ABSTRACT
Localization of mRNAs can involve multiple steps, each with its own cis-acting localization signals and transport factors. How is the transition between different steps orchestrated? We show that the initial step in localization of Drosophila oskar mRNA − transport from nurse cells to the oocyte − relies on multiple cis-acting signals. Some of these are binding sites for the translational control factor Bruno, suggesting that Bruno plays an additional role in mRNA transport. Although transport of oskar mRNA is essential and robust, the localization activity of individual transport signals is weak. Notably, increasing the strength of individual transport signals, or adding a strong transport signal, disrupts the later stages of oskar mRNA localization. We propose that the oskar transport signals are weak by necessity; their weakness facilitates transfer of the oskar mRNA from the oocyte transport machinery to the machinery for posterior localization.
KEY WORDS: mRNA localization, mRNA transport, Oskar, Bruno, TLS, Oogenesis
Summary: Multiple cis-acting signals direct the first step in localization of oskar mRNA. These signals are collectively strong but individually weak, a property essential for later steps of localization.
INTRODUCTION
Analysis of early stages of animal development highlighted the phenomenon of mRNA localization. Proteins responsible for patterning certain insect and amphibian embryos are distributed asymmetrically within oocytes, and the patterns of protein distribution can be traced to the prior localization of the mRNAs that encode them (St Johnston, 1995). More recently, mRNA localization was shown to be surprisingly widespread. In Drosophila melanogaster embryos the majority of mRNAs display some degree of localization, with a wide variety of patterns (Lécuyer et al., 2007). Similarly, many vertebrate mRNAs exhibit subcellular localization (Holt and Bullock, 2009). How the diverse patterns of mRNA localization are programmed remains uncertain.
Localization of mRNAs typically relies on cis-acting signals, which serve to associate the mRNAs with factors involved in directed transport and anchoring (Lipshitz and Smibert, 2000). Thus far, the complexity of the localization program correlates with the complexity of the localization signal. For more-complex examples, such as the Drosophila bicoid (bcd) and oskar (osk) mRNAs, localization involves multiple steps and the mRNAs are delivered to mRNA-specific destinations (St Johnston et al., 1989; Kim-Ha et al., 1991; Ephrussi et al., 1991). Signals that localize these mRNAs are elaborate, with different elements mediating different steps (Macdonald, 1990; Macdonald et al., 1993; Macdonald and Kerr, 1997; Ferrandon et al., 1997; Kim-Ha et al., 1993; Ghosh et al., 2012).
Some other types of mRNA localization, such as apical localization in early Drosophila embryos (Davis and Ish-Horowicz, 1991) or localization to the distal regions of leading lamellae and filopodia of cultured chicken embryonic fibroblasts (Lawrence and Singer, 1986), entail a single step and are thus comparatively simple. Likewise, the signals directing such patterns of localization tend to be simple, comprising a single discrete element that may bind a single localization factor (Ross et al., 1997; Dienstbier et al., 2009). Notably, the simple localization signals are versatile: they can be used in different cell types and for multiple mRNAs (Bullock and Ish-Horowicz, 2001; Snee et al., 2005), and they can be incorporated into more-complex localization signals (Macdonald et al., 1993). The transport and localization signal (TLS) element from the Drosophila fs(1)K10 mRNA is a paradigm for these features (Serano and Cohen, 1995).
Synthesis of the fs(1)K10 mRNA occurs in the nurse cells of the ovary, with the mRNA then efficiently transported to the oocyte and enriched at the anterior margin. The TLS resides in the fs(1)K10 mRNA 3′UTR and consists of an AU-rich stem-loop (Serano and Cohen, 1995) that adopts an unusual helical structure (Bullock et al., 2010). Egalitarian (Egl) protein binds the TLS to mediate association with dynein for transport along microtubules (Dienstbier et al., 2009). Deletion of the TLS eliminates localization of fs(1)K10 mRNA (Serano and Cohen, 1995), while addition of a minimal TLS (44 nucleotides) to other mRNAs robustly confers the same program of oocyte transport and anterior localization (Serano and Cohen, 1995; Cohen et al., 2005; Bullock and Ish-Horowicz, 2001). The TLS is also active in an ectopic setting, the early Drosophila embryo. Notably, RNAs bearing the TLS move to the apical region of early embryos, dependent on the same proteins needed for transport to the oocyte in the ovary (Bullock and Ish-Horowicz, 2001).
Transport mediated by the TLS/Egl transport system can be useful in the context of more-complex localization programs. The bcd and osk mRNAs must be transported to the oocyte, and at least the bcd mRNA appears to use a version of this system. Within the bcd localization signal a stem-loop structure mediates transport to the oocyte, and is also sufficient to direct apical localization when injected into an early embryo. Both bcd localization programs − the natural process in the ovary and ectopic transport in the embryo − are sensitive to the same point mutation in the signal (Macdonald et al., 1993; Bullock and Ish-Horowicz, 2001).
A more-nuanced situation applies to the osk mRNA. Just as for mRNAs with the TLS, the osk mRNA is very strongly enriched in the oocyte. This property relies on a localization element within the 3′ UTR (Kim-Ha et al., 1993; Kim et al., 2014). Sequences contributing to the function of this element have been more-precisely mapped to part of a stem-loop structure which resembles the TLS in having an AU-rich stem. Mutation of the distal part of the stem disrupts transport of a reporter mRNA and compensatory changes restore activity, defining the oocyte entry signal (OES) (Jambor et al., 2014). Despite some structural similarity between the TLS and the OES, the OES is depauperate by comparison: it is not active in isolation and directs transport of a reporter mRNA only in the context of larger regions of the osk 3′UTR (Bullock and Ish-Horowicz, 2001; Jambor et al., 2014). Furthermore, although association of Egl and osk mRNA was detected by coimmunoprecipitation, the degree of association was less than for other localized mRNAs tested (Sanghavi et al., 2016).
Here we describe experiments on localization of osk mRNA that provide insights into the mechanisms and logic of mRNA localization. First, we show that regulatory elements used to control translation of osk mRNA also mediate mRNA transport from the nurse cells to the oocyte. By extension, we suggest that diversity in patterns of mRNA localization can be achieved by use of binding factors with other primary functions, obviating the need for a large class of previously unrecognized factors with dedicated roles in mRNA localization. Second, we demonstrate that multiple signals contribute to oocyte transport of osk mRNA. Although the use of multiple signals for mRNA localization has precedents (Kim-Ha et al., 1993; Macdonald and Kerr, 1997; Gautreau et al., 1997; Arn et al., 2003), we now show how this feature can be critical for the overall localization pathway. Notably, the individual signals most important for transport are individually weak in comparison to the action of the TLS, even though their collective activity is potent. We provide evidence that the use of weak oocyte transport signals allows the subsequent steps in osk mRNA localization, in which the mRNA is positioned at the posterior pole of the oocyte. We argue that the use of weak RNA localization signals provides one solution to a complication of complex localization programs: how to transition between different steps that rely on different mechanisms. Tight and persistent association of factors for an early step could interfere with subsequent steps, while weaker interactions would enable remodeling of localization complexes.
RESULTS
The AB or C regions of the osk 3′ UTR each confer oocyte enrichment on reporter mRNAs
Translational regulation of osk mRNA relies on Bruno (Bru), which binds to multiple sites in the osk 3′ UTR. These sites are clustered in the AB and C regions, positioned near the 5′ and 3′ ends of the osk 3′ UTR, respectively (Fig. 1A) (Kim-Ha et al., 1995; Reveal et al., 2010, 2011). GFP reporter mRNAs bearing either AB or C regions (Fig. 1A) are translationally repressed during oogenesis (Reveal et al., 2010). Notably, the presence of the osk AB or C regions also affected the distribution of these mRNAs. While the GFP mRNA was present at similar levels in the oocyte and nurse cells, the GFP-AB and GFP-C mRNAs were enriched substantially in the oocyte (Fig. 1B,D). Thus, the AB and C regions provided RNA localization activity similar to that of the TLS, although not as efficient. However, neither region is predicted to adopt a stem-loop structure, a characteristic of TLS and TLS-like signals (Serano and Cohen, 1995; Bullock et al., 2003, 2010). We conclude that the AB and C regions contain transport signals that are distinct in mechanism from the TLS paradigm.
Fig. 1.
