ABSTRACT
IRE1 mediates the unfolded protein response (UPR) in part by regulating XBP1 mRNA splicing in response to endoplasmic reticulum (ER) stress. In cultured metazoan cells, IRE1 also exhibits XBP1-independent biochemical activities. IRE1 and XBP1 are developmentally essential genes in Drosophila and mammals, but the source of the physiological ER stress and the relative contributions of XBP1 activation versus other IRE1 functions to development remain unknown. Here, we employed Drosophila to address this question. Explicitly, we find that specific regions of the developing alimentary canal, fat body and the male reproductive organ are the sources of physiological stress that require Ire1 and Xbp1 for resolution. In particular, the developmental lethality associated with an Xbp1 null mutation was rescued by transgenic expression of Xbp1 in the alimentary canal. The domains of IRE1 that are involved in detecting unfolded proteins, cleaving RNAs and activating XBP1 splicing were all essential for development. The earlier onset of developmental defects in Ire1 mutant larvae compared to in Xbp1-null flies supports a developmental role for XBP1-independent IRE1 RNase activity, while challenging the importance of RNase-independent effector mechanisms of Drosophila IRE1 function.
KEY WORDS: Unfolded protein response, Physiological ER stress, Xbp1, Ire1, Midgut, Gastric caeca, Proventriculus, Accessory gland, Ejaculatory duct, Male sterility
Summary: The stress-response proteins IRE1 and XBP1 are developmentally essential in metazoans. We identify the source of the physiological stress in Drosophila and determine the molecular requirements for stress response signaling.
INTRODUCTION
Endoplasmic reticulum (ER) is a subcellular organelle in eukaryotes where most secretory and membrane proteins are synthesized and undergo folding. Conditions that disrupt or overwhelm the protein-folding capacity of the ER must be resolved to maintain cellular function, and not surprisingly, eukaryotic cells have evolved robust quality control mechanisms that help cells cope with such ER stress. Among those quality control mechanisms is the unfolded protein response (UPR), which refers to signaling pathways that are activated in response to ER stress to regulate gene expression (Walter and Ron, 2011).
A particularly well-characterized branch of the UPR is mediated by IRE1 (also known as ERN1 in mammals) and XBP1, which are conserved across phyla from mammals to S. cerevisiae (Walter and Ron, 2011). This pathway is initiated by IRE1, which detects imbalances between unfolded proteins and chaperones through its stress-sensing luminal domain (Aragón et al., 2009; Credle et al., 2005; Gardner and Walter, 2011; Zhou et al., 2006) to activate its cytoplasmic RNase (Korennykh et al., 2011; Lee et al., 2008b). The best-characterized and most important substrate of IRE1 is the XBP1 mRNA, which undergoes an unconventional splicing reaction as part of the UPR. Spliced XBP1 mRNA encodes an active transcription factor isoform, which after translation, promotes the expression of various ER quality control genes (Calfon et al., 2002; Shen et al., 2001; Yoshida et al., 2001).
In addition to this well-established axis of IRE1 signaling, a number of additional regulatory mechanisms associated with IRE1 have been uncovered in cultured cells. These include additional substrates for the RNase activity of IRE1. Unlike XBP1 mRNA, IRE1 cleavage of these other substrates leads to their degradation, a process which is referred to as regulated IRE1-dependent decay (RIDD) (Coelho et al., 2013; Hollien et al., 2009; Hollien and Weissman, 2006). RIDD is believed to lessen the burden of new protein synthesis and folding in the ER, but its functional significance has not been rigorously tested. IRE1 also has an RNase-independent activity in stimulating JNK signaling (Urano et al., 2000). These various cytoplasmic activities of IRE1 are not only activated by misfolded peptides, but also in response to perturbation of lipid balance. Interestingly, IRE1 activation under these conditions occurs even without the luminal domain responsible for detecting misfolded peptides, in yeast as well as in cultured mammalian cells (Promlek et al., 2011; Volmer et al., 2013).
While these mechanisms have been uncovered in cultured cells exposed to exogenously imposed conditions of stress, their roles in dealing with physiological ER stress in metazoan tissues remain poorly understood. It is notable that IRE1α, one of the two mammalian IRE1 genes (also known as ERN1), and XBP1 are developmentally essential in mice (Iwawaki et al., 2009; Lee et al., 2008a; Reimold et al., 2001). However, the reported IRE1-knockout phenotype is different from that of XBP1 knockouts in mice: whereas the developmental lethality of XBP1-knockout embryos has been attributed to liver failure (Lee et al., 2005), the lethality of IRE1α-knockout mice is attributed to a different organ – specifically, the placenta (Iwawaki et al., 2009). Such difference may be due to as yet inexplicable tissue specificity of the two genes, and the functional significance of IRE1-mediated XBP1 activation during normal animal development remains unclear.
Here, we employed the molecular genetic tools of Drosophila to understand the precise role of IRE1 and XBP1 signaling during normal metazoan development. The basic mechanism of UPR is conserved in Drosophila, and the homologous genes in Drosophila are also essential for normal development (Coelho et al., 2013; Ryoo, 2015; Ryoo et al., 2007, 2013). By employing robust stress reporters, we report the identity of tissues that are impacted by the loss of Ire1 or Xbp1 during normal development. In addition, we examined the functional significance and the in vivo roles of various IRE1 domains through molecular genetic tools. The results indicate that specific tissues require Ire1 and Xbp1 during normal development, and the domains of IRE1 involved in the detection of misfolded peptides and splicing of XBP1 mRNA are particularly essential. We also find that developmental phenotypes found upon Ire1 mutation are more severe than those for Xbp1, supporting the idea that IRE1 has XBP1-independent roles. On the other hand, we found no significant evidence that the RNase-independent activities of IRE1 have a role during normal development.
RESULTS
Visualization of cells impacted by the loss of Ire1 or Xbp1 through a stress-responsive reporter
In Drosophila, Xbp1-null mutants (Xbp1ex7−/−) fail to develop beyond the second-instar larval stage (Ryoo et al., 2007, 2013). To better understand the basis of this defect, we sought a reporter that could mark cells suffering from the loss of IRE1 and/or XBP1 signaling. We specifically employed a reporter for the PERK (also known as PEK in flies and EIF2AK3 in mammals) and ATF4 (crc in flies) pathway, a branch of the UPR that is parallel to that of IRE1 and XBP1. In cells that require IRE1 and XBP1 signaling for ER homeostasis, it is predicted that the loss of IRE1 or XBP1 would aggravate stress in the ER and thereby hyper-activate PERK and ATF4 signaling. In fact, it had been observed that the loss of one branch of the UPR results in the hyper-activation of the other in mouse tissues that have high secretory burden (Harding et al., 2001). In worms, single mutants of perk or Xbp1 are viable, but the knockdown of both genes result in synthetic lethality, further supporting the compensatory activities of the two pathways (Shen et al., 2001). The PERK and ATF4 reporter that we used is 4E-BPintron dsRed, which has a cluster of predicted ATF4-binding sites in the 4E-BP intron sequence driving dsRed (Fig. 1A) (Kang et al., 2017).
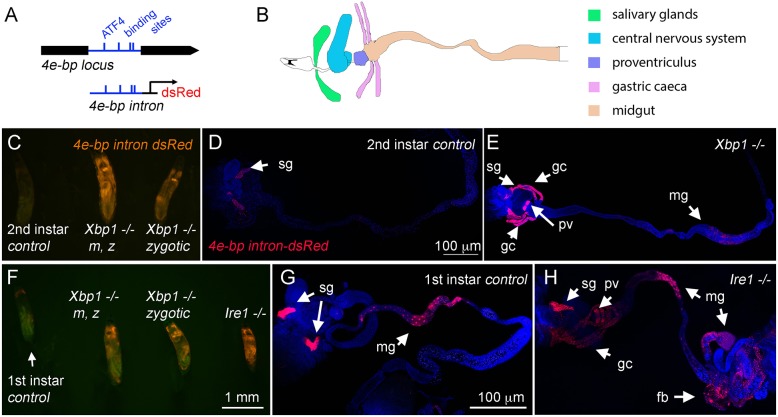
Fig. 1.
