Abstract
Duchenne muscular dystrophy (DMD) is an X-chromosome-linked disorder and the most common monogenic disease in people. Affected boys are diagnosed at a young age, become non-ambulatory by their early teens, and succumb to cardiorespiratory failure by their thirties. Despite being a monogenic condition resulting from mutations in the DMD gene, affected boys have noteworthy phenotypic variability. Efforts have identified genetic modifiers that could modify disease progression and be pharmacologic targets. Dogs affected with golden retriever muscular dystrophy (GRMD) have absent dystrophin and demonstrate phenotypic variability at the functional, histopathological, and molecular level. Our laboratory is particularly interested in muscle metabolism changes in dystrophin-deficient muscle. We identified several metabolic alterations, including myofiber type switching from fast (type II) to slow (type I), reduced glycolytic enzyme expression, reduced and morphologically abnormal mitochondria, and differential AMP-kinase phosphorylation (activation) between hypertrophied and wasted muscle. We hypothesize that muscle metabolism changes are, in part, responsible for phenotypic variability in GRMD. Pharmacological therapies aimed at modulating muscle metabolism can be tested in GRMD dogs for efficacy.
Keywords: Duchenne, golden retriever muscular dystrophy, dystrophin, muscle, metabolism, phenotype, AMPK
Introduction
Duchenne muscular dystrophy (DMD) occurs in approximately 1 in 5,000 boys, resulting from out-of-frame mutations in the DMD gene that lead to loss of dystrophin protein and its glycoprotein complex [1]. Disruption of these scaffolding and signaling molecules leads to myofiber instability and progressive muscle wasting and weakness, with patients losing ambulation by their early teens and succumbing to the condition by their thirties. A milder, allelic form of dystrophinopathy, Becker muscular dystrophy (BMD), results from in-frame DMD gene mutations that allow production of a truncated, partially functional protein [2,3]. Becker patients usually live and ambulate for several additional decades [4]. In addition to the pathologic changes directly caused by dystrophin deficiency, so-called genetic modifiers and secondary effects contribute to further phenotypic variation. Studies in the DMD animal models, X-linked muscular dystrophy (mdx) mouse [5] and golden retriever muscular dystrophy (GRMD) dog [6], have played an important role in defining the potential basis for this variability. Based on gene and protein studies, reduced glycolytic and oxidative enzyme expression and activity, a so-called “metabolic crisis,” contributes to differential involvement of both individuals and muscles [7-11]. These metabolic changes may be more pronounced in DMD and GRMD versus the mdx mouse, perhaps contributing to the more severe phenotype of dystrophin deficient boys and dogs. As such, the GRMD canine model appears to be a suitable animal model for muscle metabolism changes and response to respective therapy.
In this study, we evaluated the metabolic changes that occur in the GRMD dog. We hypothesized that GRMD muscle undergoes similar metabolic changes seen in DMD and mdx, including fiber type switching, reduced glycolytic and oxidative metabolism, defective mitochondria, and molecular aberrations. We show for the first time a molecular compensatory mechanism involving AMP-kinase (AMPK), which may drive hypertrophy versus wasting with regards to muscle metabolism.
Methods
Animals. The dogs were used and cared for according to principles outlined in the National Research Council’s Guide for the Care and Use of Laboratory Animals. They were maintained at North Carolina State University, University of Missouri, University of North Carolina, and then at Texas A&M University. Historical data for each dog in the colony were stored in Breeder’s Assistant (Tenset Technologies, Ltd; United Kingdom), including genotype, date of birth, date of death, and any other defining information.
Surgical Muscle Biopsies. Biopsy samples from the cranial sartorius (CS), long digital extensor (LDE), and vastus lateralis (VL) muscles were collected surgically, snap frozen in liquid nitrogen-cooled SUVA34A (a Freon analog; DuPont Fluorochemicals, Wilmington, DE), and subsequently archived at -80°C until analysis.
Functional Outcome Measures. Surgically measured CS muscle circumference, a directly measure of muscle size was performed, as previously described [8].
Light Microscopy. Muscle sample processing, hematoxylin, and eosin (H&E) staining, and light microscopic analysis were performed, also as previously described [8].
Immunofluorescence Microscopy. Muscle samples were stored at -80°C prior to processing. For immunofluorescence, serial muscle cryosections were cut at 7 μm, thawed on Superfrost Plus slides (Fisher Scientific, Pittsburgh, PA), rehydrated and permeabilized in physiological buffered saline (PBS) containing 0.2 percent fish skin gelatin (FSG) and 0.1 percent Triton X-100 for 10 minutes at room temperature (~20°C). Samples were washed two times with PBS. Sections were fixed in cold, 100 percent acetone for 10 minutes and then washed three times with PBS (5 minutes each). Samples were blocked for 1 hour at ~20°C with 5 percent normal goat serum, PBS+0.3 percent Triton X-100. Primary antibody incubation occurred overnight (~17 hours) at 4°C with the following antibodies: mouse monoclonal myosin heavy chain (fast twitch) (NCL-MHCF; Leica Biosystems, Buffalo Grove, IL) and mouse monoclonal myosin heavy chain (slow twitch) (NCL-MHCS; Leica Biosystems, Buffalo Grove, IL). Sections were then washed two times in PBS-FSG-Triton (5 minutes each) and then once with PBS-FSG. Secondary antibody incubation was for 1 hour at ~20°C with goat anti-mouse Alexa Fluor® 594 (A11005; Thermo Fisher, Waltham, MA). Samples were washed two times with PBS-FSG-Triton and then incubated with DAPI (for DNA) for 5 minutes at 20°C. Prolong® Gold Anti-fade reagent (Life Technologies, Carlsbad, CA) was placed on the sections followed by coverslips. Images of the sections were viewed on a Nikon Eclipse 80i microscope and collected for analysis with NIS-Elements Basic Research software (Laboratory Imaging, Version 3.22.14). Further imaging processing was performed in ImageJ software (National Institute of Health, Version 1.48).
