Abstract
Nitrogenase reduces dinitrogen (N2) to ammonia in biological nitrogen fixation. The nitrogenase Fe protein cycle involves a transient association between the reduced, MgATP-bound Fe protein and the MoFe protein and includes electron transfer, ATP hydrolysis, release of Pi, and dissociation of the oxidized, MgADP-bound Fe protein from the MoFe protein. The cycle is completed by reduction of oxidized Fe protein and nucleotide exchange. Recently, a kinetic study of the nitrogenase Fe protein cycle involving the physiological reductant flavodoxin reported a major revision of the rate-limiting step from MoFe protein and Fe protein dissociation to release of Pi. Because the Fe protein cannot interact with flavodoxin and the MoFe protein simultaneously, knowledge of the interactions between flavodoxin and the different nucleotide states of the Fe protein is critically important for understanding the Fe protein cycle. Here we used time-resolved limited proteolysis and chemical cross-linking to examine nucleotide-induced structural changes in the Fe protein and their effects on interactions with flavodoxin. Differences in proteolytic cleavage patterns and chemical cross-linking patterns were consistent with known nucleotide-induced structural differences in the Fe protein and indicated that MgATP-bound Fe protein resembles the structure of the Fe protein in the stabilized nitrogenase complex structures. Docking models and cross-linking patterns between the Fe protein and flavodoxin revealed that the MgADP-bound state of the Fe protein has the most complementary docking interface with flavodoxin compared with the MgATP-bound state. Together, these findings provide new insights into the control mechanisms in protein–protein interactions during the Fe protein cycle.
Keywords: electron transfer, MS, protein complex, protein cross-linking, protein—protein interaction, proteolysis
Introduction
Nitrogenase is the enzyme that reduces dinitrogen (N2) to ammonia (NH3) in a process known as biological nitrogen fixation (1–3). The MoFe protein and the Fe protein are the two catalytic components of the molybdenum-dependent nitrogenase found in diazotrophs, such as Azotobacter vinelandii (4). The MoFe protein is a α2β2 heterotetramer, where each αβ unit contains two unique metal clusters: the [8Fe-7S] P cluster, which is involved in electron transfer to the [7Fe-9S-Mo-C-homocitrate] FeMo-cofactor at the active site (5–9).
The Fe protein is a member of a large class of proteins that couple nucleoside triphosphate (ATP or GTP) binding and hydrolysis to protein conformational changes that are transduced within a macromolecular protein complex (10–13). Two sets of consensus amino acid sequence motifs, called the Walker A (GXXXXGKS) and Walker B motifs (DXXG), located along the nucleotide-binding regions, are used to identify members within this class (10, 11, 14–16). A unique feature that distinguishes the nitrogenase Fe protein is that it exists as a homodimer, where a [4Fe-4S] cluster bridges the two identical subunits with a Walker A motif (also known as the phosphate-binding loop or P-loop) and Walker B motif present in each subunit (10, 11, 13–15, 17, 18).
The Fe protein cycle involves transient association between the reduced, MgATP-bound Fe protein with each αβ half of the MoFe protein to deliver one electron to the MoFe protein (19–21). Complex formation between the MoFe protein and the Fe protein is followed by a conformationally gated one-electron transfer from the P cluster to the FeMo cofactor (22, 23). The reduced Fe protein then transfers one electron from its [4Fe-4S] cluster to the oxidized P cluster (P+1) in what has been described as a “deficit-spending” electron transfer process (5). Hydrolysis of MgATP to MgADP and Pi occurs after the electron transfer, and the dissociation of the oxidized, MgADP-bound Fe protein from the MoFe protein occurs after the release of two Pi molecules (24). Nucleotide exchange and re-reduction of the oxidized nucleotide-bound Fe protein must occur before another cycle of catalysis can begin. The order of events for re-reduction of the oxidized nucleotide-bound Fe protein and nucleotide exchange of MgADP for MgATP has not been established (24–27).
Recently, it was determined that, with the physiological reductant flavodoxin (Fld),3 the rate-limiting step in the Fe protein cycle is actually the release of the two Pi molecules and not the dissociation of the MoFe protein from the Fe protein, as shown previously using the chemical reductant sodium dithionite (25). It was implicated that Fld binds to the same face of the Fe protein that interacts with the MoFe protein and verified that Fld cannot reduce the Fe protein while it is in complex with the MoFe protein (25).
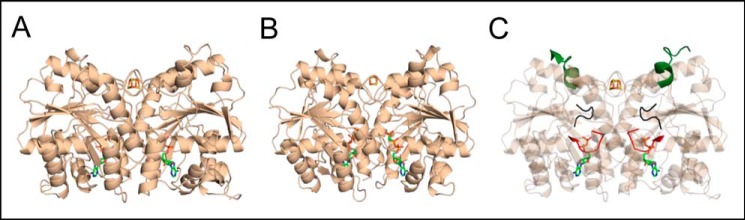
The conformation of the Fe protein is modulated by nucleotide-dependent conformational differences in the MgATP- and MgADP-bound states (28–31). The structures of the nucleotide-free and MgADP-bound states of the Fe protein are known (13, 15), but the structure of the MgATP state has remained elusive. Circular dichroism, 1H NMR and EPR spectroscopic studies have shown that regions of the Fe protein undergo significant conformational changes during nucleotide binding and that the MgADP and MgATP states are conformationally distinct (28–31). Although the structure of the MgATP-bound state of the Fe protein is unknown, there are several structures of the Fe protein–MoFe protein complex in which the Fe protein structure from these complexes is significantly distinct from the structure of the free MgADP-bound state (32). The majority of the conformational difference in the Fe protein structures observed in the complexes, relative to the MgADP-bound Fe protein conformation, is a rigid body reorientation of the subunits with respect to one another (Fig. 1, A and B) (32). The most notable changes are associated with alternative sets of inter- and intrasubunit salt bridges and different conformations of the nucleotide-dependent switch regions (switch I and II) and the P-loop (15, 17, 32). These key regions transduce conformational changes between the nucleotide-binding site and both the MoFe protein docking interface (switch I) and the [4Fe-4S] cluster (switch II) (Fig. 1C). It is attractive to consider that the MgATP-bound state of the Fe protein resembles the structure of the Fe protein observed in the stabilized nitrogenase Fe protein–MoFe protein complex (Fig. 1). This would establish a catalytic cycle in which conformational gating dictates that the MgATP-bound Fe protein is more complementary with the MoFe protein docking interface, whereas the MgADP-bound Fe protein is more complementary with the Fld docking interface.
