Abstract
A model has been proposed in which JIL-1 kinase-mediated H3S10 and H2Av phosphorylation is required for transcriptional elongation and heat shock-induced chromatin decondensation. However, here we show that although H3S10 phosphorylation is indeed compromised in the H2Av null mutant, chromatin decondensation at heat shock loci is unaffected in the absence of JIL-1 as well as of H2Av and that there is no discernable decrease in the elongating form of RNA polymerase II in either mutant. Furthermore, mRNA for the major heat shock protein Hsp70 is transcribed at robust levels in both H2Av and JIL-1 null mutants. Using a different chromatin remodeling paradigm that is JIL-1 dependent, we provide evidence that ectopic tethering of JIL-1 and subsequent H3S10 phosphorylation recruits PARP-1 to the remodeling site independently of H2Av phosphorylation. These data strongly suggest that H2Av or H3S10 phosphorylation by JIL-1 is not required for chromatin decondensation or transcriptional elongation in Drosophila.
KEY WORDS: JIL-1 kinase, Chromatin structure, Histone H3S10 phosphorylation, Drosophila, H2Av, His2Av
Highlighted Article: Histone H2Av and JIL-1 kinase are not required in Drosophila for chromatin decondensation associated with transcriptional activation, instead implicating a role for PARP-1 downstream of JIL-1-mediated H3S10 phosphorylation.
INTRODUCTION
The JIL-1 kinase localizes specifically to euchromatic interband regions of polytene chromosomes and is the kinase responsible for histone H3S10 phosphorylation at interphase in Drosophila (Jin et al., 1999; Wang et al., 2001). Furthermore, JIL-1 is enriched ∼2-fold on the male X chromosome and is implicated in transcriptional regulation as well as dosage compensation (Jin et al., 1999; Lerach et al., 2005, 2006). In a recent study, Cai et al. (2014) determined the genome-wide relationship of JIL-1 kinase-mediated H3S10 phosphorylation with gene expression and the distribution of the epigenetic H3K9me2 mark. The results showed that the H3S10ph mark in wild-type salivary gland cells is predominantly enriched at active genes, whereas the H3K9me2 mark is largely associated with inactive genes. However, mutation in JIL-1 resulted in 2-fold or greater changes in salivary gland expression of 1539 genes, with approximately half showing increased expression while the other half were downregulated. Notably, H3K9me2 marking also changed and was inversely correlated with expression level: genes showing decreased expression in the JIL-1 mutant were found to have acquired the H3K9me2 mark, whereas genes showing increased expression had either no or reduced levels of H3K9me2 marking as compared with wild type. These results are consistent with a model whereby the H3S10ph mark itself is not essential for gene transcription but rather that gene expression levels are modulated by the levels of the H3K9me2 mark independently of the state of the H3S10ph mark (Wang et al., 2011a,b, 2012; Girton et al., 2013; Cai et al., 2014). Thus, H3S10 phosphorylation acts indirectly to maintain active transcription by counteracting H3K9 dimethylation and gene silencing.
Recently, partly based on the finding that H3S10 phosphorylation is impaired in the absence of the H2Av (His2Av – FlyBase) histone variant, an alternative model has been proposed in which JIL-1 is required for gene expression by activating poly(ADP-ribose) polymerase 1 (PARP-1; also known as Parp – FlyBase) through phosphorylation of the C-terminus of H2Av (Thomas et al., 2014). In the model this leads to loosening of nucleosome structure, facilitating the subsequent H3S10 phosphorylation by JIL-1 that is required for transcription by the RNA polymerase II (Pol II) machinery (Thomas et al., 2014; Ivaldi et al., 2007). In particular, Thomas et al. (2014) claim that JIL-1 kinase activity is required for transcriptional elongation during the heat shock response as well as for PARP-1-dependent chromatin decondensation (puffing) at heat shock loci. Since these results are incompatible with those of Cai et al. (2014) described above and the demonstration by Cai et al. (2008) that JIL-1 is not enriched at developmental or heat shock-induced polytene chromosome puffs, we have re-examined some of the key findings of Thomas et al. (2014).
Although our results confirm that H3S10 phosphorylation is indeed compromised in the H2Av null mutant, we find that chromatin decondensation at heat shock loci is unaffected in the absence of JIL-1 as well as of H2Av and that there is no discernable decrease in the elongating form of Pol II in either mutant. These results, along with our previous studies (Deng et al., 2007, 2008; Cai et al., 2008, 2014; Wang et al., 2011a,b, 2012), provide further evidence that redistribution of the epigenetic H3K9me2 mark that occurs in the absence of H3S10 phosphorylation leads to transcriptional defects and argue against the model of Thomas et al. (2014) and Ivaldi et al. (2007) that JIL-1-mediated H3S10 phosphorylation is required for Pol II-dependent transcription. Furthermore, in a different chromatin decondensation paradigm that is JIL-1 dependent (Deng et al., 2008; Li et al., 2013; Wang et al., 2013), we provide evidence that ectopic tethering of JIL-1 and subsequent H3S10 phosphorylation recruits PARP-1 to the remodeling site independently of H2Av and of H2Av phosphorylation.