Transmission Electron Microscopy (TEM). Cranial sartorius and VL muscle samples were collected at necropsy from 6- to 12-month old GRMD and normal dogs and routinely prepared for TEM. Samples were fixed immediately in 3 percent glutaraldehyde + 2 percent paraformaldehyde buffered with 0.1 M sodium cacodylate (pH 7.4) for 1 hour. Individual muscle fascicles were dissected from each muscle sample and placed in fresh fixative overnight at 4°C. Muscle fascicles were then removed from the fixative and washed with 0.1 M sodium cacodylate solution. Following fixation, tissues were treated with 2 percent OsO4+1.5 percent K Ferrocyanide in 0.1 M sodium cacodylate for 5 hours at room temperature. Next, tissues were washed three times with water for 5 minutes each and then incubated with 2 percent uranyl acetate in distilled water for 3 hours. Following uranyl acetate treatment, the tissue was washed overnight in water at room temperature and three times with water prior to dehydration. Tissues were dehydrated using a graded series of absolute ethanol solutions, finishing with absolute acetone, and then incubated overnight with 1/3 EMbed 812 Resin (Electron Microscopy Sciences, Hatfield, PA) and 2/3 absolute acetone at room temperature. Next, samples were incubated with 2/3 Resin and 1/3 absolute acetone at room temperature for 8 hours and then overnight in 100 percent Resin. Finally, tissues were placed in molds, embedded in Resin, and baked at 60°C for ~72 hours. Thin sections (80 nm) were placed on formvar-coated mesh grids and copper mesh grids for staining with uranyl acetate and lead citrate. Images were collected from an FEI Transmission Electron Microscope in the Image Analysis Laboratory at Texas A&M University and digitally captured for analysis. Further imaging processing was performed in ImageJ.
Protein Extraction and Western Blot Analysis. Stain-free BioRad gels (12 percent, 1.5mm) were prepared using the TGX Stain-Free Fast Cast Acrylamide Starter Kit. Muscle tissue from normal and GRMD CS and LDE samples at 6 months of age were homogenized and quantified, as previously described [12]. One hundred μg of protein was mixed with Laemmli two times electrophoresis buffer until the total volume reached (20 μL). The samples were heated at 95°C for 4 minutes, and then each sample was loaded into the gel along with 10 μL of Novex Sharp prestained ladder (Invitrogen; Carlsbad, CA). Duplicate samples were loaded into two separate gels, which were electrophoresed for 35 minutes at 200 V and then a picture was taken using BioRad ImageLab 4.1 and Gel Doc EZ (Hercules, CA). The proteins were transferred to two separate PVDF membranes for 1 h at 100 V. The membranes were blocked in 5 percent bovine serum albumin (BSA) and 0.1 percent tween TBST at 4°C overnight. Membranes were then vigorously washed three times with 0.1 percent tween in TBST for 5 minutes at room temperature. AMPKα-rabbit antibody (detects total AMPKα1 and α2 subunits; Cell Signaling Technology #2532; Danvers, MA), at a dilution of 1:1,000 in 5 percent BSA/TBST, was applied to one membrane. PhosphoAMPKα-rabbit antibody (detects both phosphorylated AMPKα1 and α2 subunits at threonine 172; Cell Signaling Technology #2535) at a dilution of 1:500 in 5 percent BSA/TBST, was applied to the other membrane. Membranes incubated overnight at 4°C and then vigorously washed three times with 0.1 percent tween in TBST for 5 min at room temperature. Membranes were treated with Jackson ImmunoResearch (West Grove, PA) goat anti-rabbit IgG horseradish peroxidase secondary antibody at a dilution of 1:5,000 in 5 percent BSA/TBST for 1 hour under dark conditions at room temperature. Membranes were washed three times in 0.1 percent tween TBST for 5 minutes, washed one time with TBS for 5 minutes, incubated in Western Blotting Luminol Reagent: sc-2048 (Dallas, TX), and finally, exposed to radiographic film. Blots were quantified with BioRad ImageLab 4.1 and Gel Doc EZ. PhosphoAMPKα was normalized to total AMPKα for normal and GRMD CS and LDE samples.
Proteomic Profiling. Label-free, quantitative mass spectrometry was performed in CS muscle samples from three each GRMD and normal dogs at age 4 to 9 weeks and 6 months of age, as previously described [8].
Statistical Analysis. Prism software version 6 (GraphPad Software, La Jolla, CA) was used for statistical testing and schematic graph generation. A nonparametric t-test was used to test significance between GRMD and normal dog expression values. A p-value cut-off of 0.05 was considered statistically significant.
Results
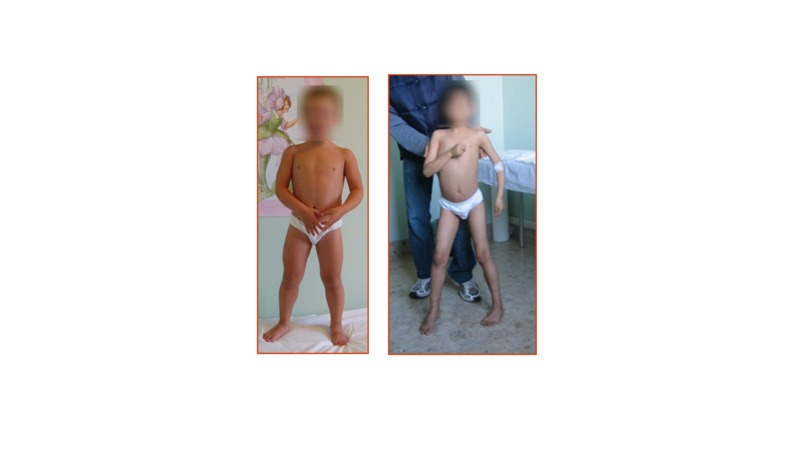
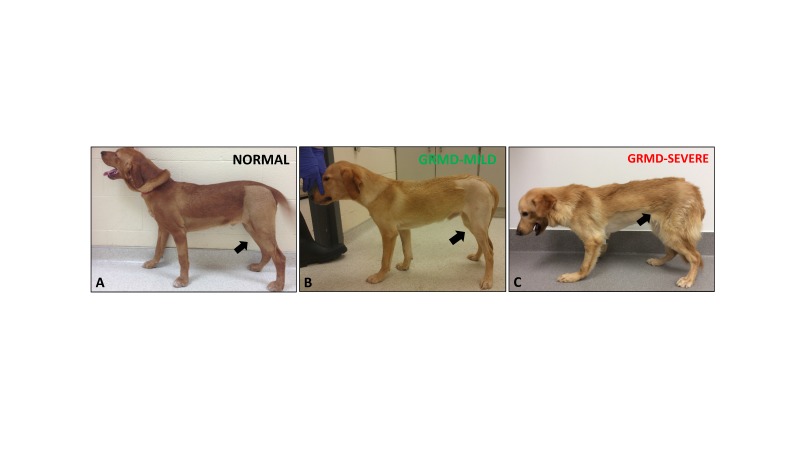
Phenotypic variability in dystrophin deficiency is associated with genetic modifiers. Over and above phenotypic variation due to in-frame vs. out-of-frame mutations, resulting in variable levels of dystrophin expression, the secondary effects of dystrophin deficiency can lead to striking functional differences in DMD boys. For instance, time to wheelchair can vary markedly from 7 to 17 years of age in the era of glucocorticoid therapy with some boys exhibiting severe muscle wasting, weakness, and postural deficits, while other boys have a milder phenotype (Figure 1) [13]. With regards to animal models, dystrophin-deficient GRMD dogs and DMD boys share pathogenetic mechanisms. Affected dogs also show phenotypic variability, including muscle weakness and postural abnormalities, despite having the same DMD gene mutation (Figure 2) [8].
