Abstract
To prolong residence on their hosts, ticks secrete many salivary factors that target host defense molecules. In particular, the tick Rhipicephalus sanguineus has been shown to produce three salivary glycoproteins named “evasins,” which bind to host chemokines, thereby inhibiting the recruitment of leukocytes to the location of the tick bite. Using sequence similarity searches, we have identified 257 new putative evasin sequences encoded by the genomes or salivary or visceral transcriptomes of numerous hard ticks, spanning the genera Rhipicephalus, Amblyomma, and Ixodes of the Ixodidae family. Nine representative sequences were successfully expressed in Escherichia coli, and eight of the nine candidates exhibited high-affinity binding to human chemokines. Sequence alignments enabled classification of the evasins into two subfamilies: C8 evasins share a conserved set of eight Cys residues (four disulfide bonds), whereas C6 evasins have only three of these disulfide bonds. Most of the identified sequences contain predicted secretion leader sequences, N-linked glycosylation sites, and a putative site of tyrosine sulfation. We conclude that chemokine-binding evasin proteins are widely expressed among tick species of the Ixodidae family, are likely to play important roles in subverting host defenses, and constitute a valuable pool of anti-inflammatory proteins for potential future therapeutic applications.
Keywords: bioinformatics, chemokine, fluorescence anisotropy, host-pathogen interaction, inhibitor, recombinant protein expression, evasin, tick protein
Introduction
Ticks are hematophagous arachnids that parasitize humans, livestock, and both domestic and wild animals. Many ticks are vectors of pathogenic microorganisms that cause disease when transmitted to the host, including anaplasmosis, ehrlichiosis, babesiosis, and Lyme disease (1–3). In particular, several species of hard tick (family Ixodidae) from the genera Amblyomma, Ixodes, and Rhipicephalus have been studied extensively due to their medical and veterinary significance (4).
Mammals respond to tick bites and other injuries or infections by mounting a complex inflammatory response, a key feature of which is the recruitment of circulating leukocytes to the site of the affected tissues. Leukocyte recruitment is mediated by chemokine proteins, which are secreted into the vasculature at the site of the injury. There, they bind to chemokine receptors on circulating leukocytes and thereby stimulate leukocyte trafficking to the damaged tissue (5, 6). Humans and other mammals produce an array of chemokines and chemokine receptors, which collectively orchestrate the migration of different types of leukocytes in response to different inflammatory stimuli (7, 8).
As an evolutionary strategy to prolong blood feeding and survival, ticks have developed an extensive repertoire of secreted, biologically active salivary factors that compromise host defenses, including vasodilators, blood coagulation inhibitors, and immunomodulatory proteins (9–12). In particular, the brown dog tick (Rhipicephalus sanguineus) has been found to produce three salivary glycoproteins, named “evasins,” which are secreted during a blood meal and bind to host chemokines, thus inhibiting the host inflammatory response (13, 14). Evasin-1 and evasin-4 are homologous with 27% amino acid sequence identity and a conserved core of eight Cys residues that form four disulfide bonds (15, 16). Evasin-3 has an unrelated sequence (16). The gene encoding a fourth protein, evasin-2, has reportedly been cloned but has not been described in detail (16). In addition, five transcripts encoding proteins with sequence similarity to either evasin-1 and -4 or evasin-3 have been reported in the salivary gland transcriptome of R. sanguineus (17). Each evasin characterized to date binds with high affinity to a different but overlapping set of chemokines, thus inhibiting their receptor binding and activation (15, 16, 18).
Prior to the current study, there have been no evasins characterized from other tick species. However, the salivary gland transcriptomes of some tick species have been reported to encode evasin homologues, including several in the genera Amblyomma (Amblyomma americanum (19, 20), Amblyomma cajennense, Amblyomma parvum, Amblyomma triste (21), and Amblyomma maculatum (22)) and Rhipicephalus (Rhipicephalus pulchellus (23) and Rhipicephalus appendiculatus (24)). Furthermore, saliva from several species of Ixodidae contains chemokine-binding activity suggested to be similar to that of evasin-3 (25). Thus, it is likely that evasins are ubiquitously expressed by a wide variety of tick species.
In addition to their functions in host defense, the interactions of chemokines with their receptors also contribute to excessive leukocyte recruitment in many inflammatory diseases (7, 8). Thus, chemokine receptors and to a lesser extent chemokines themselves are popular targets for the development of anti-inflammatory therapeutics (26, 27). To this end, Proudfoot and co-workers (16, 28–30) have demonstrated that R. sanguineus evasins are efficacious anti-inflammatory agents in animal models of psoriasis (16), lung injury (16, 28), colitis (29), atherosclerosis (30), and ischemic stroke (30).
Motivated by the potential of evasins to inhibit chemokine-mediated inflammation in other diseases, we have explored tick genomic and transcriptomic databases to identify homologues of evasin-1 and evasin-4. Here we describe the identification of more than 250 putative tick evasins, spanning three genera of hard ticks (Rhipicephalus, Amblyomma, and Ixodes), and the validation of representative members of this protein family as binders and inhibitors of human chemokines. We discuss the implications of our findings for evasin evolution and for future development of chemokine-targeted anti-inflammatory therapeutics.
Results
Identification of evasin candidates from gene and transcript databases
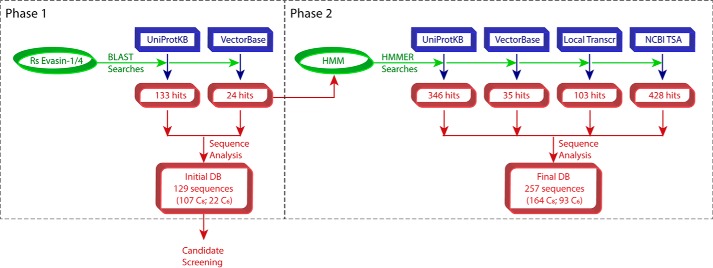
To identify predicted tick proteins with high sequence identity to R. sanguineus evasin-1 and -4, we initially searched the UniProtKB protein sequence database as well as the tick genomic and transcriptomic data in VectorBase (Fig. 1, Phase 1). UniProtKB searches yielded 133 protein sequences with 9–42% sequence identity to R. sanguineus evasin-1 and -4. These sequences were derived from one Rhipicephalus species (R. pulchellus) and five Amblyomma species (A. americanum, A. cajennense, A. maculatum, A. parvum, and A. triste). VectorBase searches yielded 24 sequences from the species Ixodes ricinus and one putative evasin from Ixodes scapularis with relatively low (9–20%) sequence identity to R. sanguineus evasin-1 and -4. To identify additional evasin candidates, we used the aligned sequences of the initial hits to generate a hidden Markov model (HMM)4 with which we then interrogated four databases: the UniProtKB protein sequence database; the genomic, transcriptomic, and peptide data in VectorBase; data from the NCBI Transcriptome Shotgun Sequence Assembly Database; and salivary gland and viscera transcriptomes from the cattle tick Rhipicephalus microplus and the Australian paralysis tick Ixodes holocyclus (Fig. 1, Phase 2). In combination with the initial search results, these searches identified a total of 428 sequences, which were curated for conserved features (see below) to yield a final database of 257 putative evasins (accession numbers are listed in supplemental Table S1) with 9–46% sequence identity to R. sanguineus evasin-1 and -4.
Figure 1.
Schematic workflow for identification of evasins by sequence similarity searches. DB, database; Local Transcr, locally obtained I. holocyclus and R. microplus transcriptome data sets; Rs, R. sanguineus; TSA, Transcriptome Shotgun Assembly.