The ATF4-responsive 4E-BPintron dsRed reporter is induced in Xbp1 and Ire1 mutant larvae. (A) A schematic diagram of the 4E-BP locus. The intron is depicted in blue, and the predicted ATF4-binding sites are indicated by vertical lines. The dsRed reporter expression is driven by this intron element (Kang et al., 2017). (B) A schematic diagram of the salivary glands (green), central nervous system (blue), proventriculus (purple), gastric caeca (pink) and midgut (yellow) in the second-instar Drosophila larva. Compare this diagram with images in D,E,G and H. (C–H) 4E-BPintron dsRed expression (red) in the indicated genetic backgrounds. TO-PRO-3 (blue) was used to show the outline of tissues. (C) An intact control of wild-type Xbp1 (left), an Xbp1 maternal zygotic (m, z) mutant (center) and an Xbp1 zygotic mutant (right) at the second-instar stage juxtaposed to each other without dissection. (D) Dissected wild-type second-instar larva with 4E-BPintron dsRed expression primarily restricted to the salivary glands. (E) In the Xbp1ex79−/− background, 4E-BPintron dsRed is strongly induced in tissues beyond the salivary glands, which include the gastric caeca and the proventriculus (arrows). (F) Intact wild-type (left), Xbp1 maternal zygotic (m, z) mutant (left center), Xbp1 zygotic mutant (right center) and Ire1f02170−/− (right) first-instar larvae are juxtaposed. (G) Dissected wild-type first-instar larva showing 4E-BPintron dsRed expression most prominently in the salivary glands. (H) In the Ire1−/− larva, the reporter is also strongly expressed in the proventriculus, gastric caeca, midgut and fat body (indicated with arrows). The scale bar in D applies to D and E; the scale bar in F applies to panels C and F, and the scale bar in G applies to images in G and H. sg, salivary glands; gc, gastric caeca; pv, proventriculus; mg, midgut; fb, fat body.
A schematic diagram of a few relevant organs in the second-instar larva is shown in Fig. 1B. Specifically, the mouth hook and the salivary glands are at anterior end. They are connected to the esophagus, which passes between the brain lobes to the proventriculus, a bulb-like structure where the ingested food passes through to reach the midgut. Around the proventriculus–midgut junction are four long tubes called the gastric caeca. While the activity of the 4E-BPintron dsRed reporter is low in the Xbp1+/+ control second-instar larva, the reporter signal was readily visible in the equivalent stage of Xbp1ex79−/− larvae even without dissection (Fig. 1C, right larva). We also generated a maternal zygotic Xbp1 mutant larva (Fig. 1C, middle) and found that they also survived to the second-instar larval stage with a similar degree of 4E-BPintron dsRed reporter activation as in the zygotic mutant. In the Xbp1+/+ background, the reporter activity was mostly confined to the salivary glands, but the dissected Xbp1 mutants reveal specific 4E-BPintron dsRed reporter induction in the gastric caeca, proventriculus and certain parts of the posterior intestine (Fig. 1D,E). Fat bodies also induced 4E-BPintron dsRed, but with lesser intensity (see below in Fig. 2D). Notably, these are the tissues with reportedly high levels of Xbp1 transcripts (Ryoo et al., 2007, 2013). On the other hand, there were many other tissues that did not show signs of stress in the Xbp1 mutant background, which included certain parts of the midgut, the larval brain and imaginal discs, indicating that the role of Xbp1 in suppressing the activation of 4E-BPintron dsRed reporter in the context of normal fly development is specific to the cell type.
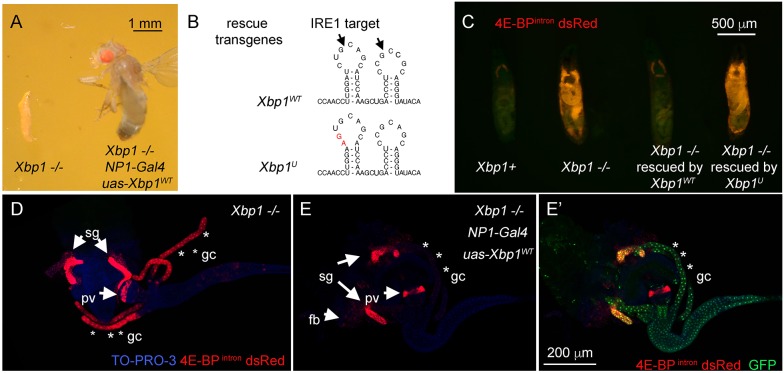
Fig. 2.
Rescue of Xbp1 mutant-associated lethality through the expression of Xbp1 transgene. (A) Xbp1−/− larvae survive only up to the second-instar larval stage (left), whereas Xbp1 mutants expressing transgenic Xbp1 with the NP1-Gal4 driver survive to adulthood (right). (B) The IRE1 target cleavage sequence within the XBP1 mRNA. Arrows point to the sites that are spliced. The wild-type sequence (upper image) and the mutated sequence within the rescue transgene (lower image, in red) are shown. (C–E) The effects of Xbp1 transgene expression driven by NP1-Gal4 on the 4E-BPintron dsRed reporter levels in Xbp1−/− second-instar larvae (red). (C) Non-dissected second-instar larvae of the indicated genotypes. (D,E) 4E-BPintron dsRed signal (red) from dissected second-instar larval tissues. TO-PRO-3 (blue) was used to show the outline of tissues. The strong expression of 4E-BPintron dsRed signal in the Xbp1−/− background (D) is suppressed by the wild-type Xbp1 transgene expression through the NP1-Gal4 driver (E). UAS-GFP was co-expressed with the Xbp1 transgene, and GFP signal (green) marks the NP-Gal4 active tissues, which includes the gastric caeca (asterisks) and the midgut (E′), and 4E-BPintron dsRed signal is specifically suppressed in those domains. The scale bar in E′ applies to D and E. sg, salivary glands; gc, gastric caeca; pv, proventriculus; fb, fat body.
We also examined the effect of Ire1 loss-of-function mutation on the 4E-BPintron dsRed reporter. Ire1 mutants develop only up to the first-instar larval stage, and strong induction of the 4E-BPintron dsRed in these mutants was also obvious without dissection. The dsRed signal from Ire1 mutant larvae were visibly more intense than that of zygotic Xbp1 null mutants as well as maternal zygotic Xbp1 mutants of the same developmental stage (Fig. 1F). Dissected Ire1 mutant first-instar larvae showed similar patterns of 4E-BPintron dsRed induction to that of second-instar Xbp1 mutants, which included the gastric caeca, several parts of the intestine and the fat body (Fig. 1G,H). These results indicate that the same tissues of the developing larvae are impacted by the loss of Ire1 or Xbp1 but the consequences of Ire1 loss are more severe.
Rescue of the larval lethality with transgenic Xbp1
We found that the larval lethality of the Drosophila Xbp1 mutants could be rescued by the ectopic expression of Xbp1 through tubulin-Gal4 (tub-Gal4) and UAS-Xbp1. The surviving adults eclosed with the predicted Mendelian ratio (approximately one-third of all progeny, due to the use of balancer chromosomes in the parents that are recessive lethal; Fig. S1). To determine the specific tissue types that require Xbp1 during development, we expressed UAS-Xbp1 with tissue-specific Gal4 drivers in Xbp1 mutants. Various tissue-specific Gal4 drivers were used (see Materials and Methods), but aside from tub-Gal4, only NP1-Gal4 rescued the larval lethality (Fig. 2A). NP1-Gal4 is a Gal4 enhancer trap in the Myosin 1A gene (also known as Myo31DF), which encodes a protein that localizes to the brush borders of the gut enterocytes (Morgan et al., 1995). The Gal4 activity is likewise restricted to the enterocytes of the alimentary canal and the salivary gland in the larva (Jiang and Edgar, 2009) (Fig. S2B). In adults, NP1-Gal4 also marks the intestine, but is not active in tissues such as the ovary (Fig. S2D). Transgenic Xbp1 expression using this Gal4 driver suppressed signs of ER stress as evidenced by the loss of the 4E-BPintron dsRed signal (Fig. 2C).
