Abstract
Bacterial copper (Cu+) homeostasis enables both precise metallation of diverse cuproproteins and control of variable metal levels. To this end, protein networks mobilize Cu+ to cellular targets with remarkable specificity. However, the understanding of these processes is rather fragmented. Here, we use genome-wide transcriptomic analysis by RNA-Seq to characterize the response of Pseudomonas aeruginosa to external 0.5 mm CuSO4, a condition that did not generate pleiotropic effects. Pre-steady-state (5-min) and steady-state (2-h) Cu+ fluxes resulted in distinct transcriptome landscapes. Cells quickly responded to Cu2+ stress by slowing down metabolism. This was restored once steady state was reached. Specific Cu+ homeostasis genes were strongly regulated in both conditions. Our system-wide analysis revealed induction of three Cu+ efflux systems (a P1B-ATPase, a porin, and a resistance-nodulation-division (RND) system) and of a putative Cu+-binding periplasmic chaperone and the unusual presence of two cytoplasmic CopZ proteins. Both CopZ chaperones could bind Cu+ with high affinity. Importantly, novel transmembrane transporters probably mediating Cu+ influx were among those largely repressed upon Cu+ stress. Compartmental Cu+ levels appear independently controlled; the cytoplasmic Cu+ sensor CueR controls cytoplasmic chaperones and plasma membrane transporters, whereas CopR/S responds to periplasmic Cu+. Analysis of ΔcopR and ΔcueR mutant strains revealed a CopR regulon composed of genes involved in periplasmic Cu+ homeostasis and its putative DNA recognition sequence. In conclusion, our study establishes a system-wide model of a network of sensors/regulators, soluble chaperones, and influx/efflux transporters that control the Cu+ levels in P. aeruginosa compartments.
Keywords: copper, copper transport, metal homeostasis, Pseudomonas aeruginosa (P. aeruginosa), transcriptomics
Introduction
Copper is a prosthetic group of redox enzymes (1–3). However, Cu+ is also toxic to cells, disrupting Fe–S centers and participating in harmful Fenton reactions (4, 5). Thus, cells must mobilize the metal ion to specific protein targets while preventing the presence of potentially toxic free Cu+. Our goal is to understand how bacterial cells accomplish Cu+ homeostasis, shifting the attention from understanding how cells tolerate this redox active metal to how physiological needs for this ion are satisfied.
The toxicity of free Cu+ has been employed in many studies to pinpoint genes responsible for conferring tolerance to external Cu2+ (6–8). Various cytoplasmic Cu+-sensing transcriptional regulators (CueR, CsoR, and CopY) (9–11) as well as periplasmic Cu+-sensing two-component systems (CopR/S, CusR/S, PcoR/S, and CinR/S) (7, 12–14) have been identified. Biochemical analyses have shown that cytoplasmic Cu+-sensing transcriptional regulators bind Cu+ with KD in the 10−17 to 10−21 m range (10, 15), suggesting the virtual absence of free Cu+ inside cells (16). In most bacteria, these might only regulate the expression of a single Cu+ efflux ATPase and a cytoplasmic chaperone (9, 17, 18). On the other hand, the periplasmic two-component sensor CusR/S regulates expression of the multicomponent trans-periplasmic Cu+ transporter CusCFBA, a resistance-nodulation-division (RND)2-type transport system (12, 19). In any case, studies have not yet provided a clear description of the coordinated work of the independent periplasmic and cytoplasmic metal sensors.
Transmembrane P1B-type ATPases (CopA) responsible for cytoplasmic Cu+ efflux are extensively characterized elements of Cu+ tolerance systems. They receive Cu+ from the cytoplasmic chaperone (CopZ) and transfer it to periplasmic chaperones (CusF) via protein–protein interactions (20, 21). In some bacterial species, an outer membrane porin (PcoB) and the RND-type CusCBA system appear to contribute to periplasmic Cu+ efflux (22, 23). Overall, these molecular studies have showed that high binding affinities and metal transfer via protein–protein interactions are key mechanisms to confer directional specificity while maintaining the absence of free Cu+. However, the integrated interplay among various transporters present in a given system is not understood. The role of Cu+ chaperone proteins mobilizing the metal within compartments has been characterized (24, 25). In some organisms, a periplasmic chaperone (CusF) associated with the RND-type efflux system is also present (26, 27). On the other hand, organisms apparently lacking cytoplasmic or periplasmic chaperones have been identified (27). Consequently, identification of the complete set of chaperones (Cu+-binding soluble proteins) participating in compartmental metal levels homeostasis would significantly contribute to establish Cu+ distribution mechanisms.
Beyond Cu+ tolerance, metallation of cuproenzymes seems similarly mediated by Cu+ transport and chaperone proteins mobilizing and targeting Cu+ via protein–protein interactions (28, 29). For instance, SenC, a membrane protein that binds Cu+ in its periplasmic domain, acts as a chaperone transferring the metal to cytochrome c oxidase in the plasma membrane (28). However, this process requires the routing of Cu+ across the cytoplasm (i.e. influx into the cytoplasm via the CcoA transporter (30) and efflux to the periplasm by a dedicated P-type ATPase (CopA2) (31)). Thus, there is a Cu+ distribution network devoted to the process and cytoplasmic control of the events as the targeted Cu+ ion is mobilized through this compartment.
As a result of Cu+ tolerance studies, a preponderant model composed of a metal-sensing transcriptional regulator, a chaperone, and a plasma membrane efflux ATPase has emerged to explain the fate of Cu+ in bacterial cells. Microarray analyses of transcriptional responses to high, or even lethal in some cases, Cu2+ levels have further contributed to this rudimentary model (13, 32–37). Consequently, there is still a lack of an integrated vision of a system-wide Cu+ homeostatic network in a given organism. We have chosen Pseudomonas aeruginosa to search for, identify, and describe novel elements involved in this role. This opportunistic pathogen is a major cause of infection and mortality among immunosuppressed patients and it is among the microorganisms for which there is an urgent need for novel antibiotics (38). BLAST searches in the P. aeruginosa genome using known homologues reveal the presence of Cu+-sensing transcriptional regulators, cuproenzymes, putative Cu+-binding chaperone/storage proteins, and likely Cu+ transmembrane transporters (supplemental Table S1). However, the connections among these proteins and their participation in alternative Cu+ pathways (metallation of cuproenzymes or tolerance) are not self-evident.
Previous studies have shown numerous genes activated during the P. aeruginosa transcriptomic response to lethal copper concentrations (exposure to 10 mm Cu2+ for 45 min or 6 h) (13). A complex response was described, including the overexpression of various porins, several P-type ATPases, ABC ATPases, RND transporters, and other genes that are probably part of a general response to cell damage. These observations clearly include pleiotropic changes associated with the lethal conditions employed in these studies. More recently, it was reported that P. aeruginosa CueR was activated by the LasR/LasI quorum-sensing system (17). The transcriptomic response of the ΔcueR mutant strain to 10 μm Cu2+ stress during 3 h under anaerobic conditions revealed a regulon that included copA1, an RND transporter, putative chaperones PA3520 and PA3574.1, and genes PA3515 to PA3519 coding for lyases, methyl transferases, and hypothetical proteins. The functional role or integrated interplay of the coding proteins was not considered.
Here we show the genome-wide transcriptomic response of P. aeruginosa to Cu2+ stress under selected treatment conditions designed to prevent nonspecific pleiotropic effects. RNA-Seq analysis was performed at short treatment times and after Cu+ fluxes reached steady-state conditions. Our studies showed the specific up-regulation of Cu+ transporters, the uncommon occurrence of two cytoplasmic Cu+ chaperones, soluble periplasmic proteins that probably mediate the trafficking to and from Cu+ efflux transporters, and the previously unobserved repression of putative Cu+ influx transporters. Comparison of the response in ΔcueR and ΔcopR mutant strains reveals two independent regulons, controlling the distribution networks in the cytoplasmic and periplasmic compartments, respectively. Departing from copper tolerance studies, our transcriptomic and functional data allowed us to generate a model of compartmentally defined and independently regulated Cu+ distribution networks. The framework presented below as a summary of these studies suggests that Cu+ fluxes and pool sizes should be described considering multicomponent functional parameters beyond the sensor affinities or plasma membrane transport rates.