Figure 1.
Structures of the Fe protein show conformational differences during relevant stages of nitrogenase catalysis. A, the crystal structure of the MgADP-bound Fe protein (tan, PDB code 1FP6). B, the Fe protein structure stabilized with an MgATP analog, MgAMPPCP, in the nitrogenase complex (tan, PDB code 4WZB), which displays rigid body movement of the subunits with respect to one another that closes the dimer interface and poises the region around the active site to promote interactions with the MoFe protein. The MoFe protein is not shown. C, key regions in the Fe protein that transduce conformational changes between the nucleotide-binding site and the MoFe protein docking interface: part of switch I that interacts with the MoFe protein (green), part of switch I that binds Mg2+ (black), and the P-loop, which interacts with the phosphates of the nucleotides (red).
In this work, time-resolved limited proteolysis and chemical cross-linking in combination with LC-MS and MALDI-TOF mass spectrometry are used to gain insight into differences between the MgADP- and MgATP-bound conformations of the Fe protein. This then allows for investigation of protein regions involved in complex formation with Fld. Time-resolved limited proteolysis allows the identification of proteolytic cleavage sites that become more dynamic because of conformational differences and protein–protein interactions (33, 34). Additionally, covalent cross-linking followed by mass analysis of proteolyzed peptides allows protein domains in close proximity to be identified based on the ability of a short chemical linker to bridge side chains. Chemical cross-linking and proteolysis patterns are consistent with the conformation of the Fe protein observed in the stabilized nitrogenase complex being the best structural mimic for the MgATP-bound state of the Fe protein in solution. In silico docking models and chemical cross-linking data show that more complementary docking interactions occur between Fld and the MgADP-bound Fe protein compared with the MgATP-bound Fe protein conformation. The results reveal that the nucleotide state of the Fe protein influences interactions with Fld and suggest mechanisms for control of protein–protein interactions during the Fe protein cycle.
Results and discussion
Distinguishing the Fe protein in different nucleotide-bound states
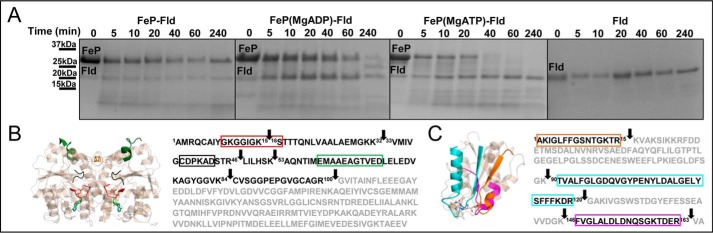
Time-resolved limited proteolysis and chemical cross-linking experiments were used to examine differences in Fe protein nucleotide-dependent conformational states and probe the hypothesis that the structural conformation of the MgATP-bound Fe protein resembles the Fe protein in the nitrogenase complex structures (32). A series of trypsin proteolysis reactions of the Fe protein with and without nucleotides produced a distinct banding pattern over the course of a 240-min reaction (Fig. 2A). The proteolysis pattern for the MgADP-bound Fe protein and nucleotide-free Fe protein are similar in that there is still some intact Fe protein after 240 min (Fig. 2A). The main difference between these two forms is in the gradual appearance of a band at 20 kDa over the course of proteolysis (Fig. 2A). Mass spectrometry analysis indicates that this 20-kDa band is a C-terminal fragment arising from cleavage at Arg100 of the Fe protein (Fig. 2B).
Figure 2.
Chemical cross-linking and proteolysis patterns can be used to distinguish the Fe protein in the different nucleotide states. A, differences in the proteolytic rates of the Fe protein band at 32 kDa are seen in the SDS-PAGE gel of the time-resolved limited proteolysis reactions and depend on the presence of nucleotide. B, mapping limited proteolysis results on the amino acid sequence display Fe protein tryptic cleavage sites (arrows) that are involved in the largest differences because of nucleotide binding. These include the P-loop (Gly9-Ser16, boxed in red), the part of the switch I region that coordinates the bound Mg2+ of the nucleotide (Cys38-Asp43, boxed in black), the part of the switch I region that interacts with the MoFe protein (Glu59-Asp69, boxed in green) and Arg100 (arrow). C, in the presence of MgATP, the Fe protein shows a more complex cross-linking pattern in comparison with the MgADP-bound state. Several types of dimers and higher-molecular-weight aggregates (arrows) are formed during the same time of exposure to glutaraldehyde (GA). In addition, when the Fe protein is separated on a gel, two differentially intra-cross-linked monomers are revealed (arrows). D, proteolysis and cross-linking patterns show structural changes in the P-loop region (red) and switch I region (green) because of MgATP binding. Rearrangements are consistent with what is observed in the Fe protein structure from the MgAMPPCP-stabilized nitrogenase complex (PDB code 4WZB).