Neither mouse nor fly studies has yet determined the functional significance of IRE1-mediated XBP1 mRNA splicing during development. IRE1 activates XBP1 by cleaving two evolutionarily conserved positions in the double stem-loop structure within the XBP1 mRNA (Calfon et al., 2002; Yoshida et al., 2001). To determine whether IRE1-mediated XBP1 splicing is required for survival, we introduced silent mutations in the Xbp1 transgene, changing the sequence of a conserved double stem-loop IRE1 target sequence within the mRNA (Fig. 2B). The targeted sequence is conserved from yeast to mammals, and mutation in those sequences impair IRE1-mediated XBP1 mRNA cleavage across phyla (Calfon et al., 2002; Shen et al., 2001; Yoshida et al., 2001). At the same time, the mutations do not alter the encoded amino acid residues, and precede the splice site. Unlike the wild-type transgene, the UAS-Xbp1 transgene containing such mutated stem-loop (referred to as UAS-Xbp1U) failed to suppress the 4E-BPintron dsRed signal in the Xbp1−/− background (Fig. 2C). Consistently, Xbp1U expression failed to rescue the larval lethality. Upon dissection of these larvae, we found that the strong 4E-BPintron dsRed signal in the gastric caeca of Xbp1 mutants was suppressed by the wild-type Xbp1 transgene expression through the NP1-Gal4 driver (Fig. 2D,E). On the other hand, the lower intensity 4E-BPintron dsRed signal from the fat body was not suppressed, a tissue where NP1-Gal4 is not active (Fig. 2E). This observation suggests that IRE1 and XBP1 signaling in the alimentary canal is essential for survival during development.
Many other Gal4 drivers that are active in other cell types could not rescue the Xbp1 lethality when driving UAS-Xbp1 expression. These included dilp2-Gal4 driving gene expression in the insulin-producing cells of the larval brain, prospero-Gal4 that drives gene expression in entero-endocrine cells that specialize in peptide secretion within the midgut, escargot-Gal4, which is expressed in salivary glands, imaginal discs and the adult midgut precursor cells (Fig. S2), the neuronal-specific Elav-Gal4, the glia-specific Repo-Gal4 and the fat body-specific cg-Gal4. These observations indicate that Xbp1 in the nervous system, insulin-producing cells or entero-endocrine cells alone are not sufficient to rescue the developmental lethality associated with the loss of Xbp1.
Requirement for IRE1 during normal development
To better understand the developmental role of Ire1, we performed a structure function study of IRE1. First, we established that the developmental lethality of the Ire1f01270 allele can be specifically attributed to the loss of Ire1 by rescuing this lethality with an 8.8 kb genomic transgene from the Ire1 locus (Fig. 3A,B). The Drosophila Ire1-coding sequence is largely conserved with that of yeast and mammalian IRE1, with an ER luminal domain that detects misfolded peptides, and cytoplasmic domains that encode a kinase and an RNase. Through BAC-recombineering of this Ire1 genomic transgene, we mutated sequences that would abolish specific functions of encoded IRE1 protein (Fig. 3C). Specifically, we expressed an Ire1LD Del transgene with a deletion in the luminal domain that detects misfolded peptides in the ER lumen. Structure and function analysis of yeast and mammalian IRE1 have shown that deletion of a portion of its luminal domain blocks its ability to respond to misfolded peptides, while not affecting its activation by lipid imbalances (Promlek et al., 2011; Volmer et al., 2013). As deleting a large segment of a protein can compromise overall protein stability, we deleted precisely the part of the Drosophila IRE1 that is equivalent to what was deleted in the aforementioned yeast and mammalian studies. To abolish the RNase activity of IRE1, we introduced an H890A mutation, also based on the fact that the equivalent amino acid residue in yeast and mammalian IRE1 is critical for the RNase activity (Korennykh et al., 2009; Lee et al., 2008b). To test whether these mutants abolish IRE1 activity in the larva, we introduced the XBP1–EGFP reporter in which the GFP epitope is designed to be expressed when XBP1 mRNA undergoes IRE1-mediated splicing (Sone et al., 2013). Ire1f02170−/− with wild-type Ire1 genomic transgenes readily activated this XBP1–EGFP reporter when exposed to tunicamycin treatment (Fig. S3A,B), but the mutant Ire1 transgenes failed to do so (Fig. S3C,D). Consistent with these results, these mutant genomic transgenes rescued neither the first-instar larval lethality of Ire1 nor the strong 4E-BPintron dsRed signal associated with Ire1 mutations (Fig. 3D).
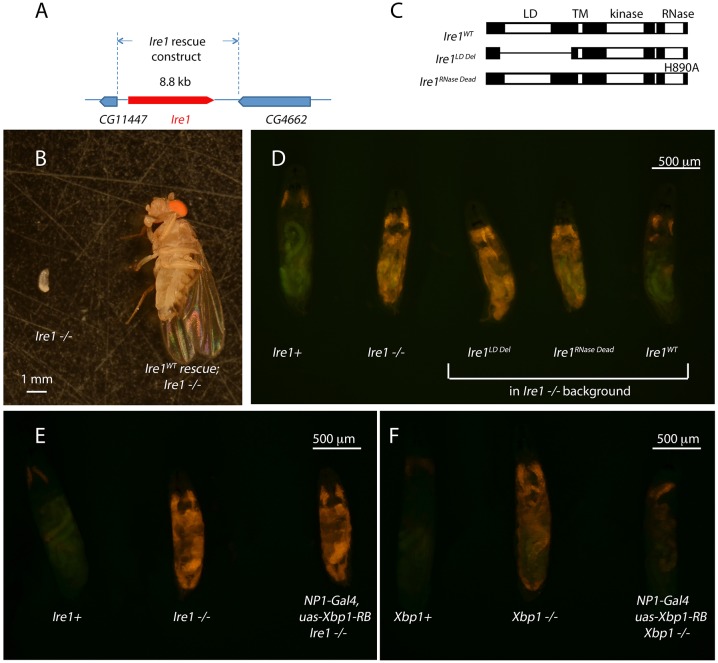
Fig. 3.
The roles of specific IRE1 domains and spliced XBP1 in development. (A) A schematic diagram of the Ire1 rescue genomic construct. (B) Ire1−/− larva at its latest stage of survival (left), juxtaposed to an Ire1−/− fly rescued with the wild-type Ire1 genomic transgene. (C) A diagram of wild-type and mutant Ire1 primary structures that were tested for rescue. While the mutations were made in the context of the 8.8 kb genomic rescue transgene, only the changes in the resulting coding sequence are shown here. The mutant constructs are designed to specifically impair misfolded peptide sensing in the ER by the deletion of the luminal domain (Ire1LD Del) and RNase activity (Ire1RNase Dead). The latter has a missense mutation that changes H890 to an alanine residue. (D) The effect of the wild-type and mutant Ire1 transgenes on the 4E-BPintron dsRed signal of the Ire1−/− first-instar larvae. The genotypes are indicated below each larva. (E,F) The effect of expressing Xbp1-RB (spliced Xbp1 isoform) through the NP1-Gal4 driver in the Ire1 (E), or Xbp1 (F) mutant backgrounds. All larvae contain the 4E-BPintron dsRed (red) reporter in the background, and the genotypes are indicated below each larva.
If the sole function of IRE1 during development was to splice and activate XBP1 mRNA, the larval lethality of Ire1 mutants would be rescued by the transgenic expression of the spliced XBP1 isoform, Xbp1-RB. We tested this possibility by driving Xbp1-RB expression through the NP1-Gal4 driver, which neither rescued the lethality nor suppressed the strong induction of the 4E-BPintron dsRed signal in Ire1 mutants (Fig. 3E). To make sure that the Xbp1-RB transgene is functional, we expressed this transgene in the Xbp1 mutant larva using the NP1-Gal4 driver, which strongly reduced the 4E-BPintron dsRed signal (Fig. 3F). These experiments indicate that while IRE1-mediated XBP1 splicing is essential for normal Drosophila development, XBP1-independent IRE1 functions are also functionally important.