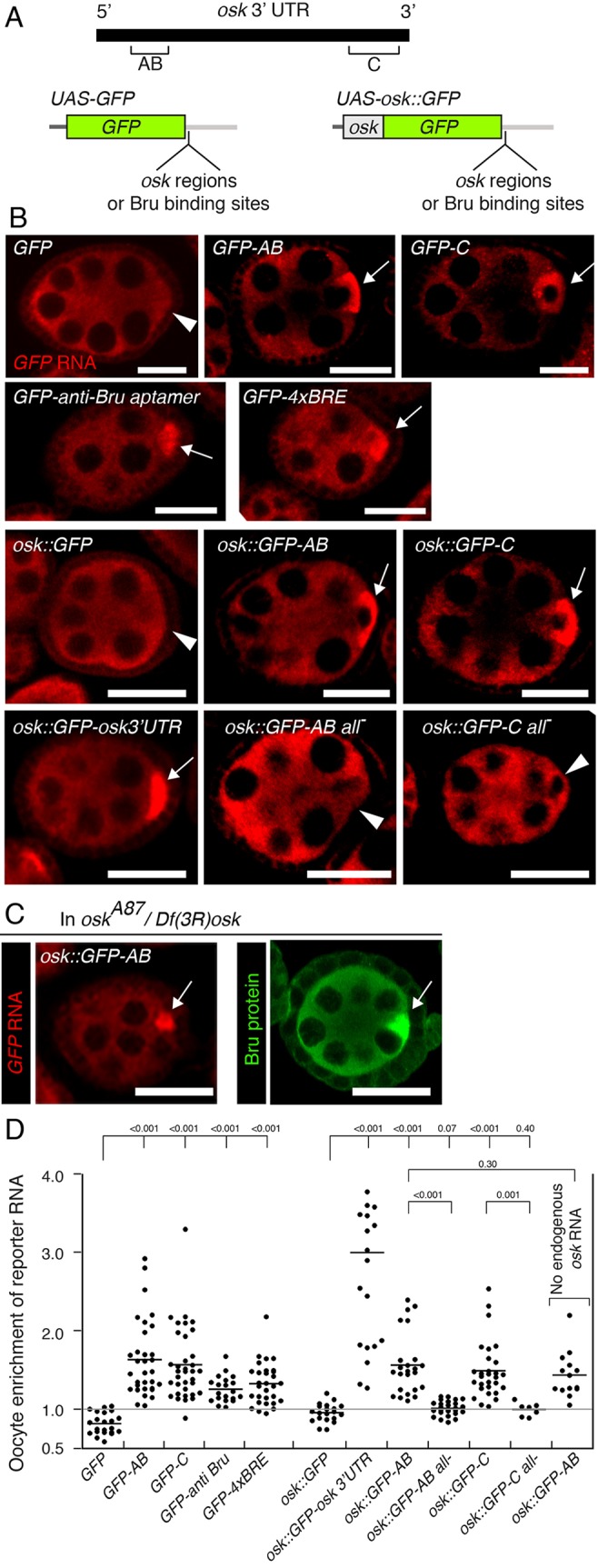
Bru-binding sites confer oocyte transport on GFP reporter mRNAs. (A) The diagram at top shows the osk mRNA 3′ UTR, with the two regions containing Bru-binding sites (AB and C) indicated. Below are two types of GFP transgene tested for localization, one with a 5′ part of the osk mRNA. The segments added to the transgenes (additions) are as indicated for the image panels below. (B) In situ hybridization to detect GFP mRNAs. For each panel the type of transgene and the segment added (if any) are indicated. The designation ‘all−’ indicates that all identified Bru-binding sites in the segment of osk RNA being tested have been mutated. For each egg chamber the position of the oocyte is indicated with an arrowhead (if no enrichment of the mRNA) or an arrow (if the mRNA is enriched). All egg chambers in B and C are previtellogenic stages. (C) GFP reporter mRNA and Bru distributions in ovaries lacking osk mRNA (oskA87/Df(3R)osk). The osk::GFP-AB mRNA remains concentrated in the oocyte, as does Bru, indicating that neither requires osk mRNA for enrichment in the oocyte. (D) Quantification of RNA distributions. Oocyte enrichment represents the ratio of signal in the oocyte cytoplasm to that in nurse cell cytoplasm. Each data point represents a single egg chamber, with the mean indicated by a horizontal line. P values from t-tests are indicated above the plot. Where multiple P values are shown above a single horizontal line, the comparisons are all relative to the sample indicated by the longest vertical line. The gray line at the ratio of 1 indicates the value for equal RNA levels in oocyte and nurse cells. RNAs with the GFP reporter are grouped at the left and those with the osk::GFP reporter at the right. The graph is truncated at the upper limit of 4.0 for oocyte enrichment ratio, although several data points for one sample (osk::GFP-osk 3′UTR) are above that value. Statistical analysis for that sample is based on all data points, including those omitted from the graph. All scale bars: 25 µm.
Bru-binding sites alone confer oocyte enrichment on reporter mRNAs
Because both AB and C regions conferred enrichment of the mRNAs in the oocyte, any feature shared by these regions would be a likely candidate for the transport signal. The AB and C regions were initially defined by their ability to bind Bru (Kim-Ha et al., 1995). To ask whether Bru-binding sites are required for oocyte enrichment, we made use of reporter transgenes that had originally been constructed to study translational regulation by Bru. The UAS-osk1-534::GFP reporter includes the first 534 bp of the osk transcription unit (most of the first exon and encoding the amino-terminal 173 amino acids) (Kim et al., 2015). Versions of this reporter transgene (referred to as UAS-osk::GFP for simplicity) include the osk AB or C regions, in wild-type form or with Bru-binding sites mutated (Fig. 1A). The UAS-osk::GFP reporter alone, without osk 3′ UTR sequences, produced an mRNA that was not enriched in the oocyte (Fig. 1B,D). Just as for the GFP reporter mRNA, addition of the osk 3′ UTR AB or C regions conferred substantial oocyte enrichment on the osk::GFP mRNA (Fig. 1B,D). Notably, mutation of the Bru sites largely eliminated oocyte enrichment of the mRNAs (Fig. 1B,D).
Although these results showed that Bru-binding sites are required for enrichment of the reporter mRNAs in the oocyte, other sequences in the AB and C regions might also be required. To ask whether Bru-binding sites are sufficient to confer oocyte enrichment, two additional transgenes were tested. Each has multiple copies of Bru-binding sites inserted in the reporter mRNA 3′ UTR. Importantly, each set of Bru-binding sites is synthetic, not derived from the osk mRNA and, thus, highly unlikely to fortuitously include the sites for any other proteins that bind the osk AB or C regions.
The GFP-anti-Bru aptamer mRNA includes multiple copies of the bru.4 and bru.18 aptamers. These anti-Bru aptamers were selected in vitro for their ability to bind recombinant Bru protein, and they confer translational repression on the GFP reporter mRNA (Reveal et al., 2011). The GFP-anti-Bru aptamer mRNA was enriched in the oocyte (Fig. 1B,D).
The GFP-4xBRE mRNA includes four copies of the UGUUUUAUAUGU motif, which corresponds to a BRE-type Bru-binding site. Inclusion of these Bru-binding sites confers Bru-dependent translational repression on the GFP-4xBRE mRNA (Reveal et al., 2011). These binding sites also resulted in enrichment of the mRNA in the oocyte (Fig. 1B,D).
We conclude that the presence of Bru-binding sites in an mRNA is sufficient for oocyte enrichment. For both the GFP-anti-Bru aptamer and GFP-4xBRE mRNAs, oocyte enrichment was less than conferred by the osk AB or C regions (Fig. 1D). However, less enrichment was expected if Bru were to mediate transport, because the synthetic Bru-binding sites are not as effective as the osk AB or C regions in conferring translational repression on the reporter mRNA, and the synthetic binding sites have lower affinity for Bru than the AB and C region RNAs (Reveal et al., 2011).
Bru-binding sites mediate oocyte mRNA enrichment independent of endogenous osk mRNA
A possible mechanism for Bru-binding site-dependent oocyte mRNA enrichment is hitchhiking, in which the reporter mRNAs would be transported to the oocyte via association with endogenous osk mRNA. Such a mechanism has been established for the localization of reporter mRNAs to the posterior of the oocyte later in oogenesis, with Polypyrimidine tract-binding protein (PTB, also known as Heph) mediating association of osk mRNA and the hitchhiking mRNAs (Hachet and Ephrussi, 2004; Besse et al., 2009). By analogy, Bru or another binding factor might bridge between the reporter and endogenous osk mRNAs to allow hitchhiking.
Two results argue against hitchhiking. First, concentration of Bru in the early-stage oocyte (Webster et al., 1997) persists in the absence of osk mRNA, showing that Bru does not require osk mRNA for its localization (Fig. 1C; Kanke et al., 2015). Second, we monitored the distribution of the osk::GFP-AB reporter mRNA in ovaries lacking endogenous osk mRNA. Even in the absence of osk mRNA, substantial oocyte enrichment of the osk::GFP-AB mRNA was detected (Fig. 1C,D). Thus, Bru-binding site-dependent RNA enrichment in the oocyte did not require hitchhiking on osk mRNA.
Degenerate cis-acting signals mediate transport of osk mRNA to the oocyte
We have shown that the AB and C regions of the osk mRNA 3′ UTR are each sufficient to direct transport into the oocyte. A different transport signal in the osk 3′ UTR was mapped initially by deletion analysis (Kim-Ha et al., 1993), with a recent study showing that a highly conserved part of that region is important for transport (Kim et al., 2014). Another recent report also addressed the function of this signal, using reporter mRNAs bearing parts of the osk 3′ UTR to implicate one domain of a stem-loop structure, SL2b (Jambor et al., 2014). Mutation of either stem strand in the distal portion of SL2b impairs oocyte transport of the GFP reporter mRNA, while mutation of both strands to restore base pairing restores transport. These mutations define the OES, which has some similarity to the TLS, being an AU-rich region of double-stranded RNA.