Figure 1.
Phenotypic variation was observed in DMD boys. Left: Mildly affected DMD patient. Note the apparent leg musculature enlargement. Right: Severely affected DMD boy with muscle wasting and weakness in the arms and legs. Note the boy needed assistance to stand. Photos courtesy of L.B.
Figure 2.
GRMD dogs showed significant variation in phenotype. A) Normal dog (Winthrop) at 6 months of age. B) Mildly affected GRMD dog (Lunes) at 6 months of age. Note the mild muscle atrophy of the pelvic limbs (black arrows) and mild hyperextension of the carpal joints. C) Severely affected GRMD dog (Kermit) at 6 months. Note the prominent joint angle changes and postural deficits, particularly at the carpal, tibiotarsal, stifle, and hip joints, which were due to severe muscle weakness/wasting and joint contractures.
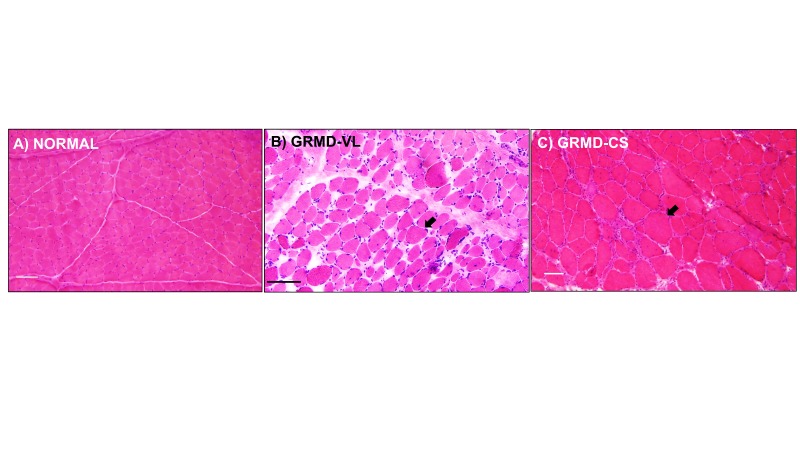
Phenotypic variability is observed among different dystrophin deficient muscles. In DMD, GRMD, and mdx, histopathological variability occurs at the individual muscle level. Most muscles, such as the VL head of the quadriceps, undergo progressive dystrophy, with initial necrosis followed by immune cell infiltration and fibrosis (Figure 3B) [8]. On the other hand, the sartorius muscle of DMD boys is spared. Similarly, after a period of early necrosis, the CS of GRMD dogs regenerates and may undergo paradoxical hypertrophy by 6 months of age. (Figure 3C) [14,15].
Figure 3.
H&E staining: Muscles from GRMD dogs were variably affected. A) Normal muscle. Note the similar-sized myofibers, minimal connective tissue, and lack of immune cell infiltration. B) VL muscle of a GRMD dog showed degeneration and regeneration (myofiber size variation, black arrow), increased fibrosis and connective tissue, and increased immune cell infiltration. C) The CS muscle and its myofibers in GRMD were hypertrophied (black arrow), as noted by their increased myofiber diameter compared to GRMD VL and normal muscle. Marker = 50 microns.
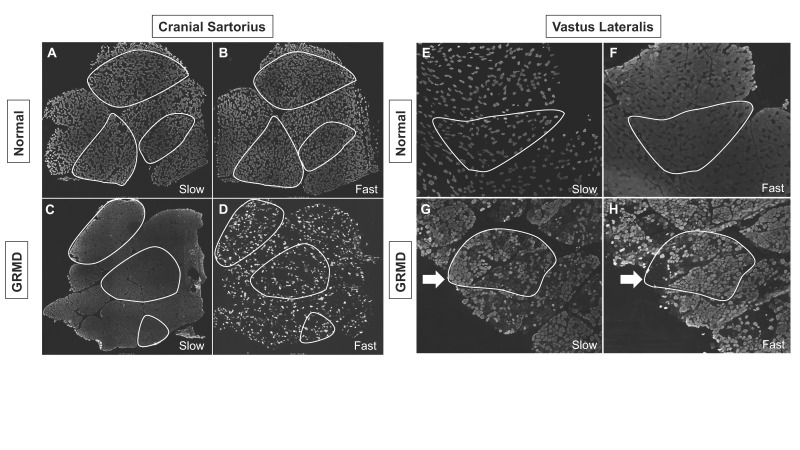
Dystrophin-deficient skeletal muscle undergoes myofiber type switching. In further evaluating the molecular signatures associated with phenotypic variability, we evaluated the well characterized switching of fiber types, as detected by fiber-type-specific myosin heavy chain (MHC) staining [16]. Indeed, we detected reduced fast twitch (type II) and increased slow twitch (type I) staining in both hypertrophied GRMD CS and wasted VL muscles compared to normal (Figure 4).
Figure 4.