Requirements for Xbp1 beyond the alimentary canal
The rescue of Xbp1−/− with NP1-Gal4 and wild-type UAS-Xbp1 was partially incomplete in a number of ways: the NP1-Gal4 active domain does not include the proventriculus and the fat body (Fig. 2E′, in green), and consistent with this, stress, as evidenced by a strong 4E-BPintron dsRed signal, was not resolved in those tissues (Fig. 2E). It was also noticeable that most of the progeny that survived to adulthood were males (Fig. S1). While the males survived close to the Mendelian ratio at 25°C, rescue became very rare at 22°C or lower temperatures. The underlying reasons for such dependence on temperature and sex remain unclear.
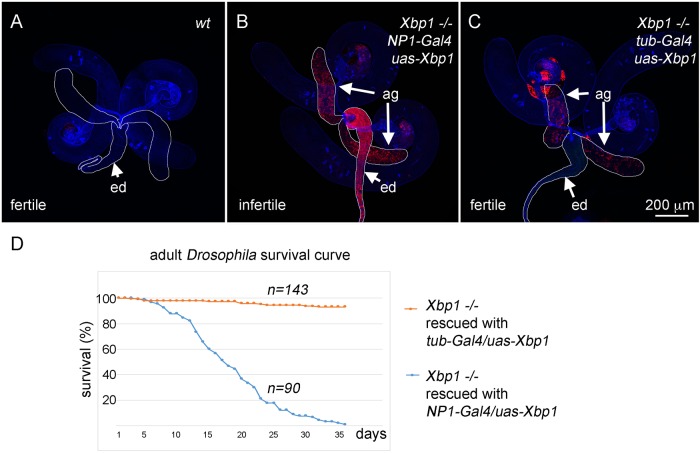
In adults, we found that the rescued males were completely sterile. To determine the specific tissues affected in these adults, we examined 4E-BPintron dsRed expression in the male reproductive system. The reporter was largely inactive in the wild-type background, whereas strong signs of stress were detected through the reporter in the accessory glands and ejaculatory ducts of Xbp1 mutants that were rescued to adulthood with NP1-Gal4 and UAS-Xbp1 (Fig. 4A,B). When the ubiquitous tub-Gal4 driver was used instead of NP1-Gal4, such conditions rescued not only larval lethality, but also male infertility. Correlating with the restoration of fertility, tub-Gal4-mediated rescue suppressed 4E-BPintron dsRed induction in the ejaculatory duct (Fig. 4C). In addition to these defects, Xbp1 mutants that had been rescued with NP1-Gal4/UAS-Xbp1 had significantly shorter lifespans (Fig. 4D). These observations suggest that Xbp1 plays functionally significant roles in tissues beyond the alimentary canal.
Fig. 4.
Requirements for Xbp1 beyond the alimentary canal. To examine the effects of Xbp1 beyond the alimentary canal, Xbp1 mutants were either rescued to adulthood with an alimentary canal-specific NP1-Gal4 driver expressing UAS-Xbp1 (B, and blue line in D), and the phenotypes were compared with mutants that were rescued with a ubiquitous tub-Gal4 driver (C, orange line in D). (A–C) 4E-BPintron dsRed (red) in the male reproductive system. Phalloidin labeling is shown in blue. Ejaculatory ducts (ed) and accessory glands (ag) are outlined and indicated with arrows. (A) In the wild-type background, the reporter expression level is insignificant (genotype, 4E-BPintron dsRed/+). (B) NP1-Gal4-rescued mutants have male infertility, and correlating with this, strong reporter signals are detected in the ejaculatory ducts and accessory glands. (C) Xbp1 mutants rescued with tub-Gal4 are fertile, and correlating with this, there is reduced reporter activity in the ejaculatory duct. (D) Lifespan of Xbp1 mutant adult flies that were rescued with either tub-Gal4 (red) or NP1-Gal4 (blue) driving UAS-Xbp1. The number of flies examined are indicated as n on the graph. The scale bar in C also applies to panels A and B.
Xbp1 mutant larvae show hyperactive innate immune response
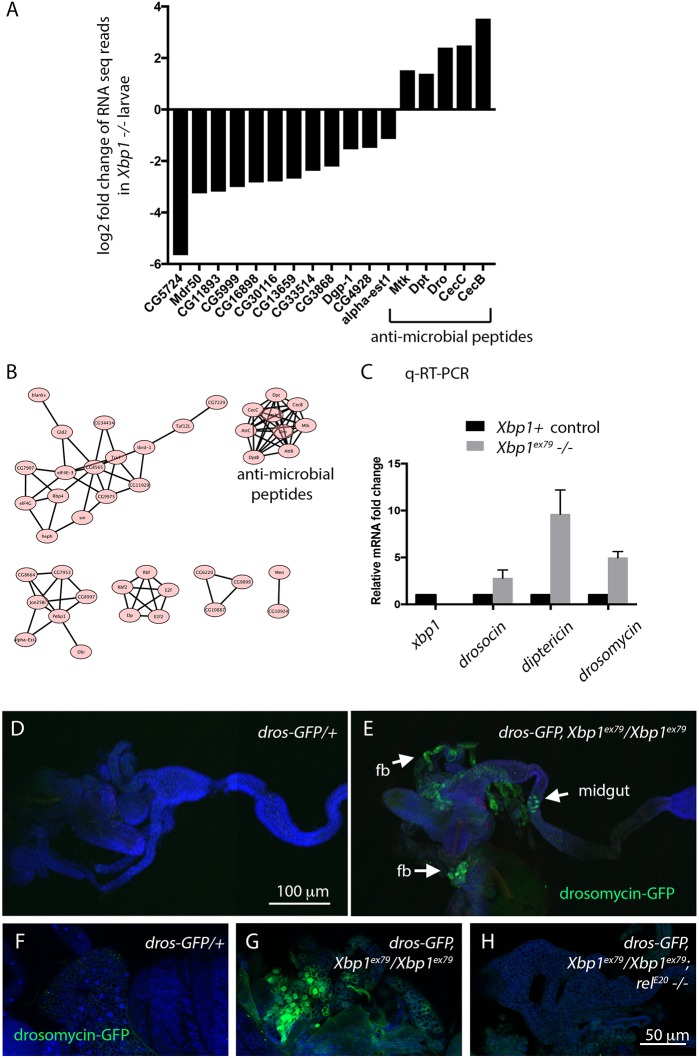
To obtain an unbiased view of the changes in gene expression that occurs in Xbp1 mutant larvae, we performed RNAseq analysis of transcripts from Xbp1+/+ control and Xbp1ex79−/− second-instar larvae. The full list of analyzed RNAs, and their relative levels in mutant versus wild-type are shown in Table S1. The entire sequence data is accessible through NCBI's Gene Expression Omnibus (GEO accession number GSE99676). The transcripts from the RNAseq data were ranked by the adjusted P values (<0.05) in order to select significantly upregulated or downregulated genes in Xbp1 mutants for further analysis. According to this criteria, 79 genes were downregulated in Xbp1 mutants, while 75 genes were upregulated (see Table S1). The gene ontology (GO) terms with P<0.05 are listed in Tables 1 and 2. Specifically, the downregulated gene GO terms included those involved in dephosphorylation (GO 0016791, P value of 3.5×10−3), oxidation-reduction (GO 0055114, P value of 4.2×10−3) and iron ion binding (GO 0005506, P value of 4.3×10−2). Some of the genes were previously reported as tunicamycin-inducible genes in flies (Chow et al., 2013; Fig. 5A), consistent with the idea that their expression depends on Xbp1. Prominent among the genes whose expression increased significantly were those encoding anti-microbial peptides (AMPs) (GO 0019731, P value of 2.40×10−5), which included cecB, cecC, mtk, dro and dipt (also known as DptA) (Fig. 5A, Table 2). Induction of AMP transcripts in Xbp1−/− larvae were further validated through quantitative real-time PCR (qRT-PCR; Fig. 5C). To visualize the pattern of AMP induction, we utilized drosomycin-GFP, a reporter that is driven by the upstream regulatory element of one of the AMPs (Ferrandon et al., 1998). Xbp1 mutant larvae had strong drosomycin-GFP induction, most prominently in the fat body, but also occasionally in the midgut epithelium (Fig. 5D,E). There is no evidence that these innate immune response genes are under direct transcriptional control by XBP1. Rather, innate immune response activation may be a secondary effect of tissue damage and impairment due to the loss of Xbp1. In fact, drosomycin-GFP induction was strongly suppressed when the canonical innate immune response was blocked in a rel mutant background (Fig. 5F–H), which disrupts a Drosophila NF-κB homolog that is a key mediator of the innate immune pathway (Hedengren et al., 1999). The association between the loss of Xbp1 and innate immune response had been reported in other organisms, such as mice (Adolph et al., 2013; Kaser et al., 2008). In C. elegans, Xbp1 mutants become susceptible to Pseudomonas aeruginosa infection (Richardson et al., 2010). Thus, these observations reveal a correlative relationship between impaired UPR and active innate immune response.