Results
Cells maintain normal growth rates under a high-intracellular copper steady-state condition
Our goal was to identify specific mechanisms of Cu+ homeostasis in P. aeruginosa. We hypothesized that exposure to non-toxic external Cu2+ stress would selectively activate homeostatic mechanisms rather than pleiotropic responses to metal toxicity, such as redox, permeability, osmotic, and metabolic imbalances. Fig. 1A shows the effect of 2-h exposure to high external CuSO4 concentrations on P. aeruginosa viability. As expected, 1–4 mm CuSO4 led to significant cell death, and no survival was observed after a 2-h exposure to 4–6 mm CuSO4. Alternatively, 0.5 mm CuSO4 had no effect on cell viability (Fig. 1A). Importantly, P. aeruginosa survived in the presence of 0.5 mm CuSO4, although the levels of intracellular copper increased >5-fold compared with untreated samples (Fig. 1B). Thus, the not only does the cell achieves homeostasis by extruding intracellular Cu+ via efflux transporters, but other mechanisms are also in play.
Figure 1.
P. aeruginosa response to external CuSO4. A, cell survival after 2-h exposure to CuSO4. B, intracellular copper level after 2-h exposure to CuSO4. C and D, copper uptake into cells exposed to 0.5 mm CuSO4. 100% = 0.30 amol of copper/cfu. Data are the mean ± S.E. (error bars), n = 3.
Based on these observations, it is reasonable to hypothesize a multifaceted cellular response to external copper, with the expression of alternative sets of genes immediately following exposure and upon stabilization of intracellular metal levels (i.e. pre-steady-state and steady-state conditions). Surprisingly, no information could be found in the literature on the kinetics of whole-cell Cu+ fluxes leading to a steady state in bacterial cells. Defining these metabolic states, Fig. 1C shows the rate of Cu+ uptake into P. aeruginosa cells upon exposure to 0.5 mm CuSO4. The influx of metal was relatively fast, and a maximum level of intracellular Cu+ was reached after 10 min. Importantly, in the presence of 0.5 mm CuSO4, the intracellular Cu+ level remained constant for at least 6 h (Fig. 1D). This implies that the system stabilizes with equal Cu+ influx and efflux rates. Moreover, this remarkable result indicates that regulation of Cu+-responsive genes occurs before the maximum Cu+ level is reached (i.e. within 10 min after copper exposure).
Copper stress triggers a metabolic shift
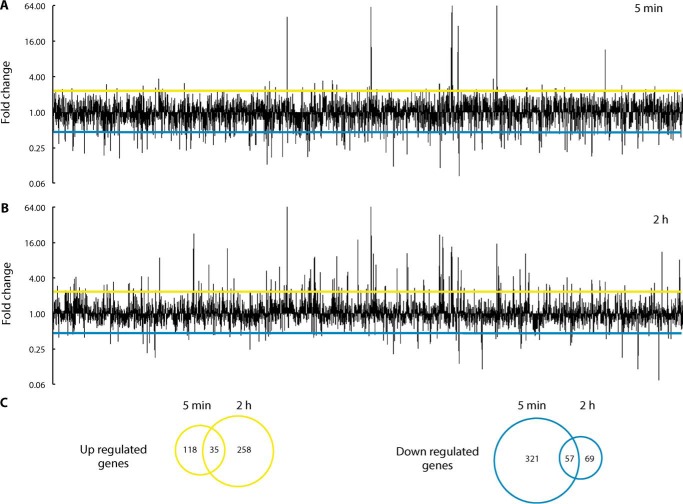
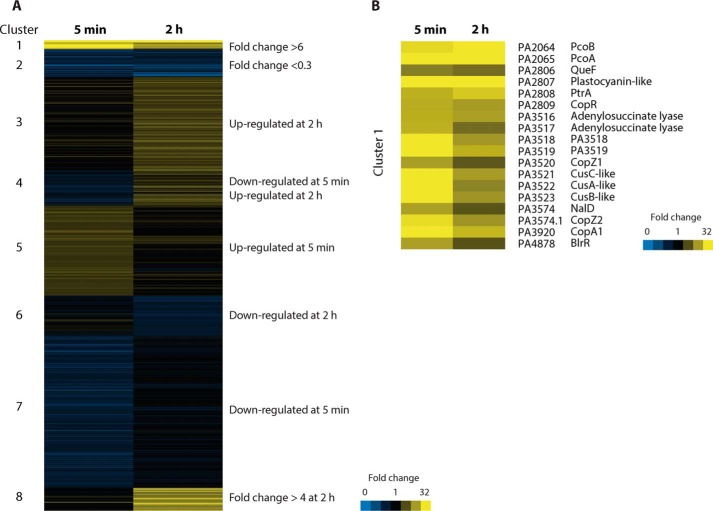
Toward capturing this initial response, as well as the distinct landscape under steady-state conditions, genome-wide transcriptomic analysis at 5 min (pre-steady state) and 2 h (steady state) of exposure to 0.5 mm CuSO4 was performed using RNA-Seq. A first examination of the genome-wide expression profile clearly showed distinct responses under each condition (Fig. 2 and supplemental Data Set S1). Cluster analysis of the differentially expressed genes (DEGs) (>2-fold change) was employed to define groups of genes with similar expression patterns. As an immediate response after 5 min of treatment (pre-steady state), a relatively small number of DEGs were induced, whereas a much larger number were repressed (Figs. 2 (A and C) and 3A (clusters 2, 4, and 7)). Examination of the functions of repressed genes points out the slowdown of cell metabolism. Genes related to fatty acid and amino acid degradation; pyruvate, nitrogen, and purine metabolism; and chemotaxis and biofilm formation pathways were all significantly down-regulated (supplemental Fig. S1) (i.e. the cell slows its metabolism, diverting resources toward the immediate response to metal stress). It was noticeable that the cluster of genes up-regulated only at 5 min included genes coding for σ factors associated with iron homeostasis (femR, femI, fiuR, fiuI, foxR, and foxI), the Fe2+ transporter FeoB, and PA3530, a bacterioferritin-associated ferredoxin. These findings are not surprising, considering the likely disruption of Fe–S centers by the high copper levels in these cells (4). Alternatively, after 2 h of exposure to external Cu2+, many DEGs were up-regulated, including some that were repressed at 5 min (Figs. 2 (B and C) and 3A (clusters 3, 4, and 8)). The functional analysis of up-regulated DEGs at 2 h showed the activation of various metabolic pathways, predominantly genes involved in the metabolism and transport of sulfur and amino acids (supplemental Fig. S2). These data indicate that a new metabolic status is established once a steady state for Cu+ levels has been achieved.
Figure 2.
Transcriptome profiling of P. aeruginosa after exposure to copper. Changes in P. aeruginosa transcriptome after 5-min (A) and 2-h (B) exposure to 0.5 mm CuSO4. Values are relative to expression levels at time 0. C, Venn diagrams of unique and overlapping DEGs between the two treatment times. Genes represented have -fold change ≥ 2 or ≤ 0.5 and corrected p value ≤ 0.05.
Figure 3.
Identification of signature genes involved in the response to copper. A, heat map of P. aeruginosa gene expression profiles upon exposure for 5 min or 2 h to 0.5 mm CuSO4. DEGs were clustered into eight groups (clusters 1–8) representing different expression patterns. Genes within clusters 1 and 2 have -fold change values > 6 or < 0.3. B, cluster of DEGs with >6-fold change after exposure to 0.5 mm CuSO4.
Compartmentally defined protein pools maintain copper levels
A relatively small subset of genes was strongly up-regulated (>6-fold change) by the exposure to Cu2+, indicating the presence of a core adaptive regulon (Fig. 3A, cluster 1). Considering first those largely up-regulated genes (Fig. 3B), five code for membrane transporters: pcoB (PA2064), an outer membrane porin; copA1 (PA3920), the Cu+-ATPase; and PA3521 (cusC), PA3522 (cusA), and PA3523 (cusB), an RND system similar to the Cu+ efflux Cus system (referred to as CusCAB in this study) (12, 39). We have previously shown Cu+ efflux by P. aeruginosa CopA1 (31). Although establishing the role of the RND-type system is beyond the scope of the present report, the abundance of Met residues in CusA and its induction by Cu+ suggest a role in extrusion of Cu+ to the extracellular milieu. Genes coding for proteins probably targeted to the periplasm, PA2807 (putative Cu+-binding protein with cupredoxin folding), PcoA (PA2065), and PtrA (PA2808), were also significantly up-regulated. copR (PA2809), encoding part of the periplasmic Cu+ two-component sensor CopR/S, belonged to this cluster of highly induced genes, whereas copS expression increased 4.8-fold at 5 min and 3.7-fold at 2 h. Moreover, an array of genes coding for cytoplasmic proteins fall into this cluster (i.e. queF (PA2806), PA3520, and PA3574.1, which encode the putative cytoplasmic Cu+ chaperones CopZ1 and CopZ2, and PA3516, PA3517, PA3518, and PA3519, which encode adenylsuccinate lyases and hypothetical proteins, respectively). Intriguingly, nalD (PA3574), a member of the TetR family of repressors, and the transcriptional regulator brlR (PA4878) were among the highly expressed genes. Some of these largely overexpressed genes (copA1, cusCAB, copZ1, copZ2, PA3516, PA3517, PA3518, and PA3519) make up the CueR regulon (17). However, cueR (PA4778) appeared in a different cluster, with a -fold change of 2.1 at 5 min and non-induction at 2 h. pcoB, pcoA, queF, PA2807 (cupredoxin-like), ptrA, and the transcriptional regulators nalD and brlR do not respond to CueR (see below). Interestingly, except for the transcriptional regulators and QueF, these remaining proteins reside in the outer membrane or in the periplasm. Moreover, soluble proteins PcoA, PA2807 (cupredoxin-like), and PtrA are probably cuproproteins with potential redox/chaperone roles. PtrA has been proposed to play a role in Cu2+ tolerance (40). A more detailed analysis of how these highly up-regulated genes might work together is presented under “Discussion.”