Dystrophin-deficient GRMD dog muscle underwent fiber type switching. In GRMD cranial sartorius (CS) and vastus lateralis (VL) muscle, there was reduction in the number of fast staining fibers, while nearly every fiber stained with the myosin heavy chain (MHC) slow twitch antibody (gray colors). A: Normal CS: Slow twitch MHC staining. B: Normal CS: Fast twitch MHC staining. C: GRMD CS: Slow twitch MHC staining. D: GRMD CS: Fast twitch MHC staining. E: Normal VL: Slow twitch MHC staining. F: Normal VL: Fast twitch MHC staining. G: GRMD VL: Slow twitch MHC staining. H: GRMD VL: Fast twitch MHC staining. All dogs shown at ~6 months of age. Images A-D were 3X3 stitched 4X objective images displaying the entire muscle biopsy sections. Images E-H were single images collected with a 4X objective. Outlined white areas highlight representative serially sectioned muscle fascicles used for identifying slow and fast twitch fiber type variation. White arrows indicate serially sectioned area in the GRMD VL that had increased slow twitch staining.
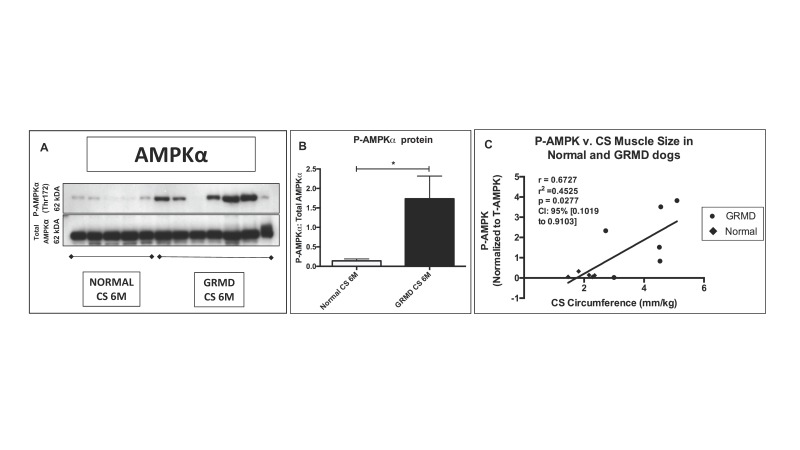
AMP-kinase (AMPK) modifies the dystrophic process at the molecular level. Several studies in the mdx mouse have demonstrated a beneficial effect of AMPK activation [17-19]. In an analogous way, we found that AMPKα phosphorylation (activation) was increased in the spared/hypertrophied GRMD CS muscle compared to normal dogs (Figures 5A and 5B), while GRMD and normal levels were comparable in the LDE muscle (wasted and measuring half the size of normal in GRMD dogs) [14]. Interestingly, we observed a positive correlation between phosphoAMPKα and surgically measured CS muscle size in GRMD dogs at 6 months of age (Figure 5C).
Figure 5.
The hypertrophied CS muscle had increased phosphorylated (P) AMPKα compared to normal. A) Note the variability in western blot expression of P-AMPKα in GRMD dog samples. B) P-AMPKα was normalized to Total AMPKα protein and increased compared to normal samples. * = p < 0.05. C) P-AMPKα was positively correlated with CS circumference.
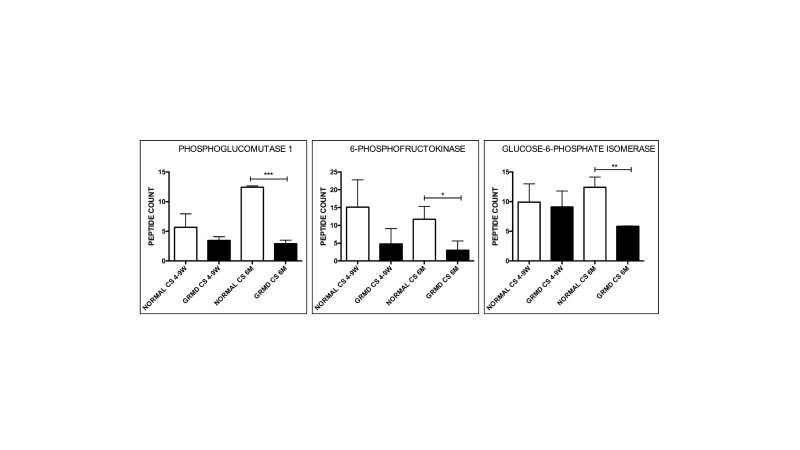
Dystrophin-deficient muscle has reduced glycolytic and oxidative capacity. Regardless of the molecular pathway(s) responsible, dystrophic muscle is under a higher metabolic demand due to a change in glycolytic and oxidative metabolism. The Hoffman laboratory performed genome-wide mRNA expression profiling on DMD muscle and found reduced glycolytic and oxidative enzyme expression compared to normal, suggesting a secondary metabolic defect [7]. Using proteomic profiling via mass spectrometry [8], we also detected similar changes in glycolytic enzyme expression in the GRMD CS muscle at 6 months of age, specifically a reduction in phosphoglucomutase 1, 6-phosphofructokinase, and glucose-6-phosphate isomerase (Figure 6).
Figure 6.
Glycolytic enzymes were reduced in hypertrophied GRMD CS muscle at 6 months of age compared to normal. Phosphoglucomutase-1, 6-phophofructokinase, and glucose-6-phosphate isomerase were reduced at 6 months (M) in GRMD cranial sartorius (CS) (black bars) compared to normal (white bars). There was not a significant change at 4 to 9 weeks (W). N = 3 for each group. Total spectral counts were used to determine relative abundance. * = P < 0.05; ** p < 0.01; *** p < 0.001.
