Abstract
The organic anion transporters OAT1 (SLC22A6) and OAT3 (SLC22A8) have similar substrate specificity for drugs, but it is far from clear whether this holds for endogenous substrates. By analysis of more than 600 metabolites in the Oat3KO (Oat3 knockout) by LC/MS, we demonstrate OAT3 involvement in the movement of gut microbiome products, key metabolites, and signaling molecules, including those flowing through the gut–liver–kidney axis. Major pathways affected included those involved in metabolism of bile acids, flavonoids, nutrients, amino acids (including tryptophan-derivatives that are uremic toxins), and lipids. OAT3 is also critical in elimination of liver-derived phase II metabolites, particularly those undergoing glucuronidation. Analysis of physicochemical features revealed nine distinct metabolite groups; at least one member of most clusters has been previously validated in transport assays. In contrast to drugs interacting with the OATs, endogenous metabolites accumulating in the Oat1KO (Oat1 knockout) versus Oat3KO have distinct differences in their physicochemical properties; they are very different in size, number of rings, hydrophobicity, and molecular complexity. Consistent with the Remote Sensing and Signaling Hypothesis, the data support the importance of the OAT transporters in inter-organ and inter-organismal remote communication via transporter-mediated movement of key metabolites and signaling molecules (e.g. gut microbiome–to–intestine–to–blood–to–liver–to–kidney–to–urine). We discuss the possibility of an intimate connection between OATs and metabolite sensing and signaling pathways (e.g. bile acids). Furthermore, the metabolomics and pathway analysis support the view that OAT1 plays a greater role in kidney proximal tubule metabolism and OAT3 appears relatively more important in systemic metabolism, modulating levels of metabolites flowing through intestine, liver, and kidney.
Keywords: drug transport, kidney, multidrug transporter, renal physiology, xenobiotic, OAT3, kidney drug transport, organic anion, solute carrier, uremic toxin
Introduction
There has been a recent surge of interest in “drug” transporters, in large part because of new regulatory guidelines from the Food and Drug Administration indicating the need to test new drugs for interaction with and/or transport by drug transporters (1). These transporters, sometimes classified as phase 3 drug-metabolizing enzymes (DMEs),2 are generally considered to be critical for absorption, distribution, and elimination of a wide range of drugs (2–5).
Drug transporters belong to both the solute carrier (SLC) and ATP-binding cassette (ABC) superfamilies; however, two of the seven transporters highlighted by the Food and Drug Administration are the SLC22 transporters known as the organic anion transporters (OATs) (1). These are OAT1 (SLC22A6, first described as NKT, which was identified using codon-optimized differential display) (6–8) and OAT3 (SLC22A8, also known as Roct) (9). Despite their importance in the transport of drugs and toxins, which is now well-supported by in vivo data from murine knockouts of each gene (10–27), detailed evolutionary analyses indicate that these transporters are highly conserved and are present in organisms such as fly, worm, and sea urchin (28–30). These multispecific transporters are abundantly expressed on the basolateral membrane of proximal tubule (PT) cells of the kidney (as well as several other epithelial tissues). Together these two transporters represent an important rate-limiting step in the uptake from the plasma of a wide variety of drugs, exogenous toxins, nutrients, and endogenous metabolites (including uremic toxins) (2, 3, 27, 31).
Moreover, although their normal adult expression is in the kidney, choroid plexus, and a few other tissues, during embryogenesis OAT1 and OAT3 are highly expressed in a number of developing tissues, which has raised the question of a functional role in the transport of small molecules involved in morphogenesis (22, 32). In addition, OAT family members that are closely related to OAT1 and OAT3, such as OAT6 (SLC22A20) and URAT1 (SLC22A12; originally identified as Rst in mice), are, respectively, known to bind and/or transport odorant molecules and uric acid (30, 33–35).
All of this raises the question as to what is the endogenous physiological function of OAT1, OAT3, and other multispecific drug transporters (2). Because OAT1 and OAT3 are critical for the movement of endogenous organic anion compounds from the plasma and into the renal PT cell, deletion of these transporters should therefore result in the accumulation of such compounds in the serum of knock-out animals (and/or their deficiency in the urine). Thus, metabolomics analyses of the serum and/or urine of such Oat knock-out animals would provide a means to link these transporters into normal physiological pathways.
Based on metabolomics analyses (as well as metabolic reconstruction studies) using data from the OAT1 knockouts, which indicate a broad role for OAT1 in the transport of key metabolites and signaling molecules, it has been proposed that these transporters regulate systemic and local physiology through the transfer within and between organs, body fluids, and organisms of small molecules regulating metabolism, signaling, and redox state. The more fully developed theory is called the Remote Sensing and Signaling Hypothesis (2, 3, 21, 25, 31, 33, 36–39). There is recent support for this idea in murine pathophysiological models and in human disease (2, 3, 37, 40–43).
Despite the very clear implication of OAT1 in regulating a wide variety of metabolic and signaling processes in vivo, and despite the fact that OAT3, by and large, appears to transport very similar (organic anion) common drugs in vitro and in vivo (39, 44) (with the exception that OAT3 has a greater propensity to interact with a limited number of organic cation drugs than OAT1 (44, 45)), the data on the role of OAT3 in regulating metabolism and signaling in vivo is comparatively sparse. In part, this is the result of the fact that early targeted and untargeted metabolomics studies that were successful in identifying endogenous OAT1 substrates in the Oat1KO were less successful in the Oat3KO (11, 17, 20, 25). Nevertheless, the limited early metabolomics data from the Oat3KO, as well as in vitro transport data (3), indicated differences in the ligand/substrate preferences between the two OATs, particularly in the case of metabolites and signaling molecules.
We have now analyzed and performed pathway analysis of robust metabolomics data from the Oat3KO mouse, which indicate a role for OAT3 in the movement of metabolites flowing through the “gut–liver–kidney” axis, especially gut microbiome metabolites, bile acids, and nutrients that have undergone modification by phase 2 liver DMEs involved in sulfation and glucuronidation reactions. Furthermore, a large number of these metabolites are potentially involved in “metabolite sensing and signaling” via GPCRs throughout the body.
Surprisingly, in contrast to drug transport studies that indicate OAT1 and OAT3 (drug) ligands have extremely similar physicochemical characteristics, endogenous metabolites altered in the Oat3KO tend to be much larger, more hydrophobic, and much more complex than those seen in the Oat1KO. The data seem to reflect distinct roles in endogenous physiology for the two OATs, in that OAT3 appears to be more important in regulating the levels of metabolites flowing through a gut–liver–kidney axis, but less important than OAT1 in regulating kidney PT energy metabolism.
Results
From a physiological standpoint, both of the OAT knockouts appear to have no growth abnormalities and have survival comparable with the wild type (10, 11). In the Oat3KO, there are no clear histological abnormalities in the liver, choroid plexus, or kidney (10). Detailed physiological analysis of renal function revealed no abnormalities (17, 22), although the Oat3KO has a slightly lower blood pressure than the wild type (17).
Much is known about drug transport capacity by OAT1 and OAT3 (2, 3, 31). The Oat3KO (like the Oat1KO) has been extensively characterized from a pharmacological standpoint, and the in vivo data are quite consistent with the in vitro data (11–26). For example, the Oat3KO has diminished uptake of the classical substrate estrone sulfate in renal slices (10), a diminished diuretic response due to the inability to transport diuretics into the renal PT lumen (13, 17), alterations in the renal handling of the known OAT3 substrate urate (12), diminished excretion of antibiotics (14, 15), as well as diminished uptake of antivirals in renal and choroid plexus organ/tissue cultures (18, 19, 23). All of these are known in vitro substrates for OAT3 in cell-based transport assays (3). In summary, the Oat3KO has normal growth and survival, normal histology, and the expected defects in transport of pharmaceuticals based on in vitro transport data in cells expressing OAT3.
Metabolomics data from the Oat1KO has led to the identification of many metabolites, many reflecting altered kidney proximal tubule metabolism, and its involvement in aspects of systemic metabolism and signaling (11, 20, 21, 37). Until now, similar metabolomics data from the Oat3KO has not been available. In this study, robust metabolomics data from the serum of adult Oat3KO mice have been obtained and indicate a role for OAT3 in the “handling” of a wide range of metabolites, including gut microbiome metabolites, bile acids, and nutrients, many of which flow through the gut–liver–kidney axis.
Of note, metabolites accumulating in serum of the Oat3KO may be present because they are not transported out (cleared) of the blood due to the deletion of the transporter or because of indirect cascade effects resulting from the lack of transport of a key metabolite in a biological pathway (20, 34). In addition, although in some cases, OAT3-mediated metabolite transport has been directly tested (using a labeled metabolite), such instances are relatively few; in many cases, and as with drugs, transport is assumed based on assays involving inhibition of uptake of a labeled probe, an assumption that may not always hold (40). For these reasons, we prefer the term handling by OAT3 when generally referring to metabolites accumulating in the knockout to avoid the implication that each metabolite is proven to be directly transported.
By analysis of molecular features and the use of agglomerative hierarchical clustering tools, we have been able to group likely (and partly validated in vitro) OAT3 ligands into nine groups with very distinct characteristics. Despite the extreme similarity of OAT1 and OAT3 drug ligands (44), quite surprisingly, the endogenous metabolites with altered concentrations in the two knock-out animals are structurally and functionally very different (see below); endogenous metabolites with altered concentrations in the Oat3KO are larger, more hydrophobic, and more complex metabolites (with more ring structures) than those altered in the Oat1KO (see below), supporting a very different role in systemic physiology for OAT3 than OAT1, particularly with respect to inter-organ and inter-organismal communication involving metabolites and signaling molecules (e.g. gut microbiome–gut–blood–liver–kidney–urine). Furthermore, renal OAT3 appears closely tied to UDP-glucuronosyltransferases-mediated liver phase II glucuronidation and may, indeed, be the primary route of excretion for glucuronidated molecules. Below, we detail these molecules, the metabolomics, and signaling pathways modulated by OAT3, and we arrive at a novel physiological perspective on the roles of OAT1 and OAT3 in systemic and kidney (proximal tubule) physiology.
