Abstract
An ever-expanding body of evidence in both humans and animal models demonstrates the influence of the resident gut microbiota on host health and disease susceptibility. However, as unwanted bacterial, viral, protozoal, and parasitic agents have gradually been eliminated from colonies of purpose-bred laboratory mice, the resident microbiota has lost richness and complexity. Recent studies have shown that the ultra-hygienic environment of traditional laboratory mice and lack of antigenic exposure during development results in mice with an immune system more akin to that of a neonate than an adult human. In contrast, wild mice or mice purchased from pet stores are exposed to much greater antigen burdens and their immune system reflects this with significantly greater numbers of memory T cells and more robust vaccine responses. The current review explores the use of alternative sources for research rodents, with an emphasis on the differences in resident gut microbiota and pathogen burden between wild mice, pet store-origin mice, and traditional laboratory mice. Specifically, the literature is compared and contrasted to our own data reflecting the endogenous gut microbiota and pathogen load of wild and pet store mice, as well as the changes in both during and after procedures intended to eliminate certain zoonotic agents present in pet store mice. These data demonstrate that, while alternative sources of research rodents will likely provide models that are more translatable to the human condition, there are also several real-world considerations for scientists including contamination of research facilities and human health risks such as zoonotic diseases.
Keywords: mouse, microbiota, microbiome, translatability, comparative medicine
Background & Introduction
The numbers are now familiar; bacteria in our gut are, conservatively, equivalent to the number of somatic cells in our body [1]. The fact that our own eukaryotic genome encodes only around 1 percent of the total genetic diversity in our entire “metagenome” highlights our emerging appreciation of the influence of the gut microbiota on host health. At its core, such a concept is not at all novel and it is largely due to the development of innovative culture-independent molecular techniques to characterize gut microbial communities that the field has seen a resurgence in interest. Using data generated from the seminal Human Microbiome Project as a foundation, researchers quickly began studies comparing individuals affected with various conditions and healthy controls. Perhaps not surprisingly, a multitude of associations were found between characteristics of the gut microbiota (GM) and a wide range of health conditions and entire review papers are now available describing disease-associated changes in the GM and cardiovascular health [2], cognition and mental health [3], obesity and endocrine disorders [4,5], and neoplasia [6].
Despite the undeniable links between the GM and human health, a careful reading of the literature reveals that, with rare exception, those links are correlative rather than definitively causative [7]. The reason for this is multifold. First, most of these conditions develop unexpectedly and over the course of years or decades, obviating the ability to collect samples before and after the development of disease. Second, studies comparing affected individuals and healthy controls are impossible to control for environmental and genetic variables which can influence the composition of the GM. Lastly, prospective studies to test causality in humans are often simply unethical. The comparative medicine approach mitigates all of these considerations and allows for well-controlled, prospective, and longitudinal studies designed to test the nature of these associations. While it is well-recognized that, at a species or strain level, the microbes colonizing the mouse gut are distinct from those colonizing humans [8], it is logical to assume that the resident microbes perform many of the same functions within their cognate hosts. With that in mind, the present review explores the translatability of GM-related research performed in murine models, and presents important considerations and caveats for scientists working with novel model systems aimed at enhancing translatability.
The GM of research mice has changed substantially from that of the original wild and fancy mice used as founders of the first inbred mice established by pioneers such as Clarence Little (1888-1971), Leonell Strong (1894-1982), and Halsey Baggs (1889-1947), founders of the DBA and C57BL/6, A, and BALB/c strains respectively [9]. As those researchers first strived to generate strains of isogenic mice via repeated brother/sister matings, they were also faced with outbreaks of infectious disease brought about through both the gradual loss of allelic heterozygosity or “hybrid vigor”, and sharing of infectious agents via intensive breeding. After making it through the genetic bottleneck successfully, these early inbred strains were precious and there was a need to keep these mice as disease-free as possible. Thus, the Jackson Laboratory and Harlan Sprague Dawley (now Envigo), the first commercial producers of research rodents established in 1929 and 1931 respectively, were faced with the formidable task of eliminating unwanted pathogens and eventually developing exclusion lists of those microbes. Several decades later and following the establishment of Charles River Laboratories and Taconic in 1947 and 1952 respectively (the other two primary producers of research rodents in the U.S. marketplace), these exclusion lists have grown dramatically. That growth was fueled by multiple factors including the development of extremely sensitive molecular methods of identifying and screening research colonies for novel agents, as well as the inherently competitive business practice of being able to offer the “cleanest” mice and rats to scientists. While certain bacterial pathobionts such as Helicobacter spp. were placed on those lists based on strong empirical evidence of adverse effects on animal health [10,11], others such as Klebsiella oxytoca and Pseudomonas aeruginosa are opportunistic organisms that have been added based on scant evidence of any untoward or confounding effects in the absence of secondary factors [12,13]. Clearly, such lists are a necessary component of biomedical research using animal models, but a growing body of evidence suggests that there may be unappreciated consequences of the ongoing quest to keep research mice “clean” and standardize their GM.
