfd− mutant than in WT cells because of the availability of more UvrA, -B, and -C proteins to act on the NTS (52). Thus, in highly transcribed genes, the mfd− mutation causes ∼20-fold change in the repair ratio of TS/NTS in favor of the NTS (Fig. 3C). Second, the map reveals that in a significant number of annotated genes the NTS appears to be repaired more efficiently than the TS in WT cells. In most cases, this is caused by the presence of a few hot spots of repair in the NTS of these genes or the presence of antisense transcription in these genes (80, 83–85). Some of the antisense transcripts are those of overlapping genes, and some are caused by spurious transcripts by RNAP from sequences of sufficient similarities to the E. coli consensus promoter sequence, which is not very stringent (80). Further studies are required to explain these apparent exceptions, which reveal that at the genome level the transcription and TCR in E. coli are rather complex.
DNA damage and repair maps of the human genome at single-nucleotide resolution
Genome-wide analyses of damage formation and repair have enabled us to make the following conclusions.
Damage formation
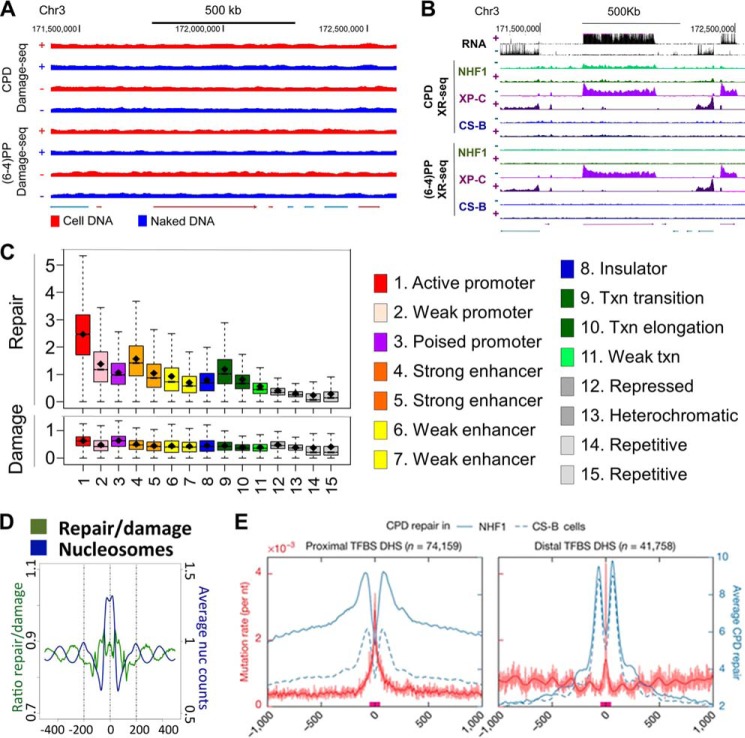
In general, within the resolutions of our assays we do not observe major effects of genomic location or chromatin states on damage formation, with a few exceptions. As shown in Fig. 4A, the damage distribution of either (6-4)PP or CPD in cellular DNA is similar with the pattern seen in UV-irradiated naked DNA, indicating that damage formation is mainly determined by sequence context. The same is also true for cisplatin and oxaliplatin damage (12). However, transcription factor (TF) binding can affect damage formation at specific sites within or around the binding motifs (73). For UV damage, TF binding can enhance or inhibit damage formation or have no effect, depending on the specific TFs, damage types, strands, and positions with no universal rule. This phenomenon is likely due to DNA structural changes induced by the particular TF binding. In contrast, for cisplatin damage, TF binding generally inhibits damage formation with few exceptions (73). This is probably because TF binding blocks the access of cisplatin molecule to DNA.
Figure 4.