Metabolomics analysis of the Oat3KO
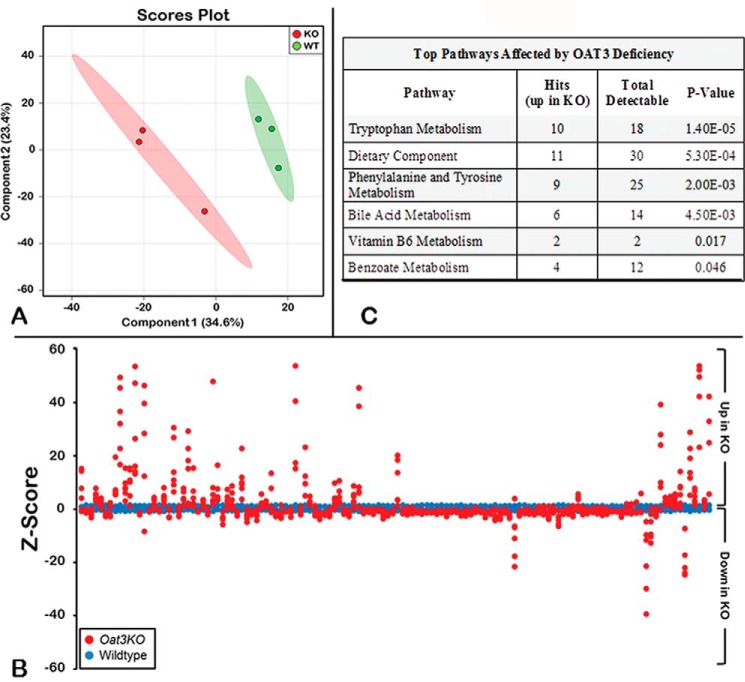
In the metabolomics analysis of the plasma collected from wild-type and Oat3KO mice, a total of 611 metabolites of known identity were detected across both groups. Partial least squares discriminant analysis revealed a clear separation of knock-out and wild-type groups based on their metabolite profile (Fig. 1). Overall, in the plasma of the Oat3KO mouse, a total of 137 metabolites (∼22% of all the metabolites detected) were found to have significantly altered serum concentrations (either increased or decreased).
Figure 1.
Metabolomics profiling of serum distinguishes Oat3KOs from wild type. A, metabolomic profiling reveals separation between serum metabolites from Oat3KO (KO; red) and wild-type control mice (WT; green) by partial least squares discrimination analysis. B, Z-score plot of Oat3KOs metabolite intensities (red) against wild-type metabolites (blue). Each dot represents one metabolite observation for one sample, and positive changes in metabolite concentration (up in the plasma of the Oat3KO) are seen above the x axis, and negative changes in metabolite concentration (down in the plasma of the Oat3KO) are seen below the x axis. The distance from the x axis is the magnitude of the alteration in plasma concentration. As can be seen, although many metabolites show a decrease in their serum concentration in the Oat3KO, the metabolites displaying the largest changes in concentration are overwhelmingly those that accumulate in the plasma of the Oat3KO. C, table of pathway analysis of metabolites accumulating in the serum of the Oat3KO. The top pathways affected are shown and are ranked by p value (determined by hypergeometric test). Hits refer to the number of endogenous metabolites within each pathway that were found to significantly accumulate in the serum of the Oat3KO. Total detectable refers to the number of all metabolites in each pathway that are actually detectable on the metabolomics platform.
In addition to these 137 significantly altered metabolites, another 60 metabolites exhibited trends toward significant differences between the two groups (0.05 < p < 0.10). Of this total of 197 endogenous metabolites, 80 were increased, and 117 metabolites were decreased in the serum of the Oat3KO compared with the wild-type controls.
The intensities of the samples altered in the Oat3KO serum were also plotted relative to the intensities of the samples in wild type in a Z-score plot (Fig. 1). Here, the distance of each dot from the x axis is reflective of the number of standard deviations above (positive value; up) or below (negative value; down) the average value of the wild-type observation. From the Z-score plot, it is evident that the endogenous metabolites displaying the greatest magnitude of change are those that accumulate in the serum of the Oat3KO; in contrast, those with significant decreases in concentration of the serum of the Oat3KO display much smaller changes (Fig. 1). Thus, in the analysis that follows, we focused our attention on those 80 endogenous metabolites that are accumulating in the serum of the Oat3KO mouse (supplemental Table 1).
Oat3KO accumulates gut microbiome-derived metabolites involved in gut–liver–kidney axis
To investigate the functional role(s) of the metabolites accumulating in the plasma of Oat3KO mice, a pathway set enrichment analysis was performed (Fig. 1C). The affected metabolite/signaling pathways included the metabolism of amino acids (tryptophan, phenylalanine, and tyrosine), lipids (bile acid metabolism), cofactors, and vitamins (vitamin B6), as well as xenobiotics (dietary components, including benzoate) (Fig. 1C). The most highly represented metabolic pathways are shown and ranked according to the p value determined by hypergeometric analysis.
What is perhaps most striking about these various pathways is the degree to which they are comprised, at least in part, of metabolites that are presumed to originate from or are dependent on the gut microbiome. For example, tryptophan and phenylalanine are essential amino acids absorbed into the circulation via the gut, whereas the level of bile acids (which are synthesized in the liver as primary bile acids, secreted into the bile, and reabsorbed in the intestine as secondary bile acids) (46) depends upon gut microbial metabolism (47). In this regard, it is worth noting that secondary bile acids were significantly altered in the plasma of the Oat3KO (p < 0.01). In addition, vitamin B6 (pyridoxine) and benzoate (a simple carboxylic acid produced in the gut from the digestion of dietary aromatic compounds (e.g. polyphenols, purines, and aromatic organic acids and amino acids), as well as other dietary components (48–50) are all also derived from the gut.
In fact, of those metabolites that were significantly increased (p ≤ 0.05) in the Oat3KO, a number are derived from the microbiome (51); of the more than 40 metabolites that have a gut microbiome origin among the 611 detectable (Table 1), 12 were found to be significantly increased (p < 0.05) in the plasma of the Oat3KO, which represents a significant enrichment for these types of metabolites (p ≤ 2.53E-05).
Table 1.
Metabolites of microbiome origin detected on the metabolomics platform
A list of 41 metabolites found on the Metabolon platform which have presumed microbiome origin (51). A check mark in column 4 indicates that this metabolite significantly accumulated in the plasma of the Oat3KO, and no mark indicates that significant accumulation was not detected. Please see Ref. 51 for details.
| Metabolite | KEGG | HMDB | Significant plasma accumulation in Oat3KO | -Fold change | p value |
|---|---|---|---|---|---|
| 2-Hydroxyhippurate | C07588 | HMDB00840 | |||
| 3-(3-Hydroxyphenyl)propionate | C11457 | HMDB00375 | |||
| 3-(4-Hydroxyphenyl)lactate | C03672 | HMDB00755 | |||
| 3-(4-Hydroxyphenyl)propionate | C01744 | HMDB02199 | |||
| 3-Indoxyl sulfate | HMDB00682 | √ | 3.83 | 8.00E-04 | |
| 3-Phenylpropionate | C05629 | HMDB00764 | |||
| 4-Hydroxycinnamate | C00811 | HMDB02035 | |||
| 4-Hydroxyhippurate | HMDB13678 | ||||
| 4-Hydroxyphenylpyruvate | C01179 | HMDB00707 | |||
| Betaine | C00719 | HMDB00043 | |||
| Cholate | C00695 | HMDB00619 | √ | 158.95 | 1.80E-03 |
| Creatine | C00300 | HMDB00064 | |||
| Creatinine | C00791 | HMDB00562 | |||
| Daidzein | C10208 | HMDB03312 | |||
| Deoxycholate | C04483 | HMDB00626 | √ | 8.59 | 2.55E-02 |
| Dimethylglycine | C01026 | HMDB00092 | |||
| Genistein | C06563 | HMDB03217 | |||
| Glucose | C00031 | HMDB00122 | |||
| Hippurate | C01586 | HMDB00714 | |||
| Hyocholate | C17649 | HMDB00760 | |||
| Indole-3-carboxylic acid | C19837 | HMDB03320 | √ | 2.85 | 6.00E-03 |
| Indoleacetate | C00954 | HMDB00197 | √ | 2.8 | 8.80E-03 |
| Indole lactate | C02043 | HMDB00671 | √ | 4.63 | 1.03E-07 |
| Indole propionate | HMDB02302 | ||||
| Isovalerate | C08262 | HMDB00718 | |||
| Ketoisovalerate | C00141 | HMDB00019 | |||
| Lactate | C00186 | HMDB00190 | |||
| Monacylglycerol | C01885 | HMDB11561 | |||
| N-Acetyltryptophan | C03137 | HMDB13713 | √ | 2.74 | 1.75E-02 |
| p-Cresol sulfate | HMDB11635 | √ | 3.83 | 1.54E-02 | |
| Phenol sulfate | C02180 | HMDB60015 | √ | 2.84 | 3.50E-03 |
| Phenylacetylglycine | C05598 | HMDB00821 | |||
| Phenyl lactate | C01479 | HMDB00779 | √ | 3.2 | 4.95E-05 |
| p-Hydroxybenzaldehyde | C00633 | HMDB11718 | |||
| Putrescine | C00134 | HMDB01414 | |||
| Spermidine | C00315 | HMDB01257 | |||
| Succinate | C00042 | HMDB00254 | |||
| Tauroursodeoxycholate | HMDB00874 | ||||
| Trimethylamine-N-oxide | C01104 | HMDB00925 | √ | 4.54 | 2.00E-04 |
| Urea | C00086 | HMDB00294 | |||
| Ursodeoxycholate | C07880 | HMDB00946 | √ | 14.6 | 8.60E-03 |
Another striking feature about the list of accumulating metabolites is that they generally reflect the movement of molecules from the gut microbiome through the gut–liver–kidney axis. For example, many of these gut microbiome-derived metabolites are metabolized in the liver (i.e. indoxyl sulfate being formed as a result of phase 2 DME modification of indole in the liver), but, based on their accumulation in the plasma of the Oat3KO, they are excreted largely via the kidney. The data indicate a central role for OAT3 in the flow of metabolites within this gut–liver–kidney axis involved in the absorption, metabolism, and excretion of endogenous metabolites (as discussed in more detail below, this is different from OAT1, which appears to have a more substantial role in proximal tubule energy metabolism). Because these gut microbiome-derived/contributed metabolites participate in a variety of biological pathways, including pathways aberrant in chronic kidney disease (27), they will also be discussed below in the context of the pathways (Fig. 2).
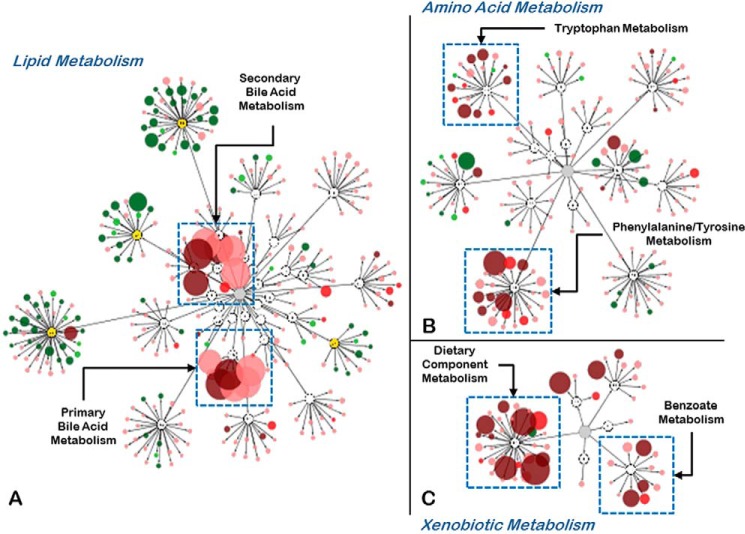
Figure 2.
