Abstract
The Gram-negative bacterium Sphingomonas sp. A1 incorporates alginate into cells via the cell-surface pit without prior depolymerization by extracellular enzymes. Alginate import across cytoplasmic membranes thereby depends on the ATP-binding cassette transporter AlgM1M2SS (a heterotetramer of AlgM1, AlgM2, and AlgS), which cooperates with the periplasmic solute-binding protein AlgQ1 or AlgQ2; however, several details of AlgM1M2SS-mediated alginate import are not well-understood. Herein, we analyzed ATPase and transport activities of AlgM1M2SS after reconstitution into liposomes with AlgQ2 and alginate oligosaccharide substrates having different polymerization degrees (PDs). Longer alginate oligosaccharides (PD ≥ 5) stimulated the ATPase activity of AlgM1M2SS but were inert as substrates of AlgM1M2SS-mediated transport, indicating that AlgM1M2SS-mediated ATP hydrolysis can be stimulated independently of substrate transport. Using X-ray crystallography in the presence of AlgQ2 and long alginate oligosaccharides (PD 6–8) and with the humid air and glue-coating method, we determined the crystal structure of AlgM1M2SS in complex with oligosaccharide-bound AlgQ2 at 3.6 Å resolution. The structure of the ATP-binding cassette transporter in complex with non-transport ligand-bound periplasmic solute-binding protein revealed that AlgM1M2SS and AlgQ2 adopt inward-facing and closed conformations, respectively. These in vitro assays and structural analyses indicated that interactions between AlgM1M2SS in the inward-facing conformation and periplasmic ligand-bound AlgQ2 in the closed conformation induce ATP hydrolysis by the ATP-binding protein AlgS. We conclude that substrate-bound AlgQ2 in the closed conformation initially interacts with AlgM1M2SS, the AlgM1M2SS–AlgQ2 complex then forms, and this formation is followed by ATP hydrolysis.
Keywords: ABC transporter, ATPase, bacteria, crystal structure, X-ray crystallography
Introduction
ATP-binding cassette (ABC)3 transporters are present in cytoplasmic membranes of all species, and the translocation of substrates by transporters is coupled with ATP hydrolysis (1). ABC transporters comprise two transmembrane domains and two nucleotide-binding domains. The transmembrane domains form the translocation pathway, and the nucleotide-binding domains hydrolyze ATP to generate energy for transport (2). ABC transporters are classified as exporters and importers (3), and whereas ABC exporters are ubiquitous from bacteria to humans, ABC importers are mainly found in prokaryotes (4) and require accessory solute-binding proteins (5). Several ABC importers, such as MalFGK2 (6), the maltose transporter in Escherichia coli, and the E. coli vitamin B12 transporter BtuCD (7) have been well-characterized. When importing substrate, ABC importers change conformations from the inward-facing to outward-facing states (5). However, the mechanisms that regulate this conformational change and ATP hydrolysis of ABC importers are not fully understood.
Interactions between the ABC transporter MalFGK2 and the periplasmic maltose-binding protein MalE induce ATP hydrolysis (8). Moreover, a mutant MalE was shown to bind sucrose (non-transport ligand) and enhance the ATPase activity of MalFGK2, suggesting that ATP hydrolysis is induced by ligand-bound MalE but not by the translocated substrate (i.e. maltose). Recently, two models of solute-binding protein MalE conformations in contact with MalFGK2 were described as closed (6) and open states (9). However, these findings were based on in vitro assays, and the structure of the ABC transporter in complex with the solute-binding protein with a non-transport ligand has not been directly characterized.
Alginate is a linear acidic polysaccharide that comprises β-d-mannuronate (M) and its C-5-epimer α-l-guluronate (G). The Gram-negative and alginate-assimilating bacterium Sphingomonas sp. strain A1 incorporates polysaccharides into cells through cell-surface pits (10). Genes that are involved in alginate import and degradation form a cluster on the genome of strain A1 and encode the periplasmic alginate-binding proteins AlgQ1 and AlgQ2; the cytoplasmic membrane-bound ABC transporter (AlgM1M2SS); the cytoplasmic endotype alginate lyases A1-I, A1-II, and A1-III; the exotype alginate lyase (A1-IV); and the alginate-dependent transcription factor AlgO (11, 12). Alginate concentrated in the cell-surface pit is incorporated into the periplasm and is delivered to the ABC transporter by alginate-binding proteins. The ABC transporter for alginate import is a heterotetramer comprising transmembrane domains (heterodimer of AlgM1 and AlgM2) and an ATP-binding domain (homodimer of AlgS). After import into the cytoplasm by the ABC transporter, alginate is depolymerized to monosaccharides by endo- and exotype alginate lyases. Moreover, the genetic cluster for import and degradation of alginate is expressed in the presence of the polysaccharide under the control of the transcription factor AlgO (13).
Characteristics of ATPase and transport activities of reconstituted AlgM1M2SS in liposomes and crystal structures of the transporter have mainly been determined using alginate oligosaccharides with polymerization degrees (PDs) of <4 (14–16). In contrast with the E. coli maltose ABC transporter, interactions of AlgM1M2SS with AlgQ2 result in the formation of an alginate-binding tunnel that is accessible to solvent. In the present study, in vitro assays of the ATPase and transport activities of AlgM1M2SS were performed using long alginate oligosaccharides (PD ≥ 5), and non-transport ligand–bound AlgQ2 in the closed conformation interacted with the ABC transporter in the inward-facing conformation. These X-ray crystallography structure analyses were performed using the humid air and glue-coating method and demonstrated that the solute-binding protein in the closed conformation induces ATP hydrolysis in this bacterial ABC transporter.
Results
Purification of alginate oligosaccharides
M-rich polysaccharides (M-block) were obtained from alginate, and unsaturated M oligosaccharides with C=C bonds in their non-reducing ends were prepared by treatment of M-block with alginate lyase. Saturated M oligosaccharides were then prepared by acid hydrolysis of M-block, and unsaturated M oligosaccharides were further fractionated by anion exchange chromatography into unsaturated M disaccharide (Δ2M), unsaturated M trisaccharide (Δ3M), unsaturated M tetrasaccharide (Δ4M), unsaturated M pentasaccharide (Δ5M), unsaturated M hexasaccharide (Δ6M), and a mixture of unsaturated M hepta- and octasaccharides (Δ7-8M). Saturated M oligosaccharides were also divided into mixtures of saturated M hexa-, hepta-, and octasaccharides (6–8M) and of saturated M hepta-, octa-, ennea-, and decasaccharides (7–10M) using anion exchange chromatography. PDs in each oligosaccharide were determined using fluorophore-assisted carbohydrate electrophoresis (FACE) (17) (Fig. 1A), and the alginate oligosaccharides and their pyridylamino (PA)-saccharides fluorescence-labeled with 2-aminopyridine were used for subsequent experiments.
Figure 1.
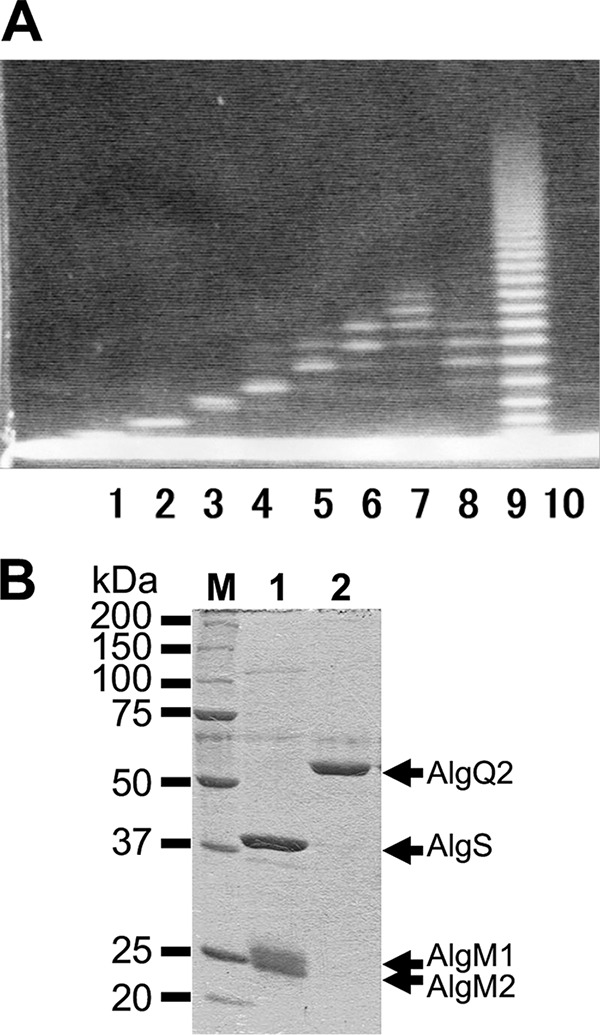
Electrophoretic profiles of oligosaccharides and proteins. A, FACE profiles of alginate oligosaccharides. Lane 1, Δ2M; lane 2, Δ3M; lane 3, Δ4M; lane 4, Δ5M; lane 5, Δ6M; lane 6, Δ7-8M; lane 7, 7–10M; lane 8, 6–8M; lane 9, saturated M oligosaccharides before purification; lane 10, negative control treated with 8-amino-1,3,6-naphthalenetrisulfonic acid in the absence of oligosaccharide. B, SDS-PAGE profiles of AlgM1M2SS (lane 1) and AlgQ2 (lane 2). Three protein bands indicated by arrows correspond to AlgS, AlgM1, and AlgM2 in the purified AlgM1M2SS (lane 1). An arrow indicates the position of AlgQ2 (lane 2). M, mass.