Damage formation and repair maps of the human genome. A, comparison of UV damage distribution in cellular DNA and naked DNA. Screen shots of the indicated coordinates of chromosome 3 are shown (73). B, screen shots of the XR-seq data for the same chromosome 3 coordinates shown in A. Note the transcribed strand-specific repair for CPD both in WT and XP-C mutant cells and in XP-C mutant cells only for (6-4)PP and the absolute dependence of repair of both photoproducts on transcription and only in the transcribed strand. In CS-B mutant there is no effect of transcription on repair of either lesion (11), and transcription does not inhibit repair of template strand lesions as is seen in mfd− E. coli (Fig. 3C). C, chromatin states affect repair rates of cisplatin damage (upper panel) but not cisplatin damage formation (lower panel). Open chromatin states and transcriptionally active regions are repaired more efficiently compared with weakly transcribed regions and heterochromatin (12). D, effect of nucleosomes. Repair efficiency is anti-phase with the nucleosome center in agreement with the in vitro data showing inhibition of cisplatin damage in the nucleosome core (12). E, “volcano pattern” of excision repair of CPDs around active transcription factor-binding sites (TFBS, red bars on x axis), which overlap with DNase I-hypersensitive sites (DHS). Repair is strongly inhibited at the center of the transcription factor-binding sites and is flanked by two peaks of repair in the DNase I-hypersensitive sites, whereas mutation rate “erupts” at the center of the repair “crater” where repair is inhibited (93).
Damage repair
The excision repair map, in contrast to the damage map, revealed a rather interesting landscape, exhibiting repair peaks, valleys, and canyons (11, 12, 73). This heterogeneity in repair landscape is caused by transcription, chromatin states (86), nucleosomes, and transcription factors. Naturally, these various factors are interconnected with one another and with histone modifications (87, 88) that affect nearly every DNA transaction in eukaryotes.
Transcription by RNA pol II has a strong effect on repair rate (transcription-coupled repair)
In wild type cells, transcription, specifically RNA pol II stalled at a damage site, strongly enhances the rate of repair of the transcribed strand as shown for human chromosome 7 in Fig. 4B. As a general rule, the enhancement of repair rate is greater for DNA lesions poorly recognized by the core excision repair nuclease such as CPDs and Pt-d(GpG) adducts and is modest for well-recognized damages such as the (6-4)PPs and BPDE-dG (11, 59, 89). The coupling of transcription to repair is mediated by the CSA and CSB proteins and is independent of XPC, which is required for transcription-independent “global” excision repair (50). The repair map from CS-B mutant cells shows the absolute dependence of TCR on CSB (11), whereas the absolute confinement of excision repair of all types of lesions to the transcribed strand of an actively transcribed gene and total lack of repair in the NTS of the same gene are observed in XP-C mutant cells (11). It is noteworthy that in most promoters and enhancers there is a switch of preferential repair to the opposite strand from the transcribed (template) strand upstream of the promoter (11), because it has been found that nearly 80% of promoters and enhancers are bi-directional (90, 91) and hence TCR follows this pattern of transcribed strand switching.
Chromatin states
The effect of chromatin states (86) on repair is shown in Fig. 4C. As a general rule, those states associated with open chromatin are repaired at a faster rate for all the lesions tested. Heterochromatic and repetitive/repressed states are repaired rather slowly, and at a physiological dose of UV light, CPD repair in these regions can be detected for more than 2 days after damage formation (72, 73).
Nucleosomes
In contrast to damage formation, excision repair exhibits periodicity that is antiphase with the nucleosome center (Fig. 4D) (72). This is in accord with the in vitro finding that nucleosomal DNA is repaired less efficiently than naked DNA (14, 15). However, the inhibition observed in vivo is rather modest compared with what is seen in vitro presumably because under in vivo conditions access to damage is facilitated by nucleosome remodeling factors such as the SWI/SNF1 complex (92).
Transcription factors
In contrast to no effect or only a modest effect on damage formation, TFs have a significant effect on excision repair. As a rule, TF with high affinity to the target sequence strongly inhibit repair (Fig. 4E). As a consequence, TF-binding sites are hot spots for mutation (93, 94). Analysis of mutation signatures (93–97) in some of the most common cancers by The Cancer Genome Atlas initiative (97) revealed that mutations that are associated with damage repaired by nucleotide excision repair (UV damage in skin cancer and polyaromatic hydrocarbon damage in lung cancer) are found at higher frequencies at transcription factor-binding sites relative to control sequences (Fig. 4E). Thus, it was concluded that inhibition of repair by transcription factors is a causative factor in skin and lung cancers (93, 94).