Mapping of the tryptic peptides from the digestion of the nucleotide-free and MgADP-bound states of Fe protein produced nearly identical patterns, indicating that they adopt similar conformations in solution. This is consistent with nucleotide-free and MgADP-bound Fe protein crystal structures (13, 15). The proteolysis pattern was strikingly different for the MgATP-bound state of the Fe protein compared with the MgADP-bound state. The MgATP-bound Fe protein is more susceptible to proteolysis, as the intact protein band is no longer present after 60 min of digestion (Fig. 2A). In addition, the 20-kDa C-terminal Fe protein band appears at a faster rate. This indicates that the region around Arg100, near the [4Fe-4S] cluster, is more exposed, increasing susceptibility to proteolysis. Furthermore, cleavage in the switch I region (Glu59-Asp69, Fig. 2B, green) is observed after only 5 min, whereas it does not appear in the MgADP-bound state until 20 min. Fe protein tryptic peptides identified in time-resolved limited proteolysis experiments with highlighted peptides that change because of the presence of nucleotides are listed in supplemental Table S1. Furthermore, representative MALDI-TOF mass spectra for digestion of the Fe protein (supplemental Fig. S1) are shown to supplement peptide identification.
There are also differences in cross-linking patterns for the nucleotide-bound Fe protein (Fig. 2C). In general, the MgATP-bound Fe protein cross-links more readily than the MgADP-bound state. This is evidenced by more extensive band broadening because of an increase in both intersubunit cross-links (above 50 kDa) between the Fe protein subunits and intrasubunit cross-links (around 32 kDa) within Fe protein monomers (Fig. 2C). Mapping of the peptides that were cross-linked confirm this, where cross-linking in the P-loop region only occurs in the MgATP-bound state, and may suggest that the Fe protein subunits are in closer proximity, as documented in previous reports (32). Fe protein tryptic and non-tryptic peptides observed to appear and disappear during the chemical cross-linking experiments with highlighted peptides that change because of the presence of nucleotides are listed in supplemental Table S2.
The increased diversity of cross-links together with increased proteolytic susceptibility of MgATP-bound Fe protein compared with the MgADP-bound state indicates that the conformations are distinct. Compared with the MgADP-bound Fe protein structure, the Fe protein in the nitrogenase complexes displays a more closed dimer interface in which part of the switch I region as well as the protein region around the [4Fe-4S] cluster form the docking site for the MoFe protein (Fig. 1B) (32). The increased proteolytic susceptibility of switch I (Fig. 2D, green) and cleavage of Arg100, near the [4Fe-4S] cluster, as well as increased cross-linking in the P-loop region (Fig. 2D, red) all suggest a MgATP-bound state that resembles the conformation of the Fe protein observed in the stabilized nitrogenase complex structures (32).
Defining Fe protein–flavodoxin interactions
In silico docking studies using ClusPro 2.0 show that the MgADP-bound structure of the Fe protein has a more complementary docking interface with Fld compared with the MgATP-bound Fe protein (Fig. 3) (25). A distance of 6.4 Å between the [4Fe-4S] cluster of the Fe protein and FMN cofactor of Fld was measured for the docking model between the MgADP-bound Fe protein with Fld (Fig. 3A). For the docking model between Fld and the Fe protein from the nitrogenase complexes, the distance between the cofactors was determined to be 9.4 Å, a 3 Å larger distance between the [4Fe-4S] cluster of the Fe protein and FMN cofactor of Fld (Fig. 3B) than observed for the MgADP-bound state docked to Fld. Furthermore, the solvent-excluded surface area for the docking model between the MgADP-bound Fe protein and Fld was calculated in Chimera to be 1128 Å2 compared with 456 Å2 for the docking model between the MgATP-bound Fe protein and Fld, translating to 2.5 times more buried surface area with the MgADP-bound state than the MgATP-bound state. Previously, electrostatic interactions were implicated in Fe protein interactions with the MoFe protein and Fld (25, 35, 36). Fe protein residues Arg100 and Arg140, implicated previously in Fe protein–MoFe protein interactions (35, 36), are also key in Fe protein–Fld interactions. In our docking model with the MgADP-bound Fe protein, Arg100 from one subunit interacts with the phosphate moiety of the FMN cofactor, and the other Arg100 from the second subunit forms a salt bridge with Glu104 (Fig. 3C). Additionally, Arg140 from both subunits forms individual salt bridge interactions with the Fld acidic residues Asp154 and Glu59 (Fig. 3C). There are fewer complementary electrostatic interactions observed with the MgATP-bound Fe protein and Fld (Fig. 3D). Arg100 interacts with Glu61 of Fld on one subunit and with both Fld acidic residues, Asp68 and Glu70, on the other subunit (Fig. 3D). Arg140, however, is positioned so that salt bridge interactions are not observed with any Fld acidic residues (Fig. 3D). Together, the differences in distances between the FeS cluster and the FMN cofactor, the buried solvent-excluded surface area, and the extent of electrostatic interactions indicate that MgADP-bound Fe protein shows the most complementary binding interface with Fld.
Figure 3.
Interactions between Fld and the Fe protein analyzed in silico docking models suggest more complementary docking between the MgADP-bound Fe protein and Fld. A, the docking model between the MgADP-bound Fe protein (light blue, PDB code 1FP6) and Fld (violet, PDB code 1YOB) displays a distance of 6.4 Å between the [4Fe-4S] cluster and the FMN cofactor measured in PyMOL. B, a distance of 9.4 Å between the [4Fe-4S] cluster and the FMN cofactor was measured in PyMOL for the docking model between the Fe protein from the nitrogenase complexes (tan, PDB code 4WZB) and Fld (violet, PDB code 1YOB). C, interactions for the docking model between the MgADP-bound Fe protein basic amino acid residues Arg100 and Arg140 and docked Fld acidic residues Glu104, Asp154, and Glu59. D, interactions between Arg100 of the Fe protein from the MgAMPPCP-stabilized nitrogenase complex and docked Fld acidic residues Glu61, Asp68, and Glu70. Arg140 is positioned so that it can no longer form salt bridge interactions with any Fld acidic residues.