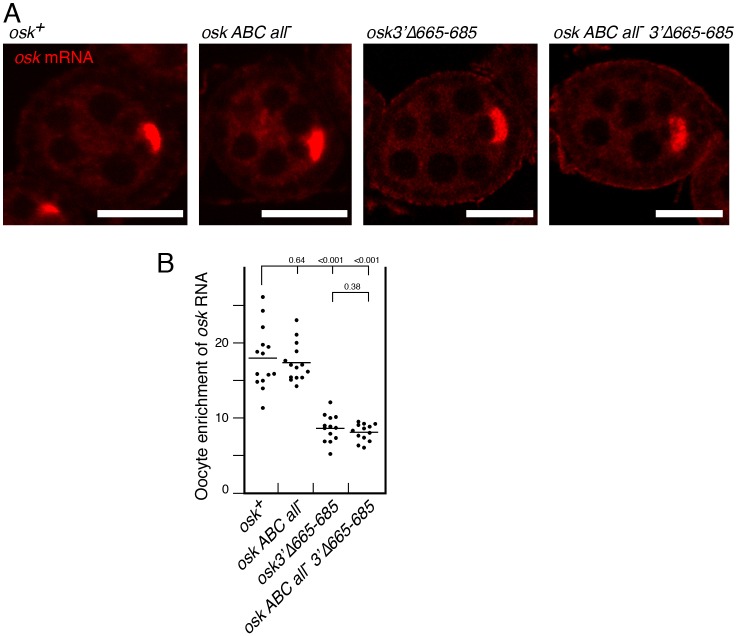
The experiments with the OES and the Bru-binding sites demonstrated that multiple elements in the osk 3′ UTR can direct transport of reporter mRNAs to the oocyte. These experiments did not, however, test whether either individual type of transport element is essential for oocyte transport of the osk mRNA (as opposed to a reporter mRNA containing only parts of the osk mRNA). To address this issue we mutated the different elements in osk transgenes, which were then tested in flies lacking endogenous osk mRNA to eliminate the possibility of hitchhiking. The osk ABC all− mRNA has all the identified Bru-binding sites in both AB and C regions mutated (as indicated by all−) (Reveal et al., 2010). These mutations abolished the ability of the AB and C regions to confer oocyte transport of the GFP-AB and GFP-C mRNAs (Fig. 1B,D) but had no discernible effect on transport of osk mRNA to the oocyte: the osk ABC all− mRNAs showed strong oocyte enrichment, similar to that of wild type osk mRNA (Fig. 2A,B). To inactivate the OES in the osk mRNA, we used a deletion mutant, osk3'Δ665-685, that lacks one strand of the distal stem of SL2b shown to be required for OES function. The osk3'Δ665-685 mutant was strongly enriched in the oocyte, although to a lesser extent than for wild-type osk RNA (Fig. 2A,B). Thus, mutating either Bru sites or the OES did not eliminate transport of osk mRNA to the oocyte, suggesting degeneracy in this process. To assess the consequences of eliminating both Bru-binding site- and OES-dependent transport, an osk transgene bearing both the Bru site mutations and the OES deletion (osk ABC all− 3'Δ665-685) was tested. Despite the loss of both types of oocyte transport signal, the mRNA was still strongly enriched in the oocyte (Fig. 2A,B). Thus, additional signals for oocyte transport of osk mRNA must exist.
Fig. 2.
Persistent oocyte transport of osk mRNA when both Bru-binding sites and the OES are mutated. (A) In situ hybridization to detect osk mRNAs. All transgenes were tested in an osk RNA null background, with ovaries from at least five flies used for imaging. All egg chambers are previtellogenic stages. Scale bars: 25 µm. (B) Quantification of RNA distributions as in Fig. 1. P values from t-tests are indicated above the plot. Where multiple P values are shown above a single horizontal line, the comparisons are all relative to the sample indicated by the longest vertical line. See Fig. S1 for transgene RNA levels.
Identification of sequences critical for oocyte transport of osk mRNA
To search for the predicted additional oocyte transport signal or signals, we used a collection of osk transgenes bearing deletions in the 3′ UTR. As a sensitive assay of reduced oocyte transport we monitored the effects of the deletions on osk noncoding RNA (ncRNA) activity. Although osk is best known for encoding a protein that is essential for embryonic development (Lehmann and Nüsslein-Volhard, 1986), the osk 3′ UTR has a separate and distinct noncoding function that was revealed by the discovery of osk RNA null mutants (Jenny et al., 2006; Kanke et al., 2015). In the absence of osk mRNA, oogenesis is arrested well before the stage at which Osk protein normally begins to accumulate, and no eggs are laid. Partial rescue of this phenotype allows oogenesis to progress further and restores some egg laying, with the rate of egg laying providing a measure of osk ncRNA activity (Jenny et al., 2006; Kanke et al., 2015). Because osk ncRNA function depends on transport of osk RNA to the oocyte (Jenny et al., 2006; Kanke et al., 2015), mutations that impair transport to the oocyte also impair osk ncRNA activity.
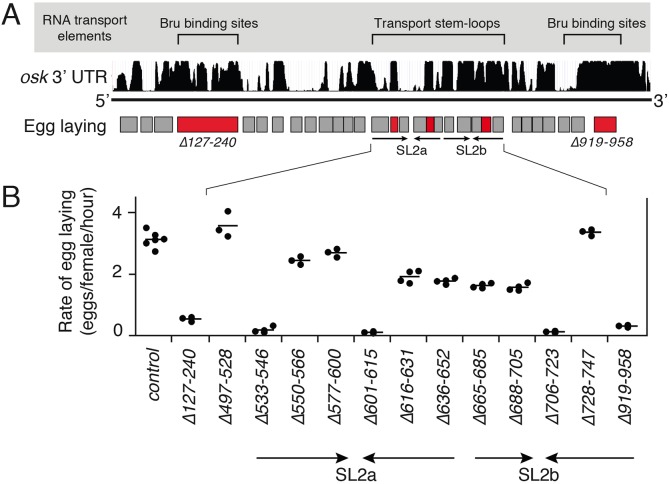
Among osk transgenes with 3′ UTR deletions, only five had dramatically reduced rates of egg laying (Fig. 3A) [the deletions do not cover the extreme 3′ end of the mRNA, which contains sequences not required for oocyte transport but critical for osk ncRNA activity (Kanke et al., 2015)]. Two of the deletions remove the Bru-binding sites, which are required for osk ncRNA function in a manner that is independent of their degenerate and nonessential contributions to osk mRNA transport (Kanke et al., 2015). Of the remaining three deletion mutants with very low egg laying, one lacks part of SL2b, while the others are missing parts of the predicted structure SL2a, which is adjacent to SL2b. All deletion mutants from the region encompassing both SL2a and SL2b (i.e. SL2) were examined in more detail by using quantitative egg laying assays (Fig. 3B). For the strongly affected mutants in SL2, egg laying rates were reduced to <10% of wild type. Deletions removing other parts of SL2b also caused a substantial, though less extreme, reduction in egg laying. Reductions in osk ncRNA activity and egg laying could be due to either weak oocyte transport or reduced levels of osk mRNA, but the mutants with egg-laying defects did not have substantially lower mRNA levels than unaffected mutants (Fig. S1).
Fig. 3.
Identification of osk mRNA 3′ UTR regions required for osk ncRNA activity. (A) Diagram of the osk mRNA 3′ UTR, showing a summary of all signals contributing to oocyte transport of the RNA (top; within gray area), sequence conservation among Drosophila species (middle; height of black bars indicates extent of sequence conservation) and deletions (rectangles) introduced into otherwise intact osk transgenes (bottom). Deletion mutants with dramatically reduced egg laying (an indicator of osk ncRNA activity) are in red. The arrows indicate the positions of SL2a and SL2b, as indicated in B. (B) Rates of egg laying for relevant deletion mutants, including all those in the SL2a and SL2b regions. Each transgene was tested as a single copy in the osk0/osk0 background (osk RNA null). Each data point represents a separate experiment (at least three for each genotype), with the mean indicated by a line. The control osk transgene has a small deletion in the 3′ UTR (Δ370-390) that causes no detectable change in osk mRNA properties or function. Data for the control transgene, as well as key deletion mutants, are reproduced in Figs 5 and 6 to facilitate comparisons. See Fig. S1 for transgene RNA levels.