RESULTS
JIL-1 kinase localizes to chromatin in H2Av null mutants at wild-type levels but euchromatic H3S10 phosphorylation is decreased
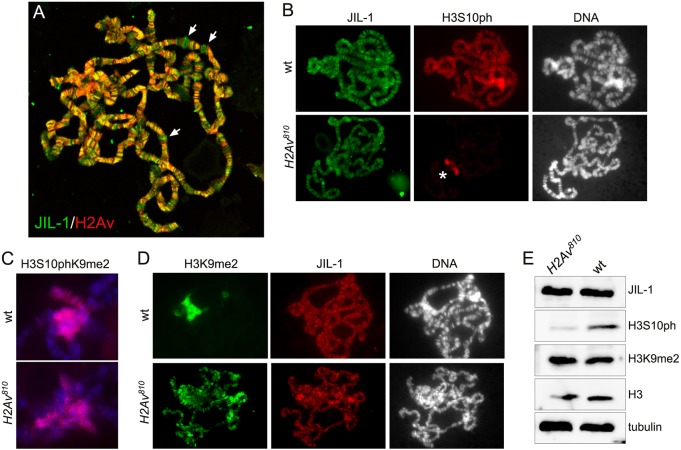
Thomas et al. (2014) reported that JIL-1 colocalizes with H2Av and that H3S10 phosphorylation is completely absent in H2Av null mutant larvae (H2Av810/810) (Van Daal and Elgin, 1992). To verify these claims we performed double labelings of polytene squash preparations with antibodies to JIL-1 and H2Av. We found that although H2Av and JIL-1 do colocalize at many locations, as indicated by the yellow/orange coloring in Fig. 1A, this colocalization was not universal as many interband locations were only positive for either JIL-1 or H2Av. This distribution is similar to that shown in figure 2B of Thomas et al. (2014). However, although JIL-1 and H2Av do not show complete colocalization, H3S10 phosphorylation was greatly diminished throughout the chromosome arms in H2Av null polytene chromosomes (Fig. 1B). Interestingly, the H3S10 phosphorylation by JIL-1 associated with the H3S10phK9me2 composite mark on pericentric heterochromatin and on the fourth chromosome (Wang et al., 2014) was unaffected (Fig. 1B,C).
Fig. 1.
Immunocytochemical and immunoblot characterization of JIL-1, H3S10ph and H3K9me2 in wild-type and H2Av null mutant backgrounds. (A) Confocal image of a polytene squash preparation double labeled with antibodies to JIL-1 (green) and H2Av (red). Although largely colocalized, as indicated by the yellow/orange coloring, many interband locations were only positive for either JIL-1 (arrows) or H2Av. (B) Polytene squash preparations from wild-type (wt) and homozygous H2Av810 null Drosophila larvae double labeled with antibodies to JIL-1 (green) and H3S10ph (red). The asterisk indicates H3S10ph labeling at the chromocenter in the H2Av810 null mutant background. DNA labeling by Hoechst is in gray. (C) Chromocenters from polytene squash preparations of wild-type and homozygous H2Av810 null larvae labeled with antibody to the H3S10phK9me2 double mark (red). DNA labeling by Hoechst is in blue. (D) Polytene squash preparations from wild-type and homozygous H2Av810 null mutant larvae double labeled with antibodies to H3K9me2 (green) and JIL-1 (red). DNA labeling by Hoechst is in gray. (E) Immunoblots of protein extracts from salivary glands from wild-type and homozygous H2Av810 null larvae labeled with antibodies to JIL-1, H3S10ph, and H3K9me2. Labeling with histone H3 and tubulin antibodies were used as loading controls.
Fig. 1D shows that the H3K9me2 mark spreads to ectopic locations on the chromosome arms in the H2Av null mutant background, as predicted by the decrease in euchromatic H3S10 phosphorylation (Zhang et al., 2006). Furthermore, JIL-1 antibody labelings and immunoblotting of salivary gland protein extracts show that JIL-1 is localized to chromatin (Fig. 1B,D) and present at wild-type levels in the H2Av null mutant although H3S10ph levels are substantially reduced (Fig. 1E). In order to quantify this reduction we determined the H3S10ph levels on immunoblots of salivary gland protein extracts from H2Av mutant larvae as a percentage of the levels in wild-type larvae. The data from six independent biological replicates indicate that the average reduction was ∼5-fold (19.8±7.2%, n=6). Moreover, the immunoblot indicates that H3K9me2 levels are indistinguishable from wild-type levels in the H2Av null mutant as well (Fig. 1E). This result is contrary to that of Swaminathan et al. (2005), who reported that H3K9me2 is not present in H2Av null mutants. However, we confirm the finding of Thomas et al. (2014) that H3S10ph levels are reduced at euchromatic sites in H2Av null mutants. Furthermore, we show that this reduction occurs despite JIL-1 protein levels being unaffected and that JIL-1 localizes to chromatin in the absence of H2Av.
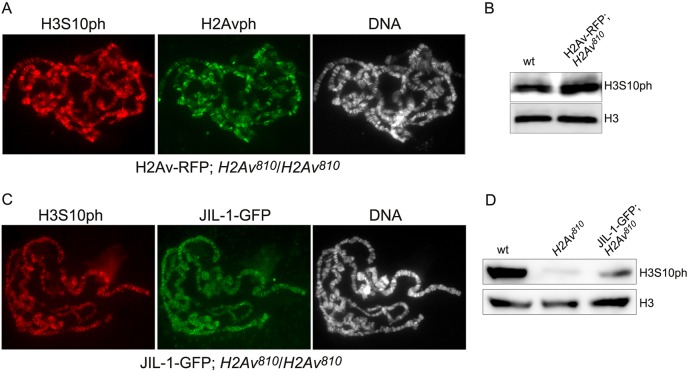
In order to verify that changes to H3S10 phosphorylation in the H2Av810 homozygous mutant were caused by the absence of H2Av we expressed an H2Av-RFP transgene (Deng et al., 2005) in the mutant. This restored H3S10ph to wild-type levels, indicating rescue of H2Av function (Fig. 2A,B). We also expressed a JIL-1-GFP transgene under heat shock promoter control (Jin et al., 1999) in the H2Av null mutant background. Interestingly, overexpressing JIL-1 substantially restored H3S10 phosphorylation levels, as indicated by both the immunofluorescence in polytene chromosome squash preparations (Fig. 2C) and immunoblot analysis (Fig. 2D). This suggests that H2Av is not required for JIL-1-mediated H3S10 phosphorylation, but rather facilitates JIL-1 kinase activity when both proteins are present at wild-type levels.
Fig. 2.
Immunocytochemical and immunoblot analysis of H2Av-RFP and JIL-1-GFP transgene expression in the H2Av null mutant background. (A) Polytene squash preparation from a homozygous H2Av810 null larvae expressing the H2Av-RFP transgene double labeled with antibodies to H3S10ph (red) and H2Avph (green). DNA labeling by Hoechst is in gray. (B) Immunoblots of protein extracts from salivary glands from wild-type and homozygous H2Av810 null larvae expressing the H2Av-RFP transgene labeled with antibody to H3S10ph. Labeling with histone H3 antibody provided a loading control. (C) Polytene squash preparation from a homozygous H2Av810 null larva expressing the JIL-1-GFP transgene double labeled with antibodies to H3S10ph (red) and GFP (green). DNA labeling by Hoechst is in gray. (D) Immunoblots of protein extracts from salivary glands of wild-type, homozygous H2Av810 null, and homozygous H2Av810 null larvae expressing the JIL-1-GFP transgene labeled with antibody to H3S10ph. Labeling with tubulin antibody provided a loading control.