Modeling Antigen-experienced Adults versus Naïve Neonates
Recent studies assessing the influence of the GM on the development of the immune system and response to vaccination or challenge with an infectious agent suggest that the use of standard purpose-bred laboratory mice could result in misleading data, particularly in research involving the immune system. Specifically, Beura et al. demonstrated that, like neonatal (but not adult) humans, laboratory mice largely lack differentiated CD8+ memory T cell subsets in the periphery [14]. In contrast, wild and pet-store origin mice were found to harbor higher numbers of such T cell subsets, akin to what is found in adult humans and presumably reflecting a greater degree of antigen exposure. Notably, pet store mice evinced the greatest numbers of CD8+ memory T cells while wild mice were intermediate to the traditional laboratory mice and pet store mice. Support for these differences being driven by components of the microbiota was provided by the fact that co-housing laboratory mice with pet store mice resulted in significant increases in numbers of peripheral memory T cells and constitutive antibody production. Similarly, Reese et al. reported that a history of intentional antigen exposure led to differences in basal expression of several genes involved in interferon production (type I and II) and T cell responses, and an enhanced response to subsequent vaccine challenge [15]. Again, the differences in gene expression observed between antigen-exposed and control (mock-infected) laboratory mice mirror the differences seen between human adult and cord blood peripheral blood mononuclear cells. These data are in agreement with an earlier report that wild Mus musculus mount greater immune responses than C57BL/6 mice, as measured by IFN-γ and antibody production and leukocyte expression of several activation markers [16]. Collectively, these data have brought into question the translatability of many findings made with traditional laboratory mice and support the need for interdisciplinary approaches such as eco-immunology or “wild immunology” [17,18]. In this paradigm, host immune responses in feral animals, presumably with a history of exposure to a broad array of antigens, are viewed as a valuable complement to data generated in purpose-bred, highly “sanitized” animals. One of the primary difficulties in such an approach is that, unless one intends to capture and confine the animals for the duration of the study, collection of repeat or post-treatment samples is problematic. Thus, as it is unlikely that many researchers and laboratories are going to abandon their traditional barrier-housed model species (accompanied by decades of robust historical data) in favor of feral animals living in the wild, wild-caught Mus musculus offer an attractive compromise. However, their introduction to a biomedical research facility is complicated by the high potential of carrying one or more pathogenic organisms. Moreover, it is unclear as to whether the aforementioned differences in the development or function of the immune system are due solely to prior exposure to recognized pathogens, or if, as in the observed differences between mice from different commercial producers, the commensal resident GM are involved.
Differences in the Composition of the GM among Research Mice
Before comparing the GM of laboratory mice and mice obtained from alternative sources, it is important to clarify what is meant by “laboratory mice”, and describe GM-dependent differences within that group. Clearly, there is a large amount of variability within the GM of laboratory mice purchased from the primary producers of research rodents. While the dominant phyla found within mice from these sources do not vary, there are differences in the average richness and diversity [19], as well as the presence of specific bacteria known to induce changes in the host physiology such as segmented filamentous bacteria (SFB; Candidatus Savagella) [20]. This highlights the fact that the term “specific pathogen-free” (SPF) is a rather nebulous term with no universal meaning. That said, the influence of a complex GM on host metabolism and immunity is most evident when comparing germ-free (GF) mice to traditional SPF research mice [21,22]. Notably, mice colonized with a limited “defined” microbiota such as altered Schaedler flora (ASF) have an intermediate phenotype with regard to immunophenotype [23]. That said, mice obtained from different commercial producers, while all SPF, possess substantial differences in various characteristics of the GM [19,24], and these differences have the potential to result in altered model phenotypes [25-27]. This is an important consideration when reviewing the literature or performing meta-analyses and should be investigated when published studies cannot be reproduced.
Indeed, the NIH laments the lack of reproducibility in pre-clinical studies involving rodent models [28,29] and myriad factors are known to affect the GM of mice. This suggests that differences in the GM owing to the different origins of colony founders and institutional differences in husbandry may contribute to poor reproducibility. Considering the number of different institutions performing research using animal models, each faced with its own set of environmental influences and budgetary constraints, the likelihood of normalizing or controlling the composition of the GM across all biomedical research is an untenable goal. However, an equally important consideration is the translatability of animal model-based research to the human condition being studied. As humans are an outbred population, exposed on a daily basis to a wide array of antigenic stimuli including microbial pathobionts and pathogens, how informative are the physiological and immunological responses of inbred mice raised in controlled ultra-hygienic conditions?
Methods
Animals. Six-week-old BALB/cJ (n = 10) and BALB/cAnNHsd (n = 10) mice were purchased from the Jackson Laboratory and Envigo respectively. Adult pet store mice were purchased in two groups (n = 10 and n = 20) from a pet store located in Columbia, MO, or bred on site (n = 29). Adult wild mice (n = 9) were captured using “live capture” traps placed near a livestock facility on the University of Missouri (MU) campus in Columbia, MO. Laboratory mice were housed under barrier conditions in microisolator cages with compressed pelleted paper bedding and nestlets, on ventilated racks (Thoren, Hazleton, PA) with ad libitum access to irradiated chow (LabDiet 5058, LabDiet, St. Louis, MO) and acidified, autoclaved water, under a 14:10 light/dark cycle. Pet store and wild mice were housed under conventional conditions in microisolator cages with corncob bedding and nestlets, on ventilated racks (Allentown, Allentown, NJ) with ad libitum access to irradiated chow (LabDiet 5008) and acidified, autoclaved water, under a 14:10 light/dark cycle. Pet store mice received fenbendazole-supplemented feed (Purina, St. Louis, MO) as indicated in the text.
DNA extraction. Following collection of a freshly evacuated fecal pellet into a 2 mL round bottom tube containing 800 µL lysis buffer [30] and a 0.5 cm-diameter stainless steel ball bead, samples were mechanically disrupted for 3 minutes using a TissueLyser II (Qiagen, Venlo, Netherlands), incubated at 70°C for 20 minutes with periodic vortexing, and then centrifuged at 5000×g for five minutes at room temperature. Supernatant was collected and mixed with 200 µL of 10 mM ammonium acetate, and then allowed to incubate on ice for 5 minutes. Following centrifugation as above, 750 µL of supernatant was mixed with an equal volume of chilled isopropanol and allowed to incubate on ice for 30 minutes. Following centrifugation at 16000×g for 15 minutes at 4°C, supernatant was aspirated and discarded, and the DNA pellet washed several times with 70 percent ethanol. Following resuspension in 150 µL of Tris-EDTA, 15 µL of proteinase-K and 200 µL of Buffer AL (DNeasy Blood and Tissue kit, Qiagen) were added and samples were incubated at 70°C for 10 minutes. 200 µL of 100 percent ethanol was added and the contents of each tube were transferred to a spin column from the DNeasy kit. DNA was then purified according to the manufacturer’s instructions and eluted in 200 µL of EB buffer (Qiagen). DNA yield was determined via fluorometry (Qubit, Life Technologies, Carlsbad, CA) using quant-iT BR dsDNA reagent kit (Invitrogen, Carlsbad, CA).