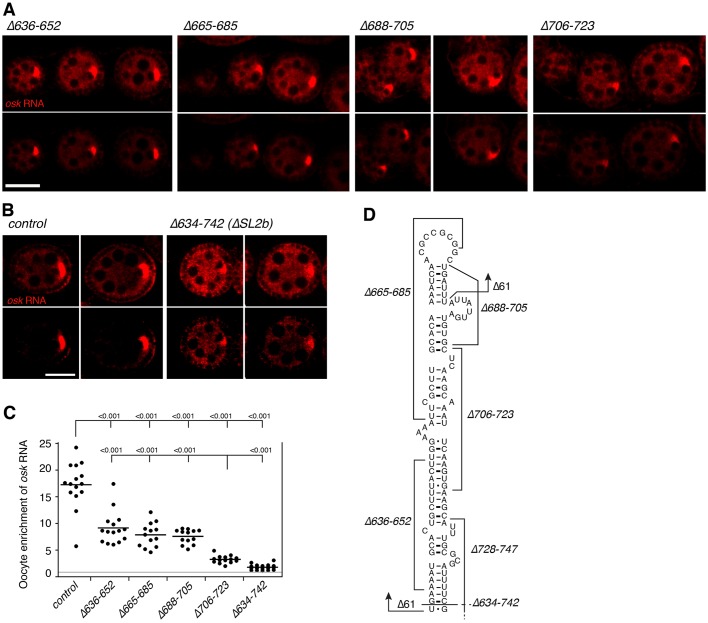
Mutants of SL2b with reduced egg laying were tested for effects on transport of osk mRNA to the oocyte, revealing a correlation between disruption of egg laying (osk ncRNA activity) and oocyte transport (Fig. 4A-C). The small deletion mutant that was most defective in egg laying, i.e. osk3′Δ706-723, was also most defective in oocyte transport. The SL2b mutants with intermediate levels of egg laying also had intermediate levels of oocyte transport. A likely explanation for why mutant osk3′ Δ706-723 is most defective is suggested by the positions of the deletions within SL2b (Fig. 4D). Mutation Δ706-723 alters a central part of the SL2b stem and disrupts both proximal and distal regions. By contrast, the other mutations alter either distal or proximal regions of the SL2b stem, but not both. Thus, multiple parts of the stem appear to contribute to oocyte transport and deletion of the entire SL2b could be even more damaging. This is, indeed, the case: a mutant lacking all of SL2b (osk3′Δ634-742) (Fig. 4D) retains only a very low level of oocyte transport (Fig. 4B,C) and is completely defective in egg laying (below).
Fig. 4.
Contributions of parts of stem-loop 2b to transport of osk mRNA to the oocyte. (A) Effects of small deletions in SL2b on osk mRNA transport to the oocyte. Each transgene was tested as a single copy in the osk RNA null background, with the transgene RNA detected by in situ hybridization. The upper and lower panels are identical, except that the signal intensity was increased in images shown in the upper panel in order to always display the entire egg chamber (this also resulted in saturation of signal intensities for some samples; lower panel provides a better display of the efficiency of RNA transport). See Fig. S1 for transgene RNA levels. All egg chambers in A and B are previtellogenic stages. (B) Effect of deleting the entire SL2b on osk mRNA transport to the oocyte. The panels are as in A. (C) Quantification of RNA distributions as in Fig. 1. The gray line at the ratio of 1 indicates the value for equal RNA levels in oocyte and nurse cells. P values from t-tests are indicated above the plot. Where multiple P values are shown above a single horizontal line, the comparisons are all relative to the sample indicated by the longest vertical line. The lower set of pairwise comparisons of means indicates that the RNA transport defect of mutant Δ706-723 is intermediate between those of mutants Δ636-652, Δ665-685 and Δ688-705 (all with stronger transport) and mutant Δ634-742 (with weaker transport). (D) Proposed structure of SL2b (Jambor et al., 2011) with the positions of deletions indicated. The Δ61 mutant is GFP-oskΔ61, which is largely defective in oocyte transport (Kim et al., 2014). All scale bars: 25 µm; all images in panel A are at the same magnification. Likewise, all images in panel B are at the same magnification.
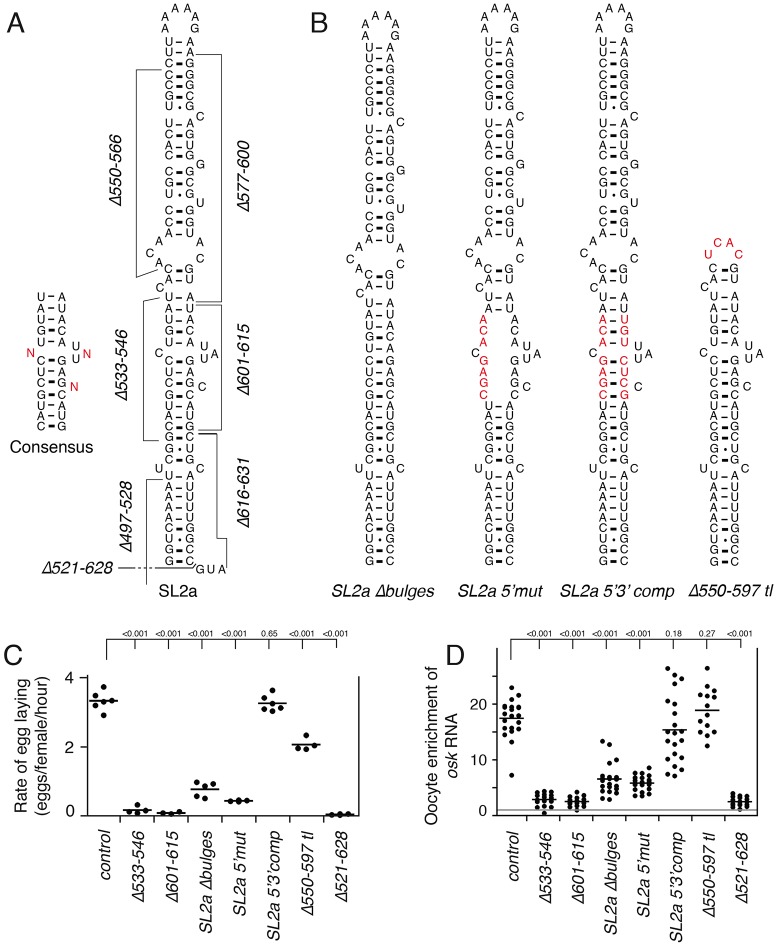
Sequences adjacent to SL2b are also predicted to form an extended stem-loop structure, SL2a (Fig. 5A), although only part of this region is conserved across multiple Drosophila species (Fig. 3A). Strikingly, the deletions that affected the highly conserved sequences in SL2a most strongly disrupted egg laying (Δ533-546 and Δ601-615; Figs 3B and 5C) and also dramatically reduced RNA transport to the oocyte (Fig. 5D).
Fig. 5.
Structure of stem-loop 2a and its role in transport of osk mRNA to the oocyte. (A) Predicted structure of the entire SL2a, with deletion mutants (from Fig. 3) indicated. The consensus structure for the highly conserved domain (left), was obtained from comparison of multiple Drosophila species. (B) Mutants used to demonstrate that the highly conserved region of SL2a does form a stem, in which the bulges are important but the identities of base pairs within the stem are not critical. Mutant bases are shown in red. For Δ550-597 tl, the deletion was replaced by UCAC, which forms a tetraloop to stabilize the adjacent stem region (Cheong et al., 1990). (C) Rates of egg laying for mutants affecting SL2a. Each transgene was tested as a single copy in the osk0/osk0 background (osk RNA null). Each data point represents a separate experiment (at least three for each genotype), with the mean indicated by a line. P values from t-tests are indicated above the plot. The comparisons are all relative to the sample indicated by the longest vertical line. The control osk transgene is as in Fig. 3. See Fig. S1 for transgene RNA levels. (D) Quantification of RNA distributions as in Fig. 1. P values from t-tests are indicated above the plot. The comparisons are all relative to the sample indicated by the longest vertical line. The gray line at the ratio of 1 indicates the value for equal RNA levels in oocyte and nurse cells.
Unlike for SL2b, there have been no studies to confirm the predicted structure of SL2a. To ask whether the critical part of the predicted stem does, indeed, form, we made additional mutants (Fig. 5B). They were tested for effects on egg laying and oocyte enrichment, with the two assays providing similar results (Fig. 5C,D). Altering the bases in one strand of this region (mutation SL2a 5′mut) strongly reduced both egg laying and oocyte transport (Fig. 5C,D), with defects largely rescued by the introduction of compensatory mutations into the other strand of the predicted stem (in mutant SL2a 5′3′comp) (Fig. 5C,D). These results provide strong evidence that the part of the stem required for osk ncRNA activity does form, although a particular sequence within that part of the stem is not essential. We also made a mutant in which all nucleotides predicted to be bulged out of the helix of the critical region were removed (SL2a Δbulges). Egg laying and oocyte transport were substantially reduced (Fig. 5C,D), further indicating that the structure of this region is important for function. Finally, because none of the initial set of deletion mutants had the loop of SL2a removed, we tested a mutant lacking the loop as well as the poorly conserved distal stem region (Δ550-597 tl), which retained strong osk ncRNA activity and oocyte transport (Fig. 5B-D). For mutants with reduced osk ncRNA activity and oocyte transport, these effects cannot be explained by lower mRNA levels (Fig. S1).