Conserved features of evasin sequences: C8 and C6 subfamilies
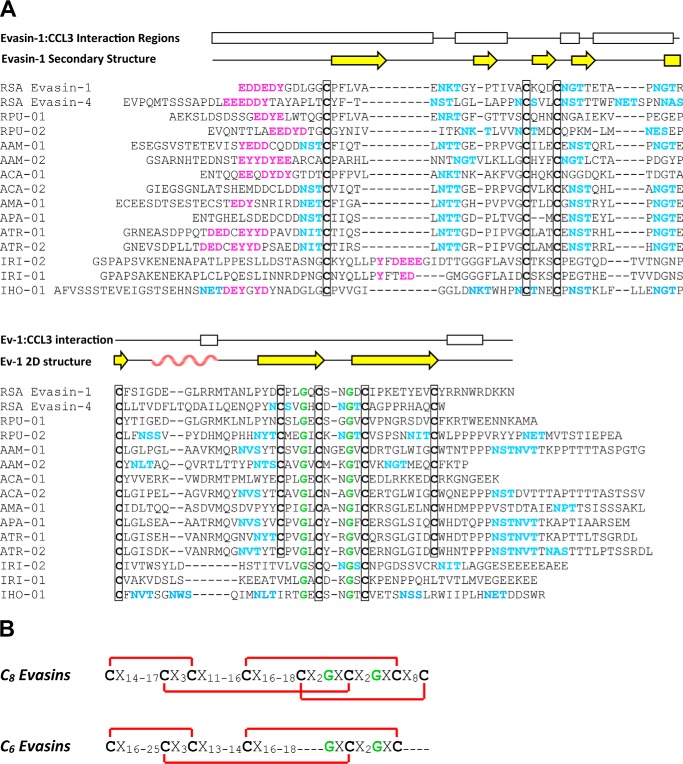
R. sanguineus evasin-1 and -4 contain several sequence features that are expected to be conserved in orthologous proteins. Eight conserved Cys residues form four disulfide bonds in the structure of evasin-1, most of which should be retained in other evasins. Because evasins are secreted proteins, N-terminal signal sequences are also anticipated to be present. In addition, R. sanguineus evasin-1 and -4 contain three conserved potential N-linked glycosylation sites (evasin-4 contains four additional sites) and have previously been shown to be glycosylated when expressed in both HEK293 cells and TN5 insect cells (15). Finally, the N-terminal regions of both evasin-1 and -4 contain tyrosine residues adjacent to several acidic residues (sequences EDDEDY and EEEDDY, respectively), which we identified as likely to be post-translationally sulfated (12, 31, 32). This feature of evasin sequences has not been identified previously.
All of these sequence features are present in the majority of putative evasin sequences retrieved in the above searches as illustrated for 13 selected sequences in Fig. 2. Although the pattern of Cys residues was completely conserved in putative evasin sequences from Rhipicephalus and Amblyomma species, all Ixodes sequences were missing the fifth and/or eighth Cys, which are disulfide-bonded to each other in evasin-1. Interestingly, the Ixodes sequences retain the putative tyrosine sulfation site, but it is located after the first conserved Cys in these sequences in contrast to being before the first conserved Cys in the Rhipicephalus and Amblyomma sequences. Although the effects of these sequence differences on evasin structure and function remain to be determined, the sequence comparisons led us to propose that there are two distinct subfamilies of evasins homologous to R. sanguineus evasin-1 and -4, namely the C8 evasins (found in Rhipicephalus and Amblyomma) and the C6 evasins (found in Ixodes species). Both subfamilies are distinct from the evasin-3 type evasins.
Figure 2.
Features of the evasin protein family. A, multiple sequence alignment (obtained using MUSCLE) of selected evasin candidates with R. sanguineus evasin-1 and -4. Features indicated are conserved cysteines (bold and boxed) and glycines (green), potential N-linked glycosylation sites (cyan), and putative tyrosine sulfation sites (magenta). The secondary structure and regions of evasin-1 that interact with CCL3 in the crystal structure are shown above the alignment; yellow arrows indicate β-strands and the rose wavy line indicates the α-helix. Abbreviations use for tick species are: RSA, R. sanguineus; RPU, R. pulchellus; AAM, A. americanum; ACA, A. cajennense; AMA, A. maculatum; APA, A. parvum; ATR, A. triste; IRI, I. ricinus; IHO, I. holocyclus. B, conserved sequence motifs of the C8 and C6 evasins showing (in red) the disulfide bond connectivity observed in the structure of R. sanguineus evasin-1.
Bacterial expression of evasins
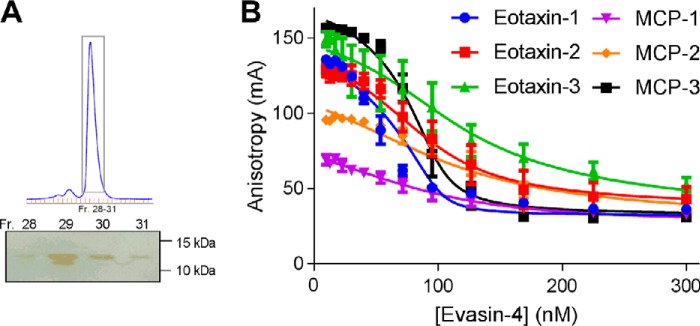
Previous studies of recombinant evasin-1 and -4 have used mammalian or insect cell expression systems. To evaluate whether evasins expressed in a bacterial system would be folded and functional, we expressed N-terminally His6-tagged R. sanguineus evasin-1 and -4 in Escherichia coli. Evasin-4 was expressed in inclusion bodies but could be renatured under disulfide exchange conditions and purified chromatographically (Fig. 3A). Evasin-1 was expressed poorly and was not pursued further.
Figure 3.
Purification and characterization of recombinant R. sanguineus evasin-4. A, size exclusion chromatogram for purification of evasin-4 (after His6 tag removal) with non-reducing SDS-PAGE of fractions (Fr.) spanning the main peak (boxed on the chromatogram); the molecular weight marker used was Bio-Rad Precision Plus ProteinTM unstained standards. B, fluorescence anisotropy-binding curves showing displacement of a fluorescent CCR2-derived sulfopeptide from each of six chemokines using purified recombinant evasin-4. Data plotted are the averages of three independent experiments, each recorded in duplicate, and error bars represent the S.E. Solid lines are fitted binding displacement curves.
Binding of purified evasin-4 to each of several chemokines was detected using a competitive fluorescence anisotropy-binding assay (Fig. 3B) (33). In this assay, a fluorescein-labeled peptide derived from the N-terminal region of a chemokine receptor is displaced from the chemokine, resulting in decreased fluorescence anisotropy with increased concentration of evasin. As shown by simulated displacement curves (supplemental Fig. S1), the shape of the curve is indicative of the equilibrium dissociation constant (Kd) between evasin and chemokine, although the concentration of evasin required for 50% displacement of the fluorescent peptide is substantially higher than the Kd; this difference is related to the concentrations of the chemokine and fluorescent peptide used in the assay and their affinity for each other. We observed that His6-tagged evasin-4 bound to each of six chemokines tested with Kd values ranging from ∼1 to ∼20 nm (Fig. 3B and Table 1). These results indicated that glycosylation and/or tyrosine sulfation is not essential for chemokine binding, consistent with previous results showing that deglycosylation of evasin-1 did not affect activity (15). We concluded that the E. coli system was appropriate for screening novel evasin candidates.
Table 1.