Library preparation and sequencing. All library preparation and sequencing were performed at the MU DNA Core facility as previously reported. Briefly, amplicon libraries of the 16S rRNA gene were generated with universal primers (U515F/806R) previously developed against the V4 region, flanked by Illumina standard adapter sequences [31,32]. A single forward primer and reverse primers with a unique 12-base index were used in all reactions. PCR cycling parameters were as reported previously [19]. Amplified product (5 µL) from each reaction was combined and thoroughly mixed; pooled amplicons were purified by addition of Axygen AxyPrep MagPCR Clean-up beads to an equal volume of 50 µL of amplicons and incubated at room temperature for 15 minutes. Products were washed multiple times with 80 percent ethanol and the dried pellet resuspended in Qiagen EB Buffer (32.5 µL), incubated at room temperature for 2 minutes, and then placed on the magnetic stand for 5 minutes. The final amplicon pool was evaluated using the Advanced Analytical Fragment Analyzer automated electrophoresis system, quantified with the Qubit fluorometer using the quant-iT HS dsDNA reagent kit (Invitrogen), and diluted according to Illumina’s standard protocol for sequencing on the MiSeq.
Informatics. Assembly, binning, and annotation of DNA sequences was performed at the MU Informatics Research Core Facility. Briefly, contiguous sequences of DNA were assembled using FLASH software [33], and contigs were culled if found to be short after trimming for a base quality less than 31. QIIME v1.8 was used to perform de novo and reference-based chimera detection and removal, and remaining contiguous sequences were assigned to operational taxonomic units (OTUs) using a criterion of 97 percent nucleotide identity. Taxonomy was assigned to selected OTUs using BLAST [34] against the Greengenes database [35] of 16S rRNA sequences and taxonomy.
Statistical analysis. Differences in β-diversity were determined via permutational multivariate analysis of variance (PERMANOVA) of Bray-Curtis distances using Past 3.15 [36] software. Differences in richness were determined via traditional ANOVA using SigmaPlot 13.0 (Systat software, San Jose, CA). The threshold for significance in all cases was p ≤ 0.05.
Pathogen testing. All pathogen testing was performed by IDEXX BioResearch (Columbia, MO) including serological testing for Clostridium piliforme, Mycoplasma spp., cilia-associated respiratory (CAR) bacillus, ectromelia, EDIM, Hantaan, K virus, LCMV, LDEV, MAV1, MAV2, MCMV, MHV, MNV, MPV, MTV, MVM, Polyoma, PVM, REO3, Sendai, TMEV, and Encephalitozoon cuniculi; and PCR testing for Aspiculuris tetraptera, Cryptosporidium sp., Helicobacter spp., Hymenolepis diminuta, Myocoptes sp., Pneumocystis carinii, Radfordia/Myobia sp., Rodentolepis nana, Salmonella sp., and Syphacia obvelata.
Differences in the Composition of the GM in Wild Mice, Pet Store Mice, and Research Mice
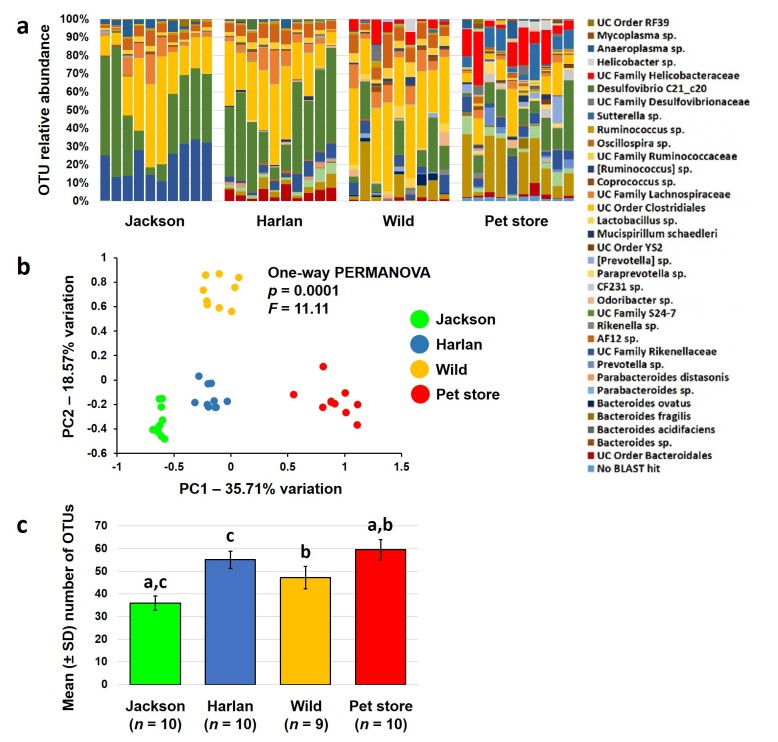
In the context of the GM, there are marked differences between laboratory mice and wild mice or mice purchased from a pet store. Studies performed using molecular, culture-independent methods have revealed that the GM of laboratory mice is very different from that of wild or pet store mice with regard to both pathogens [14] and resident microbes [37,38]. Our own survey of the GM of wild-caught Mus musculus, pet store-origin Mus musculus, and colonies of laboratory mice from standard mouse producers revealed significant differences between all groups (Figure 1A and 1B). As previously reported, the GM of substrains of BALB/c from the Jackson Laboratory and Harlan differ significantly with a greater proportion of unclassified (UC) microbes in family Rikenellaceae, and several genera of Bacteroides and Parabacteroides, detected in the two groups respectively. Table 1 lists the mean relative abundance of all OTUs detected at greater than 0.50 percent relative abundance in at least one group. In addition to overall compositional differences, the gut microbial richness of mice from standard producers was substantially lower than that of pet store mice (Figure 1C), largely reflecting the relative paucity of Proteobacteria (e.g., family Helicobacteraceae and Sutterella sp.) found in laboratory mice. Interestingly, the microbial richness of wild mice captured on the MU campus did not differ significantly from that of laboratory mice. Considering the myriad reported associations between increased bacterial richness and diversity in the resident GM and protection from intestinal inflammation [39-43], it is plausible that the GM of traditional research mice renders them artificially susceptible to such conditions. While this may be a positive model attribute in the evaluation of therapeutic modalities, it could be considered a negative attribute in terms of translation to a human population. An argument could be made that treatments, be they genetic or pharmaceutical, should be applied in parallel to colonies of mice harboring more than one of these GM profiles. To do so in a controlled fashion would require introduction of these differentially colonized mice to a research institution and, just as animal producers abide by pathogen exclusion lists, so too do academic and industry institutions. Thus, such studies are faced with very real-world concerns regarding contamination with pathogenic organisms.
