ABSTRACT
The 2 main degradative pathways that contribute to proteostasis are the ubiquitin-proteasome system and autophagy but how they are molecularly coordinated is not well understood. Here, we demonstrate an essential role for an effector caspase in the activation of compensatory autophagy when proteasomal activity is compromised. Functional loss of Hsp83, the Drosophila ortholog of human HSP90 (heat shock protein 90), resulted in reduced proteasomal activity and elevated levels of the effector caspase Dcp-1. Surprisingly, genetic analyses showed that the caspase was not required for cell death in this context, but instead was essential for the ensuing compensatory autophagy, female fertility, and organism viability. The zymogen pro-Dcp-1 was found to interact with Hsp83 and undergo proteasomal regulation in an Hsp83-dependent manner. Our work not only reveals unappreciated roles for Hsp83 in proteasomal activity and regulation of Dcp-1, but identifies an effector caspase as a key regulatory factor for sustaining adaptation to cell stress in vivo.
KEYWORDS: apoptosis, caspase, compensatory autophagy, Dcp-1, Drosophila, heat-shock protein, Hsp83, ubiquitin-proteasome system
Introduction
Proteostasis, or protein homeostasis, is a term that describes how the proteome is regulated and maintained to protect cellular integrity. Loss of proteostasis is a common feature of aging and diseases such as cancer and neurogenerative disorders as it causes inappropriate protein aggregate formation in various tissues.1 This aggregation is routinely managed by the proteostasis network, a network defined by macromolecular machines that contribute to maintenance of proteome integrity by coordinating protein synthesis, folding, disaggregation, and degradation.1
The 2 main degradative pathways that contribute to proteostasis are the ubiquitin-proteasome system (UPS) and autophagy. The proteasome consists of a core 20S cylindrical complex containing proteolytic sites with 2 19S regulatory complexes at each terminus to form the functional 26S proteasome. It serves as a major contributor to proteostasis by degrading misfolded, damaged proteins and regulatory proteins that have been targeted with ubiquitin, along with a few exceptional ubiquitin-free proteins that can also be degraded by the proteasome.2,3 Macroautophagy (hereafter referred to as autophagy) is a ubiquitous cellular process responsible for the degradation of long-lived proteins and turnover of organelles. It is a multistep process involving the formation of a double-membrane phagophore that engulfs cytoplasmic cargo and delivers it to the lysosome for degradation. In instances when the UPS is dysfunctional, it has been shown in multiple systems that compensatory autophagy contributes to protein degradation in an effort to maintain healthy cellular proteostasis.2-5 However, the molecular mechanisms that regulate compensatory autophagy are not well understood.
Heat-shock proteins play a central role in proteostasis by controlling protein expression, acting as chaperones, and assisting with protein disaggregation and degradation.6 One of the most conserved, ubiquitous and highly-expressed heat-shock proteins is HSP90. HSP90 is a chaperone that alters protein configurations using ATP hydrolysis in a homodimer conformation, and has a large and growing “clientele” of greater than 200 proteins including numerous nuclear receptors, protein kinases, and transcription factors.7 The wide associations of HSP90 play a pivotal role in cell signaling and regulation of diverse cellular processes in normal biology and its dysregulation can have a marked effect on disease. HSP90 has emerged as a molecule of interest for cancer therapeutics as its upregulation, mislocalization and stabilization of proteins involved in metastasis, evasion of apoptosis and proliferation make it a prime target.8-10 While HSP90 is known to be an important hub for signaling and proteostasis, there are many HSP90 relationships and related pathways yet to be discovered.
Investigations into the regulation of autophagy have led to the discovery of a Drosophila effector caspase, Dcp-1, that promotes starvation-induced autophagy in Drosophila oogenesis.11,12 Although caspases are well known for their role in apoptosis, it is becoming increasingly evident that caspases have nonapoptotic functions in diverse processes such as immunity, differentiation, compensatory proliferation and autophagy.12-14 In an effort to elucidate the molecular mechanisms underlying Dcp-1-mediated autophagy regulation, an immune-affinity purification (IAP) and tandem mass spectrometry (MS/MS) assay has identified sesB, an adenine nucleotide translocase, as a downstream regulator of autophagy.11 Upstream factors and other downstream pathway components and their relationship to Dcp-1-mediated autophagy still remain largely unknown.
In this paper we report 24 candidate Dcp-1-interacting proteins identified in the IAP-MS/MS screen, 13 of which were found to negatively regulate autophagic flux in vitro. We focused further on one of the identified interactors, Hsp83, since its human ortholog HSP90 has links to disease, proteostasis and a current ambiguous role in autophagy.15-17 In vivo analyses revealed that loss-of-function Hsp83 mutants induced autophagy and cell death during Drosophila mid-oogenesis. Biochemical analyses showed that Hsp83 binds to the zymogen pro-Dcp-1 and that the loss of Hsp83 led to elevated levels of cleaved and pro-Dcp-1 that were not due to transcriptional regulation. As an explanation for elevated levels of Dcp-1, we investigated the functionality of the UPS, and found that Hsp83 mutants had decreased proteasomal activity. The levels of Dcp-1 were increased in flies with suppressed proteasomal activity supporting that Dcp-1 itself is affected by the proteasome. Analysis of Dcp-1;Hsp83 double mutants indicated that Dcp-1 was responsible for autophagy induced in mid-stage egg chambers (MSECs) and larval fat bodies, female fertility and organism viability when Hsp83 function is compromised. Additionally, in vitro double RNAi experiments revealed that Dcp-1 is needed to induce autophagy when Hsp83 or the proteasomal subunit Rpn11 is knocked down. These findings indicate that Hsp83 is important for basal proteasomal activity and that loss of it causes an induction of Dcp-1-mediated compensatory autophagy.
Results
Proteomic analysis and RNAi screen identify candidate Dcp-1-interacting proteins that modulate autophagy
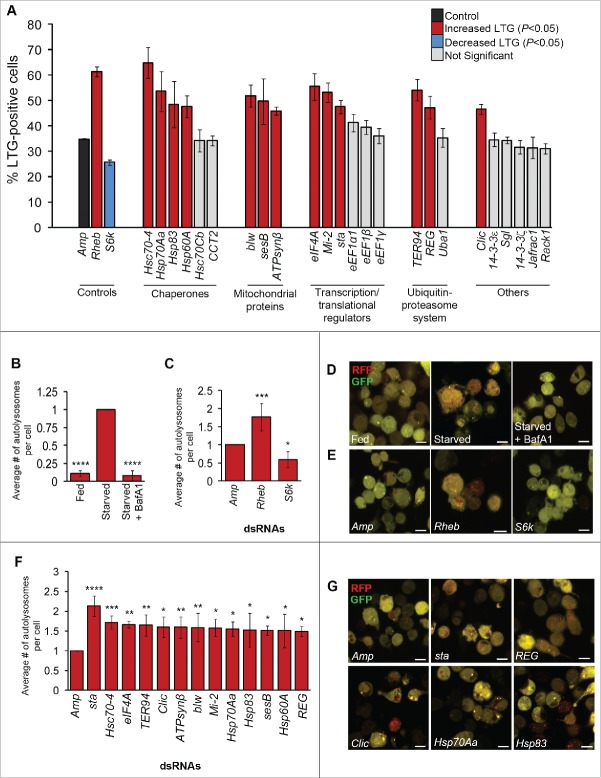
To identify candidate substrates and interactors of Dcp-1 that regulate starvation-induced autophagy, we took an IAP-MS/MS-based approach.11 Catalytically inactive Dcp-1 (Dcp-1C<A) was overexpressed in Drosophila l(2)mbn cells to stabilize the interactions that would normally be transient if there was proteolytic activity. Following immuno-affinity purification, cell lysates were analyzed by LC-MS/MS to identify copurifying proteins. A subset of 24 high-confidence candidate interacting proteins was identified that met the selection threshold detailed in Materials and methods (Table 1, S1). Using this method, we have previously reported sesB as an interacting partner of Dcp-1.11 We selected all 24 candidates for subsequent autophagy analyses, initially using a RNAi and LysoTracker® Green (LTG) flow cytometry strategy that we described previously.12 Of the 24 candidates, 13 showed a statistically significant increase in LTG fluorescence following RNAi and starvation treatment, indicating that these candidates act as potential negative regulators of autophagy (Fig. 1A). The 24 candidates were compared with control RNAi-treated cells: Rheb, a negative regulator of autophagy that significantly increased LTG fluorescence following starvation, and S6k, a positive regulator of autophagy that significantly reduced LTG fluorescence (Fig. 1A). A second set of nonoverlapping dsRNAs were designed and used to validate the LTG findings (Fig. S1). The 13 validated hits included the heat shock proteins Hsc70–4, Hsp70Aa, Hsp60A and Hsp83, translation initiation factor eIF4A, the chromatin remodeler Mi-2, the ribosomal constituent sta, the AAA+ ATPase TER94, the chloride intracellular channel protein Clic, the proteasome activator REG, and the mitochondrial proteins ATPsynβ, blw and sesB.
Table 1.
Candidate Dcp-1 interactors and substrates identified by mass spectrometry. Symbols, CG numbers and molecular functions are from FlyBase.
| FlybaseSymbol | CG Number | Gene Ontology molecular function | Mean log(e) | Mean unique peps | UniProt human gene name | # of Expts observed |
|---|---|---|---|---|---|---|
| 14–3–3ζ | CG17870 | Protein binding, protein heterodimerization activity, protein homodimerization activity | −9.6 | 1.75 | YWHAZ | 4 |
| eEF1α1 | CG8280 | Translation elongation factor activity | −27 | 3.75 | EEF1A1 | 4 |
| Hsc70–4 | CG4264 | Chaperone binding | −70 | 8.50 | HSPA8 | 4 |
| Hsp83 | CG1242 | ATPase activity, coupled | −32 | 4.75 | HSP90AA1 | 4 |
| Jafrac1 | CG1633 | Thioredoxin peroxidase activity | −30 | 4.00 | PRDX1 | 4 |
| 14–3–3ε | CG31196 | Protein binding; protein heterodimerization activity | −23 | 3.33 | YWHAE | 3 |
| blw | CG3612 | Hydrogen exporting ATPase activity; phosphorylative mechanism | −16 | 2.67 | ATP5A1 | 3 |
| CCT2 | CG7033 | Unfolded protein binding; ATP binding | −6.5 | 1.67 | CCT2 | 3 |
| Clic | CG10997 | Calcium ion binding; chloride channel activity; lipid binding | −7.2 | 1.67 | CLIC2 | 3 |
| eEF1β | CG6341 | Translation elongation factor activity | −10 | 2.00 | EEF1B2 | 3 |
| Rack1 | CG7111 | Protein kinase c binding | −3.8 | 1.33 | RACK1 | 3 |
| sesBa | CG16944 | ATP:ADP antiporter activity | −27 | 4.00 | ANT2 | 3 |
| sgl | CG10072 | UDP-glucose 6-dehydrogenase activity | −8.1 | 1.67 | UGDH | 3 |
| TER94 | CG2331 | ATPase activity; golgi & ER organization | −16 | 2.67 | VCP | 3 |
| Uba1 | CG1782 | Ubiquitin activating enzyme activity | −43 | 5.67 | UBA1 | 3 |
| ATPsynβ | CG11154 | Hydrogen exporting ATPase activity; phosphorylative mechanism | −3.1 | 1.00 | ATP5B | 2 |
| eEF1γ | CG11901 | Translation elongation factor activity | −5.6 | 1.50 | EEF1G | 2 |
| eIF4a | CG9075 | Translation initiation factor activity; RNA helicase activity | −17 | 3.00 | EIF4A1 | 2 |
| Hsc70Cb | CG6603 | Chaperone binding | −19 | 3.00 | HSPA4 | 2 |
| Hsp60A | CG12101 | Unfolded protein binding | −4.7 | 1.50 | HSPD1 | 2 |
| Hsp70Aa | CG31366 | ATP binding, response to hypoxia | −19 | 1.00 | HSPA1A/1B/1L | 2 |
| Mi-2 | CG8103 | Protein binding; nucleosome-dependent ATPase activity; chromatin binding | −6.1 | 1.50 | CHD3 | 2 |
| REG | CG1591 | Endopeptidase inhibitor activity; endopeptidase activator activity | −3.4 | 1.00 | PSME3 | 2 |
| sta | CG14792 | Structural constituent of ribosome | −5.8 | 1.50 | RPSA | 2 |
Notes. The mean number of unique peptides that corresponded to each gene and the mean X!Tandem log (e) score for the peptides identified are listed. The human gene names were determined from a BLAST analysis of the Drosophila genes against the human UniProt database. The list is ordered by number of observations made from 4 independent immuno-affinity purifications of the Dcp-1 protein (Expts) with the most being 4 out of 4 experiments and the least being 2. See Table S1 for all raw values.
reported in.11
Figure 1.