CopZ1 and CopZ2 are two cytoplasmic copper chaperones
The presence of two genes, PA3520 (CopZ1) and PA3574.1 (CopZ2), coding for proteins highly homologous to well-described Cu+ chaperones Atox1, Atx1, and CopZ (41–43), is striking. CopZ1 and CopZ2 are similar proteins, sharing 37% identity (51% similarity) with the invariant CXXC Cu+-binding motif close to their N-terminal ends. The CD spectra of apo-CopZ1 and apo-CopZ2 were comparable and in agreement with the βαββαβ-fold characteristic of Cu+ chaperones (Fig. 4A). The content of secondary structure elements was 42% α-helix, 17% β-strand, 17% turns, and 23% disordered in CopZ1 and 56% α-helix, 16% β-strand, 9% turns, and 17% disordered in CopZ2. Similar to other chaperones, Cu+ binding did not drastically change CopZ1 and CopZ2 secondary structures (Fig. 4B) (44). Both CopZ1 and CopZ2 bound Cu+ with 1:1 stoichiometry (CopZ1 1.00 ± 0.07 Cu+ ions/protein; CopZ2 1.04 ± 0.02 Cu+ ions/protein) and high affinity (CopZ1 KD = 3.5 ± 0.3 × 10−15 m; CopZ2 KD = 8.4 ± 0.8 × 10−17 m) (Fig. 4, C and D). These subfemtomolar KD values are comparable with those reported for other Cu+ chaperones (45). We have shown that cytoplasmic Cu+ chaperones deliver the metal to membrane Cu+-ATPases upon electrostatically driven protein–protein interactions (46, 47). In this direction, the surface of CopZ1 probably involved in the putative interaction is considerably more electronegative than that of CopZ2 (Fig. 4, E and 4F), suggesting that these two Cu+ chaperones might exchange the cation with different targets.
Figure 4.
Characterization of PA3520 (CopZ1) and PA3574. 1 (CopZ2). A and B, far-UV circular dichroism spectra of the apo-form (A) and holo-form (B) of CopZ1 (blue) and CopZ2 (red). C and D, dissociation constant KD (Cu+) of CopZ1 (C) and CopZ2 (D) determined using CuBCS2 as a competitive probe. The data were fit to KD = 3.5 ± 0.3 × 10−15 m for CopZ1 and KD = 8.4 ± 0.8 × 10−17 m for CopZ2. E and F, CopZ1 (E) and CopZ2 (F) structural models colored according to electrostatic surface potential. Cysteines involved in Cu+ binding are highlighted in yellow. Circles indicate the areas that might interact with the platform structures of P1B-ATPases based on Archaeoglobus fulgidus CopZ–CopA interaction (46).
Copper influx transporters participate in copper homeostasis
The occurrence of cell influx transporters is a prerequisite for homeostasis of a given cellular constituent. Considering that Cu+ efflux proteins are up-regulated under Cu+ stress, genes coding for Cu+ influx proteins should be repressed. We focused our attention on strongly down-regulated DEGs (<0.3-fold change) to screen for putative participants in Cu+ homeostasis (Fig. 3A, cluster 2). Three interesting proteins were identified among these: OprC (PA3790), an outer membrane porin with a proposed role in Cu+ influx; PA3789, an inner membrane protein; and PA5030, a member of the major facilitator superfamily (MFS) of transporters with an abundance of Met and His in its primary sequence. qPCR analysis confirmed these observations (Fig. 5A). Alternatively, genes encoding previously proposed Cu+ importers ccoA and hmtA (30, 48) did not change their expression levels (Fig. 5).
Figure 5.
Identification of putative copper importers. Copper-dependent expression of PA3789, oprC, PA5030, hmtA, and ccoA in WT strain after a 2-h exposure to CuSO4 (black). Values are relative to expression levels at time 0 (white). Expression values were normalized against rpsL (PA4268). Data are the mean ± S.E. (error bars), n = 3. Asterisks indicate statistically significant differences (unpaired t test; *, p ≤ 0.05; ***, p ≤ 0.001).
Metallation of cuproenzymes is regulated independently of copper levels
Contrary to the highly regulated genes involved in the control of Cu+ levels, genes associated with cuproproteins and their metallation (senC, copA2, ccoA, ccoN, nozZ, cotA, and azu; see supplemental Table S1) were not regulated. This indicates that independent Cu+ distribution networks are involved in these functions. These observations were further confirmed by qPCR of selected genes (csp1, copA2, and azu) (data not shown). The induction of genes that might participate in producing Cu+ storage pools was also explored. No changes in the expression of PA2140 (metallothionein-like) or glutathione biosynthesis genes were observed. Recently, the high Cu+-binding capability of Methylosinus trichosporium OB3b and Bacillus subtilis Csp1s has led to a suggested role for these molecules in Cu+ homeostasis (49). However, P. aeruginosa csp1 was not induced in any condition, suggesting perhaps an alternative role for Csp1.
Compartmental copper levels are independently controlled by the CopR/S and CueR regulons
Studies have shown the role of the cytoplasmic regulator CueR and the two-component system CusR/S in sensing periplasmic copper and regulating the Cus RND-type transport system (9, 12, 17). The RNA-Seq analysis showed that a number of P. aeruginosa genes strongly up-regulated during Cu2+ exposure were not part of the described CueR regulon (17). RNA-Seq analysis of ΔcueR and ΔcopR mutant strains after 5-min and 2-h exposure to 0.5 mm CuSO4 was performed to seek an integrated view of the different roles of each transcriptional regulator (supplemental Data Set S1). Both strains were fully viable in medium supplemented with this level of Cu2+ (Fig. 6A). As expected, cluster analysis of the DEGs from wild type, ΔcopR, and ΔcueR revealed two distinct subsets of genes whose Cu+-dependent induction was abolished in each one of the mutant strains (Fig. 6B, clusters 2 and 6). Importantly, expression of copR in the ΔcueR mutant background and expression of cueR in the ΔcopR mutant background were not affected (Fig. 7A). Mutation of cueR abolished expression of copA1, cusCAB, copZ1, copZ2, PA3516, PA3517, PA3518, and PA3519, confirming the described CueR regulon (17). This observation was further verified by qPCR analysis of selected genes (Fig. 7B). Alternatively, expression of pcoB, pcoA, queF, PA2807 (cupredoxin-like), and ptrA was suppressed in the ΔcopR mutant strain (Fig. 7C). These genes are organized in three operons (pcoAB, PA2808, and PA2807/06) and appear to constitute the full CopR/S regulon. Analysis of their 5′-untranslated regions revealed that the CopR putative DNA-binding region sequence is homologous to the reported Escherichia coli CusR-binding site (32) (Fig. 7D). A BLAST search of the P. aeruginosa genome did not show the additional presence of the CopR DNA-binding motif (TGACXXXTGTAAT). These results are consistent with the idea that CueR and CopR exert different regulatory roles (i.e. CueR regulates the expression of genes implicated in cytoplasmic Cu+ homeostasis, whereas control of the periplasmic metal level relies on CopR).
Figure 6.
Comparative analysis of CopR and CueR-dependent gene expression in response to copper. A, copper sensitivity after 2 h of exposure to CuSO4 in P. aeruginosa PAO1 (black), mutant strains ΔcopR (yellow), and ΔcueR (gray). Data are the mean ± S.E. (error bars) (n = 3). B, transcriptomic profiles in WT and the mutant strains ΔcopR and ΔcueR upon exposure for 5 min or 2 h to 0.5 mm CuSO4. DEGs were clustered into 10 groups (clusters 1–10) representing different expression patterns.
Figure 7.