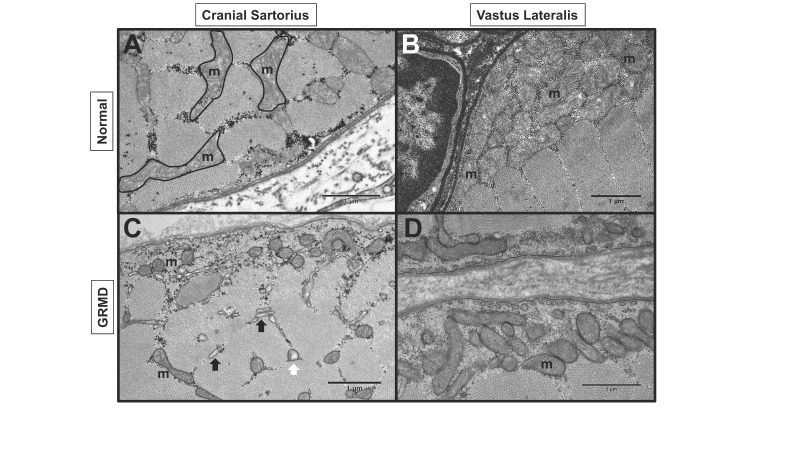
Dystrophic myofibers have reduced mitochondrial numbers and activity. Given the changes in metabolic expression, mitochondrial structure and function was a logical area for further inquiry in evaluating oxidative capacity. In mdx muscle, down-regulation of key metabolic enzymes was associated with reduced mitochondria number and ATP-producing activity; furthermore, mitochondria were reduced in areas where dystrophin was absent from the myofiber membrane [20]. In GRMD muscle, we also observed fewer than normal and swollen mitochondria in both the hypertrophied CS and wasted VL muscle (Figure 7).
Figure 7.
GRMD dog muscle had abnormal mitochondria (m). Transmission electron microscopy (TEM) of normal and GRMD cranial sartorius (CS) and vastus lateralis (VL) at 6 to 12 months of age. A: Normal CS with intermyofibrillar mitochondria. B: Normal VL with numerous subsarcolemmal mitochondria C: GRMD CS with reduced size and density of subsarcolemmal and intermyofibrillar mitochondria, swollen (white arrow) mitochondria and dilated sarcoplasmic reticulum (black arrows). D: GRMD VL with reduced subsarcolemmal mitochondria.
Discussion
DMD boys and GRMD dogs have progressive muscle weakness, but phenotypic variability is observed among individuals and within different muscle groups [2,8]. Although some phenotypic variation can be explained by the amount of dystrophin expression in muscle [2-4], genes outside the DMD gene may modify the disease process. Giacopelli et al. described a polymorphism in the promoter of the SPP1 gene, encoding the cytokine osteopontin (OPN), which reduced OPN expression at baseline [21]. Pegoraro et al. described the same, minor allele, which was associated with a more severe phenotype in DMD [22,23]. Given that the genotype seemed to alter longitudinal, functional changes in DMD (i.e. a faster deterioration of ambulatory function was observed with the minor allele) and also increase eccentric contraction damage by affecting Sp1 binding sites [24], we recommended OPN be considered as a covariate in DMD clinical trials [25]. A draft guidance for drug development in DMD, initiated by patient advocates and submitted to the FDA, suggested that genetic modifiers should be used in post-hoc analyses of clinical trial data [26]. Flanigan and colleagues described an additional DMD modifier, a coding haplotype in LTPB4, which segregated with age at loss of ambulation [27]. A more recent exome-chip based association study found that a single nucleotide polymorphism in the CD40 gene, encoding a T helper costimulatory protein, affected function in DMD [28]. Definition of complex molecular mechanisms that explain the effects of modifier polymorphisms should improve understanding of disease pathophysiology and allow modifier proteins to be manipulated pharmacologically. As an example, pharmacological blockade of LTBP4 was suggested as a promising therapeutic strategy for fibrotic disorders such as DMD [29]. Genome-wide mRNA/microRNA expression (Affymetrix chip) and proteomic profiling (mass spectrometry) in GRMD dogs from our colony demonstrated that SPP1 was associated with several key molecular pathways and could serve as a biomarker for progression [11,30]. Although the SPP1 promoter has not been evaluated in a similar fashion in GRMD dogs, we believe osteopontin significantly modifies the disease process. Taken together, these studies strongly suggest that genetic modifiers influence the phenotype of dystrophin deficient individuals.
This phenotypic variation confounds assessment of outcome parameters in preclinical trials, which are typically conducted between 3 and 6 months of age [10,29], a period that corresponds to ages 5 to 10 years in DMD [31,32]. Dogs enrolled in these studies are humanely euthanized at 6 months of age. However, a number of mildly affected GRMD dogs are maintained beyond 6 months for use as breeders. We retrospectively evaluated our entire GRMD colony perpetuated over years and observed phenotypes ranging from near normal appearance and muscle strength to a fulminating deadly neonatal disorder [33]. In particular, mildly affected GRMD dogs that have lived 3 years of age or greater were characterized. With regard to genetic modifiers, a recent study from a Brazilian GRMD colony identified a Jagged1 polymorphism in so-called “escaper” GRMD dogs that lived to 9 years of age and contrasted their genetic profile with a group of dogs that died before 5 years [34]. The study concluded that the Jagged1 polymorphism contributed to the mild phenotype, presumably because of enhanced muscle regeneration. However, none of the mildly affected GRMD dogs from our colony for which DNA samples were available had the Jagged1 polymorphism. This is not surprising in that the Jagged1 mutation identified in the Brazilian colony traced to an outcross and would not be expected in other colonies. As indicated by our earlier studies, other genetic modifiers such as osteopontin, myostatin, dystroglycan, and myotrophin are probably associated with phenotypic variability in GRMD dogs [8,10,30]. Proof of concept that modifiers can cross species barriers and influence the DMD phenotype was provided by identification of the LTBP4 locus in a delta-sarcoglycan deficient mouse strain, carrying a 36 bp insertion/deletion [35], before a different modifier haplotype was identified in the homologous human gene [27].
In DMD and its animal models, phenotypic variability occurs at the histopathological level. In the GRMD dog, this differential muscle involvement is thought to arise at least partially from the opposing of flexors versus extensor muscles, with the former being affected early in life, as the animal crawls, and the latter spared [36]. As the individual begins to ambulate, extensors become more affected. An alternative, not mutually exclusive explanation might be that specific muscles have peculiar fiber type composition, innervation, and gene expression patterns. This is intriguing from a therapeutic standpoint; identifying molecular signatures associated with muscle sparing/hypertrophy could provide molecular targets for therapeutic development [8,37].