Thirteen candidate Dcp-1 interactors modify LysoTracker® Green and autolysosomes in vitro (A) RNAi-treated l(2)mbn cells stained with LysoTracker® Green (LTG) and starved to measure autophagy-associated activity via flow cytometry. Error bars represent ± SEM(n = 3). Statistical significance was determined using one-way ANOVA with a Dunnet post-test. Knockdown of targets that significantly increased LTG levels are indicated in red (P < 0.05), and knockdown of targets that significantly decreased LTG levels are indicated in blue (P < 0.05). All samples were compared with the negative Amp control dsRNA (ampicillin resistance gene) that is shown in black. (B to G) Analysis of RFP-GFP-Atg8a puncta in RNAi-treated Drosophila S2 cells. At least 50 cells were counted per treatment (n = 3), and graphs represent the average number of autolysosomes per cell relative to the Amp control. Error bars represent the average ± SEM, and statistical significance was determined using one-way ANOVA with a Dunnet post-test. (B) Cells were subjected to nutrient rich or deprived conditions for 7 h in the presence or absence of 0.1 μM bafilomycin A1 (BafA1). ****P < 0.0001. (C) Cells were treated with Amp, Rheb, or S6k dsRNA and subjected to 7 h of starvation, *P < 0.05, ***P < 0.001. (D) Representative images of S2-RFP-GFP-Atg8a cells subjected to fed, starved, or starved + BafA1 conditions. (E) Representative images of S2-RFP-GFP-Atg8a cells treated with the indicated dsRNAs. (F) Cells were treated with the indicated dsRNAs and subjected to starvation conditions for 7 h. *P < 0.05, **P < 0.01, ***P < 0.001 ****P < 0.0001. (G) Representative images of S2-RFP-GFP-Atg8a cells treated with the indicated dsRNAs. Scale bars: 10 μm.
Candidate Dcp-1 interacting partners regulate autophagic flux in vitro
To investigate the role of candidate Dcp-1 substrates and interacting partners in autophagic flux in vitro, we monitored autolysosome formation using a Drosophila S2 cell line stably expressing RFP-GFP-Atg8a. Following autophagy induction, Atg8 becomes lipidated and inserted into the autophagosomal membrane. Here, the overlap of RFP and GFP fluorescence results in the formation of yellow puncta that signify autophagosomes. In the acidic environment of the autolysosome, GFP fluorescence is quenched, whereas RFP fluorescence remains, and the resulting red puncta indicate that flux through the lysosomal compartment has occurred.18,19 As expected, we observed a significant increase in the number of autolysosomes per cell following starvation and this was blocked following the addition of the late-stage autophagy inhibitor bafilomycin A1 (BafA1) (Fig. 1B, D). As an additional control, we monitored the formation of autolysosomes following RNAi-mediated knockdown of Rheb and S6k and found a significant increase and decrease, respectively, in the number of autolysosomes per cell (Fig. 1C, E). Candidate Dcp-1-interacting partners that modified LTG following starvation were next tested for their ability to mediate autophagic flux in vitro. We found that RNAi-mediated knockdown of all 13 candidates significantly increased the number of autolysosomes per cell following starvation compared with the control (Fig. 1F and G), indicating that these proteins negatively regulate starvation-induced autophagic flux in vitro. Knockdown of all 13 candidates in fed conditions also led to a statistically significant, although modest, increase in autolysosomes, with the greatest effects observed for the 4 heat-shock proteins identified (Fig. S1).
Loss of Hsp83 increases TUNEL staining, the percentage of mid-stage degenerating egg chambers (MSDECs) and autophagic flux
Our in vitro autophagy assays revealed that Dcp-1 may regulate, or may be regulated by, one or more of the 13 identified negative regulators of starvation-induced autophagy. Of the interacting partners identified, we chose to focus on Hsp83. To further confirm our in vitro findings that Hsp83 acts as a negative regulator of autophagy we designed additional Hsp83 dsRNAs and analyzed LTG levels by flow cytometry (Fig. S2). Two RNAi constructs used for further experiments were tested for efficacy and consistently achieved 65% to 80% knockdown (Fig. S2).
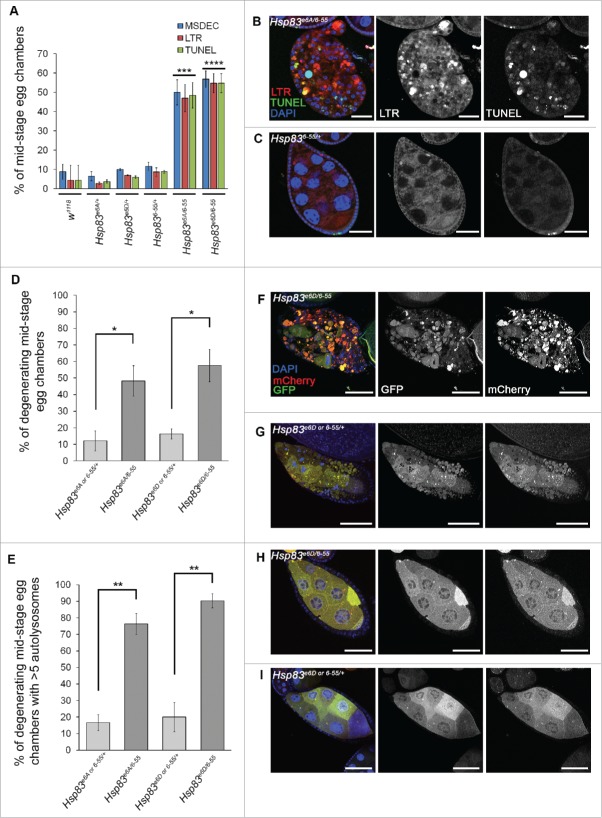
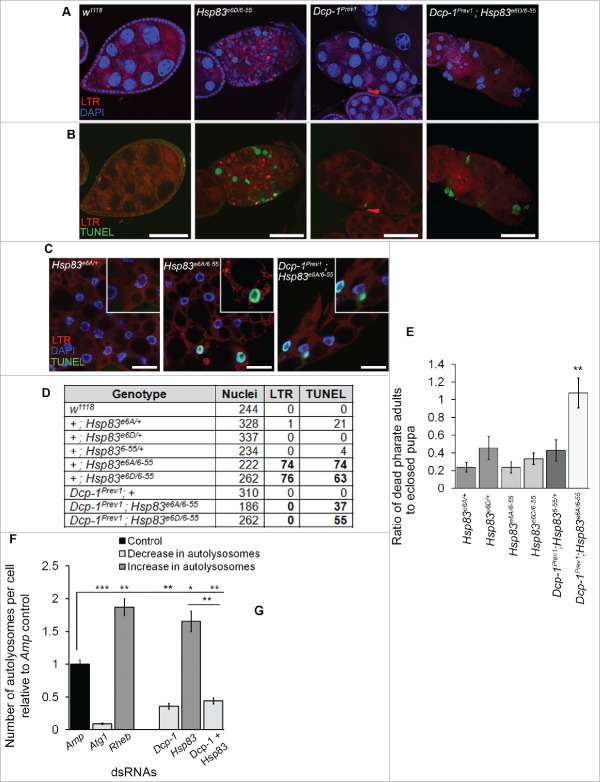
For in vivo autophagy analyses, we chose to analyze Drosophila ovaries as nutrient deprivation leads to both Dcp-1 dependent cell death and autophagy.11,20 Loss-of-function studies were performed using transheterozygous (transhet) combinations of Hsp83 alleles because Hsp83 homozygous mutant flies are lethal.21,22 We examined the role of 2 transhet combinations with decreased Hsp83 function of Hsp83 alleles, Hsp83e6A with Hsp836–55 (Hsp83e6A/6–55) and Hsp83e6D with Hsp836–55 (Hsp83e6D/6–55), for their role in autophagy during Drosophila oogenesis by analyzing mid-stage egg chambers (MSECS). These alleles carry missense point mutations and the combinations were functionally screened by genetic complementation tests.21 Ovaries from Hsp83e6A/6–55 and Hsp83e6D/6–55 flies contained an increase in the percentage of degenerating MSECs (MSDECS) that stained positively for LysoTracker® Red (LTR) and TUNEL relative to control flies and balanced monoallelic Hsp83 mutants (Fig. 2A to C). MSECs undergoing cell death are characterized by nurse cell nuclear condensation and fragmentation, uptake of the germline by follicle cells and follicle cell death.23 These findings suggest that there is an increase in lysosomal activity, a key feature of autophagy, in addition to cell death.
Figure 2.
Loss of Hsp83 function leads to an increase in autophagy and cell death features in vivo. (A) Mid-stage egg chambers (MSECS) scored as being TUNEL positive, LysoTracker® Red (LTR)-positive or as having condensed degenerating nurse cell nuclei by DAPI (MSDEC) with percentages reported according to their genotype. At least 50 MSECs were counted per genotype (n = 3). Error bars represent ± SEM and statistical significance was determined using one-way ANOVA with a Bonferroni post-test and compared with w1118, ***P < 0.001, ****P < 0.0001. (B, C) Representative MSDECs and nondegenerating MSECs stained with LTR, TUNEL and DAPI, scale bars: 25 μm. (B) MSDEC from an Hsp83e6A/6–55 ovariole that scored positive for LTR and TUNEL staining. (C) Nondegenerating MSEC from Hsp836–55/+ scored as negative for LTR and TUNEL (D to I) MSECs were scored from flies expressing GFP-mCherry-Atg8a in the germline (UAsp-GFP-mCherry-DrAtg8a with single copies of the drivers otu-GAL4 and NGT-GAL4). Hsp83e6D/+ and Hsp836–55/+, Hsp83e6A/+ and Hsp836–55/+ from the same cross were analyzed together. (D) Percentage of MSDECs for the indicated genotypes is represented on the graph and reflects the mean of at least 100 MSECs scored per genotype (n = 3). Error bars represent ± SEM and statistical significance was determined using a 2-tailed Student t test, *P < 0.05. (E) Flies expressing GFP-mCherry-Atg8a in the germline were scored as either having more than 5 autolysosomes or less than or equal to 5 autolysosomes. The percentages shown reflect the mean of at least 100 MSECs scored per genotype (n = 3). Error bars represent ± SEM and statistical significance was determined using a 2-tailed Student t test, **P < 0.01. (F) to I) Representative images of MSECs expressing the construct GFP-mCherry-Atg8a, scale bar = 25μm. MSDECS found in (F) Hsp83e6D/6–55 flies and (G) Hsp83e6D or 6-55/+. (H, I) Examples of nondegenerating MSECs from (H) Hsp83e6D/6–55 flies and (I) Hsp83e6D or 6-55/+.
