Abstract
One central goal in molecular evolution is to pinpoint the mechanisms and evolutionary forces that cause an enzyme to change its substrate specificity; however, these processes remain largely unexplored. Using the glycolytic ADP-dependent kinases of archaea, including the orders Thermococcales, Methanosarcinales, and Methanococcales, as a model and employing an approach involving paleoenzymology, evolutionary statistics, and protein structural analysis, we could track changes in substrate specificity during ADP-dependent kinase evolution along with the structural determinants of these changes. To do so, we studied five key resurrected ancestral enzymes as well as their extant counterparts. We found that a major shift in function from a bifunctional ancestor that could phosphorylate either glucose or fructose 6-phosphate (fructose-6-P) as a substrate to a fructose 6-P-specific enzyme was started by a single amino acid substitution resulting in negative selection with a ground-state mode against glucose and a subsequent 1,600-fold change in specificity of the ancestral protein. This change rendered the residual phosphorylation of glucose a promiscuous and physiologically irrelevant activity, highlighting how promiscuity may be an evolutionary vestige of ancestral enzyme activities, which have been eliminated over time. We also could reconstruct the evolutionary history of substrate utilization by using an evolutionary model of discrete binary characters, indicating that substrate uses can be discretely lost or acquired during enzyme evolution. These findings exemplify how negative selection and subtle enzyme changes can lead to major evolutionary shifts in function, which can subsequently generate important adaptive advantages, for example, in improving glycolytic efficiency in Thermococcales.
Keywords: archaea, crystal structure, phosphofructokinase, protein evolution, substrate specificity, ADP-dependent kinase, ancestral enzymes, glucokinase
Introduction
Deciphering the biochemical and genetic mechanisms by which new protein functions appear is a central goal in molecular evolution (1). In enzyme families, functionally diverse members share a common mechanism of catalysis (2), but have structural determinants that dictate different specificities and therefore different functions in metabolic pathways. Many observations support the hypothesis that different functions or specificities evolve through either promiscuous activities or bifunctionality, although promiscuity has also been proposed as an evolutionary vestige of ancestral function (3). In this respect, it is important to distinguish between promiscuity and bifunctionality. The former results from a secondary catalytic activity that is not physiologically relevant, whereas bifunctionality refers to the simultaneous existence of two catalytic activities both of which have physiological relevance (4). On the other hand, the mechanism and evolutionary modes whereby a bifunctional enzyme for substrates “A” and “B” becomes specific for “A” have been poorly explored. In principle, the specialization for “A” can be either the result of a positive selection for “A” or a negative selection against “B.” Directed evolution experiments indicate that negative selection induces selectivity much more effectively (5) and two modes of substrate discrimination have been proposed: ground-state (loss of substrate affinity) and transition-state (loss of catalysis) and that such discrimination imposes an accuracy rate tradeoff (6). Furthermore, experimental microbial evolution has shown that new functionalities that enable adaptation often involve a small number of genetic alterations, in which a single change causing a large effect on the phenotype or fitness tends to appear first, followed by subsequent mutations that generate small enhancing effects (7). Nonetheless, a question that remains open is whether this mechanism could also act at the level of substrate specificity in enzymes.
Traditional biochemical studies regarding functional specificity in extant homologous enzymes often focus on the identification of key amino acid changes that would account for different functions in different families (8). However, during evolution most changes are expected to have a small or null effect on enzyme specificity, making the identification of truly critical residues a difficult problem to solve. This difficulty can be overcome if one considers the evolutionary history of extant members by employing ancestral sequence reconstruction (ASR)5 (8). The availability of current methodologies for the reconstruction of ancestral sequences with low uncertainty, and specifically those of key ancestors responsible for the appearance of new functions, enable us to explore the mechanisms and relevant changes that gave rise to the functions of current proteins (9).
In this work we study ADP-dependent kinases, which are key enzymes in the glycolytic pathway of Euryarchaeotas in archaea. The members of this family have only two types of catalytic activity; as phosphofructokinases (PFK) and glucokinases (GK), both of which participate in fundamental steps in the glycolytic pathway. In archaea, these enzymes have been described mainly in the orders Thermococcales, Methanosarcinales, and Methanococcales as well as in higher eukaryotes (10). According to the Kyoto Encyclopedia of Genes and Genomes (62),6 the genomes of Thermococcales have two copies of these genes (glkA and pfkC), which have been characterized experimentally as glucokinases (ADP-GK), highly specific for glucose (11), and phosphofructokinases (ADP-PFK), highly specific for fructose-6-P (12). Methanosarcinales genomes also have two genes pfkC and glkA, coding ADP-K1 and ADP-K2, respectively, but thus far there are no reports concerning the specificities of these enzymes. By contrast, the Methanococcales genomes have only one gene (pfkC) and the corresponding enzyme has been characterized to be bifunctional, suggesting that it satisfies both functions in vivo (13, 14). These bifunctional enzymes have been described to be phylogenetically closest to the specific PFKs from Thermococcales (15) (Fig. 1).
Figure 1.

Dendrogram of the ADP-dependent kinases family and Euryarcheotas organisms. A, consensus phylogenetic tree of the ADP-dependent sugar kinases family obtained by Bayesian inference using protein sequences. Nodes of resurrected ancestral enzymes are indicated as colored circles: ancGK/PFK in black, ancMMT in green, ancPFK-MT in pink, ancPFK-M in purple, and ancPFK-T in red. The following current enzymes are shown in their respective groups: glucose-specific from T. litoralis (TlGK), the specific by fructose-6-P from P. horikoshii (PhPFK), and the bifunctional from M. jannaschii (MjPFK/GK). B, species tree adapted from TimeTree (19) and Petitjean et al. (18), dotted lines show possible horizontal gene transfer events.
Several structures of specific ADP-dependent kinases have been reported. Among them there are the specific ADP-GK from Thermococcus litoralis, Pyrococcus horikoshii, Pyrococcus furiosus, and mouse (PDB codes 1GC5, 1L2L, 1UA4, and 5CK7, respectively) and the specific ADP-PFK from P. horikoshii (PDB code 1U2X).
Inspection of the active site of the ADP-PFK from P. horikoshii highlights the importance of Ala-71 in substrate specificity, since replacement of this residue for glutamic acid enables the catalysis with Glc as substrate (16). However, to date it is not known if the ancestral trait was bifunctionality or fructose-6-P-specific. Moreover, whether the change from bifunctional to specific (or the opposite) was acquired through positive or negative selection and whether it involved a ground-state or transition-state mechanism are still unanswered questions.
We studied five key ancestral enzymes of increasing historical age from the ADP-dependent kinase family to compare their activities with contemporary enzymes. This allows us to explore the mechanism underlying the change of protein function during natural evolution, illuminating the evolutionary basis for substrate recognition. Also, we were able to reconstruct the evolutionary history of the substrate-binding affinity using a simple evolutionary model of discrete binary characters and also to identify the large-effect mutation that is the structural determinant responsible for the emergence of fructose-6-P specificity in the ADP-PFKs enzymes from Thermococcales.
Results
Phylogeny of ADP-dependent enzymes and ancestral sequence reconstruction
We reconstructed the phylogeny of the ADP-dependent kinase family and performed ancestral protein resurrection to characterize the substrate specificity of key ancestors. The phylogeny of the ADP-dependent kinases was inferred using Bayesian inference on the basis of amino acid and nucleotide sequences (Fig. 1 and supplemental Fig. S1). Furthermore, a maximum likelihood phylogeny based on amino acid sequences was also constructed for comparison (supplemental Fig. S2). All the methodologies employed resulted in the same general topology for the phylogeny (Fig. 1A), which is consistent with previous studies (14, 15, 17). Six groups can be identified: PFK from Thermococcales, GKs from Thermococcales, and GKs from Eukaryotas, bifunctional enzymes PFK/GK from Methanococcales as well as homologous proteins from the order Methanosarcinales here denominated ADP-K1 and ADP-K2. All of these groups, as well as the key nodes employed to perform the inference of ancestral sequences, present robust statistical support (supplemental Figs. S1 and S2). Five ancestral enzymes from the family were selected for ancestral inference and reconstruction. Each of them is located at a major branch point and positioned progressively deeper within the phylogeny; these ancestors are indicated in Fig. 1A and their corresponding sequences are shown in supplemental Fig. S3. AncMT was considered particularly interesting because it is the ancestor of both the bifunctional enzymes from Methanococcales and the specific fructose-6-P enzymes from Thermococcales. Characterization of this ancestor should prove whether bifunctionality or fructose-6-P specificity is the ancestral trait.