Affinities for binding of purified evasins to human CC chemokines
Affinities are reported as pKd values (−log10 of the Kd; in m) ±S.E. The corresponding Kd values (in nm) are in parentheses. The affinity of RSA evasin-4 for MCP-3 (not shown in the table) was pKd = 9.21 ± 0.2 (Kd = 0.62 nm).
| Evasin | Chemokine |
||||
|---|---|---|---|---|---|
| Eotaxin-1 | Eotaxin-2 | Eotaxin-3 | MCP-1 | MCP-2 | |
| RSA evasin-4 | 9.25 ± 0.20 (0.57) | 8.15 ± 0.10 (7.1) | 8.00 ± 0.10 (10) | 7.71 ± 0.09 (20) | 7.68 ± 0.07 (21) |
| RPU-01 | 7.88 ± 0.08 (13) | 6.81 ± 0.05 (150) | 7.44 ± 0.04 (36) | 7.39 ± 0.07 (40) | 7.78 ± 0.06 (17) |
| ACA-01 | 7.94 ± 0.07 (12) | 6.05 ± 0.07 (890) | 6.16 ± 0.05 (690) | 6.45 ± 0.04 (360) | 8.09 ± 0.10 (8.1) |
| IRI-01 | 6.76 ± 0.06 (170) | a | 6.46 ± 0.04 (350) | 5.95 ± 0.05 (1100) | 5.97 ± 0.04 (1100) |
| IHO-01 | 6.90 ± 0.06 (130) | 6.48 ± 0.05 (330) | 7.08 ± 0.03 (83) | 6.04 ± 0.08 (920) | 6.10 ± 0.06 (800) |
a Kd >1 mm (pKd + S.E. <6).
To determine whether the evasin candidates identified above are able to bind chemokines, we expressed 12 selected sequences in E. coli, two each from the species R. pulchellus, A. americanum, A. cajennense, A. triste, and I. ricinus and one each from A. maculatum and A. parvum; evasin candidates are identified by the names listed in Fig. 2 (e.g. ACA-01 and ACA-02 for the two candidates from A. cajennense). Initial expression trials indicated that the majority of candidates were sequestered in inclusion bodies, which were therefore harvested and denatured to allow the His6-tagged proteins to be partially purified. After renaturation by rapid dilution and subsequent immobilized metal affinity purification, nine of the 12 candidates (all except APA-01, ATR-01, and IRI-02) were isolated in sufficient quantities to screen for chemokine binding.
Evasin candidates bind to chemokines
The competitive fluorescence anisotropy-binding assay was used to assess the binding of the nine isolated evasin candidates to each of five CC chemokines previously expressed and purified in our laboratory (supplemental Fig. S2). With the exception of ACA-02 (from A. cajennense), all evasin candidates bound to at least one of the chemokines tested with Kd values of <50 nm (Table 2). We conclude that these eight proteins are chemokine-binding evasins and, by extension, that the majority of evasin candidates identified in our sequence-based searches are also likely to be chemokine binders.
Table 2.
Affinities for binding of nine evasin candidates to six human CC chemokines
Affinities are reported as pKd values (−log10 of the Kd; in m) ±S.E. The corresponding Kd values (in nm) are in parentheses.
| Evasin | Chemokine |
|||||
|---|---|---|---|---|---|---|
| Eotaxin-1 | Eotaxin-2 | Eotaxin-3 | MCP-1 | MCP-2 | MCP-3 | |
| RPU-01 | 7.26 ± 0.09 (55) | 7.06 ± 0.09 (86) | 7.28 ± 0.10 (53) | 6.74 ± 0.08 (180) | 7.11 ± 0.10 (77) | 7.70 ± 0.10 (20) |
| RPU-02 | 7.33 ± 0.06 (74) | 6.88 ± 0.09 (130) | 7.63 ± 0.09 (23) | 6.33 ± 0.04 (470) | 6.27 ± 0.10 (540) | 7.09 ± 0.06 (81) |
| AAM-01 | 7.66 ± 0.03 (22) | 6.88 ± 0.04 (130) | 7.73 ± 0.05 (19) | 6.06 ± 0.10 (870) | 5.94 ± 0.09 (1100) | 6.99 ± 0.04 (100) |
| AAM-02 | 7.72 ± 0.09 (19) | 6.87 ± 0.09 (130) | 7.77 ± 0.07 (170) | 5.99 ± 0.10 (1000) | a | 6.75 ± 0.05 (180) |
| ACA-01 | 7.93 ± 0.10 (12) | 7.16 ± 0.09 (70) | 7.26 ± 0.10 (55) | 6.79 ± 0.07 (160) | 8.24 ± 0.10 (5.8) | 7.76 ± 0.05 (17) |
| ACA-02 | 6.73 ± 0.03 (180) | 5.94 ± 0.10 (1100) | 6.65 ± 0.04 (220) | a | a | a |
| AMA-01 | 7.60 ± 0.05 (25) | 6.94 ± 0.05 (120) | 7.66 ± 0.05 (22) | 6.38 ± 0.10 (420) | 6.33 ± 0.06 (470) | 6.97 ± 0.03 (110) |
| ATR-02 | 7.41 ± 0.09 (39) | 6.87 ± 0.09 (140) | 7.35 ± 0.07 (44) | 5.99 ± 0.20 (1000) | 6.22 ± 0.08 (610) | 7.01 ± 0.07 (98) |
| IRI-01 | 7.28 ± 0.10 (53) | 6.95 ± 0.10 (110) | 6.80 ± 0.10 (160) | 6.70 ± 0.06 (200) | 6.85 ± 0.09 (140) | 7.39 ± 0.09 (41) |
a Kd >1 mm (pKd + S.E. <6) or binding not detected.
We selected one newly identified evasin from each genus (ACA-01, RPU-01, and IRI-01) and an additional putative evasin (IHO-01; Fig. 2) from the Australian paralysis tick I. holocyclus for detailed comparison with R. sanguineus evasin-4 in chemokine-binding and inhibition assays. These proteins were expressed, refolded, and purified by size exclusion chromatography (Fig. 4). Non-reducing SDS-PAGE (Fig. 4) indicated that the purified proteins did not contain intermolecular disulfide bonding, suggesting that they were predominantly correctly folded. These four evasins bound to five CC chemokines tested (Fig. 4) with the Kd values listed in Table 1). Notably, the two C6 evasins (from Ixodes species) bind with lower affinity than the C8 evasins (from Rhipicephalus and Amblyomma species) to the chemokines tested here. This may be an indication of differences in target chemokine selectivity, the requirement for post-translational modifications, or simply lower intrinsic affinity.
Figure 4.