As an additional measure to examine autophagic flux, we used the tandem-tagged Atg8a reporter system expressing GFP-mCherry-Atg8a in the germline of Hsp83 transhets in nutrient-rich conditions. Due to the nature of the cross, Hsp836–55/+ and either Hsp83e6A/+ or Hsp83e6D/+ were pooled and analyzed together. First, we confirmed that expression of the Atg8a reporter construct did not affect the frequency of MSDECs (Fig. 2D). Hsp83e6A/6–55 and Hsp83e6D/6–55 had very distinct MSDECs that had high levels of autolysosomes in comparison to the control Hsp83 heterozygotes (Fig. 2E-G). It was found that 75% to 90% of MSDECs had greater than 5 autolysosomes compared with only 15% to 20% when carrying one mutated Hsp83 allele (Fig. 2E). The nondegenerating MSECs in the mutant transhets and the controls looked morphologically normal and did not appear to have high levels of autophagic flux (Fig. 2H, I). From these studies we can conclude that loss of Hsp83 enhances the percentage of MSDECs undergoing autophagic flux. To determine if the phenotype is affected by the stress of nutrient deprivation, the flies were fed a sucrose-only diet for 4 to 5 d. Both the control Hsp83 heterozygotes and the mutant Hsp83e6A/6–55 transhets showed an increase in autolysosomal activity in the early stage egg chambers. However, in the mutant Hsp83 transhets only, we observed an exacerbated phenotype of nearly obliterated mCherry-positive mid-stage egg chambers in contrast to the degenerating mid-stage egg chambers observed in fed conditions (Fig. S3).
Hsp83 interacts with pro-Dcp-1 and regulates its levels
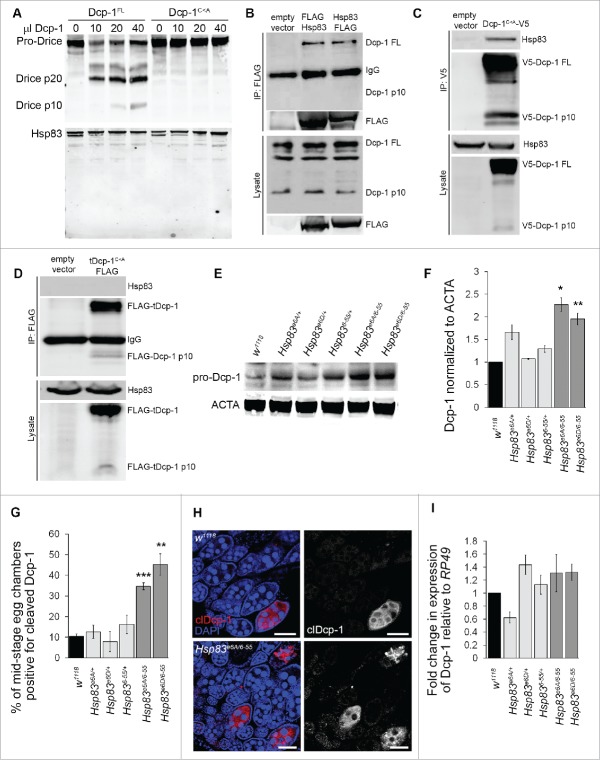
To further understand the nature of the interaction between Hsp83 and Dcp-1, we tested if Hsp83 is cleaved by Dcp-1. In vitro cleavage assays were performed using catalytically active Dcp-1 (Dcp-1FL) and catalytically inactive Dcp-1 (Dcp-1C<A) that were purified from l(2)mbn cells. Whereas Dcp-1FL was able to cleave in vitro translated Drice, a known target of Dcp-1 proteolytic activity,24 there were no detectable Hsp83 cleavage products (Fig. 3A).
Figure 3.
Hsp83 interacts with pro-Dcp-1 and suppresses its levels in a manner independent of transcriptional regulation. (A) Purified catalytically active Dcp-1 (Dcp-1FL) and catalytically inactive Dcp-1 (Dcp-1C<A) were incubated with in vitro translated Drice or Hsp83. Dcp-1FL cleaved Drice but not Hsp83. (B) N and C-terminal FLAG-tagged constructs of Hsp83 were expressed in l(2)mbn cells and immunoprecipitated with anti-FLAG antibodies. A representative western blot shows that both N- and C-FLAG-tagged constructs immunoprecipitated endogenous pro-Dcp-1 (Dcp-1 FL). No processed Dcp-1 was detected following immunoprecipitation. Similar results were observed in 3 independent experiments. (C) Dcp-1C<A-V5 was expressed in l(2)mbn cells and immunoprecipitated with anti-V5 antibodies. A representative western blot shows that Dcp-1C<A-V5immunoprecipitated endogenous Hsp83. Similar results were observed in 3 independent experiments. (D) Truncated Dcp-1C<A-FLAG (tDcp-1C<A-FLAG) was expressed in l(2)mbn cells and immunoprecipitated with anti-FLAG antibodies. A representative western blot (from n = 2 independent experiments) shows that tDcp-1 was unable to immunoprecipitate endogenous Hsp83. (E) Western blot of whole body lysates from females of the specified genotypes; pro-Dcp-1 = 35 kDa, ACTA/actin = 42 kDa. (F) Quantification of levels of pro-Dcp1 was determined by densitometry and normalized to levels of ACTA/actin. The average relative levels of pro-Dcp-1 were determined with 8 females per lysate (n = 3). Error bars represent ± SEM and statistical significance was determined by comparison to the w1118 control using one-way ANOVA with a Dunnet post-test, *P < 0.05, **P < 0.01. (G) MSECs were scored as being positive or negative for cleaved Dcp-1 (clDcp-1). The average percentage was determined by analyzing over 50 MSECs per genotype (n = 3). Error bars represent ± SEM and statistical significance was determined by comparison to w1118 using one-way ANOVA with a Dunnet post-test, **P < 0.01,***P < 0.001. (H) Representative images of Drosophila ovarioles that were stained with cleaved Dcp-1 antibody and DAPI for w1118 control and Hsp83e6A/6–55, scale bars: 50 μm. (I) QRT-PCR was performed on mRNA extracts from 8 animals each to determine levels of Dcp-1 mRNA relative to the RP49 control mRNA (n = 3). The relative fold change was normalized to wild-type w1118 with error bars representing ± SEM. There was no significant difference in Dcp-1 mRNA expression between genotypes.
Because Dcp-1 does not cleave Hsp83, we reasoned that Hsp83 might be affecting Dcp-1 since chaperones stabilize proteins or facilitate their turnover. First, we investigated whether Hsp83 interacted with the full-length zymogen and/or the truncated active form of Dcp-1. Hsp83 and full-length Dcp-1 were confirmed to associate via affinity isolation assays using either Hsp83 or full-length Dcp-1C<A as bait (Fig. 3B, C). However, the truncated form of Dcp-1 did not pull down Hsp83, indicating that the prodomain of Dcp-1 is required for its association with Hsp83 (Fig. 3D). We next quantified levels of the zymogen pro-Dcp-1 in Hsp83 flies by western blot analysis. Hsp83e6A/6–55 and Hsp83e6D/6–55 flies showed a significant increase in the levels of pro-Dcp-1 in comparison to wild type and the single alleles of Hsp83 (Fig. 3E, F). To determine if Hsp83 loss also led to elevated levels of catalytically active Dcp-1, we analyzed levels of cleaved Dcp-1 by scoring immunofluorescence. We found a statistically significant increase in the percentage of MSECS that stained positively for cleaved Dcp-1 in Hsp83 mutants compared with controls (Fig. 3G, H). The elevated levels of both pro-Dcp-1 and processed Dcp-1 indicate that loss of Hsp83 does not impair pro-Dcp-1 processing into its active form.
To test the possibility of transcriptional regulation as a reason for the elevated Dcp-1, we performed qRT-PCR on RNA extracts from Hsp83 flies. We found that there was no significant difference in Dcp-1 transcript levels between Hsp83 transhets, Hsp83 heterozygote controls and wild type (Fig. 3I). These data suggest that loss of functional Hsp83 leads to increased Dcp-1 that is independent of transcriptional regulation. Next, we explored the potential role of the death-associated inhibitor of apoptosis 1 (Diap-1) protein. Diap1 was shown previously to ubiquitinate and inhibit the activity of cleaved Dcp-1 without affecting its levels,25 but the consequences on pro-Dcp-1 levels are unknown. We overexpressed Diap1 in vivo and found that Diap1 overexpression did not have a significant effect on the protein levels of pro-Dcp-1 (Fig. S4A, B). We also overexpressed Diap1 in the Hsp83 transhet background and found no statistically significant difference in levels compared with the Hsp83 transhets (Fig. S4C, D). These results, together with the association of Hsp83 with only the zymogen form of Dcp-1, indicate that the regulation of pro-Dcp-1 levels in this context is largely independent of Diap-1.
Hsp83 mutants have reduced proteasomal activity
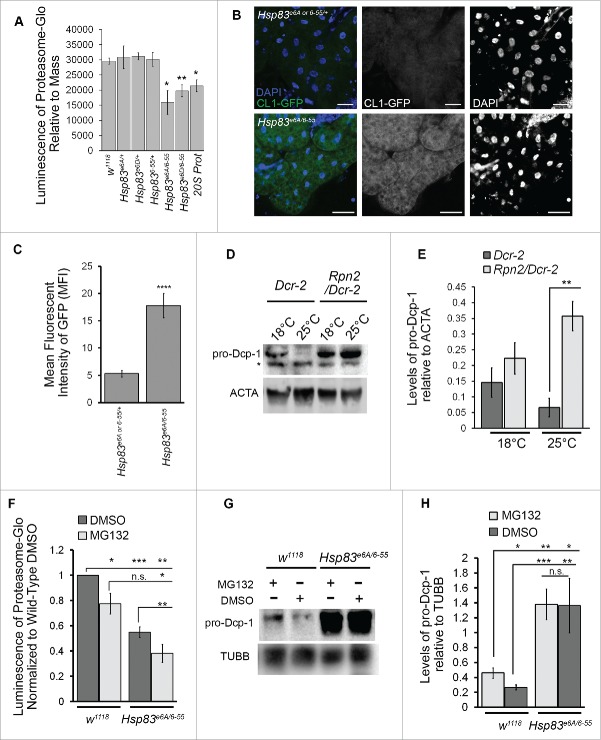
Orthologs of Hsp83, yeast Hsc82 and Hsp82 (orthologs of mammalian HSP90α and HSP90β, respectively), and human HSP90α, are needed for 26S proteasomal assembly respectively in yeast and in humans for antigen processing.26,27 To test whether altered proteasomal activity could be a possible explanation for the elevated levels of Dcp-1 observed in Hsp83 mutants, we used a luminescence assay with proteasome-targeted substrates that react when cleaved. Luminescence was significantly decreased in flies harboring RNAi targeting Prosα1, a subunit of the 20S proteasome, and in Hsp83 transhets when compared with Hsp83 heterozygote controls and wild-type flies (Fig. 4A). To confirm the decreased proteasome activity in Hsp83 transhets, we expressed the proteasome activity reporter CL1-GFP, a fusion protein with a degradation signal introduced into the otherwise stable GFP molecule,28 in the larval fat body using the larval midgut specific driver cg-GAL4. The Hsp83 transhets had a marked increase in GFP expression relative to controls, indicating inefficient turnover of CL1-GFP (Fig. 4B, C). The observed reduction in proteasomal activity could explain why Dcp-1 levels are elevated in the Hsp83 transhet mutants.
Figure 4.