Conclusion
DNA repair has been the focus of intensive research in recent years. The enzymes carrying out the repair reactions have been purified; the repair pathways have been reconstituted in vitro, and reasonably detailed models for all the major repair mechanisms have been developed (3–5). In addition, a variety of in vivo imaging methods have been used to follow the repair reactions in vivo to complement the in vitro studies (98). Moreover, structural (99) and single molecule (100) studies have added additional insights into the mechanistic and kinetic aspects of various repair pathways. The high-resolution DNA damage formation and repair mapping methods we described here and results for certain damages we have obtained with these methods further complement and extend existing in vitro and in vivo information on nucleotide excision repair over short DNA segments (7–10). The damage-seq and XR-seq methods provide genome damage and repair landscapes that can be superimposed on various genome-wide structure and function landscapes and thus help integrate DNA repair into the overall genomic context and cellular reaction networks.
This work was supported by National Institutes of Health Grants GM118102 and ES027255. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- nt
- nucleotide
- BPDE
- benzo[α]pyrene diol epoxide
- DHS
- DNase I-hypersensitive site
- TCR
- transcription-coupled repair
- TS
- transcribed strand
- NTS
- non-template strand
- TRCF
- transcription-repair coupling factor
- RNAP
- RNA polymerase
- CPD
- cyclobutane pyrimidine dimer
- pol
- polymerase
- Exo
- exonuclease
- (6-4)PP
- (6-4) photoproduct
- TF
- transcription factor
- seq
- sequencing
- IP
- immunoprecipitation
- ssDNA
- single-stranded DNA.
References
- 1. Crick F. (1974) The double helix: a personal view. Nature 248, 766–769 [DOI] [PubMed] [Google Scholar]
- 2. Rupert C. S., Goodgal S. H., and Herriott R. M. (1958) Photoreactivation in vitro of ultraviolet-inactivated Hemophilus influenzae transforming factor. J. Gen. Physiol. 41, 451–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lindahl T. (2016) The intrinsic fragility of DNA (Nobel Lecture). Angew. Chem. Int. Ed. Engl. 55, 8528–8534 [DOI] [PubMed] [Google Scholar]
- 4. Modrich P. (2016) Mechanisms in E. coli and human mismatch repair (Nobel Lecture). Angew. Chem. Int. Ed. Engl. 55, 8490–8501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sancar A. (2016) Mechanisms of DNA repair by photolyase and excision nuclease (Nobel Lecture). Angew. Chem. Int. Ed. Engl. 55, 8502–8527 [DOI] [PubMed] [Google Scholar]
- 6. Sancar A., Lindsey-Boltz L. A., Unsal-Kaçmaz K., and Linn S. (2004) Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73, 39–85 [DOI] [PubMed] [Google Scholar]
- 7. Denissenko M. F., Pao A., Tang M., and Pfeifer G. P. (1996) Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science 274, 430–432 [DOI] [PubMed] [Google Scholar]
- 8. Besaratinia A., and Pfeifer G. P. (2012) Measuring the formation and repair of UV damage at the DNA sequence level by ligation-mediated PCR. Methods Mol. Biol. 920, 189–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zavala A. G., Morris R. T., Wyrick J. J., and Smerdon M. J. (2014) High-resolution characterization of CPD hotspot formation in human fibroblasts. Nucleic Acids Res. 42, 893–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bryan D. S., Ransom M., Adane B., York K., and Hesselberth J. R. (2014) High resolution mapping of modified DNA nucleobases using excision repair enzymes. Genome Res. 24, 1534–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hu J., Adar S., Selby C. P., Lieb J. D., and Sancar A. (2015) Genome-wide analysis of human global and transcription-coupled excision repair of UV damage at single-nucleotide resolution. Gene Dev. 29, 948–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hu J., Lieb J. D., Sancar A., and Adar S. (2016) Cisplatin DNA damage and repair maps of the human genome at single-nucleotide resolution. Proc. Natl. Acad. Sci. U.S.A. 113, 11507–11512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sancar A. (1996) DNA excision repair. Annu. Rev. Biochem. 65, 43–81 [DOI] [PubMed] [Google Scholar]