Co-immunoprecipitation of AlgQ2 by AlgM1M2SS
Co-immunoprecipitation assays were conducted to analyze interactions between AlgM1M2SS and AlgQ2. In these analyses, recombinant AlgM1M2SS and AlgQ2 were expressed in E. coli cells and were purified to homogeneity (Fig. 1B). After interactions of anti-His tag antibody conjugated Dynabeads Protein G (Veritas) and AlgM1M2SS containing a histidine tag at the C terminus of AlgM2, AlgQ2 was added in the presence or absence of ATP and Δ3M. Dynabeads Protein G was then collected, and bound proteins were subjected to SDS-PAGE, followed by Western blotting using anti-AlgQ2 antiserum (18) (Fig. 2). AlgQ2 was observed only in the presence of both ATP and Δ3M (Fig. 2, lanes 2 and 3), and no interactions between AlgM1M2SS and AlgQ2 occurred in the absence of Δ3M (Fig. 2, lane 1). No AlgQ2 was detected in the immunoprecipitation assay using other His tag protein as a negative control in place of AlgM1M2SS (data not shown). Because AlgQ2 comprises N- and C-terminal domains and adopts substrate-free open and substrate-bound closed conformations through domain dynamics (16, 19), these co-immunoprecipitation assays suggested that Δ3M-bound AlgQ2 interacts with AlgM1M2SS in the closed conformation. However, the substrate-free AlgQ2 in the open conformation did not interact with AlgM1M2SS. Moreover, following the addition of vanadate, the intensity of the AlgQ2 band increased slightly in the presence of ATP and Δ3M (Fig. 2, lane 3). The phosphate analogue vanadate bound the ATP-binding site with ADP and inhibited ATP hydrolysis. Hence, the increase in AlgQ2 band intensity probably reflects inhibited dissociation of AlgQ2 from AlgM1M2SS, suggesting that this dissociation event is coupled with ATP hydrolysis.
Figure 2.
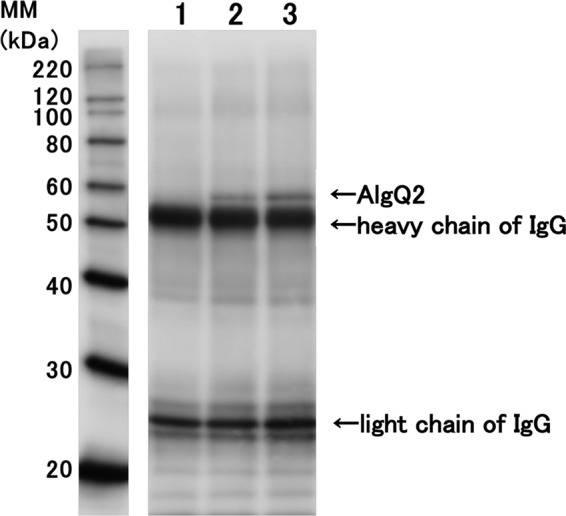
Co-immunoprecipitation of AlgM1M2SS and AlgQ2. Lane 1, no ligand; lane 2, 2 mm Mg-ATP and 0.2 mm Δ3M; lane 3, 2 mm Mg-ATP, 0.2 mm Δ3M, and 5 mm vanadate. MM, molecular mass.
ATPase and transport activities of AlgM1M2SS
ATPase and transport activities of reconstituted AlgM1M2SS in liposomes were measured in the presence of various alginate oligosaccharides and AlgQ2 (Fig. 3). Specific ATPase activity of AlgM1M2SS in the absence of saccharide was determined to around 30 nmol/min/mg. In these experiments, all alginate oligosaccharides enhanced ATPase activity, and saccharide chain lengths, structures of non-reducing ends (saturated or unsaturated), and modifications at reducing saccharide residues with PA in alginate oligosaccharides had limited effects on ATPase activities. In contrast, transport activities varied with saccharide chain lengths, with the highest transport activity in the presence of alginate tetrasaccharide as a substrate, and almost no transport of alginate oligosaccharides with PD of >5. In addition, transport of these long oligosaccharides was comparable with that of the non-alginate oligosaccharide chitotetraose, indicating that long alginate oligosaccharides with PD of >5 were not transport substrates of AlgM1M2SS in this assay system.
Figure 3.
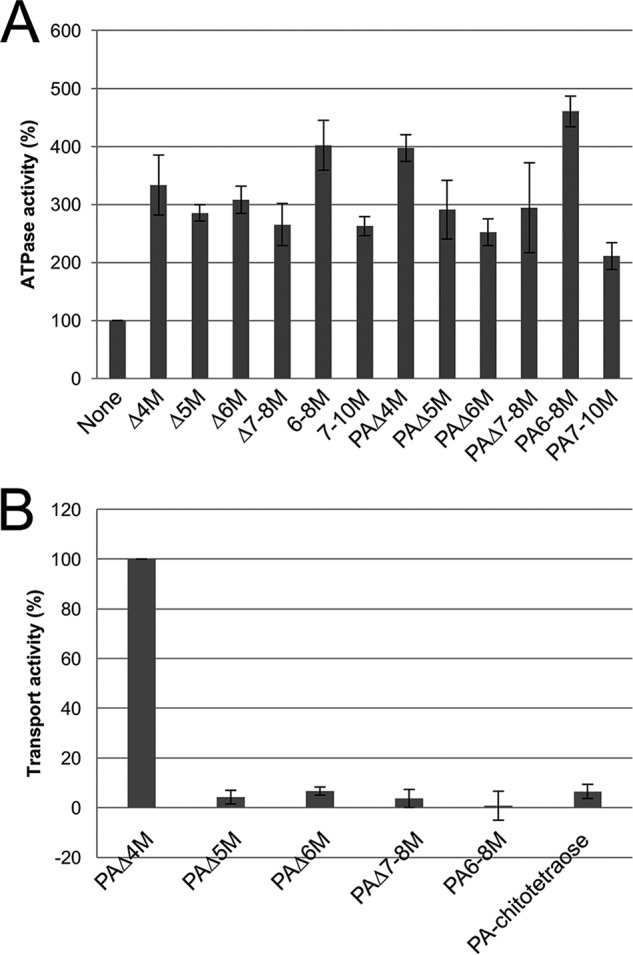
In vitro assay of AlgM1M2SS. A, ATPase activity; B, alginate transport activity; activities are shown as relative values. Activities in the absence of oligosaccharides (A) and with Δ4M (B) were taken as 100%. Assays were conducted three times, and data are presented as means with error bars representing S.E.