Loss of Hsp83 decreases proteasomal activity resulting in elevated Dcp-1 levels. (A) Proteasomal activity was measured in females from the indicated genotypes using luminescent output (RLU) produced from cleavage of proteasomal substrates (ProteasomeGlo kit) and made relative to mass. Error bars represent ± SEM and statistical significance was determined using one-way ANOVA with a Bonferroni post-test and compared with w1118, *P < 0.05, **P < 0.01. (n = 5) (B, C) UAS-CL1-GFP expressed in the larval fat body using the driver cg-GAL4 and visualized with nonadjusted GFP channel images taken in the same experiment with identical confocal microscope settings (n = 3). (B) Representatives images, scale bars: 50 μm and (C) quantification of fluorescence intensity using mean fluorescent intensity (MFI). Error bars represent ± SEM and statistical significance was determined using a 2-tailed Student t test, ****P < 0.0001. (D, E) Females with the maternal driver nosGAL4 were collected after 2 d of exposure to 18°C or 25°C with the genotypes +/UAspDcr-2; nosGAL4/+ (Dcr-2) and Rpn2-RNAI/UAspDcr-2;nosGAL4/+ (Rpn2/Dcr-2). (D) Representative western blot of pro-Dcp-1 and ACTA/actin levels in Dcr-2 and Rpn2/Dcr-2 pro-Dcp-1 = 35 kDa, ACTA/actin = 42 kDa, * represents a nonspecific band detected by the Dcp-1 antibody. (E) Quantification of pro-Dcp-1 levels was performed by densitometry and normalized to levels of ACTA/actin. The average relative levels of pro-Dcp-1 were determined with 8 females per lysate (n = 3). Error bars represent ± SEM and statistical significance was determined using a 2-way Student t test, **P < 0.01. (F) to H) Hsp83e6A/6–55 and w1118 flies were fed proteasomal inhibitor MG132 or the control DMSO for 4 d. (F) Proteasomal activity was measured and normalized to w1118 flies fed with DMSO (n = 3). Error bars represent ± SEM and statistical significance was determined using a Student t test, *P < 0.05, **P < 0.01, ***P < 0.001. (G) Representative image of a western blot probed for pro-Dcp-1(35 kDa) and TUBB/tubulin (55 kDa). (H) Quantification of pro-Dcp-1 levels was performed by densitometry and normalized to levels of TUBB/tubulin. The average relative levels of pro-Dcp-1 were determined with 8 females per lysate (n = 3). Error bars represent ± SEM and statistical significance was determined using a 2-way Student t test, *P < 0.05, **P < 0.01, ***P < 0.001.
Reduction in proteasomal activity leads to an increase in Dcp-1
To determine if Dcp-1 levels are affected by the proteasome, we quantified levels of Dcp-1 in an RNAi line targeted against Rpn2, a regulatory subunit of the 26S proteasome. RNAi knockdown was enhanced by expressing one copy of Dcr-2/Dicer2 using the oogenesis-specific driver nosGAL4. An increase in temperature enhances expression of GAL4-driven Dcr-2 under the UAsp promoter and thus the knockdown is more efficacious. Rpn2 RNAi flies (Rpn2/Dcr-2) or control flies (Dcr-2), were incubated at 18°C or 25°C for 2 d before collection. The enhanced knockdown of Rpn2 at 25°C correlated with a greater increase in pro-Dcp-1 levels in comparison to the Dcr-2 flies (Fig. 4D, E). In addition to genetic suppression of the proteasome, we tested pharmacological inhibition by feeding flies the proteasome inhibitor MG132. A significant decrease in proteasomal activity was observed in both wild type and Hsp83 transhets following MG132 treatment (Fig. 4F). The levels of pro-Dcp-1 increased in wild type with the addition of MG132, consistent with the decrease in proteasomal activity (Fig. 4G, H). There was no further increase in the levels of pro-Dcp-1 in Hsp83 transhets treated with MG132 compared with the DMSO control despite reduced proteasomal activity. Together, these results show that Dcp-1 levels are affected by the proteasome and that the reduction of Dcp-1 is dependent on functional Hsp83.
Hsp83 loss induces Dcp-1-dependent compensatory autophagy that is required to maintain female fertility and larval viability
We previously found that Dcp-1 levels are increased and induce autophagic flux in response to starvation.11,12 To understand if the increased levels of autophagy in Hsp83 mutants are dependent on Dcp-1, we performed an epistasis experiment. We constructed double mutants consisting of Hsp83 transhets and the Dcp-1Prev1 loss-of-function alleles. Ovaries from Dcp-1Prev1 flies contain persisting nurse cell nuclei and premature loss of follicle cells.20 We found the MSECs of double mutants Dcp-1Prev1; Hsp83e6A/6–55 and Dcp-1Prev1; Hsp83e6D/6–55 flies have a phenotype similar to Dcp-1Prev1 but with several differences (Fig. 5A, B). The MSECs appear to have a premature loss of follicle cells, similar to Dcp-1Prev1 flies, but the nurse cell nuclei are only partially condensed relative to those observed in MSDECs from Hsp83 transhet mutants. The nurse cell nuclei in Hsp83; Dcp-1Prev1 double-mutant MSECs also stain positively for TUNEL similar to Hsp83, however, fail to stain for LTR.
Figure 5.
Dcp-1 is required for autophagic flux but not cell death resulting from loss of Hsp83. (A,B) Representative images of MSECs for the genotypes w1118, Hsp83e6D/6–55, Dcp-1Prev1 and Dcp-1Prev1; Hsp83e6D/6–55, stained with (A) LTR and DAPI or (B) LTR and TUNEL; experiments were performed on at least 8 females per genotype (n = 3), scale bars: 50 μm. (C)Representative images of first in-star larval fat bodies stained with LTR, DAPI and TUNEL; scale bars:10 μm. (D) Quantification of the total number of cells, determined by DAPI staining, that stained positively for LTR and/or TUNEL in larval fat bodies from listed genotype. Experiments were performed in triplicate with the total number of cells assessed listed in the table. (E) The ratio of dead pharate adult pupae to eclosed pupae was counted in vials containing different combinations of mutant Hsp83 and Dcp-1 alleles. Vials were incubated at room temperature for 14 d past first fly eclosion and then ratios were counted for at least 80 animals (n = 3). Error bars represent ± SEM and statistical significance was determined using one-way ANOVA with a Dunnet post-test (**P < 0.01). (F, G) The number of autolysosomes per cell was quantified in S2 cells stably expressing GFP-RFP-Atg8a and treated with the indicated dsRNAs. (F) All counts were normalized to the Amp dsRNA control. Atg1 and Rheb dsRNA's served as controls for decreasing and increasing the number of autolysosomes, respectively (n = 3). Error bars represent ± SEM and statistical significance was determined using one-way ANOVA with a Dunnet post-test, *P < 0.05, **P < 0.01,***P < 0.001. More than 100 cells were analyzed per treatment. (G) Representative images of GFP-RFP-Atg8a S2 cells following treatment with the indicated dsRNAs; scale bars:10 μm.
To determine if the relationship between Hsp83 and Dcp-1 occurs in other tissues we analyzed the larval fat body. The larval fat body is also a good model for monitoring autophagy as it is sensitive to oxidative stress and starvation.29,30 Fat bodies from Hsp83 transhets had increased TUNEL and LTR staining, similar to the ovaries. In addition, fat bodies from Dcp-1;Hsp83 double mutants had only TUNEL staining whereas control Hsp83 alleles, Dcp-1Prev1 and wild-type flies had no significant LTR or TUNEL (Fig. 5C, D).
The Hsp83;Dcp-1Prev1 double mutants also affected fecundity as both females and males were sterile whereas only male Hsp83 transhets are sterile and Dcp-1Prev1 flies are fertile. In addition, the double mutants showed a reduction in viability with a significant increase in the fraction of animals dying in the pharate adult stage and not being able to eclose, as quantified by the ratio of dead pharate adults to eclosed pupae (P < 0.01) (Fig. 5E). These results suggest that loss of Hsp83 can lead to activation of cell death through a pathway independent of Dcp-1, but that loss of Hsp83 induces compensatory autophagy through a pathway that requires Dcp-1.
To further validate the relationship between Hsp83 and Dcp-1, we conducted RNAi studies in the S2-GFP-RFP-Atg8a cell line as a more specific autophagy assay. As expected, Atg1 and Rheb showed a relative decrease and increase in the numbers of autolysosomes, respectively, compared with the control (Fig. 5F, G). The decrease in autolysosomes seen in the Dcp-1 RNAi-treated cells was reiterated in the double knockdown of Hsp83 and Dcp-1 and was in stark contrast to the increase observed in Hsp83 RNAi-treated cells.
Proteasome disruption induces Dcp-1-dependent compensatory autophagy
To determine if Dcp-1 is required for compensatory autophagy following proteasome disruption specifically we used RNAi targeted against Rpn11, a Drosophila proteasome regulatory subunit, that was shown to compromise proteasomal activity.31 Similar to Hsp83-RNAi, the Rpn11-RNAi resulted in enhanced levels of autolysosomes that were significantly reduced in combination with Dcp-1 RNAi (Fig. 6A, B). Since mutant Hsp83 transhets showed an increase in pro-Dcp-1, cleaved Dcp-1, and LTR staining, we investigated whether there was also an increase in cleaved Dcp-1 and LTR staining in proteasome-compromised flies in addition to the elevated pro-Dcp1 levels already observed (Fig. 4D, E). Rpn2/Dcr-2 flies kept at 25°C showed a significant increase in the number of MSDECs expressing cleaved Dcp-1 that were LTR positive (Fig. 6C, D). To determine if LTR levels correlated with Dcp-1 levels, we quantified staining at both 18°C and 25°C, and found a significant increase (P < 0.05) in LTR staining, along with increased Dcp-1 levels, at the higher temperature (Fig. 6E). Together, these findings support that the increase in autophagy detected in vitro and in vivo in response to reduced proteasome function is regulated by Dcp-1.
Figure 6.
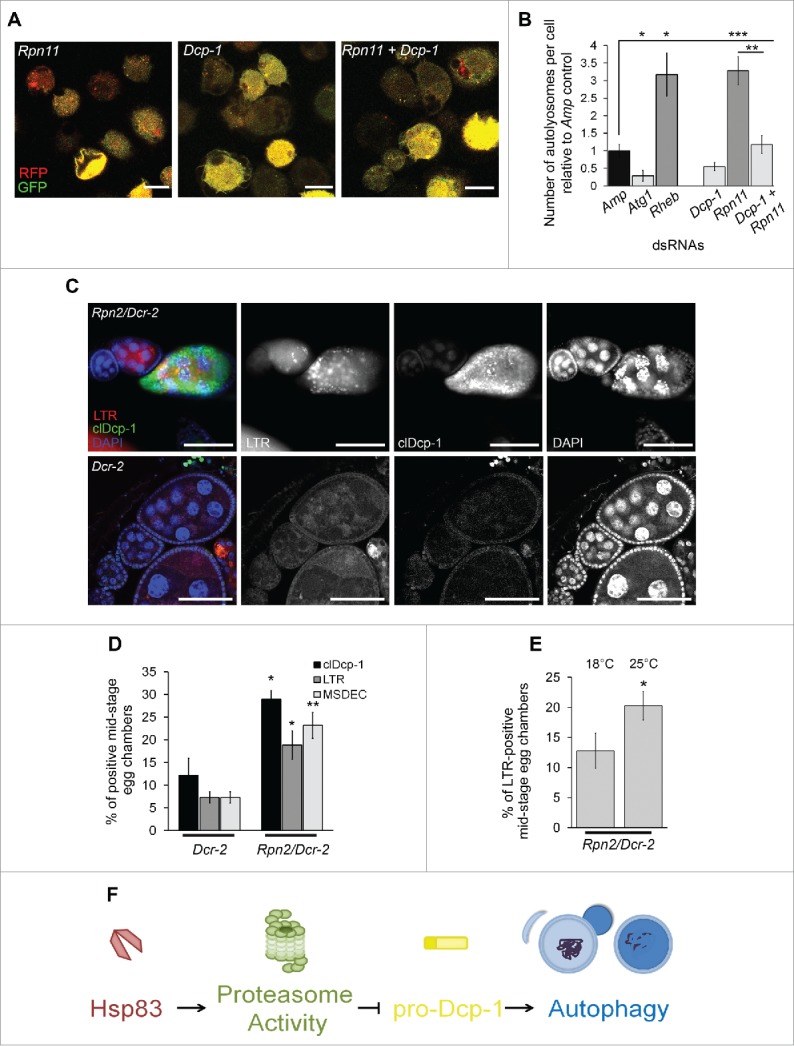
Proteasomal subunit loss results in Dcp-1-dependent compensatory autophagy. (A, B) The number of autolysosomes per cell was quantified in S2 cells stably expressing GFP-RFP-Atg8a and treated with the indicated dsRNAs. (A) Representative images of GFP-RFP-Atg8a S2 cells following treatment with the indicated dsRNAs; scale bars: 10 μm. (B) All counts were normalized to the Amp dsRNA control. Atg1 and Rheb dsRNA's served as controls for decreasing and increasing the number of autolysosomes, respectively. Error bars represent ± SEM and statistical significance was determined using one-way ANOVA with a Dunnet post-test, *P < 0.05, **P < 0.01,***P < 0.001 (C, D) Females were collected from flies with the UAS maternal driver kept at 25°C with the genotypes UAspDcr-2/+; nosGAL4/+ (Dcr-2) and Rpn2-RNAI/UAspDcr-2;nosGAL4/+ (Rpn2/Dcr-2). (C) Representative images of MSECs from Rpn2/Dcr-2 and Dcr-2 kept at 25°C. Ovaries were imaged with clDcp-1 antibody, LTR and DAPI; scale bars: 50 μm. (D) MSECS from Dcr-2 and Rpn2/Dcr-2 flies were scored for cleaved Dcp-1 (clDcp-1), LTR and DAPI. The graph represents the average percentage of MSECs that scored positive for LTR, MSDEC or clDcp-1. Experiments were performed with at least 8 females per genotype (n = 3). Error bars represent ± SEM and statistical significance was determined using the 2-way Student t test, *P < 0.05, **P < 0.01 (E) MSECS in Rpn2/Dcr-2 flies were scored for LTR positivity at 18°C and 25°C. At least 50 MSECS were analyzed per temperature (n = 4). Error bars represent ± SEM and statistical significance was determined using the 2-way Student t test, *P < 0.05 (F) A proposed pathway indicating that Hsp83 functions in basal conditions to contribute to proteasomal activity which suppresses pro-Dcp-1 levels and thus prevents activation of autophagy or cell death.