Neither osk 3′ UTR stem-loop transport signal is sufficient for transport
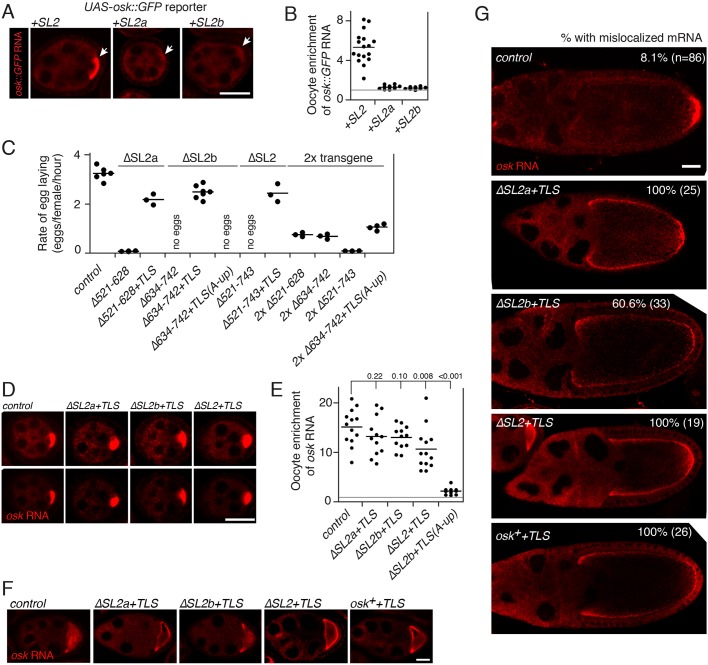
Deletion of either SL2a (in mutant Δ521-628) or SL2b (in mutant Δ634-742) dramatically reduces transport of osk mRNA to the oocyte (Figs 4B,C and 5C,D). Jambor et al. showed that SL2 conferred oocyte transport on a GFP reporter mRNA, but SL2b did not (Jambor et al., 2014). We repeated these experiments using a GFP reporter mRNA containing a 5′ part of the osk coding region and extended the analysis to include SL2a. The complete SL2 supported robust mRNA transport to the oocyte, but neither SL2a nor SL2b was sufficient for transport (Fig. 6A,B). Thus, neither of the osk stem-loop structures on their own have the robust transport activity of the TLS stem-loop transport signal.
Fig. 6.
A strong oocyte transport signal restores both oocyte transport and ncRNA activity of SL2 deletion mutants, but alters patterns of osk mRNA localization. (A) Oocyte transport activity of SL2 and its subdomains. The osk 3′ UTR parts shown above each panel were added to an osk::GFP reporter mRNA and tested for transport activity by in situ hybridization. SL2 is osk 3′ UTR sequences 521-743, SL2a is 521-628 and SL2b is 634-742. The position of the oocyte in each early-stage egg chamber is indicated with an arrow. (B) Quantification of RNA distributions as in Fig. 1. (C) Rates of egg laying for mutants affecting SL2a and SL2b, and rescue of the defects from addition of the TLS. Each transgene was tested as a single copy in the osk0/osk0 background (osk RNA null). In some cases (at right) two copies of the transgene were present; these are labeled ‘2x’. Each data point represents a separate experiment (at least three for each genotype), with the mean indicated by a line. The control osk transgene (panels C-G) is as in Fig. 3. See Fig. S1 for transgene RNA levels. (D) The TLS rescues transport of mutant osk mRNAs to the oocyte. Each transgene was tested as a single copy in the osk RNA null background, with the transgene RNA detected by in situ hybridization. The upper and lower panels are identical, except that the signal intensity was increased for the upper panels to display the entire egg chambers. (E) Quantification of RNA distributions as in Fig. 1. The gray line at the ratio of 1 indicates the value for equal RNA levels in oocyte and nurse cells. P values from t-tests are indicated above the plot. The comparisons are all relative to the sample indicated by the longest vertical line. (F) Enhanced cortical enrichment of RNAs as the oocyte begins to expand in stage 8 of oogenesis. In wild type the RNA is dispersed throughout the oocyte, with either no strong cortical enrichment, or enrichment just at the anterior (left side of the oocyte in these images). Cortical association, in which the RNA outlines the oocyte, due to the presence of the TLS is stronger than for the control osk mRNA in all cases, but is the least pronounced with ΔSL2b. (G) Persistent cortical enrichment and reduced posterior localization following addition of the TLS. Transgenes were present in two copies for all genotypes and RNAs detected by in situ hybridization. The fraction of late stage 9 and stage 10 oocytes with cortical osk mRNA not correctly localized to the posterior is indicated. To be scored as mislocalized, the in situ hybridization signal had to appear along most of the length of the oocyte at levels clearly above background (as seen in all genotypes except the control osk RNA). All scale bars: 25 µm. All images within a single lettered panel are at the same magnification. All egg chambers in A and D are previtellogenic stages.
A potent oocyte transport signal disrupts posterior localization of osk mRNA
Given the importance of efficient transport of osk mRNA to the oocyte for its ncRNA function, it is intriguing that the individual osk stem-loop transport signals are depauperate relative to the TLS. Among several possible explanations for the relative weakness of the osk mRNA transport signals, two seem most plausible.
First, the transport elements in SL2a and SL2b might play dual roles: one in transport and a separate, and additional, role in osk ncRNA function. If so, constraints on the sequence imposed by the ncRNA role could limit the effectiveness of transport. To ask whether SL2a and SL2b are required for osk ncRNA activity independent of their contributions to mRNA transport, we made osk transgenes in which these regions were deleted and the TLS was added. We also tested mutant Δ521-743 (ΔSL2), in which both SL2a and SL2b were deleted, as well as the companion mutant with the TLS added. In all cases the presence of the TLS dramatically improved egg laying relative to the respective stem-loop deletion (Fig. 6C). Consistent with the rescue of egg laying, oocyte transport of each SL2 deletion mutant was strongly rescued by the TLS: all previtellogenic egg chambers had a high concentration of the mRNA in the oocyte (Fig. 6D,E). Thus, it appears that both SL2a and SL2b contribute to osk ncRNA function primarily via mRNA transport, although we cannot rule out a minor contribution to osk ncRNA function that is independent of transport.
A second possible explanation for the relative weakness of oocyte transport directed by individual osk transport signals is suggested by the complex localization program of osk mRNA. After the initial transport from the nurse cells to the oocyte, osk mRNA concentrated at the anterior margin of the oocyte. This pattern is shared with the fs(1)K10 mRNA, as well as other mRNAs containing TLS-like transport signals. The osk mRNA differs in having additional steps in localization. First, osk mRNA is displaced from the anterior and appears in particles throughout the oocyte. As development proceeds, the particles are enriched at the posterior of the oocyte. Finally, osk mRNA becomes confined to the narrow crescent at the posterior (Fig. 6G; Kim-Ha et al., 1991; Ephrussi et al., 1991). Perhaps, the need to transfer osk mRNA between distinct localization machines − one for transport to the oocyte and another to deliver the mRNA to the posterior pole − places constraints on the interaction with the oocyte transport machine. While persistent association of certain other mRNAs with the transport machine is acceptable, the osk mRNA may need to be released to allow the later steps of localization.
Notably, replacement of part or all of SL2 with the TLS did alter patterns of osk mRNA localization. During the initial expansion of the oocyte, osk mRNAs bearing the TLS were noticeably enriched at cortical regions (Fig. 6F). This cortical localization persisted: by stage 10, when wild-type osk mRNA is largely restricted to the posterior of the oocyte, the TLS-containing mRNAs remained arrayed along the periphery of the oocyte with only modest posterior localization (Fig. 6G). This aberration is consistent with impaired transfer of the mRNA between the different localization machineries. Alternatively, disruption of these stages of localization could be due to loss of elements within SL2a and SL2b that help direct posterior localization. Two additional osk transgenes were made to distinguish between these interpretations.
First, we asked whether adding the TLS to an osk mRNA, while leaving SL2 intact, also interferes with osk mRNA localization. This was, indeed, the case, with the osk++TLS mRNA showing the same cortical enrichment and reduction of posterior localization (Fig. 6F,G).
Second, we asked whether weakening the TLS of osk3′Δ634-742+TLS, in which SL2b is deleted and the TLS added, alleviates the mRNA localization defects. TLS-dependent apical transport in early embryos is impaired by the A-up mutation, which alters nucleotides in the TLS stem (Bullock et al., 2010). Similarly, this mutation [osk3′Δ634-742+TLS(A-up)] reduced, but did not eliminate, transport of the modified osk mRNA to the oocyte (Fig. 7A, quantification in Fig. 6F). Egg laying of osk3′Δ634-742+TLS(A-up) (Fig. 6C) was dramatically reduced (no eggs with only one copy of the transgene), reinforcing the notion that the noncoding function of osk mRNA requires strong oocyte transport. To evaluate the later stages of localization for the osk3′Δ634-742+TLS(A-up) mRNA, two copies of the transgene were used, thereby increasing osk ncRNA activity and allowing a fraction of egg chambers to proceed through oogenesis. Notably, the cortical mislocalization of the mRNA containing the intact TLS was largely corrected by weakening the TLS (Fig. 7B). The posterior localization was still less than for wild-type osk mRNA, but this might be due to the weaker transport of the mRNA into the oocyte, which makes less mRNA available for localization.
Fig. 7.
