Abstract
Hedgehog signaling plays crucial roles in the development of calvarial bone, relying on the activation of Gli transcription factors. However, the molecular mechanism of the role of regulated Gli protein level in osteogenic specification of mesenchyme still remains elusive. Here, we show by conditionally inactivating Suppressor of Fused (Sufu), a critical repressor of Hedgehog signaling, in Wnt1-Cre–mediated cranial neural crest (CNC) or Dermo1-Cre–mediated mesodermal lineages that Sufu restraint of Hedgehog activity level is critical for differentiation of preosteogenic mesenchyme. Ablation of Sufu results in failure of calvarial bone formation, including CNC-derived bones and mesoderm-derived bones, depending on the Cre line being used. Although mesenchymal cells populate to frontonasal destinations, where they are then condensed, Sufu deletion significantly inhibits the proliferation of osteoprogenitor cells, and these cells no longer differentiate into osteoblasts. We show that there is suppression of Runx2 and Osterix, the osteogenic regulators, in calvarial mesenchyme in the Sufu mutant. We show that down-regulation of several genes upstream to Runx2 and Osterix is manifested within the calvarial primordia, including Bmp2 and its downstream genes Msx1/2 and Dlx5. By contrast, we find that Gli1, the Hedgehog activity readout gene, is excessively activated in mesenchyme. Deletion of Sufu in CNC leads to a discernible decrease in the repressive Gli3 form and an increase in the full-length Gli2. Finally, we demonstrate that simultaneous deletion of Gli2 and Sufu in CNC completely restores calvarial bone formation, suggesting that a sustained level of Hedgehog activity is critical in specification of the osteogenic mesenchymal cells.
Keywords: craniofacial development, gene knockout, Hedgehog signaling pathway, mouse, osteoblast, Gli2/3, Sufu, intramembranous ossification, neural crest-derived mesenchyme, skull vault
Introduction
The mammalian skull vault (top of neurocranium), which is composed of the paired frontal bones and parietal bones, an interparietal bone, and junctions between calvarial bones called the sutures, is formed from cranial osteogenic mesenchyme derived from two distinct embryonic tissue sources, the cranial neural crest (CNC)4 and paraxial mesoderm (1–4).
Calvarial bones form by intramembranous ossification, which in mice begins with the condensation of mesenchymal progenitors at E12.5 when the frontal and parietal bone primordia become evident. Condensed mesenchymal progenitor (osteoprogenitor) cells then undergo vigorous proliferation and differentiate into osteoblast precursor cells and express the early osteoblast marker Runx2. As the next 2 days of development proceed, these progenitor cells progress to preosteoblasts and mature osteoblasts expressing Runx2, Osterix, type 1 collagen, bone sialoprotein (BSP), osteocalcin (OC), and osteopontin (OPN) and secreting bone matrix. The ossification centers are formed by direct bone matrix deposition forming bone plates between the brain and the epidermis. Osteoblast differentiation occurs in the margins of osteogenic centers (the osteogenic front), where osteoblasts invade into, and progenitor cells are recruited from, the surrounding mesenchyme. Calvarial bone plates grow until they are nearly approximated but remain unfused at the sutures (5, 6).
Hedgehog (Hh) signaling plays a pivotal role in calvarial patterning and ossification (5, 7, 8). The mutation and disruption of the Hh signaling network result in a variety of genetic disorders associated with the growth defects in the calvaria and the sutures (craniosynostosis), both in humans and in mice (5, 9, 10). Combinatorial data of null mutation phenotypes and distinct expression patterns suggest that Sonic hedgehog (Shh) may prevent suture fusion, whereas Indian hedgehog (Ihh) activates the intramembranous ossification and sutural fusion, although there is disputability regarding whether Ihh and Shh function as negative or positive regulators of intramembranous bone development (8, 11–14). The Hh signaling is mediated mostly through Gli family transcription factors upon Hh activation (15). Ligand binding to the receptor Patched (Ptc) results in release of the transmembrane protein Smoothened (Smo) from Ptc inhibition, so that Smo transduces the signal intracellularly through interaction with Gli family of transcription factors, Gli1, Gli2, and Gli3 (5). Gli1 is transcriptionally regulated by Gli2 and Gli3 and functions as a transcription activator (16, 17), although it is dispensable during mouse embryogenesis (18, 19). However, it is involved in osteoblast differentiation during endochondral ossification (20, 21). Gli2 and Gli3 both have an N-terminal repressor domain and a C-terminal activator domain flanking the zinc fingers that can function as both activator and repressor, depending on Hh signal presence. In general, Gli2 remains as a transcription activator due to inefficient protein processing, whereas Gli3 mostly functions as a transcription repressor because of efficient degradation. In both human and mice, loss of function of GLI3 causes Greig cephalopolysyndactyly syndrome, which causes the premature fusion of metopic (interfrontal) and lambdoidal sutures and abnormal frontal bone morphology (9, 22–24). Deletion of repressor Gli3 results in excessive osteoprogenitor proliferation and differentiation, strongly implicating GLI activity and regulation in the control of calvarial bone development. Moreover, in vitro studies have shown that Gli2 mediates the Ihh control of osteoblast differentiation of mesenchymal cells through direct interaction with Runx2 expression (25, 26), suggesting the importance of Gli in the regulation of osteogenesis. However, it remains unclear whether there is a genetic requirement of GLI activity level for early development of the calvarial bone development and how the on/off switch of the activator/repressor Gli2 and Gli3 is conducted in osteogenic mesenchymal cell proliferation and differentiation of osteoprogenitor cells.
In mammals, Suppressor of Fused (Sufu) is a major repressor of Hh signaling by modulation of Gli2 and Gli3. In the absence of Hh signal activation, Sufu can sequester the full-length Gli2 and Gli3 proteins (GliA) to the cytoplasm, facilitating the formation of repressive Gli2/3 (GliR) and thereby regulating Gli at the protein levels (27–32). By contrast, full-length Gli2 and Gli3 remain as activators in the presence of high Hh levels, probably by dissociation with Sufu (33, 34). The Sufu null mutation in mice causes a global increase in up-regulation of Ptc1 (35, 36). Conditional deletion of Sufu in the limb bud mesenchyme resulted in the alteration of anterior/posterior patterning and polydactyly, associated with the reduction in repressive Gli2 and Gli3 (37, 38). The fact that expression of Shh and Ihh in the calvarial primordia region is detected in a relatively late stage around E16.5 raises the possibility that activity of Gli2R and Gli3R is required for regulation of osteoprogenitor differentiation and maturation in intramembranous bone development (5, 8, 39). In this study, by ablation of Sufu in neural crest-derived mesenchymal cells, we show the requirement of Sufu regulation of Gli2 and Gli3 for calvarial bone formation during development. Sufu deletion in CNC lineage causes activation of Hh signaling in the mesenchyme for the calvarial bone primordia, resulting in the failure of developmental progression of osteogenesis beyond initial condensations in the calvarial mesenchyme via dysregulation of downstream molecules.
Results
Functional Sufu is required for skull vault development
In situ hybridization showed that Sufu is globally expressed in the calvarial mesenchyme (Fig. 1A and supplemental Fig. S1) that was consistent with immunofluorescence analysis results using the Sufu antibody (Fig. 1B), suggesting a role for this gene in the presumptive skull vault primordia. To investigate the potential function of Sufu in calvarial bone development, we examined mice with CNC-specific conditional deletion of Sufu using the Wnt1-Cre mouse (Sufufx/fx;Wnt1-Cre or SufuWnt1Cre) and mice with mesoderm-specific Sufu deletion using the Dermo1-Cre strain (Sufufx/fx;Dermo1-Cre). Inactivation of Sufu with Wnt1-Cre or Dermo1-Cre caused immediate postnatal lethality. Immunofluorescence and Western blot analyses demonstrated removal of Sufu in the calvarial mesenchyme of Sufufx/fx;Wnt1-Cre mutants (Fig. 1, C and D). Skeletal preparations with Alcian blue/Alizarin red staining showed that the Wnt1-Cre–mediated Sufu deletion resulted in specific loss of CNC-derived calvarial bones, such as the frontal bones and the central portion of the interparietal bone, as well as the facial bones (Fig. 1 (E–L) and supplemental Fig. S2); by contrast, ablation of Sufu with Dermo1-Cre caused specific defects in mesoderm-derived calvarial bones, including parietal bones and the lateral part of the interparietal bone (Fig. 1, M–T). These data suggest that an intrinsic function of Sufu is required for the calvarial bone formation. We thus focused our studies on the CNC-derived calvarial bone formation.
Figure 1.