Figure 1.
Differences in the gut microbiota of laboratory, wild, and pet store mice. Stacked bar charts showing relative abundance of operational taxonomic units (OTUs) detected in the feces of adult BALB/c mice purchased the Jackson Laboratory or Harlan (Envigo), adult mice trapped on the University of Missouri campus, or adult mice purchased from a pet store in Columbia, MO. Legend at right shows identity of OTUs detected at greater than 0.50% mean relative abundance in at least one group (a); principal component analysis plot showing differences in β-diversity of fecal communities in the same groups of mice (b); and bar charts showing mean (± standard deviation) number of OTUs detected in feces from each group. Like letters indicate significant (p < 0.05) differences as determined via Kruskal-Wallis one-way ANOVA on ranks (c).
Table 1. Relative abundance of dominant operational taxonomic units (OTUs) in feces of mice purchased from two commercial suppliers (Jackson and Harlan), wild mice, and pet store mice.
| OTU | Jackson (n = 10) | Harlan (n = 10) | Wild (n = 9) | Pet St. (n = 10) | ||||
| mean | SD | mean | SD | mean | SD | mean | SD | |
| No BLAST hit | 0.00% | 0.00% | 0.01% | 0.01% | 0.00% | 0.00% | 1.91% | 0.58% |
| UC order Bacteroidales | 0.01% | 0.00% | 4.87% | 2.76% | 0.79% | 0.80% | 1.67% | 2.10% |
| Bacteroides sp. | 0.01% | 0.00% | 3.15% | 2.36% | 7.79% | 10.25% | 18.05% | 12.70% |
| B. acidifaciens | 0.03% | 0.05% | 2.81% | 2.34% | 0.00% | 0.00% | 4.32% | 2.92% |
| B. fragilis | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.96% | 1.78% |
| B. ovatus | 0.00% | 0.00% | 0.05% | 0.04% | 1.71% | 2.01% | 0.00% | 0.00% |
| Parabacteroides sp. | 0.00% | 0.00% | 0.12% | 0.10% | 0.59% | 0.81% | 0.12% | 0.12% |
| P. distasonis | 0.00% | 0.00% | 0.96% | 0.96% | 0.11% | 0.20% | 0.00% | 0.01% |
| Prevotella sp. | 0.00% | 0.00% | 0.30% | 0.21% | 0.15% | 0.20% | 2.30% | 3.13% |
| UC family Rikenellaceae | 22.80% | 8.95% | 4.76% | 3.13% | 4.05% | 3.56% | 6.99% | 5.48% |
| AF12 sp. | 0.00% | 0.00% | 0.60% | 0.48% | 0.29% | 0.29% | 0.25% | 0.23% |
| Rikenella sp. | 0.00% | 0.00% | 0.00% | 0.00% | 0.67% | 1.22% | 0.50% | 0.78% |
| UC family S24-7 | 33.02% | 20.77% | 34.13% | 16.23% | 11.00% | 10.21% | 12.43% | 10.20% |
| Odoribacter sp. | 0.00% | 0.00% | 0.58% | 0.47% | 1.82% | 2.39% | 0.83% | 1.28% |
| CF231 sp. | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 1.58% | 2.38% |
| Paraprevotella sp. | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 1.65% | 1.87% |
| [Prevotella] sp. | 0.00% | 0.00% | 0.05% | 0.12% | 0.42% | 0.79% | 2.85% | 4.55% |
| UC order YS2 | 0.00% | 0.00% | 0.06% | 0.07% | 0.01% | 0.04% | 0.71% | 0.94% |
| Mucispirillum schaedleri | 0.00% | 0.00% | 0.69% | 0.61% | 0.00% | 0.00% | 1.06% | 1.24% |
| Lactobacillus sp. | 0.10% | 0.12% | 0.43% | 0.51% | 2.84% | 2.26% | 0.14% | 0.13% |
| UC order Clostridiales | 26.01% | 16.65% | 26.72% | 12.25% | 35.31% | 10.12% | 9.32% | 8.26% |
| UC family Lachnospiraceae | 5.48% | 5.41% | 6.25% | 5.32% | 7.43% | 3.94% | 1.98% | 1.64% |
| Coprococcus sp. | 0.36% | 0.29% | 0.36% | 0.30% | 1.16% | 0.99% | 0.24% | 0.47% |
| [Ruminococcus] sp. | 0.00% | 0.00% | 0.00% | 0.00% | 1.34% | 1.24% | 0.00% | 0.00% |
| UC family Ruminococcaceae | 1.75% | 0.83% | 2.68% | 1.09% | 4.47% | 2.02% | 0.95% | 0.61% |
| Oscillospira sp. | 4.15% | 2.33% | 5.97% | 2.07% | 8.18% | 4.32% | 1.19% | 1.01% |
| Ruminococcus sp. | 0.94% | 0.40% | 0.82% | 0.36% | 1.18% | 0.84% | 0.16% | 0.15% |
| Sutterella sp. | 0.00% | 0.00% | 0.16% | 0.11% | 0.24% | 0.48% | 7.03% | 6.15% |
| UC family Desulfovibrionaceae | 0.00% | 0.00% | 0.00% | 0.00% | 1.04% | 2.89% | 1.62% | 1.83% |
| Desulfovibrio C21_c20 | 0.00% | 0.00% | 0.57% | 1.32% | 0.00% | 0.00% | 0.00% | 0.00% |
| UC family Helicobacteraceae | 0.00% | 0.00% | 0.00% | 0.00% | 4.57% | 4.04% | 11.76% | 9.15% |
| Helicobacter sp. | 0.00% | 0.00% | 0.00% | 0.00% | 0.88% | 2.33% | 1.38% | 1.88% |
| Anaeroplasma sp. | 3.30% | 2.74% | 0.67% | 1.48% | 0.09% | 0.17% | 1.72% | 2.94% |
| Mycoplasma sp. | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.00% | 0.53% | 0.60% |
| UC order RF39 | 0.76% | 1.19% | 0.34% | 0.17% | 0.05% | 0.05% | 0.33% | 0.83% |
Table showing mean and standard deviation (SD) relative abundance of all operational taxonomic units (OTUs) detected at greater than 0.50% relative abundance in any group, in feces collected from adult BALB/c mice purchased from the Jackson Laboratory, Harlan Laboratories (Envigo), adult wild mice captured on the MU campus, and adult pet store mice (all Mus musculus).