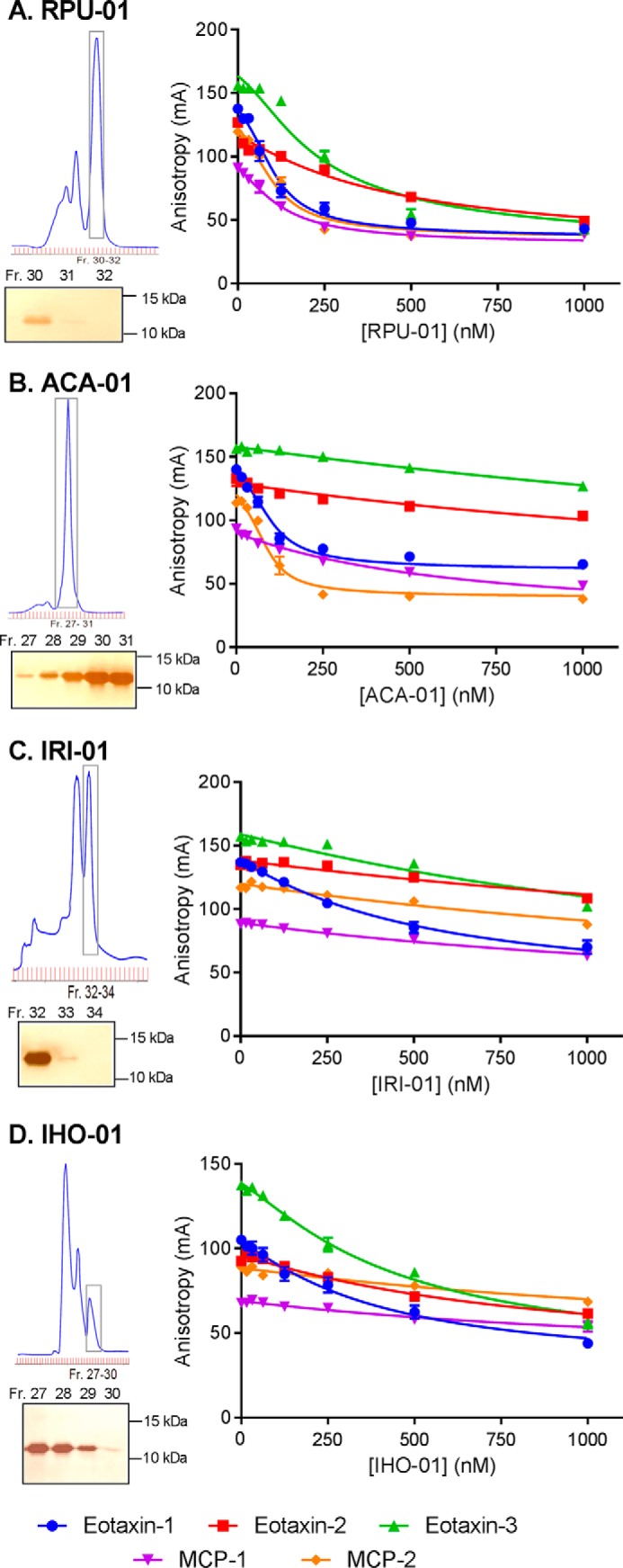
Purification and characterization of representative evasins from three genera. For evasins, ACA-01 from A. cajennense (A), RPU-01 from R. pulchellus (B), IRI-01 from I. ricinus (C), and IHO-01 from I. holocyclus (D) are shown. Top left, the size exclusion chromatogram for purification of the evasin protein (with a C-terminal His6 tag); bottom left, non-reducing SDS-PAGE of fractions (Fr.) spanning the main peak (boxed on the chromatogram); right, competitive fluorescence anisotropy curves for binding of the purified evasin to each of five CC chemokines. The molecular weight marker used for SDS-PAGE was Bio-Rad Precision Plus Protein unstained standards. Binding data represent the average ± S.E. (error bars) of values from three independent experiments, each recorded in duplicate. Solid lines are fitted binding displacement curves.
Evasin candidates inhibit chemokine activity
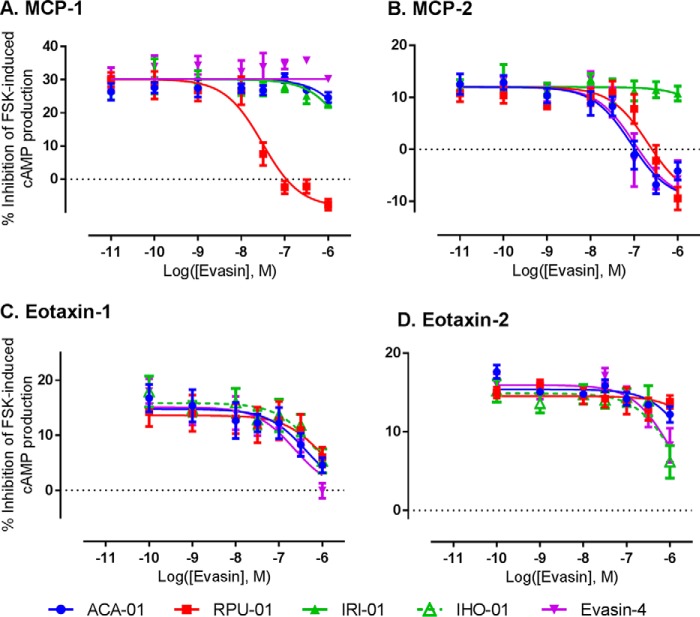
Chemokines activate their receptors to initiate a variety of G protein-dependent (e.g. inhibition of cAMP synthesis) and G protein-independent (e.g. β-arrestin recruitment) signaling pathways. To determine whether the newly identified evasins inhibit chemokine signaling via their cognate receptors, we determined the effects of evasins on the inhibition of cAMP production induced by treatment of CCR2-expressing HEK293 cells with the chemokines monocyte chemoattractant protein-1 (MCP-1/CCL2) and MCP-2/CCL8 or by treatment of CCR3-expressing HEK293 cells with the chemokines eotaxin-1/CCL11 and eotaxin-2/CCL24. As shown in Fig. 5 and Table 3, three of the novel evasins (RPU-01, ACA-01, and IHO-01) were able to inhibit the activity of one or more chemokine tested, although again the evasin from the Ixodes species exhibited relatively low potency. Notably, the selectivities for chemokine inhibition clearly differ among the evasins, suggesting that it may be possible to identify or engineer evasins for desired inhibitory selectivity against target chemokines.
Figure 5.
Inhibition of chemokine activity by purified evasins. Shown are concentration-response curves for inhibition of the chemokines MCP-1 (10 nm) (A) and MCP-2 (100 nm) (B) acting at the receptor CCR2 and for inhibition of the chemokines eotaxin-1 (100 nm) (C) and eotaxin-2 (100 nm) (D) acting at the receptor CCR3. Chemokine activity was detected as the ability of the chemokine to inhibit forskolin (FSK)-induced production of cAMP as detected via a BRET sensor (see “Experimental procedures” for details); thus, evasins inhibit the cAMP-inhibitory activity of the chemokines. Data represent the average ± S.E. (error bars) of values from three independent experiments, each recorded in duplicate.
Table 3.
Inhibition constants for inhibition of chemokine activity by purified evasins
Inhibition constants are reported as pIC50 values (−log10 of the IC50; in m) ±S.E. The corresponding IC50 values (in nm) are in parentheses.
| MCP-1 | MCP-2 | Eotaxin-1 | Eotaxin-2 | |
|---|---|---|---|---|
| RSA evasin-4 | a | 7.00 ± 0.2 (100) | 6.65 ± 0.2 (220) | 5.99 ± 0.1 (1000) |
| RPU-01 | 7.47 ± 0.3 (34) | 6.68 ± 0.1 (210) | 6.03 ± 0.3 (940) | a |
| ACA-01 | a | 7.07 ± 0.1 (84) | 6.39 ± 0.2 (400) | a |
| IHO-01 | b | b | 6.20 ± 0.2 (630) | 5.95 ± 0.2 (1100) |
a IC50 >1 μm (pIC50 + S.E. <6) or no inhibition detected.
b Not determined.
Discussion
We have shown here that the genomes or transcriptomes of several species of hard ticks, spanning the three genera Rhipicephalus, Amblyomma, and Ixodes, encode numerous proteins with sequences similar to the previously characterized evasin-1 and -4 from R. sanguineus. Among 12 of these proteins screened, nine were expressed well in E. coli and were sufficiently renatured for binding measurements. Of these nine candidate evasins, eight bound to one or more chemokine(s) tested with affinities of ∼50 nm or tighter. Moreover, three of the four fully purified evasins tested were found to inhibit chemokine signaling. These results strongly suggest that many of the proteins identified by our bioinformatics approach are indeed chemokine-inhibitory evasins.
During the final revisions of the current manuscript, a study by Singh et al. (34) describing the application of yeast surface display to identify several evasins from ticks in the genera Rhipicephalus and Amblyomma was published. Among the 10 evasins characterized in that study, two were the same as evasins we have characterized, namely ACA-01 (designated by Singh et al. (34) as P974_AMBCA) and RPU-01 (P467_RHIPU).