Discussion
In this study we identified 13 putative negative autophagy regulators that could be involved in Dcp-1-mediated regulation of autophagy via an IAP and MS-MS, and flow cytometry screen. Of those interactors, we further characterized the role of Hsp83 and found that it negatively regulates autophagy and cell death in Drosophila in vivo and in vitro. Hsp83 was found to interact with the zymogen pro-Dcp1, and loss of Hsp83 function led to a decrease in proteasomal activity and an increase in Dcp-1 that was not transcriptionally regulated. Analysis of proteasome-compromised flies also showed an increase in levels of Dcp-1 and LTR staining supporting that Dcp-1 is regulated by the proteasome. The Dcp-1 observed in Hsp83 mutants was found to be required for the ensuing compensatory autophagy, and contributed to both female fertility and development to adulthood. These findings highlight a novel role for Hsp83 in the proteasome-dependent regulation of pro-Dcp-1 that functions normally to prevent activation of autophagy (Fig. 6F).
Putative Dcp-1 interactors identified
It is possible that the 11 additional proteins identified in the IAP-MS/MS study may also positively or negatively regulate autophagy, but due to potential incomplete knockdown by RNAi or functional redundancy did not show an autophagy phenotype. Further in vivo studies to validate the remaining genes identified in the RNAi screen for their autophagy-regulatory role during Drosophila oogenesis or in other tissues will be valuable. It will be important to identify potential Dcp-1 cleavage substrates as it has been found that Dcp-1 catalytic activity is required to activate autophagy.11
pro-Dcp-1 levels are regulated by the proteasome
Caspases require constitutive regulation to prevent unwanted activation and cell death. Inhibitor of apoptosis proteins are one way caspases can be regulated, acting either through baculovirus inhibitor of apoptosis protein repeat (BIR) domain binding or E2-E3 ubiquitin ligase activity which can be degradative or nondegradative.32 While the active (cleaved) form of Dcp-1 is regulated by Diap1 through nondegradative ubiquitination,25 it was unknown how or if the zymogen form of Dcp-1 is regulated. We found that the elevated levels of pro-Dcp-1 in Hsp83 mutants neither correspond to Diap1 levels nor to transcript levels. Instead, we found that pro-Dcp-1 levels were elevated following the loss of function of Hsp83, 2 different proteasomal subunits or pharmacological inhibition of the proteasome. While other Drosophila caspases, Drice and Dronc, were found not to be stabilized by proteasome inhibition,25,33,34 there is precedence for proteasome regulation of the cleaved form, but not the proform, of CASP8 and CASP3 in human cell lines.35,36 Based on our protein interaction studies that showed Hsp83 only interacts with the proform of Dcp-1, we propose a model where Hsp83 acts at a relatively early stage, before Diap1, to regulate levels of pro-Dcp-1 via the proteasome in basal conditions. Functional loss of Hsp83 results in elevated pro-Dcp-1 that is available for the subsequent cleavage into active Dcp-1 and the induction of autophagy. Further studies are required to determine if pro-Dcp-1 itself is being directly processed by the proteasome or is instead indirectly regulated by the proteasome through as of yet unknown molecular components. Another outstanding question is the threshold of Dcp-1 required for autophagy induction.
Hsp83 mediates proteasomal degradation
It has been shown previously that Hsp83 plays a role in promoting protein degradation involving a degradative complex function. Loss of Hsp83 leads to a build-up of proteins targeted for degradation in the cell cycle due to inhibition of the anaphase-promoting complex/cyclosome.21 In a Drosophila neurodegenerative disease model, Hsp83 is required for targeting the mutant protein TPIsugarkill for proteasomal degradation,37 and, similarly, the human ortholog HSP90 is required for the delivery of substrates to the proteasome.38 While proteasomal activity was further decreased in Hsp83 transhets treated with MG132, the level of pro-Dcp-1 was not further increased, suggesting that Hsp83 specifically is a rate-limiting step in the proteasomal processing of pro-Dcp-1. However, it is still unknown if the physical interaction between pro-Dcp-1 and Hsp83 is ultimately important for proteasomal regulation of pro-Dcp-1, or if Hsp83 acts indirectly to control pro-Dcp-1 levels by promoting proteasomal activity. Since Hsp83 and pro-Dcp-1 could have been immuno-affinity purified in a complex involving the proteasome, further work is required to understand if pro-Dcp-1 requires Hsp83 for delivery or targeting to the proteasome or if Hsp83 might play a more direct role at the proteasome to facilitate degradation of pro-Dcp-1.
Dcp-1 regulates compensatory autophagy
Proteostasis requires a balance between the 2 major degradative UPS and autophagy pathways. When proteasome activity is inhibited, as is the case in Hsp83 mutants, there is increased autophagic flux known as compensatory autophagy.39 Previous to this study, Dcp-1 is reported to be a positive regulator of autophagy only in the context of starvation although overexpression also induces autophagy.11,12,40 Here, we observed a Dcp-1 dependent increase in the number of autolysosomes or lysosomal staining with loss of Hsp83 or Rpn11, and thus reduced proteasomal activity, even in nutrient-rich conditions. In fly models compensatory autophagy occurs when the UPS is inhibited,4,5 but this is the first time an effector caspase has been reported to be involved. The discovery that Dcp-1 is required for compensatory autophagy when proteasomal activity is compromised begs further exploration into whether there are other forms of stresses or contexts, such as hypoxia, that also rely on Dcp-1 for autophagy induction.
Hsp83;Dcp-1 double mutants have effects on cell death, cell division, autophagy, fertility and development
Loss of Dcp-1 prevents the autophagy that is otherwise induced in loss-of-function Hsp83 mutants, indicating a Dcp-1-dependent role in the regulation of autophagy in this context. In addition to autophagy dysregulation, double Dcp-1;Hsp83 mutants had other surprising phenotypes involving cell death and division. Dcp-1;Hsp83 flies had an abnormal number of persisting nurse cell nuclei that showed partial condensation and stained positively for TUNEL. This suggests that the cell death associated with Hsp83 loss is Dcp-1-independent and that Dcp-1 contributes at least partially to the pyknosis observed. The double mutants also rendered females sterile and did not rescue the male sterility observed in Hsp83 mutants, indicating an increasingly impaired phenotype. Furthermore, there was an increase in animals with developmental defects as more flies mutant with Hsp83 and Dcp-1 flies died in the pharate adult stage. This divergence between control of cell death and autophagy with Dcp-1 and Hsp83 is a unique and surprising finding. Discovery of this relationship provides a potential avenue for further understanding the complex cross-talk between autophagy and apoptosis, as well as for exploitation of promoting cell death without autophagy by counterintuitively targeting an effector caspase in the presence of Hsp83/HSP90 genetic or pharmacologic inhibition.
In summary, our Dcp1-related IAP-MS/MS strategy followed by an in vitro RNAi screen resulted in the identification of several novel regulators of autophagy and provides a foundation for further in vivo analyses. Further investigation into one of the interactors, Hsp83, led to the identification of its role not only in maintaining proteasomal activity but also suppressing autophagy through the maintenance of low levels of pro-Dcp-1. Understanding the role Hsp83 and its orthologs play is important as HSP90 plays a prominent role in cancer and neurodegeneration.10,41,42 The knowledge that inhibition of HSP90 function can affect proteasomal activity and induce compensatory autophagy is particularly pertinent as HSP90 is currently a drug target under trial for these diseases.41,42 Additionally, the proteasome is inhibited by the drug bortezomib for treatment of cancers such as multiple myeloma and mantle cell lymphoma and has been shown to induce autophagy in melanomas.43,44 The identification of a caspase being responsible for the regulation of compensatory autophagy could be conserved in humans and is an important avenue for future investigation. This study highlights the discovery of a previously unidentified proteostatic relationship in Drosophila between Hsp83 and Dcp-1 in vivo.
Materials and methods
Fly strains
w1118 was used as the wild-type control strain in this study. A complete list of stocks can be found in Table 2. All flies were collected in nutrient-rich conditions at room temperature unless otherwise stated.
Table 2.
Drosophila stock list.
| Genotype | Description | Source |
|---|---|---|
| Hsp83e6A/TM6B | point mutation in Hsp83 gene causing amino acid replacement: S592F | Bloomington Stock Center (Stock number 36576) |
| Hsp83e6D/TM6B | point mutation in Hsp83 gene causing amino acid replacement: E317K | Bloomington Stock Center (Stock number 5696) |
| Hsp836–55/TM6B | point mutation in Hsp83 gene causing amino acid replacement: P380S | B. Edgar (Heidelberg University, Heidelberg, Germany).21 |
| Dcp-1Prev1 | contains a 40-bp partial P element insertion in the coding region of Dcp-1, resulting in an in-frame stop | K. McCall (Boston University, Boston, MA)20 |
| UASp-GFP-mCherry-Atg8a | Expression of GFP-mCherry-Atg8a under the UASp promoter | T.E. Rusten (Center for Cancer Biomedicine, Oslo University Hospital, Montebello, Oslo, Norway)18 |
| UAS-Dcr-2; nosGAL4 | Female germline driver and construct of Dcr-2/Dicer-2 driven under the UAS promoter | Bloomington Stock Center (Stock number 25751) |
| 20ProtS-GD | RNAi strain targeted against the gene encoding proteasomal subunit Prosα1 | Vienna Drosophila Resource Center (transformant ID 49681) |
| Rpn2-KK | RNAi strain targeted against the gene encoding regulatory proteasomal subunit Rpn2 | Vienna Drosophila Resource Center (transformant ID 106457) |
| MTD-Gal-4 | Maternal triple driver used for crosses | Bloomington Stock Center (Stock number 31777) |
| sco/CyO;TM6B/MKRS | Balancer stock used for crosses | Bloomington Stock Center (Stock number 3703) |
| UASt-CL1-GFP | Expression of CL1-GFP under the UASt promoter | U. Pandey (Children's Hospital of Pittsburgh, University of Pittbursgh Medical Center, Pittsburgh, PA)5 |
| Cg-GAL4 | GAL4 driver in hemocytes, fat body and lymph gland used for larval fat body experiments | Bloomington Stock Center (Stock number 7011) |
| UASp-Diap1.P | Expression of Diap1 under the UASp promoter | Bloomington Stock Center (Stock number 63820) |
Cell culture
Drosophila l(2)mbn cells were grown in Schneider medium (Invitrogen, 11720–034) supplemented with 10% fetal bovine serum in 25-cm2 suspension cell flasks (Sarstedt, 83.1810.502) at 25°C. Drosophila S2-GFP-RFP-Atg8a cells were grown in ESF921 medium (Expression Systems, 96–001–01) in 25-cm2 suspension cell flasks at 25°C and treated with 50 μg/mL Zeocin (Invitrogen, R250–01). All experiments were performed 3 to 4 d after passage of cells.