- 14. Wood R. D. (1997) Nucleotide excision repair in mammalian cells. J. Biol. Chem. 272, 23465–23468 [DOI] [PubMed] [Google Scholar]
- 15. Reardon J. T., and Sancar A. (2005) Nucleotide excision repair. Prog. Nucleic Acid Res. Mol. Biol. 79, 183–235 [DOI] [PubMed] [Google Scholar]
- 16. Truglio J. J., Croteau D. L., Van Houten B., and Kisker C. (2006) Prokaryotic nucleotide excision repair: the UvrABC system. Chem. Rev. 106, 233–252 [DOI] [PubMed] [Google Scholar]
- 17. Goosen N., and Moolenaar G. F. (2008) Repair of UV damage in bacteria. DNA Repair 7, 353–379 [DOI] [PubMed] [Google Scholar]
- 18. Naegeli H., and Sugasawa K. (2011) The xeroderma pigmentosum pathway: decision tree analysis of DNA quality. DNA Repair 10, 673–683 [DOI] [PubMed] [Google Scholar]
- 19. Sancar A., and Rupp W. D. (1983) A novel repair enzyme–uvrabc excision nuclease of Escherichia coli cuts a DNA strand on both sides of the damaged region. Cell 33, 249–260 [DOI] [PubMed] [Google Scholar]
- 20. Svoboda D. L., Taylor J. S., Hearst J. E., and Sancar A. (1993) DNA-repair by eukaryotic nucleotide excision nuclease–removal of thymine dimer and psoralen monoadduct by HeLa cell-free extract and of thymine dimer by Xenopus laevis oocytes. J. Biol. Chem. 268, 1931–1936 [PubMed] [Google Scholar]
- 21. Huang J. C., Svoboda D. L., Reardon J. T., and Sancar A. (1992) Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5` and the 6th phosphodiester bond 3` to the photodimer. Proc. Natl. Acad. Sci. U.S.A. 89, 3664–3668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guzder S. N., Habraken Y., Sung P., Prakash L., and Prakash S. (1995) Reconstitution of yeast nucleotide excision-repair with purified Rad proteins, replication protein-A, and transcription factor TFIIH. J. Biol. Chem. 270, 12973–12976 [DOI] [PubMed] [Google Scholar]
- 23. Canturk F., Karaman M., Selby C. P., Kemp M. G., Kulaksiz-Erkmen G., Hu J., Li W., Lindsey-Boltz L. A., and Sancar A. (2016) Nucleotide excision repair by dual incisions in plants. Proc. Natl. Acad. Sci. U.S.A. 113, 4706–4710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sancar A., and Hearst J. E. (1993) Molecular matchmakers. Science 259, 1415–1420 [DOI] [PubMed] [Google Scholar]
- 25. Orren D. K., Selby C. P., Hearst J. E., and Sancar A. (1992) Post-incision steps of nucleotide excision repair in Escherichia coli--disassembly of the Uvrbc-DNA complex by helicase-II and DNA-polymerase-I. J. Biol. Chem. 267, 780–788 [PubMed] [Google Scholar]
- 26. Hopfield J. J. (1974) Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc. Natl. Acad. Sci. U.S.A. 71, 4135–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ninio J. (1975) Kinetic amplification of enzyme discrimination. Biochimie 57, 587–595 [DOI] [PubMed] [Google Scholar]
- 28. Brutlag D., and Kornberg A. (1972) Enzymatic synthesis of deoxyribonucleic acid. 36. A proofreading function for the 3` leads to 5` exonuclease activity in deoxyribonucleic acid polymerases. J. Biol. Chem. 247, 241–248 [PubMed] [Google Scholar]
- 29. Bar-Ziv R., Tlusty T., and Libchaber A. (2002) Protein-DNA computation by stochastic assembly cascade. Proc. Natl. Acad. Sci. U.S.A. 99, 11589–11592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Larson M. H., Zhou J., Kaplan C. D., Palangat M., Kornberg R. D., Landick R., and Block S. M. (2012) Trigger loop dynamics mediate the balance between the transcriptional fidelity and speed of RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 109, 6555–6560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Burgess S. M., and Guthrie C. (1993) A mechanism to enhance mRNA splicing fidelity: the RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell 73, 1377–1391 [DOI] [PubMed] [Google Scholar]