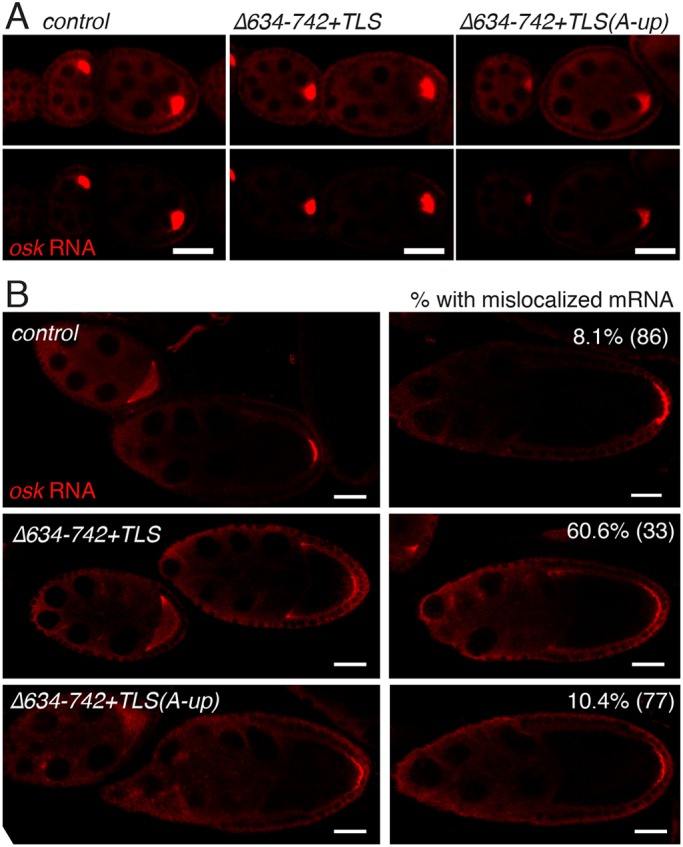
A weak TLS does not alter patterns of osk mRNA localization. (A) The A-up mutation reduces TLS-dependent oocyte transport (experiments as those described for Fig. 6D; quantification is shown in Fig. 6E). See Fig. S1 for transgene RNA levels. The control osk transgene is as described in Fig. 3. All egg chambers are previtellogenic stages. (B) The A-up mutation suppresses the RNA localization defects associated with presence of the TLS. Transgenes were present in two copies for all genotypes and RNAs detected by in situ hybridization. The fraction of late stage 9 and stage 10 oocytes with anterior or lateral cortical osk mRNA is indicated, and was scored as noted in Fig. 6. Egg chambers in the left column are stage 9, while those in the right column are stage 10A. All scale bars: 25 µm.
We conclude that strengthening oocyte transport of osk mRNA, by adding a strong transport signal or by replacing a weaker signal with a strong signal, enhances cortical enrichment and disrupts posterior localization.
DISCUSSION
Three main conclusions emerge from these studies. First, our results suggest that Bru, a translational regulator, plays an additional role in mRNA transport. The ability of a factor with a primary function − distinct from mRNA localization − to also direct localization of a bound mRNA could explain how the remarkable diversity of mRNA localization patterns can be achieved. Second, we show that the strong and essential transport of osk mRNA to the oocyte relies not on a strong cis-acting signal but, instead, on multiple weak signals. Third, we find that when osk mRNA is transported to the oocyte under control of a strong signal, then the subsequent steps in osk mRNA localization are disrupted, providing a rationale for why osk relies on multiple weaker forms of transport.
Evidence that Bru can act in RNA transport suggests a model for generating diverse patterns of mRNA localization
Bru was identified originally because of its role in translational repression (Kim-Ha et al., 1995). Subsequent studies also implicated Bru in translational activation (Reveal et al., 2010). Now we have found that the presence of Bru-binding sites in mRNAs confers a specific form of mRNA localization, i.e. transport to the oocyte during early stages of oogenesis. Most likely, these binding sites mediate RNA transport by binding of Bru, although an unidentified protein with the same binding specificity could be responsible. The distribution of Bru in the ovary fits well with the possible new role. At early stages Bru protein is highly concentrated in the oocyte, despite a more-uniform distribution of the mRNA (Webster et al., 1997). Bru must, therefore, be moved to this destination, a process that would allow colocalization of any associated RNAs. Despite this evidence for a novel role for Bru, it is important to emphasize that the contribution of the Bru-binding sites to RNA transport is modest relative to that of the transport stem-loops. Removing all identified Bru sites has little effect on oocyte enrichment of osk mRNA, while mutating one of the stem-loops has a measurable effect (Fig. 2). Thus, we focus here on the significance of these results in terms of strategies for achieving mRNA localization.
It is noteworthy that a large fraction of all mRNAs display some form of localization. Within the ovary ∼10% are concentrated in early-stage oocytes (Dubowy and Macdonald, 1998), a pattern similar to the localization conferred by Bru-binding sites. This high incidence of RNA transport to the oocyte could represent, at least in part, colocalization with proteins that function primarily in some other role but facilitate a form of mRNA localization as they are themselves moved to the oocyte. Recent studies have highlighted secondary roles in RNA binding for various proteins with other more-primary functions (Castello et al., 2012). We propose a related form of secondary function: RNA-binding proteins acting in regulation or metabolism can have the secondary function of RNA transport. When a regulatory protein is stably bound to an mRNA, that mRNA can be carried along with the protein if it is itself enriched at a particular subcellular location. This scenario does not address the question of how a specific distribution pattern is achieved, as it simply shifts the question to one of protein localization rather than RNA localization. However, the significant difference is that the instructions for localization reside in the protein, not the RNA. This paradigm could apply to the remarkable prevalence and diversity of mRNA localization found for embryonic mRNAs in Drosophila, of which ∼70% exhibit some form of localization (Lécuyer et al., 2007). For some, or even many, of these mRNAs localization could rely on association with proteins whose primary role is not mRNA transport but can, nevertheless, confer localization as a consequence of their own subcellular distributions. Because these proteins have enormous diversity in their distributions, associated RNAs could have comparable diversity in their localization patterns. Notably, the degree to which a particular mRNA is localized is often low (Dubowy and Macdonald, 1998; Lécuyer et al., 2007), implying lesser efficiency − which would be consistent with our model. If our model is correct, an extremely large and diverse set of RNA-binding sites serve, indirectly, as mRNA localization signals. This situation would stymie any effort to identify localization signals, based on the assumption that there is only a modest number of transport factors and cognate binding sites.
Multiple cis-acting signals contribute to transport of osk mRNA to the oocyte
The work presented here highlights the value of combining different types of assay to explore mechanisms of mRNA localization. Testing the ability of an RNA signal to confer localization on reporter mRNAs reveals what the signal can do. Testing the effects of removing the signal from its natural setting, the host RNA, reveals whether that function is essential. By using both approaches, we have shown that different types of signal share the ability to direct the osk mRNA to the oocyte, but removing any single signal does not eliminate transport.
The system for transport of osk mRNA to the oocyte displays both degeneracy and redundancy. Degeneracy refers to the ability of structurally distinct elements to perform the same function (Tononi et al., 1999). The two types of osk mRNA oocyte transport signal − the Bru sites and the stem-loop structures − have degenerate roles in RNA transport. Redundancy involves the use of multiple iterations of the same element, each performing the same function (Tononi et al., 1999). For osk mRNA transport the different Bru-binding sites act redundantly, with either of two clusters of sites conferring oocyte localization on a reporter mRNA. Similarly, the two stem-loop structures may act redundantly. Although determining how these structures mediate transport is beyond the scope of this report, their structural similarities to the TLS suggests that they, too, are bound by Egl. For SL2a it is only one part of the stem-loop that is important for transport; this region could represent an Egl-binding site, one with a lower affinity for Egl than the TLS, given the inability of SL2a to direct transport on its own. The SL2b transport signal is more complex. The previous mapping of OES activity within SL2b was based on testing mutations in the distal portion of the SL2b stem-loop within an osk 3′ UTR fragment included in a reporter transgene mRNA (Jambor et al., 2014). We have now shown that mutations that disrupt this distal part of SL2b have only a modest effect on transport of otherwise intact osk mRNA. Instead, a larger proportion of SL2b, including both distal and proximal parts of the stem-loop, had to be deleted to dramatically reduce transport. Therefore, SL2b may provide multiple, redundant binding sites for Egl, with loss of most or all of the sites required to strongly disrupt mRNA transport to the oocyte.
Robust transport is integrated with additional steps in mRNA localization
The different roles of the osk mRNA impose different demands on its localization. For the ncRNA function of osk, our results strongly argue that robust transport of osk mRNA to the oocyte is critical. For the protein-coding role of osk mRNA, it is essential that the protein accumulates only at the posterior pole of the oocyte, a distribution that relies on additional steps in mRNA localization.
Although robust transport of osk mRNA to the oocyte occurs, how this is achieved is intriguing. Instead of a single strong transport system, like that mediated by the TLS, osk uses a combination of weaker systems: Bru-binding sites and the various parts of SL2. The Bru sites are weak in the sense that, although they clearly confer oocyte enrichment of the mRNA, transport is not strong enough for osk ncRNA activity or to deplete the mRNA from the nurse cells. The SL2 parts are weak by comparison to the TLS. The TLS is largely insensitive to sequence context: a short segment containing just the TLS stem-loop has been shown to confer strong oocyte transport on multiple different mRNAs (Serano and Cohen, 1995; Cohen et al., 2005). This robustness suggests that formation of the TLS structure, which is critical for its function and for binding Egl (Dienstbier et al., 2009; Bullock et al., 2010), is highly favored. By contrast, neither SL2a nor SL2b alone support oocyte transport of a reporter mRNA.