Immuno-affinity purification (IAP) and MS/MS analysis
A detailed protocol for the IAP and MS/MS is described in a previous paper.11 Putative interacting proteins were identified by having an X!Tandem log(e) score less than −3 and were identified in at least 2 experimental samples (V5–Dcp-1C<A) and not in the negative control (V5 vector only). Protein sequences corresponding to the identified peptides were obtained from FlyBase. A subset of high-confidence candidates that met the criteria of unique peptides ≥ 1, log(E) <−3 and observed in at least 2 of 4 experiments from both fed and 2-h starvation conditions were selected.
dsRNA synthesis and RNAi
Each PCR primer for RT-PCR was designed to contain a 5’ T7 RNA polymerase-binding site (TAATACGACTCACTATAGG) followed by sequences specific for the target gene. The ampicillin resistance gene was used as a control dsRNA. The PCR products were generated by RT-PCR using Superscript one-step RT-PCR with Platinum Taq (Invitrogen). RT-PCR products were ethanol precipitated and used as a template for in vitro transcription reactions using T7 RiboMax Express RNAi systems (Promega). Quality of the RNA was analyzed by gel electrophoresis. dsRNA was quantified using PicoGreen and adjusted to 200 to 400 ng/μL with nuclease-free water. PCR primers for dsRNA synthesis are listed in Table 3.
Table 3.
Sets of primers used for dsRNA synthesis.
| Gene | Forward Primer Sequence (5′-3′) | Reverse Primer Sequence (5′-3′) |
|---|---|---|
| 1st set | ||
| 14–3–3ε | TAATACGACTCACTATAGGGACAGGTGGAGAAGGAGCTG | TAATACGACTCACTATAGGTCAGTGTATCCAACTCGGCA |
| 14–3–3ζ | TAATACGACTCACTATAGGGTCACAGAGACTGGCGTTGA | TAATACGACTCACTATAGGCGTAGCAGATTTCCCTCAGC |
| ATPsynβ | TAATACGACTCACTATAGGCGTCGATGTCCAGTTTGATG | TAATACGACTCACTATAGGTGATGCGTCCTAGTGTTTCG |
| blw | TAATACGACTCACTATAGGTGTGTTCTACCTGCATTCGC | TAATACGACTCACTATAGGCACGTACTGACCCTGCTTGA |
| CCT2 | TAATACGACTCACTATAGGGTGGACAACATCATCCGTTG | TAATACGACTCACTATAGGCAGCACTCATCCTCGAATCA |
| Clic | TAATACGACTCACTATAGGTTCCGTACCAATTTTGAGGC | TAATACGACTCACTATAGGATCAGCTCACAGTCGAAGCA |
| eEF1α1 | TAATACGACTCACTATAGGGTGACTCCAAGGCTAACCCC | TAATACGACTCACTATAGGTTAGAGGGCACCAGGTTGAC |
| eEF1β | TAATACGACTCACTATAGGGAAGTCTAAGAAACCCGCCC | TAATACGACTCACTATAGGTGCTTAGTTCGCTTTGCTCA |
| eEF1γ | TAATACGACTCACTATAGGGGTGTTCATGTCGTGCAATC | TAATACGACTCACTATAGGGAAGATCTTGCCCTGGTTGA |
| eIF4A | TAATACGACTCACTATAGGTTACGTCAACGTGAAGCAGG | TAATACGACTCACTATAGGAATGTAGTTCTCGCGGTTCG |
| Hsp60A | TAATACGACTCACTATAGGCACCCTCACCGATATGGC | TAATACGACTCACTATAGGGGGTGTCGTCCTTGGTGA |
| Hsc70–4 | TAATACGACTCACTATAGGATCTGACCACCAACAAGCGT | TAATACGACTCACTATAGGATGACCGACTTGTCCAGCTT |
| Hsp70Aa | TAATACGACTCACTATAGGGGGAGGATTTGGAGGCTACT | TAATACGACTCACTATAGGTCGATCGAAACATTCTTATCAGTC |
| Hsc70Cb | TAATACGACTCACTATAGGGAAAAACACAGTTGGCGGAT | TAATACGACTCACTATAGGGGTCTTGGCGTTGATCTTGT |
| Hsp83(1) | TAATACGACTCACTATAGGGAGCTGAACAAGACCAAGCC | TAATACGACTCACTATAGGTTGCGGATCACCTTTAGGAC |
| Jafrac1 | TAATACGACTCACTATAGGATGGAGTGCTCGATGAGGAG | TAATACGACTCACTATAGGTACTCCTTGGACTTGGTGGG |
| Mi-2 | TAATACGACTCACTATAGGCGCAAGTACGACATGGAAGA | TAATACGACTCACTATAGGTCGACCTTGAGCTTGGACTT |
| Rack1 | TAATACGACTCACTATAGGACCTCAATGACGGCAAGAAC | TAATACGACTCACTATAGGATTTGACGCCCGTTACAAAG |
| REG | TAATACGACTCACTATAGGCCATTCAAGAGGACACGCTT | TAATACGACTCACTATAGGACAAGAATTGCTGACCGTCC |
| sesB | TAATACGACTCACTATAGGCTGATACTGGCAAGGGTGGT | TAATACGACTCACTATAGGCCCAGCTGATGTAGATGGGT |
| sgl | TAATACGACTCACTATAGGAATCTCCAGCATCAATTCGC | TAATACGACTCACTATAGGAACAAACTCATCCCACTCCG |
| sta | TAATACGACTCACTATAGGAGTTCGCCAAGTACACCGAC | TAATACGACTCACTATAGGGGATCGCGGTAGAAGAACAG |
| TER94 | TAATACGACTCACTATAGGCATGGGAGCCAAGAAGAATG | TAATACGACTCACTATAGGGTCACCTTGGCGATGTAGGT |
| Uba1 | TAATACGACTCACTATAGGGATTTCGCAAAGCTGGACTC | TAATACGACTCACTATAGGTAGGCTTCTGCACATCATGC |
| 2nd set | ||
| ATPsynβ | TAATACGACTCACTATAGGCTCCTGGCTCCATACGC | TAATACGACTCACTATAGGATATGGCCTGAACAGAAGTAAT |
| blw | TAATACGACTCACTATAGGGTACTGCATCTACGTCGCCA | TAATACGACTCACTATAGG ACGTTGGTTGGAATGTAGGC |
| Clic | TAATACGACTCACTATAGGATCAGCCTGAAGGTGACGAC | TAATACGACTCACTATAGGACAGGTTCTCGATCAGGGTG |
| eIF4A | TAATACGACTCACTATAGGTCGATTGCTATCCTTCAGCA | TAATACGACTCACTATAGGGATCTGATCCTTGAAACCGC |
| Hsp60A | TAATACGACTCACTATAGGGGGAGGGAGATGTGATGAGA | TAATACGACTCACTATAGGGCGAAGCAAAACAAAGTTCC |
| Hsc70–4 | TAATACGACTCACTATAGGGGCTGACAAGGAGGAGTACG | TAATACGACTCACTATAGGTGTCGTTTGACCCGTTTGTA |
| Hsp70Aa | TAATACGACTCACTATAGGCCCACTTTCATTGGGAATTG | TAATACGACTCACTATAGGAATGCATTGTTGTCCTTCGTC |
| Hsp83(2) | TAATACGACTCACTATAGGATTGCTCAGCTGATGTCCCT | TAATACGACTCACTATAGGGGAGTAGAAACCCACACCGA |
| Mi-2 | TAATACGACTCACTATAGGATTTGCGTGGTAAATCGGAG | TAATACGACTCACTATAGGGTTCTTGCTTCACCTCGCTC |
| REG | TAATACGACTCACTATAGGGTTGATCCTCAAGGCAGAGC | TAATACGACTCACTATAGGTCCTCCACAAGCTTCCTGAT |
| sesB | TAATACGACTCACTATAGGGCAAGAACCCTTCCTTCCTC | TAATACGACTCACTATAGGTTCGGAGGCGAAAGAATCTA |
| sta | TAATACGACTCACTATAGGTTTCCACGTTAACATGTCGG | TAATACGACTCACTATAGGCCCAGGTTGAGGATGTTGAC |
| TER94 | TAATACGACTCACTATAGGGCATGATGATGTTGACCTGG | TAATACGACTCACTATAGGCTGCATGCCAAACTTCAAGA |
| Additional primer sets | ||
| Hsp83A | TAATACGACTCACTATAGGAAATCCCTGACCAACGACTG | TAATACGACTCACTATAGGTTGCGGATCACCTTTAGGAC |
| Hsp83B | TAATACGACTCACTATAGGGAGCTGAACAAGACCAAGCC | TAATACGACTCACTATAGGGAGTCGACCACACCCTTCAT |
| Rpn11 | TAATACGACTCACTATAGGCTGCTACGTCTTGGAGGTGCTATGCCACAGG | TAATACGACTCACTATAGGACAGTGCTCATTGTAGTCGGACAACGTGAGGC |
l(2)mbn cells and S2-GFP-RFP-Atg8a were washed and resuspended in ESF921 medium (Expression Systems, 96–001–01) to a concentration of 2 × 106 cells/mL. Cells (333 μL for 24-well plates or 1 mL for 6-well plates) were plated. dsRNAs were added at 10 μg (for 24-well plates) or 30 μg (for 6-well plates) per well and incubated at 25°C for 1 h. Following incubation, 667 μL (for 24-well plates) or 2 mL (for 6-well plates) of Schneider medium + 10% FBS was added back to each well and incubated for an additional 72 h at 25°C. S2-GFP-RFP-Atg8a cells were transferred to an 8-well Chamber Slide (Thermo Fisher Scientific, 154941) overnight and the next d treated with a second dose of dsRNAs in E2F921 for 7 h before fixation. Puncta were either counted manually from saved microscopy images or automatically by using a contours discovery algorithm in OpenCV Python package as described in,45 was applied to images preprocessed by filtering colors and applying a Gaussian filter and adaptive threshold.
For LTG (Invitrogen, L-7526) experiments for flow cytometry, 66 μL of 2 × 106 cells/mL were plated in triplicate in a 96-well plate and analyzed on a BD FACSCalibur™ (BD Biosciences, San Jose, CA USA). dsRNA (10 μg) was added per well and incubated for 1 h at room temperature. Schneider medium + 10% FBS (134 μL) was added back to each well and incubated for 72 h at 25°C. Cells were then treated with a second dose of dsRNAs in ESF921 for 7 h before analysis.
Immunofluorescence studies and microscopy
Flies were conditioned on wet yeast paste for 2 d before dissection. Ovaries were dissected in phosphate-buffered saline (PBS; Sigma, P3813) and fixed with 4% paraformaldehyde. Flies expressing fluorescent reporters were then mounted with Slowfade Gold Antifade Reagent with DAPI (Invitrogen, S36939). Fluorescence intensity for the CL1-GFP reporter was measured by outlining the fat body observed in the DAPI channel and measuring the mean gray value from the unadjusted images in the GFP channel and subtracting the mean background using ImageJ 1.45s (https://imagej.nih.gov/ij/).