According to a variety of protein markers (18) and the tree time published for archaea (19) the divergence of Methanococcales from Thermococcales occurred about 3594 million years ago, whereas that of Methanococcales from Methanosarcinales occurred later, about 3468 million years ago (Fig. 1B). This inconsistency between the tree topology of the ADP-dependent enzymes and the phylogeny described for archaeal organisms can be explained by horizontal gene transfer (HGT) events. The appearance of the Thermococcales PFKs close to the bifunctional Methanococcales enzymes is the result of an HGT event that occurred relatively recently (despite the fact that Thermococcales organisms were the first to diverge). Such an event would explain the phylogenetic closeness between theses enzymes and allow us to hypothesize that ancMT was a bifunctional enzyme that gave rise to the fructose-6-P-specific enzymes present today in Thermococcales. Another HGT event could have occurred from Thermococcales toward Methanosarcinales explaining the presence of two genes in the latter group. However, in this case it is difficult to be certain given the lack of biochemical information concerning enzymes from the Methanosarcinales. Considering these likely HGT events it can be hypothesized that ancestors ancGK/PFK and ancMMT existed about 3594 and 3468 million years ago, respectively (19).
Robustness analysis of the five reconstructed ancestral sequences showed that the probability distribution for ancGK/PFK resulted in 54% of its residues with a posterior probability greater than 0.5. By comparison, the analogous posterior probability values for ancMMT, ancMT, ancT, and ancM were 86, 95, 96, and 95%, respectively (supplemental Fig. S4). According, with the objective of elucidating the evolutionary history of substrate usage, which led to the existence of extant bifunctional and fructose-6-P-specific enzymes, we also analyzed the robustness of residue positions that reside within or near the active site. All positions within 12 Å of the sugar were examined (supplemental Table S1). Indeed, 12 Å is a generously set threshold because it incorporates all positions that make up the active site including catalytic residues and their second-shell neighbors. The analysis shows that residues of the first-shell (out to 5 Å from the substrate) and second-shell (from 5 to 12 Å) have a high robustness in all the reconstructed sequences except for ancGK/PFK. The latter was analyzed using three different methodologies; maximum likelihood using protein sequences (20) and a Bayesian approach employing either protein or gene sequences (21), revealing that the reconstructed first-shell residues are not dependent on the method employed (supplemental Table S2). Although this ancestor shows a relatively weak robustness, we decided to analyze it because it represents the deepest node in the phylogeny and might give light about the evolutionary trajectory if it is considered along with the results obtained for the other ancestors, which have very robust statistics support. In the case of ancMMT, ancMT, ancT, and ancM, high robustness was observed for both the first and second shell residues, supporting the notion that the kinetic characteristics of the enzymes are not affected by uncertainties in the reconstructed sequences.
Kinetic characterization of extant and ancestral enzymes
In the order Thermococcales the extant GK and PFK ADP-dependent kinases are highly specific for their substrates (Table 1). We have evaluated the GK from T. litoralis (TlGK), which has no detectable activity with fructose-6-P, and the PFK from P. horikoshii (PhPFK), which has no activity with glucose. In addition, TlGK and PhPFK were not significantly inhibited by fructose-6-P or glucose, respectively, even at concentrations of 50 mm, which emphasizes that these enzymes are neither able to bind nor perform catalysis on the inappropriate substrate. On the other hand, bifunctionality is present in both the Methanococcus jannaschii (MjPFK/GK) (Table 1) (14) and Methanococcus maripaludis enzymes (13). To gain insight into changes in substrate specificity during the evolutionary history of this family we characterized the enzyme kinetics of the resurrected ancestral enzymes employing both potential substrates (Table 1). Because ancT saturation with glucose was not reached, only rough estimates for kcat and Km were obtained (see “Experimental procedures”), to have at least an approximate order of magnitude for these values. The results show that the ancestral family trait is bifunctionality, whereas the trait of fructose-6-P specificity in PFK from Thermococcales is the evolutionary novelty. Fructose-6-P specificity emerged in the transitions between ancMT and ancT (Fig. 2A), whereas bifunctionality is retained in ancM (the last common ancestor of Methanococcales) (Fig. 2B). It is important to note that in the ancMT to ancT transition the enzyme suffers a large loss in glucose affinity (Km 31 mm to >1 m) without a corresponding change in catalysis (kcat 4 to ∼15 s−1) (Fig. 2A). Concomitantly, there appears an increase in fructose-6-P affinity and catalysis. This analysis leads to two possibilities: (i) a mixture of two mechanisms acting in series with negative selection via a ground-state discrimination mode (loss of affinity for glucose) and then a positive selection increasing both the catalytic rate and affinity for fructose-6-P or (ii) the loss of affinity for glucose is a side-product of an increase in catalytic efficiency for fructose-6-P occurring only by positive selection. Another important result is that even though ancT is specific for fructose-6-P, it is also able to phosphorylate glucose at very high concentrations (Km> 1 m). This means that the GK activity has become a promiscuous activity and a vestige of its evolutionary past.
Table 1.
Kinetic constants of extant and ancestral enzymes
The mean ± S.E. were calculated from three independent measurements.
| Enzyme | Fructose-6-P |
Glucose |
|||||
|---|---|---|---|---|---|---|---|
| Km | kcat | kcat/Km | Km | kcat | Ki | kcat/Km | |
| μm | s−1 | m−1s−1 | μm | s−1 | mm | m−1s−1 | |
| TlGK | NDa | ND | ND | 0.44 ± 0.09 | 42 ± 6 | 14 ± 2 | 9.5 × 104 |
| PhPFK | 56 ± 4 | 21 ± 1 | 3.9 × 105 | ND | ND | ND | |
| MjPFK/GK | 6.3 ± 0.2 | 2.70 ± 0.06 | 4.2 × 105 | 1.1 ± 0.3 | 5.0 ± 0.7 | 97 ± 23 | 4.5 × 103 |
| ancGK/PFK | 119 ± 8 | 0.33 ± 0.02 | 2.8 × 103 | 4 ± 1 | 0.24 ± 0.04 | 6.0 × 101 | |
| ancMMT | 540 ± 20 | 0.53 ± 0.05 | 9.7 × 102 | 12 ± 1 | 0.35 ± 0.07 | 3.3 × 101 | |
| ancMT | 2900 ± 600 | 1.0 ± 0.2 | 3.5 × 102 | 31 ± 5 | 4.1 ± 0.2 | 1290 ± 75 | 1.3 × 102 |
| ancT | 60 ± 2 | 43 ± 3 | 7.2 × 105 | >1 × 103b | ∼15b | 1.1 × 101c | |
| ancM | 370 ± 60 | 8.5 ± 0.6 | 2.3 × 104 | 5.7 ± 0.4 | 15 ± 1 | 121 ± 4 | 2.6 × 103 |
| ancMT-E72A | 2800 ± 600 | 0.38 ± 0.02 | 1.4 × 102 | >1 × 103b | ∼ 0.3b | 8.1 × 10−2c | |
a ND, no activity detected.
b Estimated from double-reciprocal plot.
c Determined from initial slope of saturation curve.
Figure 2.
Trace of the kinetic parameters of PFKs specificity en route to Thermococcales (A) and Methanococcales (B) using glucose or fructose-6-P as substrates. Divergence times were estimated by the RelTime method implemented in Mega7, using the speciation events shown in Fig. 1B (ancGK/PFK 3.59 Ga and ancMMT 3.47 Ga) as calibration points. The estimated times were: ancMT 1.98, ancM 1.51, and ancT 0.93 Ga. Evolution of Km and kcat shows that the main change toward fructose-6-P specificity in Thermococcales occurred in the transition between ancestors ancMT (1.98 Ga) and ancT (0.93 Ga), and was mainly caused by a loss in glucose affinity together with a concomitant increase in fructose-6-P affinity, leading to a 6 × 104-fold increment in fructose-6-P specificity.