- 32. Hopfield J. J., Yamane T., Yue V., and Coutts S. M. (1976) Direct experimental evidence for kinetic proofreading in amino acylation of tRNAIle. Proc. Natl. Acad. Sci. U.S.A. 73, 1164–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thompson R. C., and Karim A. M. (1982) The accuracy of protein biosynthesis is limited by its speed: high fidelity selection by ribosomes of aminoacyl-tRNA ternary complexes containing GTP[γS]. Proc. Natl. Acad. Sci. U.S.A. 79, 4922–4926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cochella L., and Green R. (2005) An active role for tRNA in decoding beyond codon:anticodon pairing. Science 308, 1178–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Blanchard S. C., Gonzalez R. L., Kim H. D., Chu S., and Puglisi J. D. (2004) tRNA selection and kinetic proofreading in translation. Nat. Struct. Mol. Biol. 11, 1008–1014 [DOI] [PubMed] [Google Scholar]
- 36. Kurland C. G. (1992) Translational accuracy and the fitness of bacteria. Annu. Rev. Genet. 26, 29–50 [DOI] [PubMed] [Google Scholar]
- 37. Moore P. B. (2013) Ribosomal ambiguity made less ambiguous. Proc. Natl. Acad. Sci. U.S.A. 110, 9627–9628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McKeithan T. W. (1995) Kinetic proofreading in T-cell receptor signal transduction. Proc. Natl. Acad. Sci. U.S.A. 92, 5042–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. MacGlashan D., Jr. (2001) Signaling cascades: escape from kinetic proofreading. Proc. Natl. Acad. Sci. U.S.A. 98, 6989–6990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kirschner M. W. (1980) Implications of treadmilling for the stability and polarity of actin and tubulin polymers in vivo. J. Cell Biol. 86, 330–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lu Y., Wang W., and Kirschner M. W. (2015) Specificity of the anaphase-promoting complex: a single-molecule study. Science 348, 1248737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kesseler K. J., Kaufmann W. K., Reardon J. T., Elston T. C., and Sancar A. (2007) A mathematical model for human nucleotide excision repair: damage recognition by random order assembly and kinetic proofreading. J. Theor. Biol. 249, 361–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Reardon J. T., and Sancar A. (2004) Thermodynamic cooperativity and kinetic proofreading in DNA damage recognition and repair. Cell Cycle 3, 141–144 [PubMed] [Google Scholar]
- 44. Reardon J. T., and Sancar A. (2003) Recognition and repair of the cyclobutane thymine dimer, a major cause of skin cancers, by the human excision nuclease. Gene Dev. 17, 2539–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mu D., Hsu D. S., and Sancar A. (1996) Reaction mechanism of human DNA repair excision nuclease. J. Biol. Chem. 271, 8285–8294 [DOI] [PubMed] [Google Scholar]
- 46. Mu D., Wakasugi M., Hsu D. S., and Sancar A. (1997) Characterization of reaction intermediates of human excision repair nuclease. J. Biol. Chem. 272, 28971–28979 [DOI] [PubMed] [Google Scholar]
- 47. Wakasugi M., and Sancar A. (1998) Assembly, subunit composition, and footprint of human DNA repair excision nuclease. Proc. Natl. Acad. Sci. U.S.A. 95, 6669–6674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mao P., Wyrick J. J., Roberts S. A., and Smerdon M. J. (2017) UV-induced DNA damage and mutagenesis in chromatin. Photochem. Photobiol. 93, 216–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mao P., Smerdon M. J., Roberts S. A., and Wyrick J. J. (2016) Chromosomal landscape of UV damage formation and repair at single-nucleotide resolution. Proc. Natl. Acad. Sci. U.S.A. 113, 9057–9062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hanawalt P. C., and Spivak G. (2008) Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 9, 958–970 [DOI] [PubMed] [Google Scholar]