In further evaluating the molecular signatures associated with muscle hypertrophy versus wasting in dystrophin deficiency, changes in muscle metabolism are of great interest to our lab. As such, evaluating myofiber type composition would be a logical first step. Type II (fast-twitch) fibers seem to be preferentially affected (and reduced in number) in dystrophin-deficient muscle, most likely due to reduced oxidative-phosphorylative and increased glycolytic capacity of these fiber types. On the other hand, type I (slow twitch) fibers have increased oxidative capacity, are relatively spared and even increased in number compared to type II, suggesting a metabolic switch from glycolytic to oxidative metabolism [16]. We showed in this study that GRMD dogs undergo fiber switching from fast to slow twitch, but no obvious differences between hypertrophied and wasted muscle. In the GRMD (CXMDJ) Japanese colony, perpetuated from a descendent of our colony, researchers also showed type II to I fiber type switching in dystrophic muscle [38]. In healthy animals, cross-innervation of muscles with alpha motor neurons of an opposing nerve resulted in fiber type switching [39]. In dystrophic muscle, the molecular signatures responsible for the fast to slow switch are mediated, in part, by the folliculin interacting protein-1 (fnip-1) – AMPK pathway [40].
In the context of phenotypic variability at the muscle level, we showed that phosphorylated AMPKα1- and -α2 are increased in hypertrophied muscle. Moreover, P-AMPK was directly correlated with surgically measured CS muscle size, suggesting the energy sensor kinase may promote muscle hypertrophy. These data are supported by the fact that the wasted LDE, a muscle measuring half its normal size in GRMD dogs [14], showed similar phosphoAMPKα expression compared to normal muscle. Pharmacological activation of the kinase was associated with neuronal nitric oxide synthase (nNOS) activation and increased nitric oxide production in mdx mouse cardiomyocytes [17]. The authors suggested that the mechanical stretch-mediated AMPK phosphorylation of nNOS was dependent on an intact DGC; pharmacological activation of AMPK in dystrophin-deficient mdx muscle restored this function [17]. Indeed, another study in mdx mice showed that administration of the AMPK activator, metformin, increased downstream expression of several muscle proteins, including peroxisome proliferator-activated receptor Ɣ Co-activator 1α and the dystrophin homologue utrophin [18]. Providing further evidence of the importance of AMPKα1, mdx mice treated with the AMPK agonist, 5-aminoimidazole-4-carboxamide-1-β-D-ribofuranoside (AICAR) had improved function, reduced creatine kinase levels, and increased oxidative metabolism proteins [19]. The previous mdx studies and the GRMD data presented here suggest that AMPKα activation may have a protective and even beneficial effect in dystrophic muscle. Further studies are underway to activate AMPK in the GRMD dog and evaluate changes in phenotype and downstream molecular signatures.
The accumulating data outline the increased metabolic demand that occurs secondarily to dystrophin deficiency. Chen and colleagues evaluated microarray profiles of DMD muscle and found reduced glycolytic and oxidative metabolism genes [7]. In mdx mice, there was a reduction in lactate dehydrogenase (marker for glycolytic enzyme) activity [9]. We also showed in this study several glycolytic enzymes reduced at the protein level in GRMD muscle, as detected by proteomic profiling and mass spectrometry [8]. Terrill and colleagues revealed this glycolytic/oxidative phosphorylative enzyme reduction was due to protein thiol oxidation [41]. A quantitative proteomic profile of GRMD versus normal muscle confirmed a defect of metabolic proteins, many regulated by PGC-1α [42].
In further evaluating metabolic capacity, we evaluated mitochondria localization in GRMD muscles. The ATP producing organelles are normally dispersed throughout myofibers and interface with the sarcolemmal membrane [43]. A recent study by Percival and colleagues showed a reduction in mitochondria in mdx mice and suggested the metabolic defect was directly related to dystrophin deficiency [20]. We also observed reduced and swollen mitochondria in both dystrophin-deficient hypertrophied and wasted muscle. A recent study evaluated muscle damage induced by cardiotoxin injection and showed similar histopathological, ultrastructural, and metabolic profiles compared to dystrophic muscle [44]. Indeed, the reduced glycolytic enzyme expression and mitochondria observed in our GRMD dogs may be due to the inflammation and muscle degeneration that occurs during the dystrophic process. Nevertheless, our lab is interested in studying mitochondrial activity and pharmacologic approaches that can enhance mitochondrial function.
Conclusion
The role of genetic modifiers in altering muscle metabolism and its association with phenotypic variability is an area of active investigation. In addition to well-documented changes in the mdx mouse model, we have identified analogous GRMD abnormalities, including fiber type switching, glycolytic enzyme reduction, mitochondrial defects, and AMPKα activation in hypertrophied muscle. Given the similar progressive phenotype observed between GRMD and DMD, affected dogs appear to be a suitable model for the metabolic crisis occurring in dystrophic muscle. Pharmacological approaches to enhance muscle metabolism in dystrophin-deficient individuals should be explored using the GRMD dog for efficacy.
Acknowledgments
We acknowledge Dr. Jack Guo from the Kornegay laboratory for providing images of affected dogs; Dr. Jyoti Jaiswal for experiment-related discussions. Experiments in this study were funded by a startup package (PPN) from the Department of Veterinary Integrative Biosciences and Texas A&M University.
Glossary
- AMPK
AMP kinase
- CS
cranial sartorius muscle
- DMD
Duchenne muscular dystrophy
- GRMD
golden retriever muscular dystrophy
- LDE
long digital extensor muscle
- MHC
myosin heavy chain
- PCR
polymerase chain reaction
- SPP1
secreted phosphoprotein 1 or osteopontin
- VL
vastus lateralis muscle
Author Contributions
PPN directed the experimental studies, developed hypotheses, and wrote the manuscript. LB and JNK contributed valuable insight in hypotheses and manuscript content. WBS, AHV, SML, BVH, KNH, TRG, AKB, CJBA, & HHB performed experiments, animal husbandry, and/or contributed to the manuscript content.