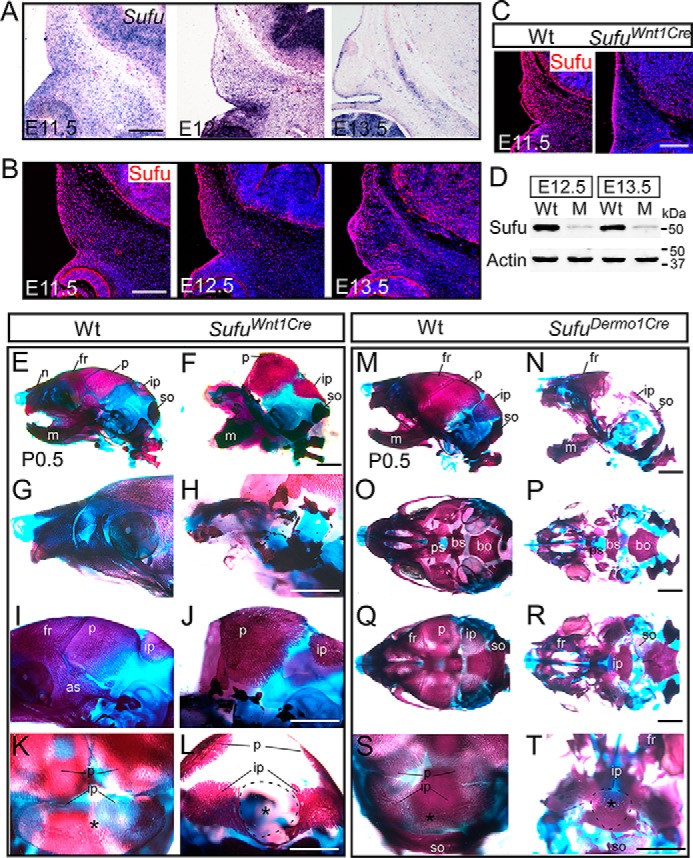
Sufu is required for the development of calvarial bones. A and B, in situ hybridization and immunohistochemical analyses showing the expression of Sufu in the development of presumptive frontal primordium. C and D, immunohistochemistry and Western blot analyses showing the mesenchymal cell–specific removal of Sufu with Wnt1-Cre in the presumptive frontal primordium, respectively. E–L, skeletal staining with Alcian blue and Alizarin red shows the missing CNC-derived craniofacial bones, including the frontal bones and the central portion of the interparietal bone in neonatal Sufufx/fx;Wnt1-Cre (SufuWnt1Cre) mice compared with littermate wild type control mice (n = 8). (Wild-type control mice in this study did not carry the Cre allele). M–T, Alcian blue/Alizarin red staining showing mesoderm lineage–specific developmental defects of calvarial bones in neonatal Sufufx/fx;Dermo1-Cre (SufuDermo1Cre) mutant compared with littermate wild-type mice (n = 3). O and P, ventral view of skull base bones. Q and R, dorsal view of calvarial bones. K, L, S, and T, back view of skull bones. Asterisks, central portion of the interparietal bone. C57B6/J mice were used for A and B. Wt, wild type; M, Sufufx/fx;Wnt1-Cre; as, alisphenoid bone; bo, basioccipital bone; bs, basisphenoid bone; fr, frontal bone; ip, interparietal bone; m, mandible; n, nasal bone; p, parietal bone; ps, presphenoid bone; so, supraoccipital bone. Scale bars, 200 μm (A–C) and 1 mm (E–T).
Mutation of Sufu in CNC does not affect migration but impedes condensation of CNC-derived mesenchymal cells
Formation of mesenchymal condensations at E12.5 is an early sign of intramembranous bone development in mice. However, scanning electron microscopy analysis showed that developmental hypoplasia of the facial/nasal processes occurred before E11.5 in Sufufx/fx;Wnt1-Cre (supplemental Fig. S3). To determine whether the deletion of Sufu interferes with CNC cell migration and population to the calvarial bone primordia, we traced the CNC population using Sufufx/fx;Wnt1-Cre mice carrying the R26R reporter allele (R26R;Sufufx/fx;Wnt1-Cre) between E11.5 and E13.5. X-gal staining on sections of the embryonic head showed that the CNC cells could normally populate to the presumptive calvarial sites in the Sufu mutant (Fig. 2, A–H), indicating that Wnt1-Cre–mediated Sufu deletion does not affect CNC cell migration. To determine whether Sufu deletion impedes the formation of the mesenchymal condensations, we performed a series of histological analyses on the calvarial tissues between E12.5 and E14.5 throughout the process of the calvarial bone development (Fig. 2, I–N). Hematoxylin and eosin (H&E) staining on the embryonic head sections showed that the mesenchyme within the presumptive calvarial bone primordium appeared condensed in E12.5 Sufufx/fx;Wnt1-Cre, comparable with the wild type (Fig. 2, I and J). The mesenchymal condensation became evident at E13.5 in wild type but was much less in Sufufx/fx;Wnt1-Cre mutants (Fig. 2, K and L). Calvarial bone primordium underwent ossification in the wild type at E14.5, but it did not develop in Sufufx/fx;Wnt1-Cre mutants (Fig. 2, M and N). In the wild type, calvarial bone development was progressing toward differentiation after E13.5 and mineralization of mature osteoblasts at E14.5, as exhibited by von Kossa staining and the expression of OPN, the osteoblast marker involved in bone matrix mineralization (Fig. 2, O–T). However, neither of these markers was detectable in Sufufx/fx;Wnt1-Cre (Fig. 2, O–T), suggesting that there is no sign of mineralization of mature osteoblasts in the absence of Sufu. In summary, these data suggest that the failure of the calvarial bone formation in Sufufx/fx;Wnt1-Cre was attributed to interruption of mesenchymal cell condensation and differentiation to osteoblasts.
Figure 2.
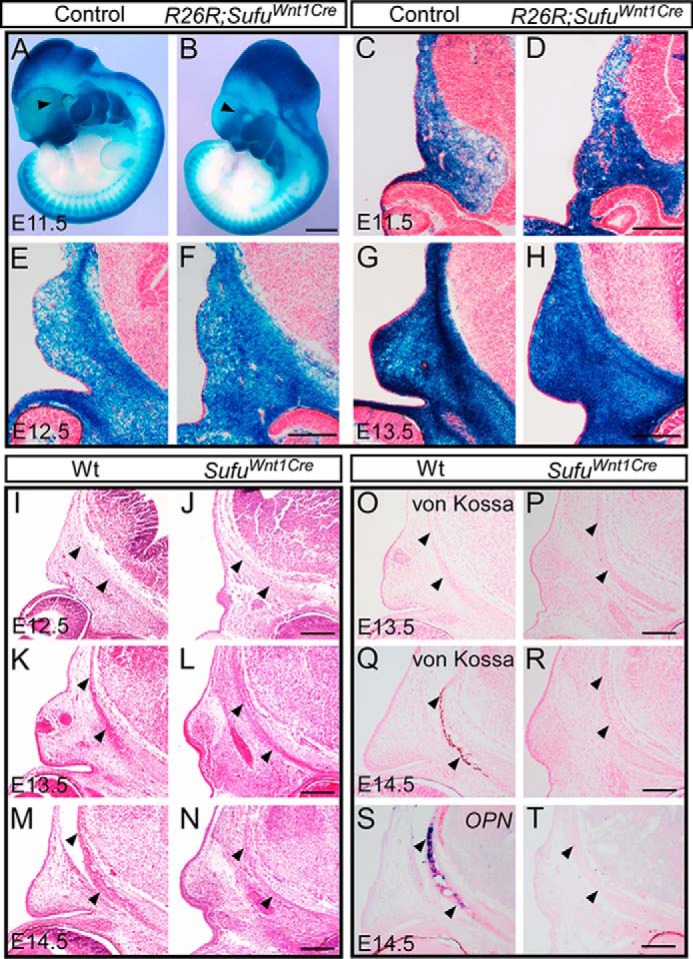
Removal of Sufu in CNC does not affect migration, but does affect condensation and ossification of CNC-derived mesenchymal cells. A–H, X-gal staining showing the migration of CNC-derived mesenchymal cells to the presumptive frontal primordium in R26R;Sufufx/fx;Wnt1-Cre (R26R;SufuWnt1Cre) embryos and littermate control mice (R26R;Sufufx/+;Wnt1-Cre, n = 3). I–N, H&E staining showing the condensation of mesenchymal cells in the frontal primordium both in littermate wild type and Sufufx/fx;Wnt1-Cre (SufuWnt1Cre) mutant (n = 3). O–R, von Kossa staining showing the mineralization of frontal bones in E14.5 in littermate wild type versus Sufufx/fx;Wnt1-Cre (SufuWnt1Cre) samples (n = 3). S and T, In situ hybridization showing the expression of mineralization marker OPN in the wild-type frontal bone but not in the littermate Sufu mutant (n = 2). Arrowhead, the presumptive frontal primordium. Scale bars, 1 mm (A and B) and 200 μm (C–T).