Mapping Fe protein–Fld interactions
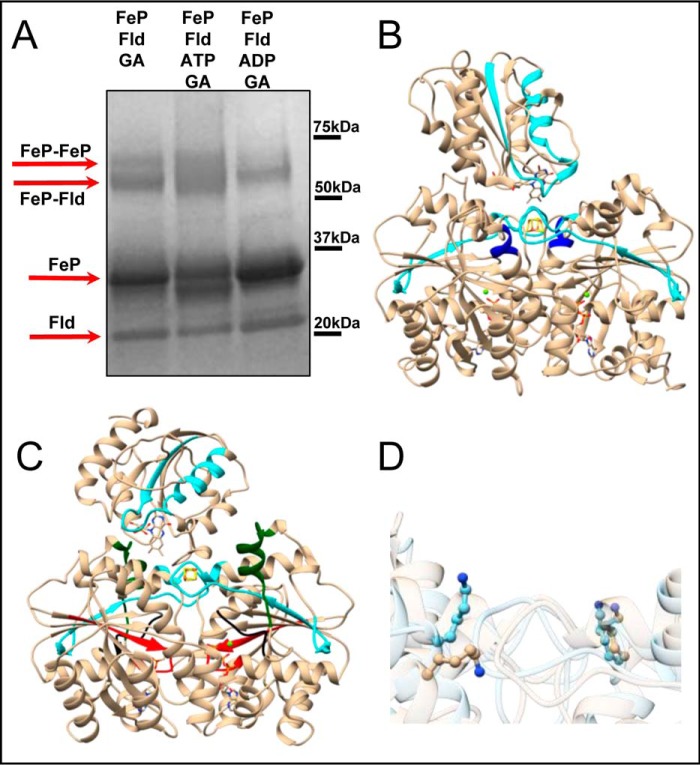
The nature of MgATP-bound Fe protein and MgADP-bound Fe protein interactions with Fld was further probed using time-resolved limited proteolysis (Fig. 4) and chemical cross-linking experiments (Fig. 5). Interestingly, the MgADP-bound Fe protein was more susceptible to proteolysis in the presence of Fld than when digested alone (Figs. 2A and 4A). This was confirmed by cleavage that mapped to part of the switch I region (Fig. 4B, green) and appeared in the earliest time point for the digestion of the MgADP-bound Fe protein with Fld. This region was not observed until 20 min in the digestion of the MgADP-bound Fe protein alone (Fig. 2A). However, the significantly higher degree of proteolytic susceptibility of the MgATP-bound Fe protein relative to the MgADP-bound Fe protein suggests that the MgADP-bound state forms a more productive complex with Fld (Fig. 4A). This is a key difference and supports the docking models, which suggest that the MgATP-bound Fe protein docking interface is not as complementary for interactions with Fld compared with the MgADP-bound state. The limited proteolysis mapping verify this by comparing the digestion of Fld alone versus in the presence of the nucleotide-bound Fe protein to distinguish different Fe protein–Fld interaction sites. Digestion of purified Fld alone leads to cleavage in regions at the Fe protein–Fld interaction site near the FMN cofactor, producing Ala1-Arg15 (Fig. 4C, orange), Thr90-Arg120 (Fig. 4C, cyan), and Phe146-Arg163 (Fig. 4C, magenta). All three Fld tryptic cleavage sites are protected from proteolysis in the MgADP-bound Fe protein, indicating that these regions are involved in Fe protein–Fld interactions. The Arg15 cleavage site (Fig. 4C, orange) was not protected in the presence of the MgATP-bound Fe protein, which is consistent with the MgATP-bound Fe protein having a less complementary docking site with Fld. Fld and the Fe protein tryptic peptides identified in the time-resolved limited proteolysis experiments with highlighted peptides that change because of complex formation and/or the presence of nucleotides are listed in supplemental Table S1. Representative MALDI-TOF mass spectra are also shown for the digestion of Fld (supplemental Fig. S2) to further support peptide identification.
Figure 4.
Time-resolved limited proteolysis reactions display differences in interactions between Fld and the Fe protein based on the presence of nucleotide. A, differences in the proteolytic rates of the Fe protein with Fld depend on the presence of nucleotide for the proteolysis patterns analyzed by SDS-PAGE. B, limited proteolysis peptide mapping of the amino acid sequence of Fe protein shows tryptic cleavage sites (arrows) that report on interactions with Fld and/or nucleotide binding. These are the P-loop (Gly9-Ser16, boxed in red), the part of the switch I region that coordinates the bound Mg2+ of the nucleotide (Cys38-Asp43, boxed in black), the part of the switch I region that interacts with the MoFe protein (Glu59-Asp69, boxed in green), and Arg100 (arrow). These regions are mapped onto the Fe protein (tan, PDB code 1FP6) for structural context. C, Fld tryptic cleavage sites (arrows) that change because of interactions with both nucleotide-bound states of the Fe protein observed in limited proteolysis experiments are placed on the Fld amino acid sequence for structural context (tan, PDB code 1YOB; Ala1-Arg15 (boxed in orange), Thr90-Arg120 (boxed in cyan), and Phe146-Arg163 (boxed in magenta).
Figure 5.
Differences in cross-linking patterns are observed based on the presence or absence of nucleotides. A, differences in cross-linking patterns based on migration rate occur for the Fe protein in complex with Fld (arrows) and depend on the presence or absence of nucleotides as analyzed by SDS-PAGE. B and C, protein regions showing differences in the cross-linking pattern were mapped onto docking models between Fld (PDB code 1YOB) and the MgADP-bound Fe protein (PDB code 1FP6, B) and the Fe protein from the MgAMPPCP-stabilized nitrogenase complex (PDB code 4WZB, C) for structural context. Cross-linking occurred in protein regions around the [4Fe-4S] cluster for the Fe protein (Tyr80-Arg100, cyan) and the FMN cofactor of Fld (Thr90-Lys123, cyan), around residue Lys170 (blue), the P-loop (red), and both switch I regions (green and black) of the Fe protein. D, different orientations of the side chain for Fe protein residue Lys170, depending on the type of bound nucleotide presented on superimposed Fe protein structures from the nitrogenase complex stabilized with MgAMPPCP (tan, PDB code 4WZB) and MgADP-bound (light blue, PDB code 1FP6).