Considering that the current experiments were limited to a small subset of human chemokines, it seems likely that some of the evasins tested may be more potent inhibitors of different human chemokines or chemokines from other species, such as the natural hosts of the relevant tick species. In addition, it is possible that evasins may inhibit binding of chemokines to not only chemokine receptors but also glycosaminoglycans, which are important for establishment of chemokine gradients required for effective leukocyte trafficking in vivo (35). Of course, it also remains possible that evasins may bind to other host proteins in addition to chemokines. Future studies will be needed to investigate these hypotheses.
The new evasins identified here bound to a panel of related CC chemokines with Kd values in the range ∼5 nm to ∼1 μm. In comparison, R. sanguineus evasin-1 and -4 have been reported to bind various chemokines with Kd values ranging from ∼30 pm to ∼200 nm, although the reported values varied somewhat depending on the experimental methods used (13, 16) and may also be dependent on expression systems used due to variable post-translational modifications. In addition, the new evasins exhibited distinct binding selectivities to the various chemokines. For example, ACA-01 binds to the chemokines eotaxin-1 and MCP-2 substantially more tightly than to eotaxin-2, eotaxin-3, or MCP-1, whereas RPU-01 has similar affinities for eotaxin-1, eotaxin-3, MCP-1, and MCP-2 and lower affinity for eotaxin-2 (supplemental Fig. S3 and Table 1). The existence of such affinity and selectivity differences is consistent with the sequence variation among the evasins and chemokines tested. In particular, the evasin sequences differ substantially in the regions that interact with the chemokine in the structure of R. sanguineus evasin-1 bound to human CCL3 (Fig. 2A) and in the few residues of evasin-4 previously found to contribute to binding of CCL5 (36, 37). Similarly, there are numerous differences in the evasin-binding regions of the chemokines, such as the flexible N-terminal region. Future studies will be required to more fully elucidate the structural basis of the selectivity differences.
Despite the sequence variation in chemokine-binding residues, the evasins identified here share several highly conserved features, represented by the sequence motifs in Fig. 2. Like R. sanguineus evasin-1 and -4, most of these proteins (dubbed C8 evasins) share a distinctive pattern of eight core Cys residues, which form four disulfide bonds in R. sanguineus evasin-1. Ixodes sequences (dubbed C6 evasins) lack one of these disulfide bonds, suggesting that the missing disulfide may not be critical for folding and stability, but the C6 evasins also bind more weakly to the chemokines tested here, so the additional disulfide may enhance binding affinity. Additional conserved features are glycine residues, glycosylation sites, and an apparent tyrosine sulfation site, which has not been identified previously.
Future studies will be required to determine whether the conserved post-translational modifications occur in vivo and influence the properties of evasins. Previous studies of the R. sanguineus evasin-1 and -4 in animal models (16, 28, 29) have utilized proteins produced in eukaryotic cell lines and therefore presumably decorated heterogeneously with various post-translational modifications. We have shown here that homogeneous evasins produced in E. coli retain the ability to bind chemokines with high affinity, suggesting that the post-translational modifications are not essential for chemokine binding. However, it remains possible that these modifications influence such properties as binding affinity, selectivity, stability, or pharmacokinetics, thereby affecting the efficacy of evasins in disease models.
Previously, the only tick evasins shown to bind and inhibit chemokines were the three R. sanguineus evasins. Our results indicate that evasins are likely to be broadly expressed by many hard tick species and therefore that chemokine inhibition has been an advantageous evolutionary strategy for ticks. As all hard ticks are obligatory blood feeders, this may reflect positive selection pressure to maintain immunomodulatory evasin proteins and to evade immune recognition and remain attached to hosts for extended periods to enable continued blood feeding.
Among the 257 putative evasins identified in our bioinformatics searches, there is substantial sequence variation within each tick species and between different species, both common characteristics of tick protein families (11). Within a single species, we identified numerous different evasin-like sequences. For example, we found 13 R. pulchellus sequences with pairwise sequence identities ranging from 14 to 50%. Considering the current and previous observations that chemokine selectivity can vary substantially among evasins (15, 16, 18), it seems likely that the expression of several evasins by the same species may be a strategy to more effectively suppress the activities of multiple host chemokines and thereby inhibit the recruitment of their target leukocytes.
Comparison of sequences from different species also indicated a high level of sequence divergence. The average ± S.D. of cross-species pairwise identities was 20.1 ± 6.7%, and pairwise identities were <50% for 99% of cross-species comparisons and <30% for 95% of cross-species comparisons. The maximum identity found to R. sanguineus evasin-1 or -4 was 46% (to sequence JAA60818.1 from R. pulchellus). These sequence variations suggest that there has been substantial evolutionary pressure for evasin sequences to diverge after tick speciation events. Because speciation of ticks would typically be associated with a new mammalian host, the evolutionary pressure may arise from the different arrays of chemokines expressed by the hosts for the different tick species.
Many tick species are of medical or agricultural importance because they function as vectors for microbial pathogens. Within the genus Ixodes, I. ricinus and the related I. scapularis are of particular interest as vectors of the Lyme disease spirochete Borrelia burgdorferi (38–40). Within the genus Rhipicephalus, R. microplus (previously known as Boophilus microplus) is considered the most economically important tick for the United States and Australian cattle industries as it can cause weight loss in host animals and transmit agents of several cattle diseases (41, 42). Based on the prevalence of evasin sequences in the currently available databases, we anticipate that ongoing genome projects for I. scapularis and R. microplus are likely to reveal additional evasins as a mechanism of host immune evasion by these important tick species.
The numerous tick evasins identified in the current study represent a rich “library” of chemokine-binding proteins from which to identify proteins that selectively inhibit chemokines of interest in specific human or animal diseases. Notably, several of the newly identified evasins have high affinity for the chemokine eotaxin-1, a major chemoattractant for eosinophils and basophils, which play a key role in allergic responses and reactions to parasitic infestations. This is consistent with the role of evasins being to suppress host responses to ticks. However, it also suggests that evasins may be useful therapeutics in human diseases in which these types of leukocytes are involved as observed in a previous study of experimental colitis (29).
Conclusion
Our sequence similarity searches have identified more than 250 predicted tick protein sequences with features similar to R. sanguineus evasin-1 and -4 but substantial sequence variation in chemokine recognition residues. Among these sequences, we have identified two distinct subfamilies of evasins (C8 and C6). Among nine candidates screened for chemokine binding, eight exhibited nanomolar affinities for one or more chemokine tested. Our results suggest that most hard ticks express chemokine-binding evasins and that these evasins play important roles in evasion of host immune responses. The evasins identified here provide a valuable pool of potential inhibitors for development of chemokine-targeted anti-inflammatory therapies.
Experimental procedures
Media and reagents
Dulbecco's modified Eagle's medium (DMEM) and Hanks' balanced salt solution were from Invitrogen. Fetal bovine serum (FBS) was from In Vitro Technologies (Noble Park, Victoria, Australia). Polyethyleneimine (PEI) was from Polysciences, Inc. (Warrington, PA). Coelenterazine h was from NanoLight (Pinetop, AZ). All other reagents were purchased from Sigma-Aldrich.
Bioinformatics and evasin identification
Sequences of the canonical R. sanguineus evasin-1 and evasion-4 were retrieved from UniProt (accession numbers P0C8E7 and P0C8E9). These were used as queries against UniProtKB (restricted to Arthropoda; July 20, 2015 release) using the Basic Local Alignment Search Tool (BLAST) (43) with the BLASTp algorithm and against VectorBase (I. scapularis and I. ricinus transcriptome and genome sequences) (44) using tBLASTn with default parameters. Hits were ranked based on highest pairwise identity to the known evasin sequences using a pairwise identity matrix generated by Clustal Omega (45). The presence of the cysteines conserved in the canonical evasins was assessed. Putative evasin sequences were then analyzed using SignalP (46) to predict signal peptide cleavage sites. The putative evasin sequences with highest pairwise identity to the known evasins for each tick species were selected as candidates for cloning and expression. These candidate sequences were analyzed for the presence of putative sulfation sites using Sulfinator (47) and N-linked glycosylation sites by the motif N*(S/T).