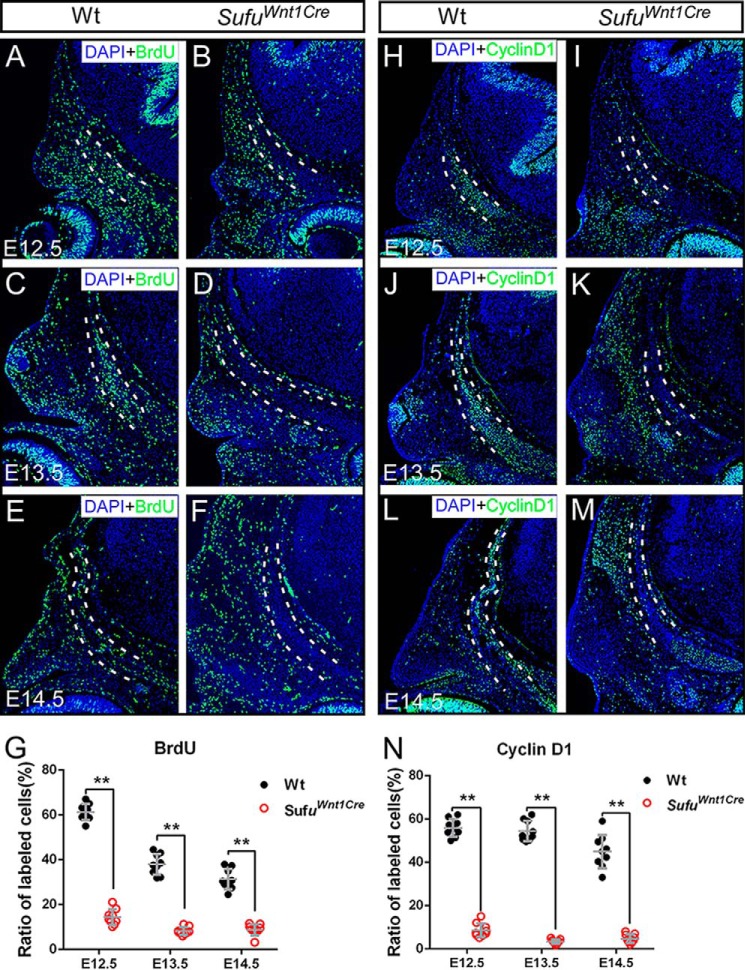
Sufu deletion disturbs proliferation but not apoptosis of osteogenic mesenchymal cells in the calvarial bone primordium
For intramembranous bone formation, initially condensed mesenchymal cells undergo vigorous proliferation prior to differentiation and mineralization (5, 6) (Fig. 3). To understand the cellular mechanism causing a defect in the differentiation of preosteogenic mesenchymal cells in the Sufu mutant, we examined cell proliferation using BrdU labeling. Immunofluorescence and statistical analyses showed a significant decrease in the rate of the BrdU-labeled cells within the presumptive calvarial bone primordia in Sufufx/fx;Wnt1-Cre between E12.5 and E14.5, compared with wild type (Fig. 3, A–G). We also applied immunofluorescence staining using antibody against cyclin D1, a marker for proliferation, on the sections. We found that the rate of cyclin D1-positive cells was significantly decreased in the Sufufx/fx;Wnt1-Cre mice (Fig. 3, H–N) through all detected stages, consistent with the BrdU labeling results. To determine whether the failure of calvarial bone formation resulted from an increase of abnormal cell death within the primordium, we performed a TUNEL assay on the histological sections. Cell apoptosis signal of mesenchymal cells within the calvarial bone primordium was indistinguishable between the wild-type and Sufufx/fx;Wnt1-Cre embryos (supplemental Fig. S4). Together, these data suggest that the function of Sufu is critical for cell proliferation of CNC-derived mesenchymal cells during osteogenesis but not for cell survival. Decreased mesenchymal cell proliferation may account for the developmental defect of the calvarial bone in the Sufu mutant.
Figure 3.
Cell proliferation analysis of mesenchymal cells within the presumptive frontal primordium. A–G, BrdU labeling analysis showing the reduction of cell proliferation rate of mesenchymal cells within the presumptive frontal primordium in Sufufx/fx;Wnt1-Cre (SufuWnt1Cre) mutants compared with littermate control mice during E12.5–E14.5 (n = 3 animals, three serial sections for each). H–N, immunofluorescence staining using cyclin D1 antibody showing the reduction of cell proliferation within presumptive frontal primordium in Sufufx/fx;Wnt1-Cre (SufuWnt1Cre) mutants compared with littermate control mice (n = 3 animals, three serial sections for each). Dashed lines outline the presumptive frontal primordium. The cell proliferation rate was calculated with the number of BrdU or cyclin D1 marked cells divided by the DAPI-labeled cells within the presumptive frontal primordium. Statistical evaluation of the differential of cell proliferation rate between wild type and mutants was performed with Student's t test. Error bars, S.D. **, p < 0.01. Scale bars, 200 μm.
Sufu is required for specification of osteo-mesenchyme within calvarial bone rudiment
To understand whether and how the aberrant differentiation of preosteoblasts contributes to the failure of calvarial bone formation in Sufufx/fx;Wnt1-Cre embryos, we examined expression of Runx2 and Osterix (Sp7) in E12.5 and E13.5 embryos. Expression of Runx2, a core transcription factor and specific marker for preosteoblast (40), was initiated at late E12.5 in the osteogenic condensations and enhanced in preosteoblasts at E13.5 in wild type (Fig. 4, A and C). Osterix is a zinc-finger-containing transcription factor that is necessary for osteoblast differentiation and bone formation, and acts downstream of Runx2 (41). Osterix transcripts appeared in the preosteoblasts at E13.5 in the wild type (Fig. 4, E and G). However, neither Runx2 nor Osterix expression was recognized in the mesenchymal cells within the presumptive calvarial bone primordium at both stages in the Sufu mutant (Fig. 4, B, D, F, and H). Immunofluorescence analyses with antibodies against Runx2 and Osterix did not show the protein products specific to the mesenchymal condensations of the presumptive calvarial bone primordia in Sufufx/fx;Wnt1-Cre mice (Fig. 4, I–P), consistent with our in situ hybridization data. Taken together, these data suggest that Sufu is essential for the specification and differentiation of the initially condensed mesenchyme to Runx2/Osterix-expressing osteoblast precursors, and the dysregulation of these osteo-marker genes in conditional ablation of Sufu accounts for the failure of calvarial bone formation.
Figure 4.
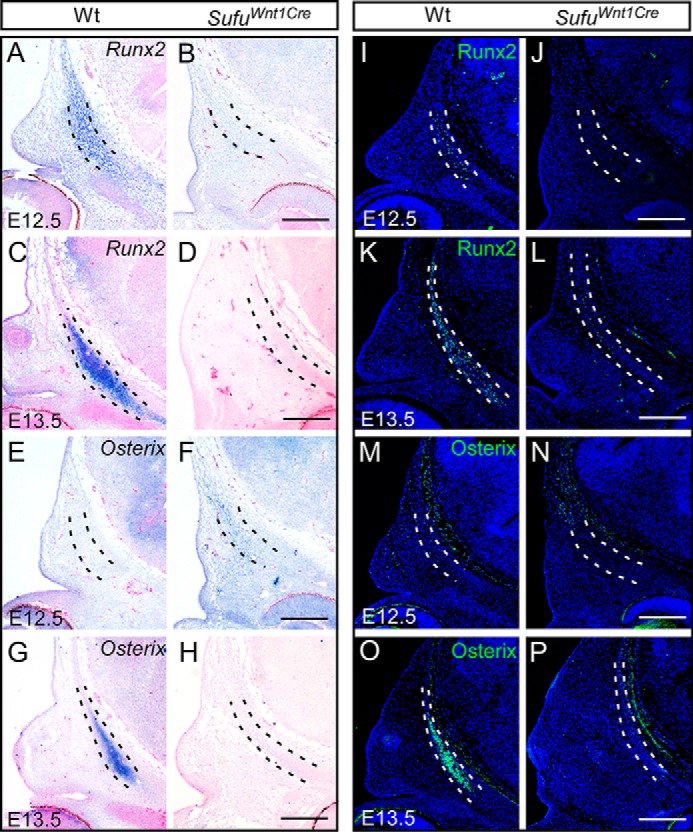
Expression of the osteogenic markers in development of the frontal bone primordium with in situ hybridization and immunofluorescence analyses. A–H, in situ hybridization showing the expression of Runx2 and Osterix during the frontal bone development in Sufufx/fx;Wnt1-Cre (SufuWnt1Cre) mutants and littermate wild-type embryos (n = 3). I–P, immunofluorescence analysis showing the reduction of Runx2 and Osterix production between E12.5 and E13.5 in Sufufx/fx;Wnt1-Cre (SufuWnt1Cre) embryos compared with littermate wild-type mice (n = 3). Dashed lines outline the presumptive frontal primordium. Scale bars, 200 μm.
Altered gene expression by loss of Sufu within calvarial mesenchyme
We next examined by in situ hybridization the expression of potential downstream targets of Sufu-dependent regulation in the calvarial mesenchyme of Sufufx/fx;Wnt1-Cre mice, in particular for several signaling molecules and transcription factors previously known for regulating the expression of Runx2 and Osterix in osteogenic differentiation. Dlx5 is an important mediator of calvarial osteoblast differentiation by positive regulation of Runx2 (42). Expression of Dlx5 was detected by in situ hybridization in the osteogenic mesenchyme at E12.5 and significantly up-regulated at E13.5 within the calvarial bone rudiment of the wild type (Fig. 5, A and C). However, expression of Dlx5 was faintly detected in the calvarial mesenchyme of the Sufu mutant at E12.5 and decreased in E13.5 Sufufx/fx;Wnt1-Cre compared with the wild-type control (Fig. 5, A–D). Previous studies have shown that homeobox genes Msx1 and Msx2 are essential for the differentiation and proliferation of osteogenic cells within the calvarial rudiments (43). In a combination of Msx1/2 mutants, skull vault components, including the frontal bones, fail to form (44), indicating the requirement for regulation of CNC cell differentiation during calvarial bone development (44). At E13.5, both Msx1 and Msx2 were expressed in the calvarial osteogenic mesenchyme in wild type, but Msx2 transcripts within the preosteoblast rudiment were much stronger than those of Msx1 (Fig. 5, E and G). In contrast, no Msx1 transcript was detected by in situ hybridization in the Sufu mutant (Fig. 5F), whereas Msx2 expression was significantly decreased in the Sufu mutant (Fig. 5, G and H). In addition, in the calvarial mesenchyme of the Sufufx/fx;Wnt1-Cre mice, we observed dramatic down-regulation in expression of Bmp2 (Fig. 5, I and J), which is a well-known signaling molecule associated with Msx2 in calvarial bone development (39, 45). Together, these data suggest that Sufu-dependent Hh signaling may control osteogenic differentiation of calvarial mesenchyme through regulation of transcription factions upstream of Runx2 and Osterix.
Figure 5.