Deficiency of OAT3 affects the concentration of metabolites found in a number of biochemical pathways. A–C, metabolites altered in the Oat3KO visualized using the Metabolon Metabolync Pathway application for Cytoscape. Metabolites are arranged by their associations with the various pathways (Lipid Metabolism (A); Amino Acid Metabolism (B); Xenobiotic Metabolism (C)). The size and color of each node (metabolite) is based on the magnitude of the change (size) and whether its concentration was increased (dark red, p ≤ 0.05; bright red, 0.05 < p < 0.10) or decreased (dark green, p ≤ 0.05; bright green, 0.05 < p < 0.10) in the plasma of the Oat3KO. Metabolites with concentrations that were trending up (pale red) or down (pale green), but did not reach levels of significance (p ≥ 0.1) are also shown.
Xenobiotics and dietary components
Of the 611 metabolites detectable in the metabolomics analysis, 55 of them (∼9%) fall into the xenobiotic superpathway category (Figs. 2 and 3). Of these 55 metabolites, 14 of them (∼25%) were found to significantly accumulate in the plasma of the Oat3KO (supplemental Table 1), which represents a statistically significant enrichment (p < 0.01) in such metabolites in the population of 80 metabolites accumulating in the plasma of the Oat3KO. These metabolites fell into a number of subpathways, including those involved in the metabolism of benzoate and food components, as well as drugs and chemicals.
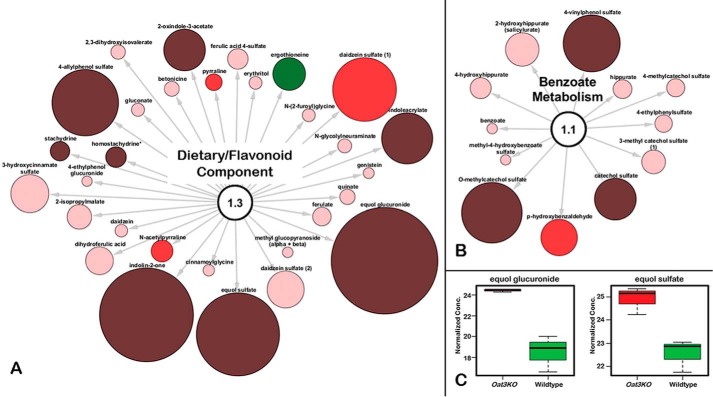
Figure 3.
Endogenous metabolites accumulating in the serum of the Oat3KO are derived from metabolism of xenobiotics/dietary components. A, metabolites associated with the dietary/food component; B, metabolites associated with benzoate metabolism. Metabolites are colored and sized based upon their magnitude of change (size) and direction of change (color; red, accumulated in plasma of Oat3KO; green, decreased in plasma of Oat3KO). In addition, the color is also indicative of the significance of the change (i.e. dark red and dark green (p ≤ 0.05); bright red or bright green (0.05<p < 0.10); pale red or pale green (trending toward significance, p > 0.1). C, Whisker-Boxplots of two selected metabolites increasing in dietary/food component (equol glucuronide and equol sulfate).
The food component metabolite displaying the greatest level of accumulation was equol glucuronide, which was significantly increased more than 42-fold in the Oat3KO (p ≤ 0.05; q ≤ 0.05) (Fig. 3). Equol, a metabolite of the soy isoflavone daidzein, is produced by the gut microbiome and metabolized to equol glucuronide by UDP-glucuronosyltransferases, phase II drug-metabolizing enzymes in the liver (52). Circulating equol metabolites are generally present as glucuronidated conjugates; however, monosulfated forms can also be present (53), and here equol sulfate was also increased more than 7-fold in the Oat3KO (Fig. 3).
In addition to equol, other dietary metabolites, including those belonging to pathways involved in the metabolism of benzoate, were also significantly increased in the plasma of the Oat3KO (supplemental Table 1) (Fig. 3). In the liver and kidneys, dietary benzoate is conjugated to glycine to form hippurate (54), which is the most abundant amino acid conjugate excreted in the urine (55), which is yet another link to the gut–liver–kidney axis.
Amino acid metabolism, tryptophan
Deletion of Oat3 also resulted in changes in the plasma levels for a number of metabolites belonging to several different pathways involved in the metabolism of amino acids (supplemental Table 1) (Fig. 4). Among those metabolites demonstrating high levels of plasma accumulation in the Oat3KO were those metabolites arising from the metabolism of tryptophan.
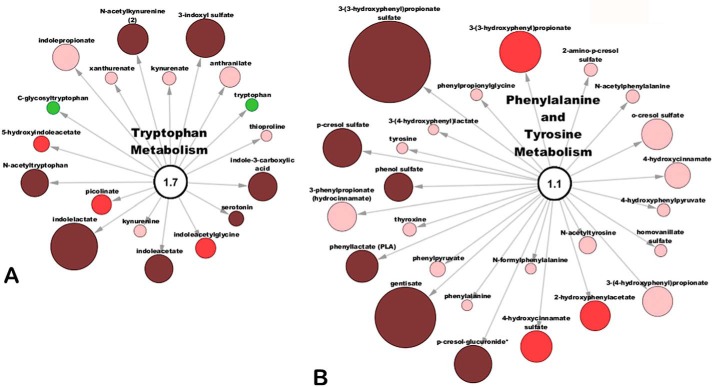
Figure 4.
Metabolites involved in amino acid metabolism accumulate in the serum of the Oat3KO. A, metabolites associated with the metabolism of tryptophan; B, metabolites associated with the metabolism of phenylalanine and tyrosine. As above, metabolites are colored and sized based upon their magnitude of change (size) and direction of change (color; red, accumulated in plasma of Oat3KO; green, decreased in the serum of Oat3KO).
Tryptophan, an essential amino acid, is absorbed from the gut and enters the circulation via transporters expressed on the enterocytes (56). A number of metabolites arising from the metabolism of tryptophan were found to accumulate in the plasma of the Oat3KO, including N-acetylkynurenine, serotonin, indoleacetate, indole lactate, 3-indoxyl sulfate, indole-3-carboxylic acid, and N-acetyltryptophan (supplemental Table 1). Among these metabolites, indole lactate, which had an ∼5-fold increase in plasma concentration in the Oat3KO (p ≤ 0.05; q ≤ 0.05), had the largest change in concentration for metabolites in this pathway (Fig. 4).
Essential amino acids, such as tryptophan, are also part of this gut–liver–kidney axis. For example, tryptophan is absorbed from the gut, and much of this ingested tryptophan will be metabolized in the liver via the kynurenine pathway (57). Some of this absorbed tryptophan will also be converted to the neurotransmitter, serotonin, either in enterochromaffin cells of the gut or by the pineal gland (56, 58–60), whereas a small fraction of ingested and absorbed tryptophan is excreted unchanged in urine. Unabsorbed tryptophan in the gut will undergo degradation by the microbiome yielding indole, indican, and indole acid derivatives (56, 58, 59). Indole is absorbed by the gut and metabolized in the liver to indoxyl sulfate, which is excreted by the kidney.
Amino acid metabolism, phenylalanine and tyrosine
About a quarter of the metabolites comprising the metabolism of phenylalanine and tyrosine pathway were significantly increased in the Oat3KO (supplemental Table 1), including gentisate, phenyllactate (PLA), p-cresol sulfate, p-cresol-glucuronide, phenol sulfate, and 3-(3-hydroxyphenyl)propionate sulfate (Fig. 4). For example, a gut microflora-derived metabolite of caffeic acid, 3-(3-hydroxyphenyl)propionate sulfate (61) (the sulfated form of 3-(3-hydroxyphenyl)propionate, which was also increased and trending toward significance (Fig. 4)), displayed the largest increase in plasma concentration with a >8-fold change (p ≤ 0.05; q ≤ 0.05). Similar to tryptophan, phenylalanine is an essential amino acid absorbed from dietary sources in the gut, and it is converted into tyrosine in the liver by phenylalanine hydrolase (62, 63). Both of these amino acids also share a common pathway of degradation in the liver (64). Moreover, taken together with the metabolomics data on tryptophan above, a number of metabolites comprising tryptophan metabolism, as well as the phenylalanine and tyrosine metabolism pathway are subsequently excreted by the kidney, providing another link to the gut–liver–kidney axis.
Lipid metabolism
The majority of altered lipid metabolites were actually up in the plasma of wild-type mice compared with Oat3KOs, especially those lipids belonging to the phospholipid and sphingolipid metabolic pathways (Fig. 2). Phospholipids, sphingolipids and lysolipids are the principal components of lipid bilayers in cell membranes, and these findings may reflect reductions in cell membrane remodeling in the Oat3KO compared with the wild type. Nevertheless, endogenous metabolites associated with lipid metabolism (particularly primary and secondary bile acids) were also found to accumulate in the plasma of Oat3KO mice.
Bile acid metabolism
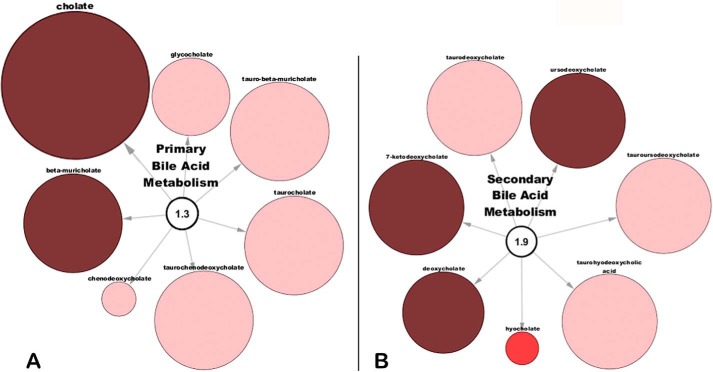
Cholate, a primary bile acid metabolite, was increased almost 160-fold in the plasma of the Oat3KO (p ≤ 0.05; q ≤ 0.05) (Fig. 5; supplemental Table 1), whereas another primary bile acid metabolite, β-muricholate, was increased over 24-fold (p ≤ 0.05; q ≤ 0.05) (Fig. 5; supplemental Table 1). Interestingly, several other primary bile acids, including taurocholate, glycocholate, chenodeoxycholate, tauro-β-muricholate, and taurochenodeoxycholate were also increased (almost 30-fold in the case of taurochenodeoxycholate), although the increases did not reach statistical significance but were trending toward significance (Fig. 5).
Figure 5.
Primary and secondary bile acids accumulate in the serum of the Oat3KO. A, metabolites associated with primary bile acid metabolism; B, metabolites associated with the metabolism of secondary bile acids. As above, metabolites are colored and sized based upon their magnitude of change (size) and direction of change (color; dark red (p ≤ 0.05); bright red (0.05 < p < 0.10); pale red (p < 0.10)).
A number of secondary bile acid metabolites were also significantly increased, including 7-ketodeoxycholate, deoxycholate, and ursodeoxycholate (again, several other secondary bile acid metabolites were increased over 10-fold and trending toward significance, including tauroursodeoxycholate, taurodeoxycholate, and taurohyodeoxycholic acid) (Fig. 5).