Multiple sequence alignments of initial hits were generated using MUSCLE v3.8.31 (48). HMMs were constructed from these MUSCLE alignments using HMMER v3.1b1 (49). The HMMs were used with hmmsearch (default parameters) to identify more distantly related evasin sequences in UniProtKB (version January 11, 2016), NCBI Transcriptome Shotgun Assembly databases (downloaded February 10, 2016), VectorBase (I. scapularis and I. ricinus transcriptome sets translated with TransDecoder (50) and six-frame translated genome sequences), and I. holocyclus and R. microplus transcriptome data sets provided by Manuel Rodriguez Valle. Sequences were retained if they contained conserved cysteine residues equivalent to the canonical evasins and a predicted signal sequence and were less than 200 residues long. Hits were compiled into a final non-redundant set of putative evasins.
Cloning, expression, and purification of putative evasins for screening
Plasmids containing the coding sequences of evasin candidates plus a C-terminal His6 tag in the background expression vector pET28a were ordered from GenScript; codon usage was optimized for expression in E. coli. BL21(DE3) E. coli cells transformed with these plasmids were grown in 1 liter of LB medium supplemented with kanamycin sulfate (30 μg/ml) until the optical density at 600 nm reached 0.6. Protein expression was initiated by addition of isopropyl β-d-1-thiogalactopyranoside (1 mm), and the culture was grown for either 4 h or overnight. Cells were harvested by centrifugation; resuspended in 15 ml of 20 mm Tris·HCl, 500 mm NaCl, 5 mm imidazole, 0.02% (w/v) NaN3, pH 8.0; sonicated; and centrifuged to separate the soluble from insoluble (inclusion body) fraction. Inclusion bodies were washed in 20 mm Tris·HCl, 500 mm NaCl, 5 mm imidazole, 0.02% (w/v) NaN3, 2 mm DTT, 0.5% (v/v) Triton X-100, pH 8.0, and denatured in 20 mm Tris·HCl, 6 m guanidine·HCl, 20 mm imidazole, 20 mm β-mercaptoethanol, pH 8.0, before being added to 15 ml of loose nickel-nitrilotriacetic acid resin (Qiagen) to purify recombinant protein by batch immobilized metal affinity chromatography. Denatured protein was then refolded via rapid dilution (flow rate, 0.1 ml/min) in 2 liters of 20 mm Tris·HCl, 400 mm NaCl, 2 mm reduced glutathione, 0.5 mm oxidized glutathione, 0.02% (w/v) NaN3, pH 8.0. This was followed by further purification by immobilized metal affinity chromatography (HisTrap HP 5-ml column, GE Healthcare). After initial screening, expression of selected evasins was repeated at 4-liter scale, including a final purification step of size exclusion chromatography (HiLoad 16/60 Superdex 75 prep grade column, GE Healthcare). The molecular weights of pure proteins were confirmed by mass spectrometry using an Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific) in intact protein mode.
Chemokine-binding assay
Competition-binding assays, measuring the displacement of a fluorescent receptor peptide from each of six CC chemokines (CCL2/MCP-1, CCL7/MCP-3, CCL8/MCP-2, CCL11/eotaxin-1, CCL24/eotaxin-2, and CCL26/eotaxin-3), were conducted for each candidate evasin to screen for chemokine-binding ability as described (33). Briefly, a solution of the candidate evasin was prepared in MOPS buffer (50 mm MOPS, pH 7.4), and a serial 2-fold dilution was conducted on a 96-well plate (final concentration range from 1 μm to 15.6 nm). A mixed solution of the fluorescent peptide and chemokine (final concentrations of 10 nm and 100 nm, respectively) was added such that the final volume in each well was 200 μl. Competition-binding assays were also performed using the purified evasins to measure the displacement of a fluorescent receptor peptide from each of five CC chemokines (MCP-1, MCP-2, eotaxin-1, eotaxin-2, and eotaxin-3) under similar conditions. Briefly, a solution of the purified evasin was prepared in MOPS buffer, and a serial 2-fold dilution (1.33-fold dilution for evasin-4) was conducted on a 384-well plate coated with a 0.001% (w/v) solution of poly-l-lysine (final concentration range from 1 μm to 15.6 nm or 300 nm to 9.5 nm for evasin-4). A mixed solution of the fluorescent peptide and chemokine (final concentrations of 10 and 100 nm, respectively) was added such that the final volume in each well was 20 μl. In both cases, fluorescence anisotropy was measured 5 min after plating with excitation and emission wavelengths of 485 and 520 nm, respectively, using a BMG Labtech PHERAstar FS plate reader. Experiments were conducted in duplicate three times independently, and the mean anisotropy was fitted by non-linear regression analysis using GraphPad Prism v.6.0 software to the equation for a 1:1 competitive displacement curve, described previously (51), transformed to enable −log Kd (pKd) values ±S.E. to be determined. Multiple t tests were conducted to assess significance of the differences in pKd.
Inhibition of chemokine signaling
Chemokine signaling via the receptor CCR2 or CCR3 was measured using a cAMP biosensor assay developed previously (52). The assay measures the ability of chemokines to inhibit forskolin-induced cAMP production in c-Myc-FLAG-CCR2 or c-Myc-FLAG-CCR3 FlpIn TREx HEK293 cells transiently transfected with the CAMYEL cAMP bioluminescence resonance energy transfer (BRET) biosensor (53). Cells were grown overnight in 10-cm dishes using DMEM + 5% FBS before transfection using 6:1 (w/w) PEI:DNA. Tetracycline (10 μg/ml) was added 24 h after transfection to induce expression of c-Myc-FLAG-CCR2 or c-Myc-FLAG-CCR3, and cells were seeded (25,000 cells/well) in a white, poly-d-lysine-coated 96-well plate (CulturPlates, PerkinElmer Life Sciences) and incubated overnight at 37 °C in 5% CO2. The following day, cells were washed and equilibrated in Hanks' balanced salt solution for 30 min at 37 °C. Cells were then incubated with the Rluc substrate coelenterazine h (final concentration, 5 μm) for 5 min followed by a further 5-min incubation with a solution containing a fixed concentration of chemokine (10 nm MCP-1, 100 nm MCP-2, 100 nm eotaxin-1, or 100 nm eotaxin-2, corresponding to approximately the EC80, as shown in supplemental Fig. S4) with various concentrations of an evasin candidate that had been preincubated for ∼15 min to allow binding of the evasin to the chemokine. A final 5-min incubation step was conducted after addition of forskolin (final concentration,10 μm), which directly stimulates the production of cAMP via adenylyl cyclase. The Rluc and yellow fluorescent protein (YFP) emissions were then measured at 475 and 525 nm, respectively, using a BMG Labtech PHERAstar FS plate reader. Data are presented as a BRET ratio, calculated as the ratio of YFP to Rluc signals and expressed as the percentage of the forskolin-induced signal.
Author contributions
J. H. and J. S. participated in research design, performed experiments, and conducted data analyses. A. P. and J. H. conducted bioinformatics searches and analyses. C. H. performed and interpreted mass spectrometry experiments. M. R. V. collected transcriptomics data for Australian tick species and contributed to bioinformatics analyses/interpretation. M. C. designed and supervised cell-based assays. R. J. P. and M. J. S. conceived the study and designed and interpreted the experiments. J. H., J. S., and M. J. S. drafted the manuscript, and all authors read and critically reviewed the manuscript.
Supplementary Material
Acknowledgments
We thank Professor Catherine Hill (Purdue University) for helpful discussions and Natalya Stone for technical assistance.