CopR and CueR regulon. A, copper-dependent expression of copR, copS, and cueR. B, copper-dependent expression of genes belonging to the CueR regulon. C, copper-dependent expression of genes belonging to the CopR regulon. Black, WT; yellow, ΔcopR; gray, ΔcueR. Expression values were normalized against rpsL (PA4268). D, in silico analysis of DNA binding motif of CopR. The consensus sequence between E. coli cusCFBA and P. aeruginosa CopR targets is boxed. Error bars, S.E.
Discussion
The goal of this work was to obtain a global description of the bacterial mechanisms that control cellular Cu+ levels. Transcriptomic analysis, under carefully selected stress conditions minimizing pleotropic responses to Cu+ toxic effects, revealed key components of Cu+ homeostasis. The data unveil the overall cellular physiology under pre-steady-state and steady-state conditions, discriminate between elements involved in controlling compartmental Cu+ levels from others related to cuproprotein metallation, and uncover novel putative periplasmic chaperones, metal influx transporters, and the presence of two cytoplasmic chaperones. In addition, the identified regulons of both periplasmic and cytoplasmic Cu+ sensors demonstrate that Cu+ levels are controlled separately according to subcellular compartments. These findings led us to propose a testable model for Cu+ compartmental distribution in the whole organism (Fig. 8).
Figure 8.
Copper homeostasis networks in P. aeruginosa. Model depicting key components of P. aeruginosa Cu+ distribution mechanisms. The candidates for Cu+ influx transporters (green, down-regulated genes), the CopR and CueR regulon (red, up-regulated genes), and the proteins responsible for metallation of cuproproteins (non-induced) are shown. Blue, transmembrane transporters; orange, transcriptional regulators; green, cuproproteins; yellow, chaperones; gray, proteins of unknown role. Hexagonal shapes highlight redox activity.
Copper levels reach steady state in P. aeruginosa
Limited information is available on the kinetics of Cu+ distribution among bacterial cellular compartments and the sizes of the various metal pools. This is relevant in transcriptomic response studies to identify Cu+ distribution proteins while neglecting those involved in the general cellular response to a homeostatic imbalance. Nevertheless, substantial progress can be made by analyzing the rate of cellular Cu+ change upon homeostasis disruption and how fast an eventual steady state is reached. In bacterial systems, steady state implies not only equal influx/efflux rates but, more importantly, cell integrity (i.e. the absence of toxic effects leading to cell death). We observed that in the presence of sublethal external Cu2+ levels (<0.5 mm Cu2+), P. aeruginosa was in a steady state, because intracellular Cu+ level and cell viability remained constant for as long as the experiments were extended. In addition, our determinations showed that under aerobiosis, P. aeruginosa reaches steady state between 5 and 10 min following the change in external Cu2+ concentration. Notably, we did not observe an excess Cu+ load that was later decreased to basal or lower steady-state levels upon expression of homeostatic proteins. This implies that transcriptional regulation and expression/repression of relevant proteins is achieved before Cu+ equilibria. This regulation is continued, albeit slightly attenuated, maintaining a steady-state Cu+ level higher than the basal (in the absence of external Cu+). Importantly, the normal growth with high intracellular Cu+ levels implies that an unidentified intracellular Cu+-binding molecular pool should be present.
Metabolic changes occur in response to copper stress
Our approach led us to observe that the most strongly up-regulated or repressed (6–70-fold) genes were those responsible for Cu+ distribution, whereas differential expression of all others genes was significantly lower (2–3-fold). Among the latter, the pre-steady-state repression of genes participating in general metabolism was observed, suggesting the need for energy economy or reassignment to essential tasks specific to the Cu+ stress response. As cells entered steady state, there was an increase in the number of genes with a low degree of up-regulation, with the consequent restoration of primary metabolic functions.
Subsets of genes involved in Cu+ metallation of cytochrome c oxidases and denitrifying enzymes, NosZ and NirS, as well as genes coding for possible Cu+ storage pools were also distinctly unresponsive to Cu+ stress. It has been proposed that CcoA, SenC, and CopA2 are involved in metallation of cytochrome c oxidases (30, 31, 50). No induction of their coding genes was observed, suggesting separate regulatory networks for metallation of cytochrome c oxidases. However, this might be quite complex. Consider, for instance, that there are three cytochrome c oxidase systems in P. aeruginosa, the cbb3-1 oxidase (Cbb3-1), the cbb3-2 oxidase (Cbb3-2), and the aa3 oxidase (Aa3) (51), but only one set of CcoA/SenC/CopA2 proteins. Is this protein set participating in the metallation of all cytochrome c oxidases? Or are alternative unidentified transporters part of this network? Among missing elements, a cytoplasmic molecule carrying Cu+ from CcoA to CopA2 should be taken into account, because both chaperone CopZs are up-regulated by Cu+ independently of CopA2. Assembly and copper metallation of the periplasmic NosZ and NirS oxidoreductases are poorly understood, preventing further consideration. A single report indicated that the NosF and NosY, subunits of an ABC ATPase, and the periplasmic NosD, but not OprC, are required for function (52).
Among possible mechanisms contributing to Cu+ homeostasis, the presence of cytoplasmic molecules buffering a Cu+ pool has been proposed (53, 54). We observed that there is a large increase in the intracellular Cu+ under stress conditions. Early studies showed that glutathione influences the metal tolerance of E. coli cells lacking metal efflux systems; however, wild-type cells do not use glutathione to cope with metal stress (53). In agreement with these reports, we did not observe changes in the expression of enzymes involved in glutathione metabolism. Recently, a small tetrameric protein, Csp1, with high Cu+-binding capability was described in M. trichosporium OB3b and B. subtilis (49). However, no changes in expression levels in the csp1 gene were observed. Thus, whereas a Cu+-buffering cellular function appears necessary, no evidence of likely responsible molecules emerged from our studies.
Defined transport mechanisms control compartmental copper levels
P. aeruginosa-specific response to Cu+ stress could be described in the form of three components: genes up-regulated by CueR that handle cytoplasmic Cu+ levels; genes under control of the two-component CopR/S system affecting Cu+ periplasmic distribution; and genes largely down-regulated, probably involved in copper influx (Fig. 8). Previous studies suggested that P. aeruginosa OprC functions as an outer membrane porin involved in copper uptake into the periplasm (55). Our transcriptomic data support this idea as oprC is strongly down-regulated in the presence of external Cu2+. oprC is part of an operon with PA3789, coding for a protein probably located in the plasma membrane. However, the structure of PA3789 does not suggest a transport function but rather a peptidase with a PepSy domain (56). Alternatively, the gene PA5030, coding for an MFS transporter localized in the plasma membrane, was also strongly down-regulated. The abundance of Met and His in its sequence suggests metal-binding/transport capabilities. More importantly, deletion of either oprC or PA5030 conferred significant tolerance to external Cu2+. It has been suggested that CcoA, also a MFS transporter, and the P1B-type ATPase HtmA might function as plasma membrane copper uptake systems (30, 48). Rhodobacter capsulatus CcoA is required for metallation of cytochrome c oxidase, and deletion of ccoA does not confer Cu+ tolerance (30). In agreement with a role in protein metallation, external Cu2+ did not affect the expression of P. aeruginosa ccoA. Similarly, P. aeruginosa htmA does not appear to respond to Cu+ stress. HmtA increases Cu2+ and Zn2+ levels but does not alter Ag+ and Cd2+ when heterologously expressed in E. coli (48). This surprising result appears contrary to the transport mechanism of P-type ATPases, where the transported ion is required in the phosphorylation step, when metal sites face the cell cytoplasm (57). Moreover, the putative metal-coordinating residues present in HtmA are distinct from Cu+/2+- or Zn2+-ATPases (58), and similar selectivity of biomolecules for elements in the same periodic group (i.e. electronic structure (group 10 Cu+/Ag+/Au+ and group 11 Zn2+/Cd2+)) has been described (59). Thus, considering the data shown in this report, OprC and PA5030 are likely to work in tandem providing Cu+ to the cytoplasm. In this direction, we believe that an unidentified periplasmic chaperone is likely to mediate the metal movement between both transporters.