Continuing backwards in the evolutionary history of the use of glucose as a substrate it becomes clear that the affinity for glucose dates back to ancGK/PFK. As we mentioned before, this ancestor may raise some concerns about its reconstruction robustness. However, the kinetic parameters obtained for this ancestor fit perfectly well with the evolutive trajectory described by the other ancestors and then we included it in the analysis. From this ancestor onwards the Km for glucose increased up to ancMT, as did that for fructose-6-P. However, this decrease in affinity is accompanied by an increase in kcat from ancGK/PFK to ancMT for both substrates (Fig. 2). On the other hand, during the evolutionary trajectory from ancGK/PFK to ancMT the efficiency constant (kcat/Km) shows a slight decrease for fructose-6-P, whereas that for glucose decreases slightly between ancGK/PFK and ancMMT and then increases from ancMMT to ancMT, achieving approximately the same efficiency as for fructose-6-P (Fig. 2). However, after the dramatic loss in glucose affinity observed during the ancMT to ancT transition (Fig. 2A), the kinetic parameters for fructose-6-P, kcat, and kcat/Km increased, reaching the same order of magnitude as those reported for the current PFK enzyme from P. horikoshii (PhPFK) (Fig. 2A).
Interestingly, the transition from ancMT to ancT correlates with the possible event of horizontal gene transfer from Methanococcales to the Thermococcales organism. This HGT event would have provoked a GK redundancy in the ancestor of Thermococcales, giving freedom for ancMT to specialize in fructose-6-P catalysis. On the other hand, during the evolutive pathway from ancMT to ancM and then to extant Methanococcales enzymes (Fig. 2B), bifunctionality was maintained. The kcat values of ancM and MjPFK/GK enzymes for both substrates are very similar, whereas the specificity constant (kcat/Km) of the ancestor in respect to the extant enzyme differ only by 18 or 2 times to fructose-6-P or glucose, respectively (Fig. 2B).
Structural bases for loss of glucose affinity
We determined the crystal structure of ancMT to shed light on the structural determinants involved in the dramatic loss of glucose affinity, which occurred en route to ancT. The ancestor was crystallized and the structure was determined to a resolution of 2.6 Å. The data collection and refinement statistics are given in Table 2. The structure shows that, like contemporary ADP-dependent enzymes, the ancestor is a monomer with a ribokinase-like fold (22). The overall structure shows the same organization seen in extant ADP-dependent kinases; the large domain has a Rossmann-fold (α/β/α) architecture with a central 10-stranded β-sheet surrounded by 12 α-helices and three 310 helices and the small domain consists of seven β-strands and four α-helices. The asymmetric unit contains two monomers, chains A and B, which have open and semiclosed conformations, respectively (Fig. 3A). These two conformations probably imply that the kinetically relevant ligand-induced conformational changes reported for other members of the family are also applicable here (17, 23). Although, ancMT crystallized in the presence of fructose-6-P and the product AMP, electron density was observed only for the latter (Fig. 3C). In this case, the phosphate group of AMP appears in two different positions, perhaps due to the lack of interactions resulting from the absence of the β-phosphate of ADP.
Table 2.
Data collection and refinement statistics for ancMT and E72A mutant
Statistics for the highest resolution shell are shown in parentheses.
| ancMT | E72A | |
|---|---|---|
| Data collection | ||
| X-ray source | DLS I04-1 | DLS I04-1 |
| Detector | PILATUS 2 m | PILATUS 2 m |
| Wavelength (Å) | 0.92 | 0.92 |
| Resolution Range (Å) | 53.42 − 2.58 (2.69 − 2.58) | 59.27 − 2.61 (2.72 − 2.61) |
| Space group | P41212 | P41212 |
| Cell dimensions (Å) a, b, c | 132.35, 132.35, 116.77 | 132.53, 132.53, 117.44 |
| I/σ(I) | 14.8 (2.1) | 11.8 (2.2) |
| Rmerge | 0.073 (0.853) | 0.122 (0.937) |
| CC1/2 | 0.998 (0.720) | 0.995 (0.747) |
| Rpim | 0.033 (0.398) | 0.044 (0.360) |
| Completeness (%) | 99.5 (99.2) | 97.7 (97.4) |
| Multiplicity | 5.8 (5.4) | 8.5 (7.6) |
| Wilson B factor | 59.46 | 54.81 |
| Reflections | 190,416 (21,462) | 271,462 (28,619) |
| Unique reflections | 33,041 (3,943) | 31,763 (3,090) |
| Refinement | ||
| Rwork/RFree | 0.2144 / 0.2568 | 0.2234 / 0.2576 |
| Number of atoms | ||
| Protein | 6838 | 6918 |
| Ligands | 74 | 53 |
| B-factors | ||
| Protein | 64.81 | 65.10 |
| Ligands | 69.23 | 65.71 |
| Water | 58.66 | 56.19 |
| Coordinate error (ML based) (Å) | 0.41 | 0.33 |
| Phase error (°) | 30.27 | 26.40 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.007 | 0.002 |
| Bond angles (°) | 1.009 | 0.56 |
| Ramachandran plot | ||
| Favored (%) | 97.77 | 97.88 |
| Allowed (%) | 2.12 | 1.90 |
| Outliers (%) | 0.11 | 0.22 |
| All atom clashscore | 12.07 | 6.22 |
| PDB code | 5K27 | 5KKG |
Figure 3.
Crystallographic structure of ancMT. A, open and semi-closed conformation of the ancMT structure, the open conformation corresponds to chain A (in red) and the semi-closed conformation to chain B (in blue). The r.m.s. deviation values for the whole protein and for the small domain are 1.2 and 2.5 Å, respectively. B, ancMT colored according to secondary structure: α-helices in red, β-sheet strands in yellow, and loops in green. Surface of the small domain is colored in gray and that of the large domain in white. C, AMP-binding site, the phosphate group of AMP appears in two positions due to conformational heterogeneity. Water molecules are labeled as w and the mFo − DFc omit map is contoured at 3σ for AMP. Yellow dotted lines indicate h-bonds calculated with PyMOL.
To identify the key residues responsible for sugar binding we modeled the ancestors ancMT, ancT, and ancM in the closed conformation in complex with Mg-ADP, using glucokinase from Pyrococcus furiosus (PDB code 1UA4) (23) and the crystal structure of ancMT reported here as templates. Docking assays with fructose-6-P and glucose were performed, which were subjected to an energy minimization protocol to allow for the proper accommodation of the substrates (Fig. 4). Minimized structures show that the ancMT and ancM ancestors bound both substrates practically identically. In ancT, fructose-6-P is stabilized in a manner similar to that observed for the previous two ancestors involving several hydrogen bonds through its hydroxyl groups and a salt bridge between the phosphate moiety and Arg-198. However, the binding of glucose to ancT is stabilized by only three hydrogen bonds mediated by its hydroxyl groups, compared with interactions involving all such groups in the ancMT and ancM ancestors. The almost null affinity of ancT for glucose (Km > 1 m) can be explained by the absence of these interactions, which is due to the presence of residue Ala-76 instead of a glutamic acid (Glu-72 in ancMT and Glu-79 in ancM). This glutamic acid receives a hydrogen bond from the C1 hydroxyl of glucose but does not interact directly with fructose-6-P. It may also play a second role by stabilizing Arg-192 (in ancMT) in an appropriate conformation for its interaction with glucose. This is expected to be unnecessary in the case of fructose-6-P because of the formal charge on the phosphate moiety (Fig. 4). It is noteworthy that this single change is the main conserved difference found between the sugar-binding sites of bifunctional and specific ancestors. Moreover, reconstruction of ancestral proteins using gene sequences shows that only one base change in the second position of the codon is required to mutate from glutamic acid (GAA and GAG) to alanine (GCC, GCT, GCA, and GCG) (supplemental Table S3).
Figure 4.
Molecular modeling of fructose-6-P and glucose in the active site of the ancMT, ancM, and ancT ancestors. Oxygen atoms are colored in red, carbons in green, nitrogens in blue, and polar hydrogens are showed in white. The only change found at the active site of these ancestors (E to A) is highlighted in bold. Yellow dotted lines indicate h-bonds calculated with PyMOL.