X-ray diffraction experiments using the humid air and glue-coating (HAG) method
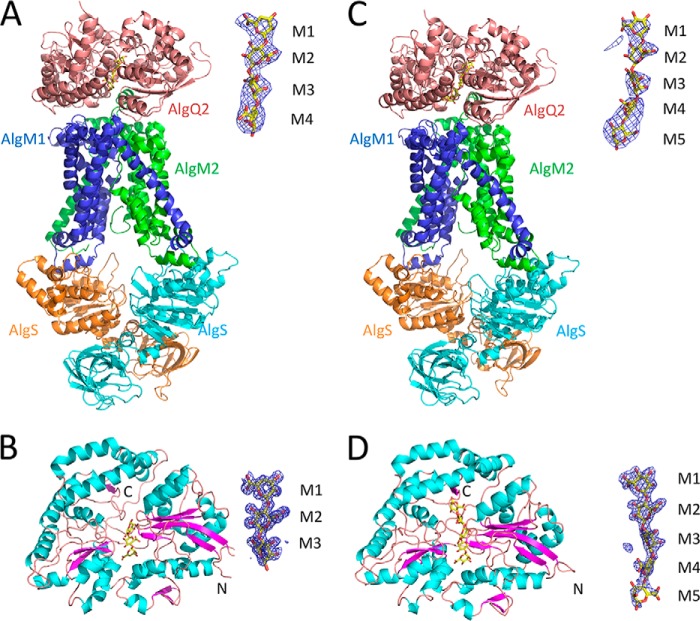
Previously, the crystal structure of AlgM1M2SS was determined in complex with Δ3M-bound AlgQ2 at a resolution of 3.2 Å (14), and similar crystals were frozen using the HAG method (20) to improve X-ray diffraction images. The HAG method is a novel technique for crystal mounting, in which crystals are coated with a water-soluble polymer and are then exposed to controlled humid air using humidifier HUM-1F (RIGAKU). AlgM1M2SS crystals in complex with Δ3M-bound AlgQ2 were obtained in drop solutions comprising 22% PEG 3000, 0.1 m ADA-NaOH (pH 6.6), 0.15 m NaCl, and 16 mm CHAPSO. Crystals were then picked up using a loop covered with 10% polyvinyl alcohol (PVA) 4500 and were subjected to X-ray diffraction experiments with air controlled at 86–92% relative humidity. In these experiments, the crystals had optimal mosaicity (0.607) at 90% relative humidity, and after freezing at 90% relative humidity, crystals were exposed to X-rays and provided diffraction data at 3.4 Å resolution, allowing determination of crystal structure by molecular replacement. The overall structure of AlgM1M2SS adopted the inward-facing conformation (Fig. 4A), and the electron density map that was obtained using the HAG method included an additional electron density at C-terminal regions of AlgM1 and AlgM2. In contrast with the previous structure, three residues (from Thr-321 to Val-323) in AlgM1 and seven residues (from Gly-285 to Val-291) in AlgM2 were accurately traced in the electron density map, indicating improved X-ray diffraction data after crystal freezing of AlgM1M2SS in complex with AlgQ2 using the HAG method.
Figure 4.
Overall structure. The images on the right show the Fo − Fc map contoured at 2.8σ around the alginate oligosaccharide. A, AlgM1M2SS/AlgQ2 + Δ3M; B, AlgQ2 + Δ3M; C, AlgM1M2SS/AlgQ2 + 6–8M; D, AlgQ2 + 7–10M.
To obtain a snapshot of the structure of the ABC transporter in complex with the non-transport ligand-bound solute-binding protein, AlgM1M2SS and AlgQ2 were crystallized in the presence of long alginate oligosaccharides. In these experiments, crystals of AlgM1M2SS, AlgQ2, and 6–8M were obtained in a drop solution comprising 22% PEG 3000, 0.1 m ADA-NaOH (pH 6.6), 0.15 m sodium formate, 8 mm CHAPSO, and 4 mm ATP and were picked up using a loop covered with 10% PVA 4500 and 5% ethylene glycol for X-ray diffraction experiments with controlled humid air. Dehydration of the crystal at 85% relative humidity for 10 min significantly improved mosaicity (1.714–0.732), which was reduced to 0.846 at 83% relative humidity. The crystal was finally frozen and subjected to X-ray diffraction experiments at 85% relative humidity. Subsequently, the crystal structure of AlgM1M2SS in complex with long alginate oligosaccharide-bound AlgQ2 was determined at 3.6 Å using molecular replacement. In the resulting complex, AlgM1M2SS and AlgQ2 adopted the inward-facing and closed conformations, respectively (Fig. 4C). However, although crystallization was conducted in the presence of ATP, no density map of ATP was observed. Diffraction data and refinement statistics are shown in Table 1.
Table 1.
Crystallization conditions, data collection, and structure refinement
| AlgM1M2SS/AlgQ2 + 6–8M | AlgQ2 + 7–10M | AlgM1M2SS/AlgQ2 + Δ3M | AlgQ2 + Δ3M | |
|---|---|---|---|---|
| Crystallization conditions | ||||
| Sample solution | 7 mg/ml AlgM1(d24) M2(H10)SS(E160Q) | 7 mg/ml AlgM1(d24) M2(H10)SS(E160Q) | ||
| 3.5 mg/ml AlgQ2 | 3 mg/ml AlgQ2 | 3.5 mg/ml AlgQ2 | 3 mg/ml AlgQ2 | |
| 2 mm 6–8M | 1 mm 7–10M | 2 mm Δ3M | 1 mm Δ3M | |
| 8 mm CHAPSO | 16 mm CHAPSO | |||
| Reservoir solution | 22% PEG 3000 | 25% PEG 4000 | 22% PEG 3000 | 30% PEG 4000 |
| 0.1 m ADA-NaOH (pH 6.6) | 0.1 m Tris-HCl (pH 8.5) | 0.1 m ADA-NaOH (pH 6.6) | 0.1 m Tris-HCl (pH 8.5) | |
| 0.15 m HCOONa | 0.2 m CaCl2 | 0.15 m NaCl | 0.2 m Li2SO4 | |
| Data collection | ||||
| Beam line | SPring-8 BL38B1 | SPring-8 BL26B2 | SPring-8 BL38B1 | SPring-8 BL38B1 |
| Detector | MX225HE (Rayonix) | MX225 (Rayonix) | Quantum315r (Area Detector Systems Corp.) | MX225HE (Rayonix) |
| Detector–crystal distance (mm) | 250 | 175 | 350 | 130 |
| HAG conditions | ||||
| Glue | 10% PVA 4500, 5% ethylene glycol | 10% PVA 4500 | ||
| Optimized humidity (%) | 85 | 90 | ||
| Wavelength (Å) | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| Resolution range (Å) | 50.0–3.60 (3.66–3.60)a | 50.0–2.00 (2.03–2.00) | 50.0–3.40 (3.46–3.40) | 30–1.55 (1.58–1.55) |
| Space group | P212121 | P1 | P212121 | P21 |
| Unit-cell parameters | ||||
| a, b, c (Å) | 71.6, 133.0, 273.0 | 46.1, 59.2, 82.7 | 72.2, 133.4, 272.7 | 76.3, 53.2, 127.5 |
| α, β, γ (degrees) | 84.7, 90.2, 88.0 | 90, 93.6, 90 | ||
| No. of molecules/asymmetric unit | 1 | 2 | 1 | 2 |
| Total observations | 144,544 | 173,757 | 266,184 | 539,873 |
| Unique reflections | 30,118 | 56,452 | 37,120 | 143,400 |
| Completeness (%) | 95.6 (98.0) | 97.4 (96.1) | 99.8 (100) | 96.8 (94.4) |
| I/σ(I) | 21.4 (3.2) | 31.4 (5.6) | 36.9 (4.1) | 25.9 (3.2) |
| Rmerge | 0.042 (0.461) | 0.053 (0.157) | 0.046 (0.536) | 0.084 (0.420) |
| CC½ in outer shell (%) | 80.2 | 95.2 | 85.3 | 85.8 |
| Mosaicity (degrees) | 1.014 | 1.510 | 0.634 | 0.524 |
| Refinement | ||||
| Resolution range (Å) | 37.4–3.60 (3.72–3.60) | 35.7–2.01 (2.04–2.01) | 37.6–3.40 (3.49–3.40) | 29.9–1.55 (1.57–1.55) |
| Rwork/Rfree | 0.238/0.289 (0.286/0.338) | 0.201/0.261 (0.264/0.325) | 0.236/0.290 (0.271/0.327) | 0.167/0.196 (0.214/0.228) |
| No. of atoms | ||||
| Protein | 14,207 | 8012 | 14,222 | 8130 |
| Alginate | 61 | 122 | 48 | 72 |
| Water | 0 | 564 | 0 | 481 |
| Calcium ion | 0 | 6 | 0 | 2 |
| Chloride ion | 0 | 0 | 0 | 2 |
| PEG | 0 | 0 | 0 | 64 |
| RMSD | ||||
| Bond lengths (Å) | 0.003 | 0.010 | 0.003 | 0.007 |
| Bond angles (degrees) | 0.781 | 1.041 | 0.820 | 1.126 |
| Ramachandran plot | ||||
| Most favored (%) | 93.32 | 97.94 | 92.26 | 97.97 |
| Allowed (%) | 6.23 | 1.96 | 7.34 | 2.03 |
| Outlier (%) | 0.45 | 0.10 | 0.39 | 0 |
a Data in the highest-resolution shell are shown in parentheses.