This work was supported in part by Australian Research Council Discovery Grant DP120100194 (to M. J. S. and R. J. P.), the Monash Institute of Pharmaceutical Sciences Large Grant Support Scheme (to M. C.), and ANZ Trustees Grant 12-3831 (to M. J. S. and M. C.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S4 and Table S1.
- HMM
- hidden Markov model
- AAM
- A. americanum
- ACA
- A. cajennense
- AMA
- A. maculatum
- APA
- A. parvum
- ATR
- A. triste
- BRET
- bioluminescence resonance energy transfer
- BLAST
- Basic Local Alignment Search Tool
- CCL
- CC chemokine ligand
- CCR
- CC chemokine receptor
- EC80
- 80% maximal effective concentration
- IRI
- I. ricinus
- Kd
- equilibrium dissociation constant
- MCP
- monocyte chemoattractant protein
- RPU
- R. pulchellus
- RSA
- R. sanguineus
- Rluc
- Renilla luciferase
- IHO
- I. holocyclus.
References
- 1. Dantas-Torres F., Chomel B. B., and Otranto D. (2012) Ticks and tick-borne diseases: a One Health perspective. Trends Parasitol. 28, 437–446 [DOI] [PubMed] [Google Scholar]
- 2. Becker C. A., Bouju-Albert A., Jouglin M., Chauvin A., and Malandrin L. (2009) Natural transmission of zoonotic Babesia spp. by Ixodes ricinus ticks. Emerging Infect. Dis. 15, 320–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. de la Fuente J., Ayoubi P., Blouin E. F., Almazán C., Naranjo V., and Kocan K. M. (2006) Anaplasmosis: focusing on host–vector–pathogen interactions for vaccine development. Ann. N.Y. Acad. Sci. 1078, 416–423 [DOI] [PubMed] [Google Scholar]
- 4. Fivaz B., Petney T., and Horak I. (1992) Tick Vector Biology, Springer-Verlag, Berlin [Google Scholar]
- 5. Moser B., Wolf M., Walz A., and Loetscher P. (2004) Chemokines: multiple levels of leukocyte migration control. Trends Immunol. 25, 75–84 [DOI] [PubMed] [Google Scholar]
- 6. Zlotnik A., and Yoshie O. (2000) Chemokines: a new classification system and their role in immunity. Immunity 12, 121–127 [DOI] [PubMed] [Google Scholar]
- 7. Baggiolini M. (2001) Chemokines in pathology and medicine. J. Intern. Med. 250, 91–104 [DOI] [PubMed] [Google Scholar]
- 8. Gerard C., and Rollins B. J. (2001) Chemokines and disease. Nat. Immunol. 2, 108–115 [DOI] [PubMed] [Google Scholar]
- 9. Kazimírová M., and Štibrániová I. (2013) Tick salivary compounds: their role in modulation of host defences and pathogen transmission. Front. Cell. Infect. Microbiol. 3, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rodriguez-Valle M., Moolhuijzen P., Piper E. K., Weiss O., Vance M., Bellgard M., and Lew-Tabor A. (2013) Rhipicephalus microplus lipocalins (LRMs): genomic identification and analysis of the bovine immune response using in silico predicted B and T cell epitopes. Int. J. Parasitol. 43, 739–752 [DOI] [PubMed] [Google Scholar]
- 11. Rodriguez-Valle M., Xu T., Kurscheid S., and Lew-Tabor A. E. (2015) Rhipicephalus microplus serine protease inhibitor family: annotation, expression and functional characterisation assessment. Parasit. Vectors 8, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Thompson R. E., Liu X., Ripoll-Rozada J., Alonso-García N., Parker B. L., Pereira P. J. B., and Payne R. J. (2017) Tyrosine sulfation modulates activity of tick-derived thrombin inhibitors. Nat. Chem. 10.1038/nchem.2744 [DOI] [PubMed] [Google Scholar]
- 13. Bachelerie F., Ben-Baruch A., Burkhardt A. M., Combadiere C., Farber J. M., Graham G. J., Horuk R., Sparre-Ulrich A. H., Locati M., Luster A. D., Mantovani A., Matsushima K., Murphy P. M., Nibbs R., Nomiyama H., et al. (2014) International Union of Basic and Clinical Pharmacology. [corrected]. LXXXIX. Update on the extended family of chemokine receptors and introducing a new nomenclature for atypical chemokine receptors. Pharmacol. Rev. 66, 1–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bonvin P., Power C. A., and Proudfoot A. E. (2016) Evasins: therapeutic potential of a new family of chemokine-binding proteins from ticks. Front. Immunol. 7, 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Frauenschuh A., Power C. A., Déruaz M., Ferreira B. R., Silva J. S., Teixeira M. M., Dias J. M., Martin T., Wells T. N., and Proudfoot A. E. (2007) Molecular cloning and characterization of a highly selective chemokine-binding protein from the tick Rhipicephalus sanguineus. J. Biol. Chem. 282, 27250–27258 [DOI] [PubMed] [Google Scholar]
- 16. Déruaz M., Frauenschuh A., Alessandri A. L., Dias J. M., Coelho F. M., Russo R. C., Ferreira B. R., Graham G. J., Shaw J. P., Wells T. N., Teixeira M. M., Power C. A., and Proudfoot A. E. (2008) Ticks produce highly selective chemokine binding proteins with antiinflammatory activity. J. Exp. Med. 205, 2019–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Anatriello E., Ribeiro J. M., de Miranda-Santos I. K., Brandão L. G., Anderson J. M., Valenzuela J. G., Maruyama S. R., Silva J. S., and Ferreira B. R. (2010) An insight into the sialotranscriptome of the brown dog tick, Rhipicephalus sanguineus. BMC Genomics 11, 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Déruaz M., Bonvin P., Severin I. C., Johnson Z., Krohn S., Power C. A., and Proudfoot A. E. (2013) Evasin-4, a tick-derived chemokine-binding protein with broad selectivity can be modified for use in preclinical disease models. FEBS J. 280, 4876–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Radulovic Ž. M., Kim T. K., Porter L. M., Sze S. H., Lewis L., and Mulenga A. (2014) A 24–48 h fed Amblyomma americanum tick saliva immuno-proteome. BMC Genomics 15, 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Karim S., and Ribeiro J. M. (2015) An insight into the sialome of the lone star tick, Amblyomma americanum, with a glimpse on its time dependent gene expression. PLoS One 10, e0131292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Garcia G. R., Gardinassi L. G., Ribeiro J. M., Anatriello E., Ferreira B. R., Moreira H. N., Mafra C., Martins M. M., Szabó M. P., de Miranda-Santos I. K., and Maruyama S. R. (2014) The sialotranscriptome of Amblyomma triste, Amblyomma parvum and Amblyomma cajennense ticks, uncovered by 454-based RNA-seq. Parasit. Vectors 7, 430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Karim S., Singh P., and Ribeiro J. M. (2011) A deep insight into the sialotranscriptome of the gulf coast tick, Amblyomma maculatum. PLoS One 6, e28525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tan A. W., Francischetti I. M., Slovak M., Kini R. M., and Ribeiro J. M. (2015) Sexual differences in the sialomes of the zebra tick, Rhipicephalus pulchellus. J. Proteomics 117, 120–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. de Castro M. H., de Klerk D., Pienaar R., Latif A. A., Rees D. J., and Mans B. J. (2016) De novo assembly and annotation of the salivary gland transcriptome of Rhipicephalus appendiculatus male and female ticks during blood feeding. Ticks Tick Borne Dis. 7, 536–548 [DOI] [PubMed] [Google Scholar]
- 25. Vancová I., Hajnická V., Slovák M., Kocáková P., Paesen G. C., and Nuttall P. A. (2010) Evasin-3-like anti-chemokine activity in salivary gland extracts of ixodid ticks during blood-feeding: a new target for tick control. Parasite Immunol. 32, 460–463 [DOI] [PubMed] [Google Scholar]