H2Av phosphorylation at S137 is indistinguishable from wild type in JIL-1 null mutants in vivo
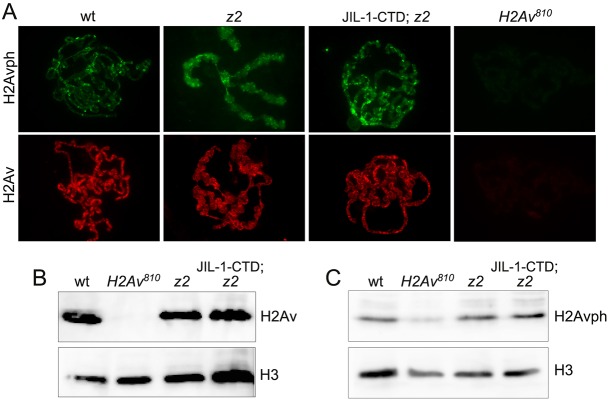
A key feature of the model of Thomas et al. (2014) is that phosphorylation of H2AvS137 (H2Avph) by JIL-1 is required for chromatin decondensation and subsequent H3S10 phosphorylation. However, this conjecture is mainly based on in vitro phosphorylation assays (Thomas et al., 2014), and kinases are well known for promiscuity in such assays (Peck, 2006; Mohamed and Hollfelder, 2012; Xue et al., 2012). We examined H2Av and H2Avph levels in wild-type and JIL-1 null (JIL-1z2/z2) mutant chromosome squash preparations as well as on immunoblots of protein extracts from salivary glands (Fig. 3). For H2Avph labeling we used a previously validated monoclonal antibody raised specifically to the Drosophila phosphorylated H2AvS137 residue (Lake et al., 2013). Since chromosome morphology is grossly perturbed in the JIL-1 null mutant (Deng et al., 2005), we expressed a CFP-tagged JIL-1 C-terminal construct (JIL-1-CTD) in the JIL-1 null mutant background that rescues chromosome morphology to near wild type without JIL-1 kinase activity (Bao et al., 2008). This allows for easier comparison of mutant squash preparations with those of wild type (Wang et al., 2013). We found that the H2Av and H2Avph distribution in JIL-1 mutants was indistinguishable from that of wild-type polytene squash preparations (Fig. 3A), as were the levels of H2Av (Fig. 3B) and H2Avph (Fig. 3C). As a control, H2Av and H2Avph antibody labeling was undetectable or at very low levels in the H2Av null mutant background (Fig. 3A-C). Thus, these results indicate that JIL-1 is not a major kinase, if at all, for H2AvS137 phosphorylation in vivo.
Fig. 3.
Immunocytochemical and immunoblot analysis of H2Av and H2Avph in the JIL-1 null mutant background. (A) Polytene squash preparation from wild-type, homozygous JIL-1z2 null (z2), homozygous JIL-1z2 null expressing the CFP-tagged JIL-1-CTD transgene (JIL-1-CTD; z2), and homozygous H2Av810 null larvae labeled with antibodies to H2Avph (green) or H2Av (red). (B,C) Immunoblots of protein extracts from salivary glands of wild-type, homozygous H2Av810 null, homozygous JIL-1z2 null (z2), and homozygous JIL-1z2 null larvae expressing the CFP-tagged JIL-1-CTD transgene (JIL-1-CTD; z2) labeled with antibody to H2Av (B) or H2Avph (C). Labeling with histone H3 antibody provided a loading control.
Decreased levels of PARP-1 activity do not affect H3S10 phosphorylation by JIL-1
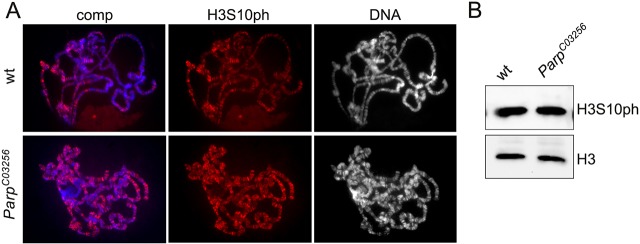
In the model of Thomas et al. (2014), PARP-1 activation following H2Av phosphorylation by JIL-1 leads to loosening of chromatin structure, allowing JIL-1-mediated H3S10 phosphorylation to occur. A prediction of this model is that H3S10 phosphorylation would be decreased in mutants with reduced PARP-1 activity. We examined H3S10ph levels and distribution in ParpC03256 homozygous salivary glands (Fig. 4). ParpC03256 is a strong hypomorphic allele with only low levels of residual polyADP-ribosylation activity (Kotova et al., 2010). As illustrated in Fig. 4A, the distribution and levels of H3S10ph were indistinguishable in wild-type and ParpC03256 homozygous mutant polytene chromosome squash preparations. That the levels of H3S10ph were undiminished in the ParpC03256 homozygous mutant compared with wild type was confirmed by immunoblotting (Fig. 4B). These results suggest that PARP-1 activity and polyADP-ribosylation are not required for JIL-1-mediated H3S10 phosphorylation.
Fig. 4.
Decreased levels of PARP-1 activity do not affect H3S10 phosphorylation by JIL-1. (A) Polytene squash preparations from wild-type and hypomorphic homozygous ParpC03256 mutant larvae labeled with antibody to H3S10ph (red), with DNA (Hoechst) in blue or gray. (B) Immunoblots of protein extracts from salivary glands of wild-type and hypomorphic homozygous ParpC03256 mutant larvae labeled with antibody to H3S10ph. Labeling with histone H3 antibody provided a loading control.