Alginate recognition by AlgQ2 in the complex
In addition to AlgM1M2SS in complex with long alginate oligosaccharide-bound AlgQ2, crystal structures of AlgQ2 complexed with Δ3M and 7–10M were determined to analyze configurations of oligosaccharides bound to AlgQ2. The crystal of Δ3M-bound AlgQ2 was then generated in drop solution comprising 30% PEG 4000, 0.1 m Tris-HCl (pH 8.5), and 0.2 m LiSO4. AlgQ2 was also co-crystallized with 7–10M in drop solution comprising 25% PEG 4000, 0.1 m Tris-HCl (pH 8.5), and 0.2 m calcium chloride. Finally, crystal structures of Δ3M- and 7–10M-bound AlgQ2 were determined by molecular replacement at 1.55 and 2.0 Å resolution, respectively, and overall structures of both were shown to adopt the closed conformation (Fig. 4, B and D).
The present structures of AlgM1M2SS in complex with Δ3M and 6–8M-bound AlgQ2 were essentially the same as the previously determined structure (Protein Data Bank entry 4TQU), with root mean square deviations (RMSDs) of <0.6 Å for 1641 Cα atoms. Moreover, no significant differences between structures of AlgM1M2SS in complex with Δ3M and 6–8M-bound AlgQ2 were observed at the AlgM1M2SS–AlgQ2 interface, suggesting that saccharide chain lengths had no effects on structures and conformations of AlgM1M2SS and AlgQ2.
The electron density map corresponded to four saccharide residues of AlgQ2 in the complex of AlgM1M2SS and Δ3M-bound AlgQ2, whereas three saccharide residues in Δ3M were bound to AlgQ2 in a monomer form (Fig. 4, A and B). These observations suggest two binding modes of Δ3M in the AlgQ2 interaction with AlgM1M2SS. However, the present data were insufficient for analyses of occupancy and conformations of the two trisaccharides in the complex of AlgM1M2SS and Δ3M-bound AlgQ2, and the coordinates corresponding to a tetrasaccharide were included for convenience. AlgQ2 recognizes the first to fourth saccharide residues from the non-reducing end of the substrate at subsites 1–4 (15, 16, 19). In the structure of AlgM1M2SS complexed with Δ3M-bound AlgQ2, Δ3M was accommodated at subsites 1–3 or subsites 2–4 in AlgQ2, and in the latter scenario, Δ3M was considered a translocation product of Δ3M at subsites 1–3 with the slide of one residue. This translocation of Δ3M was probably carried out through docking of Δ3M-bound AlgQ2 and the ABC transporter AlgM1M2SS.
In structures of the AlgM1M2SS and AlgQ2 complex and AlgQ2 alone, the electron density map corresponded to only five saccharide residues of AlgQ2, despite the addition of 6–8M to the crystallization drop solution (Fig. 4, C and D). Hence, AlgQ2 may strictly recognize five saccharide residues of alginate, and the sixth or later saccharide residue is flexible due to limited interactions with AlgQ2.
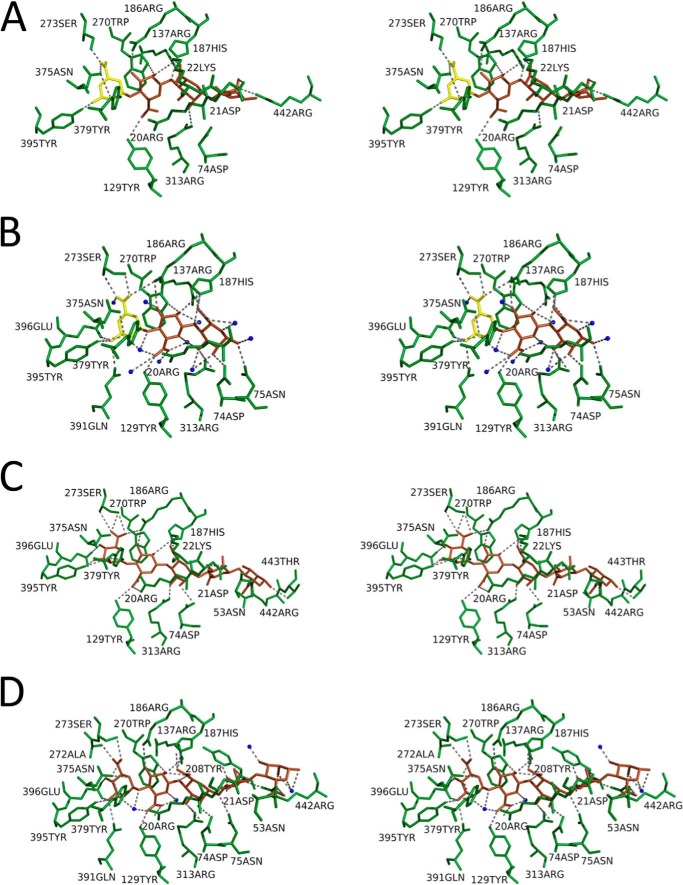
Structures of the substrate-binding site of AlgQ2 in four kinds of crystals are presented in Fig. 5. Tables 2 and 3 show amino acid residues of AlgQ2 interacting with saccharide residues via hydrogen bonds (<3.5 Å) and C–C interactions (<4.4 Å), respectively. The interaction of AlgQ2 with AlgM1M2SS produced little effect on the structure of the subsite in AlgQ2 (Fig. 5) or on the substrate-binding mode (Tables 2 and 3). Saccharide residues are represented as M1, M2, M3, M4, and M5 in that order from the non-reducing end. As reported previously (16), the number of hydrogen bonds between residues of amino acids and saccharides was greatest in M1, confirming that AlgQ2 strongly recognizes the saccharide of the non-reducing end. M5 also formed C–C interactions with multiple amino acid residues, suggesting that AlgQ2 recognizes alginate at subsites 1–5.
Figure 5.
Cross-eyed stereo views of the alginate-binding site of AlgQ2. A, AlgM1M2SS/AlgQ2 + Δ3M; B, AlgQ2 + Δ3M; C, AlgM1M2SS/AlgQ2 + 6–8M; D, AlgQ2 + 7–10M. ΔM and M are shown as yellow and brown saccharides, respectively. Water molecules are shown as blue spheres. Dotted lines, hydrogen bonds.
Table 2.