To evaluate the dynamics of protein ligand interactions, we performed a molecular dynamics simulation. The ancMT ancestor was simulated in complex with each sugar substrate and Mg-ADP to analyze the Van der Waals plus electrostatic interactions (nonbonding interaction energy) between active site residues and fructose-6-P or glucose. Molecular dynamics trajectories show that the sugar-binding site of ancMT displays mainly attractive interactions with fructose-6-P where Lys-159 and Arg-192 display the strongest interactions and that with Glu-72 the only repulsive one (Fig. 5). In stark contrast, the latter represents one of the strongest attractive interaction energies when glucose is the substrate (Fig. 5), whereas Arg-192 has a significant repulsive contribution. Although Arg-192 in docking assays was shown to be able to interact with glucose by a hydrogen bond, the molecular dynamics showed that over time the interaction is mostly repulsive. These results point to the fact that residue Glu-72 is essential for glucose, but not for fructose-6-P, binding. Therefore, it seems reasonable to assume that the E72A mutation alone is responsible for the transition from bifunctionality to fructose-6-P specificity, resulting in an evolutionary novelty in the ADP-dependent kinase family. Although this replacement has no major effect on the protein structure or on the ligand-binding site itself, it leads to an alteration of the hydrogen-bonding network that determines ligand binding and, as a consequence, to the overall energy landscape of these interactions. It is important to note that these results are based on theoretical homology modeling, docking, and classic molecular dynamics, so experimental evidence should be added to support these results.
Figure 5.
Nonbonding energy between ancMT and β-d-fructose-6-P or β-d-glucose calculated from molecular dynamic simulations. Simulations of 30 ns were performed for the Mg-ADP complex. The average nonbonding energy (Van der Waals plus electrostatic interactions) for all residues with at least one atom within 5 Å from the substrate was calculated. Only residues with over 1 kcal/mol are shown. The averaged interactions with positive energy values are shown in red and negative values are shown in black. Both simulations were performed with the force-field CHARMM 27 and NAMD 2.8 software. Error bars represent S.D. from 1500 frames. Residue conservation in sequence alignment of current enzymes (bifunctional and fructose-6-P-specific enzymes from Methanococcales and Thermococcales, respectively) is shown as sequence logo.
Recreating the evolutionary pathway toward fructose-6-P specificity
Because Glu-72 has been pointed out to be the main factor responsible for glucose binding to the ancMT ancestral enzyme, it was a logical next step to generate a mutant (E72A) of ancMT, and to determine its structure (Table 2) and kinetic properties. The structure of the mutant protein, crystallized in the presence of fructose-6-P and AMP, presents no significant structural modifications beyond the mutation itself, presenting an r.m.s. deviation on Cα atoms of 0.29 Å when compared with the wild type (supplemental Fig. S5). As in the case of the wild-type enzyme, the structure of the E72A mutant also shows an open and a semiclosed conformation for the two molecules in the asymmetric unit. Kinetic characterization of the E72A mutant shows no significant changes with respect to the kinetic parameters for fructose-6-P. Mainly, the catalytic efficiency decreases almost 2.5 times, implying that the increase in selectivity imposes a slight accuracy rate tradeoff. However, although the mutation did not affect significantly the kinetic parameters for fructose-6-P (Table 1) the mutation leads to a huge loss of glucokinase activity. Nonetheless, enzyme activity can be observed at very high glucose concentrations, exhibiting a behavior similar to that displayed by the ancT ancestor (Fig. 6). The linear portion of the initial velocity curve versus glucose concentration for the E72A mutant allowed us to determine a catalytic efficiency (kcat/Km) of 8.1 × 10−2 m−1 s−1 (see “Experimental procedures”), a value that is 1,600 times lower than that observed for the wild-type ancMT (1.3 × 102 m−1 s−1). A rough estimation of the kcat and glucose Km values from a double-reciprocal plot, showed a minor change in kcat, which dropped from 4.1 to ∼0.3 s−1, whereas the Km increased from 31 mm to over 1 m, showing that the loss of glucokinase activity is mainly due to a dramatic loss in glucose affinity caused by the E72A mutation, in agreement with the structural analysis described above (Figs. 4 and 5). However, the kcat of the E72A mutant is still lower than that of ancT indicating that additional changes en route from ancMT to ancT must have led to an improved catalytic rate. Taken together, the data suggest a mixture of negative and positive selection mechanisms in which the two effects are uncoupled. Feasibly, glucose affinity loss (negative selection with ground-state discrimination) occurred prior to the increase in affinity for fructose-6-P and the rate of catalysis in general (positive selection), which discards the loss of affinity for glucose as a side product of the increase in catalytic efficiency toward fructose-6-P during the transition from ancMT to ancT.
Figure 6.

Glucose saturation curves for ancMT and its E72A mutant. Saturation curve for ancMT was fitted to the substrate inhibition equation. For the E72A mutant saturation was not observed even at glucose concentrations higher than 600 mm. Inset, activity of the E72A mutant with glucose as substrate in a different velocity scale. The activity shows a linear dependence on glucose concentration and the kcat/Km value was obtained from the slope, considering that glucose concentrations are below the Km value. Error bars represent S.E. from three independent measurements.
Evolutionary history of enzyme substrate affinity reconstructed as a discrete trait
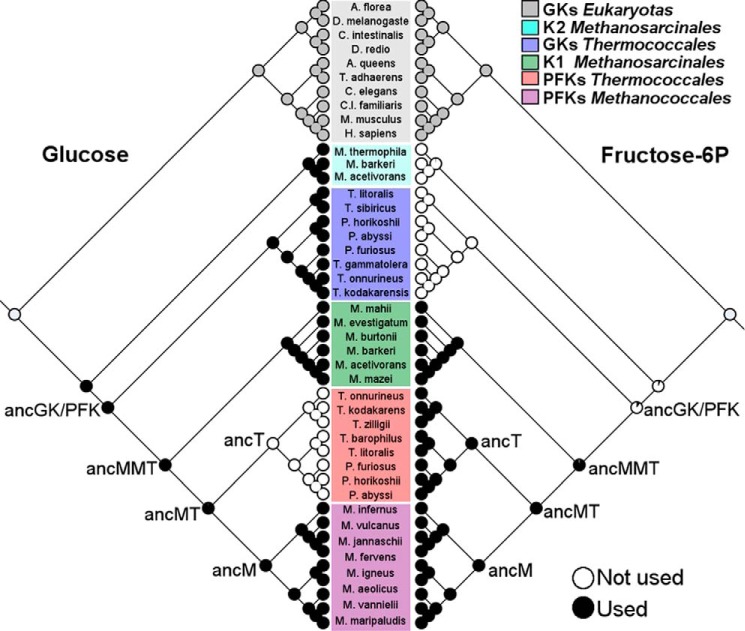
Because fructose-6-P specificity is caused by a single mutation, the evolutionary history of substrate affinity could be reconstructed using a simple evolutionary model such as the Markov k-state 1 parameter model (Mk1) of discrete binary characters (24). We performed the analysis using this model as implemented in the Mesquite software employing the maximum likelihood approach. We used the consensus tree obtained from the Bayesian phylogeny reconstruction based on protein sequences (Fig. 1A) together with the binary character defined as the ability to bind (or not) either substrate, at concentrations considered physiological (<50 mm). Specific GK and PFK activities of the Thermococcales group were assigned based on extant experimentally characterized enzymes, whereas Methanococcales enzymes were assigned as bifunctional according to that reported in the literature. In the Methanosarcinales group, there is no experimentally characterized enzyme to date. However, protein sequence alignment shows that the active site residues of the bifunctional PFK/GKs from Methanococcales are also conserved in ADP-K1s from Methanosarcinales, whereas those of the GKs from Thermococcales are conserved in ADP-K2s, suggesting that the former should be classified as bifunctional and the latter glucose specific. The data obtained from this analysis (Fig. 7) showed that the ancMT ancestor has a probability of 98.9% to bind glucose and 99.9% to bind fructose-6-P, in total agreement with the experimental bifunctional trait obtained from the kinetic characterization of this ancestor. This analysis also perfectly reconstructs the use of substrates by other ancestors; ancT (Fru-6-P specific), ancM (bifunctional), and ancMMT (bifunctional). However, ancGK/PFK showed a probability of 99.9% to bind glucose and a probability of only 10% for binding of fructose-6-P. In this case, the discrete binary character model presents a discrepancy with respect to the experimental characterization, but it should be borne in mind that this ancestor also presents the lowest posterior probabilities in the ASR procedure. To analyze the robustness of the reconstruction based on the maximum likelihood of discrete characters, we also employed Bayesian inference with BayesTrait version 2.0 (25) using one rate of change (q10 = q01). This analysis showed essentially the same results as those obtained by maximum likelihood, with the only difference being the value of the posterior probability of ancGK/PFK for fructose-6-P, whose mean value was 0.44 (supplemental Table S4). These analyses show the uncertainty in the inference of the use of fructose-6-P in this ancestor, which can be attributed to the node-depth. In any case, the analysis of discrete traits is in full agreement with the robust posterior probabilities obtained for the reconstruction of the ancestor using ASR and with the experimental data obtained for the characterized ancestors.