A final chemical cross-linking experiment was completed on the Fe protein—Fld complex with and without nucleotides. The presence of a sharper band above 50 kDa in the MgADP-bound state relative to the MgATP-bound state suggests that, in the presence of MgADP, a complex is formed more effectively (Fig. 5A) and is consistent with the docking studies. For both nucleotide-bound states, the observed cross-linking suggests interactions near the [4Fe-4S] cluster of the Fe protein (Fig. 5, B and C, cyan) and FMN cofactor of Fld (Thr90-Lys123; Fig. 5, B and C, cyan), consistent with the docking models. Mapping shows cross-linking in the P-loops and both switch I regions only with MgATP (Fig. 5C). However, with MgADP, another Fe protein domain involved in interactions with Fld around Lys170 (Fig. 5B, blue) is observed. In the MgADP-bound state, the side chains of Lys170 of both Fe protein subunits interact with Fld (Fig. 5D), whereas, in the MgATP-bound state, only one Lys170 side chain interacts with Fld (Fig. 5D). Therefore, mapping of proteolytic fragments identified more protein regions that are cross-linked between the MgADP-bound Fe protein and Fld. This is in agreement with the docking models and proteolysis patterns showing that the MgADP-bound state has a more complementary docking interface with Fld than the MgATP-bound state. Fld and the Fe protein tryptic and non-tryptic peptides observed to appear and disappear during chemical cross-linking experiments with highlighted peptides that change because of complex formation and/or the presence of nucleotides are listed in supplemental Table S2.
Conclusions
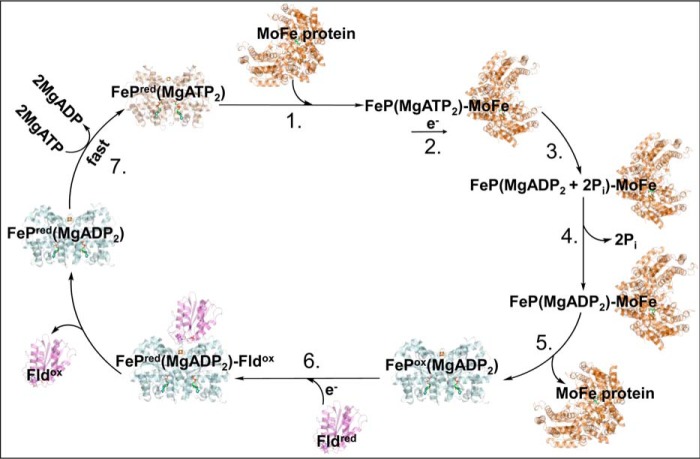
It has been reported that Fld, as a reductant, stimulates the reaction rate of the Fe protein cycle by ∼2-fold compared with sodium dithionite (25). This is consistent with the increased dissociation rate between the MgADP-bound Fe protein and MoFe protein (>760 s−1), rapid re-reduction of the nucleotide-bound Fe protein (>1200 s−1), and fast nucleotide exchange (70 s−1) compared with the relatively slow rate for release of Pi (25–27 s−1) with Fld as the source of reductant (25, 37). However, the rate of reduction for the oxidized Fe protein by Fld is fast regardless of which nucleotide is bound, and the oxidized Fe protein has similar affinities for both nucleotides (25, 38). These factors make it difficult to distinguish between the order of re-reduction for the Fe protein and nucleotide exchange for the final steps in the Fe protein cycle. A key result is that reduction of the MgADP-bound Fe protein, while bound to the MoFe protein, is very slow, indicating that dissociation is obligatory for reduction to occur with Fld (25, 39). This is important because it indicates that interactions between Fld, the nucleotide-bound Fe protein, and the MoFe protein are intrinsically controlled. Limited proteolysis and cross-linking patterns in combination with the docking models can be used to differentiate further patterns of control of interactions between the nucleotide-bound Fe protein and Fld. The decrease in observed Fld tryptic cleavage sites (increased protection) and different cross-linking patterns of Fld and the MgADP-bound Fe protein show that Fld has a more complementary docking site with the MgADP-bound state of the Fe protein. In contrast, the increase in proteolytic susceptibility (decreased protection) of Fld, absence of cross-links to the Fe protein near Lys170, and decrease in salt bridge interactions indicate that the Fld docking site is not as complementary to the MgATP-bound state. This difference in complementarity would result in less competition between the MoFe protein and Fld binding to the MgADP-bound Fe protein. An advantage of this situation is that the MgATP-bound Fe protein would be free to transiently associate with the MoFe protein, directing the flow of electron transfer toward substrate reduction. From this, a rational model of the Fe protein cycle can be proposed, where the MgATP-bound Fe protein delivers an electron to the MoFe protein, followed by MgATP hydrolysis, release of Pi, and complex dissociation between the MgADP-bound Fe protein and MoFe protein (Fig. 6). Fld then delivers an electron to the MgADP-bound Fe protein, followed by nucleotide exchange for MgATP to end the Fe protein cycle. This model is consistent with reports that demonstrate that re-reduction of the Fe protein should occur before nucleotide exchange because of the rapid re-reduction rate of the nucleotide-bound Fe protein by Fld (Fig. 6) (25, 37, 40). These results provide a deeper mechanistic understanding of the Fe protein cycle.
Figure 6.
Model of the Fe protein cycle with reduction preceding nucleotide exchange. 1, the MgATP-bound Fe protein (tan, PDB code 4WZB) transiently associates with one-half of the MoFe protein (orange, PDB code 2NIP). 2, the MgATP-bound Fe protein transfers an electron to the MoFe protein upon complex formation. 3, nucleotide hydrolysis of 2MgATP to 2MgADP and 2Pi occurs. 4, inorganic phosphate is released from the Fe protein (MgADP2)–MoFe protein complex. 5, the MoFe protein and MgADP-bound Fe protein dissociate. 6, Fld (violet, PDB code 1YOB) delivers an electron to the MgADP-bound Fe protein (light blue, PDB code 1FP6). 7, MgADP is exchanged for MgATP, and this induces structural changes in the MgATP-bound Fe protein to promote interactions with the MoFe protein.