Pathogen Exposure and Burden in Wild and Pet Store Mice
Regarding such pathogens and pathobionts, one of the most consistent differences is the presence of Helicobacter spp. in wild-caught mice [38,44]. While still problematic in some institutions, Helicobacter spp. are rarely, if ever, present in mice obtained from the large producers. The fact that the majority of inbred laboratory mice are resistant to helicobacter-induced inflammation underscores the notion that, while these microbes can serve as triggers of inflammation, they do so primarily in the context of genetic susceptibility or secondary insult. Moreover, their presence in wild mice is not associated with any lesions. The use of mice endemically (or experimentally) colonized with Helicobacter sp. is not necessarily problematic in a research setting as it does not transmit from cage to cage, assuming standard barrier husbandry procedures are in place. Of greater concern in wild mice is the abundance of pathogenic viral and parasitic microbes found at high prevalence. In a survey of wild mice captured on the University of Pennsylvania campus [45], mites and mite eggs were found via direct exam on 31 percent (13/55) and 24 percent (17/55) of mice respectively. Serologic or PCR-based evidence of multiple viruses including parvoviridae (MPV, MVM), adenoviridae (MAV2), betaherpesviridae (MCMV), caliciviridae (MNV), picornaviridae (TMEV), coronaviridae (MHV), and reoviridae (EDIM) was also found sporadically, often suggesting co-infections. Notably, two pathogens of particular concern to laboratory animal veterinarians, Mycoplasma pulmonis and pinworms were detected in 0/55 and 1/55 mice via serology and wet mount examination, respectively. A similar study from Australia published 16 years earlier [46] reported comparably low prevalence of M. pulmonis, but much higher serologic evidence of MCMV, MHV, and EDIM in wild mice. Aside from the obvious geographical contrast, these differences could reflect purely serological testing in the former report, and a combination of serology and PCR testing in the latter. This is supported by the fact that three of the agents found in the 2009 study were detected only by serology, with negative PCR results. This is an important consideration as this “antigen experience” could ostensibly provide a more appropriate immunological context for research animals, assuming there is no risk of contamination with viable pathogens within a facility. Our own screening of wild adult Mus musculus on the MU campus in Columbia, MO detected both Helicobacter hepaticus and H. ganmani in 100 percent (9/9), pinworms (S. obvelata) in 22 percent (2/9), fur mites (Radfordia/Myobia sp.) in 33 percent (3/9), and MCMV in 56 percent (5/9) of mice (Table 2). Surprisingly, no serological evidence of exposure to M. pulmonis, Cryptosporidium sp., or any other adventitious viruses, bacteria, protozoa, or parasites included on the most comprehensive diagnostic panel offered by IDEXX BioResearch was detected in any mice.
Table 2. Results of pathogen testing in wild mice and pet store mice before and after quarantine.
| Agent | Wild (n = 9) | Pet store | Pet store | Pet store |
| arrival (n = 20) | 6w-post (n = 19) | 10w-post (n = 19) | ||
| E. cuniculi | 0/9 (0%) | 15/20 (75%) | 0/19 (0%) | 0/19 (0%) |
| Ectromelia | 0/9 (0%) | 6/20 (30%) | 0/19 (0%) | 0/19 (0%) |
| EDIM | 0/9 (0%) | 11/20 (55%) | 0/19 (0%) | 0/19 (0%) |
| M. pulmonis | 0/9 (0%) | 17/20 (85%) | 0/19 (0%) | 0/19 (0%) |
| LDEV | 0/9 (0%) | 1/20 (5%) | 0/19 (0%) | 0/19 (0%) |
| MAV1 | 0/9 (0%) | 2/20 (10%) | 7/19 (37%) | 5/19 (26%) |
| MAV2 | 0/9 (0%) | 15/20 (75%) | 18/19 (95%) | 14/19 (74%) |
| MCMV | 5/9 (56%) | 1/20 (5%) | 0/19 (0%) | 0/19 (0%) |
| MHV | 0/9 (0%) | 20/20 (100%) | 14/19 (74%) | 12/19 (63%) |
| MNV | 0/9 (0%) | 20/20 (100%) | 1/19 (5%) | 2/19 (11%) |
| MPV | 0/9 (0%) | 20/20 (100%) | 17/19 (89%) | 15/19 (79%) |
| MVM | 0/9 (0%) | 20/20 (100%) | 13/19 (68%) | 15/19 (79%) |
| Polyoma | 0/9 (0%) | 2/20 (10%) | 0/19 (0%) | 0/19 (0%) |
| PVM | 0/9 (0%) | 17/20 (85%) | 0/19 (0%) | 0/19 (0%) |
| Reo3 | 0/9 (0%) | 5/20 (25%) | 0/19 (0%) | 0/19 (0%) |
| TMEV | 0/9 (0%) | 17/20 (85%) | 14/19 (74%) | 16/19 (84%) |
| Helicobacter sp. | 9/9 (100%) | 2/2 pools (100%) | 2/2 pools (100%) | 2/2 pools (100%) |
| H. bilis | 0/9 (0%) | n/a | n/a | n/a |
| H. ganmani | 9/9 (100%) | n/a | n/a | n/a |
| H. hepaticus | 9/9 (100%) | n/a | n/a | n/a |
| H. mastomyrinus | 0/9 (0%) | n/a | n/a | n/a |
| H. rodentium | 0/9 (0%) | n/a | n/a | n/a |
| H. typhlonius | 1/9 (11%) | n/a | n/a | n/a |
| Mites | 3/9 (33%) | 2/2 pools (100%) | 0/19 (0%) | 0/19 (0%) |
| Pinworms | 2/9 (22%) | 2/2 pools (100%) | 0/19 (0%) | 0/19 (0%) |
| Cryptosporidium | 0/9 (0%) | 1/2 pools (50%) | 19/19 (100%) | 18/19 (95%) |
| R. nana | 0/9 (0%) | 3/20 (15%) | 3/19 (16%) | 1/19 (5%) |
Table showing results of comprehensive pathogen testing of adult wild mice captured on the MU campus, and adult pet store mice (all Mus musculus) upon arrival in our facility and after 6 and 10 weeks (6w- and 10w-post, respectively) of quarantine procedures including topical cydectin and fenbendazole-treated chow. Testing of pet store mice for Helicobacter sp., mites, and pinworms via PCR was performed on pooled samples. No evidence of any other infectious agent including C. piliforme, cilia-associated respiratory (CAR) bacillus, Hymenolepis diminuta, Hantaan virus, K virus, LCMV, LDEV, MTV, or Sendai virus was detected in any sample. Materials and Documentation: All experiments and procedures were performed according to guidelines put forth in the Guide for the Care and Use of Laboratory Animals and were approved by the University of Missouri Institutional Animal Care and Use Committee, under protocol #8524.