Figure 7.
Maximum likelihood reconstruction of substrate utilization as discrete binary characters. The left tree of the figure shows the results for the use of glucose, and on the right those corresponding to the use of fructose-6-P. The circles on leaf nodes and on ancestors represent a pie chart where black is the probability of use of the particular substrate (glucose or fructose-6-P) and white the probability of it not being used. Gray circles indicate out-group sequences used in the analysis.
Discussion
Directed evolution experiments performed with enzymes presenting promiscuous activities have provided important hints regarding evolutionary, structural, and mechanistic relationships within enzyme superfamilies (3). However, changes in function along natural lineages employing ASR have been less thoroughly explored. Here we present such a study in which we focus on enzyme substrate specificity in a family of key enzymes involved in glycolytic metabolism in archaea.
The ASR field is still developing and there is still no overall agreement about optimal reconstruction methods. Sometimes many sequence positions have alternative predictions, all with relatively high probabilities, in which case the posterior probability difference between the first and the second most likely amino acid at that position can be considered a measure of robustness. Alternatively, a robust reconstruction may be considered to be one that was independent of the method used. Regarding the sequence reconstructions presented in this study, ancMMT, ancM, and mainly ancMT and ancT showed a high level of robustness (supplemental Table S1) providing confidence that the experimental results obtained for them represent a genuine description of these ancestors. On the other hand, ancGK/PFK although showing robust results for residues that form the first shell of contacts to the sugar substrate showed uncertainties mainly within the second shell (supplemental Table S2), which may be relevant for enzymatic activity (26). Nevertheless, experimental results on ancGK/PFK are consistent with both the other ancestors and extant enzymes.
By characterizing ancestral enzymes we have been able to show that the fructose-6-P-specific enzymes (PFKs) found in Thermococcales originated from a PFK/GK bifunctional ancestor. Hence, the bifunctional activity of enzymes from the extant Methanococcales represents the preservation of this ancestral trait to the present day, whereas the evolutionary novelty is the appearance of fructose-6-P specificity that emerges during the transition from ancMT to ancT. The resolution of the crystallographic structure of ancMT together with molecular modeling and molecular dynamics of the ancestral proteins showed that a single mutation (E72A) at the active site of ancMT is the principal continuant responsible for the loss in glucose affinity. It is evident that discrete mutations, in this case just one, can cause large changes in enzyme function, which may affect the fitness of an organism and thus provide certain metabolic advantages in particular ecological environments. A similar situation, where a shift in function was driven primarily by only two amino acid changes, was found in the study by Harms et al. (27) on ligand specificity in the steroid receptors. Furthermore, Heilbron et al. (28) using a hypermutator strain of the bacterium Pseudomonas aeruginosa showed that rare mutations of large effect have a strong influence on fitness changes.
The topology of the tree describing ADP-dependent kinase evolution, and particularly regarding why PFKs and GKs from Thermococcales are not phylogenetically close, can be better understood looking at the change in substrate specificity observed between the ancMT and ancT ancestors. It has been previously suggested (15) that HGT of an ADP-dependent gene from a Methanococcales to a Thermococcales occurred at some point during their evolutionary history. Redundancy of ancMT GK activity in Thermococcales (which already have a glkA gene) would have given the required freedom for a bifunctional gene to specialize toward Fru-6-P specificity, presumably starting the mutational pathway via the E72A mutation. This would explain, on the one hand, the presence of specific PFKs in Thermococcales and on the other, the distant phylogenetic relationship of these genes with the GKs from the same group. Nonetheless, there is no direct evidence of HGT from Methanococcales to Thermococcales, either by organization of neighboring genes or codon usage, which may be due to the long time elapsed because these events occurred. However, there is evidence that HGT has occurred on other occasions (15) within the ADP-dependent kinase family; the gene of ADP-K2 from Methanothrix thermophile (Methanosarcinal) has a large difference in codon usage compared with its own genome, but presents a great similarity with that of GKs from Thermococcales. Furthermore, evidence shows the importance and high rate of HGT events occurring during the evolution of prokaryotes in general (29).
The kinetic parameters of the resurrected enzymes are consistent with those for extant enzymes, showing an increase in catalytic efficiency on evolving toward the current forms. This is in overall agreement with other studies of ancestral enzyme reconstruction (30), although there are examples in which ancestral enzymes show greater efficiency than their extant counterparts (31). Both cases agree with current theories for the evolution of enzyme catalysis (32) and recently reviewed by Newton et al. (33), which propose that changes in the kinetic parameters depend on the function and metabolic context in which the enzyme works. Considering the above, ADP-dependent kinases from Methanococcales are bifunctional enzymes with low catalytic efficiencies, in accordance with the fact that these organisms obtain their energy mainly from methanogenesis, whereas glycolysis/gluconeogenesis participates in glycogen metabolism, which could be an environmental advantage when carbon sources are scarce (34). On the other hand, in Thermococcales, glycolysis would be a major source of energy, in accordance with the presence of ADP-dependent specific enzymes with higher catalytic efficiencies.
The emergence of a specialist enzyme (ancT) from a bifunctional one (ancMT) by negative selection is due to the loss of glucose affinity resulting from the E72A mutation (ground-state discrimination mode). It is likely that the loss of glucokinase activity by the E72A mutation provided the required freedom for the appearance of subsequent mutations that increased both the affinity and catalytic rate when using fructose-6-P as substrate. From these results it is not possible to deduce if the specialist ancT was fixed in the population by genetic drift or positive selection. However, the latter would make more sense if we consider a positive selective pressure toward more efficient glycolysis in these organisms leading to an increase in catalysis and affinity for fructose-6-P after the appearance of the E72A mutation.
Our findings are also consistent with studies of experimental microbial evolution, which reveal that a modest number of mutations have a great effect on the phenotype or fitness (7), shedding light on the classic debate about the size distribution of the mutational effects. Here we present a clear example where the evolutionary shift in function was driven by only a single amino acid substitution through negative selection, which caused a 1600-fold change in the ancestral specificity of the protein by a ground-state discrimination mode. This in turn, could provide the freedom for the specialization and increased efficiency of the enzyme and possibly provide an adaptive advantage through a gain in glycolytic efficiency in Thermococcales. Although there is agreement regarding the role of promiscuity as a starting point for the appearance of new protein function, there have been few insights into the mechanisms and mutational paths that lie behind the process of functional divergence. Here we were able to reconstruct the evolutionary history of substrate affinity using a simple evolutionary model of discrete binary characters and identify a large-effect mutation responsible for a critical event that is characteristic in the transformation of a specialized enzyme from a broad specificity progenitor. This allows substrate usage under physiological conditions in the family of ADP-dependent kinases to be considered as traits that are lost or acquired in a discreet manner, which can be considered as a punctuated equilibrium mechanism operating at the enzymatic function level.
Experimental procedures
Alignment and inference of phylogeny
The Protein BLAST Server was used to extract sequences present in the non-redundant protein sequence database (nr) using a PSI-BLAST algorithm with 3 iterations and ADP-GK from T. litoralis as template (UNIPROT codes of corresponding sequences used are shown in supplemental Table S5). A multiple sequence alignment was constructed based on three-dimensional and secondary structure constraints using Promals3D (35), and then misaligned positions were corrected by visual inspection in Multiseq from VMD (36). To obtain the multiple sequence alignment of the gene sequences, the protein sequence alignment was fixed and back-translated into DNA using the genes of each protein using the RevTrans server (37). The Bayesian inference of phylogeny was performed with MrBayes version 3.1.2 (38). For the protein sequences we used Cprev+ I+G as a fixed rate model, which showed a posterior probability of 1.0. For the gene sequences, a GTR+I+G model was used according to JMoldelTest (39). For Bayesian inference, the number of generations was set to 1 × 106, sample frequency to 100, with 2 runs and 4 chains for each run, and a temperature parameter of 0.2. The average standard deviation of the split frequencies was less than 1 × 10−4. The maximum likelihood inference was performed with PhyML 3.0, using protein sequences with a Cprev+I+G model.