- 51. Selby C. P. (2017) Mfd protein and transcription-repair coupling in Escherichia coli. Photochem. Photobiol. 93, 280–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Selby C. P., and Sancar A. (1990) Transcription preferentially inhibits nucleotide excision repair of the template DNA strand in vitro. J. Biol. Chem. 265, 21330–21336 [PubMed] [Google Scholar]
- 53. Selby C. P., Drapkin R., Reinberg D., and Sancar A. (1997) RNA polymerase II stalled at a thymine dimer: footprint and effect on excision repair. Nucleic Acids Res. 25, 787–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lindsey-Boltz L. A., and Sancar A. (2007) RNA polymerase: the most specific damage recognition protein in cellular responses to DNA damage? Proc. Natl. Acad. Sci. U.S.A. 104, 13213–13214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tremeau-Bravard A., Riedl T., Egly J. M., and Dahmus M. E. (2004) Fate of RNA polymerase II stalled at a cisplatin lesion. J. Biol. Chem. 279, 7751–7759 [DOI] [PubMed] [Google Scholar]
- 56. Selby C. P., and Sancar A. (1993) Molecular mechanism of transcription-repair coupling. Science 260, 53–58 [DOI] [PubMed] [Google Scholar]
- 57. Roberts J., and Park J. S. (2004) Mfd, the bacterial transcription-repair coupling factor: translocation, repair and termination. Curr. Opin. Microbiol. 7, 120–125 [DOI] [PubMed] [Google Scholar]
- 58. Selby C. P., and Sancar A. (1997) Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 94, 11205–11209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Li W., Hu J., Adebali O., Adar S., Yang Y., Chiou Y. Y., and Sancar A. (2017) Human genome-wide repair map of DNA damage caused by the cigarette smoke carcinogen benzo[a]pyrene. Proc. Natl. Acad. Sci. U.S.A. 114, 6752–6757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Aravind L., Walker D. R., and Koonin E. V. (1999) Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 27, 1223–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Verhoeven E. E., van Kesteren M., Moolenaar G. F., Visse R., and Goosen N. (2000) Catalytic sites for 3` and 5` incision of Escherichia coli nucleotide excision repair are both located in UvrC. J. Biol. Chem. 275, 5120–5123 [DOI] [PubMed] [Google Scholar]
- 62. Lin J. J., and Sancar A. (1992) Active-site of (a)Bc excinuclease. 1. Evidence for 5` incision by Uvrc through a catalytic site involving Asp399, Asp438, Asp466, and His538 residues. J. Biol. Chem. 267, 17688–17692 [PubMed] [Google Scholar]
- 63. Husain I., Van Houten B., Thomas D. C., Abdel-Monem M., and Sancar A. (1985) Effect of DNA-polymerase-I and DNA helicase-II on the turnover rate of uvrabc excision nuclease. Proc. Natl. Acad. Sci. U.S.A. 82, 6774–6778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Adebali O., Chiou Y. Y., Hu J., Sancar A., and Selby C. P. (2017) Genome-wide transcription-coupled repair in Escherichia coli is mediated by the Mfd translocase. Proc. Natl. Acad. Sci. U.S.A. 114, E2116–E2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kemp M. G., Reardon J. T., Lindsey-Boltz L. A., and Sancar A. (2012) Mechanism of release and fate of excised oligonucleotides during nucleotide excision repair. J. Biol. Chem. 287, 22889–22899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hu J., Choi J. H., Gaddameedhi S., Kemp M. G., Reardon J. T., and Sancar A. (2013) Nucleotide excision repair in human cells fate of the excised oligonucleotide carrying DNA damage in vivo. J. Biol. Chem. 288, 20918–20926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sibghat-Ullah, Sancar A., and Hearst J. E. (1990) The repair patch of Escherichia coli (a) Bc excinuclease. Nucleic Acids Res. 18, 5051–5053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lehmann A. R. (2011) DNA polymerases and repair synthesis in NER in human cells. DNA Repair 10, 730–733 [DOI] [PubMed] [Google Scholar]