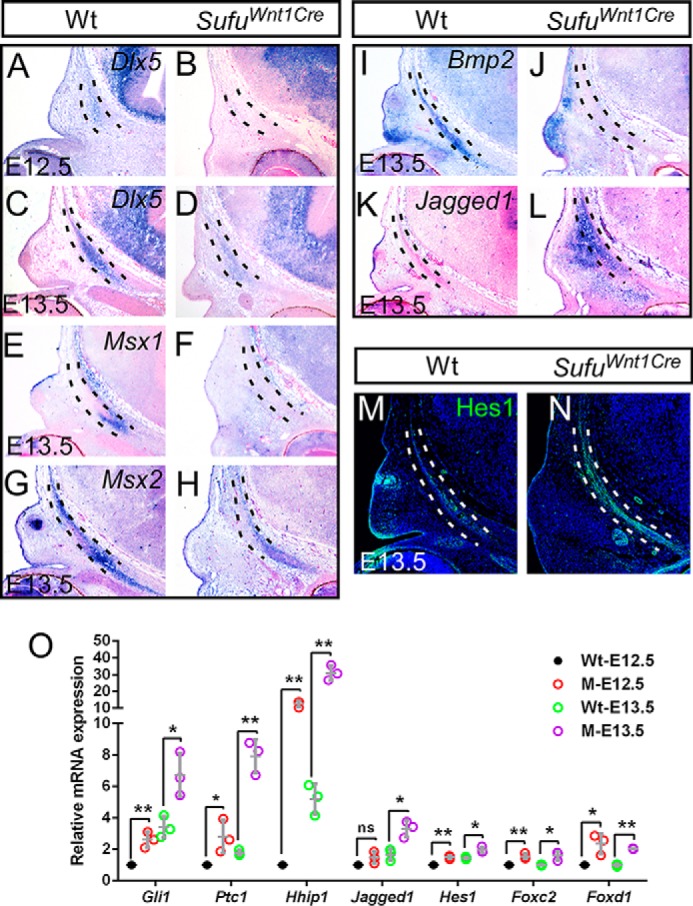
Mutation of Sufu in CNC results in expression alteration of downstream genes. A–L, in situ hybridization showing the down-regulation of Dlx5, Msx1/2, and Bmp2 and up-regulation of Notch signaling ligand Jagged1 in the frontal primordium of Sufufx/fx;Wnt1-Cre (SufuWnt1Cre) mutants compared with littermate wild type controls (n = 3). M and N, immunofluorescence showing the activation of Hes1 in the frontal primordium of Sufufx/fx;Wnt1-Cre (SufuWnt1Cre) versus the littermate wild type (n = 3). O, qRT-PCR analysis showing the up-regulation of Hh signaling target or downstream genes in Sufu mutants at E12.5 and E13.5. Three sets of mutant and littermate wild-type control at each stage were analyzed with independent triplicate experiments. Dashed lines outline the presumptive frontal primordium. Wt, wild type; M, Sufufx/fx;Wnt1-Cre.
If Sufu were directly involved in the regulation of calvarial mesenchyme differentiation to preosteoblasts by inhibitory modulation of Hh activity, we would expect up-regulation of genes downstream of the Hh pathway concomitant with elevated Shh activity within the calvarial mesenchyme in the absence of Sufu. Previously, studies have demonstrated that a number of genes encoding transcription factors and signal molecules are associated with the craniofacial development under the guidance of the SHH signaling pathway. To characterize Sufu functional contributions to the regulation of preosteoblast differentiation through Shh cascades, we examined by quantitative RT-PCR the expression of several targets of Hh signaling, including Gli1, Ptc1, Hhip, Jagged1, Foxc2, and Foxd1 (5, 46–48). Significant increases in expression of these genes were demonstrated using calvarial RNA samples of Sufufx/fx;Wnt1-Cre mice at E12.5 and E13.5 (Fig. 5O), suggesting up-regulation of the Hh signaling cascade in the osteogenic mesenchymal cells for calvarial bone development.
Jagged1, the molecule in the Notch signaling pathway, is one of the downstream genes of Hh signaling (48–50). Transcripts of Jagged1 were not present in the osteogenic mesenchyme within or surrounding the frontal bone in E13.5 wild-type mice (Fig. 5K). However, we found by in situ hybridization that Jagged1 expression was highly activated in CNC-derived mesenchymal cells in the Sufu mutant (Fig. 5L). Intriguingly, Hes1, the target gene of Notch signaling, was specifically increased within calvarial primordia, suggesting that Sufu-dependent up-regulation of Notch signaling is associated with dysregulation of preosteoblast differentiation (Fig. 5, M and N). Overall, these results suggest that Sufu-dependent Hh signaling is critical for regulation of osteogenic differentiation of CNC-derived mesenchymal cells during calvarial bone development, probably through regulation of multiple Shh signaling downstream genes.
Deletion of Sufu in CNC-derived mesenchymal cells perturbs the regulation of SHH activity via changing the protein stability of Gli2/3
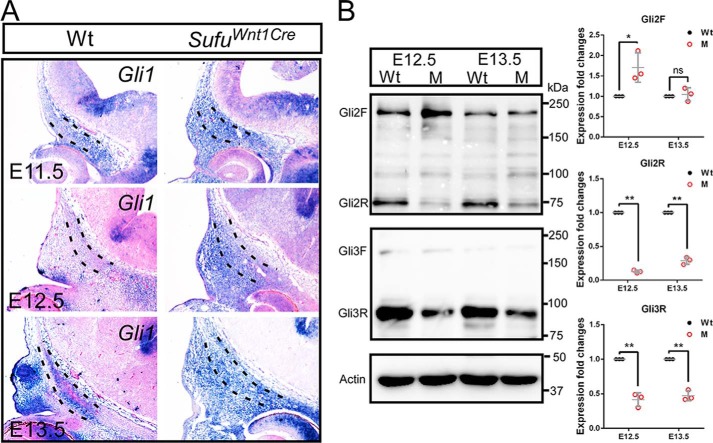
Sufu is a critical negative regulator of Hh signaling by binding to the transcription factor GLIs. To investigate the effect of Sufu deletion on the Hh activity, we next detected the expression of Gli1, the target gene and readout of Hh signaling. During development of the skull vault in wild-type mice, expression of Gli1 was extensive in the CNC-derived cranial mesenchyme in E11.5 (Fig. 6A) and was dramatically down-regulated in the condensed mesenchyme for the calvarial primordia at E12.5. Interestingly, the transcripts of Gli1 were obviously detected in preosteoblasts at E13.5 (Fig. 6A). This dynamic expression pattern may suggest that the regulatory level of Hedgehog activity is critical for the condensed mesenchyme toward the preosteogenic fate. However, expression of Gli1 was retained and broadly detected in the CNC-derived mesenchyme throughout stages in the absence of Sufu (Fig. 6A), indicating that the level of Shh activity is abnormally up-regulated in the calvarial mesenchyme of Sufufx/fx;Wnt1-Cre. This is consistent with previously reported data for Sufu null mutation (35), suggesting that Sufu inhibitory regulation of Shh activity is essential for preosteogenic differentiation of the mesenchyme during intramembranous ossification.
Figure 6.
Sufu mutation results in activation of Hh signaling and protein stability variation of Gli2/3. A, in situ hybridization showing up-regulation of Gli1 within the presumptive frontal primordium in Sufufx/fx;Wnt1-Cre (SufuWnt1Cre) mutants compared with littermate wild-type control (n = 3) during calvarial bone development. Dashed lines outline the presumptive frontal primordium. B, Western blot showing a discernable increase of full-length Gli2 (Gli2F) and significant decrease of truncated repressor forms Gli2 (Gli2R) and Gli3 (Gli3R) in mesenchymal cells within the frontal primordium in the Sufu mutant versus the wild type (n = 3). Each image shows a representative result of independent triplicated experiments. Expression levels were quantified from the band intensity as relative values of the target protein/actin expression ratios. *, p < 0.05; **, p < 0.01; ns, non-significant; Student's t test. Error bars, S.D. Wt, wild type; M, Sufufx/fx;Wnt1-Cre.
We sought to identify the means by which Sufu modulates Hh signaling within the calvarial mesenchyme by clarifying the expression of Gli2/3 at the protein level. Gli2 primarily functions as a full-length activator, and Gli3 mainly acts as a repressor by an N-terminal truncated form in Hh signaling, and binding of Sufu to Gli2 or Gli3 can inhibit their processing and keep the stability of the full-length forms. To distinguish the effect of Sufu deletion on these processes, Western blot analyses were performed with protein extracted from embryonic calvarial samples. We found that the Gli2 full-length form was intensified, concomitant with a significant decrease in its truncated repressor form, in the Sufu mutant compared with the wild type (Fig. 6B). By contrast, Gli3 existed primarily in the truncated repressor form, and the Gli3 repressor (Gli3R) in the CNC-derived mesenchyme was significantly decreased in E12.5 and E13.5 Sufufx/fx;Wnt1-Cre mutant samples, in comparison with wild type (Fig. 6B). We did not detect a concomitant increase of Gli3 full-length form (Fig. 6B). These findings, together with in situ hybridization data showing the abnormal activation of Gli1 expression in calvarial mesenchyme, suggest that within the calvarial mesenchyme, the inactivation of Sufu reduces Gli2 and Gli3 repressor forms and increases the activator form of Gli2, resulting in an abnormal high Hh activity.