Ancestral sequence reconstruction
For the Bayesian approach we used a modified method described by Hall (21) using gene sequences. Ancestral sequences were inferred through a hierarchical Bayesian approach implemented in MrBayes 3.1.2 with model and parameters used for phylogeny inference (40). Each target node was constrained and the probabilities for each nucleotide (or amino acid) were calculated at each position of the alignment, where the nucleotide (or amino acid) with highest posterior probability was selected. The maximum likelihood ASR inference was performed with PALM 4.4 (41), using the Lazarus tool according to that described by Hanson-Smith et al. (42). Sequence gaps were inferred using a model of binary evolutionary traits with one rate of change, considering only the presence of gaps or characters in the alignment (21). The positions that were most likely to be a gap for each ancestor were eliminated from the sequence, which produced ancestral sequences with different residue numbers (e.g. Glu-72 in ancMT is found at the same alignment position as Glu-79 in ancM and Ala-76 in ancT).
Gene synthesis, protein expression, and purification
The protein sequences for the ancestors inferred by the hierarchical Bayesian approach using gene sequences were codon-optimized for expression in Escherichia coli and genes were synthesized by GENSCRIPT (Piscataway, NJ), then cloned into a modified pET-28b vector and verified by DNA sequencing. E. coli BL21(DE3) cells were transformed and grown in LB broth containing 35 μg/ml of kanamycin at 37 °C until the A600 reached ∼0.8. Expression of the recombinant proteins was induced with 1 mm isopropyl β-d-thiogalactopyranoside overnight. Cells were harvested by centrifugation, re-suspended in binding buffer (50 mm Tris-HCl, pH 7.6, 500 mm NaCl, 20 mm imidazole, and 5 mm MgCl2), and disrupted by sonication. After centrifugation (18,514 × g for 45 min), ancestors ancMT, ancM, and ancT were heated at 70 °C during 10 min, chilled on ice, and centrifuged at 18,514 × g for 45 min before loaded onto a nickel-nitrilotriacetic acid affinity column (HisTrap HP, GE Healthcare, UK). Proteins were eluted with a linear gradient between 20 and 500 mm imidazole and fractions presenting enzyme activity were pooled. After the nickel-nitrilotriacetic acid affinity column, ancMMT and ancGK/PFK were dialyzed against buffer 25 mm Tris-HCl, pH 7.8, and 50 mm NaCl and chromatographed in an anionic exchange column HiTrapQ (GE Healthcare, UK). Purified proteins were stored at 4 °C in the presence of 5% glycerol. Enzyme purity was analyzed by SDS-PAGE stained with Coomassie Blue. Before crystallization, ancMT and its mutant were dialyzed against 25 mm Hepes/NaOH, pH 7.8, 5 mm MgCl2, concentrated by filtration and supplemented with ligand.
Reagent concentrations
Glucose and fructose-6-P were titrated with ADP-dependent glucokinase from T. litoralis and ADP-dependent PFK from P. horikoshii, respectively. ADP was titrated with the GK assay using ADP-GK from T. litoralis. Glucose-6-P was titrated with glucose-6-P-dehydrogenase from Leuconostoc mesenteroides and NAD+ under conditions in which the co-substrate concentration was at least 20-fold over the compound titrated. The protein concentration was determined using the Bio-Rad protein assay (Bio-Rad) with bovine serum albumin (BSA) standard (Thermo Scientific) as reference.
Determination of enzyme activity and kinetic parameters
PFK activity was assayed spectrophotometrically as described previously (43). The enzyme preparation was mixed with reaction buffer containing 10 mm ADP, 15 mm MgCl2, 25 mm PIPES, pH 6.5, 0.2 mm NADH and variable concentrations of fructose-6-P. α-Glycerophosphate dehydrogenase, triose-phosphate isomerase, and aldolase from rabbit (purchased from Sigma) were used as auxiliary enzymes.
GK activity was measured spectrophotometrically by following NAD+ reduction at 340 nm coupled with glucose-6-P oxidation. The assay was carried out in the presence of 10 mm ADP, 15 mm MgCl2, 25 mm Hepes, pH 7.8, 0.5 mm NAD+ and glucose. Glucose-6-P dehydrogenase from L. mesenteroides was used as an auxiliary enzyme.
All enzyme activities were determined at 40 °C using a photodiode array spectrophotometer (Hewlett Packard/Agilent 8453, Santa Clara, CA). An extinction coefficient of 6.22 mm−1 cm−1 for NADH was used for specific activity (U/mg) determination (U = μmol/min). The data were analyzed using nonlinear regression with GraphPad Prism version 5.0 (GraphPad Software Inc., San Diego, CA), and fitted to the Michaelis-Menten or substrate inhibition equations.
In the event that saturation was not reached, the kcat/Km value was determined from the initial linear portion of the saturation curve. In the Michaelis-Menten equation at low substrate concentrations ([S] ≪ Km) the velocity exhibits a linear dependence on substrate concentration whose slope is (kcat/Km) × [E], so the kcat/Km value can be obtained by dividing the slope by the enzyme concentration. A rough estimation of the kcat and Km values for glucose of the ancT and E72A mutant were obtained by double-reciprocal plot.
Crystallization, data collection, and structure determination
Crystallization screening was performed using a Honeybee963 robot (genomic Solution) in Greiner CrystalQuick plates using a 1:1 ratio of protein to precipitant. Optimization rounds were carried out manually by the hanging drop method resulting in good quality crystals of ancMT and its E72A mutant. These were obtained for both proteins using a drop of 2 μl of protein stock solution (8 mg/ml of protein in 25 mm Hepes/NaOH, pH 7.8, buffer, 30 mm MgCl2, 20 mm fructose-6-P, and 20 mm AMP) plus 2 μl of the reservoir solution comprising 18% PEG 3350 and 0.15 m NaI. Crystals appeared after 3 days at 18 °C, the crystals were briefly transferred to glycerol as cryoprotective solution and flash-frozen in liquid nitrogen. The X-ray diffraction data were collected at the Diamond Light Source using the beamline I04-1. The data were indexed, integrated, and scaled with XDS (44), and merged using Aimless from the CCP4 package (45). The structure of the ancMT in complex with AMP was solved by molecular replacement using the program Phaser (46), with the PFK structure from P. horikoshii split into two domains (major and minor) as searches models (PDB code 3DRW). The structure of the E72A mutant was solved using the structure of ancMT as the search model. Structure refinement was carried out using Phenix (47) and COOT for model building (48). The AMP ligands were automatically placed using COOT, and water molecules were located with a combination of Phenix and COOT. The quality of the model was evaluated using Molprobity (49). Full statistics for data collection and refinement are summarized in Table 2.
Molecular modeling, docking, and molecular dynamics
For homology modeling two template structures were employed. One was the GK from P. furiosus in its closed conformation (PDB code 1UA4), which presents AMP and glucose in its active site. The second template was the structure for ancMT solved here. In the latter case the structure was first divided into its two domains, which were then structurally aligned independently to their homologues in PDB code 1UA4. These two structures were then used as templates for modeling the enzyme in the closed conformation. Fifty models were constructed with MODELLER 8 (50). From these, the best 10 models based on the DOPE potential were chosen, and their quality evaluated with PROSA2003 (51), Procheck (52), and VERIFY_3D (53).