Why does osk mRNA rely on multiple weak transport signals, rather than a single strong signal such as the TLS? We propose that the need to switch between different localization systems − one for transport to the oocyte and one for the subsequent redistribution within the oocyte − places constraints on the initial transport mechanisms. In particular, for the TLS-like form of transport mediated by SL2a and SL2b it is critical that the transport machine is not tenaciously associated with the mRNA. Replacing these osk transport signals with the TLS, or simply adding the TLS to osk, led to persistent retention of the mRNA at the anterior of the oocyte and reduction in posterior localization, fully consistent with this model. We speculate that this principle − weak by necessity − is applied to other mRNAs with complex programs of localization, in which different localization systems must work together.
MATERIALS AND METHODS
Flies and transgenes
Mutants of the osk locus
oskA87 (Jenny et al., 2006), Df(3R)osk (Reveal et al., 2010) and osk0 (Kanke et al., 2015). For experiments in which an osk transgene was tested as the only source of osk mRNA, the background was an osk RNA null combination: oskA87/Df(3R)osk for Figs 1 and 2, and osk0/osk0 for all others.
Genomic osk transgenes
These were based on a genomic fragment that fully rescues osk null mutants (Kim-Ha et al., 1991). All, with the exception of osk ABC all− (Reveal et al., 2010; Kanke et al., 2015), have three copies of the (human influenza hemagglutinin) HA epitope tag at position T140 (Kanke and Macdonald, 2015) and were made by P element transgenesis (those in Fig. 2) or by phiC31 transgenesis (all others) to the attP site on chromosome II at 51C (in Bloomington stock center strain 24482).
UAS transgenes
All UAS-GFP transgenes have been described previously (Reveal et al., 2010, 2011). The osk 3′ UTR AB and C regions included in some of these correspond to coordinates 3R 8937486-8937612 and 8938214-8938372, respectively, of R6.13 within the Drosophila genome sequence. Most of the UAS-osk::GFP transgenes, which include a 5′ sequence of the osk gene and coding region (osk mRNA coordinates 1−534, including the first 173 amino acids of Long Osk), have been described previously (Kim et al., 2015). The novel UAS-osk::GFP transgenes are those with the osk 3′ UTR C region and include the osk 3′ UTR C region segment (above) in wild-type form or with the Bru-binding site mutations of osk ABC all−. The matalpha4-GAL-VP16 driver (Martin and St Johnston, 2003) was used for expression of UAS transgenes, all made by P element transgenesis.
Mutations to transgenes were introduced by PCR or from synthetic DNA fragments (gBlocks; Integrated DNA Technologies). Mutations of Bru-binding sites were as described (Reveal et al., 2010, 2011). The TLS (44 n ucleotides) (Serano and Cohen, 1995) was introduced into genomic osk transgenes at one of two positions. For osk ΔSL2b transgenes the TLS was inserted at the site of the deletion. For all others, the TLS replaced sequences 57−78 of the osk 3′ UTR (an osk 3′Δ57-78 transgene behaves like wild type).
Detection of RNAs and proteins
RNA levels were determined by RNase protection assays, using rp49 for normalization of sample amounts (Reveal et al., 2010). At least three assays were carried out for each mRNA. RNA distributions were detected by in situ hybridization (Reveal et al., 2010; Jambor et al., 2014). The probes for in situ hybridization were the complete osk 3′ UTR or the complete GFP gene.
Proteins were detected by fluorescence (GFP) or immunofluorescence (Bru). For the latter, rat anti-Bru antibodies (Kanke et al., 2015) were used at 1:2000 dilution. Samples were imaged by using a Leica TCS-SP confocal microscope or a Zeiss LSM 710 confocal microscope. For all imaging experiments the samples were obtained from at least five flies, typically many more. Imaging experiments in order to detect mRNAs by in situ hybridization were performed in groups, with all samples from a single comparison set imaged in the same session and by using identical confocal settings. Quantification of in situ hybridization data was done using FIJI (Schindelin et al., 2012). Parts of nurse cells and oocytes were traced by using a Wacom Intuos tablet to define regions of interest (ROIs). Because the signal is dramatically lower in nuclei, only regions within the cytoplasm were traced. For oocytes, the ROIs correspond to almost the complete oocyte (except the nucleus). For nurse cells, a single representative nurse cell was used to define an ROI, again tracing almost the complete nurse cell (except the nucleus). Egg chambers in which signal levels clearly varied between different nurse cells were excluded from the analysis. This is an issue that primarily occurs when RNA is expressed by using the UAS/GAL4 expression system, as with the reporter transgenes of Figs 1 and 6A. In a small fraction of egg chambers the activity of the UAS/GAL4 system is markedly lower in a subset of nurse cells. Egg chambers in which the oocyte boundary was difficult to define were also excluded. Average signal intensities were obtained for the ROIs and the ratio of oocyte to nurse cell signal for each individual egg chamber was calculated. Trial quantifications testing multiple nurse cells for each chamber, rather than just one, revealed no substantial change in the results. Each egg chamber from this analysis corresponds to a single data point in the quantification graphs, showing the ratio of oocyte:nurse cell signal.
RNA structure predictions
Predictions of RNA folding were performed in the Mfold Web Server using the default settings (http://unafold.rna.albany.edu/?q=mfold/RNA-Folding-Form) (Zuker, 2003).
Egg laying assays
Newly eclosed flies were collected, aged for 2−3 days (when the first eggs appeared) and placed in cages with yeasted apple juice plates. Collections were performed over 60−75 h, during which plates were changed twice a day and the total number of eggs scored to determine egg laying rates (eggs/hour/female). The results of each such experiment (i.e. multiple collections over several days) are presented as a single value in the figures, with at least three such experiments for each genotype. The females were osk0/osk0 with one copy of an osk transgene, unless otherwise indicated.
Statistical methods
The egg laying and imaging assays can be divided into two groups − those with dramatically different outcomes and those with more graded differences that can be small. For results regarding the first group, such as those shown in Fig. 3, no statistical analysis is included. For the second group, statistical tests were used to evaluate the significance of any observed differences. Student's unpaired t-tests were used to compare the means of two samples and means were calculated using Microsoft Excel. Reproducibility was confirmed by performing independent experiments. Biochemical experiments were repeated at least three times. Imaging experiments, which were performed at least twice, involve examination of multiple individual egg chambers in each experiment. Here, repetition serves to reveal any technical problems, and the large number of individual egg chambers scored in each experiment addresses issues of consistency and reproducibility. Egg laying experiments were performed at least three times. Rates of egg laying often show circadian variation but, because each experiment consists of egg collections over several days, this source of variation is largely eliminated. The experiments were not randomized and no statistical method was used to predetermine sample size.
Acknowledgements
We thank members of the Fischer, Macdonald and Stein labs for comments during the course of the work, and Janice Fischer for comments on the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: Y.H.R., P.M.M.; Investigation: Y.H.R., A.K., Y.G., M.S., P.M.M.; Resources: P.M.M.; Writing - original draft: Y.H.R., P.M.M.; Writing - review & editing: Y.H.R., A.K., P.M.M.; Supervision: P.M.M.; Project administration: P.M.M.; Funding acquisition: P.M.M.