Table 1.
GO terms of genes downregulated in Xbp1 mutant larvae
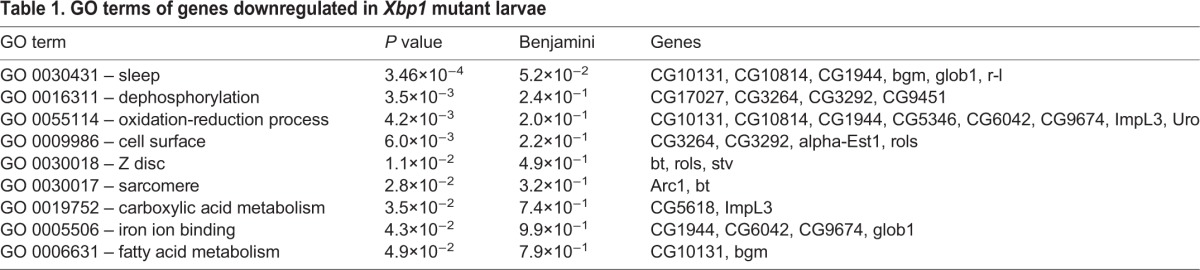
Table 2.
GO terms of genes upregulated in Xbp1 mutant larvae
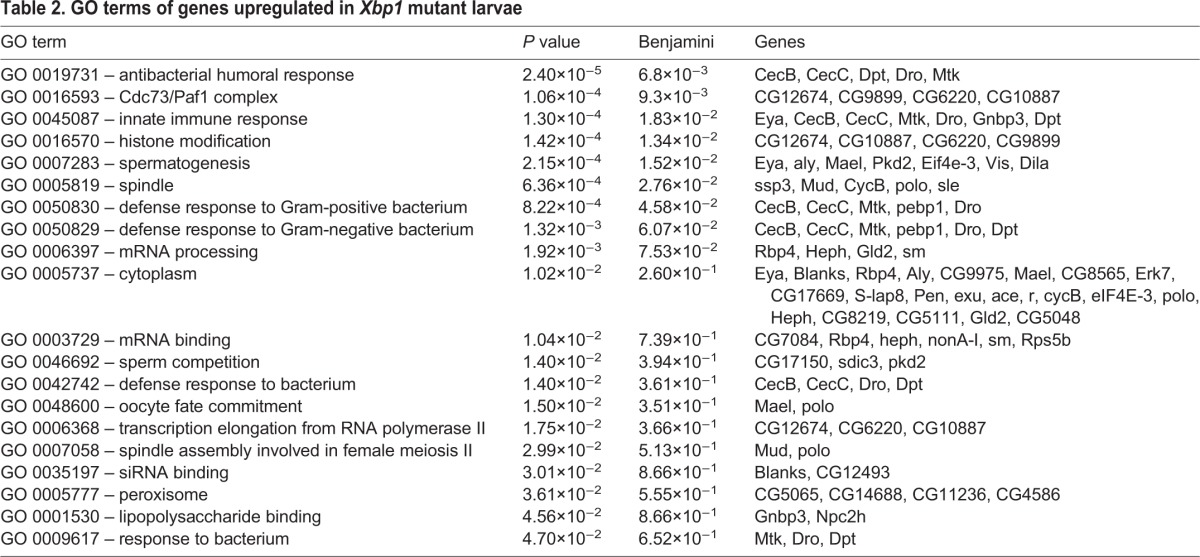
Fig. 5.
The innate immune response is activated in Xbp1 mutant larvae. (A) A select set of genes with altered gene expression (adjusted P value <0.05) in Xbp1 mutant larvae. The full list is in Table S1. The y-axis shows the log2 fold change of RNAseq reads in Xbp1 mutants. Genes shown here are those previously reported to be tunicamycin-inducible genes in the fly (reported in Chow et al., 2013; left 12 bars), and those encoding anti-microbial peptides (right five bars). (B) Network analysis of the genes that are induced in Xbp1 mutants. Those that encode anti-microbial peptides form a major cluster. (C) Validation of AMP transcript induction through qRT-PCR. qRT-PCR against Xbp1 was performed to confirm that the samples were from Xbp1ex79−/− null larvae. As representative AMPs, drosocin, diptericin and drosomycin transcripts were examined. (D,E) drosomycin-GFP, a reporter for an anti-microbial peptide expression (green) in wild-type (D) and Xbp1 mutant (E) second-instar larvae. TO-PRO-3 (blue) was used to show the outline of tissues. D and E are composite images. The scale bar for D and E is shown in D. (F–H) Induction of drosomycin-GFP in the fat body of Xbp1 mutants (wild-type in F and Xbp1−/− in G) is abolished in the mutant background of rel20 (H), which encodes a Rel family of immune responsive transcription factor. The scale bar for F–H is shown in H.
DISCUSSION
Here, we report evidence that IRE1 plays essential developmental roles through its XBP1-dependent and -independent activities during normal Drosophila development and tissue homeostasis. While it was known that IRE1α and XBP1 are developmentally essential in mice (Iwawaki et al., 2009; Kaser et al., 2008; Lee et al., 2008a; Reimold et al., 2001), our study is the first to determine the functional significance of IRE1-mediated XBP1 splicing versus XBP1 independent function during metazoan development. The results show that this branch of UPR is engaged in resolving physiological stress associated with normal development, and the ability of IRE1 to splice XBP1 mRNA plays a particularly prominent role during this process.
In addition to the IRE1 and XBP1 axis, several cell culture studies have revealed XBP1-independent roles of IRE1. One of these is a role for IRE1 in degrading mRNAs other than XBP1 (Hollien et al., 2009; Hollien and Weissman, 2006). Recently, the significance of such a mechanism has been validated in vivo, specifically in the developing Drosophila eye (Coelho et al., 2013). In addition, IRE1 regulates JNK signaling that is independent of the RNase activity of IRE1 (Urano et al., 2000); however, the significance of that signaling axis during metazoan development has not yet been determined. Consistent with the XBP1-independent role of IRE1, we also find that the Ire1 loss-of-function phenotype is generally stronger in the larva than that of Xbp1 mutants, specifically in terms of earlier lethality and stronger induction of the 4E-BPintron dsRed reporter. Our structure–function study provides further hints regarding the two XBP1-independent roles of IRE1: an RNase-dependent process of degrading other mRNAs (RIDD) and an RNase-independent process of JNK signaling. Our data particularly highlights the importance of RNase-dependent IRE1 function, as the RNase-dead allele of Ire1 fails to rescue the first-instar larval lethality and as the degree of 4E-BPintron dsRed induction is indistinguishable from that of Ire1 nulls.