Neither JIL-1 nor H2Av is required for chromatin decondensation or transcriptional elongation during heat shock
Thomas et al. (2014) reported that chromatin decondensation and transcriptional elongation during the heat shock response require JIL-1 and H2Av phosphorylation. However, in a Su(var)3-9 mutant with reduced H3K9 histone methyltransferase activity, chromosome defects and the lethality associated with the JIL-1 null phenotype are substantially rescued (Deng et al., 2007). Furthermore, comparing global transcription profiles from wild-type and JIL-1 null mutant salivary glands, Cai et al. (2014) found that overall levels of transcription were unchanged, with about half of the altered genes upregulated and the other half downregulated. This indicates that Pol II transcription can occur even in the complete absence of JIL-1 kinase activity and the associated loss of interphase H3S10 phosphorylation. In addition, we have previously shown that transcription of heat shock loci still occurs in the JIL-1 null mutant background (Cai et al., 2008).
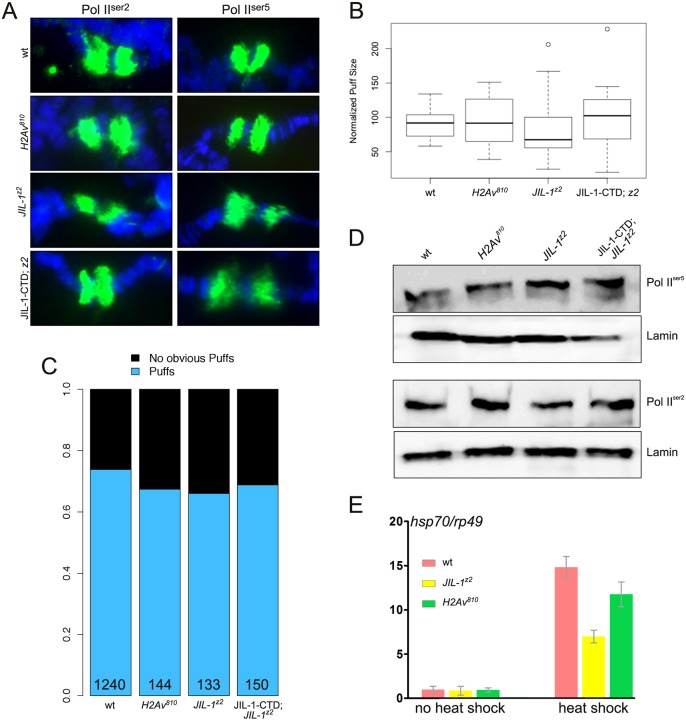
In order to reaffirm the previous observation that loss of JIL-1 reduces but does not eliminate heat shock-induced transcription, as well as to compare this effect with results observed after loss of H2Av, we investigated the distribution of Pol II0ser2 and Pol II0ser5 labeling and measured heat shock transcript levels in both JIL-1 null and H2Av null mutant backgrounds (Fig. 5). The antibody to Pol II0ser2 recognizes the elongating form of RNA polymerase II, which is phosphorylated at Ser2 in the C-terminal domain and which serves as a marker for active transcription, whereas antibody to Pol II0ser5 recognizes the paused form (Weeks et al., 1993; Boehm et al., 2003; Ivaldi et al., 2007). Fig. 5A shows labeling of heat shock puffs by Pol II0ser2 and Pol II0ser5 antibodies in polytene chromosome squashes from salivary glands of wild type, H2Av null, JIL-1 null, and a JIL-1 null expressing the JIL-1-CTD only (in order to restore chromosome morphology). In all mutant genotypes there was robust labeling of heat shock puffs indistinguishable from wild type. To quantify this aspect we measured the area of the 87A/C heat shock puffs labeled by Pol II0 antibody and normalized it to the width of the adjacent unpuffed region in order to adjust for different degrees of chromosome spreading during the squash process. As illustrated in Fig. 5B, there was no statistical difference (P>0.15, ANOVA test) between the normalized puff size from the various genotypes and wild type. Measurements were obtained from more than 30 salivary gland nuclei from at least five different larvae for each genotype. In addition, we determined the number of nuclei with clearly recognizable puffs among the total number of nuclei examined and found a frequency close to 70% for all the genotypes (Fig. 5C). Furthermore, on immunoblots of extracts from wild-type, H2Av null, JIL-1 null, and JIL-1-CTD-expressing JIL-1 null salivary glands, there was no detectable difference in Pol II0ser2 or Pol II0ser5 levels (Fig. 5D).
Fig. 5.
Neither JIL-1 nor H2Av is required for chromatin decondensation or transcriptional elongation during heat shock. (A) 87 A/C heat shock puffs from polytene squash preparations labeled with Pol II0ser2 and Pol II0ser5 antibodies (green) from wild-type, homozygous H2Av810 null, homozygous JIL-1z2 null (z2), and homozygous JIL-1z2 null larvae expressing the CFP-tagged JIL-1-CTD transgene (JIL-1-CTD; z2). DNA (Hoechst) is in blue. (B) Normalized 87 A/C puff size from wild-type, homozygous H2Av810 null, homozygous JIL-1z2 null (z2), and homozygous JIL-1z2 null larvae expressing the CFP-tagged JIL-1-CTD transgene (JIL-1-CTD; z2). Measurements were obtained from more than 30 salivary gland nuclei from at least five different larvae for each genotype. The box plot representation defines 25th to 75th percentiles (boxes), 50th percentile (lines in boxes), ranges (whiskers, 1.5 times the interquartile range extended from both ends of the box or the maximal/minimal value), and outliers more than 3/2 times the upper quartile (circles). There was no statistically significant difference in puff size between the four genotypes (P>0.15; ANOVA test). (C) The distribution of salivary gland nuclei with clearly recognizable puffs among the total number of nuclei examined from wild-type, homozygous H2Av810 null, homozygous JIL-1z2 null (z2), and homozygous JIL-1z2 null larvae expressing the CFP-tagged JIL-1-CTD transgene (JIL-1-CTD; z2). The total number of nuclei examined is indicated for each genotype. (D) Immunoblots of protein extracts labeled with Pol II0ser2 and Pol II0ser5 antibody from wild-type, homozygous H2Av810 null, homozygous JIL-1z2 null (z2), and homozygous JIL-1z2 null larvae expressing the CFP-tagged JIL-1-CTD transgene (JIL-1-CTD; z2). Labeling with lamin antibody provided a loading control. (E) Transcript levels of Hsp70 mRNA in wild-type and homozygous H2Av810 and JIL-1z2 null mutant backgrounds in response to heat shock treatment. Hsp70 transcript levels were determined by qRT-PCR and normalized to the mRNA levels for the control non-heat shock protein Rp49 both without and after heat shock treatment. The data shown are the average from three independent experiments in which each determination of transcript levels was performed in duplicate. The error bars indicate s.d.