Experimental procedures
General procedures
Argon and dinitrogen gases were passed through an activated copper catalyst to remove dioxygen contamination prior to use. Proteins and buffers were handled anaerobically in septum-sealed serum vials under an inert atmosphere (argon) on a Schlenk vacuum line or in an anaerobic glove box (Teledyne Analytical Instruments, MO-10-M, Hudson, NH). The transfers of gases and liquids were done with gastight syringes.
Purification of Fe protein and Fld from A. vinelandii
Growth, expression, and purification for both the Fe protein and Fld were carried out as described previously (25, 41).
Time-resolved limited proteolysis of Fe protein and Fld
Time-resolved limited proteolysis of Fld and Fe protein was carried out as described previously using a 1:1 ratio of Fe protein:Fld and a 1:1000 ratio of trypsin (Promega) to total protein (25). All reactions were carried out in sealed vials under an argon atmosphere to maintain anaerobic conditions. Samples were taken at 0, 5, 10, 20, 40, 60, and 240 min and quenched with 10% formic acid (Sigma). MgATP and MgADP were at final concentrations of 10 mm. Proteolysis reactions with MgATP were quenched by boiling the reactions for 5 min at 95 °C to avoid protein precipitation. Proteolysis patterns were analyzed by SDS-PAGE (4–20% linear gradient gel, Bio-Rad) and stained with Coomassie Brilliant Blue (Thermo Fisher). Tryptic peptides were identified with a maXis Impact QTOF instrument (Bruker Daltonics) interfaced with Dionex 3000 nano-HPLC (Thermo Fisher) and Autoflex III MALDI-TOF/TOF (Bruker Daltonics) mass spectrometers as described previously (25). Identification and mapping of the observed proteolytic cleavage sites were carried out using the Protein Analysis Worksheet (PAWS) software package (ProteoMetrics, LLC) and Peptide Shaker, as described previously (25). Identified limited proteolysis tryptic peptides for the Fe protein and Fld are listed in supplemental Table S1. Tryptic peptides with a confidence score reported by Peptide Shaker of ≥95% were considered for analysis. Representative MALDI-TOF spectra for Fe protein and Fld tryptic peptides are shown in supplemental Figs. S1 and S2.
In silico docking study between Fld and the Fe protein in its different nucleotide states
In silico protein–protein docking simulations were performed using the computational docking program ClusPro 2.0 to derive the structural models as described previously (25). Fe protein and Fld (flavodoxin II, PDB code 1YOB) were used in each case as the ligand and the Fe protein in three states: nucleotide-free (PDB code 2NIP), MgADP-bound (PDB code 1FP6), and nitrogenase complex stabilized with MgAMPPCP (PDB code 4WZB) were used as the respective receptors. The difference in molecular surface (solvent-excluded surface) between MgATP/MgADP-bound Fe protein alone and in complex with flavodoxin was calculated in Chimera, implementing the Lee–Richards molecular surface definition (42), using a water molecule as a probe of a radius of 1.4 Å.
Chemical cross-linking
Chemical cross-linking and LC-MS analysis were performed as described previously (25). Briefly, Fe protein at 20 μm concentration was exposed to 10 mm MgATP (Sigma) or MgADP (Sigma), mixed with Fld in a 1:1 molar ratio, and immediately reacted with 10 mm glutaraldehyde (Sigma). Reactions were quenched with 100 mm Tris buffer (pH 8) after 10 min. All control reactions were set up in the same fashion. Cross-linked Fe protein samples were separated on SDS-PAGE (4–20% linear gradient gel, Bio-Rad) and stained with Coomassie Brilliant Blue (Thermo Fisher). Protein bands of interest were excised from the gel and digested with a trypsin/chymotrypsin (Promega) mixture. Proteolyzed peptides were identified using a maXis Impact QTOF instrument (Bruker Daltonics) interfaced with a Dionex 3000 nano-HPLC (Thermo Fisher), followed by data analysis in Peptide Shaker. Identified cross-linking tryptic and non-tryptic peptides for the Fe protein and Fld are listed in supplemental Table S2. Tryptic peptides with a confidence score reported by Peptide Shaker of ≥95% were considered for analysis. All reactions were carried out under strictly anaerobic conditions and in primary amine-free buffer at room temperature.
Author contributions
Purification of Fe protein and Fld was performed by N. P., R. N. L., and Z. Y. Y. Time-resolved limited proteolysis experiments, MALDI-TOF data collection and analysis, LC-MS analysis, and chemical cross-linking experiments were performed by M. T. L. and N. P. Docking calculations were performed by N. P. Data interpretation was conducted by N. P., M. T. L., L. C. S., B. B., and J. W. P. All coauthors contributed to manuscript preparation.
Supplementary Material
Acknowledgments
We thank the Montana State University Microfabrication Facility for help with the preparation of gold-coated borosilica capillaries for non-covalent mass spectrometry. The Mass Spectrometry Facility at Montana State University is supported in part by the Murdock Charitable Trust and National Institutes of Health Institutional Development Award (IDeA) Program Grant P20GM103474.
This work was supported as part of the Biological Electron Transfer and Catalysis Energy Frontier Research Center funded by the Department of Energy, Office of Science, Basic Energy Sciences under Award DE-SC0012518. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Figs. S1 and S2 and Tables S1 and S2.
- Fld
- flavodoxin.