Funding
This work was funded by the National Institutes of Health, grants R01GM54409 and R01GM096730. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.202069.supplemental
References
- Arn E. A., Cha B. J., Theurkauf W. E. and Macdonald P. M. (2003). Recognition of a bicoid mRNA localization signal by a protein complex containing Swallow, Nod, and RNA binding proteins. Dev. Cell 4, 41-51. 10.1016/S1534-5807(02)00397-0 [DOI] [PubMed] [Google Scholar]
- Besse F., Lopez de Quinto S., Marchand V., Trucco A. and Ephrussi A. (2009). Drosophila PTB promotes formation of high-order RNP particles and represses oskar translation. Genes Dev. 23, 195-207. 10.1101/gad.505709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock S. L. and Ish-Horowicz D. (2001). Conserved signals and machinery for RNA transport in Drosophila oogenesis and embryogenesis. Nature 414, 611-616. 10.1038/414611a [DOI] [PubMed] [Google Scholar]
- Bullock S. L., Zicha D. and Ish-Horowicz D. (2003). The Drosophila hairy RNA localization signal modulates the kinetics of cytoplasmic mRNA transport. EMBO J. 22, 2484-2494. 10.1093/emboj/cdg230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock S. L., Ringel I., Ish-Horowicz D. and Lukavsky P. J. (2010). A′-form RNA helices are required for cytoplasmic mRNA transport in Drosophila. Nat. Struct. Mol. Biol. 17, 703-709. 10.1038/nsmb.1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B. M., Strein C., Davey N. E., Humphreys D. T., Preiss T., Steinmetz L. M. et al. (2012). Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393-1406. 10.1016/j.cell.2012.04.031 [DOI] [PubMed] [Google Scholar]
- Cheong C., Varani G. and Tinoco I. J. (1990). Solution structure of an unusually stable RNA hairpin, 5'GGAC(UUCG)GUCC. Nature 346, 680-682. 10.1038/346680a0 [DOI] [PubMed] [Google Scholar]
- Cohen R. S., Zhang S. and Dollar G. L. (2005). The positional, structural, and sequence requirements of the Drosophila TLS RNA localization element. RNA 11, 1017-1029. 10.1261/rna.7218905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis I. and Ish-Horowicz D. (1991). Apical localization of pair-rule transcripts requires 3′ sequences and limits protein diffusion in the Drosophila blastoderm embryo. Cell 67, 927-940. 10.1016/0092-8674(91)90366-7 [DOI] [PubMed] [Google Scholar]
- Dienstbier M., Boehl F., Li X. and Bullock S. L. (2009). Egalitarian is a selective RNA-binding protein linking mRNA localization signals to the dynein motor. Genes Dev. 23, 1546-1558. 10.1101/gad.531009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubowy J. and Macdonald P. M. (1998). Localization of mRNAs to the oocyte is common in Drosophila ovaries. Mech. Dev. 70, 193-195. 10.1016/S0925-4773(97)00185-8 [DOI] [PubMed] [Google Scholar]
- Ephrussi A., Dickinson L. K. and Lehmann R. (1991). oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell 66, 37-50. 10.1016/0092-8674(91)90137-N [DOI] [PubMed] [Google Scholar]
- Ferrandon D., Koch I., Westhof E. and Nüsslein-Volhard C. (1997). RNA-RNA interaction is required for the formation of specific bicoid mRNA 3′ UTR-STAUFEN ribonucleoprotein particles. EMBO J. 16, 1751-1758. 10.1093/emboj/16.7.1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautreau D., Cote C. A. and Mowry K. L. (1997). Two copies of a subelement from the Vg1 RNA localization sequence are sufficient to direct vegetal localization in Xenopus oocytes. Development 124, 5013-5020. [DOI] [PubMed] [Google Scholar]
- Ghosh S., Marchand V., Gáspár I. and Ephrussi A. (2012). Control of RNP motility and localization by a splicing-dependent structure in oskar mRNA. Nat. Struct. Mol. Biol. 19, 441-449. 10.1038/nsmb.2257 [DOI] [PubMed] [Google Scholar]
- Hachet O. and Ephrussi A. (2004). Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature 428, 959-963. 10.1038/nature02521 [DOI] [PubMed] [Google Scholar]
- Holt C. E. and Bullock S. L. (2009). Subcellular mRNA localization in animal cells and why it matters. Science 326, 1212-1216. 10.1126/science.1176488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambor H., Brunel C. and Ephrussi A. (2011). Dimerization of oskar 3′ UTRs promotes hitchhiking for RNA localization in the Drosophila oocyte. RNA 17, 2049-2057. 10.1261/rna.2686411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambor H., Mueller S., Bullock S. L. and Ephrussi A. (2014). A stem-loop structure directs oskar mRNA to microtubule minus ends. RNA 20, 429-439. 10.1261/rna.041566.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny A., Hachet O., Závorszky P., Cyrklaff A., Weston M. D., Johnston D. S., Erdélyi M. and Ephrussi A. (2006). A translation-independent role of oskar RNA in early Drosophila oogenesis. Development 133, 2827-2833. 10.1242/dev.02456 [DOI] [PubMed] [Google Scholar]
- Kanke M. and Macdonald P. M. (2015). Translational activation of oskar mRNA: reevaluation of the role and importance of a 5′ regulatory element. PLoS ONE 10, e0125849 10.1371/journal.pone.0125849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanke M., Jambor H., Reich J., Marches B., Gstir R., Ryu Y. H., Ephrussi A. and Macdonald P. M. (2015). oskar RNA plays multiple noncoding roles to support oogenesis and maintain integrity of the germline/soma distinction. RNA 21, 1096-1109. 10.1261/rna.048298.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Lee J., Lee S., Lee B. and Kim-Ha J. (2014). Phylogenetic comparison of oskar mRNA localization signals. Biochem. Biophys. Res. Commun. 444, 98-103. 10.1016/j.bbrc.2014.01.021 [DOI] [PubMed] [Google Scholar]
- Kim G., Pai C.-I., Sato K., Person M. D., Nakamura A. and Macdonald P. M. (2015). Region-specific activation of oskar mRNA translation by inhibition of bruno-mediated repression. PLoS Genet. 11, e1004992 10.1371/journal.pgen.1004992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Ha J., Smith J. L. and Macdonald P. M. (1991). oskar mRNA is localized to the posterior pole of the Drosophila oocyte. Cell 66, 23-35. 10.1016/0092-8674(91)90136-M [DOI] [PubMed] [Google Scholar]
- Kim-Ha J., Webster P. J., Smith J. L. and Macdonald P. M. (1993). Multiple RNA regulatory elements mediate distinct steps in localization of oskar mRNA. Development 119, 169-178. [DOI] [PubMed] [Google Scholar]
- Kim-Ha J., Kerr K. and Macdonald P. M. (1995). Translational regulation of oskar mRNA by bruno, an ovarian RNA-binding protein, is essential. Cell 81, 403-412. 10.1016/0092-8674(95)90393-3 [DOI] [PubMed] [Google Scholar]
- Lawrence J. B. and Singer R. H. (1986). Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell 45, 407-415. 10.1016/0092-8674(86)90326-0 [DOI] [PubMed] [Google Scholar]
- Lécuyer E., Yoshida H., Parthasarathy N., Alm C., Babak T., Cerovina T., Hughes T. R., Tomancak P. and Krause H. M. (2007). Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131, 174-187. 10.1016/j.cell.2007.08.003 [DOI] [PubMed] [Google Scholar]
- Lehmann R. and Nüsslein-Volhard C. (1986). Abdominal segmentation, pole cell formation, and embryonic polarity require the localized activity of oskar, a maternal gene in Drosophila. Cell 47, 141-152. 10.1016/0092-8674(86)90375-2 [DOI] [PubMed] [Google Scholar]
- Lipshitz H. D. and Smibert C. A. (2000). Mechanisms of RNA localization and translational regulation. Curr. Opin. Genet. Dev. 10, 476-488. 10.1016/S0959-437X(00)00116-7 [DOI] [PubMed] [Google Scholar]
- Macdonald P. M. (1990). bicoid mRNA localization signal: phylogenetic conservation of function and RNA secondary structure. Development 110, 161-171. [DOI] [PubMed] [Google Scholar]
- Macdonald P. M. and Kerr K. (1997). Redundant RNA recognition events in bicoid mRNA localization. RNA 3, 1413-1420. [PMC free article] [PubMed] [Google Scholar]
- Macdonald P. M., Kerr K., Smith J. L. and Leask A. (1993). RNA regulatory element BLE1 directs the early steps of bicoid mRNA localization. Development 118, 1233-1243. [DOI] [PubMed] [Google Scholar]
- Martin S. G. and St Johnston D. (2003). A role for Drosophila LKB1 in anterior-posterior axis formation and epithelial polarity. Nature 421, 379-384. 10.1038/nature01296 [DOI] [PubMed] [Google Scholar]
- Reveal B., Yan N., Snee M. J., Pai C.-I., Gim Y. and Macdonald P. M. (2010). BREs mediate both repression and activation of oskar mRNA translation and act in trans. Dev. Cell 18, 496-502. 10.1016/j.devcel.2009.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reveal B., Garcia C., Ellington A. and Macdonald P. M. (2011). Multiple RNA binding domains of Bruno confer recognition of diverse binding sites for translational repression. RNA Biol. 8, 1047-1060. 10.4161/rna.8.6.17542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross A. F., Oleynikov Y., Kislauskis E. H., Taneja K. L. and Singer R. H. (1997). Characterization of a β-actin mRNA zipcode-binding protein. Mol. Cell. Biol. 17, 2158-2165. 10.1128/MCB.17.4.2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanghavi P., Liu G., Veeranan-Karmegam R., Navarro C. and Gonsalvez G. B. (2016). Multiple roles for Egalitarian in polarization of the Drosophila egg chamber. Genetics 203, 415-432. 10.1534/genetics.115.184622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676-682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serano T. L. and Cohen R. S. (1995). A small predicted stem-loop structure mediates oocyte localization of Drosophila K10 mRNA. Development 121, 3809-3818. [DOI] [PubMed] [Google Scholar]
- Snee M. J., Arn E. A., Bullock S. L. and Macdonald P. M. (2005). Recognition of the bcd mRNA localization signal in Drosophila embryos and ovaries. Mol. Cell. Biol. 25, 1501-1510. 10.1128/MCB.25.4.1501-1510.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D. (1995). The intracellular localization of messenger RNAs. Cell 81, 161-170. 10.1016/0092-8674(95)90324-0 [DOI] [PubMed] [Google Scholar]
- St Johnston D., Driever W., Berleth T., Richstein S. and Nüsslein-Volhard C. (1989). Multiple steps in the localization of bicoid RNA to the anterior pole of the Drosophila oocyte. Development 107, 13-19. [DOI] [PubMed] [Google Scholar]
- Tononi G., Sporns O. and Edelman G. M. (1999). Measures of degeneracy and redundancy in biological networks. Proc. Natl. Acad. Sci. USA 96, 3257-3262. 10.1073/pnas.96.6.3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster P. J., Liang L., Berg C. A., Lasko P. and Macdonald P. M. (1997). Translational repressor bruno plays multiple roles in development and is widely conserved. Genes Dev. 11, 2510-2521. 10.1101/gad.11.19.2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406-3415. 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]