These results indicate that the Pol II complex is transcriptionally active in both JIL-1 and H2Av null mutant backgrounds. However, in order to obtain a more direct measure of the level of heat shock gene transcription, we performed quantitative RT-PCR (qRT-PCR) assays on the six nearly identical Hsp70 genes, which encode the major heat shock protein in Drosophila (Gong and Golic, 2004), before and after heat shock in the H2Av null mutant background and compared the results with those obtained from the JIL-1 null mutant background. qRT-PCR results from primers that amplify transcripts from all six Hsp70 genes were normalized to qRT-PCR results from a primer pair specific to the non-heat shock-sensitive rp49 (RpL32) as in Cai et al. (2008). Three independent experiments with total RNA isolated from wild-type, H2Av null, and JIL-1 null third instar larvae were performed; in each case, qRT-PCR determination of transcript levels was performed in duplicate. As illustrated in Fig. 5E, very low levels of Hsp70 mRNA in wild-type, JIL-1 and H2Av mutant backgrounds were detected under non-heat shock conditions. However, a robust increase in Hsp70 transcript levels was detected in response to heat shock treatment in all three genotypes relative to rp49 transcript levels (Fig. 5E). The increase in JIL-1 and H2Av null mutant larvae was at least two orders of magnitude greater than under non-heat shock conditions. Thus, although total transcript levels were reduced (1/3 to 2/3) compared with wild type, a strong heat shock response was clearly observed. The results shown in Fig. 5E for transcript levels in the JIL-1 null mutant background replicate and reaffirm the previous findings shown in figure 8C of Cai et al. (2008).
PARP-1, but neither H2Av nor H2Avph, is upregulated at LacI-JIL-1 targeting sites
Previously, we showed that ectopic tethering of LacI-JIL-1 to lacO repeats inserted into a condensed, heterochromatic-like polytene chromosome band resulted in robust H3S10 phosphorylation and a more open euchromatic state at the targeting region (Deng et al., 2008). LacI tethering affords an excellent experimental system to assess whether targeting of JIL-1 could induce H2Av phosphorylation and/or the recruitment of H2Av to the targeting site. As illustrated in Fig. 6A, tethering of LacI-JIL-1 to 96C1-2 resulted in band ʻopening', as previously reported (Deng et al., 2008). However, labeling of the insertion site with H2Av or H2Avph antibody (Fig. 6A) revealed no signal above background levels, suggesting that there is no direct involvement of H2Av or H2Av phosphorylation in this process. For comparison, Fig. 6B shows control anti-H2Av and anti-H2Avph antibody labelings when LacI-GFP was tethered instead of LacI-JIL-1. In contrast to LacI-JIL-1, tethering of LacI-GFP induced no opening of the band and both H2Av and H2Avph antibody labelings coincided with the targeting site. In order to further exclude a role for H2Av in this chromatin remodeling we tethered LacI-JIL-1 to the targeting site in a homozygous H2Av810 null mutant background. This resulted in robust H3S10 phosphorylation at the targeting site and ʻopening' of the band (Fig. 6C).
Fig. 6.
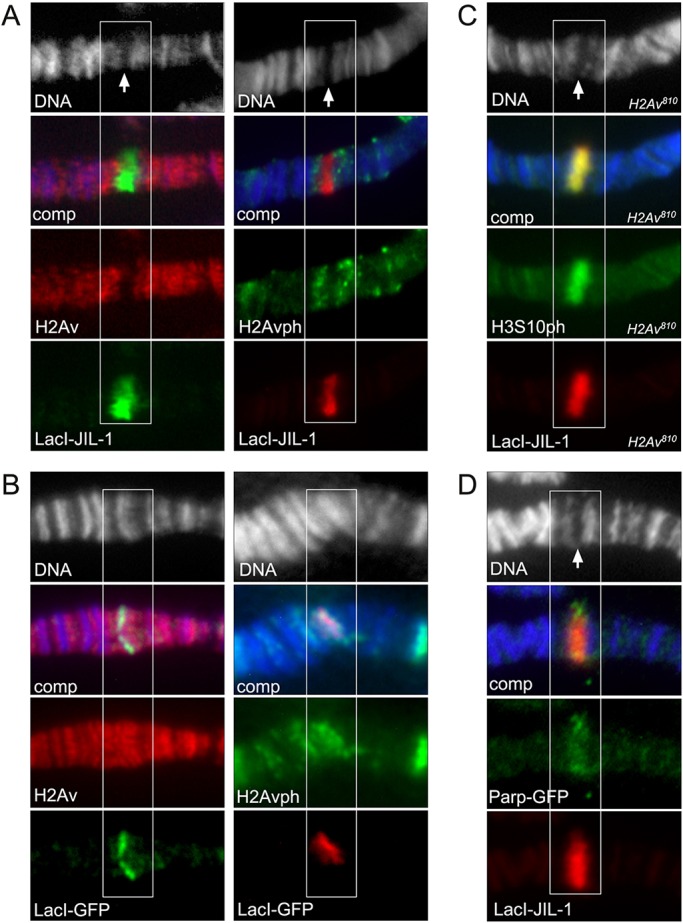
Tethering of LacI-tagged JIL-1 constructs to a polytene chromosome band lacO insertion site. Antibody (anti-LacI, anti-H2Av, anti-H2Avph, anti-H3S10ph or anti-GFP) and Hoechst (blue or gray) labelings of polytene squash preparations from third instar larvae homozygous for the lacO repeat line P11.3, which is inserted into the middle of a polytene band in region 96C1-2. The white boxes indicate the location of the polytene band with the lacO repeat insertion. Arrows indicate the ‘split’ in the polytene bands reflecting decondensation of the chromatin when LacI-JIL-1 is tethered to the band. (A) Tethering of full-length LacI-JIL-1. (B) Tethering of LacI-GFP. In contrast to when LacI-JIL-1 is tethered, there is no opening of the band. (C) Tethering of full-length LacI-JIL-1 in a homozygous H2Av810 null mutant background. (D) Tethering of full-length LacI-JIL-1 from a preparation co-expressing GFP-tagged PARP-1 (Parp-GFP).