CueR-controlled genes are primarily involved in maintaining lower cytoplasmic Cu+ levels. The key players of this system are the well-characterized plasma membrane CopA1 (31) and a CusCAB transport system spanning the outer and inner membranes (39). Previous studies indicated that several P. aeruginosa RND systems might participate in the response to Cu+ stress (13). However, we observed that P. aeruginosa CusCAB is the sole RND system probably exerting this function. This, however, has a distinctive characteristic, the regulation by CueR the cytoplasmic sensor, rather than a periplasmic sensing two-component system as occurs in E. coli (17). It is interesting that the periplasmic chaperone CusF, observed in E. coli for instance, is not present in P. aeruginosa. These two observations might suggest that CusCAB acquires Cu+ from the cytoplasm. Consider that alternative transport mechanisms have been proposed for this system, acquiring Cu+ either from the cytoplasm (60) or the periplasm (61). Moreover, considering the proximity of the copZ1 (PA3520) in the genome and the presence of an extra chaperone, CopZ2, it is tempting to speculate that CopZ1 delivers Cu+ not to an ATPase but to CusCAB instead. The data suggest that both CopZ1 and CopZ2, albeit with higher affinity, are cytoplasmic Cu+ chaperones. The presence of two Cu+-ATPases in P. aeruginosa and the demonstrated role of chaperones delivering Cu+ to these transporters might suggest that each chaperone delivers metal to a distinct ATPase. However, the independent regulation and participation of CopA2 in the metallation of cytochrome c oxidase make this possibility less appealing.
We have shown that CopA delivers Cu+ to the periplasmic chaperone CusF in E. coli (21). Because CusF is not present in P. aeruginosa, it appears unlikely that CopA1 works in tandem with CusCAB. Alternatively, CopA1 might operate in conjunction with PcoB, a porin in the outer membrane under the control of CopR. However, the need for a periplasmic chaperone is still pertinent (see below). In any case, the possibility of cytoplasmic Cu+ extrusion via CopA1/PcoB and CusCAB presents the question of when the cell uses one or the other system.
The relevance of periplasmic Cu+-sensing two-component systems has been shown previously (62–64). However, a complete description of a CopR regulon was lacking. The comparative analysis of the wild-type, ΔcueR, and ΔcopR strain transcriptomes in response to external Cu2+ uncovered a small set of genes that were exclusively induced in wild-type and ΔcueR and thus likely to constitute the CopR regulon. All members of the regulon shared a conserved operator sequence that matches the E. coli CusR DNA-binding site (CusR box). Curiously, among the genes controlled, only ptrA appears to affect copper tolerance, as evaluated by bacterial survival in high metal levels (40). PcoA and PcoB were initially described as part of a copper resistance Cop operon in E. coli and Pseudomonas syringae (7, 65). As mentioned, pcoB codes for a copper-binding outer membrane porin (65). E. coli PcoA has multicopper oxidase activity and requires interaction with the periplasmic chaperone PcoC for full activity (66, 67). In P. aeruginosa, this role might be exerted by the cupredoxin-like protein (PA2807), also part of the CopR regulon. Alternatively, we hypothesize that PA2807 might participate in the copper trafficking from CopA1 to PcoB as well as in the delivery of copper to periplasmic cuproproteins. PA2807 along with ptrA and queF form the gene cluster next to the copRS operon. P. aeruginosa PtrA, a periplasmic protein of unknown function, has been associated with Cu+ tolerance (40). PA2807 and QueF are induced by Cu+, but deletion of these genes in P. putida does not lead to an increase in Cu2+ sensitivity (40). QueF is a nitrile reductase that participates in the synthesis of the tRNA-modified nucleoside queosine, which in turn improves translational accuracy (68). Consequently, QueF might be necessary to fine-tune the activation of the Cu+ homeostatic machinery.
In summary, selective copper stress conditions revealed specific homeostasis mechanisms enabling compartmental metal sensing and distribution coupled to transmembrane influx and efflux (Fig. 8). This model, although it does not include components required for cuproenzyme metallation, provides a testable system to model copper fluxes and pools under various environmental conditions, such as those encountered during bacterial virulence in eukaryotic hosts.
Experimental procedures
P. aeruginosa strains
Bacterial strains, plasmids, and oligonucleotides used in this study are listed in supplemental Table S2. P. aeruginosa PAO1 served as wild-type strain. Mutant strains PW5704 (ΔcopR) and PW9026 (ΔcueR) were obtained from the P. aeruginosa PAO1 transposon mutant library (University of Washington) (69, 70). Strains were grown at 37 °C in LB medium supplemented with Irgasan (25 μg/ml) (wild-type strain) and tetracycline (30 μg/ml) (mutant strains).
Copper sensitivity
Overnight cultures of P. aeruginosa PAO1, ΔcopR, and ΔcueR mutant strains were diluted 1:30 and grown to mid-log phase in antibiotic-free LB medium, followed by the addition of 0, 0.5, 1, 2, 4, 6, or 8 mm CuSO4. Cells were grown for 2 h, 1:10 serial dilutions were plated and incubated overnight, and colonies were counted.
Copper uptake assays
Copper uptake was measured using cells in mid-log phase in antibiotic-free LB medium. Upon the addition of 0.5 mm CuSO4, aliquots were taken at the indicated times, treated with 1 mm DTT and 1 mm bathocuproinedisulfonic acid (BCS), and harvested by centrifugation. Pellets were washed twice with 150 mm NaCl. Cells were mineralized with 15.6 m HNO3 (trace metal grade) for 1 h at 80 °C and overnight at 20 °C. Digestions were completed with 2 m H2O2. Copper levels were measured by atomic absorption spectroscopy (AAS). When required, samples were diluted with 300 mm HNO3 before analysis by AAS.
RNA isolation and sequencing
P. aeruginosa PAO1, ΔcopR, and ΔcueR mutant strains were grown to mid-log phase in antibiotic-free LB medium and treated with 0.5 mm CuSO4 for 0 min, 5 min, and 2 h. At each point, 7.5 × 108 cells were collected for RNA extraction, and 2× RNAprotect Bacteria Reagent (Qiagen) was added to stabilize RNA. Total RNA extraction was performed using an RNeasy® Plus kit (Qiagen), following the manufacturer's instructions. To minimize biological variation within samples, RNA from three biological replicates was pooled to constitute single RNA sequencing samples. Furthermore, two independent RNA samples were sequenced for each strain and condition (71).
Library construction and sequencing were performed at the Biopolymers Facility, Harvard Medical School (Boston, MA). Briefly, mRNA was enriched using the Ribo-Zero rRNA removal kit (Illumina). Strand-specific cDNA libraries were synthesized with the Apollo324TM system (WaferGen). Transcriptome sequencing was carried out to generate 150-bp pair end reads on a Next Seq 500 Mid Output platform. The number of raw reads ranged 5–10 million/sample (average = 8,429,273), ensuring optimal sequencing depth (72).
Gene expression analysis and functional categorization
Workflows were built with the Galaxy platform (73). Clean reads (average quality ≥25) were mapped against the P. aeruginosa PAO1 genome using TopHat (mean inner distance between mate pairs = 50; S.D. for distance between mate pairs = 20) (74). Count files were generated from aligned reads with HTSeq (mode to handle reads that overlap more than one feature = intersection-strict; minimum alignment quality = 20) (75). Screening of DEGs in pairwise comparisons (treated versus untreated) was performed using DEseq2 (76). We defined differential expression as the ratio between expression values ≥ ±2 (p value ≤ 0.01 and corrected p value ≤ 0.05) at 5 min and 2 h versus time 0. -Fold change values and statistical significance for each gene are provided in supplemental Data Set S1. Putative function of DEGs was manually assigned based on PseudoCap annotation (77) and literature search. Pathway annotation of genes down-regulated at 5 min and up-regulated at 2 h was performed and visualized using ClueGO App within Cytoscape (78). DEGs were clustered using Cluster 3.0, and the corresponding heat maps were generated with Java Tree View (79).
Quantitative PCR analysis was performed on samples obtained under the same conditions as those used for RNA-Seq. Total RNA was isolated as described above. Following DNase I treatment (New England Biolabs), RNA was purified with phenol/chloroform extraction and ethanol precipitation. cDNA was synthesized from 1 μg of total RNA using the ProtoScript II first strand DNA synthesis kit (New England Biolabs) in accordance with the manufacturer's protocol. All reactions were prepared in a 20-μl final volume with 10 μl of 2× Fast Essential DNA Green Master (Roche Applied Science), 0.4 μm each primer, and 25 ng of cDNA. Amplification, after a 10-min preincubation at 95 °C, consisted of 40 cycles of 95 °C for 10 s and 60 °C for 30 s, followed by a melting stage. To compare the transcript levels between different samples, we used the comparative Ct (2−ΔΔCt) method (80).
Identification of DNA-binding motif of CopR
200-bp sequences upstream of the putative CopR targets (PA2808, PA2807/PA2806, and PA2064/PA2065) were aligned with the promoter regions of target genes of orthologous regulators (E. coli CusCFBA) (63). The consensus motif obtained with MEME suit (GTCACAN5GTAAT) was used as input for a DNA motif search within the entire PAO1 chromosome (http://www.pseudomonas.com/replicon/setmotif)3 (90).