Perhaps not surprisingly, pet store mice possess a richer microbiota and much more antigen-experienced immune system. In agreement with a recently published report of pathogen testing performed on pet store mice [14], our own survey of pet store mice found them to carry large pathogen burdens, despite their outward appearance of good health. Specifically, Helicobacter sp., mites, pinworms, and antibodies to MHV, MNV, MPV, and MVM were ubiquitous, and M. pulmonis was detected in 85 percent (17/20) of mice. Moreover, the zoonotic agents Cryptosporidium sp. and Rodentolepis nana were detected in 100 percent (19/19) and 15 percent (3/20) of mice, respectively. Thus, our diagnostic testing suggests that pet store mice experience significantly greater pathogen exposure than wild mice, both of which experience greater pathogen burdens than standard laboratory mice. This paradigm dove-tails perfectly with the number of CD8+ memory T cells found in similar cohorts; pet store mice have the greatest number, wild mice are intermediate, and laboratory mice have the least [14].
Elimination of Pathogens Reduces Richness of Resident Gut Microbiota
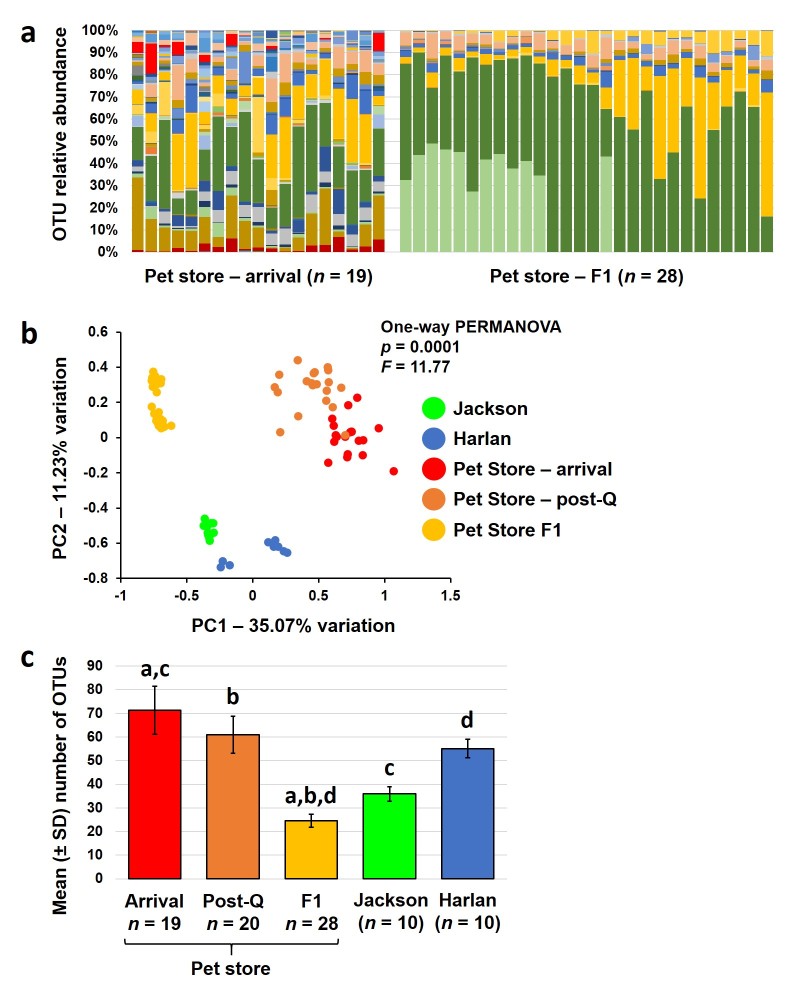
The two main concerns with introduction of wild or pet store mice to a standard rodent vivarium are spread of infectious agents to other rodents, and exposure of personnel to zoonotic agents. The transmission of pathogens can also occur indirectly as is often the case with pinworm ova which can aerosolize, adhere to surfaces for extended periods of time, and then infect naïve animals long after the infected animal is gone. Efforts in our lab to eliminate pathogens from pet store mice using traditional quarantine procedures (i.e., fenbendazole-treated food and topical cydectin treatment) in conjunction with temporary cessation of breeding resulted in elimination of mites and pinworms but was ineffective at removing Cryptosporidium sp. and R. nana (as determined via PCR) (Table 2). Even after 10 weeks in quarantine, these zoonotic pathogens persisted. Following treatment with azithromycin (intended to eliminate Cryptosporidium sp.), mice were eventually allowed to breed, in hopes of generating offspring that retained the rich GM of the original pet store mice but that were at least free of M. pulmonis, pinworms, Cryptosporidium sp., and R. nana. Comparison of the GM in first generation (F1) offspring to the parental mice revealed a stark shift in composition and reduction in richness (Figure 2A). Principal component analysis (PCA) of the parental mice (upon arrival and following an initial 6-week quarantine period), the F1 mice, and representative mice from the Jackson Laboratory and Harlan (Envigo) Laboratories results in a subtle separation in the original pet store mice before and after quarantine, and a substantial shift in the F1 mice across principal component 1 (Figure 2B). Much of this divergence is likely due to the significant reduction in taxonomic richness, with the adult F1 mice harboring fewer OTUs than traditional research mice from Harlan (Figure 2C). Collectively, the pronounced separation of F1 samples and pre- and post-quarantine samples from the parental mice, along with the significant decrease in richness in the F1 mice, suggests that treatment with azithromycin had a more substantial influence on the GM than the initial quarantine procedures. Of greatest concern however, despite daily treatment of the parental mice with azithromycin for one month, 24 percent (7/29) of the pups tested from two litters at weaning were colonized with Cryptosporidium sp. As different pet stores in different cities acquire their mice from varied sources, the pathogen burden also undoubtedly varies. That said, the above data highlight the difficulties in eliminating patent infection with certain pathogens while retaining the rich commensal microbiota. They also corroborate the notion that production practices over the last half century aimed at generating rodents free of adventitious pathogens have also led to a loss of richness and diversity in the GM of contemporary rodent colonies.