Hydrogen bond between AlgQ2 and alginate oligosaccharide
| AlgM1M2SS/AlgQ2 + 6–8M |
AlgQ2 + 7–10M |
AlgM1M2SS/AlgQ2 + Δ3M |
AlgQ2 + Δ3M |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sugar | Atom | Amino acid | Atom | Distance | Sugar | Atom | Amino acid | Atom | Distance | Sugar | Atom | Amino acid | Atom | Distance | Sugar | Atom | Amino acid | Atom | Distance |
| Å | Å | Å | Å | ||||||||||||||||
| M1 | O2 | Tyr-379 | OH | 3.0 | M1 | O2 | Asn-375 | ND2 | 3.3 | M1 | O2 | Asn-375 | ND2 | 3.3 | M1 | O2 | Asn-375 | ND2 | 3.1 |
| M1 | O2 | Tyr-395 | OH | 3.4 | M1 | O2 | Tyr-395 | OH | 3.1 | M1 | O2 | Tyr-379 | OH | 2.9 | M1 | O2 | Tyr-395 | OH | 3.1 |
| M1 | O3 | Tyr-395 | OH | 2.2 | M1 | O3 | Gln-391 | NE2 | 3.2 | M1 | O3 | Tyr-395 | OH | 2.3 | M1 | O3 | Gln-391 | NE2 | 3.3 |
| M1 | O4 | Glu-396 | OE2 | 3.5 | M1 | O3 | Tyr-395 | OH | 2.5 | M1 | O6A | Ser-273 | OG | 2.2 | M1 | O3 | Tyr-395 | OH | 2.6 |
| M1 | O6A | Ser-273 | OG | 3.0 | M1 | O3 | Glu-396 | OE1 | 3.0 | M1 | O6A | Asn-375 | ND2 | 2.7 | M1 | O3 | Glu-396 | OE1 | 2.7 |
| M1 | O6A | Ser-273 | N | 3.4 | M1 | O4 | Glu-396 | OE1 | 3.5 | M1 | O5 | Asn-375 | ND2 | 3.2 | |||||
| M1 | O6B | Ser-273 | OG | 3.2 | M1 | O4 | Glu-396 | OE2 | 3.1 | M1 | O6A | Arg-186 | NH2 | 3.5 | |||||
| M1 | O5 | Asn-375 | ND2 | 3.4 | M1 | O6A | Ser-273 | OG | 2.6 | ||||||||||
| M1 | O6A | Ser-273 | OG | 2.9 | M1 | O6A | Asn-375 | ND2 | 3.0 | ||||||||||
| M1 | O6A | Asn-375 | ND2 | 3.2 | M1 | O6B | Ser-273 | N | 2.9 | ||||||||||
| M1 | O6B | Ser-273 | N | 3.0 | M1 | O6B | Ser-273 | OG | 3.4 | ||||||||||
| M2 | O2 | His-187 | NE2 | 3.5 | M2 | O2 | Arg-137 | NH2 | 3.2 | M2 | O2 | Trp-270 | NE1 | 3.0 | M2 | O2 | Trp-270 | NE1 | 3.0 |
| M2 | O2 | Trp-270 | NE1 | 3.3 | M2 | O2 | Trp-270 | NE1 | 2.6 | M2 | O2 | Arg-137 | NH2 | 3.5 | M2 | O2 | Arg-137 | NH2 | 3.1 |
| M2 | O3 | Arg-186 | NH2 | 2.6 | M2 | O3 | Arg-186 | NH2 | 3.0 | M2 | O3 | Arg-186 | NH2 | 2.7 | M2 | O2 | His-187 | NE2 | 3.4 |
| M2 | O4 | Tyr-379 | OH | 2.9 | M2 | O3 | Trp-270 | NE1 | 3.1 | M2 | O6A | Tyr-129 | OH | 3.3 | M2 | O3 | Arg-186 | NH2 | 3.0 |
| M2 | O6B | Arg-20 | NH2 | 3.5 | M2 | O4 | Tyr-379 | OH | 3.5 | M2 | O3 | Trp-270 | NE1 | 3.3 | |||||
| M2 | O6B | Tyr-129 | OH | 3.1 | M2 | O6B | Tyr-129 | OH | 2.6 | M2 | O4 | Tyr-379 | OH | 3.5 | |||||
| M2 | O6A | Tyr-129 | OH | 2.6 | |||||||||||||||
| M3 | O2 | Asp-74 | OD2 | 3.3 | M3 | O2 | Arg-313 | NH2 | 3.3 | M3 | O1 | Asp-74 | OD2 | 3.5 | |||||
| M3 | O3 | Arg-313 | NH2 | 2.3 | M3 | O2 | Asp-74 | OD2 | 2.9 | M3 | O4 | His-187 | NE2 | 2.8 | M3 | O1 | Asn-75 | ND2 | 2.7 |
| M3 | O6A | His-187 | NE2 | 2.8 | M3 | O3 | Arg-313 | NH1 | 3.3 | M3 | O6A | Lys-22 | NZ | 2.5 | M3 | O2 | Asp-74 | OD2 | 2.8 |
| M3 | O6B | Lys-22 | NZ | 3.0 | M3 | O3 | Arg-313 | NH2 | 3.3 | M3 | O6B | Lys-22 | NZ | 3.3 | M3 | O3 | Arg-313 | NH1 | 3.2 |
| M3 | O6A | His-187 | NE2 | 3.3 | M3 | O3 | Arg-313 | NH2 | 3.2 | ||||||||||
| M3 | O4 | His-187 | NE2 | 3.4 | |||||||||||||||
| M3 | O6A | His-187 | NE2 | 2.7 | |||||||||||||||
| M4 | O3 | Asp-21 | OD2 | 3.4 | M4 | O2 | Asp-21 | OD2 | 3.2 | M4 | O2 | Asp-21 | OD2 | 3.4 | |||||
| M4 | O4 | Asp-74 | OD2 | 3.4 | M4 | O3 | Arg-20 | O | 3.5 | ||||||||||
| M4 | O4 | Asn-75 | ND2 | 2.9 | M4 | O4 | Asp-74 | OD2 | 3.4 | ||||||||||
| M4 | O6A | Arg-442 | NH2 | 3.2 | M4 | O6B | Arg-442 | NH2 | 3.0 | ||||||||||
| M5 | O1 | Thr-443 | OG1 | 2.8 | M5 | O2 | Arg-442 | NH1 | 2.4 | ||||||||||
| M5 | O2 | Arg-442 | NE | 2.5 | M5 | O6A | Asn-53 | ND2 | 3.5 | ||||||||||
| M5 | O2 | Arg-442 | NH2 | 3.4 | |||||||||||||||
| M5 | O6B | Asn-53 | ND2 | 2.5 | |||||||||||||||
Table 3.
C–C interaction between AlgQ2 and alginate oligosaccharide
| AlgM1M2SS/AlgQ2 + 6–8M |
AlgQ2 + 7–10M |
AlgM1M2SS/AlgQ2 + Δ3M |
AlgQ2 + Δ3M |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sugar | Atom | Amino acid | Atom | Distance | Sugar | Atom | Amino acid | Atom | Distance | Sugar | Atom | Amino acid | Atom | Distance | Sugar | Atom | Amino acid | Atom | Distance |
| Å | Å | Å | Å | ||||||||||||||||
| M1 | C4 | Trp-399 | CH2 | 4.4 | M1 | C1 | Trp-270 | CD2 | 4.0 | M1 | C1 | Trp-270 | CE2 | 4.4 | M1 | C1 | Trp-270 | CZ3 | 3.9 |
| M1 | C4 | Trp-399 | CZ2 | 4.3 | M1 | C1 | Trp-270 | CE3 | 4.1 | M1 | C1 | Trp-270 | CD2 | 4.3 | M1 | C1 | Trp-270 | CH2 | 4.0 |
| M1 | C5 | Trp-270 | CB | 4.3 | M1 | C1 | Trp-270 | CE2 | 4.0 | M1 | C1 | Trp-270 | CE3 | 4.0 | M1 | C1 | Trp-270 | CD2 | 3.9 |
| M1 | C5 | Trp-270 | CG | 4.2 | M1 | C1 | Trp-270 | CZ2 | 4.1 | M1 | C1 | Trp-270 | CZ3 | 3.7 | M1 | C1 | Trp-270 | CE3 | 3.9 |
| M1 | C5 | Trp-270 | CD2 | 4.0 | M1 | C1 | Trp-270 | CZ3 | 4.1 | M1 | C1 | Trp-270 | CH2 | 3.8 | M1 | C1 | Trp-270 | CE2 | 4.0 |
| M1 | C5 | Trp-270 | CE3 | 3.9 | M1 | C1 | Trp-270 | CH2 | 4.2 | M1 | C1 | Trp-270 | CZ2 | 4.2 | M1 | C1 | Trp-270 | CZ2 | 4.1 |
| M1 | C6 | Ser-273 | CB | 4.0 | M1 | C3 | Glu-396 | CD | 3.9 | M1 | C3 | Glu-396 | CD | 4.2 | M1 | C3 | Glu-396 | CD | 4.0 |
| M1 | C6 | Trp-270 | CD1 | 4.2 | M1 | C4 | Trp-399 | CH2 | 4.1 | M1 | C4 | Glu-396 | CD | 4.3 | M1 | C3 | Trp-270 | CZ3 | 4.1 |
| M1 | C6 | Trp-270 | CB | 3.3 | M1 | C4 | Trp-399 | CZ2 | 4.2 | M1 | C4 | Trp-399 | CH2 | 4.0 | M1 | C3 | Trp-270 | CE3 | 4.2 |
| M1 | C6 | Trp-270 | CG | 3.5 | M1 | C4 | Glu-396 | CD | 4.0 | M1 | C4 | Trp-399 | CZ2 | 4.2 | M1 | C4 | Glu-396 | CD | 3.7 |
| M1 | C6 | Trp-270 | CD2 | 3.8 | M1 | C5 | Trp-270 | CB | 3.8 | M1 | C5 | Trp-270 | CD2 | 4.3 | M1 | C4 | Trp-270 | CZ3 | 4.3 |
| M1 | C6 | Trp-270 | CE3 | 4.1 | M1 | C5 | Trp-270 | CG | 3.8 | M1 | C5 | Trp-270 | CE3 | 4.0 | M1 | C4 | Trp-270 | CE3 | 3.8 |