Protein expression and purification
PA3520 (copZ1) and PA3574.1 (copZ2) ORFs were amplified from genomic DNA by PCR. Amplicons were cloned in the pET-30b+ vector and transformed into E. coli BL21(DE3)pLysS cells (Invitrogen). Protein expression was induced for 3 h by the addition of 1 mm isopropyl-β-d-thiogalactopyranoside. Protein purification was carried out as described (20). Briefly, cells expressing PA3520 (CopZ1) and PA3574.1 (CopZ2) were disrupted by sonication in 25 mm Tris, pH 8, 150 mm NaCl, 100 mm sucrose, 1 mm phenylmethylsulfonyl fluoride, and 1 mm tris(2-carboxyethyl)phosphine (TCEP). Homogenates were centrifuged at 10,000 × g for 30 min at 4 °C. The resulting supernatants were loaded onto Ni2+-nitrilotriacetic acid columns, and His-tagged proteins were eluted using a 5–300 mm imidazole gradient. Buffer was exchanged using 3-kDa Centricon filters, and proteins were stored in 25 mm Hepes, pH 8, 100 mm sucrose, 150 mm NaCl, and 5 mm TCEP at −20 °C. Protein determinations were performed in accordance with Bradford (81). Protein-thiol content was measured using the Ellman method (82) and l-cysteine as a standard.
Cu+ loading to chaperone proteins
Prior to Cu+-binding studies, apo-PA3520 (CopZ1) and apo-PA3574.1 (CopZ2) were fully reduced by incubation overnight with 5 mm TCEP. Reducing agent was removed with 3-kDa Centricon filters immediately before the assay. Cu+ loading to apo-PA3520 (CopZ1) and apo-PA3574.1 (CopZ2) was performed by incubating each protein with a 2-fold molar excess of CuSO4 in 25 mm Hepes, pH 8, 150 mm NaCl, 5 mm DTT for 5 min at room temperature with gentle agitation. DTT and unbound Cu+ were removed by passage through a Sephadex G-10 column (GE Healthcare). The amount of Cu+ bound to chaperones was determined by the bicinchoninic acid method (83), using BCS as color reagent. CuSO4 was used as a standard.
Copper-binding affinity
Protein-Cu+ dissociation constants (KD) were determined by using a competition assay with BCS following the formation of the CuBCS2 complex at 483 nm (β2 formation constant for CuBCS2 = 1019.8 m−2, ϵ = 13,000 m−1 cm−1 (45)). Briefly, 10 μm Cu+, 25 μm BCS in 25 mm Hepes, pH 8, 150 mm NaCl, 10 mm ascorbic acid was titrated with 1–20 μm protein and incubated for 5 min at room temperature, and the 300–800 nm absorption spectra were recorded. KD values were calculated by curve fitting of the experimental data to the equilibrium equation (45).
| (Eq. 1) |
Reported errors are asymptotic S.E. values provided by the fitting software (KaleidaGraph, Synergy).
Circular dichroism spectroscopy
Far-UV CD spectra (190–240 nm) of apo- and holo-forms of PA3520 (CopZ1) and PA3574.1 (CopZ2) were recorded using a 1-mm quartz cuvette at 37 °C and a J-1500 spectrometer. Different protein concentrations (0.06–0.1 mg/ml) were prepared in 10 mm buffer phosphate, pH 8, 20% glycerol, 1 mm TCEP. The consensus spectrum averaged 20 individual scans. Secondary structure analyses using the CDDSTR algorithm (set 4) (84) were carried out with the DichroWeb analysis server (85, 86). The CD data sets are reported as mean residue molar ellipticity (degrees cm2 dmol−1 residue−1).
Molecular modeling
Homology modeling of P. aeruginosa PA3520 (CopZ1) and PA3574.1 (CopZ2) was performed with the Phyre2 server (87), using Salmonella enterica GolT (Protein Data Bank entry 4Y2I) as a template. Quality of the models was assessed with the ProQ2 algorithm (88). The electrostatic surface potentials were calculated using the Adaptive Poisson-Boltzmann Solver (APBS) with the PyMOL APBS tool (89).
Author contributions
J. Q. and J. M. A. designed the research; J. Q. and L. N.-A. performed the research; J. Q., L. N.-A., and J. M. A. analyzed the data; and J. Q., L. N.-A., and J. M. A. wrote the paper.
Supplementary Material
Acknowledgments
We thank James McIsaac for providing vectors for expression of PA3520 and PA3574.1, Virginia Nuñez Mir for help with the initial processing of RNA-Seq data, Drs. Scarlet Shell and Reeta Prusty for helpful discussions, and Dr. Elizabeth Bafaro for critical reading of the manuscript.
This work was supported by NIGMS, National Institutes of Health Grant R01 GM114949 (to J. M. A.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Data Set S1, Tables S1 and S2, and Figs. S1 and S2.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- RND
- resistance-nodulation-division
- ABC
- ATP-binding cassette
- DEGs
- differentially expressed genes
- MFS
- major facilitator superfamily
- AAS
- atomic absorption spectroscopy
- TCEP
- tris(2-carboxyethyl)phosphine
- BCS
- bathocuproinedisulfonic acid.
References
- 1. Fraústo da Silva J. J. R., and Williams R. J. P. (2001) The Biological Chemistry of the Elements: The Inorganic Chemistry of Life, 2nd Ed., pp. 418–425, Oxford University Press, Oxford, UK [Google Scholar]
- 2. Tavares P., Pereira A. S., Moura J. J., and Moura I. (2006) Metalloenzymes of the denitrification pathway. J. Inorg. Biochem. 100, 2087–2100 [DOI] [PubMed] [Google Scholar]
- 3. Cobine P. A., Pierrel F., and Winge D. R. (2006) Copper trafficking to the mitochondrion and assembly of copper metalloenzymes. Biochim. Biophys. Acta 1763, 759–772 [DOI] [PubMed] [Google Scholar]
- 4. Macomber L., and Imlay J. A. (2009) The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. U.S.A. 106, 8344–8349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dupont C. L., Grass G., and Rensing C. (2011) Copper toxicity and the origin of bacterial resistance: new insights and applications. Metallomics 3, 1109–1118 [DOI] [PubMed] [Google Scholar]
- 6. Argüello J. M., Raimunda D., and Padilla-Benavides T. (2013) Mechanisms of copper homeostasis in bacteria. Front. Cell Infect. Microbiol. 3, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rensing C., and Grass G. (2003) Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol. Rev. 27, 197–213 [DOI] [PubMed] [Google Scholar]
- 8. Osman D., and Cavet J. S. (2008) Copper homeostasis in bacteria. Adv. Appl. Microbiol. 65, 217–247 [DOI] [PubMed] [Google Scholar]
- 9. Outten F. W., Outten C. E., Hale J., and O'Halloran T. V. (2000) Transcriptional activation of an Escherichia coli copper efflux regulon by the chromosomal MerR homologue, CueR. J. Biol. Chem. 275, 31024–31029 [DOI] [PubMed] [Google Scholar]
- 10. Liu T., Ramesh A., Ma Z., Ward S. K., Zhang L., George G. N., Talaat A. M., Sacchettini J. C., and Giedroc D. P. (2007) CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat. Chem. Biol. 3, 60–68 [DOI] [PubMed] [Google Scholar]
- 11. Strausak D., and Solioz M. (1997) CopY is a copper-inducible repressor of the Enterococcus hirae copper ATPases. J. Biol. Chem. 272, 8932–8936 [DOI] [PubMed] [Google Scholar]
- 12. Outten F. W., Huffman D. L., Hale J. A., and O'Halloran T. V. (2001) The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J. Biol. Chem. 276, 30670–30677 [DOI] [PubMed] [Google Scholar]
- 13. Teitzel G. M., Geddie A., De Long S. K., Kirisits M. J., Whiteley M., and Parsek M. R. (2006) Survival and growth in the presence of elevated copper: transcriptional profiling of copper-stressed Pseudomonas aeruginosa. J. Bacteriol. 188, 7242–7256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Quaranta D., McCarty R., Bandarian V., and Rensing C. (2007) The copper-inducible cin operon encodes an unusual methionine-rich azurin-like protein and a pre-Q0 reductase in Pseudomonas putida KT2440. J. Bacteriol. 189, 5361–5371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Changela A., Chen K., Xue Y., Holschen J., Outten C. E., O'Halloran T. V., and Mondragón A. (2003) Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science 301, 1383–1387 [DOI] [PubMed] [Google Scholar]
- 16. Rae T. D., Schmidt P. J., Pufahl R. A., Culotta V. C., and O'Halloran T. V. (1999) Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science 284, 805–808 [DOI] [PubMed] [Google Scholar]