Primary bile acids are synthesized in the liver from cholesterol and then transformed by the gut microbiome to secondary bile acids (65). Primary and secondary bile acids can be reabsorbed from the gut and enter the circulation where most return to the liver via portal circulation. Although bile acids are largely excreted by the hepatobiliary system (66), in the setting of cholestasis, the renal excretion of bile acids increases (67), apparently mediated by OAT3 (68). In addition, in the setting of chronic kidney disease, serum bile acid levels are increased (69). Thus, OAT3 appears to be capable of mediating the renal secretion of bile acids, and this again supports the central role of renal OAT3 in regulating metabolites flowing through the gut–liver–kidney axis.
Physicochemical analysis of OAT3 interacting metabolite ligands
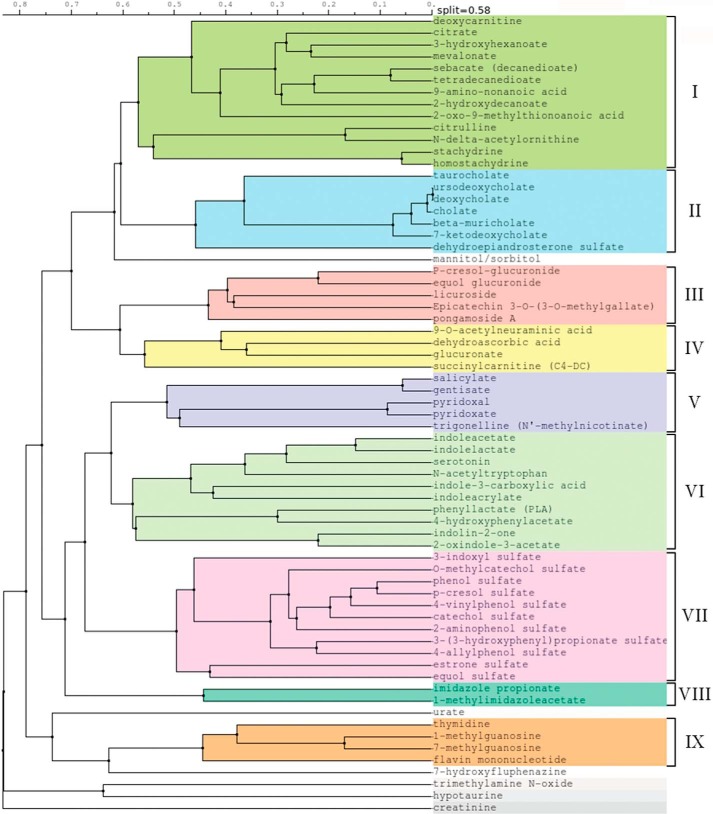
To further characterize the endogenous metabolites altered in the Oat3KO, 67 OAT3 metabolites (i.e. metabolites identified in this study, together with those from previous metabolomics analyses) (12, 17, 25) were computationally analyzed and grouped based on the similarities of their chemical structures. Although a few metabolites did not cluster into any of the groups, this analysis resulted in the generation of nine distinct groups of metabolites (Fig. 6). The number of metabolites within each group varied with the smallest group (group VIII) containing just two metabolites (i.e. imidazole propionate and 1-methylimidazole acetate) and the largest group (group I) containing 13 metabolites (including a number of short chain fatty acids known to function in signaling) (Fig. 6).
Figure 6.
Clusterization of metabolites. Endogenous metabolites found to increase in the serum of the Oat3KO in this and previous studies (17, 23) were clustered based on their physicochemical structures. The metabolites clustered into nine distinct groups; there were also several orphans that did not cluster into any group.
The physicochemical properties of the various metabolite ligands comprising each cluster were calculated and compared (Table 2). Despite structural similarity within the clusters, there were striking differences in the physicochemical properties between the clusters (Table 2). For example, the clusters displayed distinct differences in average molecular mass, ranging from a low of ∼140 Da (group VIII, imidazole-containing compounds) to a high of ∼433 Da (group III, flavonoid metabolites) (Table 2). The clusters also showed distinct differences in the ratio of polar surface area to total area and structural complexity, as well as a number of other attributes (Table 2). In addition, variations in hydrophobicity (logP) and solubility (logS) were also observed (Table 2).
Table 2.
Quantitative molecular property measurements of Oat3 knockout metabolites
The table summarizes the comparisons of the physiochemical descriptors for each of the nine clusters of OAT3 metabolite ligands. Metabolites were clustered based on their physiochemical descriptors into nine distinct groups (Fig. 6). The table shows the average value for several different descriptors for the metabolite ligands comprising each group (mean ± S.E.). MM indicates molecular mass; PSA is polar surface area; No. of rings is number of ring structures; No. of chiral is number of chiral centers; Complexity indicates a measure of structural complexity; logP indicates partition coefficient; logS indicates aqueous solubility.
| MM | Volume | PSA | Area | PSA/area | No. of atoms | No. of heavy atoms | No. of rings | No. of chiral | Complexity | logP | logS | Hydrogen bond | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acceptor | Donor | |||||||||||||
| I | 171 ± 9 | 176 ± 10 | 75.1 ± 7.0 | 209 ± 13 | 0.381 ± 0.046 | 25.9 ± 1.9 | 11.8 ± 0.6 | 0.19 ± 0.10 | 0.56 ± 0.13 | 155 ± 11 | −0.069 ± 0.628 | −0.553 ± 0.408 | 3.6 ± 0.3 | 1.9 ± 0.3 |
| II | 413 ± 18 | 451 ± 16 | 98.3 ± 9.7 | 455 ± 20 | 0.215 ± 0.015 | 67.7 ± 3.0 | 29.0 ± 1.1 | 4 ± 0 | 9.86 ± 0.67 | 682 ± 38 | 3.657 ± 0.382 | −4.381 ± 0.213 | 5.0 ± 0.4 | 3.3 ± 0.5 |
| III | 433 ± 49 | 389 ± 47 | 161 ± 20 | 420 ± 44 | 0.382 ± 0.020 | 52.6 ± 5.8 | 31.0 ± 3.5 | 3.8 ± 0.5 | 5.20 ± 1.08 | 637 ± 93 | 1.180 ± 0.493 | −3.842 ± 0.690 | 9.8 ± 1.1 | 5.6 ± 0.9 |
| IV | 235 ± 36 | 213 ± 38 | 128 ± 21 | 237 ± 44 | 0.565 ± 0.087 | 29.5 ± 6.2 | 16.0 ± 2.4 | 0.75 ± 0.29 | 3.50 ± 1.37 | 292 ± 50 | −2.225 ± 1.320 | 0.392 ± 0.462 | 7.3 ± 1.1 | 3.5 ± 1.4 |
| V | 156 ± 9 | 141 ± 9 | 68.1 ± 8.1 | 163 ± 9 | 0.417 ± 0.048 | 18.6 ± 1.2 | 11.2 ± 0.6 | 1 ± 0 | 0 ± 0 | 156 ± 12 | 1.040 ± 0.441 | −0.817 ± 0.197 | 3.6 ± 0.5 | 2.0 ± 0.5 |
| VI | 201 ± 23 | 198 ± 29 | 59.6 ± 4.1 | 226 ± 36 | 0.286 ± 0.023 | 27.2 ± 4.7 | 14.7 ± 1.7 | 1.82 ± 0.01 | 0.36 ± 0.15 | 231 ± 28 | 1.783 ± 0.598 | −2.733 ± 0.589 | 2.4 ± 0.2 | 2.2 ± 0.2 |
| VII | 227 ± 17 | 182 ± 20 | 86.3 ± 4.1 | 218 ± 19 | 0.413 ± 0.029 | 24.4 ± 2.6 | 14.8 ± 1.3 | 1.55 ± 0.31 | 0.46 ± 0.37 | 304 ± 38 | 1.273 ± 0.231 | −1.975 ± 0.329 | 4.7 ± 0.2 | 1.4 ± 0.2 |
| VIII | 140 ± 0 | 133 ± 3 | 60.6 ± 5.5 | 157 ± 1 | 0.385 ± 0.053 | 18.0 ± 0 | 10.0 ± 0 | 1 ± 0 | 0 ± 0 | 133 ± 6 | −0.4 ± 0.1 | −0.343 ± 0.564 | 3.0 ± 0 | 1.5 ± 0.5 |
| IX | 323 ± 46 | 292 ± 39 | 150.3 ± 21.1 | 311 ± 43 | 0.480 ± 0.038 | 38.8 ± 4.6 | 22.5 ± 3.0 | 2.75 ± 0.25 | 3.50 ± 0.29 | 522 ± 109 | −1.650 ± 0.343 | −2.095 ± 0.253 | 7.5 ± 1.0 | 4.3 ± 0.6 |
In vitro support for the Oat3KO metabolomics
Although it is a huge undertaking to test each of the metabolites changing in the Oat3KO for direct transport (requiring the synthesis and use of dozens of labeled substrates) by in vitro transport assays, it is possible to compare physicochemical features of known metabolites transported in vitro by OAT3 (3) to those metabolites accumulating in the Oat3KO. Therefore, we examined published wet lab data, including our own (17, 25), for the ability of metabolites within each cluster to interact with OAT3 in vitro (i.e. either transport data (Km) or inhibition data (Ki or IC50)) (3). Thus, an extensive list of endogenous metabolites for which in vitro data exist for interaction with OAT3 was compiled and compared with the metabolite ligands comprising each metabolite cluster. When all in vitro endogenous metabolite transport data from this literature search was re-clustered together with the knock-out metabolites, ∼90% of the metabolites with in vitro data fell into one of the nine groups of knock-out metabolites. For example, in the re-clustered cluster II (which contains bile acids and steroids), while three metabolites had both in vivo and in vitro support, altogether 19 metabolites with in vitro and/or in vivo support clustered together (based on common physicochemical properties, supplemental Table 2). Using this approach, nearly all clusters of endogenous metabolite ligands were found to contain at least one metabolite for which in vitro assays have shown an ability to interact with OAT3; in other cases, a structurally related metabolite (e.g. epicatechin galleate as opposed epicatechin 3-O-(3-O-methylgallate)) which clustered with the group has been shown to interact with OAT3 in in vitro assays (Table 3). Taken together, the data provide broad support for the notion that molecules with physicochemical properties unique to each group of metabolites accumulating in the Oat3KO in vivo are capable of interacting with the OAT3 transporter in cell-based transport assays.
Table 3.
Examples from each OAT3 endogenous metabolite cluster with wet lab data showing interaction with OAT3
Exemplary chemical structures for metabolites in each group are shown (Column 2). Metabolites within each cluster for which in vitro data exist are shown in Column 3. If in vitro data do not currently exist for any of the metabolites within a cluster, then a structurally related molecule for which in vitro data exists for interaction with OAT3 is shown (Column 4, related metabolites with in vitro data).