| M1 | C5 | Trp-270 | CD2 | 3.6 | M1 | C6 | Trp-399 | CH2 | 4.1 | M1 | C5 | Asn-375 | CG | 4.4 | |||||
| M1 | C5 | Trp-270 | CE3 | 3.6 | M1 | C6 | Trp-270 | CG | 4.3 | M1 | C5 | Trp-270 | CZ3 | 4.4 | |||||
| M1 | C5 | Trp-270 | CZ3 | 4.4 | M1 | C6 | Trp-270 | CD2 | 4.4 | M1 | C5 | Trp-270 | CB | 4.0 | |||||
| M1 | C6 | Ser-273 | CB | 3.6 | M1 | C6 | Trp-270 | CE3 | 4.3 | M1 | C5 | Trp-270 | CG | 4.0 | |||||
| M1 | C6 | Ser-273 | CA | 4.4 | M1 | C6 | Trp-270 | CB | 4.0 | M1 | C5 | Trp-270 | CD2 | 3.8 | |||||
| M1 | C6 | Trp-399 | CH2 | 4.1 | M1 | C6 | Ser-273 | CB | 4.0 | M1 | C5 | Trp-270 | CE3 | 3.7 | |||||
| M1 | C6 | Trp-270 | CB | 3.4 | M1 | C6 | Ser-273 | CB | 3.7 | ||||||||||
| M1 | C6 | Trp-270 | CG | 3.8 | M1 | C6 | Asn-375 | CG | 4.3 | ||||||||||
| M1 | C6 | Trp-270 | CD2 | 4.2 | M1 | C6 | Trp-399 | CH2 | 4.1 | ||||||||||
| M1 | C6 | Trp-270 | CB | 3.3 | |||||||||||||||
| M1 | C6 | Trp-270 | CG | 3.8 | |||||||||||||||
| M1 | C6 | Trp-270 | CD2 | 4.1 | |||||||||||||||
| M1 | C6 | Trp-270 | CE3 | 4.2 | |||||||||||||||
| M2 | C2 | His-187 | CE1 | 4.3 | M2 | C2 | His-187 | CE1 | 3.9 | M2 | C2 | Arg-186 | CZ | 4.4 | M2 | C2 | His-187 | CE1 | 3.7 |
| M2 | C5 | Arg-20 | CZ | 4.2 | M2 | C2 | Trp-270 | CE2 | 4.4 | M2 | C2 | His-187 | CE1 | 3.9 | M2 | C3 | Trp-270 | CE2 | 4.3 |
| M2 | C6 | Tyr-129 | CE2 | 4.3 | M2 | C3 | Trp-270 | CD1 | 4.3 | M2 | C4 | Trp-270 | CE2 | 4.3 | M2 | C4 | Trp-270 | CH2 | 4.4 |
| M2 | C3 | Trp-270 | CE2 | 4.2 | M2 | C4 | Trp-270 | CH2 | 4.3 | M2 | C4 | Trp-270 | CE2 | 3.9 | |||||
| M2 | C4 | Trp-270 | CE2 | 3.9 | M2 | C4 | Trp-270 | CZ2 | 4.0 | M2 | C4 | Trp-270 | CZ2 | 3.8 | |||||
| M2 | C4 | Trp-270 | CZ2 | 3.8 | M2 | C6 | Tyr-129 | CZ | 4.3 | ||||||||||
| M2 | C5 | Arg-20 | CZ | 4.3 | M2 | C6 | Tyr-129 | CE2 | 4.1 | ||||||||||
| M2 | C6 | Tyr-129 | CZ | 4.3 | M2 | C6 | Arg-313 | CD | 4.3 | ||||||||||
| M2 | C6 | Tyr-129 | CE2 | 4.1 | |||||||||||||||
| M3 | C2 | Arg-313 | CZ | 4.2 | M3 | C4 | His-187 | CD2 | 4.4 | M3 | C2 | Arg-313 | CZ | 4.1 | |||||
| M3 | C3 | Arg-313 | CZ | 4.1 | M3 | C5 | His-187 | CD2 | 3.9 | M3 | C3 | Arg-313 | CZ | 4.0 | |||||
| M3 | C4 | Arg-20 | CG | 4.3 | M3 | C6 | Arg-20 | CD | 4.4 | M3 | C5 | His-187 | CD2 | 4.0 | |||||
| M3 | C4 | Arg-20 | CD | 4.4 | M3 | C6 | Lys-22 | CE | 4.2 | M3 | C6 | His-187 | CD2 | 4.0 | |||||
| M3 | C5 | His-187 | CD2 | 4.3 | |||||||||||||||
| M3 | C5 | Arg-20 | CG | 4.4 | |||||||||||||||
| M3 | C6 | Arg-20 | CB | 4.2 | |||||||||||||||
| M3 | C6 | His-187 | CD2 | 4.3 | |||||||||||||||
| M3 | C6 | Arg-20 | CG | 4.0 | |||||||||||||||
| M3 | C6 | Arg-20 | CD | 4.2 | |||||||||||||||
| M4 | C2 | Asp-21 | CG | 4.3 | M4 | C2 | Asp-21 | CG | 3.7 | M4 | C2 | Asp-21 | CG | 4.0 | |||||
| M4 | C3 | Asp-21 | CG | 4.2 | M4 | C3 | Asp-21 | CG | 4.3 | M4 | C3 | Arg-20 | CA | 3.9 | |||||
| M4 | C6 | Tyr-208 | CD2 | 4.3 | M4 | C3 | Arg-20 | C | 4.4 | ||||||||||
| M4 | C5 | Asn-75 | CG | 4.3 | |||||||||||||||
| M5 | C1 | Arg-442 | CD | 4.2 | M5 | C1 | Tyr-446 | CG | 4.4 | ||||||||||
| M5 | C1 | Thr-443 | CA | 4.4 | M5 | C2 | Tyr-446 | CE1 | 4.3 | ||||||||||
| M5 | C2 | Arg-442 | CD | 4.1 | M5 | C2 | Tyr-446 | CG | 3.8 | ||||||||||
| M5 | C2 | Tyr-446 | CG | 4.2 | M5 | C2 | Tyr-446 | CD1 | 4.2 | ||||||||||
| M5 | C2 | Tyr-446 | CD2 | 4.4 | M5 | C2 | Tyr-446 | CZ | 4.1 | ||||||||||
| M5 | C2 | Tyr-446 | CB | 4.2 | M5 | C2 | Tyr-446 | CE2 | 3.7 | ||||||||||
| M5 | C3 | Tyr-446 | CG | 4.4 | M5 | C2 | Tyr-446 | CD2 | 3.6 | ||||||||||
| M5 | C3 | Tyr-446 | CD2 | 4.3 | M5 | C3 | Tyr-446 | CZ | 4.3 | ||||||||||
| M5 | C3 | Tyr-446 | CE2 | 4.1 | |||||||||||||||
| M5 | C6 | Gln-55 | CG | 4.3 | |||||||||||||||
Discussion
ABC importers are classified as type I or II based on their structures. Type I ABC transporters include the maltose transporter MalFGK2 and AlgM1M2SS, whereas the vitamin B12 transporter BtuCD is categorized as a type II ABC transporter. MalFGK2 exhibits significant ATPase activity in the presence of both maltose-binding protein (MalE) and maltose, whereas the MalE mutant (sMBP) accommodates maltose and sucrose, which is a non-transport substrate. MalFGK2 reportedly exhibited similar ATPase activity in the presence of sMBP and either maltose or sucrose (8), suggesting that ATP hydrolysis is induced by interactions between the ABC transporter and the solute-binding protein with any substrate, but not by translocation of the substrate by the ABC transporter. However, the structure of the MalFGK2 and sucrose-bound sMBP complex has not been determined, and the initial interaction mode between the ABC transporter and solute-binding protein remains unknown. In contrast, when present in excess of ATP (ATP/ABC transporter = 1:20), the ATPase activity of MalFGK2 in the presence of MalE was higher than that in the presence of both MalE and maltose (9). These observations suggest that interactions between MalE in the open conformation and MalFGK2 induce ATP hydrolysis. Accordingly, in the presence of excess ATP (ATP/ABC transporter = 1000:1), ATPase activity of MalFGK2 in the presence of MalE and maltose was higher than that in the presence of MalE alone, suggesting that MalE interacts with MalFGK2 in closed and open conformations (6). Because the complex structure of MalFGK2 and MalE has not yet been characterized at the moment of formation, the ensuing interaction scheme remains controversial. Similarly, the solute-binding protein BtuF reportedly induces ATP hydrolysis activity of the type II transporter BtuCD (21), although the details of the interaction scheme between BtuCD and BtuF are not fully understood.