Failure of calvarial bone development in Sufufx/fx;Wnt1-Cre mutant can be restored by compound mutation of Sufu and Gli2 in CNC
The evidence above suggests that loss of Sufu in the CNC-derived mesenchyme interferes with Sufu regulation of Gli2 protein, resulting in unbalanced elevation of Hh output critical for differentiation of osteoprogenitor cells. To test this proposal in vivo, we generated Wnt1-Cre–mediated compound conditional deletion of Sufu and Gli2 (Fig. 7M). Skeletal preparations revealed that the calvarial bone formation in mice carrying Sufufx/fx;Gli2fx/fx;Wnt1-Cre allele was comparable with that of wild-type control (Fig. 7, A–C), indicative of rescue of intramembranous bone ossification. von Kossa staining showed that mineralization of osteoblasts within the calvarial bone occurred in the double mutant at E14.5 of Sufufx/fx;Gli2fx/fx;Wnt1-Cre (Fig. 7, D–F). Immunofluorescence analyses showed reactivation of cyclin D1 and Runx2 in the calvarial bone in the E14.5 double mutant (Fig. 7, G–I). In situ analysis showed that the expression of Gli1 was decreased in Sufufx/fx;Gli2fx/fx;Wnt1-Cre mutants, compared with Sufufx/fx;Wnt1-Cre (Fig. 7, J–L). It was further confirmed by qRT-PCR analysis that the expression of Hh signaling targets and downstream genes in Sufufx/fx;Gli2fx/fx;Wnt1-Cre is comparable with wild-type control (Fig. 7N). In contrast, simultaneous deletion of Gli3 in Sufufx/fx;Wnt1-Cre background failed to restore the calvarial bone formation but led to more severe craniofacial defects (supplemental Fig. S5). Taken together, these data may suggest that unbalanced Hh activity from loss of Sufu can be genetically neutralized by simultaneous deletion of Gli2, providing evidence that a stabilized level of Hh activity is required for developmental progress of mesenchyme beyond condensations in the formation of calvarial bones (Fig. 8).
Figure 7.
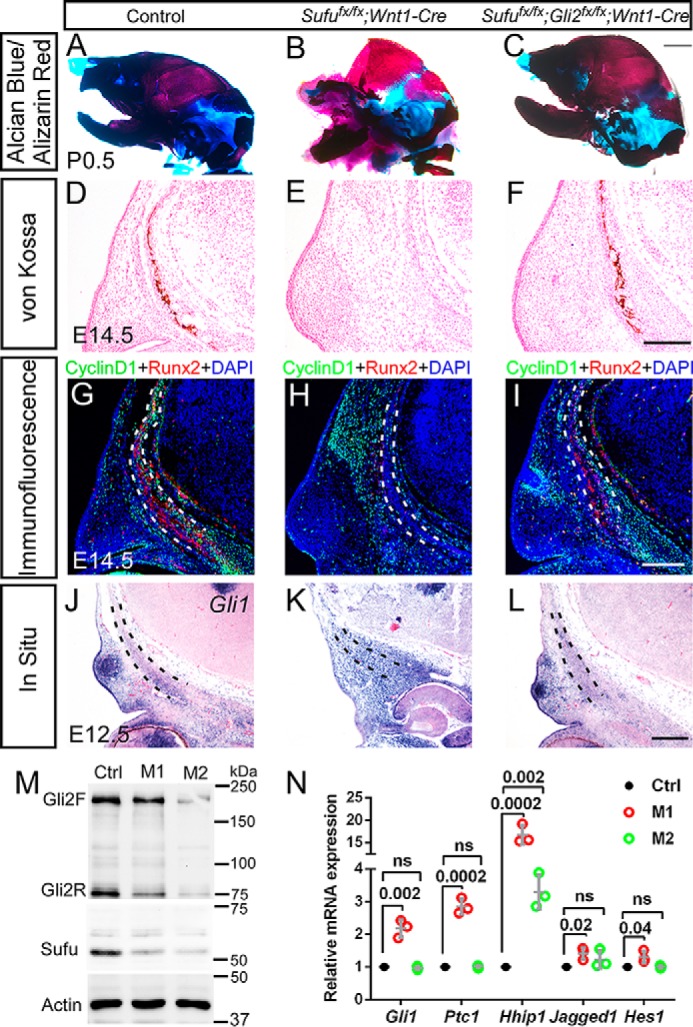
Compound mutation of Sufu and Gli2 in CNC rescues the developmental defects of calvarial bones in the Sufufx/fx;Wnt1-Cre mutant. A–C, skeletal staining with Alcian blue/Alizarin red showing the complete restoration of calvarial bone formation in Sufufx/fx;Gli2fx/fx;Wnt1-Cre mice (n = 5). D–F, von Kossa staining showing the restoration of frontal bone mineralization in Sufufx/fx;Gli2fx/fx;Wnt1-Cre at E14.5 (n = 2). G–I, immunofluorescence showing the restored expression of cyclin D1 and osteogenic marker Runx2 in the frontal primordium of Sufufx/fx;Gli2fx/fx;Wnt1-Cre at E14.5 (n = 3). J–L, in situ hybridization analysis reveals that expression of Gli1 in the frontal primordium of Sufufx/fx;Gli2fx/fx;Wnt1-Cre is comparable with wild type control at E12.5 (n = 3). Dashed lines outline the presumptive frontal primordium. Scale bars, 1 mm (A–C) and 200 μm (E–L). M, Western blot showing substantial removal of Sufu and Gli2 in calvarial mesenchyme of compound mutant Sufufx/fx;Gli2fx/fx;Wnt1-Cre at E12.5 (n = 3). N, qRT-PCR shows that the expression of the target and downstream genes of the Hh signaling pathway in Sufufx/fx;Gli2fx/fx;Wnt1-Cre at E12.5 is comparable with wild-type control (n = 3). The significance level of differential expression between wild type control and mutants was evaluated with Student's t test. p values are indicated above each pair compared. ns, non-significant. Error bars, S.D. Ctrl, wild-type control; M1, Sufufx/fx;Wnt1-Cre; M2, Sufufx/fx;Gli2fx/fx;Wnt1-Cre.
Figure 8.
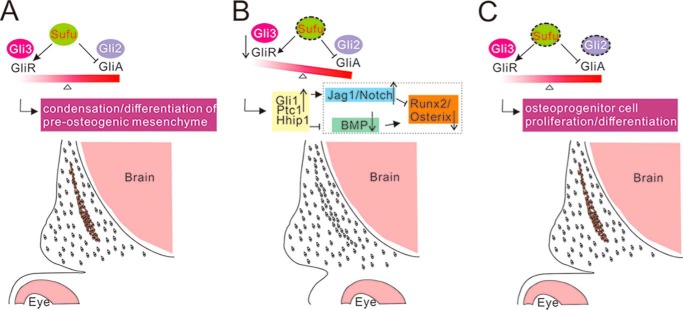
Schematic of hypothesized model for Sufu-dependent balance of Gli repressors and activators in regulating calvarial bone development. In wild-type calvarial mesenchymal cells, Sufu is required for maintaining the balance of Gli2 and Gli3 activity (A). Calvarial mesenchymal cells in Sufufx/fx;Wnt1-Cre mutant cannot undergo normal condensation and osteogenic differentiation due to the deletion of Sufu that leads to a decrease of Gli repressors and an increase of Hh signaling activity (B). This leads to activation of downstream Jag1/Notch signaling and inhibition of BMP signaling, respectively, resulting in dysregulation of osteogenic markers Runx2 and Osterix. Simultaneous removal of Sufu and Gli2 can compromise Hh activity by rebalancing the Gli repressors and full-length activators, which promotes the proliferation and differentiation of osteoprogenitor cells (C). The regulatory relationships shown in dashed lines are drawn according to previous studies from other researchers (41, 44, 45, 51–53).
Discussion
In this study, by conditional inactivation of Sufu in CNC cells, we provide genetic evidence that the level of Sufu-regulated Hh activity is critical for osteogenic specification of mesenchyme in calvarial bone formation. Moreover, inactivation of Sufu in the osteogenic mesenchyme leads to interruption of the molecular network critical for developmental progress of the initial condensed mesenchyme toward an osteoblastic lineage. Finally, we show the complete restoration of calvarial bone formation by compound deletion of Sufu and Gli2 in CNC. Our study suggests that Sufu acts as an inhibitor of full-length Gli2 activity and an activator of Gli3 repressor activity in mesenchyme differentiation of osteo-progenitors in intramembranous bone development (Fig. 8).
It is noted that Sufu-dependent Hh signaling activity probably targets the BMP2 signaling cascade during the regulation of preosteogenic mesenchyme. Previous studies have suggested that BMPs are required for commitment of CNC-derived mesenchyme to the osteogenic fate in intramembranous ossification, and Ihh negatively regulates the further differentiation of osteoblast precursors (11). We found that the osteogenic marker Bmp2 was diminished within the frontal bone-forming mesenchyme of Sufufx/fx;Wnt1-Cre mutant, suggesting that Sufu-dependent Hh signaling inhibits the commitment of mesenchymal progenitors to the osteogenic lineage by suppressing BMP signaling (11, 23). Consistently, the expression of osteogenic markers downstream of BMP signaling, such as Msx1/2 and Dlx5, is overtly suppressed in the Sufufx/fx;Wnt1-Cre mutant. Msx1 and Msx2 are required for differentiation in the CNC lineage within the frontal bone primordium by regulation of Runx2 (43, 44). In frontal bone development, Msx2 and Twist1 cooperatively control the proliferation and differentiation of the osteogenic mesenchyme (51). In addition, Msx1 and Dlx5 are both involved in the osteoblast differentiation by synergic and positive regulation of Runx2 and Osterix (42). Previous data from in vitro analyses revealed that BMP2-induced Osterix expression is mainly mediated by Dlx5 and not by Runx2 (52). After identifying the above osteogenic marker genes whose expression had diminished upon conditional ablation of Sufu in the CNC-derived mesenchyme, we recognized that the absence of Sufu activity in the mesenchyme might have changed the regulation required for specifying preosteogenic mesenchyme to osteoblastic fate, in which excessive Hh signaling inhibits the specification and proliferation of mesenchymal progenitor cells by suppressing BMP signaling during calvarial bone development.