To examine the possible involvement of PARP-1, in the absence of a suitable antibody to Drosophila PARP-1 we expressed a GFP-tagged PARP-1 construct (Parp-GFP) (Tulin et al., 2002) together with LacI-JIL-1. Interestingly, as shown in Fig. 6D, Parp-GFP accumulates at the LacI-JIL-1 targeting site suggesting a possible role for PARP-1 in the chromatin remodeling process. To investigate whether the recruitment was caused by direct interactions between JIL-1 and PARP-1 or whether it was dependent on phosphorylation of H3S10, we expressed a LacI-tagged ʻkinase dead' and two deletion constructs (CTD and ΔCTD) in a JIL-1 null background as previously described (Deng et al., 2008; Li et al., 2013). JIL-1 can be divided into four main domains: an N-terminal domain (NTD), two kinase domains (KDI and KDII) and a C-terminal domain (CTD) (Jin et al., 1999). The CTD of JIL-1 is without kinase activity but sufficient for proper chromatin localization (Deng et al., 2005; Bao et al., 2008), whereas the ΔCTD JIL-1 construct without the CTD has kinase activity for histone H3S10 despite the fact that it does not localize properly (Bao et al., 2008; Wang et al., 2011b). Fig. 7 shows ʻsmush' preparations in which these constructs were co-expressed with Parp-GFP. The smush preparation is a modified whole-mount staining technique in which nuclei from dissected salivary glands are gently compressed beneath a coverslip to flatten them before fixation (Wang et al., 2001; Cai et al., 2008). Only constructs with H3S10 phosphorylation activity (full-length and ΔCTD) recruited Parp-GFP to the targeting site (Fig. 7, arrows), whereas the ‘kinase dead' construct, which only differs from wild-type JIL-1 by two alanine mutations in the two catalytic domains, does not.
Fig. 7.
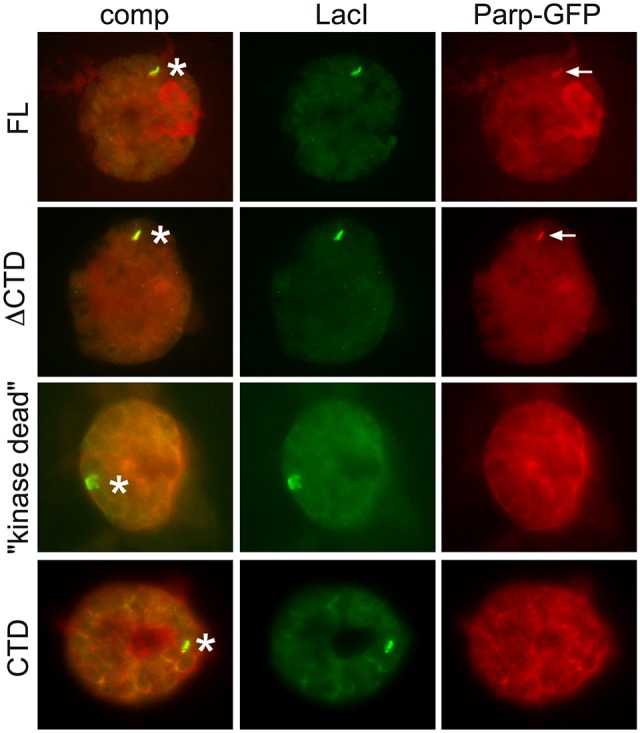
Tethering of LacI-tagged JIL-1 constructs to a polytene chromosome band lacO insertion site. Antibody labelings of smush preparations from third instar larvae homozygous for the lacO repeat line P11.3, which is inserted into the middle of a polytene band in region 96C1-2. Full-length LacI-JIL-1 (FL), LacI-JIl-1-ΔCTD (ΔCTD), LacI-JIL-1-‘kinase dead’ (“kinase dead”), and LacI-JIL-1-CTD (CTD) were tethered to the lacO repeats, in nuclei co-expressing Parp-GFP. LacI-tagged constructs were detected with anti-LacI antibody and Parp-GFP with anti-GFP antibody. Asterisks indicate the lacO repeat insertion sites. Arrows point to enhanced recruitment of Parp-GFP to the insertion sites when LacI-JIL-1 constructs with intact H3S10 phosphorylation activity are tethered (FL and ΔCTD).
Taken together, these findings indicate that PARP-1 correlates with JIL-1-induced chromatin remodeling sites and H3S10 phosphorylation independently of H2Av and H2Av phosphorylation in this paradigm.
DISCUSSION
In this study we have revisited the roles of H2Av and the JIL-1 kinase in chromatin decondensation associated with transcriptional activation, especially during the heat shock response. We provide evidence that heat shock-induced chromatin puffs occur in both H2Av and JIL-1 null mutant backgrounds and that the size and frequency of such puffs are indistinguishable from wild type. Furthermore, heat shock puffs in JIL-1 and H2Av null mutant backgrounds are strongly labeled by Pol II0ser2 antibody, indicating that Pol II0ser2 is actively involved in heat shock-induced transcription in the absence of H2Av as well as of JIL-1 kinase activity. These findings were corroborated by immunoblot analysis that showed that both Pol II0ser2 and Pol II0ser5 levels were unchanged in these mutants compared with wild type, as also previously demonstrated by Cai et al. (2008). qRT-PCR assays revealed that Hsp70 mRNA is transcribed at robust levels in H2Av and JIL-1 null mutants, confirming the results of Cai et al. (2008). Thus, these data strongly suggest that H2Av and histone H3S10 phosphorylation by JIL-1 is not required for heat shock-induced chromatin decondensation or transcriptional elongation in Drosophila. Moreover, we found that H2Av phosphorylation in vivo was indistinguishable from wild type in the absence of JIL-1 and that H3S10ph levels were unaffected in mutants with greatly reduced PARP-1 activity. These results are contrary to the model and many of the findings of Thomas et al. (2014). We cannot explain the discrepancies, apart from that they might be caused by technical issues such as the use of whole larval extracts containing a substantial mitotic H3S10ph component (Wang et al., 2001; Cai et al., 2008) and the use of hypomorphic instead of null JIL-1 alleles. Instead, the conclusions of the present study are consistent with a number of previous reports showing that polytene chromosome immunodetection of JIL-1 and H3S10ph shows no to minimal overlap with either Pol II0ser5 (paused) or Pol II0ser2 (elongating) labeling (Cai et al., 2008; Regnard et al., 2011; Wang et al., 2013). The present results are also consistent with previous studies that did not detect any JIL-1 or H3S10ph signal associated with developmental or heat shock-induced puffs (Cai et al., 2008).