The present in vitro assays indicated that long alginate oligosaccharides with PD of >5 were not transported by liposome-reconstituted AlgM1M2SS. Moreover, in alginate-importing A1 cells, AlgM1M2SS is expressed in conjunction with cell-surface pits (11), suggesting that the ABC transporter requires additional elements, such as the cell-surface pit, for import of alginate polymers. However, long alginate oligosaccharides with PD of >5 stimulated ATPase activity of AlgM1M2SS, suggesting that substrate binding by the solute-binding protein triggers ATP hydrolysis in AlgM1M2SS. Accordingly, X-ray crystallography demonstrated that long alginate oligosaccharide-bound AlgQ2 in the closed conformation interacts with AlgM1M2SS. To the best of our knowledge, this is the first structural characterization of the complex of the ABC transporter with the non-transport ligand-bound solute-binding protein.
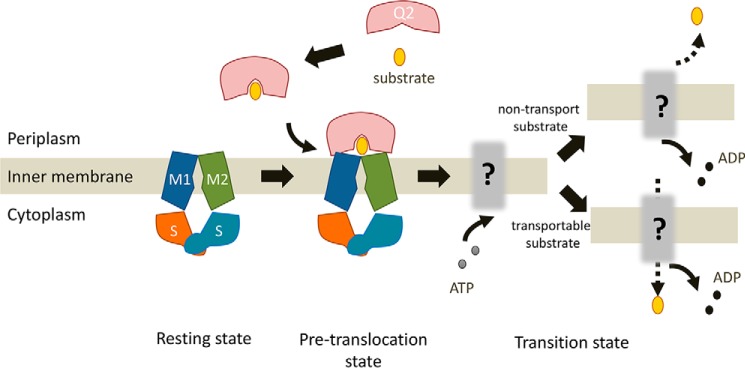
Although the structure of the AlgM1M2SS and Δ3M-bound AlgQ2 complex was determined previously, it remains unclear which step of the transport cycle was structurally characterized, because Δ3M was used as a transport substrate of AlgM1M2SS. In contrast, the complex structure of AlgM1M2SS with non-transport 6–8M-bound AlgQ2 is indicative of the initial moment of the interaction between AlgQ2 and AlgM1M2SS. This structure indicates that ATP binding and hydrolysis occur after the formation of the complex, because no ATP is included in the complex structure (Fig. 6). In conclusion, substrate-bound AlgQ2 in the closed conformation initially interacts with AlgM1M2SS, and the AlgM1M2SS–AlgQ2 complex forms subsequently, followed by ATP hydrolysis.
Figure 6.
A model for ATP hydrolysis by periplasmic solute-binding protein–dependent ABC transporter. The model shows the following steps: (i) resting state before docking of AlgM1M2SS and AlgQ2; (ii) pretranslocation state at the moment of interaction between AlgM1M2SS in the inward-facing conformation and ligand-bound AlgQ2 in the closed conformation; (iii) docking-dependent ATP binding and hydrolysis; and (iv) substrate import or return to resting state in the case of non-transport substrate.
Experimental procedures
Preparation and fluorescence labeling of alginate oligosaccharides
M oligosaccharides were prepared from alginate as reported previously (15). Briefly, alginate was hydrolyzed using HCl and was divided into M-block– and G–rich saccharides (G-block) based on differences in the solubility at pH 2.85. Treatment of neutralized M-block with alginate lyase provided unsaturated M oligosaccharides, and acid hydrolysis of M-block with HCl provided saturated M oligosaccharides. M oligosaccharides were then labeled with 2-aminopyridine as reported previously (14) and were then eluted through a HiTrap Q HP column (GE healthcare). Remaining reagents and unreacted oligosaccharides were excluded using anion exchange chromatography, and purified PA-saccharides were obtained.
FACE
FACE (17) was conducted to determine the purity of alginate oligosaccharides, which were then mixed with labeling reagent containing 75 mm 8-amino-1,3,6-naphthalenetrisulfonic acid, 0.5 m sodium cyanoborohydride, 7.5% (v/v) acetic acid, and 50% (v/v) dimethyl sulfoxide and were incubated at 37 °C for 16 h in the shade. Labeled alginate oligosaccharides were then freeze-dried and resolved in 10% glycerol and were then subjected to electrophoresis in 30% polyacrylamide gels with running buffer containing 25 mm Tris-HCl (pH 8.5) and 0.192 m glycine. Bands of labeled oligosaccharides in gels were detected using an ultraviolet lamp.
Assays of ATPase and transport activities
Expression and purification of AlgM1M2SS was conducted as reported previously (14). Briefly, E. coli BL21(DE3)Gold cells were transfected with the expression plasmid pET21b_AlgM1(d0)M2(H10)SS(WT), in which a histidine tag is attached to the C terminus of AlgM2, and AlgM1 and AlgS are encoded by wild-type sequences. The same strain was used as a host for the expression plasmid pET21b_AlgM1(d24)M2(H10)SS(E160Q), in which the encoded AlgS lacks ATPase activity and 2–24 residues of the N terminus of AlgM1 have been truncated. Membrane fractions were prepared by ultracentrifugation from cell extracts of E. coli. After solubilization with the surfactant n-dodecyl-β-d-maltoside, AlgM1M2SS was purified using a nickel-nitrilotriacetic acid column (Qiagen) and a Hi Load 16/60 Superdex 200 PG column (GE Healthcare), and expression and purification of AlgQ2 were conducted as reported previously (19). E. coli cell extracts were eluted through a HiTrap SP HP column (GE Healthcare) and a Hi Load 16/60 Superdex 200 PG column.
Proteoliposomes were prepared using l-α-phosphatidylcholine (Sigma-Aldrich) and phospholipid derived from soybean as reported previously (14). ATPase assays were performed using type II-S, whereas transport assays were performed using type IV-S. AlgM1M2SS was added to liposomes that were destabilized in n-octyl-β-d-glucoside and were diluted to less than critical micelle concentrations to produce proteoliposomes. Magnesium ions and ATP were confined in proteoliposomes for transport assays (14).
Assays of ATPase and transport activity were conducted as reported previously (14). Briefly, reactions were conducted in the presence of 0.1 μm AlgM1M2SS, 1 μm AlgQ2, 5 mm Mg-ATP, and 20 μm oligosaccharides. ATPase activity was determined as the amount of inorganic phosphate released per min, and transport activity was measured in the presence of PA-saccharides. The reaction mixture comprised 0.1 μm AlgM1M2SS, 1.3 μm AlgQ2, 2 mm ATP, 10 mm MgCl2, 20 μm PA-saccharide, and 20 mm Tris-HCl (pH 8.0). Liposomes were collected by ultracentrifugation after reactions, and transport activities were determined based on fluorescence intensities of imported PA-saccharides.
Co-immunoprecipitation assays
Interactions between AlgM1M2SS and AlgQ2 were determined using co-immunoprecipitation assays with binding/washing buffer comprising 10% glycerol, 0.1 m NaCl, 0.045% 6-cyclohexyl-hexyl-β-d-maltoside, 0.25% CHAPSO, and 20 mm Tris-HCl (pH 8.0). AlgM1M2SS exhibits significant ATPase activity in this surfactant solution. Precipitates were collected from 30 μl of Dynabeads Protein G using DynaMag-2 (Life Technologies) and were then mixed with 200 μl of binding buffer containing 0.5 μg of rabbit anti-His-tag antibody (Cosmo Bio). After incubation at 4 °C for 15 min, precipitates were washed once with wash buffer, and 200 μl of binding buffer containing 20 μg of AlgM1(d24)M2(H10)SS(WT) was added. After incubation at 4 °C for 15 min, precipitates were washed once with wash buffer, and 200-μl aliquots of binding buffers containing 20 μg of AlgQ2, 0 or 2 mm Mg-ATP, 0 or 0.2 mm Δ3M, and 0 or 5 mm sodium orthovanadate were added. After incubation at 4 °C for 15 min, precipitates were washed four times in wash buffer and were then dissolved in 10-μl aliquots of binding buffer and 10 μl of 2× SDS buffer comprising 4% SDS, 12% 2-mercaptoethanol, 20% glycerol, 0.1 m Tris-HCl (pH 6.8), and bromphenol blue. After boiling for 3 min, mixtures were centrifuged, and supernatants were subjected to SDS-PAGE analyses. AlgQ2 was then detected in Western blots using rabbit anti-AlgQ2 antiserum as a primary antibody and donkey anti-rabbit IgG as a secondary antibody (GE Healthcare).