References
- Hoffman EP, Brown RH, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. [DOI] [PubMed] [Google Scholar]
- Monaco AP, Bertelson CJ, Liechti-Gallati S, Moser H, Kunkel LM. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics. 1988;2(1):90–95. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A, Van Deutekom JC, Fokkema IF, Van Ommen GJ, Den Dunnen JT. Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve. 2006;34(2):135–144. [DOI] [PubMed] [Google Scholar]
- Emery AE. The muscular dystrophies. Lancet. 2002;359(9307):687-95. [DOI] [PubMed] [Google Scholar]
- Bulfield G, Siller WG, Wight PA, Moore KJ. X chromosome-linked muscular dystrophy (mdx) in the mouse. Proc Natl Acad Sci USA. 1984;81(4):1189–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornegay JN, Tuler SM, Miller DM, Levesque DC. Muscular dystrophy in a litter of golden retriever dogs. Muscle Nerve. 1988;11(10):1056–1064. [DOI] [PubMed] [Google Scholar]
- Chen YW, Zhao P, Borup R, Hoffman EP. Expression profiling in the muscular dystrophies: identification of novel aspects of molecular pathophysiology. J Cell Biol. 2000;151(6):1321–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nghiem PP, Hoffman EP, Mittal P, Brown KJ, Schatzberg SJ, Ghimbovschi S, Wang Z, Kornegay JN. Sparing of the Dystrophin-Deficient Cranial Sartorius Muscle Is Associated with Classical and Novel Hypertrophy Pathways in GRMD Dogs. Am J Pathol. 2013;183(5):1411–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayavarapu S, Coley W, Cakir E, Jahnke V, Takeda S, Aoki Y, Grodish-Dressman H, Jaiswal JK, Hoffman EP, Brown KJ, et al. Identification of disease specific pathways using in vivo SILAC proteomics in dystrophin deficient mdx mouse. Mol Cell Proteomics. 2013;12(5):1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornegay JN, Spurney CF, Nghiem PP, Brinkmeyer-Langford CL, Hoffman EP, Nagaraju K. Pharmacologic management of Duchenne muscular dystrophy: target identification and preclinical trials. ILAR J. 2014;55(1):119–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo CL, Soslow JH, Brinkmeyer-Langford CL, Gupte M, Smith HM, Sengsayadeth S, Sawyer DB, Benson DW, Kornegay JN, Markham LW. Translating golden retriever muscular dystrophy microarray findings to novel biomarkers for cardiac/skeletal muscle function in Duchenne muscular dystrophy. Pediatr Res. 2016;79(4):629–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nghiem PP, Bello L, Balog-Alvarez C, López SM, Bettis A, Barnett H, Hernandez B, Schatzberg SJ, Piercy RJ, Kornegay JN. Whole genome sequencing reveals a 7 base-pair deletion in DMD exon 42 in a dog with muscular dystrophy. Mamm Genome. 2017;28(3-4):106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald CM, Henricson EK, Abresch RT, Han JJ, Escolar DM, Florence JM, Duong T, Arrieta A, Clemens PR, Hoffman EP, et al. Cinrg Investigators. The cooperative international neuromuscular research group Duchenne natural history study—a longitudinal investigation in the era of glucocorticoid therapy: design of protocol and the methods used. Muscle Nerve. 2013;48(1):32–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornegay JN, Cundiff DD, Bogan DJ, Bogan JR, Okamura CS. The cranial sartorius muscle undergoes true hypertrophy in dogs with golden retriever muscular dystrophy. Neuromuscul Disord. 2003;13(6):493–500. [DOI] [PubMed] [Google Scholar]
- Marden FA, Connolly AM, Siegel MJ, Rubin DA. Compositional analysis of muscle in boys with Duchenne muscular dystrophy using MR imaging. Skeletal Radiol. 2005;34(3):140–148. [DOI] [PubMed] [Google Scholar]
- Marini JF, Pons F, Leger J, Loffreda N, Anoal M, Chevallay M, Fardeau M, Leger JJ. Expression of myosin heavy chain isoforms in Duchenne muscular dystrophy patients and carriers. Neuromuscul Disord. 1991;1(6):397–409. [DOI] [PubMed] [Google Scholar]
- Garbincius JF, Michele DE. Dystrophin-glycoprotein complex regulates muscle nitric oxide production through mechanoregulation of AMPK signaling. Proc Natl Acad Sci USA. 2015;112(44):13663–13668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljubicic V, Jasmin BJ. Metformin increases peroxisome proliferator-activated receptor γ Co-activator-1α and utrophin a expression in dystrophic skeletal muscle. Muscle Nerve. 2015;52(1):139–142. [DOI] [PubMed] [Google Scholar]
- Bueno Júnior CR, Pantaleão LC, Voltarelli VA, Bozi LH, Brum PC, Zatz M. Combined effect of AMPK/PPAR agonists and exercise training in mdx mice functional performance. PLoS One. 2012;7(9):e45699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival JM, Siegel MP, Knowels G, Marcinek DJ. Defects in mitochondrial localization and ATP synthesis in the mdx mouse model of Duchenne muscular dystrophy are not alleviated by PDE5 inhibition. Hum Mol Genet. 2013;22(1):153–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacopelli F, Marciano R, Pistorio A, Catarsi P, Canini S, Karsenty G, Ravazzolo R. Polymorphisms in the osteopontin promoter affect its transcriptional activity. Physiol Genomics. 2004;20(1):87–96. [DOI] [PubMed] [Google Scholar]
- Pegoraro E, Hoffman EP, Piva L, Gavassini BF, Cagnin S, Ermani M, Bello L, Soraru G, Pacchioni B, Bonifati MD, et al. Cooperative International Neuromuscular Research Group SPP1 genotype is a determinant of disease severity in Duchenne muscular dystrophy. Neurology. 2011;76(3):219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barp A, Bello L, Politano L, Melacini P, Calore C, Polo A, Vianello S, Sorarù G, Semplicini C, Pantic B, et al. Genetic Modifiers of Duchenne Muscular Dystrophy and Dilated Cardiomyopathy. PLoS One. 2015;10(10):e0141240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barfield WL, Uaesoontrachoon K, Wu CS, Lin S, Chen Y, Wang PC, Kanaan Y, Bond V, Hoffman EP. Eccentric muscle challenge shows osteopontin polymorphism modulation of muscle damage. Hum Mol Genet. 2014;23(15):4043–4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello L, Piva L, Barp A, Taglia A, Picillo E, Vasco G, Pane M, Previtali SC, Torrente Y, Gazzerro E, et al. Importance of SPP1 genotype as a covariate in clinical trials in Duchenne muscular dystrophy. Neurology. 2012;79(2):159–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA Draft Guidance on Duchenne. Available from: http://www.parentprojectmd.org/site/PageServer?pagename=Advocate_fdaguidance .