In this study, the use of the 4E-BPintron dsRed reporter has allowed the visualization of specific developing tissues that are impacted by the loss of Ire1 and Xbp1. The results indicate that the midgut, and in particular, the gastric caeca are particularly affected by the loss of IRE1 and XBP1 signaling. Such observations were somewhat unexpected, as there has been no literature implicating gastric caeca and proventriculus with higher physiological stress in the ER, and in adult Drosophila, knockdown of Ire1 shows a rather mild phenotype in triglyceride metabolism and a modest change in adult lifespan (Luis et al., 2016). In fact, not much is known about the precise role of the developing gastric caeca. A small number of digestive enzymes that have been characterized in Drosophila are expressed in the gastric caeca and proventriculus (Grönke et al., 2005; Matsumoto et al., 1995). Taken together, these observations raise the possibility that the gastric caeca is a major exocrine organ in Drosophila, perhaps playing a role similar to that of the mammalian pancreas in synthesizing and secreting digestive enzymes. We speculate that the essential requirements of Ire1 and Xbp1 in the alimentary canal are due to their roles in dealing with physiological levels of ER stress caused by its high secretory protein levels.
Outside the alimentary canal, we find that the accessory glands and the ejaculatory duct of male reproductive organs are particularly sensitive to the loss of Xbp1. This is consistent with our understanding that the accessory gland is a secretory organ that produces seminal fluid proteins (SFPs) (Avila et al., 2011; Monsma et al., 1990). Consistent with this, we had reported previously that this organ shows inherent IRE1 activity as evidenced by the XBP1–GFP-based splicing reporter (Sone et al., 2013). A different reporter for Xbp1 expression in Drosophila specifically marks the accessory gland and the ejaculatory duct (Ryoo et al., 2013). Furthermore, a recent study has found that the accessory gland is particularly vulnerable to conditions that perturb ER protein folding (Chow et al., 2015). The results reported in this study show that these tissues are not only vulnerable to exogenously imposed stress, but also have physiological ER stress that requires Xbp1 for resolution.
In summary, results presented in this study establish the importance of IRE1 and XBP1 signaling in a number of normally developing tissues with no experimentally imposed stress. The results suggest that cells suffer from physiological ER stress as part of the normal developmental program that needs to be resolved through the IRE1 and XBP1 pathway. On the other hand, we do not find evidence that the unconventional roles of IRE1 that are independent of its RNase and luminal domains are necessary during development.
MATERIALS AND METHODS
Constructs
UAS-Xbp1-RA was generated by cloning the entire Xbp1-RA coding sequence into the pUAST plasmid. These constructs were injected into flies using the standard P-element transgenesis technology. UAS-Xbp1U was generated through site-directed mutagenesis of the conserved splice site residues that introduced silent mutations, as shown in Fig. 4A.
Ire1 genomic rescue constructs were generated by cloning the 8.8 kb Ire1 genomic locus through recombineering. Specifically, the sequence between two adjacent genes, CG11447 and CG4662, was cloned from a BAC clone into the P[acman] plasmid using established recombineering protocols (Venken et al., 2006). Subsequently, five tandem repeats of the HA epitope tag were introduced after IRE1 residue 506 using GalK-based recombineering. The epitope tag did not interfere with IRE1 function, as this transgene was able to rescue the lethality of the Ire1−/− mutants. IRE1LD Del was made by deleting the sequence that encodes the amino acid residues from amino acids 55 to 381. IRE1RNase Dead had the H890 replaced by an alanine residue. These P[acman] constructs were injected into flies to generate transgenic lines by using the phi31-integrase, targeted to the chromosomal location 51F.
Fly genetics
Xbp1ex79 (Coelho et al., 2013), Ire1f02170 (Kang et al., 2012), tubulin-Gal4 (Lee and Luo, 1999), dilp2-Gal4 (Rulifson et al., 2002), dcg-Gal4 (Arsham and Neufeld, 2009), NP1-Gal4 (Jiang and Edgar, 2009), cadudal-Gal4 (Ryu et al., 2008), drosomycin-GFP (Ferrandon et al., 1998) and 4E-BPintron dsRed (Kang et al., 2017) were described previously. The Xbp1ex79 allele has a deletion from 260 bp upstream to 748 bp downstream of the start codon, which includes regions that encode the DNA-binding domain and the IRE1-mediated mRNA splice site (Coelho et al., 2013), and the Ire1f02170 allele has a transposon insertion within the coding sequence. The Ire1f02170 line obtained from the Bloomington Stock Center had background lethal mutations, which were cleaned up by backcrossing to the Ryoo laboratory w1118 stock. Such backcrosses resulted in the isogenization of the Ire1f02170 with other alleles generated in the Ryoo laboratory, including Xbp1ex79. The effects with UAS-Xbp1 transgenes were validated with at least two independent lines, which gave consistent results. To generate maternal Xbp1 mutants, we rescued Xbp1−/− flies by re-introducing transgenic Xbp1 through NP1-Gal4/UAS-Xbp1. As shown in Fig. S2, NP1-Gal4 is not active in the female ovary, and therefore, generates Xbp1−/− mothers that cannot deposit Xbp1 mRNA into the oocyte. These females were crossed to 4E-BPintron dsRed, Xbp1/CyO males. Half of the progeny were maternal zygotic Xbp1 mutants (the other half were CyO-containing Xbp1+/+ flies), which were distinguishable through the strong 4E-BPintron dsRed signal visible at the second-instar larval stage.
Survival assays
To assess the degree of Xbp1−/− rescue by Gal4/UAS-mediated transgene expression, crosses were set up with ten males and ten females and the cross was transferred to fresh food every 2 days. The progenies were allowed to develop at 25°C until they reached adulthood. Based on the scheme of the cross, one-third of the flies were expected to have the Xbp1−/− genotype if all flies were to survive equally. For adult survival assays, ∼20 adult flies were reared in each vial and reared at 25°C. The flies were passed to fresh food every 2 days and the number of dead flies were counted at that point.
Immunohistochemistry
The following antibodies were used: rabbit anti-GFP (Invitrogen cat. no. A-6455, 1:500). In addition, TO-PRO-3 (Fisher cat. #T3605) and phalloidin conjugated to Alexa Fluor 647 (Invitrogen cat. no. A22287) were used to visualize nuclei of cells and actin filaments. The antibody labeling was done under the standard conditions, which included 20 min of fixation in 4% paraformaldehyde solution, and subsequent incubation with antibodies in PBS that contained 0.2% Triton X-100.
RNAseq and bioinformatics analysis
RNA was isolated from second-instar larvae (48 h old) of Xbp1ex79−/− and Xbp1+/+ genotypes. As an Xbp1+/+ control, a precise excision line from Xbp1CB02061 was used. The latter is the line that was also used to derive an imprecise excision allele Xbp1ex79 (Coelho et al., 2013). Two independent extractions of RNAs from each genotype were purified through Qiagen RNeasy columns. RNAseq libraries were prepared using the Illumina TruSeq Stranded Total RNA library prep, with Ribozero Gold, starting from 500 ng of DNase I-treated total RNA, following the manufacturer's protocol, with the exception that 13 cycles of PCR were performed to amplify the libraries, to keep the duplication rate lower than with the recommended 15 cycles. The amplified libraries were purified using AMPure beads, quantified by Qubit and qPCR, and visualized in an Agilent Bioanalyzer. The libraries were pooled equimolarly, and loaded at 8 pM, on a high output HiSeq 2500 flow cell, as paired 50 nucleotide reads for sequencing at the NYU Langone Genome Technology Center. The sequencing reads were aligned to the dm6 reference genome using the Tophat/2.1.1 sequencing analysis package, and HTC count 0.6.1 was used to generate raw gene counts. The package DESeq2 was used for differential gene analysis. The full results have been deposited in NCBI's Gene Expression Omnibus (GEO) and are accessible through GEO series accession number GSE99676. The Database for Annotation, Visualization and Integrated Discovery (DAVID) was used for functional annotation (http://david.abcc.ncifcrf.gov/) of genes whose expression changed significantly in Xbp1 mutants (P<0.05). Network analysis for the selected genes was performed using The Search Tool of the Retrieval of Interacting Genes (STRING) databases (http://string-db.org). Analyzed network and expression level of individual genes were visualized by Cytoscape software (ver 3.4.0).