For immunofluorescence, ovaries were washed with PBS-T (PBS+0.3% Triton X-100 [Sigma, T8787]), permeabilized with 0.5% Triton X-100 and blocked with 2% BSA (Sigma, A2153) in PBS-T after fixation. The cleaved Dcp-1 antibody (Cell Signaling Technology, 9578) was diluted in 0.5% BSA+ PBS-T and incubated overnight at 4°C. Secondary antibodies anti-rabbit Alexa Fluor 546 or Alexa Fluor 488 (Invitrogen, A-11010, A-11008) were incubated at room temperature for 2 h and subsequently washed with PBS-T. Slides were then mounted as described above.
All microscopy images were acquired at room temperature using one of 3 confocal apparatuses. Confocal images were taken using a Nikon Confocal C1 microscope equipped with a Plan APO 60X/1.45 oil immersion objective (Nikon, Melville, NY USA) with EZ-C1 Ver 3.00 software (Nikon), Nikon A1R Eclipse Ti inverted Laser Scanning Confocal Microscope with a Plan APO 60X/1.40 oil immersion objective (Nikon, Melville, NY USA) with acquisition software NIS Elements AR 4.2 (Nikon), and Leica TCS SP8 inverted confocal microscope with a Leica HC PL APO 63x/1.40 oil objective and LAS AF software (Leica, Buffalo Grove, IL USA). The pinhole and laser brightness settings were kept constant by applying the same properties between comparable experiments. Brightness and contrast were adjusted using Photoshop (CC 2014, Adobe) and applied to the whole image.
LysoTracker® Red and TUNEL stains in ovaries and larval fat bodies
Ovaries were dissected in PBS 3 or 4 d after eclosion. Larval fat bodies were dissected in PBS from first in-star larva. Both tissues were then incubated with 50 μM LTR DND-99 (Invitrogen, L-7528) in PBS for 3 min in the dark. Tissues were washed 3 times with PBS and then fixed for 20 min with 4% paraformaldehyde in PBS. For TUNEL staining, fixed tissues were washed 3 times with PBS + 0.1% Triton-X-100. TUNEL reaction was performed using the DeadEnd Fluorimetric Tunel System (Promega, G3250). Samples were mounted as described above.
Protein extraction and western blot analysis
Cell, ovary and whole body lysates were extracted using RIPA lysis buffer (Santa Cruz Biotechnology, sc-24948) plus complete protease inhibitors (Roche, 11697498002). Proteins were quantified using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, PI23228). Proteins were separated on a 4–12% or 10% NuPAGE Bis-Tris gels (Invitrogen, NP/0335 or NP0316) and transferred to PVDF membranes (Bio-Rad, 170-5061). Membranes were blocked in milk or Odyssey blocking buffer (LI-COR Inc., 927-40003) and incubated in primary antibodies overnight at 4°C.
Primary antibodies included mouse anti-ACTA/actin (1:500, Developmental Studies Hybridoma Bank, JLA20), mouse anti-TUBB/tubulin (1:1000, Developmental Studies Hybridoma Bank, E7), rabbit anti-Dcp-1 (1:1000, a gift from K. McCall; Boston University, Boston, MA), and rabbit anti-HSP90 (1:1000, Cell Signaling Technology, 4874S). Membranes were incubated with HRP-conjugated secondary antibodies (Santa Cruz Biotech, SC2004/SC2005) or IR-labeled secondary antibodies (VWR, 610–132–121/610–132–122) and were detected using the Amersham ECL™ Enhanced Western Blotting System (VWR, RPN2106, Radnor, PA, USA) or the Odyssey System (LI-COR Biosciences, 927-40003, Lincoln, NE USA). Densitometry was performed using Image Quant 5.1 software (GE Healthcare) or Image Lab 5.1 software (BioRad).
Purification of Dcp-1 and in vitro cleavage assays
For transfection experiments, 5 μg of His-V5-Dcp-1FL or His-V5-Dcp-1C<A plasmid DNA46 was added to 1 mL of Grace's insect medium (Invitrogen, 11595–030) and vortexed to mix. 100 μl of Cellfectin II (Invitrogen, 10362–100) was added to the DNA-Grace mixture and was incubated for at least 30 min. Cells (3.75 × 106) in 4 mL of Grace medium were incubated with DNA-Grace-Cellfectin II transfection medium overnight. Schneider medium + 10% FBS (10 mL) was added back to the cells and the cells were incubated for an additional 3 d before Ni-NTA purification. Purification of His-V5-Dcp-1FL or His-V5-Dcp-1C<A was performed using HisPurTM Ni-NTA Spin Columns (Thermo Fisher Scientific, 88224). Cells were resuspended in 400 μL of Equilibration Buffer (PBS, 10 mM imidazole, pH 7.4) with 1% Triton X-100 (Sigma, T8787), incubated on a rotary shaker at 4°C for 10 min and centrifuged at 22000 g at 4°C for 15 min to remove insoluble material. Supernatants were added to the Ni-NTA beads and mixed at 4°C for 30 min. The column was centrifuged at 22000 g at 4°C for 2 min to remove the flow-through. The columns were then washed 3 times with 400 μL of wash buffer (PBS, 25 mM imidazole), and His-tagged Dcp-1 was eluted 3 times with 200 μL of elution buffer (PBS, 250 mM imidazole). The elutions were assayed for caspase activity using the Caspase-Glo 3/7 Kit (Promega, G8091). Eluate (10 μL) was added to 100 μL of Caspase-Glo 3/7 Reagent and incubated at 25°C for 30 min. Luminescence was detected using the Wallac1420 Victor plate reader (Perkin Elmer, Waltham, MA USA). The elutions were immediately used for in vitro cleavage assays. The caspase reaction buffer used for in vitro cleavage assays was as described previously in.46 A 100-μL reaction consisting of increasing volumes of Ni-NTA purified Dcp-1FL or Dcp-1C<A (10, 20, 40 μL) and 5 μL of in vitro translated Hsp83 or Drice incubated in caspase reaction buffer (10 mM Tris, pH 7.5, 150 mM NaCl, 2 mM DTT, 0.1% Triton X-100). The reaction mixture was incubated at 25°C overnight and precipitated with 400 μL of acetone for western blot analysis.
Quantitative RT-PCR (QRT-PCR) analysis
Flies were collected and ground in 1 mL TRIZOL (Ambion, 15596–026) and total RNA was extracted according to the manufacturer's instructions. RNA was treated with DNase, and QRT-PCR was performed using the One-Step SYBR green RT-PCR reagent kit (Applied Biosystems, 4389986) on a 7900 Sequence Detection System (Applied Biosystems, Foster City, CA USA). Expression levels were calculated using the comparative threshold method with Drosophila rp49 as a reference gene. QRT-PCR primers are as follows: rp49, 5’-ATACAGGCCCAAGATCGTGA-3’ and 5’-GCACTCTGTTGTCGATACCCTT-3’, and Dcp-1, 5’-CCGGAGTCTCTTGTGTTGGT-3’ and 5’-GTATTCGCTTGCATATCGTTCC-3’.
Proteasome activity assay
Animals were collected 3 to 4 d after eclosion and frozen immediately on dry ice. Microcentrifuge tubes were weighed before and after on an analytical balance to determine the mass. Whole bodies were ground and incubated for 30 min on ice in 0.5 mL of lysis buffer (50 mM HEPES, pH 7.5, 5 mM EDTA, 150 mM NaCl, 1% Triton X-100). Lysates were then separated by centrifugation at 22000 g for 15 min at 4°C. In a 96-well black plate 50 μL of lysate was added to 50 μL of chymotrypsin-like, caspase-like, trypsin-like peptide substrates from the Proteasome-Glo kit (Promega, G8531). The substrates incubated with the lysates for 10 min before the luminescence reading was measured on a Synergy H4 Hybrid (BioTek, Winooski, VT USA). Relative luminescence was then determined proportionally to total mass per sample.
MG132 treatment
Flies were fed a final concentration of 50 μM of MG132 (Cedarlane, A2585g-10mg) resuspended in DMSO and diluted with 5% sucrose (Sigma, 84097) and green food coloring. This solution was aliquoted onto wet yeast paste and fed to flies for 4 d. The control solution had the same concentrations of DMSO, sucrose and food coloring without MG132. Ingestion of the solutions was indicated by green food coloring visible in the abdomen.
Statistics
In each graph, data represent standard error of mean (SEM) of n independent experiments. As indicated in the legends, statistical significance was calculated by analysis of variance (ANOVA) plus a Dunnett or Bonferroni post test, or a 2-tailed Student t test between the indicated samples using GraphPad Prism (GraphPad Software). P values are shown in the legends.
Supplementary Material
Abbreviations
- Atg
autophagy related
- BafA1
bafilomycin A1
- Caspase
cysteine-dependent aspartate-specific protease
- CL1-GFP
fusion protein with a degradation signal in GFP
- clDcp-1
cleaved Dcp-1
- DAPI
4’6-diamidino-2-phenylindole
- Dcp-1
death caspase-1
- Dcr-2
dicer-2
- Diap-1
death-associated inhibitor of apoptosis 1
- DMSO
dimethylsulfoxide
- Drice
death related ICE-like caspase
- Dronc
Death regulator Nedd2-like caspase
- dsRNA
double-stranded ribonucleic acid
- GFP
green fluorescent protein
- Hsp
heat shock protein
- HSP90
human heat shock protein 90
- IAP
immuno-affinity purification
- l(2)mbn
lethal (2) malignant blood neoplasm
- LC
liquid chromatography
- LTG
LysoTracker® Green
- LTR
LysoTracker® Red
- mCherry
monomeric red fluorescent construct
- MG132
proteasome inhibitor
- MKRS
stubble bristles balancer chromosome
- MS
mass spectrometry
- MSDEC
mid-stage degenerating egg chamber
- MSEC
mid-stage egg chamber
- PBS
phosphate-buffered saline
- Prosα1
proteasome α1 subunit
- qRT-PCR
quantitative reverse transcription polymerase chain reaction
- RFP
red fluorescent protein
- Rheb
ras homolog enriched in brain
- RNAi
ribonucleic acid interference
- Rpn
regulatory particle non-ATPase
- S2
Schneider 2 cells
- S6K
ribosomal protein S6 kinase
- sesB
stress-sensitive B
- transhet
transheterozygote
- TUNEL
terminal deoxynucelotidyl transferase dUTP nick end labeling
- UPS
ubiquitin-proteasome system
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors would like to thank members of the Gorski laboratory and L. Megeney for helpful discussions, and S. Bortnik and V. Dreizen for assistance in puncta quantitfication. They would also like to thank K. McCall, B. Edgar, P. Meier, U. Pandey, Bloomington Stock Center and VDRC stock center for reagents, G. Sharma for flow cytometry assistance and T. Heslip, C. Soong and D. Luciani for confocal microscope access and support.
Funding
This work was supported by a CIHR Operating Grant (MOP- 78882) to SMG and GBM, and a NSERC Operating Grant (RGPIN/04982–2015) to SMG. SMG was supported in part by a CIHR New Investigator Salary Award. CC was supported in part by a NSERC Doctoral Postgraduate Scholarship.