- 69. Reardon J. T., Thompson L. H., and Sancar A. (1997) Rodent UV-sensitive mutant cell lines in complementation groups 6–10 have normal general excision repair activity. Nucleic Acids Res. 25, 1015–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lindsey-Boltz L. A., Kemp M. G., Reardon J. T., DeRocco V., Iyer R. R., Modrich P., and Sancar A. (2014) Coupling of human DNA excision repair and the DNA damage checkpoint in a defined in vitro system. J. Biol. Chem. 289, 5074–5082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Moser J., Kool H., Giakzidis I., Caldecott K., Mullenders L. H., and Fousteri M. I. (2007) Sealing of chromosomal DNA nicks during nucleotide excision repair requires XRCC1 and DNA ligase IIIα in a cell-cycle-specific manner. Mol. Cell 27, 311–323 [DOI] [PubMed] [Google Scholar]
- 72. Adar S., Hu J., Lieb J. D., and Sancar A. (2016) Genome-wide kinetics of DNA excision repair in relation to chromatin state and mutagenesis. Proc. Natl. Acad. Sci. U.S.A. 113, E2124–E2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hu J., Adebali O., Adar S., and Sancar A. (2017) Dynamic maps of UV damage formation and repair for the human genome. Proc. Natl. Acad. Sci. U.S.A. 114, 6758–6763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hu J., and Adar S. (2017) The cartography of UV-induced DNA damage formation and DNA repair. Photochem. Photobiol. 93, 199–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Choi J. H., Gaddameedhi S., Kim S. Y., Hu J., Kemp M. G., and Sancar A. (2014) Highly specific and sensitive method for measuring nucleotide excision repair kinetics of ultraviolet photoproducts in human cells. Nucleic Acids Res. 42, e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Choi J. H., Kim S. Y., Kim S. K., Kemp M. G., and Sancar A. (2015) An integrated approach for analysis of the DNA damage response in mammalian cells: nucleotide excision repair, DNA damage checkpoint, and apoptosis. J. Biol. Chem. 290, 28812–28821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Prakash S., Johnson R. E., and Prakash L. (2005) Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 74, 317–353 [DOI] [PubMed] [Google Scholar]
- 78. Witkin E. M. (1966) Radiation-induced mutations and their repair. Science 152, 1345–1353 [DOI] [PubMed] [Google Scholar]
- 79. Selby C. P., Witkin E. M., and Sancar A. (1991) Escherichia coli Mfd mutant deficient in mutation frequency decline lacks strand-specific repair–in vitro complementation with purified coupling factor. Proc. Natl. Acad. Sci. U.S.A. 88, 11574–11578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lloréns-Rico V., Cano J., Kamminga T., Gil R., Latorre A., Chen W. H., Bork P., Glass J. I., Serrano L., and Lluch-Senar M. (2016) Bacterial antisense RNAs are mainly the product of transcriptional noise. Sci. Adv. 2, e1501363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Selby C. P., and Sancar A. (1991) Gene-specific and strand-specific repair in vitro–partial purification of a transcription-repair coupling factor. Proc. Natl. Acad. Sci. U.S.A. 88, 8232–8236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Fan J., Leroux-Coyau M., Savery N. J., and Strick T. R. (2016) Reconstruction of bacterial transcription-coupled repair at single-molecule resolution. Nature 536, 234–237 [DOI] [PubMed] [Google Scholar]