The finding that heat shock protein transcription levels were attenuated somewhat in JIL-1 and H2Av mutant backgrounds, although two orders of magnitude greater than under non-heat shock conditions, is directly compatible with the model of Cai et al. (2014), which proposes that such decreases are caused by the redistribution of the H3K9me2 silencing mark that occurs in the absence of H3S10 phosphorylation (Wang et al., 2011b, 2013). This hypothesis is further supported by our demonstration in the present paper that H3K9me2 spreads to the chromosome arms and that H3K9me2 levels are indistinguishable from wild type in the H2Av null mutant.
However, our data do confirm the finding of Thomas et al. (2014) that euchromatic H3S10ph levels are severely reduced in the absence of H2Av, although pericentric and fourth chromosome H3S10 phosphorylation are unaffected. Furthermore, we show that JIL-1 is present at wild-type levels in the H2Av null mutant and that it binds to chromatin. Thus, the reduced level of H3S10ph on the euchromatic chromosome arms is not a consequence of JIL-1 degradation, as is the case in the absence of Mof acetyltransferase activity in males (Li et al., 2012), or of a failure of JIL-1 to localize to chromatin, possibilities not addressed by Thomas et al. (2014). Rather, these findings suggest a model whereby JIL-1 H3S10 phosphorylation is facilitated by a direct interaction of JIL-1 with H2Av in a complex that provides an optimal conformation for JIL-1 enzymatic activity. In this scenario, without H2Av JIL-1-mediated H3S10 phosphorylation is less efficient, leading to reduced H3S10ph levels. Alternatively, H2Av-dependent changes to nucleosome structure and histone tail alignment could serve the same function. That H2Av is not required for H3S10 phosphorylation by JIL-1 is further supported by the finding that overexpressing JIL-1 in the H2Av null mutant substantially restores H3S10ph levels.
In this study we also examined the roles of H2Av, H2Avph and PARP-1 in a different chromatin decondensation paradigm from that of heat shock-induced chromosome puffs that is JIL-1 dependent. In this paradigm, ectopic targeting of JIL-1 using a LacI-tethering system induces robust histone H3S10 phosphorylation and a change in higher order chromatin structure from a condensed heterochromatic-like state to a more open euchromatic state (Deng et al., 2008). However, as was the case for heat shock-induced chromatin decondensation, we found no evidence for a direct involvement of H2Av or H2Avph in this process. Rather, the results suggested that PARP-1 was recruited to the chromatin remodeling site by H3S10 phosphorylation itself, independently of any structural contributions from the JIL-1 protein. Thus, these data suggest a possible role for PARP-1 in chromatin remodeling downstream, not upstream (Thomas et al., 2014), of JIL-1-mediated H3S10 phosphorylation. It will be of interest in future studies to further define the role of PARP-1 and the mechanisms of chromatin decondensation caused by H3S10 phosphorylation.
MATERIALS AND METHODS
Drosophila melanogaster stocks
Drosophila lines were grown at 25°C according to standard methods (Roberts, 1998); Canton S was used for wild-type preparations. The JIL-1z2 null allele is from Wang et al. (2001) and Zhang et al. (2003), the H2Av810 null allele is described in Van Daal and Elgin (1992), and the ParpC03256 hypomorphic allele in Ji and Tulin (2009). LacI-tagged JIL-1 constructs were generated and described by Deng et al. (2008) and Li et al. (2013). These lines include LacI-JIL-1-FL, LacI-JIL-1-CTD, LacI-JIL-1-ΔCTD, and LacI-JIL-1-ʻkinase-dead'. GAL4-expression was driven by generating recombinant lines with Sgs3-GAL4 and da-GAL4 drivers obtained from the Bloomington Drosophila Stock Center. Recombinant JIL-1z2, da-GAL4 and H2Av810, Sgs3-GAL4 chromosomes were generated as described by Ji et al. (2005). Deng et al. (2008) and Li et al. (2013) describe the Lac operator insertion line P11.3. Bao et al. (2008) generated the CFP-tagged JIL-1-CTD construct. S. Heidmann (University of Bayreuth) provided the H2AvDmRFP1 transgenic line, which has been described previously (Deng et al., 2005). Jin et al. (1999) described the Hsp83 promoter-driven full-length JIL-1-GFP transgenic line GF29.1. The Parp-GFP transgenic fly line was a gift of Dr A. Tulin and is described in Tulin et al. (2002). Balancer chromosomes and markers are described in Lindsley and Zimm (2012). Heat shock experiments followed the protocols of Nowak et al. (2003) and Cai et al. (2008) with wandering third instar larvae subjected to a 25 min heat shock at 37°C.