Previous studies have shown that Jagged1, the molecule in the Notch signaling pathway, is one of the downstream genes of Hh signaling (48–50). Notch signaling is involved in regulation of osteogenesis by inhibition of endochondral bone formation and osteoblastic differentiation. Notch signaling in the bone marrow maintains mesenchymal progenitors by suppressing osteoblast differentiation through direct interaction of Hes/Hey proteins with Runx2 that down-regulates its transcriptional activity (53). Our data indicating that up-regulation of Notch ligand Jagged1 in the Sufufx/fx;Wnt1-Cre mutant, together with the increased expression of Notch signaling target gene Hes1 in the calvarial bone primordia of Sufufx/fx;Wnt1-Cre mutant, suggest that excessive Hh signaling inhibits the osteoblastic specification of mesenchymal progenitors through activing downstream Notch signaling (49).
The present results confirm and extend earlier works in long bone development suggesting that Hh signaling is required for osteogenesis in the context of temporal specificity. Hh signaling has proved an essential but transiently required component for initial specification of an osteoblast progenitor to a Runx2-expressing osteoblast precursor (54, 55), but Hh is not required once Runx2 and Osterix are expressed in the osteoblast precursor. Activation of Hh signaling in human mesenchymal stem cells at early stages, but not in differentiated osteoblasts, inhibits human osteoblast differentiation by decreasing Runx2 expression (56), suggesting that Hh signaling plays critical roles in early rather than late stages of osteogenic differentiation. Interestingly, a recent study has suggested that Runx2 is required for intramembranous ossification from the initial phase when the mesenchymal cells express Prx1 and Sca1 until the time when osteoblast precursors express Osterix (57). Our data showing the failed expression of Runx2 and Osterix in the initial stage of the osteogenic mesenchyme suggest that a Sufu-regulated level of Hh signaling activity is required for the initiation of the osteoblast progenitor.
In the absence of Hh ligands or interference with the Hedgehog signaling transduction, transcriptional repression of Hh activity and targets would result in a variety of craniofacial abnormalities (44, 55, 58–61). Gli3 null mice with excessive proliferation and differentiation of osteoblast have craniosynostosis (25). In contrast, local application of recombinant FGF2 protein can rescue loss of Gli3 as it stabilizes the increased osteoblastic proliferation observed in Gli3 mutant mice (23). Interestingly, our current study provides genetic evidence that Sufu, a crucial negative regulator of Hh signaling, acts via repressive regulation of Hedgehog activity in the preosteogenic mesenchyme to ensure osteogenesis in calvarial bone formation. Our data suggest that the function of Gli2 and Gli3 in the calvarial mesenchyme requires the presence of Sufu (Fig. 8). Sufu stabilizes full-length Gli2 and Gli3 and promotes the generation of their repressor forms that are consistent with previous studies (62, 63). We demonstrate that calvarial bone development arrests at the initial mesenchyme condensation associated with excessive activation of Gli2 and Gli3 activity by deletion of Sufu in CNC-derived mesenchyme, which leads to suppression of both osteoprogenitor proliferation and expression of osteoblastic markers. The complete restoration of calvarial bone formation in simultaneous inactivation of Sufu and Gli2 suggests that the Gli2 activator primarily contributes to maximal activation of Hh activity in CNC-derived mesenchyme of the Sufu mutant. These findings therefore provide novel evidence that a relatively low level of Hh signaling activity is required for successful progression of osteogenesis beyond the initial condensation phase in intramembranous ossification.
Experimental procedures
Animals
All mice used in this study were raised in a standard specific pathogen-free mouse facility, and the animal experiments were approved by the Committee of Laboratory Animals, Hangzhou Normal University. Sufu-floxed mice containing loxP sites flanking exon 7 were generated as we described previously (38). Wnt1-Cre (64), Dermo1-Cre (65), R26R-LacZ (66), and Gli2 (67) mice were purchased from the Jackson Laboratory, and maintained on the C57BL/6 background. The morning of vaginal plug appearance was determined as embryonic day 0.
Scanning electron microscopy analysis
Embryos were fixed overnight with 1.25% glutaraldehyde in PBS. Samples were dehydrated through a series of increasing ethanol concentration from 30 to 100%. After a 15-min treatment with isoamyl acetate, samples were critically point-dried using CO2 for 2 h. The dried samples were mounted on conductive paper and sputter-coated with gold. Images were recorded with a scanning electron microscope (Hitachi S-3000N) with a 15-kV accelerating voltage.
Skeletal preparation
Mice were fixed in 95% ethanol for 2 days after the removal of skin and viscera. They were incubated in Alcian blue staining solution (0.03% Alcian blue in 80% ethanol and 20% glacial acetic acid) for 2–3 days at room temperature. Skeletons were rehydrated and cleared in 1% KOH overnight. Samples were stained with Alizarin red solution (0.03% Alizarin red in 1% KOH) for 1–2 days followed by further clearing with 1% KOH in 20% glycerol. The stained skeletons were finally stored in 100% glycerol.
Histological analysis and von Kossa staining
Samples were fixed in 4% paraformaldehyde (PFA) overnight and embedded in paraffin. Tissue paraffin sections were cut at 7 μm for histological analysis and von Kossa staining. H&E staining was performed using a standard protocol. For von Kossa staining, the sections were flooded with 5% silver nitrate and exposed under a UV lamp for 20 min. The staining was stopped with 2% sodium thiosulfate. The stained sections were counterstained with 1% neutral red.
Immunofluorescence and in situ hybridization
Immunofluorescence analysis was carried out according to the standard protocol for paraffin section samples (68). Embryos were fixed in 4% PFA for 30 min, embedded in paraffin, and sectioned at 7 μm. Antigen epitopes were unmasked by heat-induced epitope retrieval. The sections were incubated with primary antibodies at 4 °C overnight. Secondary antibodies conjugated with Alexa Fluor 488 or 594 (1:1000; Invitrogen) were applied for 30 min to detect the primary antibody. Antibody against Sufu was from LifeSpan BioSciences (LS-C482700; 1:150). Antibodies against Hes1 (ab712559; 1:100) and Osterix (ab22552; 1:200) were from Abcam. Antibody against Runx2 (1:300) was from MBL International (D130-3). The protocol for immunofluorescence analysis with cryostat sections was described previously (68). Images were acquired using a Nikon 80i fluorescence microscope.
In situ hybridizations using whole-mount and section of embryonic samples were performed as described previously (38, 69) using digoxigenin-labeled RNA probes.
X-gal staining
Whole-mount and section X-gal staining were performed according to the standard protocols (38, 70). Embryos were incubated in fixing solution (4% PFA with 5 mm EGTA and 2 mm MgCl2 in PBS) for 1 h at room temperature. Fixed embryos were rinsed three times in washing buffer (0.014% Nonidet P-40, 0.01% sodium deoxycholate, and 2 mm MgCl2 in PBS) and stained 2–4 h at 37 °C in the dark using standard staining solution (5 mm potassium ferricyanide, 5 mm potassium ferrocyanide, 2 mm Tris (pH 7.3), 0.1% X-gal in washing buffer). The stained samples were rinsed twice in PBS and post-fixed in 4% PFA. The X-gal staining procedures for cryostat section samples were similar to that for whole-mount samples. The X-gal–stained sections were counterstained with Nuclear Fast Red.
Cell proliferation and TUNEL assays
The cell proliferation rate was evaluated by BrdU labeling and immunofluorescence staining using antibody against cyclin D1 (1:200; Abcam). BrdU labeling was performed by intraperitoneal injection of a pregnant mouse with BrdU solution (3 mg/100 g body weight) from a BrdU labeling and detection kit (Roche Applied Science) 30 min prior to harvesting embryos. The embryos were fixed in 10% neutral buffered formalin at 4 °C overnight and embedded in paraffin. Detection of BrdU-labeled cells was performed with an immunohistochemical staining method according to the manufacturer's instructions. The cell proliferation rate was calculated by dividing the positively stained cells by the total number of cells within the zone of presumptive calvarial bone primordia. TUNEL analysis was performed on 5-μm paraffin sections using the In Situ Cell Death Detection kit (Roche Applied Science) by following the manufacturer's protocol.
Real-time quantitative PCR analysis
Supraorbital mesenchyme that forms the presumptive frontal bone primordia was dissected out under a stereomicroscope and transferred to RNA stabilization reagent (Qiagen). RNA was extracted with an Ambion® RNAqueous®-4PCR kit and reverse-transcribed with an iScriptTM cDNA synthesis kit (Bio-Rad). qRT-PCR was performed using the CFX96 real-time system (Bio-Rad) with SsoFastTM EvaGreen Supermix (Bio-Rad). The relative expression level of each target gene was calculated based on a standard curve of cycle thresholds, and 18S rRNA expression was used as an internal control for normalization. qRT-PCR analysis was performed in triplicate for each set of samples.
Western blot analysis
Western blot analysis was performed according to the standard protocol (70). The primary antibodies against Sufu (1:1000; LS-C482700, LifeSpan BioSciences), Gli2 (1:2500; AF3635, R&D Systems), and Gli3 (1:1000; AF3690, R&D Systems) were used for immunoblotting to detect their expression in calvarial bone primordia. Antibody against β-actin (1:1000; catalog no. 3700, Cell Signaling Technology) was used as an internal reference. Relative quantification of protein expression was analyzed with Image-Pro Plus software (version 6.0) based on the integrated optical density of the blotting bands.
Statistical analysis
Student's t test was used to compare the differentials between data sets. The threshold for statistical significance was p < 0.05.