- 26. Proudfoot A. E. (2002) Chemokine receptors: multifaceted therapeutic targets. Nat. Rev. Immunol. 2, 106–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Proudfoot A. E., Power C. A., and Wells T. N. (2000) The strategy of blocking the chemokine system to combat disease. Immunol. Rev. 177, 246–256 [DOI] [PubMed] [Google Scholar]
- 28. Russo R. C., Alessandri A. L., Garcia C. C., Cordeiro B. F., Pinho V., Cassali G. D., Proudfoot A. E., and Teixeira M. M. (2011) Therapeutic effects of evasin-1, a chemokine binding protein, in bleomycin-induced pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 45, 72–80 [DOI] [PubMed] [Google Scholar]
- 29. Vieira A. T., Fagundes C. T., Alessandri A. L., Castor M. G., Guabiraba R., Borges V. O., Silveira K. D., Vieira E. L., Gonçalves J. L., Silva T. A., Deruaz M., Proudfoot A. E., Sousa L. P., and Teixeira M. M. (2009) Treatment with a novel chemokine-binding protein or eosinophil lineage-ablation protects mice from experimental colitis. Am. J. Pathol. 175, 2382–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Copin J. C., da Silva R. F., Fraga-Silva R. A., Capettini L., Quintao S., Lenglet S., Pelli G., Galan K., Burger F., Braunersreuther V., Schaller K., Deruaz M., Proudfoot A. E., Dallegri F., Stergiopulos N., et al. (2013) Treatment with Evasin-3 reduces atherosclerotic vulnerability for ischemic stroke, but not brain injury in mice. J. Cereb. Blood Flow Metab. 33, 490–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stone M. J., and Payne R. J. (2015) Homogeneous sulfopeptides and sulfoproteins: synthetic approaches and applications to characterize the effects of tyrosine sulfation on biochemical function. Acc. Chem. Res. 48, 2251–2261 [DOI] [PubMed] [Google Scholar]
- 32. Wang X., Sanchez J., Stone M. J., and Payne R. J. (2017) Sulfation of the human cytomegalovirus protein UL22A enhances binding to the chemokine RANTES. Angew. Chem. Int. Ed. Engl. 56, 8490–8494 [DOI] [PubMed] [Google Scholar]
- 33. Ludeman J. P., Nazari-Robati M., Wilkinson B. L., Huang C., Payne R. J., and Stone M. J. (2015) Phosphate modulates receptor sulfotyrosine recognition by the chemokine monocyte chemoattractant protein-1 (MCP-1/CCL2). Org. Biomol. Chem. 13, 2162–2169 [DOI] [PubMed] [Google Scholar]
- 34. Singh K., Davies G., Alenazi Y., Eaton J. R. O., Kawamura A., and Bhattacharya S. (2017) Yeast surface display identifies a family of evasins from ticks with novel polyvalent CC chemokine-binding activities. Sci. Rep. 7, 4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Proudfoot A. E., Handel T. M., Johnson Z., Lau E. K., LiWang P., Clark-Lewis I., Borlat F., Wells T. N., and Kosco-Vilbois M. H. (2003) Glycosaminoglycan binding and oligomerization are essential for the in vivo activity of certain chemokines. Proc. Natl. Acad. Sci. U.S.A. 100, 1885–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bonvin P., Dunn S. M., Rousseau F., Dyer D. P., Shaw J., Power C. A., Handel T. M., and Proudfoot A. E. (2014) Identification of the pharmacophore of the CC chemokine-binding proteins Evasin-1 and -4 using phage display. J. Biol. Chem. 289, 31846–31855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dias J. M., Losberger C., Déruaz M., Power C. A., Proudfoot A. E., and Shaw J. P. (2009) Structural basis of chemokine sequestration by a tick chemokine binding protein: the crystal structure of the complex between Evasin-1 and CCL3. PLoS One 4, e8514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ogden N. H., Maarouf A., Barker I. K., Bigras-Poulin M., Lindsay L. R., Morshed M. G., O'callaghan C. J., Ramay F., Waltner-Toews D., and Charron D. F. (2006) Climate change and the potential for range expansion of the Lyme disease vector Ixodes scapularis in Canada. Int. J. Parasitol. 36, 63–70 [DOI] [PubMed] [Google Scholar]
- 39. Hovius J. W. (2009) Spitting image: tick saliva assists the causative agent of Lyme disease in evading host skin's innate immune response. J. Invest. Dermatol. 129, 2337–2339 [DOI] [PubMed] [Google Scholar]
- 40. Gulia-Nuss M., Nuss A. B., Meyer J. M., Sonenshine D. E., Roe R. M., Waterhouse R. M., Sattelle D. B., de la Fuente J., Ribeiro J. M., Megy K., Thimmapuram J., Miller J. R., Walenz B. P., Koren S., Hostetler J. B., et al. (2016) Genomic insights into the Ixodes scapularis tick vector of Lyme disease. Nat. Commun. 7, 10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jonsson N. N., Davis R., and De Witt M. (2001) An estimate of the economic effects of cattle tick (Boophilus microplus) infestation on Queensland dairy farms. Aust. Vet. J. 79, 826–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jonsson N. N. (2006) The productivity effects of cattle tick (Boophilus microplus) infestation on cattle, with particular reference to Bos indicus cattle and their crosses. Vet. Parasitol. 137, 1–10 [DOI] [PubMed] [Google Scholar]
- 43. Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 44. Giraldo-Calderón G. I., Emrich S. J., MacCallum R. M., Maslen G., Dialynas E., Topalis P., Ho N., Gesing S., VectorBase Consortium, Madey G., Collins F. H., and Lawson D. (2015) VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 43, D707–D713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Petersen T. N., Brunak S., von Heijne G., and Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 47. Monigatti F., Gasteiger E., Bairoch A., and Jung E. (2002) The Sulfinator: predicting tyrosine sulfation sites in protein sequences. Bioinformatics 18, 769–770 [DOI] [PubMed] [Google Scholar]
- 48. Edgar R. C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Finn R. D., Clements J., Arndt W., Miller B. L., Wheeler T. J., Schreiber F., Bateman A., and Eddy S. R. (2015) HMMER web server: 2015 update. Nucleic Acids Res. 43, W30–W38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D., Bowden J., Couger M. B., Eccles D., Li B., Lieber M., MacManes M. D., Ott M., Orvis J., Pochet N., Strozzi F., et al. (2013) De novo transcript sequence reconstruction from RNA-Seq: reference generation and analysis with Trinity. Nat. Protoc. 8, 1494–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Huff S., Matsuka Y. V., McGavin M. J., and Ingham K. C. (1994) Interaction of N-terminal fragments of fibronectin with synthetic and recombinant D motifs from its binding protein on Staphylococcus aureus studied using fluorescence anisotropy. J. Biol. Chem. 269, 15563–15570 [PubMed] [Google Scholar]
- 52. Thompson G. L., Lane J. R., Coudrat T., Sexton P. M., Christopoulos A., and Canals M. (2015) Biased agonism of endogenous opioid peptides at the μ-opioid receptor. Mol. Pharmacol. 88, 335–346 [DOI] [PubMed] [Google Scholar]
- 53. Jiang L. I., Collins J., Davis R., Lin K. M., DeCamp D., Roach T., Hsueh R., Rebres R. A., Ross E. M., Taussig R., Fraser I., and Sternweis P. C. (2007) Use of a cAMP BRET sensor to characterize a novel regulation of cAMP by the sphingosine 1-phosphate/G13 pathway. J. Biol. Chem. 282, 10576–10584 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.