Docking assays with glucose and fructose-6-P were performed with AutodockVina 1.0 (54), with protonation states according to the pH values used in the kinetic experiments. The protonation state of the ionizable residues was calculated using the web server H++ (55). After this procedure, partial charges were derived with the Gasteiger method using AutoDockTool (56). Docking results with low interaction energies and the phosphoryl acceptor hydroxyl oriented toward the GXGD motif were selected since the aspartic acid is considered the catalytic base for the phosphate group transfer in all the Ribokinase superfamily members (57). Subsequently, the Mg-ADP complex was included into the resulting docked structures according to the procedure described by Merino et al. (58) and then both structures were subjected to energy minimization using a standard protocol in NAMD 2.7 (59) with the CHARMM27 force field. Proteins were placed in a water box of TIP3 waters that extended at least 12 Å beyond any macromolecular atom. The system was neutralized with NaCl to a final concentration of 0.1 m and 10,000 iterations were performed without restrictions. The equilibration of the system was performed in an isobaric isothermal assembly (NPT) at a temperature of 310 K, 1 atmosphere of pressure (1.01325 bar), PME cutoff of 12.0, Langevin piston pressure control, Langevin temperature control, and using a time step of 2 fs. After 10 ns of equilibration a 30-ns long simulation was produced for each system and the nonbonding energy between protein and ligand was analyzed using the NAMDenergy plugin of VMD (60).
Reconstruction of ancestral discrete traits
The reconstruction of ancestral states as discrete traits was performed by maximum likelihood using the Mesquite 2.75 software. To discriminate between the MK1 and AsymmMK models, the likelihood ratio test was used. For all traits evaluated the LR was not higher than the p parameter of the χ2 distribution with 1 degree of freedom, so the MK1 model was selected. The consensus tree obtained from the Bayesian phylogeny reconstruction using protein sequences was used for analysis.
Bayesian inference was performed using the software BayesTraits (61) using the MCMC methodology. Traits evaluated were mapped onto 500 trees obtained using Bayesian inference in MrBayes 3.1 with protein sequences. The Bayes factor was used to discriminate between a 1 (q01 = q10) or 2 rate (q01 ≠ q10) model, the former being most appropriate. Analyses were performed with 10 million iterations, with burn-in of 100,000 and a sampling frequency of 1,000. The ratedev parameter was set so that the acceptance parameter was maintained between 20 and 40%, whereas the sequence of Homo sapiens was used as out-group to root the trees. Parameter files and Bayes factor calculations were evaluated with the Tracer software.
Author contributions
V. C-F., F. M., and V. G. conceived the project and designed the research; V. C-F., A. H-M., R. Z., F. G-O., F. P-S., and V. G. performed and analyzed the enzyme kinetic experiment; V. C-F. performed the phylogenetic analysis; V. C-F. and R. Z. performed and analyzed the molecular dynamics simulations; V. C-F., H. M. P., J. B-N., and R. C. G. performed and analyzed the protein crystallography experiment; and V. C-F., R. C. G., and V. G. wrote the paper.
Supplementary Material
Acknowledgments
We thank the CeBEM/CCP4 school 2014, São Carlos, Brazil, and the Diamond Light Source beamline I04–1.
This work was supported in part by Fondo Nacional de Desarrollo Cientifico y Tecnológico from Chile Grants FONDECYT 1150460 (to V. G.) and FONDECYT Postdoctorado 3160332 (to V. C-F.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S5 and Tables S1–S5.
The atomic coordinates and structure factors (codes 5K27 and 5KKG) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- ASR
- ancestral sequence reconstruction
- PFK
- phosphofructokinase
- GK
- glucokinase
- Glc
- glucose
- HGT
- horizontal gene transfer
- TlGK
- glucokinase from T. litoralis
- PhPFK
- phosphofructokinase from P. horikoshii
- r.m.s.
- root mean square.
References
- 1. Conant G. C., and Wolfe K. H. (2008) Turning a hobby into a job: how duplicated genes find new functions. Nat. Rev. Genet. 9, 938–950 [DOI] [PubMed] [Google Scholar]
- 2. Todd A. E., Orengo C. A., and Thornton J. M. (2001) Evolution of function in protein superfamilies, from a structural perspective. J. Mol. Biol. 307, 1113–1143 [DOI] [PubMed] [Google Scholar]
- 3. Khersonsky O., and Tawfik D. S. (2010) Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu. Rev. Biochem. 79, 471–505 [DOI] [PubMed] [Google Scholar]
- 4. Allewell N. M. (2012) Thematic minireview series on enzyme evolution in the post-genomic era. J. Biol. Chem. 287, 1–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carlson J. C., Badran A. H., Guggiana-Nilo D. A., and Liu D. R. (2014) Negative selection and stringency modulation in phage-assisted continuous evolution. Nat. Chem. Biol. 10, 216–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tawfik D. S. (2014) Accuracy-rate tradeoffs: how do enzymes meet demands of selectivity and catalytic efficiency?. Curr. Opin. Chem. Biol. 21, 73–80 [DOI] [PubMed] [Google Scholar]
- 7. Orr H. A. (2005) The genetic theory of adaptation: a brief history. Nat. Rev. Genet. 6, 119–127 [DOI] [PubMed] [Google Scholar]
- 8. Harms M. J., and Thornton J. W. (2010) Analyzing protein structure and function using ancestral gene reconstruction. Curr. Opin. Struct. Biol. 20, 360–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Benner S. A., Sassi S. O., and Gaucher E. A. (2007) Molecular paleoscience: systems biology from the past. Adv. Enzymol. Relat. Areas Mol. Biol. 75, 1–132 [DOI] [PubMed] [Google Scholar]
- 10. Richter S., Richter J. P., Mehta S. Y., Gribble A. M., Sutherland-Smith A. J., Stowell K. M., Print C. G., Ronimus R. S., and Wilson W. R. (2012) Expression and role in glycolysis of human ADP-dependent glucokinase. Mol. Cell. Biochem. 364, 131–145 [DOI] [PubMed] [Google Scholar]
- 11. Koga S., Yoshioka I., Sakuraba H., Takahashi M., Sakasegawa S., Shimizu S., and Ohshima T. (2000) Biochemical characterization, cloning, and sequencing of ADP-dependent (AMP-forming) glucokinase from two hyperthermophilic archaea, Pyrococcus furiosus and Thermococcus litoralis. J. Biochem. 128, 1079–1085 [DOI] [PubMed] [Google Scholar]
- 12. Tuininga J. E., Verhees C. H., van der Oost J., Kengen S. W., Stams A. J., and de Vos W. M. (1999) Molecular and biochemical characterization of the ADP-dependent phosphofructokinase from the hyperthermophilic archaeon Pyrococcus furiosus. J. Biol. Chem. 274, 21023–21028 [DOI] [PubMed] [Google Scholar]
- 13. Castro-Fernandez V., Bravo-Moraga F., Herrera-Morande A., and Guixe V. (2014) Bifunctional ADP-dependent phosphofructokinase/glucokinase activity in the order Methanococcales: biochemical characterization of the mesophilic enzyme from Methanococcus maripaludis. FEBS J. 281, 2017–2029 [DOI] [PubMed] [Google Scholar]
- 14. Sakuraba H., Yoshioka I., Koga S., Takahashi M., Kitahama Y., Satomura T., Kawakami R., and Ohshima T. (2002) ADP-dependent glucokinase/phosphofructokinase, a novel bifunctional enzyme from the hyperthermophilic archaeon Methanococcus jannaschii. J. Biol. Chem. 277, 12495–12498 [DOI] [PubMed] [Google Scholar]
- 15. Merino F., and Guixé V. (2008) Specificity evolution of the ADP-dependent sugar kinase family: in silico studies of the glucokinase/phosphofructokinase bifunctional enzyme from Methanocaldococcus jannaschii. FEBS J. 275, 4033–4044 [DOI] [PubMed] [Google Scholar]
- 16. Currie M. A., Merino F., Skarina T., Wong A. H., Singer A., Brown G., Savchenko A., Caniuguir A., Guixé V., Yakunin A. F., and Jia Z. (2009) ADP-dependent 6-phosphofructokinase from Pyrococcus horikoshii OT3: structure determination and biochemical characterization of PH1645. J. Biol. Chem. 284, 22664–22671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rivas-Pardo J. A., Herrera-Morande A., Castro-Fernandez V., Fernandez F. J., Vega M. C., and Guixé V. (2013) Crystal structure, SAXS and kinetic mechanism of hyperthermophilic ADP-dependent glucokinase from Thermococcus litoralis reveal a conserved mechanism for catalysis. PLoS ONE PLoS ONE 8, e66687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Petitjean C., Deschamps P., López-García P., Moreira D., and Brochier-Armanet C. (2015) Extending the conserved phylogenetic core of archaea disentangles the evolution of the third domain of life. Mol. Biol. Evol. 32, 1242–1254 [DOI] [PubMed] [Google Scholar]