References
- [1].Labbadia J, Morimoto RI. The biology of proteostasis in aging and disease. Annu Rev Biochem 2015; 84:435-64; PMID:25784053; https://doi.org/ 10.1146/annurev-biochem-060614-033955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ding W-X, Ni HM, Gao W, Yoshimori T, Stolz DB, Ron D, Yin XM. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol 2007; 171:513-24; PMID:17620365; https://doi.org/ 10.2353/ajpath.2007.070188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Iwata A, Riley BE, Johnston JA, Kopito RR. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem 2005; 280:40282-92; PMID:16192271; https://doi.org/ 10.1074/jbc.M508786200 [DOI] [PubMed] [Google Scholar]
- [4].Lőw P, Varga Á, Pircs K, Nagy P, Szatmári Z, Sass M, Juhász G. Impaired proteasomal degradation enhances autophagy via hypoxia signaling in Drosophila. BMC Cell Biol 2013; 14:29; PMID:23800266; https://doi.org/ 10.1186/1471-2121-14-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, et al.. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature 2007; 447:859-63; PMID:17568747; https://doi.org/ 10.1038/nature05853 [DOI] [PubMed] [Google Scholar]
- [6].Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature 2011; 475:324-32; PMID:21776078; https://doi.org/ 10.1038/nature10317 [DOI] [PubMed] [Google Scholar]
- [7].Echeverría PC, Bernthaler A, Dupuis P, Mayer B, Picard D. An interaction network predicted from public data as a discovery tool: Application to the Hsp90 molecular chaperone machine. PLoS One 2011; 6:e26044; PMID:22022502; https://doi.org/ 10.1371/journal.pone.0026044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kang BH, Plescia J, Dohi T, Rosa J, Doxsey SJ, Altieri DC. Regulation of tumor cell mitochondrial homeostasis by an organelle-specific Hsp90 chaperone network. Cell 2007; 131:257-70; PMID:17956728; https://doi.org/ 10.1016/j.cell.2007.08.028 [DOI] [PubMed] [Google Scholar]
- [9].Pick E, Kluger Y, Giltnane JM, Moeder C, Camp RL, Rimm DL, Kluger HM. High HSP90 expression is associated with decreased survival in breast cancer. Cancer Res 2007; 67:2932-7; PMID:17409397; https://doi.org/ 10.1158/0008-5472.CAN-06-4511 [DOI] [PubMed] [Google Scholar]
- [10].Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer 2005; 5:761-72; PMID:16175177; https://doi.org/ 10.1038/nrc1716 [DOI] [PubMed] [Google Scholar]
- [11].DeVorkin L, Go NE, Hou YC, Moradian A, Morin GB, Gorski SM. The Drosophila effector caspase Dcp-1 regulates mitochondrial dynamics and autophagic flux via SesB. J Cell Biol 2014; 205:477-92; PMID:24862573; https://doi.org/ 10.1083/jcb.201303144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hou Y-CC, Chittaranjan S, Barbosa SG, McCall K, Gorski SM. Effector caspase Dcp-1 and IAP protein Bruce regulate starvation-induced autophagy during Drosophila melanogaster oogenesis. J Cell Biol 2008; 182:1127-39; PMID:18794330; https://doi.org/ 10.1083/jcb.200712091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lee REC, Brunette S, Puente LG, Megeney LA. Metacaspase Yca1 is required for clearance of insoluble protein aggregates. Proc Natl Acad Sci U S A 2010; 107:13348-53; PMID:20624963; https://doi.org/ 10.1073/pnas.1006610107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Wirawan E, Vande Walle L, Kersse K, Cornelis S, Claerhout S, Vanoverberghe I, Roelandt R, De Rycke R, Verspurten J, Declercq W, et al.. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis 2010; 1:e18; PMID:21364619; https://doi.org/ 10.1038/cddis.2009.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bandyopadhyay U, Kaushik S, Varticovski L, Cuervo AM. The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol Cell Biol 2008; 28:5747-63; PMID:18644871; https://doi.org/ 10.1128/MCB.02070-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mori M, Hitora T, Nakamura O, Yamagami Y, Horie R, Nishimura H, Yamamoto T. Hsp90 inhibitor induces autophagy and apoptosis in osteosarcoma cells. Int J Oncol 2015; 46:47-54; PMID:25351442; https://doi.org/ 10.3892/ijo.2014.2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Xu C, Liu J, Hsu LC, Luo Y, Xiang R, Chuang TH. Functional interaction of Hsp90 and Beclin 1 modulates Toll-like receptor-mediated autophagy. FASEB J 2011; 25:2700-10; PMID:21543763; https://doi.org/ 10.1096/fj.10-167676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Nezis IP, Shravage BV, Sagona AP, Lamark T, Bjørkøy G, Johansen T, Rusten TE, Brech A, Baehrecke EH, Stenmark H. Autophagic degradation of dBruce controls DNA fragmentation in nurse cells during late Drosophila melanogaster oogenesis. J Cell Biol 2010; 190:523-31; PMID:20713604; https://doi.org/ 10.1083/jcb.201002035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al.. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8:445-544; PMID:22966490; https://doi.org/ 10.4161/auto.19496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Laundrie B, Peterson JS, Baum JS, Chang JC, Fileppo D, Thompson SR, McCall K. Germline cell death is inhibited by P-element insertions disrupting the dcp-1/pita nested gene pair in Drosophila. Genetics 2003; 165:1881-8; PMID:14704173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bandura JL, Jiang H, Nickerson DW, Edgar BA. The molecular chaperone Hsp90 is required for cell cycle exit in Drosophila melanogaster. PLoS Genet 2013; 9:e1003835; PMID:24086162; https://doi.org/ 10.1371/journal.pgen.1003835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yue L, Karr TL, Nathan DF, Swift H, Srinivasan S, Lindquist S. Genetic analysis of viable Hsp90 alleles reveals a critical role in Drosophila spermatogenesis. Genetics 1999; 151:1065-79; PMID:10049923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Giorgi F, Deri P. Cell death in ovarian chambers of Drosophila melanogaster. J Embryol Exp Morphol 1976; 35:521-33; PMID:820828. [PubMed] [Google Scholar]
- [24].Song Z, Guan B, Bergman A, Nicholson DW, Thornberry NA, Peterson EP, Steller H. Biochemical and genetic interactions between Drosophila caspases and the proapoptotic genes rpr, hid, and grim. Mol Cell Biol 2000; 20:2907-14; PMID:10733594; https://doi.org/ 10.1128/MCB.20.8.2907-2914.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ditzel M, Broemer M, Tenev T, Bolduc C, Lee TV, Rigbolt KT, Elliott R, Zvelebil M, Blagoev B, Bergmann A, et al.. Inactivation of effector caspases through nondegradative polyubiquitylation. Mol Cell 2008; 32:540-53; PMID:19026784; https://doi.org/ 10.1016/j.molcel.2008.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Imai J, Maruya M, Yashiroda H, Yahara I, Tanaka K. The molecular chaperone Hsp90 plays a role in the assembly and maintenance of the 26S proteasome. EMBO J 2003; 22:3557-67; PMID:12853471; https://doi.org/ 10.1093/emboj/cdg349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yamano T, Mizukami S, Murata S, Chiba T, Tanaka K, Udono H. Hsp90-mediated assembly of the 26 S proteasome is involved in major histocompatibility complex class I antigen processing. J Biol Chem 2008; 283:28060-5; PMID:18703510; https://doi.org/ 10.1074/jbc.M803077200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science 2001; 292:1552-5; PMID:11375494; https://doi.org/ 10.1126/science.292.5521.1552 [DOI] [PubMed] [Google Scholar]
- [29].Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell 2004; 7:167-78; PMID:15296714; https://doi.org/ 10.1016/j.devcel.2004.07.009 [DOI] [PubMed] [Google Scholar]
- [30].Wu H, Wang MC, Bohmann D. JNK protects Drosophila from oxidative stress by trancriptionally activating autophagy. Mech Dev 2009; 126:624-37; PMID:19540338; https://doi.org/ 10.1016/j.mod.2009.06.1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lundgren J, Masson P, Realini CA, Young P. Use of RNA interference and complementation to study the function of the Drosophila and human 26S proteasome subunit S13. Mol Cell Biol 2003; 23:5320-30; PMID:12861018; https://doi.org/ 10.1128/MCB.23.15.5320-5330.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Vaux DL, Silke J. IAPs – the ubiquitin connection. Cell Death Differ 2005; 12:1205-7; PMID:16094398; https://doi.org/ 10.1038/sj.cdd.4401696 [DOI] [PubMed] [Google Scholar]
- [33].Lee TV, Fan Y, Wang S, Srivastava M, Broemer M, Meier P, Bergmann A. Drosophila IAP1-mediated ubiquitylation controls activation of the initiator caspase DRONC independent of protein degradation. PLoS Genet 2011; 7:e1002261; PMID:21909282; https://doi.org/ 10.1371/journal.pgen.1002261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lee TV, Kamber Kaya HE, Simin R, Baehrecke EH, Bergmann A. The initiator caspase Dronc is subject of enhanced autophagy upon proteasome impairment in Drosophila. Cell Death Differ 2016; 23:1555-64; PMID:27104928; https://doi.org/ 10.1038/cdd.2016.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Fiandalo MV, Schwarze SR, Kyprianou N. Proteasomal regulation of caspase-8 in cancer cell apoptosis. Apoptosis 2013; 18:766-76; PMID:23456622; https://doi.org/ 10.1007/s10495-013-0821-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Suzuki Y, Nakabayashi Y, Takahashi R. Ubiquitin-protein ligase activity of X-linked inhibitor of apoptosis protein promotes proteasomal degradation of caspase-3 and enhances its anti-apoptotic effect in Fas-induced cell death. PNAS 2001; 98:8662-7; PMID:11447297; https://doi.org/ 10.1073/pnas.161506698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hrizo SL, Palladino MJ. Hsp70 and Hsp90 mediated proteasomal degradation underlies TPIsugarkill pathogenesis in Drosophila. Neurobiol Dis 2010; 40:676-83; PMID:20727972; https://doi.org/ 10.1016/j.nbd.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Oura J, Tamura Y, Kamiguchi K, Kutomi G, Sahara H, Torigoe T, Himi T, Sato N. Extracellular heat shock protein 90 plays a role in translocating chaperoned antigen from endosome to proteasome for generating antigenic peptide to be cross-presented by dendritic cells. Int Immunol 2011; 23:223-37; PMID:21421737; https://doi.org/ 10.1093/intimm/dxq475 [DOI] [PubMed] [Google Scholar]
- [39].Lilienbaum A. Relationship between the proteasomal system and autophagy. Int J Biochem Mol Biol 2013; 4:1-26; PMID:23638318. [PMC free article] [PubMed] [Google Scholar]
- [40].Kim Y-I, Ryu T, Lee J, Heo YS, Ahnn J, Lee SJ, Yoo O. A genetic screen for modifiers of Drosophila caspase Dcp-1 reveals caspase involvement in autophagy and novel caspase-related genes. BMC Cell Biol 2010; 11:9; PMID:20100334; https://doi.org/ 10.1186/1471-2121-11-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Pratt WB, Gestwicki JE, Osawa Y, Lieberman AP. Targeting Hsp90/Hsp70-based protein quality control for treatment of adult onset neurodegenerative diseases. Annu Rev Pharmacol Toxicol 2015; 55:353-71; PMID:25292434; https://doi.org/ 10.1146/annurev-pharmtox-010814-124332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Trepel J, Mollapour M, Giaccone G, Neckers L. Targeting the dynamic HSP90 complex in cancer. Nat Rev Cancer 2010; 10:537-49; PMID:20651736; https://doi.org/ 10.1038/nrc2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Chen D, Frezza M, Schmitt S, Kanwar J, Dou QP. Bortezomib as the first proteasome inhibitor anticancer drug: Current status and future perspectives. Curr Cancer Drug Targets 2011; 11:239-53; PMID:21247388; https://doi.org/ 10.2174/156800911794519752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Selimovic D, Porzig BB, El-Khattouti A, Badura HE, Ahmad M, Ghanjati F, Santourlidis S, Haikel Y, Hassan M. Bortezomib/proteasome inhibitor triggers both apoptosis and autophagy-dependent pathways in melanoma cells. Cell Signal 2013; 25:308-18; PMID:23079083; https://doi.org/ 10.1016/j.cellsig.2012.10.004 [DOI] [PubMed] [Google Scholar]
- [45].Bortnik S, Choutka C, Horlings HM, Leung S, Baker JH, Lebovitz C, Dragowska WH, Go NE, Bally MB, Minchinton AI, et al.. Identification of breast cancer cell subtypes sensitive to ATG4B inhibition. Oncotarget 2016; 7:66970-88; PMID:27556700; https://doi.org/ 10.18632/oncotarget.11408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Tenev T, Zachariou A, Wilson R, Ditzel M, Meier P. IAPs are functionally non-equivalent and regulate effector caspases through distinct mechanisms. Nat Cell Biol 2005; 7:70-7; PMID:15580265; https://doi.org/ 10.1038/ncb1204 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.