Author contributions
J. L. and Zunyi Zhang designed the research. X. Z. collected animal materials. J. L., Y. C., J. X., Q. W., X. Y., Y. L., and X. Z. performed the experiments. J. L. and Zunyi Zhang analyzed the data. M. Q. and Ze Zhang helped perform the analysis with constructive discussions. J. L. and Zunyi Zhang wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank all members of the Zhang laboratory (Institute of Life Sciences, Hangzhou Normal University) for suggestions during the generation of these data.
This work was supported by National Natural Science Foundation of China Grants 81570941, 31371471, and 31201089 and Science and Technology Plan Program of Zhejiang Province Grant 2017C37163. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S5.
- CNC
- cranial neural crest
- E
- embryonic day
- qRT-PCR
- quantitative RT-PCR
- BMP
- bone morphogenetic protein
- PFA
- paraformaldehyde.
References
- 1. Jiang X., Iseki S., Maxson R. E., Sucov H. M., and Morriss-Kay G. M. (2002) Tissue origins and interactions in the mammalian skull vault. Dev. Biol. 241, 106–116 [DOI] [PubMed] [Google Scholar]
- 2. Gross J. B., and Hanken J. (2008) Review of fate-mapping studies of osteogenic cranial neural crest in vertebrates. Dev. Biol. 317, 389–400 [DOI] [PubMed] [Google Scholar]
- 3. Yoshida T., Vivatbutsiri P., Morriss-Kay G., Saga Y., and Iseki S. (2008) Cell lineage in mammalian craniofacial mesenchyme. Mech. Dev. 125, 797–808 [DOI] [PubMed] [Google Scholar]
- 4. Chai Y., Jiang X., Ito Y., Bringas P. Jr., Han J., Rowitch D. H., Soriano P., McMahon A. P., and Sucov H. M. (2000) Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development 127, 1671–1679 [DOI] [PubMed] [Google Scholar]
- 5. Pan A., Chang L., Nguyen A., and James A. W. (2013) A review of hedgehog signaling in cranial bone development. Front. Physiol. 4, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huycke T. R., Eames B. F., and Kimmel C. B. (2012) Hedgehog-dependent proliferation drives modular growth during morphogenesis of a dermal bone. Development 139, 2371–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brugmann S. A., Allen N. C., James A. W., Mekonnen Z., Madan E., and Helms J. A. (2010) A primary cilia-dependent etiology for midline facial disorders. Hum. Mol. Genet. 19, 1577–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lenton K., James A. W., Manu A., Brugmann S. A., Birker D., Nelson E. R., Leucht P., Helms J. A., and Longaker M. T. (2011) Indian hedgehog positively regulates calvarial ossification and modulates bone morphogenetic protein signaling. Genesis 49, 784–796 [DOI] [PubMed] [Google Scholar]
- 9. Naruse I., Ueta E., Sumino Y., Ogawa M., and Ishikiriyama S. (2010) Birth defects caused by mutations in human GLI3 and mouse Gli3 genes. Congenit. Anom. 50, 1–7 [DOI] [PubMed] [Google Scholar]
- 10. Dennis J. F., Kurosaka H., Iulianella A., Pace J., Thomas N., Beckham S., Williams T., and Trainor P. A. (2012) Mutations in Hedgehog acyltransferase (Hhat) perturb Hedgehog signaling, resulting in severe acrania-holoprosencephaly-agnathia craniofacial defects. PLoS Genet. 8, e1002927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Abzhanov A., Rodda S. J., McMahon A. P., and Tabin C. J. (2007) Regulation of skeletogenic differentiation in cranial dermal bone. Development 134, 3133–3144 [DOI] [PubMed] [Google Scholar]
- 12. Jacob S., Wu C., Freeman T. A., Koyama E., and Kirschner R. E. (2007) Expression of Indian Hedgehog, BMP-4 and Noggin in craniosynostosis induced by fetal constraint. Ann. Plast. Surg. 58, 215–221 [DOI] [PubMed] [Google Scholar]
- 13. Nott R. L., Stelnicki E. J., Mack J. A., Ben Y., Mitchell R., and Mooney M. P. (2002) Changes in the protein expression of hedgehog and patched-1 in perisutural tissues induced by cranial distraction. Plast. Reconstr. Surg. 110, 523–532 [DOI] [PubMed] [Google Scholar]
- 14. Murakami S., Nifuji A., and Noda M. (1997) Expression of Indian hedgehog in osteoblasts and its posttranscriptional regulation by transforming growth factor-β. Endocrinology 138, 1972–1978 [DOI] [PubMed] [Google Scholar]
- 15. Hui C. C., and Angers S. (2011) Gli proteins in development and disease. Annu. Rev. Cell Dev. Biol. 27, 513–537 [DOI] [PubMed] [Google Scholar]
- 16. Dai P., Akimaru H., Tanaka Y., Maekawa T., Nakafuku M., and Ishii S. (1999) Sonic Hedgehog-induced activation of the Gli1 promoter is mediated by GLI3. J. Biol. Chem. 274, 8143–8152 [DOI] [PubMed] [Google Scholar]
- 17. Sasaki H., Nishizaki Y., Hui C., Nakafuku M., and Kondoh H. (1999) Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development 126, 3915–3924 [DOI] [PubMed] [Google Scholar]
- 18. Bai C. B., Auerbach W., Lee J. S., Stephen D., and Joyner A. L. (2002) Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development 129, 4753–4761 [DOI] [PubMed] [Google Scholar]
- 19. Park H. L., Bai C., Platt K. A., Matise M. P., Beeghly A., Hui C. C., Nakashima M., and Joyner A. L. (2000) Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development 127, 1593–1605 [DOI] [PubMed] [Google Scholar]
- 20. Hojo H., Ohba S., Yano F., Saito T., Ikeda T., Nakajima K., Komiyama Y., Nakagata N., Suzuki K., Takato T., Kawaguchi H., and Chung U. I. (2012) Gli1 protein participates in Hedgehog-mediated specification of osteoblast lineage during endochondral ossification. J. Biol. Chem. 287, 17860–17869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hojo H., Ohba S., Taniguchi K., Shirai M., Yano F., Saito T., Ikeda T., Nakajima K., Komiyama Y., Nakagata N., Suzuki K., Mishina Y., Yamada M., Konno T., Takato T., et al. (2013) Hedgehog-Gli activators direct osteo-chondrogenic function of bone morphogenetic protein toward osteogenesis in the perichondrium. J. Biol. Chem. 288, 9924–9932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Veistinen L., Takatalo M., Tanimoto Y., Kesper D. A., Vortkamp A., and Rice D. P. (2012) Loss-of-function of Gli3 in mice causes abnormal frontal bone morphology and premature synostosis of the interfrontal suture. Front. Physiol. 3, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rice D. P., Connor E. C., Veltmaat J. M., Lana-Elola E., Veistinen L., Tanimoto Y., Bellusci S., and Rice R. (2010) Gli3Xt-J/Xt-J mice exhibit lambdoid suture craniosynostosis which results from altered osteoprogenitor proliferation and differentiation. Hum. Mol. Genet. 19, 3457–3467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tanimoto Y., Veistinen L., Alakurtti K., Takatalo M., and Rice D. P. (2012) Prevention of premature fusion of calvarial suture in GLI-Kruppel family member 3 (Gli3)-deficient mice by removing one allele of Runt-related transcription factor 2 (Runx2). J. Biol. Chem. 287, 21429–21438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shimoyama A., Wada M., Ikeda F., Hata K., Matsubara T., Nifuji A., Noda M., Amano K., Yamaguchi A., Nishimura R., and Yoneda T. (2007) Ihh/Gli2 signaling promotes osteoblast differentiation by regulating Runx2 expression and function. Mol. Biol. Cell 18, 2411–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Amano K., Densmore M., Nishimura R., and Lanske B. (2014) Indian hedgehog signaling regulates transcription and expression of collagen type X via Runx2/Smads interactions. J. Biol. Chem. 289, 24898–24910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barnfield P. C., Zhang X., Thanabalasingham V., Yoshida M., and Hui C. C. (2005) Negative regulation of Gli1 and Gli2 activator function by Suppressor of fused through multiple mechanisms. Differentiation 73, 397–405 [DOI] [PubMed] [Google Scholar]
- 28. Lin C., Yao E., Wang K., Nozawa Y., Shimizu H., Johnson J. R., Chen J. N., Krogan N. J., and Chuang P. T. (2014) Regulation of Sufu activity by p66β and Mycbp provides new insight into vertebrate Hedgehog signaling. Genes Dev. 28, 2547–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lin C., Chen M. H., Yao E., Song H., Gacayan R., Hui C. C., and Chuang P. T. (2014) Differential regulation of Gli proteins by Sufu in the lung affects PDGF signaling and myofibroblast development. Dev. Biol. 392, 324–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jia J., Kolterud A., Zeng H., Hoover A., Teglund S., Toftgård R., and Liu A. (2009) Suppressor of Fused inhibits mammalian Hedgehog signaling in the absence of cilia. Dev. Biol. 330, 452–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kise Y., Morinaka A., Teglund S., and Miki H. (2009) Sufu recruits GSK3β for efficient processing of Gli3. Biochem. Biophys. Res. Commun. 387, 569–574 [DOI] [PubMed] [Google Scholar]
- 32. Zhang Y., Fu L., Qi X., Zhang Z., Xia Y., Jia J., Jiang J., Zhao Y., and Wu G. (2013) Structural insight into the mutual recognition and regulation between Suppressor of Fused and Gli/Ci. Nat. Commun. 4, 2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Humke E. W., Dorn K. V., Milenkovic L., Scott M. P., and Rohatgi R. (2010) The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes Dev. 24, 670–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tukachinsky H., Lopez L. V., and Salic A. (2010) A mechanism for vertebrate Hedgehog signaling: recruitment to cilia and dissociation of SuFu-Gli protein complexes. J. Cell Biol. 191, 415–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Svärd J., Heby-Henricson K., Persson-Lek M., Rozell B., Lauth M., Bergström A., Ericson J., Toftgård R., and Teglund S. (2006) Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev. Cell 10, 187–197 [DOI] [PubMed] [Google Scholar]