- 19. Battistuzzia F. U., and Hedgesa S. B. (2009) Archaebacteria. in The Timetree of Life (Hedges S. B., and Kumar S., eds) pp. 101–105, Oxford University Press, New York [Google Scholar]
- 20. Yang Z., Kumar S., and Nei M. (1995) A new method of inference of ancestral nucleotide and amino acid sequences. Genetics. 141, 1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hall B. G. (2006) Simple and accurate estimation of ancestral protein sequences. Proc. Natl. Acad. Sci. U.S.A. 103, 5431–5436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ito S., Fushinobu S., Yoshioka I., Koga S., Matsuzawa H., and Wakagi T. (2001) Structural basis for the ADP-specificity of a novel glucokinase from a hyperthermophilic archaeon. Structure 9, 205–214 [DOI] [PubMed] [Google Scholar]
- 23. Ito S., Fushinobu S., Jeong J. J., Yoshioka I., Koga S., Shoun H., and Wakagi T. (2003) Crystal structure of an ADP-dependent glucokinase from Pyrococcus furiosus: implications for a sugar-induced conformational change in ADP-dependent kinase. J. Mol. Biol. 331, 871–883 [DOI] [PubMed] [Google Scholar]
- 24. Page M. (1999) The maximum likelihood approach to reconstructing ancestral character states of discrete characters on phylogenies. Syst. Biol. 48, 612–622 [Google Scholar]
- 25. Pagel M., Meade A., and Barker D. (2004) Bayesian estimation of ancestral character states on phylogenies. Syst. Biol. 53, 673–684 [DOI] [PubMed] [Google Scholar]
- 26. Bar-Rogovsky H., Stern A., Penn O., Kobl I., Pupko T., and Tawfik D. S. (2015) Assessing the prediction fidelity of ancestral reconstruction by a library approach. Protein Eng. Des. Sel. 28, 507–518 [DOI] [PubMed] [Google Scholar]
- 27. Harms M. J., Eick G. N., Goswami D., Colucci J. K., Griffin P. R., Ortlund E. A., and Thornton J. W. (2013) Biophysical mechanisms for large-effect mutations in the evolution of steroid hormone receptors. Proc. Natl. Acad. Sci. U.S.A. 110, 11475–11480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Heilbron K., Toll-Riera M., Kojadinovic M., and MacLean R. C. (2014) Fitness is strongly influenced by rare mutations of large effect in a microbial mutation accumulation experiment. Genetics 197, 981–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Treangen T. J., and Rocha E. P. (2011) Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes. PLoS Genet. 7, e1001284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Voordeckers K., Brown C. A., Vanneste K., van der Zande E., Voet A., Maere S., and Verstrepen K. J. (2012) Reconstruction of ancestral metabolic enzymes reveals molecular mechanisms underlying evolutionary innovation through gene duplication. PLos Biol. 10, e1001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hobbs J. K., Shepherd C., Saul D. J., Demetras N. J., Haaning S., Monk C. R., Daniel R. M., and Arcus V. L. (2012) On the origin and evolution of thermophily: reconstruction of functional precambrian enzymes from ancestors of Bacillus. Mol. Biol. Evol. 29, 825–835 [DOI] [PubMed] [Google Scholar]
- 32. Bar-Even A., Noor E., Savir Y., Liebermeister W., Davidi D., Tawfik D. S., and Milo R. (2011) The moderately efficient enzyme: evolutionary and physicochemical trends shaping enzyme parameters. Biochemistry 50, 4402–4410 [DOI] [PubMed] [Google Scholar]
- 33. Newton M. S., Arcus V. L., and Patrick W. M. (2015) Rapid bursts and slow declines: on the possible evolutionary trajectories of enzymes. J. R. Soc. Interface 12, 20150036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Santiago-Martínez M. G., Encalada R., Lira-Silva E., Pineda E., Gallardo-Pérez J. C., Reyes-García M. A., Saavedra E., Moreno-Sánchez R., Marín-Hernández A., and Jasso-Chávez R. (2016) The nutritional status of Methanosarcina acetivorans regulates glycogen metabolism and gluconeogenesis and glycolysis fluxes. FEBS J. 283, 1979–1999 [DOI] [PubMed] [Google Scholar]
- 35. Pei J., Kim B.-H., and Grishin N. V. (2008) PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 36, 2295–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Roberts E., Eargle J., Wright D., and Luthey-Schulten Z. (2006) MultiSeq: unifying sequence and structure data for evolutionary analysis. BMC Bioinformatics 7, 382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wernersson R., and Pedersen A. (2003) RevTrans: multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Res. 31, 3537–3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ronquist F., and Huelsenbeck J. P. (2003) MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 [DOI] [PubMed] [Google Scholar]
- 39. Posada D. (2008) jModelTest: phylogenetic averaging. Mol. Biol. Evol. 25, 1253–1256 [DOI] [PubMed] [Google Scholar]
- 40. Huelsenbeck J. P., and Bollback J. P. (2001) Empirical and hierarchical Bayesian estimation of ancestral states. Syst. Biol. 50, 351–366 [PubMed] [Google Scholar]
- 41. Yang Z. (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 [DOI] [PubMed] [Google Scholar]
- 42. Hanson-Smith V., Kolaczkowski B., and Thornton J. W. (2010) Robustness of ancestral sequence reconstruction to phylogenetic uncertainty. Mol. Biol. Evol. 27, 1988–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kengen S. W., de Bok F. A., van Loo N. D., Dijkema C., Stams A. J., and de Vos W. M. (1994) Evidence for the operation of a novel Embden-Meyerhof pathway that involves ADP-dependent kinases during sugar fermentation by Pyrococcus furiosus. J. Biol. Chem. 269, 17537–17541 [PubMed] [Google Scholar]
- 44. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Evans P. R., and Murshudov G. N. (2013) How good are my data and what is the resolution?. Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Emsley P., and Cowtan K. (2004) Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 49. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sali A., and Blundell T. L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 51. Sippl M. J. (1993) Recognition of errors in three-dimensional structures of proteins. Proteins Struct. Funct. Genet. 17, 355–362 [DOI] [PubMed] [Google Scholar]
- 52. Laskowski R. A., MacArthur M. W., Moss D. S., and Thornton J. M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26, 283–291 [Google Scholar]
- 53. Bowie J. U., Lüthy R., and Eisenberg D. (1991) A method to identify protein sequences that fold into a known three-dimensional stucture. Science 253, 164–170 [DOI] [PubMed] [Google Scholar]
- 54. Trott O., and Olson A. J. (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gordon J. C., Myers J. B., Folta T., Shoja V., Heath L. S., and Onufriev A. (2005) H++: a server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res. 33, W368–W371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., and Olson A. J. (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Guixé V., and Merino F. (2009) The ADP-dependent sugar kinase family: kinetic and evolutionary aspects. IUBMB Life 61, 753–761 [DOI] [PubMed] [Google Scholar]
- 58. Merino F., Rivas-Pardo J. A., Caniuguir A., García I., and Guixé V. (2012) Catalytic and regulatory roles of divalent metal cations on the phosphoryl-transfer mechanism of ADP-dependent sugar kinases from hyperthermophilic archaea. Biochimie 94, 516–524 [DOI] [PubMed] [Google Scholar]
- 59. Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., and Schulten K. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Humphrey W., Dalke A., and Schulten K. (1996) VMD: Visual Molecular Dynamics. J. Mol. Graph. 14, 33–38, 27–28 [DOI] [PubMed] [Google Scholar]
- 61. Pagel M., and Meade A. (2006) Bayesian analysis of correlated evolution of discrete characters by reversible-jump Markov chain Monte Carlo. Am. Nat. 167, 808–825 [DOI] [PubMed] [Google Scholar]
- 62. Kanehisa M., Sato Y., Kawashima M., Furumichi M., and Tanabe M. (2016) Kyoto Encyclopedia of Genes and Genomes. http://www.genome.jp/kegg/ (Accessed May 3, 2016)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.