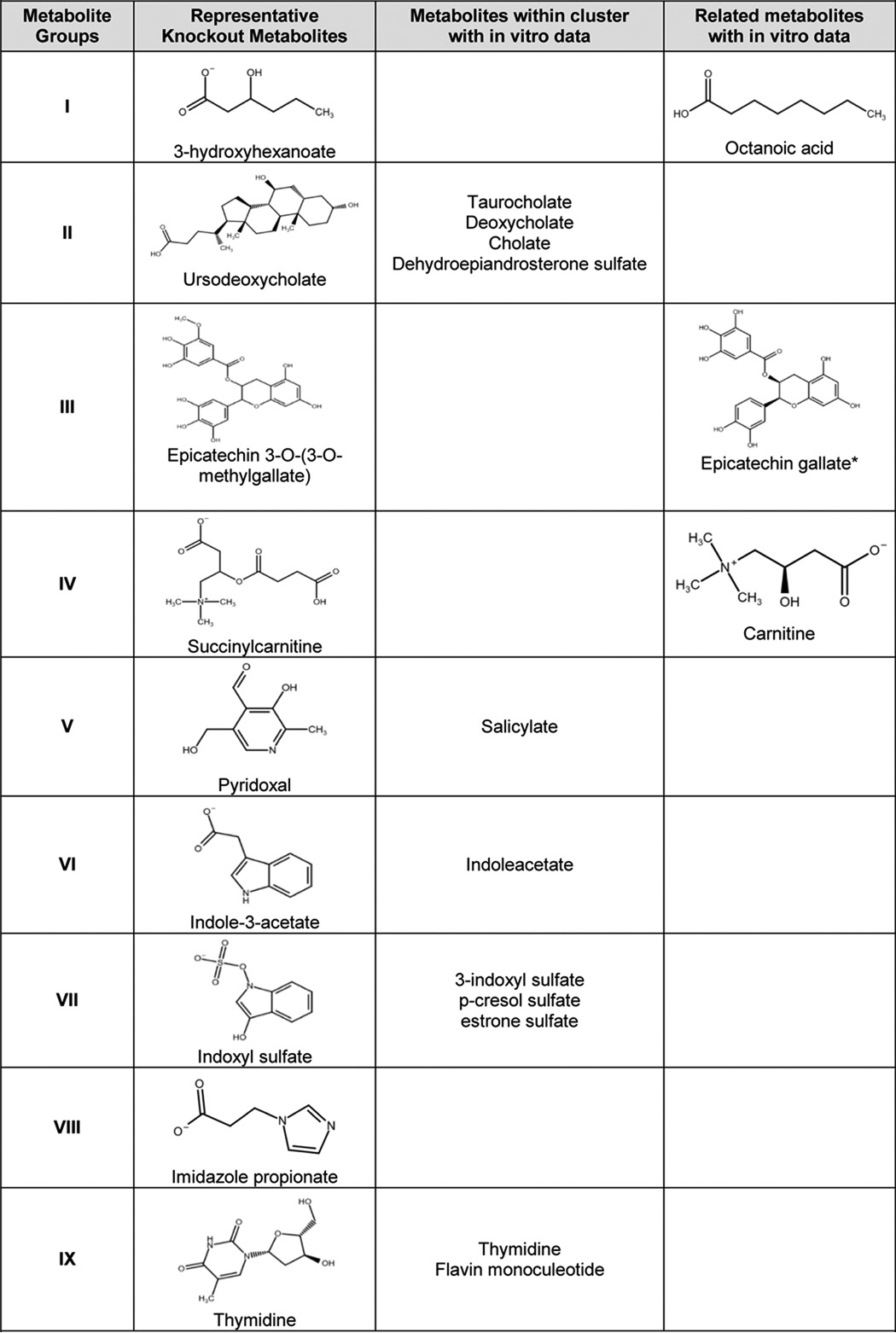
* Km, Ki, or IC50 data are not available, but it has been shown to inhibit Oat3-mediated transport activity.
Comparison of OAT3 with OAT1 metabolite and drug ligands reveals unexpected marked differences in the substrate specificity of OAT1 and OAT3
In a recent study, statistical analysis was performed on the physicochemical descriptors of ∼250 drugs known to interact with one or more SLC22 drug transporters, including OAT1 and OAT3, to investigate the structural differences of high-affinity drug ligands of the different transporters (44). In that study, it was found that although there were some subtle differences between OAT1 and OAT3 (i.e. certain OAT3 drug ligands appear to have a slightly more positive charge and longer hydrophobic chains) the drug ligands for these two organic anion transporters are remarkably similar (44). This is consistent with the conventional view in the literature as well. However, although that study provided important information about the structural features targeting drugs for interaction with a particular SLC22 transporter, the input data (i.e. physicochemical features of interacting drugs) is likely to be skewed by the approaches employed by the pharmaceutical industry (70), which could help explain why this drug ligand-based approach led to the identification of only subtle differences in the molecular characteristics targeting drugs for OAT1 or OAT3 (44).
Based on the idea that the physiological substrates of these transporters are more likely to provide better insight into the unique substrate specificities of OAT1 and OAT3, a similar structural comparison of the endogenous metabolites detected in vivo in the OAT knock-out mice was undertaken in this study. Previous untargeted and targeted metabolomics studies of the serum and/or urine of the Oat1KO resulted in an extensive list of endogenous in vivo metabolites with altered serum and/or urine concentrations (11, 20, 25, 27); similar targeted and untargeted metabolomics studies using the serum and/or urine from Oat3KOs have only described a limited number of metabolites altered in the Oat3KO (12, 17, 25, 27). By including the Oat3KO metabolomics data described here, it was possible to calculate and compare the physicochemical properties for large sets of metabolites present in the two knockouts.
ICM, a commercially available computational chemistry software (Molsoft, San Diego, CA), as well as PubChem were utilized to compare a number of physicochemical attributes of the metabolite ligands, including complexity, molecular mass, logP, atom counts, and ring counts. t test analyses were performed on pairwise comparisons of the individual attributes of OAT1 and OAT3 endogenous in vivo metabolites (Fig. 7). Unlike the drug ligands, the metabolites with altered concentrations in the Oat1KO and Oat3KO were strikingly different, and statistically significant differences were identified in the physicochemical structures of the metabolites. The attributes that had among the lowest p values for each comparison are summarized in Fig. 7. In contrast to the structurally similar drugs transported by OAT1 and OAT3 (44), metabolites accumulating in the serum of the Oat3KO were larger, more complex, more hydrophobic (e.g. higher logP), and had more rings than those found in the Oat1KO (Fig. 7). In fact, of 24 endogenous metabolites examined for which published in vitro transport or binding data exist for both OAT1 and OAT3 (supplemental Table 3), only six had similar in vitro transport or binding, a OAT1/OAT3 ratio of transport (Km) or binding (Ki or IC50) between 0.5 and 1.5. Hippurate was one of these five metabolites and has a reported Ki of 18.8 μm for OAT1 and 30.8 μm for OAT3 and thus an OAT1/OAT3 ratio of 0.61. In other words, 18 of 24 metabolites had a strong preference (≥1.5-fold) for either OAT1 or OAT3 (supplemental Table 3). Taken together, these findings not only reveal global differences in the chemical structures of the endogenous metabolites accumulating in their respective knockout (Fig. 7), but importantly, this suggests that, at least for these two OATs, the endogenous “metabolite space” is likely distinct from “drug space.”
Figure 7.
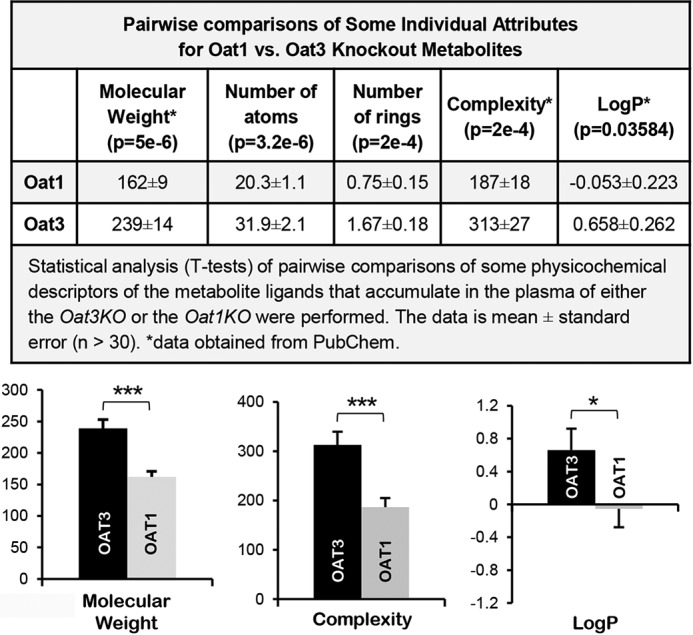
Comparisons of physicochemical descriptors reveal significant differences between metabolites accumulating in the serum of the Oat3KO versus those accumulating in the Oat1KO. Top panel, table of pairwise comparisons of some physicochemical descriptors for endogenous metabolites with altered concentrations in Oat3KOs and Oat1KOs. Metabolites were compiled from either prior metabolomics analyses of the transporters (11, 12, 17, 20, 25, 83) or from this analysis of the Oat3KO. The p value for each comparison is indicated below the name of each attribute. The data for each attribute is the mean ± S.E. Bottom panel, a series of bar graphs for some of the attributes seen in the top panel to allow for visualization of the differences. The attributes compared in each graph are shown below the graph (black bar, OAT3; gray bar, OAT1; *, p ≤ 0.05; ***, p ≤ 0.001).
Discussion
Comparison of the plasma metabolomics of the Oat3KO to the Oat1KO revealed surprisingly striking differences despite the fact that the drug molecular properties for these two transporters are largely similar (44). Metabolic differences in the Oat3KO versus the wild type included accumulation of metabolites associated with the metabolism of flavonoids, tryptophan, phenylalanine and tyrosine, and bile acids, as well as disruption of xenobiotic metabolism (e.g. dietary components and benzoate). Thus, OAT3 deletion alters normal transport functions leading to the accumulation of endogenous metabolites involved in important metabolite and signaling pathways. Many of these metabolites are linked to the gut microbiome and gut microbiome-derived metabolites, a number of which are metabolized in the liver and are ultimately excreted by the kidney. The data also point to an important association of OAT3 with flow of metabolites through the gut–liver–kidney axis.
In this context, gut–liver–kidney axis refers to the uptake of a metabolite (e.g. dietary component or gut microbiome product) from the gut, which is then metabolized in the liver and is ultimately excreted by the kidney. For example, equol glucuronide and equol sulfate, among the most elevated metabolites in the Oat3KO (increased as much as ∼40-fold), ultimately derived from metabolism of daidzein, a soy isoflavone, by the gut bacteria, is absorbed as equol from the intestinal lumen via transporters expressed on the luminal membrane of the enterocytes where it is ultimately transported into the blood (53). Equol is then picked up and transported into the liver where it is then glucuronidated or sulfated via DMEs in the hepatocytes to equol glucuronide/equol sulfate (52), which is then transported back into the blood where it has access to various organs and it is ultimately excreted by the kidney.