Quantitative real-time PCR
Total RNA was extracted from 48-h-old larvae using TRIzol reagent (Invitrogen). From this, single-stranded cDNA was synthesized by using the Superscript III reverse transcript kit (Thermo Fisher Scientific). Real-time PCR was performed by using a Power SYBR Green PCR master mix (Thermo Fisher Scientific). The following primers were used for RT-PCR: Xbp1 forward, 5′-CGTCGAACATGGATGACGATAA; Xbp1 reverse, 5′-GAGTCCAGCTTGTGGTTCTT-3′; CecA1 forward, 5′-GGGTGGCTGAAGAAAATTGG-3′; CecA1 reverse, 5′-ACATTGGCGGCTTGTTGAG-3′; CecA2 forward, 5′-TGGCAAGAAAATCGAACGTG-3′; CecA2 reverse, 5′-CTCGAGCAGTGGCTGCAA-3′; CecB forward, 5′-CGTCTTTGTGGCACTCATCC-3′; CecB reverse, 5′-CCTGGTATGCTGACCAATGC-3′; CecC forward, 5′-CCGGTTGGCTGAAGAAACTT-3′; CecC reverse, 5′-TCCCAGTCCTTGAATGGTTG-3′; Drososin forward, 5′-GTTTTCCTGCTGCTTGCTTG-3′; Drososin reverse, 5′-GGCAGCTTGAGTCAGGTGAT-3′; Drosomycin forward, 5′-TGCCTGTCCGGAAGATACAA-3′; Drosomycin reverse, 5′-CTCCTCCTTGCACACACGAC-3′; crc forward, 5′-AAAACCCGTGCTCGTAAAGG-3′; and crc reverse, 5′-CGAGCTCCTTAGCACGCATA-3′. The RT-PCR counts with these primers were normalized to that of Drosophila Ribosomal protein L15. Primer sequences to amplify the latter were as follows: Rpl15 forward, 5′-AGGATGCACTTATGGCAAGC-3′ and Rpl15 reverse, 5′-GCGCAATCCAATACGAGTTC-3′.
Statistics and bioinformatics analysis
To calculate the statistical significance of qRT-PCR results, experiments were repeated three times and the results were subjected to t-test analysis. The n number for the total number of flies subjected to lifespan examination in Fig. 4D are indicated in the graph (n=143 for tubulin-Gal4 rescued flies, and n=90 for NP1-Gal4 rescued flies).
Acknowledgements
We thank Eric Baehrecke, Claude Desplan, Tom Neufeld, Jing Wang and Kwon Yu for fly stocks, David D. Sabatini for discussions throughout the project, Won Jae Lee, David Shetlar and Marc Amoyel for various advice, Tenzin Lhakhang for help with bioinformatics, Jun Ho Chang for technical assistance and the members of the Ryoo laboratory for comments on the manuscript. RNAseq library prep, sequencing and analysis were done at the NYU Langone Genome Technology Center.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: D.R., H.R.; Methodology: D.R., H.R.; Formal analysis: T.R., H.R.; Investigation: H.H., X.Z., H.R.; Writing - original draft: H.R.; Writing - review & editing: X.Z., D.R.; Supervision: D.R., H.R.; Project administration: H.R.; Funding acquisition: D.R., H.R.
Funding
This study was supported by the National Institutes of Health (grants R01 EY020866, R01 DK047119), March of Dimes Foundation (grant FY#13-204) to H.D.R., and the Wellcome Trust (200848/Z/16/Z to D.R.). Deposited in PMC for immediate release.
Data availability
RNAseq data from this study have been deposited in the Gene Expression Omnibus (GEO) database under accession number GSE99676 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE99676).
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.203612.supplemental
References
- Adolph T. E., Tomczak M. F., Niederreiter L., Ko H. J., Böck J., Martinez-Naves E., Glickman J. N., Tschurtschenthaler M., Hartwig J., Hosomi S. et al. (2013). Paneth cells as a site of origin for intestinal inflammation. Nature 503, 272-276. 10.1038/nature12599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragón T., van Anken E., Pincus D., Serafimova I. M., Korennykh A. V., Rubio C. A. and Walter P. (2009). Messenger RNA targeting to endoplasmic reticulum stress signalling sites. Nature 457, 736-740. 10.1038/nature07641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsham A. M. and Neufeld T. P. (2009). A genetic screen in Drosophila reveals novel cytoprotective functions of the autophagy-lysosome pathway. PLoS ONE 4, e6068 10.1371/journal.pone.0006068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila F. W., Sirot L. K., LaFlamme B. A., Rubinstein C. D. and Wolfner M. F. (2011). Insect seminal fluid proteins: identification and function. Annu. Rev. Entomol. 56, 21-40. 10.1146/annurev-ento-120709-144823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M., Zeng H., Urano F., Till J. H., Hubbard S. R., Harding H. P., Clark S. G. and Ron D. (2002). IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415, 92-96. 10.1038/415092a [DOI] [PubMed] [Google Scholar]
- Chow C. Y., Wolfner M. F. and Clark A. G. (2013). Using natural variation in Drosophila to discover previously unknown endoplasmic reticulum stress genes. Proc. Natl. Acad. Sci. USA 110, 9013-9018. 10.1073/pnas.1307125110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow C. Y., Avila F. W., Clark A. G. and Wolfner M. F. (2015). Induction of excessive endoplasmic reticulum stress in the Drosophila male accessory gland results in infertility. PLoS ONE 10, e0119386 10.1371/journal.pone.0119386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho D. S., Cairrão F., Zeng X., Pires E., Coelho A. V., Ron D., Ryoo H. D. and Domingos P. M. (2013). Xbp1-independent Ire1 signaling is required for photoreceptor differentiation and rhabdomere morphogenesis in Drosophila. Cell Rep. 5, 791-801. 10.1016/j.celrep.2013.09.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Credle J. J., Finer-Moore J. S., Papa F. R., Stroud R. M. and Walter P. (2005). On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 102, 18773-18784. 10.1073/pnas.0509487102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandon D., Jung A. C., Criqui M. C., Lemaitre B., Uttenweiler-Joseph S., Michaut L., Reichhart J. M. and Hoffmann J. A. (1998). A drosomycin-GFP reporter transgene reveals a local immune response in Drosophila that is not dependent on the Toll pathway. EMBO J. 17, 1217-1227. 10.1093/emboj/17.5.1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner B. M. and Walter P. (2011). Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science 333, 1891-1894. 10.1126/science.1209126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grönke S., Mildner A., Fellert S., Tennagels N., Petry S., Müller G., Jäckler H. and Kühnlein R. P. (2005). Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab. 1, 323-330. 10.1016/j.cmet.2005.04.003 [DOI] [PubMed] [Google Scholar]
- Harding H. P., Zeng H., Zhang Y., Jungries R., Chung P., Plesken H., Sabatini D. D. and Ron D. (2001). Diabetes mellitus and exocrine pancreatic dysfunction in perk−/− mice reveals a role for translational control in secretory cell survival. Mol. Cell 7, 1153-1163. 10.1016/S1097-2765(01)00264-7 [DOI] [PubMed] [Google Scholar]
- Hedengren M., Asling B., Dushay M. S., Ando I., Ekengren S., Wihlborg M. and Hultmark D. (1999). Relish, a central factor in the control of humoral but not cellular immunity in Drosophila. Mol. Cell 4, 827-837. 10.1016/S1097-2765(00)80392-5 [DOI] [PubMed] [Google Scholar]
- Hollien J. and Weissman J. S. (2006). Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 313, 104-107. 10.1126/science.1129631 [DOI] [PubMed] [Google Scholar]