- Flanigan KM, Ceco E, Lamar KM, Kaminoh Y, Dunn DM, Mendell JR, King WM, Pestronk A, Florence JM, Mathews KD, et al. United Dystrophinopathy Project LTBP4 genotype predicts age of ambulatory loss in Duchenne muscular dystrophy. Ann Neurol. 2013;73(4):481–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello L, Flanigan KM, Weiss RB, United Dystrophinopathy Project Spitali P, Aartsma-Rus A, Muntoni F, Zaharieva I, Ferlini A, Mercuri E, Tuffery-Giraud S, Claustres M, Straub V, Lochmüller H, Barp A, Vianello S, Pegoraro E, Punetha J, Gordish-Dressman H, Giri M, McDonald CM, Hoffman EP; Cooperative International Neuromuscular Research Group. Association Study of Exon Variants in the NF-κB and TGFβ Pathways Identifies CD40 as a Modifier of Duchenne Muscular Dystrophy. Am J Hum Genet. 2016;99(5):1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceco E, Bogdanovich S, Gardner B, Miller T, DeJesus A, Earley JU, Hadhazy M, Smith LR, Barton ER, Molkentin JD, et al. Targeting latent TGFβ release in muscular dystrophy. Sci Transl Med. 2014;6(259):259ra144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornegay JN, Bogan JR, Bogan DJ, Childers MK, Li J, Nghiem P, Detwiler DA, Larsen CA, Grange RW, Bhavaraju-Sanka RK, et al. Canine models of Duchenne muscular dystrophy and their use in therapeutic strategies. Mamm Genome. 2012;23(1-2):85–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patronek GJ, Waters DJ, Glickman LT. Comparative longevity of pet dogs & humans: implications for gerontology research. J Gerontol A Biol Sci Med Sci. 1997;52:B171–B178. [DOI] [PubMed] [Google Scholar]
- Kornegay JN, Childers MK. Canine inherited dystrophinopathies and centronuclear myopathies In: Childers MK. Regenerative medicine for degenerative muscle diseases. Series title: Stem cell biology and regenerative medicine. New York: Humana Press; 2016. p. 309–329. [Google Scholar]
- Nguyen F, Cherel Y, Guigand L, Goubault-Leroux I, Wyers M. Muscle lesions associated with dystrophin deficiency in neonatal golden retriever puppies. J Comp Pathol. 2002;126(2-3):100–108. [DOI] [PubMed] [Google Scholar]
- Vieira NM, Elvers I, Alexander MS, Moreira YB, Eran A, Gomes JP, Marshall JL, Karlsson EK, Verjovski-Almeida S, Lindblad-Toh K, et al. Jagged 1 Rescues the Duchenne Muscular Dystrophy Phenotype. Cell. 2015;163(5):1204–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydemann A, Ceco E, Lim JE, Hadhazy M, Ryder P, Moran JL, Beier DR, Palmer AA, McNally EM. Latent TGF-beta-binding protein 4 modifies muscular dystrophy in mice. J Clin Invest. 2009;119(12):3703–3712. [Erratum in: J Clin Invest. 2010; 120] [2]:645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine BA, Cooper BJ. Canine X-linked muscular dystrophy: selective involvement of muscles in neonatal dogs. Neuromuscul Disord. 1991;1:31–38. [DOI] [PubMed] [Google Scholar]
- Kornegay JN, Childers MK, Bogan DJ, Bogan JR, Nghiem P, Wang J, Fan Z, Howard JF, Jr, Schatzberg SJ, Dow JL, et al. The paradox of muscle hypertrophy in muscular dystrophy. Phys Med Rehabil Clin N Am. 2012;23(1):149–172. [xii.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa K, Nakamura A, Hijikata T, Takeda S. Dystrophin deficiency in canine X-linked muscular dystrophy in Japan (CXMDJ) alters myosin heavy chain expression profiles in the diaphragm more markedly than in the tibialis cranialis muscle. BMC Musculoskelet Disord. 2008;9;9:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McComas AJ. Skeletal Muscle, Form and Function. Human Kinetics; 1996. [Google Scholar]
- Reyes NL, Banks GB, Tsang M, Margineantu D, Gu H, Djukovic D, Chan J, Torres M, Liggitt HD, Hirenallur-S DK, et al. Fnip1 regulates skeletal muscle fiber type specification, fatigue resistance, and susceptibility to muscular dystrophy. Proc Natl Acad Sci USA. 2015;112(2):424–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrill JR, Duong MN, Turner R, Le Guiner C, Boyatzis A, Kettle AJ, Grounds MD, Arthur PG. Levels of inflammation and oxidative stress, and a role for taurine in dystropathology of the Golden Retriever Muscular Dystrophy dog model for Duchenne Muscular Dystrophy. Redox Biol. 2016;9:276–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guevel L, Lavoie JR, Perez-Iratxeta C, Rouger K, Dubreil L, Feron M, Talon S, Brand M, Megeney LA. Quantitative proteomic analysis of dystrophic dog muscle. J Proteome Res. 2011;10(5):2465–2478. [DOI] [PubMed] [Google Scholar]
- Ernster L, Schatz G. Mitochondria: a historical review. J Cell Biol. 1981;91(3 Pt 2):227s–255s. [Review] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadasan-Nair R, Gayathri N, Mishra S, Sunitha B, Mythri RB, Nalini A, Subbannayya Y, Harsha HC, Kolthur-Seetharam U, Srinivas Bharath MM. Mitochondrial alterations and oxidative stress in an acute transient mouse model of muscle degeneration: implications for muscular dystrophy and related muscle pathologies. J Biol Chem. 2014;289(1):485–509. [DOI] [PMC free article] [PubMed] [Google Scholar]