The data presented above provide additional support for the relevance of the Remote Sensing and Signaling Hypothesis. This hypothesis, first formulated a decade ago (39) and subsequently elaborated (2, 3, 21, 25, 31, 36–38, 42, 43), emphasizes the central role of solute carrier and ATP-binding cassette transporters to inter-organ and inter-organismal small molecule communication throughout the body and thus the importance of the transporters to modulation of metabolism and signaling. As such, it is envisioned as a small molecule homeostatic system similar to the neuroendocrine system. A majority of the identified endogenous metabolites accumulating in the plasma of the Oat3KO are absorbed from the gut metabolized in the liver and are excreted by the kidney. This gut–liver–kidney axis can be viewed as transporter-dependent inter-organ movement of small organic anions. If we consider the gut microbiome, then this amounts to transporter-mediated inter-organismal communication; to the extent that these gut-derived molecules are involved in signaling, this amounts to remote inter-organismal signaling.
As discussed below, many of the metabolites (e.g. bile acids and lipids) are themselves capable of activating signaling pathways, and they have been implicated in what has been termed “metabolite sensing and signaling” (71, 72). For example, gentisate, an intermediate of tyrosine degradation, accumulates in the plasma of Oat3KO mice (Fig. 4; supplemental Table 1) and functions as an agonist of the G-protein-coupled receptor, GPR35 (64). GPR35 can be activated by other dietary metabolites (e.g. kynurenine and tryptophan metabolites) and belongs to a group of “metabolite-sensing” GPRs, which includes GPR43, GPR41, GPR109A, and GPR120 (73). GPR35 has been implicated in modulating synaptic transmission as well as neurogenic and inflammatory pain (74).
In addition, bile acids are now recognized as versatile signaling molecules based primarily on their ability to act as ligands for G protein-coupled receptors, such as the membrane receptor TGR5, as well as nuclear receptors like the farnesoid X receptor (65, 75, 76). Bile acid-mediated activation of these receptors leads to specific signaling pathways involved in the regulation of many physiological functions, such as lipid, glucose, and energy metabolism (65). Taken together, the data not only provide evidence supporting OAT3 as a critical component of the gut–liver–kidney axis, they also suggest a role for OAT3 in modulating local and systemic physiological pathways by regulating the uptake and excretion of intermediate metabolites and signaling molecules.
Furthermore, it has also been suggested that the OATs, which have homology to G protein-coupled receptors (such as OAT1/SLC22A6, OAT3/SLC22A8, and OAT6/SLC22A20) may also function in the sensing of odorants and/or short chain fatty acids, possibly as transceptors (18, 33–35, 39). Thus, the potentially intimate connection of SLC “drug” transporters to metabolite sensing and signaling is a vital area for further exploration in the context of the Remote Sensing and Signaling Hypothesis (2, 3).
Other small organic anions identified are also important co-factors in other biochemical pathways. For example, pyridoxal, which is converted to pyridoxal 5′-phosphate (the active form of vitamin B6) in the liver, and pyridoxic acid, the catabolic product of pyridoxal excreted in the urine, were also significantly increased in the Oat3KO (supplemental Table 1). Pyridoxal 5′-phosphate also functions as a cofactor in a number of physiological pathways, including neurotransmitter synthesis (i.e. serotonin, dopamine, epinephrine, norepinephrine and GABA). OAT3 is also involved in elimination of sulfated and acetylated metabolites, including many derived from the gut microbiome. Together with previous metabolomics analyses of the Oat1KO, these data highlight the central role of OAT3 and OAT1 in the regulation of numerous metabolites and signaling molecules, including gut microbiome-derived metabolites, many of which are also categorized as uremic toxins or uremic retention solutes (20, 27).
Clusterization based on the physicochemical descriptors of the endogenous metabolites accumulating in the serum of the Oat3KO revealed nine distinct groups (Fig. 6; Table 2). Moreover, although there is overlap of metabolites attributable to loss of OAT1 and OAT3 function, there are also striking differences (Fig. 7). For instance, examination of the physicochemical attributes of OAT1- and OAT3-interacting metabolites revealed clear differences between the groups (Fig. 7). On average, OAT3 interacting metabolites appear to be larger and more complex, possessing more ring structures (Fig. 7). OAT3 interacting endogenous metabolites also appear to be more lipophilic, having a higher logP than OAT1-interacting metabolites. Crucially, unlike drugs known to interact with OAT1 and OAT3, which were highly similar, endogenous metabolites accumulating in the Oat knockouts are physicochemically very different (Fig. 7). This finding has important consequences for metabolite-based tissue targeting of drugs, for example, to increase renal excretion.
It is widely accepted that OAT1 is an antiporter that exchanges its plasma ligand for intracellular α-ketoglutarate (77, 78). Thus, OAT1 is closely linked to proximal tubule aerobic metabolism. Consistent with this view, the Oat1KO shows a higher urinary and lower plasma concentration for α-ketoglutarate (11) (supporting the importance of OAT1-mediated regulation of the movement of α-ketoglutarate (a key TCA cycle intermediate) in the proximal tubule). The TCA cycle is noteworthy, because it includes metabolites known to be classical substrates of OAT1, such as α-ketoglutarate, citrate, fumarate, and succinate (2, 4, 39, 79); in contrast, we observed no significant change in the plasma levels of α-ketoglutarate (or other TCA cycle intermediates) in the Oat3KO. In fact, it has previously been reported that levels of these metabolites are actually decreased in the urine of Oat3KO mice (25); this is the opposite that would be expected if OAT3, like OAT1, played an important role in PT regulation of α-ketoglutarate movement. The uptake of organic anions via the OATs is believed to be driven, at least in part, by the exchange of dicarboxylates (such as α-ketoglutarate) across the basolateral membrane; however, of note in this regard, OAT1 function has been consistently shown to exchange α-ketoglutarate with organic anions taken up into the PT, although this has not been consistently demonstrated for OAT3. Taken together with the metabolomics data, as well as the comparison of the molecular properties of the endogenous metabolites, the data appear to support the idea that although OAT1 and OAT3 share many functional properties, OAT1 plays a larger role in regulation/modulation of proximal tubule metabolism, whereas OAT3 likely plays a larger role in systemic metabolism, particularly through regulating/modulating the flow of metabolites through the gut–liver–kidney axis. This raises the question of how these differing physiological roles of OAT1 and OAT3 are controlled at the transcriptional level, a question that is beginning to receive more attention (80–82).
Experimental procedures
Animals
All experimental protocols were approved by The University of California San Diego Institutional Animal Care and Use Committee (IACUC), and the animals were handled in accordance with the Institutional Guidelines on the Use of Live Animals for Research. Adult (n = 3) wild-type, Oat3KO male mice were housed separately under a 12-h light/dark cycle and were provided access to food (standard diet) and water ad libitum.
Metabolomic analysis
Serum samples (at the time of blood collection) from adult male wild-type (WT) control and Oat3KO mice were obtained, and individual unpooled samples were stored at −80 °C, and all samples were shipped together on dry-ice to Metabolon (Durham, NC) for preparation and metabolomic profiling analysis (83, 84). Samples, prepared using the automated MicroLab STAR® system (Hamilton, Reno, NV), were subjected to ultra-high performance liquid chromatography-tandem mass spectroscopy (UPLC-MS/MS) as described previously (27).
Compound identification, quantification, and data curation and statistics
A total of 611 compounds (metabolites) of known identity were detected and identified across both groups (83, 84). Metabolites were identified by automated comparison of the ion features in the experimental samples to a reference library of chemical standard entries that included retention time, molecular weight (m/z), preferred adducts, and in-source fragments as well as associated MS spectra and curated by visual inspection for quality control using software developed at Metabolon (83, 84). Metabolites with missing intensity scores, indicating low levels of the metabolite in a particular sample, were imputed with the minimum value for the study following rescaling of the metabolites to set the median equal to 1.
Peaks were quantified using area-under-the-curve, and two-way analysis of variance testing was used to calculate the p values. To take into account multiple comparisons of the data, an estimate of the false discovery rate (q value) was calculated, and a metabolite was considered to be statistically different when p < 0.05 and q < 0.10. Pathway enrichment analyses were also performed using Metabolon software.
Raw metabolomics data were also analyzed with the statistical analysis functionalities of MetaboAnalyst 3.0 (85), and partial least squares discriminant analysis was used to assess the separability of the wild-type and Oat3KO samples; because there were less than 2000 features, the missing value estimation step was skipped, and the data were not filtered, but it was normalized using log transformation. For visualization and analysis of the detected metabolites within relevant networks of metabolic pathways, the KEGG identifications for the detected metabolites were input into MetaboLync pathway analysis software, a Cytoscape plugin.
Statistical analysis of physicochemical properties of metabolites
A comprehensive list of in vivo endogenous metabolites altered in plasma or urine of either Oat1KO or Oat3KO mice was compiled from both this study and previous metabolomics analyses (supplemental Table 4) (11, 12, 17, 20, 25, 27, 86). The conditions under which the Oat1KO metabolomics were performed have been described (11, 20), and these are generally similar to the conditions of the Oat3KO analysis. The physicochemical properties of the metabolites, calculated using ICM or PubChem, were compared, and t tests were performed to determine statistically significant differences between the two groups of metabolites.
Clusterization of in vivo and in vitro OAT3 metabolite ligands
The structure data files for both in vivo and in vitro OAT3 metabolite ligands were retrieved from PubChem and input into ICM. Initially, the in vivo OAT3 metabolite ligands were hierarchically clustered based on their chemical substructure using the Unweighted Pair Group Method with Arithmetic Mean algorithm in ICM. The splitting threshold was 0.58 for the in vivo metabolites and resulted in the clusterization of Oat3KO metabolite ligands into nine structurally distinct groups. For the combined clusterization of the in vivo and in vitro Oat3KO metabolite ligands, the splitting threshold was 0.54 and resulted in the clusterization of the metabolite ligands into the nine structurally distinct groups as above.
Author contributions
K. T. B. performed experiments and data analysis and wrote the manuscript. W. W. performed experiments. C. L. performed experiments and performed analyses. S. K. N. conceived the hypothesis, supervised the experiments and analysis, and edited the manuscript. All authors reviewed the manuscript.
Supplementary Material
This work was supported by National Institutes of Health Grants R01-GM104098 and R01-GM098449 from the NIGMS, Grant R01-DK109392 from NIDDK, and by Grant U54-HD090259 from Eunice Kennedy Shriver NICHD. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Tables S1–S4.
- DME
- drug-metabolizing enzyme
- SLC
- solute carrier
- OAT
- organic anion transporter
- PT
- proximal tubule
- TCA
- tricarboxylic acid.