- Hollien J., Lin J. H., Li H., Stevens N., Walter P. and Weissman J. S. (2009). Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J. Cell Biol. 186, 323-331. 10.1083/jcb.200903014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwawaki T., Akai R., Yamanaka S. and Kohno K. (2009). Function of IRE1 alpha in the placenta is essential for placental development and embryonic viability. Proc. Natl. Acad. Sci. USA 106, 16657-16662. 10.1073/pnas.0903775106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H. and Edgar B. A. (2009). EGFR signaling regulates the proliferation of Drosophila adult midgut progenitors. Development 136, 483-493. 10.1242/dev.026955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M. J., Chung J. and Ryoo H. D. (2012). CDK5 and MEKK1 mediate pro-apoptotic signaling following endoplasmic reticulum stress in an autosomal dominant retinitis pigmentosa model. Nat. Cell Biol. 14, 409-415. 10.1038/ncb2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M.-J., Vasudevan D., Kang K., Kim K., Park J.-E., Zhang N., Zeng X., Neubert T. A., Marr M. T. II and Ryoo H. D. (2017). 4E-BP is a target of the GCN2-ATF4 pathway during Drosophila development and aging. J. Cell Biol. 216, 115-129. 10.1083/jcb.201511073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaser A., Lee A.-H., Franke A., Glickman J. N., Zeissig S., Tilg H., Nieuwenhuis E. E., Higgins D. E., Schreiber S., Glimcher L. H. et al. (2008). XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell 134, 743-756. 10.1016/j.cell.2008.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korennykh A. V., Egea P. F., Korostelev A. A., Finer-Moore J., Zhang C., Shokat K. M., Stroud R. M. and Walter P. (2009). The unfolded protein response signals through high-order assembly of Ire1. Nature 457, 687-693. 10.1038/nature07661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korennykh A. V., Korostelev A. A., Egea P. F., Finer-Moore J., Stroud R. M., Zhang C., Shokat K. M. and Walter P. (2011). Structural and functional basis for RNA cleavage by Ire1. BMC Biol. 9, 47 10.1186/1741-7007-9-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. and Luo L. (1999). Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22, 451-461. 10.1016/S0896-6273(00)80701-1 [DOI] [PubMed] [Google Scholar]
- Lee A.-H., Chu G. C., Iwakoshi N. N. and Glimcher L. H. (2005). XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. EMBO J. 24, 4368-4380. 10.1038/sj.emboj.7600903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.-H., Scapa E. F., Cohen D. E. and Glimcher L. H. (2008a). Regulation of hepatic lipogenesis by the transcription factor XBP1. Science 320, 1492-1496. 10.1126/science.1158042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. P., Dey M., Neculai D., Cao C., Dever T. E. and Sicheri F. (2008b). Structure of the dual enzyme ire1 reveals the basis for catalysis and regulation in nonconventional RNA splicing. Cell 132, 89-100. 10.1016/j.cell.2007.10.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis N. M., Wang L., Ortega M., Deng H., Katewa S. D., Li P.-W., Karpac J., Jasper H. and Kapahi P. (2016). Intestinal IRE1 is required for increased triglyceride metabolism and longer lifespan under dietary restriction. Cell Rep. 17, 1207-1216. 10.1016/j.celrep.2016.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto I., Watanabe H., Abe K., Arai S. and Emori Y. (1995). A putative digestive cysteine proteinase from Drosophila melanogaster is predominantly expressed in the embryonic and larval midgut. Eur. J. Biochem. 227, 582-587. 10.1111/j.1432-1033.1995.tb20428.x [DOI] [PubMed] [Google Scholar]
- Monsma S. A., Harada H. A. and Wolfner M. F. (1990). Synthesis of two Drosophila male accessory gland proteins and their fate after transfer to the female during mating. Dev. Biol. 142, 465-475. 10.1016/0012-1606(90)90368-S [DOI] [PubMed] [Google Scholar]
- Morgan N. S., Heintzelman M. B. and Mooseker M. S. (1995). Characterization of myosin-1A and myosin-1B, two unconventional myosins associated with the Drosophila brush border cytoskeleton. Dev. Biol. 172, 51-71. 10.1006/dbio.1995.0005 [DOI] [PubMed] [Google Scholar]
- Promlek T., Ishiwata-Kimata Y., Shido M., Sakuramoto M., Kohno K. and Kimata Y. (2011). Membrane aberrancy and unfolded proteins activate the endoplasmic reticulum stress sensor Ire1 in different ways. Mol. Biol. Cell 22, 3520-3532. 10.1091/mbc.E11-04-0295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimold A. M., Iwakoshi N. N., Manis J., Vallabhajosyula P., Szomolanyi-Tsuda E., Gravallese E. M., Friend D., Grusby M. J., Alt F. and Glimcher L. H. (2001). Plasma cell differentiation requires the transcription factor XBP-1. Nature 412, 300-307. 10.1038/35085509 [DOI] [PubMed] [Google Scholar]
- Richardson C. E., Kooistra T. and Kim D. H. (2010). An essential role for XBP-1 in host protection against immune activation in C. elegans. Nature 463, 1092-1095. 10.1038/nature08762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulifson E. J., Kim S. K. and Nusse R. (2002). Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science 296, 1118-1120. 10.1126/science.1070058 [DOI] [PubMed] [Google Scholar]
- Ryoo H. D. (2015). Drosophila as a model for unfolded protein response research. BMB Rep. 48, 445-453. 10.5483/BMBRep.2015.48.8.099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo H. D., Domingos P. M., Kang M.-J. and Steller H. (2007). Unfolded protein response in a Drosophila model for retinal degeneration. EMBO J. 26, 242-252. 10.1038/sj.emboj.7601477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo H. D., Li J. and Kang M.-J. (2013). Drosophila XBP1 expression reporter marks cells under endoplasmic reticulum stress and with high protein secretory load. PLos ONE 8, e75774 10.1371/journal.pone.0075774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu J.-H., Kim S.-H., Lee H.-Y., Bai J. Y., Nam Y.-D., Bae J.-W., Lee D. G., Shin S. C., Ha E.-M. and Lee W.-J. (2008). Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science 319, 777-782. 10.1126/science.1149357 [DOI] [PubMed] [Google Scholar]
- Shen X., Ellis R. E., Lee K., Liu C.-Y., Yang K., Solomon A., Yoshida H., Morimoto R., Kurnit D. M., Mori K. et al. (2001). Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell 107, 893-903. 10.1016/S0092-8674(01)00612-2 [DOI] [PubMed] [Google Scholar]
- Sone M., Zeng X., Larese J. and Ryoo H. D. (2013). A modified UPR stress sensing system reveals a novel tissue distribution of IRE1/XBP1 activity during normal Drosophila development. Cell Stress Chaperones 18, 307-319. 10.1007/s12192-012-0383-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano F., Wang X., Bertolotti A., Zhang Y., Chung P., Harding H. P. and Ron D. (2000). Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287, 664-666. 10.1126/science.287.5453.664 [DOI] [PubMed] [Google Scholar]
- Venken K. J. T., He Y., Hoskins R. A. and Bellen H. J. (2006). P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314, 1747-1751. 10.1126/science.1134426 [DOI] [PubMed] [Google Scholar]
- Volmer R., van der Ploeg K. and Ron D. (2013). Membrane lipid saturation activates endoplasmic reticulum unfolded protein response transducers through their transmembrane domains. Proc. Natl. Acad. Sci. USA 110, 4628-4633. 10.1073/pnas.1217611110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P. and Ron D. (2011). The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081-1086. 10.1126/science.1209038 [DOI] [PubMed] [Google Scholar]
- Yoshida H., Matsui T., Yamamoto A., Okada T. and Mori K. (2001). XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881-891. 10.1016/S0092-8674(01)00611-0 [DOI] [PubMed] [Google Scholar]
- Zhou J., Liu C. Y., Back S. H., Clark R. L., Peisach D., Xu Z. and Kaufman R. J. (2006). The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response. Proc. Natl. Acad. Sci. USA 103, 14343-14348. 10.1073/pnas.0606480103 [DOI] [PMC free article] [PubMed] [Google Scholar]