Figure 2.
Changes in the gut microbiota of pet store mice and their offspring associated with quarantine and antibiotic treatment. Stacked bar charts showing relative abundance of operational taxonomic units (OTUs) detected in the feces of a second cohort of adult mice purchased from a pet store in Columbia, MO and two litters born to those mice following 10 weeks in quarantine and treatment with azithromycin (a); Principal component analysis plot showing differences in β-diversity of fecal communities in a second cohort of adult pet store mice immediately upon arrival (n = 19), following 6 weeks of quarantine (post-Q, n = 20), adult F1 mice from two litters born to the pet store mice treated with azithromycin; and adult BALB/c mice purchased from Jackson (n = 10) or Harlan (n = 10) (b); bar chart showing the mean (± standard deviation) number of OTUs detected in feces from each group. Like letters indicate significant (p < 0.05) differences as determined via Kruskal-Wallis one-way ANOVA on ranks (c).
Directions for the Future
As discussed above, most institutions performing biomedical research maintain lists of excluded agents, making the routine use of wild or pet store mice difficult. Most investigators and facility managers would, understandably, object to pinworm- or mycoplasma-positive mice being housed in the same vivarium that houses their research colonies. However, the benefit of such mice is thought to be due to previous pathogen exposure, rather than the extant presence of said pathogens, making clean-up of such mice an attractive approach. Thus, researchers intending to work with pet store (or wild) mice would be well-advised to begin by surveying individual sources of animals for mice that are free of the most problematic agents, rather than attempting to eliminate pathogens after the fact. An alternative approach would be to allow the pathogen burden and house all mice in separate facility with dedicated staff and strict containment procedures, resulting in considerably higher per diem rates than standard SPF- or barrier-housing. Ultimately, such mice could be used as surrogate dams for surgical embryo transfer procedures as this would ostensibly allow the generation of genetically defined mice colonized with wild or pet store mouse GM. Regardless of the approach, these are very exciting times in biomedical research and we have but scratched the surface in our understanding of the relationship between host health and the invisible microbial communities that complete and surround us. Continued efforts to both standardize and diversify the GM of research animals must occur in parallel as both provide valuable pieces of the puzzle. This includes the use of rich and diverse, naturally occurring GM profiles similar to those found in humans.
Acknowledgments
The authors would like to acknowledge the University of Missouri DNA Core and Informatics Research Core facilities for their assistance with 16S rRNA amplicon library preparation and sequencing, and post-sequencing informatics respectively, IDEXX BioResearch for the pathogen diagnostic testing reported herein, and Ms. Karen Clifford for assistance formatting images.
Glossary
- GF
germ-free
- GM
gut microbiota
- OTU
operational taxonomic unit
- SFB
segmented filamentous bacteria
- SPF
specific pathogen-free
Author Contributions
ACE and CLF provided resources, designed experiments, and helped analyze data; DRM and CRS handled mice, collected and processed samples, and helped analyze data; ACE wrote the manuscript.
References
- Sender R, Fuchs S, Milo R. Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell. 2016;164(3):337–340. [DOI] [PubMed] [Google Scholar]
- Tang WH, Hazen SL. The contributory role of gut microbiota in cardiovascular disease. J Clin Invest. 2014;124(10):4204–4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13(10):701–712. [DOI] [PubMed] [Google Scholar]
- Gerard P. Gut microbiota and obesity. Cell Mol Life Sci. 2016;73(1):147–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke G, Stilling RM, Kennedy PJ, Stanton C, Cryan JF, Dinan TG. Minireview: Gut microbiota: the neglected endocrine organ. Mol Endocrinol. 2014;28(8):1221–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014;12(10):661–672. [DOI] [PubMed] [Google Scholar]
- de Vos WM, de Vos EA. Role of the intestinal microbiome in health and disease: from correlation to causation. Nutr Rev. 2012;70 Suppl 1:S45–S56. [DOI] [PubMed] [Google Scholar]
- Krych L, Hansen CH, Hansen AK, van den Berg FW, Nielsen DS. Quantitatively different, yet qualitatively alike: a meta-analysis of the mouse core gut microbiome with a view towards the human gut microbiome. PLoS One. 2013;8(5):e62578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Health NIo. Origins of Inbred Mice. Morse III HC. Bethesda, Maryland: Elsevier; 1978. 719 p . [Google Scholar]
- Ward JM, Fox JG, Anver MR, Haines DC, George CV, Collins MJ, Jr, et al. Chronic active hepatitis and associated liver tumors in mice caused by a persistent bacterial infection with a novel Helicobacter species. J Natl Cancer Inst. 1994;86(16):1222–1227. [DOI] [PubMed] [Google Scholar]
- Rao VP, Poutahidis T, Ge Z, Nambiar PR, Boussahmain C, Wang YY, et al. Innate immune inflammatory response against enteric bacteria Helicobacter hepaticus induces mammary adenocarcinoma in mice. Cancer Res. 2006;66(15):7395–7400. [DOI] [PubMed] [Google Scholar]
- Bleich A, Kirsch P, Sahly H, Fahey J, Smoczek A, Hedrich HJ, et al. Klebsiella oxytoca: opportunistic infections in laboratory rodents. Lab Anim. 2008;42(3):369–375. [DOI] [PubMed] [Google Scholar]