- 17. Thaden J. T., Lory S., and Gardner T. S. (2010) Quorum-sensing regulation of a copper toxicity system in Pseudomonas aeruginosa. J. Bacteriol. 192, 2557–2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Smaldone G. T., and Helmann J. D. (2007) CsoR regulates the copper efflux operon copZA in Bacillus subtilis. Microbiology 153, 4123–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gudipaty S. A., Larsen A. S., Rensing C., and McEvoy M. M. (2012) Regulation of Cu(I)/Ag(I) efflux genes in Escherichia coli by the sensor kinase CusS. FEMS Microbiol. Lett. 330, 30–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. González-Guerrero M., and Argüello J. M. (2008) Mechanism of Cu+-transporting ATPases: soluble Cu+ chaperones directly transfer Cu+ to transmembrane transport sites. Proc. Natl. Acad. Sci. U.S.A. 105, 5992–5997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Padilla-Benavides T., George Thompson A. M., McEvoy M. M., and Argüello J. M. (2014) Mechanism of ATPase-mediated Cu+ export and delivery to periplasmic chaperones: the interaction of Escherichia coli CopA and CusF. J. Biol. Chem. 289, 20492–20501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim E. H., Rensing C., and McEvoy M. M. (2010) Chaperone-mediated copper handling in the periplasm. Nat. Prod. Rep. 27, 711–719 [DOI] [PubMed] [Google Scholar]
- 23. Lee S. M., Grass G., Rensing C., Barrett S. R., Yates C. J., Stoyanov J. V., and Brown N. L. (2002) The pco proteins are involved in periplasmic copper handling in Escherichia coli. Biochem. Biophys. Res. Commun. 295, 616–620 [DOI] [PubMed] [Google Scholar]
- 24. Robinson N. J., and Winge D. R. (2010) Copper metallochaperones. Annu. Rev. Biochem. 79, 537–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. O'Halloran T. V., and Culotta V. C. (2000) Metallochaperones, an intracellular shuttle service for metal ions. J. Biol. Chem. 275, 25057–25060 [DOI] [PubMed] [Google Scholar]
- 26. Cubillas C., Miranda-Sanchez F., Gonzalez-Sanchez A., Elizalde J. P., Vinuesa P., Brom S., and Garcia-de Los Santos A. (2017) A comprehensive phylogenetic analysis of copper transporting P1B ATPases from bacteria of the rhizobiales order uncovers multiplicity, diversity and novel taxonomic subtypes. Microbiologyopen 10.1002/mbo3.452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hernández-Montes G., Argüello J. M., and Valderrama B. (2012) Evolution and diversity of periplasmic proteins involved in copper homeostasis in γ proteobacteria. BMC Microbiol. 12, 249–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lohmeyer E., Schröder S., Pawlik G., Trasnea P. I., Peters A., Daldal F., and Koch H. G. (2012) The ScoI homologue SenC is a copper binding protein that interacts directly with the cbb(3)-type cytochrome oxidase in Rhodobacter capsulatus. Biochim. Biophys. Acta 1817, 2005–2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Osman D., Patterson C. J., Bailey K., Fisher K., Robinson N. J., Rigby S. E., and Cavet J. S. (2013) The copper supply pathway to a Salmonella Cu,Zn-superoxide dismutase (SodCII) involves P1B-type ATPase copper efflux and periplasmic CueP. Mol. Microbiol. 87, 466–477 [DOI] [PubMed] [Google Scholar]
- 30. Ekici S., Yang H., Koch H. G., and Daldal F. (2012) Novel transporter required for biogenesis of cbb3-type cytochrome c oxidase in Rhodobacter capsulatus. MBio 10.1128/mBio.00293-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. González-Guerrero M., Raimunda D., Cheng X., and Argüello J. M. (2010) Distinct functional roles of homologous Cu+ efflux ATPases in Pseudomonas aeruginosa. Mol. Microbiol. 78, 1246–1258 [DOI] [PubMed] [Google Scholar]
- 32. Yamamoto K., and Ishihama A. (2005) Transcriptional response of Escherichia coli to external copper. Mol. Microbiol. 56, 215–227 [DOI] [PubMed] [Google Scholar]
- 33. Kershaw C. J., Brown N. L., Constantinidou C., Patel M. D., and Hobman J. L. (2005) The expression profile of Escherichia coli K-12 in response to minimal, optimal and excess copper concentrations. Microbiology 151, 1187–1198 [DOI] [PubMed] [Google Scholar]
- 34. Pontel L. B., Scampoli N. L., Porwollik S., Checa S. K., McClelland M., and Soncini F. C. (2014) Identification of a Salmonella ancillary copper detoxification mechanism by a comparative analysis of the genome-wide transcriptional response to copper and zinc excess. Microbiology 160, 1659–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shafeeq S., Yesilkaya H., Kloosterman T. G., Narayanan G., Wandel M., Andrew P. W., Kuipers O. P., and Morrissey J. A. (2011) The cop operon is required for copper homeostasis and contributes to virulence in Streptococcus pneumoniae. Mol. Microbiol. 81, 1255–1270 [DOI] [PubMed] [Google Scholar]
- 36. Schelder S., Zaade D., Litsanov B., Bott M., and Brocker M. (2011) The two-component signal transduction system CopRS of Corynebacterium glutamicum is required for adaptation to copper-excess stress. PLoS One 6, e22143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ward S. K., Hoye E. A., and Talaat A. M. (2008) The global responses of Mycobacterium tuberculosis to physiological levels of copper. J. Bacteriol. 190, 2939–2946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wagner S., Sommer R., Hinsberger S., Lu C., Hartmann R. W., Empting M., and Titz A. (2016) Novel strategies for the treatment of Pseudomonas aeruginosa infections. J. Med. Chem. 59, 5929–5969 [DOI] [PubMed] [Google Scholar]
- 39. Kim E. H., Nies D. H., McEvoy M. M., and Rensing C. (2011) Switch or funnel: how RND-type transport systems control periplasmic metal homeostasis. J. Bacteriol. 193, 2381–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Elsen S., Ragno M., and Attree I. (2011) PtrA is a periplasmic protein involved in Cu tolerance in Pseudomonas aeruginosa. J. Bacteriol. 193, 3376–3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Arnesano F., Banci L., Bertini I., Ciofi-Baffoni S., Molteni E., Huffman D. L., and O'Halloran T. V. (2002) Metallochaperones and metal-transporting ATPases: a comparative analysis of sequences and structures. Genome Res. 12, 255–271 [DOI] [PubMed] [Google Scholar]
- 42. Banci L., Bertini I., Del Conte R., Markey J., and Ruiz-Dueñas F. J. (2001) Copper trafficking: the solution structure of Bacillus subtilis CopZ. Biochemistry 40, 15660–15668 [DOI] [PubMed] [Google Scholar]
- 43. Wernimont A. K., Huffman D. L., Lamb A. L., O'Halloran T. V., and Rosenzweig A. C. (2000) Structural basis for copper transfer by the metallochaperone for the Menkes/Wilson disease proteins. Nat. Struct. Biol. 7, 766–771 [DOI] [PubMed] [Google Scholar]
- 44. Harrison M. D., Meier S., and Dameron C. T. (1999) Characterisation of copper-binding to the second sub-domain of the Menkes protein ATPase (mnkr2). Biochim. Biophys. Acta 1453, 254–260 [DOI] [PubMed] [Google Scholar]
- 45. Xiao Z., Brose J., Schimo S., Ackland S. M., La Fontaine S., and Wedd A. G. (2011) Unification of the copper(I) binding affinities of the metallo-chaperones Atx1, atox1, and related proteins: detection probes and affinity standards. J. Biol. Chem. 286, 11047–11055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Padilla-Benavides T., McCann C. J., and Argüello J. M. (2013) The mechanism of Cu+ transport ATPases: interaction with Cu+ chaperones and the role of transient metal-binding sites. J. Biol. Chem. 288, 69–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Blaby-Haas C. E., Padilla-Benavides T., Stube R., Arguello J. M., and Merchant S. S. (2014) Evolution of a plant-specific copper chaperone family for chloroplast copper homeostasis. Proc. Nat. Acad. Sc. U.S.A. 111, E5480–E5487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lewinson O., Lee A. T., and Rees D. C. (2009) A P-type ATPase importer that discriminates between essential and toxic transition metals. Proc. Natl. Acad. Sci. U.S.A. 106, 4677–4682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vita N., Landolfi G., Baslé A., Platsaki S., Lee J., Waldron K. J., and Dennison C. (2016) Bacterial cytosolic proteins with a high capacity for Cu(I) that protect against copper toxicity. Sci. Rep. 6, 39065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Trasnea P.-I. I., Utz M., Khalfaoui-Hassani B., Lagies S., Daldal F., and Koch H.-G. G. (2016) Cooperation between two periplasmic copper chaperones is required for full activity of the cbb3 -type cytochrome c oxidase and copper homeostasis in Rhodobacter capsulatus. Mol. Microbiol. 100, 345–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Comolli J. C., and Donohue T. J. (2004) Differences in two Pseudomonas aeruginosa cbb3 cytochrome oxidases. Mol. Microbiol. 51, 1193–1203 [DOI] [PubMed] [Google Scholar]