- 36. Cooper A. F., Yu K. P., Brueckner M., Brailey L. L., Johnson L., McGrath J. M., and Bale A. E. (2005) Cardiac and CNS defects in a mouse with targeted disruption of suppressor of fused. Development 132, 4407–4417 [DOI] [PubMed] [Google Scholar]
- 37. Zhulyn O., Li D., Deimling S., Vakili N. A., Mo R., Puviindran V., Chen M. H., Chuang P. T., Hopyan S., and Hui C. C. (2014) A switch from low to high Shh activity regulates establishment of limb progenitors and signaling centers. Dev. Cell 29, 241–249 [DOI] [PubMed] [Google Scholar]
- 38. Li J., Wang Q., Cui Y., Yang X., Li Y., Zhang X., Qiu M., Zhang Z., and Zhang Z. (2015) Suppressor of Fused is required for determining digit number and identity via Gli3/Fgfs/Gremlin. PLoS One 10, e0128006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kim H. J., Rice D. P., Kettunen P. J., and Thesleff I. (1998) FGF-, BMP- and Shh-mediated signalling pathways in the regulation of cranial suture morphogenesis and calvarial bone development. Development 125, 1241–1251 [DOI] [PubMed] [Google Scholar]
- 40. Ducy P., Zhang R., Geoffroy V., Ridall A. L., and Karsenty G. (1997) Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89, 747–754 [DOI] [PubMed] [Google Scholar]
- 41. Nakashima K., Zhou X., Kunkel G., Zhang Z., Deng J. M., Behringer R. R., and de Crombrugghe B. (2002) The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 108, 17–29 [DOI] [PubMed] [Google Scholar]
- 42. Chung I. H., Han J., Iwata J., and Chai Y. (2010) Msx1 and Dlx5 function synergistically to regulate frontal bone development. Genesis 48, 645–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Roybal P. G., Wu N. L., Sun J., Ting M. C., Schafer C. A., and Maxson R. E. (2010) Inactivation of Msx1 and Msx2 in neural crest reveals an unexpected role in suppressing heterotopic bone formation in the head. Dev. Biol. 343, 28–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Han J., Ishii M., Bringas P. Jr, Maas R. L., Maxson R. E. Jr, and Chai Y. (2007) Concerted action of Msx1 and Msx2 in regulating cranial neural crest cell differentiation during frontal bone development. Mech. Dev. 124, 729–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Maxson R., and Ishii M. (2008) The Bmp pathway in skull vault development. Front. Oral Biol. 12, 197–208 [DOI] [PubMed] [Google Scholar]
- 46. Jeong J., Mao J., Tenzen T., Kottmann A. H., and McMahon A. P. (2004) Hedgehog signaling in the neural crest cells regulates the patterning and growth of facial primordia. Genes Dev. 18, 937–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McGlinn E., van Bueren K. L., Fiorenza S., Mo R., Poh A. M., Forrest A., Soares M. B., Bonaldo Mde F., Grimmond S., Hui C. C., Wainwright B., and Wicking C. (2005) Pax9 and Jagged1 act downstream of Gli3 in vertebrate limb development. Mech. Dev. 122, 1218–1233 [DOI] [PubMed] [Google Scholar]
- 48. Panman L., Galli A., Lagarde N., Michos O., Soete G., Zuniga A., and Zeller R. (2006) Differential regulation of gene expression in the digit forming area of the mouse limb bud by SHH and gremlin 1/FGF-mediated epithelial-mesenchymal signalling. Development 133, 3419–3428 [DOI] [PubMed] [Google Scholar]
- 49. Ma X., Drannik A., Jiang F., Peterson R., and Turnbull J. (2017) Crosstalk between Notch and Sonic hedgehog signaling in a mouse model of amyotrophic lateral sclerosis. Neuroreport 28, 141–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Xie G., Karaca G., Swiderska-Syn M., Michelotti G. A., Krüger L., Chen Y., Premont R. T., Choi S. S., and Diehl A. M. (2013) Cross-talk between Notch and Hedgehog regulates hepatic stellate cell fate in mice. Hepatology 58, 1801–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ishii M., Merrill A. E., Chan Y. S., Gitelman I., Rice D. P., Sucov H. M., and Maxson R. E. Jr. (2003) Msx2 and Twist cooperatively control the development of the neural crest-derived skeletogenic mesenchyme of the murine skull vault. Development 130, 6131–6142 [DOI] [PubMed] [Google Scholar]
- 52. Lee M. H., Kwon T. G., Park H. S., Wozney J. M., and Ryoo H. M. (2003) BMP-2-induced Osterix expression is mediated by Dlx5 but is independent of Runx2. Biochem. Biophys. Res. Commun. 309, 689–694 [DOI] [PubMed] [Google Scholar]
- 53. Hilton M. J., Tu X., Wu X., Bai S., Zhao H., Kobayashi T., Kronenberg H. M., Teitelbaum S. L., Ross F. P., Kopan R., and Long F. (2008) Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat. Med. 14, 306–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rodda S. J., and McMahon A. P. (2006) Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development 133, 3231–3244 [DOI] [PubMed] [Google Scholar]
- 55. Long F., Chung U. I., Ohba S., McMahon J., Kronenberg H. M., and McMahon A. P. (2004) Ihh signaling is directly required for the osteoblast lineage in the endochondral skeleton. Development 131, 1309–1318 [DOI] [PubMed] [Google Scholar]
- 56. Plaisant M., Fontaine C., Cousin W., Rochet N., Dani C., and Peraldi P. (2009) Activation of hedgehog signaling inhibits osteoblast differentiation of human mesenchymal stem cells. Stem Cells 27, 703–713 [DOI] [PubMed] [Google Scholar]
- 57. Takarada T., Nakazato R., Tsuchikane A., Fujikawa K., Iezaki T., Yoneda Y., and Hinoi E. (2016) Genetic analysis of Runx2 function during intramembranous ossification. Development 143, 211–218 [DOI] [PubMed] [Google Scholar]
- 58. Sasaki T., Ito Y., Bringas P. Jr, Chou S., Urata M. M., Slavkin H., and Chai Y. (2006) TGFβ-mediated FGF signaling is crucial for regulating cranial neural crest cell proliferation during frontal bone development. Development 133, 371–381 [DOI] [PubMed] [Google Scholar]
- 59. Sun J., Ishii M., Ting M. C., and Maxson R. (2013) Foxc1 controls the growth of the murine frontal bone rudiment by direct regulation of a Bmp response threshold of Msx2. Development 140, 1034–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Iwata J., Hosokawa R., Sanchez-Lara P. A., Urata M., Slavkin H., and Chai Y. (2010) Transforming growth factor-β regulates basal transcriptional regulatory machinery to control cell proliferation and differentiation in cranial neural crest-derived osteoprogenitor cells. J. Biol. Chem. 285, 4975–4982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Iseki S., Wilkie A. O., and Morriss-Kay G. M. (1999) Fgfr1 and Fgfr2 have distinct differentiation- and proliferation-related roles in the developing mouse skull vault. Development 126, 5611–5620 [DOI] [PubMed] [Google Scholar]
- 62. Wang C., Pan Y., and Wang B. (2010) Suppressor of fused and Spop regulate the stability, processing and function of Gli2 and Gli3 full-length activators but not their repressors. Development 137, 2001–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wen X., Lai C. K., Evangelista M., Hongo J. A., de Sauvage F. J., and Scales S. J. (2010) Kinetics of hedgehog-dependent full-length Gli3 accumulation in primary cilia and subsequent degradation. Mol. Cell Biol. 30, 1910–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Danielian P. S., Muccino D., Rowitch D. H., Michael S. K., and McMahon A. P. (1998) Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr. Biol. 8, 1323–1326 [DOI] [PubMed] [Google Scholar]
- 65. Yu K., Xu J., Liu Z., Sosic D., Shao J., Olson E. N., Towler D. A., and Ornitz D. M. (2003) Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development 130, 3063–3074 [DOI] [PubMed] [Google Scholar]
- 66. Soriano P. (1999) Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21, 70–71 [DOI] [PubMed] [Google Scholar]
- 67. Corrales J. D., Blaess S., Mahoney E. M., and Joyner A. L. (2006) The level of sonic hedgehog signaling regulates the complexity of cerebellar foliation. Development 133, 1811–1821 [DOI] [PubMed] [Google Scholar]
- 68. Zhu X. J., Liu Y., Dai Z. M., Zhang X., Yang X., Li Y., Qiu M., Fu J., Hsu W., Chen Y., and Zhang Z. (2014) BMP-FGF signaling axis mediates Wnt-induced epidermal stratification in developing mammalian skin. PLoS Genet. 10, e1004687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang Z., Song Y., Zhao X., Zhang X., Fermin C., and Chen Y. (2002) Rescue of cleft palate in Msx1-deficient mice by transgenic Bmp4 reveals a network of BMP and Shh signaling in the regulation of mammalian palatogenesis. Development 129, 4135–4146 [DOI] [PubMed] [Google Scholar]
- 70. Zhu X., Zhao P., Liu Y., Zhang X., Fu J., Ivy Yu H. M., Qiu M., Chen Y., Hsu W., and Zhang Z. (2013) Intra-epithelial requirement of canonical Wnt signaling for tooth morphogenesis. J. Biol. Chem. 288, 12080–12089 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.