- Baker DG. Natural pathogens of laboratory mice, rats, and rabbits and their effects on research. Clin Microbiol Rev. 1998;11(2):231–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura LK, Hamilton SE, Bi K, Schenkel JM, Odumade OA, Casey KA, et al. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature. 2016;532(7600):512–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese TA, Bi K, Kambal A, Filali-Mouhim A, Beura LK, Burger MC, et al. Sequential Infection with Common Pathogens Promotes Human-like Immune Gene Expression and Altered Vaccine Response. Cell Host Microbe. 2016;19(5):713–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abolins SR, Pocock MJ, Hafalla JC, Riley EM, Viney ME. Measures of immune function of wild mice, Mus musculus. Mol Ecol. 2011;20(5):881–892. [DOI] [PubMed] [Google Scholar]
- Babayan SA, Allen JE, Bradley JE, Geuking MB, Graham AL, Grencis RK, et al. Wild immunology: converging on the real world. Ann N Y Acad Sci. 2011;1236:17–29. [DOI] [PubMed] [Google Scholar]
- Pedersen AB, Babayan SA. Wild immunology. Mol Ecol. 2011;20(5):872–880. [DOI] [PubMed] [Google Scholar]
- Ericsson AC, Davis JW, Spollen W, Bivens N, Givan S, Hagan CE, et al. Effects of vendor and genetic background on the composition of the fecal microbiota of inbred mice. PLoS One. 2015;10(2):e0116704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericsson AC, Hagan CE, Davis DJ, Franklin CL. Segmented filamentous bacteria: commensal microbes with potential effects on research. Comp Med. 2014;64(2):90–98. [PMC free article] [PubMed] [Google Scholar]
- Backhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci USA. 2007;104(3):979–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostman S, Rask C, Wold AE, Hultkrantz S, Telemo E. Impaired regulatory T cell function in germ-free mice. Eur J Immunol. 2006;36(9):2336–2346. [DOI] [PubMed] [Google Scholar]
- Norin E, Midtvedt T. Intestinal microflora functions in laboratory mice claimed to harbor a “normal” intestinal microflora. Is the SPF concept running out of date? Anaerobe. 2010;16(3):311–313. [DOI] [PubMed] [Google Scholar]
- Hufeldt MR, Nielsen DS, Vogensen FK, Midtvedt T, Hansen AK. Variation in the gut microbiota of laboratory mice is related to both genetic and environmental factors. Comp Med. 2010;60(5):336–347. [PMC free article] [PubMed] [Google Scholar]
- Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32(6):815–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning TL, Norris BA, Medina-Contreras O, Manicassamy S, Geem D, Madan R, et al. Functional specializations of intestinal dendritic cell and macrophage subsets that control Th17 and regulatory T cell responses are dependent on the T cell/APC ratio, source of mouse strain, and regional localization. J Immunol. 2011;187(2):733–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart ML, Ericsson AC, Franklin CL. Differing complex microbiota alter disease severity of the IL-10-/- mouse model of inflammatory bowel disease. Front Microbiol. 2017:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Tabak LA. Policy: NIH plans to enhance reproducibility. Nature. 2014;505(7485):612–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin S. Preclinical research: make mouse studies work. Nature. 2014;507(7493):423–425. [DOI] [PubMed] [Google Scholar]
- Yu Z, Morrison M. Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques. 2004;36(5):808–812. [DOI] [PubMed] [Google Scholar]
- Walters WA, Caporaso JG, Lauber CL, Berg-Lyons D, Fierer N, Knight R. PrimerProspector: de novo design and taxonomic analysis of barcoded polymerase chain reaction primers. Bioinformatics. 2011;27(8):1159–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011;108 Suppl 1:4516–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer O. Past 3.15 2016. Available from: http://folk.uio.no/ohammer/past/.
- Wilson KH, Brown RS, Andersen GL, Tsang J, Sartor B. Comparison of fecal biota from specific pathogen free and feral mice. Anaerobe. 2006;12(5-6):249–253. [DOI] [PubMed] [Google Scholar]
- Linnenbrink M, Wang J, Hardouin EA, Kunzel S, Metzler D, Baines JF. The role of biogeography in shaping diversity of the intestinal microbiota in house mice. Mol Ecol. 2013;22(7):1904–1916. [DOI] [PubMed] [Google Scholar]
- Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55(2):205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll IM, Ringel-Kulka T, Siddle JP, Ringel Y. Alterations in composition and diversity of the intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol Motil. 2012;24(6):521-30, e248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicksved J, Halfvarson J, Rosenquist M, Jarnerot G, Tysk C, Apajalahti J, et al. Molecular analysis of the gut microbiota of identical twins with Crohn’s disease. ISME J. 2008;2(7):716–727. [DOI] [PubMed] [Google Scholar]
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104(34):13780–13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa MC, Arroyo LG, Allen-Vercoe E, Stampfli HR, Kim PT, Sturgeon A, et al. Comparison of the fecal microbiota of healthy horses and horses with colitis by high throughput sequencing of the V3-V5 region of the 16S rRNA gene. PLoS One. 2012;7(7):e41484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Linnenbrink M, Kunzel S, Fernandes R, Nadeau MJ, Rosenstiel P, et al. Dietary history contributes to enterotype-like clustering and functional metagenomic content in the intestinal microbiome of wild mice. Proc Natl Acad Sci USA. 2014;111(26):E2703–E2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker SE, Malone S, Bunte RM, Smith AL. Infectious diseases in wild mice (Mus musculus) collected on and around the University of Pennsylvania (Philadelphia) Campus. Comp Med. 2009;59(5):424–430. [PMC free article] [PubMed] [Google Scholar]
- Smith AL, Singleton GR, Hansen GM, Shellam G. A serologic survey for viruses and Mycoplasma pulmonis among wild house mice (Mus domesticus) in southeastern Australia. J Wildl Dis. 1993;29(2):219–229. [DOI] [PubMed] [Google Scholar]