- 52. Wunsch P., Herb M., Wieland H., Schiek U. M., and Zumft W. G. (2003) Requirements for CuA and Cu-S center assembly of nitrous oxide reductase deduced from complete periplasmic enzyme maturation in the nondenitrifier Pseudomonas putida. J. Bacteriol. 185, 887–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Helbig K., Bleuel C., Krauss G. J., and Nies D. H. (2008) Glutathione and transition-metal homeostasis in Escherichia coli. J. Bacteriol. 190, 5431–5438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vita N., Platsaki S., Baslé A., Allen S. J., Paterson N. G., Crombie A. T., Murrell J. C., Waldron K. J., and Dennison C. (2015) A four-helix bundle stores copper for methane oxidation. Nature 525, 140–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yoneyama H., and Nakae T. (1996) Protein c (OprC) of the outer membrane of Pseudomonas aeruginosa is a copper-regulated channel protein. Microbiology 142, 2137–2144 [DOI] [PubMed] [Google Scholar]
- 56. Yeats C., Rawlings N. D., and Bateman A. (2004) The PepSY domain: a regulator of peptidase activity in the microbial environment? Trends Biochem. Sci. 29, 169–172 [DOI] [PubMed] [Google Scholar]
- 57. Rosenzweig A. C., and Argüello J. M. (2012) Toward a molecular understanding of metal transport by P1B-type ATPases. Curr. Top. Membr. 69, 113–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Argüello J. M. (2003) Identification of ion selectivity determinants in heavy metal transport P1B-type ATPases. J. Membr. Biol. 195, 93–108 [DOI] [PubMed] [Google Scholar]
- 59. Ma Z., Jacobsen F. E., and Giedroc D. P. (2009) Coordination chemistry of bacterial metal transport and sensing. Chem. Rev. 109, 4644–4681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Long F., Su C. C., Zimmermann M. T., Boyken S. E., Rajashankar K. R., Jernigan R. L., and Yu E. W. (2010) Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport. Nature 467, 484–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chacón K. N., Mealman T. D., McEvoy M. M., and Blackburn N. J. (2014) Tracking metal ions through a Cu/Ag efflux pump assigns the functional roles of the periplasmic proteins. Proc. Natl. Acad. Sci. U.S.A. 111, 15373–15378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Caille O., Rossier C., and Perron K. (2007) A copper-activated two-component system interacts with zinc and imipenem resistance in Pseudomonas aeruginosa. J. Bacteriol. 189, 4561–4568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Munson G. P., Lam D. L., Outten F. W., and O'Halloran T. V. (2000) Identification of a copper-responsive two-component system on the chromosome of Escherichia coli K-12. J. Bacteriol. 182, 5864–5871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pezza A., Pontel L. B., López C., and Soncini F. C. (2016) Compartment and signal-specific codependence in the transcriptional control of Salmonella periplasmic copper homeostasis. Proc. Natl. Acad. Sci. U.S.A. 113, 11573–11578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cha J. S., and Cooksey D. A. (1991) C Copper resistance in Pseudomonas syringae mediated by periplasmic and outer membrane proteins. Proc. Natl. Acad. Sci. U.S.A. 88, 8915–8919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Huffman D. L., Huyett J., Outten F. W., Doan P. E., Finney L. A., Hoffman B. M., and O'Halloran T. V. (2002) Spectroscopy of Cu(II)-PcoC and the multicopper oxidase function of PcoA, two essential components of Escherichia coli pco copper resistance operon. Biochemistry 41, 10046–10055 [DOI] [PubMed] [Google Scholar]
- 67. Djoko K. Y., Xiao Z., and Wedd A. G. (2008) Copper resistance in E. coli: the multicopper oxidase PcoA catalyzes oxidation of copper(I) in Cu(I)Cu(II)-PcoC. Chembiochem 9, 1579–1582 [DOI] [PubMed] [Google Scholar]
- 68. Lee B. W. K., Van Lanen S. G., and Iwata-Reuyl D. (2007) Mechanistic studies of Bacillus subtilis QueF, the nitrile oxidoreductase involved in queuosine biosynthesis. Biochemistry 46, 12844–12854 [DOI] [PubMed] [Google Scholar]
- 69. Held K., Ramage E., Jacobs M., Gallagher L., and Manoil C. (2012) Sequence-verified two-allele transposon mutant library for Pseudomonas aeruginosa PAO1. J. Bacteriol. 194, 6387–6389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Jacobs M. A., Alwood A., Thaipisuttikul I., Spencer D., Haugen E., Ernst S., Will O., Kaul R., Raymond C., Levy R., Chun-Rong L., Guenthner D., Bovee D., Olson M. V., and Manoil C. (2003) Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 100, 14339–14344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Conesa A., Madrigal P., Tarazona S., Gomez-Cabrero D., Cervera A., McPherson A., Szcześniak M. W., Gaffney D. J., Elo L. L., Zhang X., and Mortazavi A. (2016) A survey of best practices for RNA-Seq data analysis. Genome Biol. 17, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Haas B. J., Chin M., Nusbaum C., Birren B. W., and Livny J. (2012) How deep is deep enough for RNA-Seq profiling of bacterial transcriptomes? BMC Genomics 13, 734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Afgan E., Baker D., van den Beek M., Blankenberg D., Bouvier D., Čech M., Chilton J., Clements D., Coraor N., Eberhard C., Grüning B., Guerler A., Hillman-Jackson J., Von Kuster G., Rasche E., et al. (2016) The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 44, W3–W10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., and Salzberg S. L. (2013) Tophat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Anders S., Pyl P. T., and Huber W. (2015) HTSeq: a python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Love M. I., Huber W., and Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Winsor G. L., Lo R., Sui S. J. H., Ung K. S. E., Huang S., Cheng D., Ching W. K., Hancock R. E., and Brinkman F. S. (2005) Pseudomonas aeruginosa Genome Database and PseudoCAP: facilitating community-based, continually updated, genome annotation. Nucleic Acids Res. 33, D338–D343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Corbett D., Schuler S., Glenn S., Andrew P. W., Cavet J. S., and Roberts I. S. (2011) The combined actions of the copper-responsive repressor CsoR and copper-metallochaperone CopZ modulate CopA-mediated copper efflux in the intracellular pathogen Listeria monocytogenes. Mol. Microbiol. 81, 457–472 [DOI] [PubMed] [Google Scholar]
- 79. Saldanha A. J. (2004) Java treeview: extensible visualization of microarray data. Bioinformatics 20, 3246–3248 [DOI] [PubMed] [Google Scholar]
- 80. Livak K. J., and Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 81. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 82. Ellman G. L. (1959) Tissue sulfhydryl groups. Arch. Biochem. Biophys. 82, 70–77 [DOI] [PubMed] [Google Scholar]
- 83. Brenner A. J., and Harris E. D. (1995) A quantitative test for copper using bicinchoninic acid. Anal. Biochem. 226, 80–84 [DOI] [PubMed] [Google Scholar]
- 84. Sreerama N., and Woody R. W. (2000) stimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 287, 252–260 [DOI] [PubMed] [Google Scholar]
- 85. Lobley A., Whitmore L., and Wallace B. A. (2002) DICHROWEB: an interactive website for the analysis of protein secondary structure from circular dichroism spectra. Bioinformatics 18, 211–212 [DOI] [PubMed] [Google Scholar]
- 86. Whitmore L., and Wallace B. A. (2004) DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 32, W668–W673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kelley L. A., Mezulis S., Yates C. M., Wass M. N., and Sternberg M. J. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ray A., Lindahl E., and Wallner B. (2012) Improved model quality assessment using ProQ2. BMC Bioinformatics 13, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Baker N. A., Sept D., Joseph S., Holst M. J., and McCammon J. A. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Winsor G. L., Griffiths E. J., Lo R., Dhillon B. K., Shay J. A., Brinkman F. S. (2016) Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 44, D646–D653 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.