References
- 1.Food and Drug Administration and Center for Drug Evaluation and Research (2012) Guidance for industry: drug interaction studies—study design, data analysis, implications for dosing, and labeling recommendations. United States Department of Health and Human Services, Silver Spring, MD [Google Scholar]
- 2. Nigam S. K. (2015) What do drug transporters really do? Nat. Rev. Drug Discov. 14, 29–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nigam S. K., Bush K. T., Martovetsky G., Ahn S. Y., Liu H. C., Richard E., Bhatnagar V., and Wu W. (2015) The organic anion transporter (OAT) family: a systems biology perspective. Physiol. Rev. 95, 83–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. VanWert A. L., Gionfriddo M. R., and Sweet D. H. (2010) Organic anion transporters: discovery, pharmacology, regulation and roles in pathophysiology. Biopharm. Drug Dispos. 31, 1–71 [DOI] [PubMed] [Google Scholar]
- 5. You G. F., and Morris M. E. (eds) (2014) Drug Transporters: Molecular Characterization and Role in Drug Disposition, John Wiley & Sons, Inc, Hoboken, NJ [Google Scholar]
- 6. López-Nieto C. E., and Nigam S. K. (1996) Selective amplification of protein-coding regions of large sets of genes using statistically designed primer sets. Nat. Biotechnol. 14, 857–861 [DOI] [PubMed] [Google Scholar]
- 7. Lopez-Nieto C. E., You G., Barros E. J. G., Beier D. R., and Nigam S. K. (1996) Molecular cloning and characterization of a novel transport protein with very high expression in the kidney. J. Am. Soc. Nephrol. 7, 1301 (abstr.) [Google Scholar]
- 8. Lopez-Nieto C. E., You G., Bush K. T., Barros E. J., Beier D. R., and Nigam S. K. (1997) Molecular cloning and characterization of NKT, a gene product related to the organic cation transporter family that is almost exclusively expressed in the kidney. J. Biol. Chem. 272, 6471–6478 [DOI] [PubMed] [Google Scholar]
- 9. Brady K. P., Dushkin H., Förnzler D., Koike T., Magner F., Her H., Gullans S., Segre G. V., Green R. M., and Beier D. R. (1999) A novel putative transporter maps to the osteosclerosis (oc) mutation and is not expressed in the oc mutant mouse. Genomics 56, 254–261 [DOI] [PubMed] [Google Scholar]
- 10. Sweet D. H., Miller D. S., Pritchard J. B., Fujiwara Y., Beier D. R., and Nigam S. K. (2002) Impaired organic anion transport in kidney and choroid plexus of organic anion transporter 3 (Oat3 (Slc22a8)) knockout mice. J. Biol. Chem. 277, 26934–26943 [DOI] [PubMed] [Google Scholar]
- 11. Eraly S. A., Vallon V., Vaughn D. A., Gangoiti J. A., Richter K., Nagle M., Monte J. C., Rieg T., Truong D. M., Long J. M., Barshop B. A., Kaler G., and Nigam S. K. (2006) Decreased renal organic anion secretion and plasma accumulation of endogenous organic anions in OAT1 knock-out mice. J. Biol. Chem. 281, 5072–5083 [DOI] [PubMed] [Google Scholar]
- 12. Eraly S. A., Vallon V., Rieg T., Gangoiti J. A., Wikoff W. R., Siuzdak G., Barshop B. A., and Nigam S. K. (2008) Multiple organic anion transporters contribute to net renal excretion of uric acid. Physiol. Genomics 33, 180–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vallon V., Rieg T., Ahn S. Y., Wu W., Eraly S. A., and Nigam S. K. (2008) Overlapping in vitro and in vivo specificities of the organic anion transporters OAT1 and OAT3 for loop and thiazide diuretics. Am. J. Physiol. Renal Physiol. 294, F867–F873 [DOI] [PubMed] [Google Scholar]
- 14. Vanwert A. L., Srimaroeng C., and Sweet D. H. (2008) Organic anion transporter 3 (oat3/slc22a8) interacts with carboxyfluoroquinolones, and deletion increases systemic exposure to ciprofloxacin. Mol. Pharmacol. 74, 122–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vanwert A. L., Bailey R. M., and Sweet D. H. (2007) Organic anion transporter 3 (Oat3/Slc22a8) knockout mice exhibit altered clearance and distribution of penicillin G. Am. J. Physiol. Renal Physiol. 293, F1332–F1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. VanWert A. L., and Sweet D. H. (2008) Impaired clearance of methotrexate in organic anion transporter 3 (Slc22a8) knockout mice: a gender specific impact of reduced folates. Pharm. Res. 25, 453–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vallon V., Eraly S. A., Wikoff W. R., Rieg T., Kaler G., Truong D. M., Ahn S. Y., Mahapatra N. R., Mahata S. K., Gangoiti J. A., Wu W., Barshop B. A., Siuzdak G., and Nigam S. K. (2008) Organic anion transporter 3 contributes to the regulation of blood pressure. J. Am. Soc. Nephrol. 19, 1732–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Truong D. M., Kaler G., Khandelwal A., Swaan P. W., and Nigam S. K. (2008) Multi-level analysis of organic anion transporters 1, 3, and 6 reveals major differences in structural determinants of antiviral discrimination. J. Biol. Chem. 283, 8654–8663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nagle M. A., Truong D. M., Dnyanmote A. V., Ahn S. Y., Eraly S. A., Wu W., and Nigam S. K. (2011) Analysis of three-dimensional systems for developing and mature kidneys clarifies the role of OAT1 and OAT3 in antiviral handling. J. Biol. Chem. 286, 243–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wikoff W. R., Nagle M. A., Kouznetsova V. L., Tsigelny I. F., and Nigam S. K. (2011) Untargeted metabolomics identifies enterobiome metabolites and putative uremic toxins as substrates of organic anion transporter 1 (Oat1). J. Proteome Res. 10, 2842–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ahn S. Y., Jamshidi N., Mo M. L., Wu W., Eraly S. A., Dnyanmote A., Bush K. T., Gallegos T. F., Sweet D. H., Palsson B. Ø., and Nigam S. K. (2011) Linkage of organic anion transporter-1 to metabolic pathways through integrated “omics”-driven network and functional analysis. J. Biol. Chem. 286, 31522–31531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sweeney D. E., Vallon V., Rieg T., Wu W., Gallegos T. F., and Nigam S. K. (2011) Functional maturation of drug transporters in the developing, neonatal, and postnatal kidney. Mol. Pharmacol. 80, 147–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nagle M. A., Wu W., Eraly S. A., and Nigam S. K. (2013) Organic anion transport pathways in antiviral handling in choroid plexus in Oat1 (Slc22a6) and Oat3 (Slc22a8) deficient tissue. Neurosci. Lett. 534, 133–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wolman A. T., Gionfriddo M. R., Heindel G. A., Mukhija P., Witkowski S., Bommareddy A., and Vanwert A. L. (2013) Organic anion transporter 3 interacts selectively with lipophilic β-lactam antibiotics. Drug Metab. Dispos. 41, 791–800 [DOI] [PubMed] [Google Scholar]
- 25. Wu W., Jamshidi N., Eraly S. A., Liu H. C., Bush K. T., Palsson B. O., and Nigam S. K. (2013) Multispecific drug transporter Slc22a8 (Oat3) regulates multiple metabolic and signaling pathways. Drug Metab. Dispos. 41, 1825–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Torres A. M., Dnyanmote A. V., Bush K. T., Wu W., and Nigam S. K. (2011) Deletion of multispecific organic anion transporter Oat1/Slc22a6 protects against mercury-induced kidney injury. J. Biol. Chem. 286, 26391–26395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu W., Bush K. T., and Nigam S. K. (2017) Key role for the organic anion transporters, OAT1 and OAT3, in the in vivo handling of uremic toxins and solutes. Sci. Rep. 7, 4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhu C., Nigam K. B., Date R. C., Bush K. T., Springer S. A., Saier M. H. Jr, Wu W., and Nigam S. K. (2015) Evolutionary analysis and classification of OATs, OCTs, OCTNs, and other SLC22 transporters: structure-function implications and analysis of sequence motifs. PLoS ONE 10, e0140569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu W., Baker M. E., Eraly S. A., Bush K. T., and Nigam S. K. (2009) Analysis of a large cluster of SLC22 transporter genes, including novel USTs, reveals species-specific amplification of subsets of family members. Physiol. Genomics 38, 116–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Eraly S. A., Monte J. C., and Nigam S. K. (2004) Novel slc22 transporter homologs in fly, worm, and human clarify the phylogeny of organic anion and cation transporters. Physiol. Genomics 18, 12–24 [DOI] [PubMed] [Google Scholar]
- 31. Nigam S. K., Wu W., Bush K. T., Hoenig M. P., Blantz R. C., and Bhatnagar V. (2015) Handling of drugs, metabolites, and uremic toxins by kidney proximal tubule drug transporters. Clin. J. Am. Soc. Nephrol. 10, 2039–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pavlova A., Sakurai H., Leclercq B., Beier D. R., Yu A. S., and Nigam S. K. (2000) Developmentally regulated expression of organic ion transporters NKT (OAT1), OCT1, NLT (OAT2), and Roct. Am. J. Physiol. Renal Physiol. 278, F635–F643 [DOI] [PubMed] [Google Scholar]
- 33. Kaler G., Truong D. M., Sweeney D. E., Logan D. W., Nagle M., Wu W., Eraly S. A., and Nigam S. K. (2006) Olfactory mucosa-expressed organic anion transporter, Oat6, manifests high affinity interactions with odorant organic anions. Biochem. Biophys. Res. Commun. 351, 872–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Monte J. C., Nagle M. A., Eraly S. A., and Nigam S. K. (2004) Identification of a novel murine organic anion transporter family member, OAT6, expressed in olfactory mucosa. Biochem. Biophys. Res. Commun. 323, 429–436 [DOI] [PubMed] [Google Scholar]
- 35. Wu W., Bush K. T., Liu H. C., Zhu C., Abagyan R., and Nigam S. K. (2015) Shared ligands between organic anion transporters (OAT1 and OAT6) and odorant receptors. Drug Metab. Dispos. 43, 1855–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ahn S. Y., and Nigam S. K. (2009) Toward a systems level understanding of organic anion and other multispecific drug transporters: a remote sensing and signaling hypothesis. Mol. Pharmacol. 76, 481–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu H. C., Jamshidi N., Chen Y., Eraly S. A., Cho S. Y., Bhatnagar V., Wu W., Bush K. T., Abagyan R., Palsson B. O., and Nigam S. K. (2016) An organic anion transporter 1 (OAT1)-centered metabolic network. J. Biol. Chem. 291, 19474–19486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu W., Dnyanmote A. V., and Nigam S. K. (2011) Remote communication through solute carriers and ATP binding cassette drug transporter pathways: an update on the remote sensing and signaling hypothesis. Mol. Pharmacol. 79, 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kaler G., Truong D. M., Khandelwal A., Nagle M., Eraly S. A., Swaan P. W., and Nigam S. K. (2007) Structural variation governs substrate specificity for organic anion transporter (OAT) homologs. Potential remote sensing by OAT family members. J. Biol. Chem. 282, 23841–23853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Prentice K. J., Luu L., Allister E. M., Liu Y., Jun L. S., Sloop K. W., Hardy A. B., Wei L., Jia W., Fantus I. G., Sweet D. H., Sweeney G., Retnakaran R., Dai F. F., and Wheeler M. B. (2014) The furan fatty acid metabolite CMPF is elevated in diabetes and induces beta cell dysfunction. Cell Metab. 19, 653–666 [DOI] [PubMed] [Google Scholar]