Crystallization and X-ray diffraction experiments
Crystallization and crystal freezing of AlgQ2 (15) and crystallization of the AlgM1(d24)M2(H10)SS(E160Q) and AlgQ2 complex (14) were conducted as reported previously. Crystals of AlgM1(d24)M2(H10)SS(E160Q) and AlgQ2 complexes were frozen using the HAG method (20). Crystals were picked up using a loop covered with 10% PVA 4500 with or without ethylene glycol and were frozen in a cold nitrogen gas stream at optimized humidity.
Analysis of diffraction data
Diffraction data from frozen crystals were collected at a wavelength of 1.0000 Å using a BL38B1 or BL26B2 beamline in SPring-8 (Hyogo, Japan) and were processed using HKL2000 (22). Phases were solved by molecular replacement using Molrep (23) in CCP4 interface (24) with the structure of the AlgM1M2SS and AlgQ2 complex (Protein Data Bank entry 4TQU or 4XIG) or the structure of AlgQ2 as initial search models. Structures were refined using Refmac5 (25) and phenix. refine (26), and model construction was performed using WinCoot (27). The figures of structures were produced using PyMOL (28). Interactions between oligosaccharides and proteins were investigated using the program Contact in the CCP4 interface. RMSD values were calculated using PyMOL.
Author contributions
A. K., K. U., and S. B. performed the experiments. A. K., K. U., Y. M., N. M., S. B., T. K., B. M., K. M., and W. H. analyzed the data. Y. M., K. M., and W. H. designed the study. A. K., K. U., Y. M., K. M., and W. H. wrote the manuscript.
Acknowledgments
We thank Ai Matsunami, Takuya Yokoyama, and Chizuru Tokunaga for excellent technical assistance. Diffraction data for crystal were collected at the BL38B1 station of SPring-8 (Hyogo, Japan) with the approval of JASRI (Projects 2015B2058, 2014B1137, 2014A1148, 2013B1260, and 2013A1106).
This work was supported in part by Grants-in-Aid Scientific Research from the Japan Society for the Promotion of Science (to K. M., Y. M., and W. H.) and the Targeted Proteins Research Program (to W. H.) from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan. The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (codes 4XIG, 5H71, 4XTC, and 5H6U) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- ABC
- ATP-binding cassette
- M
- β-d-mannuronic acid
- G
- α-l-guluronic acid
- FACE
- fluorophore-assisted carbohydrate electrophoresis
- PA
- pyridylamino
- HAG
- humid air and glue-coating
- ADA
- N-(2-acetamido)iminodiacetic acid
- CHAPSO
- 3-((3-cholamidopropyl)dimethylammonio)-2-hydroxy-1-propane sulfonate
- PVA
- polyvinyl alcohol
- PD
- polymerization degree
- RMSD
- root mean square deviation
- strain A1
- Sphingomonas sp. strain A1.
References
- 1. Davidson A. L., Dassa E., Orelle C., and Chen J. (2008) Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 72, 317–364, table of contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jones P. M., and George A. M. (2004) The ABC transporter structure and mechanism: perspectives on recent research. Cell. Mol. Life Sci. 61, 682–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Locher K. P. (2009) Structure and mechanism of ATP-binding cassette transporters. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 239–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beis K. (2015) Structural basis for the mechanism of ABC transporters. Biochem. Soc. Trans. 43, 889–893 [DOI] [PubMed] [Google Scholar]
- 5. Oldham M. L., Davidson A. L., and Chen J. (2008) Structural insights into ABC transporter mechanism. Curr. Opin. Struct. Biol. 18, 726–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Oldham M. L., Chen S., and Chen J. (2013) Structural basis for substrate specificity in the Escherichia coli maltose transport system. Proc. Natl. Acad. Sci. U.S.A. 110, 18132–18137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Locher K. P., and Borths E. (2004) ABC transporter architecture and mechanism: implications from the crystal structures of BtuCD and BtuF. FEBS Lett. 564, 264–268 [DOI] [PubMed] [Google Scholar]
- 8. Gould A. D., and Shilton B. H. (2010) Studies of the maltose transport system reveal a mechanism for coupling ATP hydrolysis to substrate translocation without direct recognition of substrate. J. Biol. Chem. 285, 11290–11296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bao H., Dalal K., Cytrynbaum E., and Duong F. (2015) Sequential action of MalE and maltose allows coupling ATP hydrolysis to translocation in the MalFGK2 transporter. J. Biol. Chem. 290, 25452–25460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hisano T., Kimura N., Hashimoto W., and Murata K. (1996) Pit structure on bacterial cell surface. Biochem. Biophys. Res. Commun. 220, 979–982 [DOI] [PubMed] [Google Scholar]
- 11. Momma K., Okamoto M., Mishima Y., Mori S., Hashimoto W., and Murata K. (2000) A novel bacterial ATP-binding cassette transporter system that allows uptake of macromolecules. J. Bacteriol. 182, 3998–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hashimoto W., Miyake O., Momma K., Kawai S., and Murata K. (2000) Molecular identification of oligoalginate lyase of Sphingomonas sp. strain A1 as one of the enzymes required for complete depolymerization of alginate. J. Bacteriol. 182, 4572–4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hayashi C., Takase R., Momma K., Maruyama Y., Murata K., and Hashimoto W. (2014) Alginate-dependent gene expression mechanism in Sphingomonas sp. strain A1. J. Bacteriol. 196, 2691–2700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maruyama Y., Itoh T., Kaneko A., Nishitani Y., Mikami B., Hashimoto W., and Murata K. (2015) Structure of a bacterial ABC transporter involved in the import of an acidic polysaccharide alginate. Structure 23, 1643–1654 [DOI] [PubMed] [Google Scholar]
- 15. Nishitani Y., Maruyama Y., Itoh T., Mikami B., Hashimoto W., and Murata K. (2012) Recognition of heteropolysaccharide alginate by periplasmic solute-binding proteins of a bacterial ABC transporter. Biochemistry 51, 3622–3633 [DOI] [PubMed] [Google Scholar]
- 16. Mishima Y., Momma K., Hashimoto W., Mikami B., and Murata K. (2003) Crystal structure of AlgQ2, a macromolecule (alginate)-binding protein of Sphingomonas sp. A1, complexed with an alginate tetrasaccharide at 1.6-Å resolution. J. Biol. Chem. 278, 6552–6559 [DOI] [PubMed] [Google Scholar]
- 17. Jackson P. (1994) High-resolution polyacrylamide gel electrophoresis of fluorophore-labeled reducing saccharides. Methods Enzymol. 230, 250–265 [DOI] [PubMed] [Google Scholar]
- 18. Momma K., Mishima Y., Hashimoto W., Mikami B., and Murata K. (2005) Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry 44, 5053–5064 [DOI] [PubMed] [Google Scholar]
- 19. Momma K., Mikami B., Mishima Y., Hashimoto W., and Murata K. (2002) Crystal structure of AlgQ2, a macromolecule (alginate)-binding protein of Sphingomonas sp. A1 at 2.0 Å resolution. J. Mol. Biol. 316, 1051–1059 [DOI] [PubMed] [Google Scholar]
- 20. Baba S., Hoshino T., Ito L., and Kumasaka T. (2013) Humidity control and hydrophilic glue coating applied to mounted protein crystals improves X-ray diffraction experiments. Acta Crystallogr. D Biol. Crystallogr. 69, 1839–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Borths E. L., Poolman B., Hvorup R. N., Locher K. P., and Rees D. C. (2005) In vitro functional characterization of BtuCD-F, the Escherichia coli ABC transporter for vitamin B12 uptake. Biochemistry 44, 16301–16309 [DOI] [PubMed] [Google Scholar]
- 22. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 23. Vagin A., and Teplyakov A. (1997) MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 24.Collaborative Computational Project, Number 4 (1994) The CCP4 suite: progarms for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 25. Murshudov G. N., Vagin A. A., and Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 26. Adams P. D., Afonine P. V., Bunkoczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3, Schrödinger, LLC, New York [Google Scholar]