References
- 1. Hoffman B. M., Lukoyanov D., Yang Z. Y., Dean D. R., and Seefeldt L. C. (2014) Mechanism of nitrogen fixation by nitrogenase: the next stage. Chem. Rev. 114, 4041–4062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Seefeldt L. C., Hoffman B. M., and Dean D. R. (2009) Mechanism of Mo-dependent nitrogenase. Annu. Rev. Biochem. 78, 701–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Burgess B. K., and Lowe D. J. (1996) Mechanism of molybdenum nitrogenase. Chem. Rev. 96, 2983–3012 [DOI] [PubMed] [Google Scholar]
- 4. Hageman R. V., and Burris R. H. (1978) Nitrogenase and nitrogenase reductase associate and dissociate with each catalytic cycle. Proc. Natl. Acad. Sci. U.S.A. 75, 2699–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Spatzal T., Aksoyoglu M., Zhang L., Andrade S. L., Schleicher E., Weber S., Rees D. C., and Einsle O. (2011) Evidence for interstitial carbon in nitrogenase FeMo cofactor. Science 334, 940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lancaster K. M., Roemelt M., Ettenhuber P., Hu Y., Ribbe M. W., Neese F., Bergmann U., and DeBeer S. (2011) X-ray emission spectroscopy evidences a central carbon in the nitrogenase iron-molybdenum cofactor. Science 334, 974–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Einsle O., Tezcan F. A., Andrade S. L., Schmid B., Yoshida M., Howard J. B., and Rees D. C. (2002) Nitrogenase MoFe-protein at 1.16 A resolution: a central ligand in the FeMo-cofactor. Science 297, 1696–1700 [DOI] [PubMed] [Google Scholar]
- 8. Chan M. K., Kim J., and Rees D. C. (1993) The nitrogenase FeMo-cofactor and P-cluster pair: 2.2 A resolution structures. Science 260, 792–794 [DOI] [PubMed] [Google Scholar]
- 9. Kim J., and Rees D. C. (1992) Structural models for the metal centers in the nitrogenase molybdenum-iron protein. Science 257, 1677–1682 [DOI] [PubMed] [Google Scholar]
- 10. Sen S., Krishnakumar A., McClead J., Johnson M. K., Seefeldt L. C., Szilagyi R. K., and Peters J. W. (2006) Insights into the role of nucleotide-dependent conformational change in nitrogenase catalysis: structural characterization of the nitrogenase Fe protein Leu127 deletion variant with bound MgATP. J. Inorg. Biochem. 100, 1041–1052 [DOI] [PubMed] [Google Scholar]
- 11. Sen S., Igarashi R., Smith A., Johnson M. K., Seefeldt L. C., and Peters J. W. (2004) A conformational mimic of the MgATP-bound “on state” of the nitrogenase iron protein. Biochemistry 43, 1787–1797 [DOI] [PubMed] [Google Scholar]
- 12. Chiu H., Peters J. W., Lanzilotta W. N., Ryle M. J., Seefeldt L. C., Howard J. B., and Rees D. C. (2001) MgATP-Bound and nucleotide-free structures of a nitrogenase protein complex between the Leu 127 δ-Fe-protein and the MoFe-protein. Biochemistry 40, 641–650 [DOI] [PubMed] [Google Scholar]
- 13. Jang S. B., Seefeldt L. C., and Peters J. W. (2000) Insights into nucleotide signal transduction in nitrogenase: structure of an iron protein with MgADP bound. Biochemistry 39, 14745–14752 [DOI] [PubMed] [Google Scholar]
- 14. Seefeldt L. C., and Mortenson L. E. (1993) Increasing nitrogenase catalytic efficiency for MgATP by changing serine 16 of its Fe protein to threonine: use of Mn2+ to show interaction of serine 16 with Mg2+. Protein Sci. 2, 93–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schlessman J. L., Woo D., Joshua-Tor L., Howard J. B., and Rees D. C. (1998) Conformational variability in structures of the nitrogenase iron proteins from Azotobacter vinelandii and Clostridium pasteurianum. J. Mol. Biol. 280, 669–685 [DOI] [PubMed] [Google Scholar]
- 16. Bursey E. H., and Burgess B. K. (1998) Characterization of a variant iron protein of nitrogenase that is impaired in its ability to adopt the MgATP-induced conformational change. J. Biol. Chem. 273, 16927–16934 [DOI] [PubMed] [Google Scholar]
- 17. Schindelin H., Kisker C., Schlessman J. L., Howard J. B., and Rees D. C. (1997) Structure of ADP x AIF4(-)-stabilized nitrogenase complex and its implications for signal transduction. Nature 387, 370–376 [DOI] [PubMed] [Google Scholar]
- 18. Seefeldt L. C., Morgan T. V., Dean D. R., and Mortenson L. E. (1992) Mapping the site(s) of MgATP and MgADP interaction with the nitrogenase of Azotobacter vinelandii: lysine 15 of the iron protein plays a major role in MgATP interaction. J. Biol. Chem. 267, 6680–6688 [PubMed] [Google Scholar]
- 19. Seefeldt L. C., Hoffman B. M., and Dean D. R. (2012) Electron transfer in nitrogenase catalysis. Curr. Opin. Chem. Biol. 16, 19–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hageman R. V., Orme-Johnson W. H., and Burris R. H. (1980) Role of magnesium adenosine 5`-triphosphate in the hydrogen evolution reaction catalyzed by nitrogenase from Azotobacter vinelandii. Biochemistry 19, 2333–2342 [DOI] [PubMed] [Google Scholar]
- 21. Thorneley R. N., Lowe D. J., Eday R. R., and Miller R. W. (1979) The coupling of electron transfer in nitrogenase to the hydrolysis of magnesium adenosine triphosphate. Biochem. Soc. Trans. 7, 633–636 [DOI] [PubMed] [Google Scholar]