- 41. Saito H. (2010) Pathophysiological regulation of renal SLC22A organic ion transporters in acute kidney injury: pharmacological and toxicological implications. Pharmacol. Ther. 125, 79–91 [DOI] [PubMed] [Google Scholar]
- 42. Sharma K., Karl B., Mathew A. V., Gangoiti J. A., Wassel C. L., Saito R., Pu M., Sharma S., You Y. H., Wang L., Diamond-Stanic M., Lindenmeyer M. T., Forsblom C., Wu W., Ix J. H., et al. (2013) Metabolomics reveals signature of mitochondrial dysfunction in diabetic kidney disease. J. Am. Soc. Nephrol. 24, 1901–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bhatnagar V., Richard E. L., Wu W., Nievergelt C. M., Lipkowitz M. S., Jeff J., Maihofer A. X., and Nigam S. K. (2016) Analysis of ABCG2 and other urate transporters in uric acid homeostasis in chronic kidney disease: potential role of remote sensing and signaling. Clin. Kidney J. 9, 444–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu H. C., Goldenberg A., Chen Y., Lun C., Wu W., Bush K. T., Balac N., Rodriguez P., Abagyan R., and Nigam S. K. (2016) Molecular properties of drugs interacting with SLC22 transporters OAT1, OAT3, OCT1, and OCT2: a machine-learning approach. J. Pharmacol. Exp. Ther. 359, 215–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ahn S. Y., Eraly S. A., Tsigelny I., and Nigam S. K. (2009) Interaction of organic cations with organic anion transporters. J. Biol. Chem. 284, 31422–31430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chiang J. Y. (2013) Bile acid metabolism and signaling. Compr. Physiol. 3, 1191–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ridlon J. M., Kang D. J., Hylemon P. B., and Bajaj J. S. (2014) Bile acids and the gut microbiome. Curr. Opin. Gastroenterol. 30, 332–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rechner A. R., Kuhnle G., Bremner P., Hubbard G. P., Moore K. P., and Rice-Evans C. A. (2002) The metabolic fate of dietary polyphenols in humans. Free Radic. Biol. Med. 33, 220–235 [DOI] [PubMed] [Google Scholar]
- 49. Gibson D. T. (1967) Microbial degradation of aromatic compounds. Science 161, 1093–1097 [PubMed] [Google Scholar]
- 50. Nicholson J. K., and Wilson I. D. (2003) Opinion: understanding ‘global' systems biology: metabonomics and the continuum of metabolism. Nat. Rev. Drug Discov. 2, 668–676 [DOI] [PubMed] [Google Scholar]
- 51. Beebe K., and Pappan K. (2016) Metabolomics–a key technology for microbiome research. Metabolon http://metabolomics.metabolon.com/acton/media/17033/microbiome-ebook
- 52. Mazerska Z., Mróz A., Pawl̸owska M., and Augustin E. (2016) The role of glucuronidation in drug resistance. Pharmacol. Ther. 159, 35–55 [DOI] [PubMed] [Google Scholar]
- 53. Legette L. L., Prasain J., King J., Arabshahi A., Barnes S., and Weaver C. M. (2014) Pharmacokinetics of equol, a soy isoflavone metabolite, changes with the form of equol (dietary versus intestinal production) in ovariectomized rats. J. Agric. Food Chem. 62, 1294–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lennerz B. S., Vafai S. B., Delaney N. F., Clish C. B., Deik A. A., Pierce K. A., Ludwig D. S., and Mootha V. K. (2015) Effects of sodium benzoate, a widely used food preservative, on glucose homeostasis and metabolic profiles in humans. Mol. Genet. Metab. 114, 73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Badenhorst C. P., Erasmus E., van der Sluis R., Nortje C., and van Dijk A. A. (2014) A new perspective on the importance of glycine conjugation in the metabolism of aromatic acids. Drug Metab. Rev. 46, 343–361 [DOI] [PubMed] [Google Scholar]
- 56. Keszthelyi D., Troost F. J., and Masclee A. A. (2009) Understanding the role of tryptophan and serotonin metabolism in gastrointestinal function. Neurogastroenterol. Motil. 21, 1239–1249 [DOI] [PubMed] [Google Scholar]
- 57. Badawy A. A. (2017) Kynurenine pathway of tryptophan metabolism: Regulatory and functional aspects. Int. J. Tryptophan Res. 10, 1178646917691938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bender D. A. (1983) Biochemistry of tryptophan in health and disease. Mol. Aspects Med. 6, 101–197 [DOI] [PubMed] [Google Scholar]
- 59. Peters J. C. (1991) Tryptophan nutrition and metabolism: an overview. Adv. Exp. Med. Biol. 294, 345–358 [DOI] [PubMed] [Google Scholar]
- 60. Namkung J., Kim H., and Park S. (2015) Peripheral serotonin: a new player in systemic energy homeostasis. Mol. Cells 38, 1023–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Konishi Y., and Kobayashi S. (2004) Microbial metabolites of ingested caffeic acid are absorbed by the monocarboxylic acid transporter (MCT) in intestinal Caco-2 cell monolayers. J. Agric. Food Chem. 52, 6418–6424 [DOI] [PubMed] [Google Scholar]
- 62. Flydal M. I., and Martinez A. (2013) Phenylalanine hydroxylase: function, structure, and regulation. IUBMB Life 65, 341–349 [DOI] [PubMed] [Google Scholar]
- 63. Ney D. M., Blank R. D., and Hansen K. E. (2014) Advances in the nutritional and pharmacological management of phenylketonuria. Curr. Opin. Clin. Nutr. Metab. Care 17, 61–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Deng H., Hu H., and Fang Y. (2012) Multiple tyrosine metabolites are GPR35 agonists. Sci. Rep. 2, 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Martinot E., Sèdes L., Baptissart M., Lobaccaro J. M., Caira F., Beaudoin C., and Volle D. H. (2017) Bile acids and their receptors. Mol. Aspects Med. 56, 2–9 [DOI] [PubMed] [Google Scholar]
- 66. Chu L., Zhang K., Zhang Y., Jin X., and Jiang H. (2015) Mechanism underlying an elevated serum bile acid level in chronic renal failure patients. Int. Urol. Nephrol. 47, 345–351 [DOI] [PubMed] [Google Scholar]
- 67. van Berge Henegouwen G. P., Brandt K. H., Eyssen H., and Parmentier G. (1976) Sulphated and unsulphated bile acids in serum, bile, and urine of patients with cholestasis. Gut 17, 861–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chen J., Terada T., Ogasawara K., Katsura T., and Inui K. (2008) Adaptive responses of renal organic anion transporter 3 (OAT3) during cholestasis. Am. J. Physiol. Renal Physiol. 295, F247–F252 [DOI] [PubMed] [Google Scholar]
- 69. Jimenez F., Monte M. J., El-Mir M. Y., Pascual M. J., and Marin J. J. (2002) Chronic renal failure-induced changes in serum and urine bile acid profiles. Dig. Dis. Sci. 47, 2398–2406 [DOI] [PubMed] [Google Scholar]
- 70. Sahu M., Sinha S. K., and Pandey K. K. (2013) Computer aided drug design: the most fundamental goal is to predict whether a given molecule will bind to a target and if so how strongly. Comp. Engineer Intell Systems 4, 22–26 [Google Scholar]
- 71. Husted A. S., Trauelsen M., Rudenko O., Hjorth S. A., and Schwartz T. W. (2017) GPCR-mediated signaling of metabolites. Cell Metab. 25, 777–796 [DOI] [PubMed] [Google Scholar]
- 72. Tan J. K., McKenzie C., Mariño E., Macia L., and Mackay C. R. (2017) Metabolite-sensing G protein-coupled receptors-facilitators of diet-related immune regulation. Annu. Rev. Immunol. 35, 371–402 [DOI] [PubMed] [Google Scholar]
- 73. Thorburn A. N., Macia L., and Mackay C. R. (2014) Diet, metabolites, and “Western-lifestyle” inflammatory diseases. Immunity 40, 833–842 [DOI] [PubMed] [Google Scholar]
- 74. Mackenzie A. E., and Milligan G. (2017) The emerging pharmacology and function of GPR35 in the nervous system. Neuropharmacology 113, 661–671 [DOI] [PubMed] [Google Scholar]
- 75. Thomas C., Pellicciari R., Pruzanski M., Auwerx J., and Schoonjans K. (2008) Targeting bile-acid signalling for metabolic diseases. Nat. Rev. Drug Discov. 7, 678–693 [DOI] [PubMed] [Google Scholar]
- 76. Guo C., Chen W. D., and Wang Y. D. (2016) TGR5, not only a metabolic regulator. Front. Physiol. 7, 646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Pritchard J. B. (1988) Coupled transport of p-aminohippurate by rat kidney basolateral membrane vesicles. Am. J. Physiol. 255, F597–F604 [DOI] [PubMed] [Google Scholar]
- 78. Shimada H., Moewes B., and Burckhardt G. (1987) Indirect coupling to Na+ of p-aminohippuric acid uptake into rat renal basolateral membrane vesicles. Am. J. Physiol. 253, F795–F801 [DOI] [PubMed] [Google Scholar]
- 79. Pritchard J. B. (1995) Intracellular α-ketoglutarate controls the efficacy of renal organic anion transport. J. Pharmacol. Exp. Ther. 274, 1278–1284 [PubMed] [Google Scholar]
- 80. Martovetsky G., Tee J. B., and Nigam S. K. (2013) Hepatocyte nuclear factors 4a and 1a regulate kidney developmental expression of drug-metabolizing enzymes and drug transporters. Mol. Pharmacol. 84, 808–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Martovetsky G., Bush K. T., and Nigam S. K. (2016) Kidney versus liver specification of SLC and ABC drug transporters, tight junction molecules, and biomarkers. Drug Metab. Dispos. 44, 1050–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Gallegos T. F., Martovetsky G., Kouznetsova V., Bush K. T., and Nigam S. K. (2012) Organic anion and cation SLC22 “drug” transporter (Oat1, Oat3, and Oct1) regulation during development and maturation of the kidney proximal tubule. PLoS One 7, e40796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Evans A. M., DeHaven C. D., Barrett T., Mitchell M., and Milgram E. (2009) Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal. Chem. 81, 6656–6667 [DOI] [PubMed] [Google Scholar]
- 84. Ohta T., Masutomi N., Tsutsui N., Sakairi T., Mitchell M., Milburn M. V., Ryals J. A., Beebe K. D., and Guo L. (2009) Untargeted metabolomic profiling as an evaluative tool of fenofibrate-induced toxicology in Fischer 344 male rats. Toxicol. Pathol. 37, 521–535 [DOI] [PubMed] [Google Scholar]
- 85. Xia J., Sinelnikov I. V., Han B., and Wishart D. S. (2015) MetaboAnalyst 3.0–making metabolomics more meaningful. Nucleic Acids Res. 43, W251–W257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Vallon V., Eraly S. A., Rao S. R., Gerasimova M., Rose M., Nagle M., Anzai N., Smith T., Sharma K., Nigam S. K., and Rieg T. (2012) A role for the organic anion transporter OAT3 in renal creatinine secretion in mice. Am. J. Physiol. Renal Physiol. 302, F1293–F1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.