- 83. Chen H., Shiroguchi K., Ge H., and Xie X. S. (2015) Genome-wide study of mRNA degradation and transcript elongation in Escherichia coli. Mol. Syst. Biol. 11, 781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Raghavan R., Sloan D. B., and Ochman H. (2012) Antisense transcription is pervasive but rarely conserved in enteric bacteria. MBio 3, e00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Thomason M. K., and Storz G. (2010) Bacterial antisense RNAs: how many are there, and what are they doing? Annu. Rev. Genet. 44, 167–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ernst J., Kheradpour P., Mikkelsen T. S., Shoresh N., Ward L. D., Epstein C. B., Zhang X., Wang L., Issner R., Coyne M., Ku M., Durham T., Kellis M., and Bernstein B. E. (2011) Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473, 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.ENCODE Project Consortium (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Yip K. Y., Cheng C., Bhardwaj N., Brown J. B., Leng J., Kundaje A., Rozowsky J., Birney E., Bickel P., Snyder M., and Gerstein M. (2012) Classification of human genomic regions based on experimentally determined binding sites of more than 100 transcription-related factors. Genome Biol. 13, R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Tang M. S., Pao A., and Zhang X. S. (1994) Repair of benzo(a)pyrene diol epoxide- and UV-induced DNA damage in dihydrofolate reductase and adenine phosphoribosyltransferase genes of CHO cells. J. Biol. Chem. 269, 12749–12754 [PubMed] [Google Scholar]
- 90. Core L. J., Martins A. L., Danko C. G., Waters C. T., Siepel A., and Lis J. T. (2014) Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat. Genet. 46, 1311–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Core L. J., Waterfall J. J., and Lis J. T. (2008) Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Hara R., and Sancar A. (2003) Effect of damage type on stimulation of human excision nuclease by SWI/SNF chromatin remodeling factor. Mol. Cell. Biol. 23, 4121–4125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Sabarinathan R., Mularoni L., Deu-Pons J., Gonzalez-Perez A., and López-Bigas N. (2016) Nucleotide excision repair is impaired by binding of transcription factors to DNA. Nature 532, 264–267 [DOI] [PubMed] [Google Scholar]
- 94. Perera D., Poulos R. C., Shah A., Beck D., Pimanda J. E., and Wong J. W. (2016) Differential DNA repair underlies mutation hotspots at active promoters in cancer genomes. Nature 532, 259–263 [DOI] [PubMed] [Google Scholar]
- 95. Alexandrov L. B., Ju Y. S., Haase K., Van Loo P., Martincorena I., Nik-Zainal S., Totoki Y., Fujimoto A., Nakagawa H., Shibata T., Campbell P. J., Vineis P., Phillips D. H., and Stratton M. R. (2016) Mutational signatures associated with tobacco smoking in human cancer. Science 354, 618–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Nik-Zainal S., Kucab J. E., Morganella S., Glodzik D., Alexandrov L. B., Arlt V. M., Weninger A., Hollstein M., Stratton M. R., and Phillips D. H. (2015) The genome as a record of environmental exposure. Mutagenesis 30, 763–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Alexandrov L. B., Nik-Zainal S., Wedge D. C., Aparicio S. A., Behjati S., Biankin A. V., Bignell G. R., Bolli N., Borg A., Børresen-Dale A. L., Boyault S., Burkhardt B., Butler A. P., Caldas C., Davies H. R., et al. (2013) Signatures of mutational processes in human cancer. Nature 500, 415–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Katsumi S., Kobayashi N., Imoto K., Nakagawa A., Yamashina Y., Muramatsu T., Shirai T., Miyagawa S., Sugiura S., Hanaoka F., Matsunaga T., Nikaido O., and Mori T. (2001) In situ visualization of ultraviolet-light-induced DNA damage repair in locally irradiated human fibroblasts. J. Invest. Dermatol. 117, 1156–1161 [DOI] [PubMed] [Google Scholar]
- 99. Koch S. C., Simon N., Ebert C., and Carell T. (2016) Molecular mechanisms of xeroderma pigmentosum (XP) proteins. Q. Rev. Biophys. 49, e5. [DOI] [PubMed] [Google Scholar]
- 100. Uphoff S., and Sherratt D. J. (2017) Single-molecule analysis of bacterial DNA repair and mutagenesis. Annu. Rev. Biophys. 46, 411–432 [DOI] [PubMed] [Google Scholar]