- 22. Danyal K., Dean D. R., Hoffman B. M., and Seefeldt L. C. (2011) Electron transfer within nitrogenase: evidence for a deficit-spending mechanism. Biochemistry 50, 9255–9263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Danyal K., Mayweather D., Dean D. R., Seefeldt L. C., and Hoffman B. M. (2010) Conformational gating of electron transfer from the nitrogenase Fe protein to MoFe protein. J. Am. Chem. Soc. 132, 6894–6895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Duval S., Danyal K., Shaw S., Lytle A. K., Dean D. R., Hoffman B. M., Antony E., and Seefeldt L. C. (2013) Electron transfer precedes ATP hydrolysis during nitrogenase catalysis. Proc. Natl. Acad. Sci. U.S.A. 110, 16414–16419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang Z. Y., Ledbetter R., Shaw S., Pence N., Tokmina-Lukaszewska M., Eilers B., Guo Q., Pokhrel N., Cash V. L., Dean D. R., Antony E., Bothner B., Peters J. W., and Seefeldt L. C. (2016) Evidence that the π release event is the rate-limiting step in the nitrogenase catalytic cycle. Biochemistry 55, 3625–3635 [DOI] [PubMed] [Google Scholar]
- 26. Wilson P. E., Nyborg A. C., and Watt G. D. (2001) Duplication and extension of the Thorneley and Lowe kinetic model for Klebsiella pneumoniae nitrogenase catalysis using a MATHEMATICA software platform. Biophys. Chem. 91, 281–304 [DOI] [PubMed] [Google Scholar]
- 27. Thorneley R. N., and Lowe D. J. (1985) in Molybdenum Enzymes (Spiro T. G., ed.) pp 221–284, Wiley, New York [Google Scholar]
- 28. Ryle M. J., Lanzilotta W. N., Seefeldt L. C., Scarrow R. C., and Jensen G. M. (1996) Circular dichroism and x-ray spectroscopies of Azotobacter vinelandii nitrogenase iron protein. MgATP and MgADP induced protein conformational changes affecting the [4Fe-4S] cluster and characterization of a [2Fe-2S] form. J. Biol. Chem. 271, 1551–1557 [DOI] [PubMed] [Google Scholar]
- 29. Ryle M. J., Lanzilotta W. N., and Seefeldt L. C. (1996) Elucidating the mechanism of nucleotide-dependent changes in the redox potential of the [4Fe-4S] cluster in nitrogenase iron protein: the role of phenylalanine 135. Biochemistry 35, 9424–9434 [DOI] [PubMed] [Google Scholar]
- 30. Ryle M. J., Lanzilotta W. N., Mortenson L. E., Watt G. D., and Seefeldt L. C. (1995) Evidence for a central role of lysine 15 of Azotobacter vinelandii nitrogenase iron protein in nucleotide binding and protein conformational changes. J. Biol. Chem. 270, 13112–13117 [DOI] [PubMed] [Google Scholar]
- 31. Lanzilotta W. N., Holz R. C., and Seefeldt L. C. (1995) Proton NMR investigation of the [4Fe–4S]1+ cluster environment of nitrogenase iron protein from Azotobacter vinelandii: defining nucleotide-induced conformational changes. Biochemistry 34, 15646–15653 [DOI] [PubMed] [Google Scholar]
- 32. Tezcan F. A., Kaiser J. T., Mustafi D., Walton M. Y., Howard J. B., and Rees D. C. (2005) Nitrogenase complexes: multiple docking sites for a nucleotide switch protein. Science 309, 1377–1380 [DOI] [PubMed] [Google Scholar]
- 33. Hilmer J. K., Zlotnick A., and Bothner B. (2008) Conformational equilibria and rates of localized motion within hepatitis B virus capsids. J. Mol. Biol. 375, 581–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bothner B., Dong X. F., Bibbs L., Johnson J. E., and Siuzdak G. (1998) Evidence of viral capsid dynamics using limited proteolysis and mass spectrometry. J. Biol. Chem. 273, 673–676 [DOI] [PubMed] [Google Scholar]
- 35. Wolle D., Kim C., Dean D., and Howard J. B. (1992) Ionic interactions in the nitrogenase complex: properties of Fe protein containing substitutions for Arg-100. J. Biol. Chem. 267, 3667–3673 [PubMed] [Google Scholar]
- 36. Seefeldt L. C. (1994) Docking of nitrogenase iron- and molybdenum-iron proteins for electron transfer and MgATP hydrolysis: the role of arginine 140 and lysine 143 of the Azotobacter vinelandii iron protein. Protein Sci. 3, 2073–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Duyvis M. G., Wassink H., and Haaker H. (1998) Nitrogenase of Azotobacter vinelandii: kinetic analysis of the Fe protein redox cycle. Biochemistry 37, 17345–17354 [DOI] [PubMed] [Google Scholar]
- 38. Lanzilotta W. N., Parker V. D., and Seefeldt L. C. (1999) Thermodynamics of nucleotide interactions with the Azotobacter vinelandii nitrogenase iron protein. Biochim. Biophys. Acta 1429, 411–421 [DOI] [PubMed] [Google Scholar]
- 39. Thorneley R. N., and Lowe D. J. (1983) Nitrogenase of Klebsiella pneumoniae: kinetics of the dissociation of oxidized iron protein from molybdenum-iron protein: identification of the rate-limiting step for substrate reduction. Biochem. J. 215, 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Thorneley R. N., and Deistung J. (1988) Electron-transfer studies involving flavodoxin and a natural redox partner, the iron protein of nitrogenase: conformational constraints on protein-protein interactions and the kinetics of electron transfer within the protein complex. Biochem. J. 253, 587–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Christiansen J., Goodwin P. J., Lanzilotta W. N., Seefeldt L. C., and Dean D. R. (1998) Catalytic and biophysical properties of a nitrogenase Apo-MoFe protein produced by a nifB-deletion mutant of Azotobacter vinelandii. Biochemistry 37, 12611–12623 [DOI] [PubMed] [Google Scholar]
- 42. Lee B., and Richards F. M. (1971) The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol. 55, 379–400 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.