Immunohistochemistry
Wang et al. (2001) described the salivary gland nuclei smush preparations, and standard polytene chromosome squash preparations followed the methods of Cai et al. (2010). Antibody labeling protocols were as in Johansen and Johansen (2003) and Johansen et al. (2009). Primary antibodies used in this study include rabbit anti-H3S10ph (Epitomics, 1173-1, RRID:AB_732930 and Cell Signaling, 9701, RRID:AB_331535; 1:1000), mouse anti-H3K9me2 (Abcam, 1220, RRID:AB_449854; 1:1000), rabbit anti-JIL-1 (Jin et al., 1999) (1:1000), chicken anti-JIL-1 (Jin et al., 2000) (1:50), mouse anti-Pol II0Ser2 (BioLegend, 920204, RRID:AB_2616695; 1:50), mouse anti-Pol II0Ser5 (Covance, MMS-134R, RRID:AB_10119940; 1:50), mouse anti-tubulin (Sigma-Aldrich, T9026, RRID:AB_477593; 1:2000), mouse anti-H2Av which was provided by Dr R. Glaser and has been previously characterized (Madigan et al., 2012) (1:100), mouse anti-H2Avph (RRID:AB_2618077) (Lake et al., 2013) (1:3000) which was obtained from the Developmental Studies Hybridoma Bank, mouse anti-Lamin Dm0 mAb HL1203 which was provided by Drs M. Paddy and H. Saumweber and has been previously characterized (Gruenbaum et al., 1988) (1:1000), chicken anti-GFP (Aves Labs, GFP-1020, RRID:AB_10000240; 1:50), mouse anti-LacI (Millipore, 05-503, RRID:AB_11211192; 1:50), mouse anti-H3S10phK9me2 (Millipore, 05-1354, RRID:AB_11212491; 1:50) and rabbit anti-H3 (Cell Signaling, 9715, RRID:AB_331563; 1:1000). The appropriate species- and isotype-specific Texas Red-, TRITC-, and FITC-conjugated secondary antibodies (Cappel/ICN, Southern Biotech) were used (1:200) to visualize primary antibody labeling. DNA was visualized by staining with Hoechst 33258 (Molecular Probes) in PBS. Mounting of the preparations was in 90% glycerol including 0.5% n-propyl gallate. Epifluorescence optics were used to examine the preparations on a Zeiss Axioskop microscope. Images were obtained and digitized using a Spot CCD camera. A Leica confocal TCS SP5 tandem scanning microscope system equipped with separate argon-UV, argon, and krypton lasers and the appropriate filter sets for Hoechst, FITC, Texas Red and TRITC imaging was used. A Plan-Apochromat 63×/1.4 NA oil objective (Zeiss) was used to obtain a separate series of confocal images for each fluorophor of double-labeled preparations. Images were obtained simultaneously with z-intervals of ∼0.5 μm. Photoshop (Adobe) was used to pseudocolor, image process, and merge images. Non-linear adjustments were performed for some images of Hoechst labeling to obtain the best chromosomal visualization.
Immunoblot analysis
Protein extractions from third instar larval salivary glands (homogenization buffer: 20 mM Tris-HCl pH 8.0, 150 mM NaCl, 10 mM EDTA, 1 mM EGTA, 0.2% Triton X-100, 0.2% NP-40, 2 mM Na3VO4, 1 mM PMSF, 1.5 µg/ml aprotinin) were separated by SDS-PAGE and immunoblotted (Sambrook and Russell, 2001) using the Bio-Rad Mini PROTEAN III system and nitrocellulose membrane (0.2 µm pore size). Anti-mouse, anti-chicken or anti-rabbit HRP-conjugated secondary antibody (Bio-Rad; 1:3000) was used to detect primary antibody using a ChemiDoc-ItTS2 imager (UVP). Digital images were analyzed using ImageJ software (NIH) to quantify the immunolabeling as previously described (Wang et al., 2001). The ImageJ gel analysis feature was used to determine the average pixel value after the grayscale exposure of the images was adjusted so that only a few pixels in the wild-type lanes were saturated. Levels in H2Av mutant larvae were determined as a percentage relative to the level determined for wild-type control larvae after normalization to the tubulin loading control lanes.
Analysis of gene expression by qRT-PCR
The MicroPoly(A)Purist Small-Scale mRNA Purification Kit (Ambion) was used to extract total RNA from ten pooled whole third instar larvae of each genotype (wild type, JIL-1z2/z2, H2Av810/810, and JIL-1-CTD; JIL-1z2, da-GAL4/JIL-1-1z2) after heat shock; the same was performed for non-heat shock controls. SuperScript III reverse transcriptase (Invitrogen) was used to make cDNA from this RNA, which was then used as template for quantitative real-time PCR (qRT-PCR) using a Stratagene Mx4000 real-time cycler. PCR reactions included Brilliant II SYBR Green QPCR Master Mix (Stratagene) and the following primers: rp49, 5′-AACGTTTACAAATGTGTATTCCGACC-3′ and 5′-ATGACCATCCGCCCAGCATACAGG-3′; Hsp70, 5′-GTCATCACAGTTCCAGCCTACTTCAAC-3′ and 5′-CTGGGTTGATGGATAGGTTGAGGTTC-3′. A 10 min 95°C denaturation step was followed by 40 cycles of 30 s at 95°C, 60 s at 59°C, and 40 s at 72°C. The Stratagene-supplied algorithm was used to plot fluorescence intensities against the number of cycles and a calibration curve based on dilution of concentrated cDNA was used to quantify mRNA levels; rp49 results were used for normalization.
Heat shock puff size and frequency
Heat shock puffs as well as adjacent areas of chromosomes in digital images from third instar polytene squash preparations labeled with RNA Pol II0ser2 or Pol II0ser5 antibody were outlined using ImageJ, and with the outlined area determined in square pixels. Subsequently, heat shock puff areas were normalized as the ratio of the area of the puff to the width of the adjacent chromosome bands. Differences in puff size between the four genotypes analyzed were compared using an ANOVA test. The frequency of puff appearances was determined as the number of nuclei with clearly recognizable puffs divided by the total number of nuclei examined. Box plot and stacking plots were made using R (v3.0.1).
Acknowledgements
We thank members of the K.M.J. and J.J. laboratory for discussion, advice and critical reading of the manuscript. We especially thank Drs S. Heidmann, A. Tulin, R. Glaser, H. Saumweber, and M. Paddy for fly stocks and reagents.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: Y.L., J.G., J.J., K.M.J.; Methodology: Y.L., C.W., W.C., M.Z., H.D., J.G., J.J., K.M.J.; Validation: Y.L., C.W., W.C., M.Z., H.D.; Formal analysis: Y.L., C.W., W.C., S.S., M.Z., H.D., J.G., J.J., K.M.J.; Investigation: Y.L., C.W., W.C., S.S., M.Z., H.D., J.G.; Data curation: C.W., W.C.; Writing - original draft: J.J., K.M.J.; Writing - review & editing: Y.L., C.W., W.C., S.S., M.Z., H.D., J.G., J.J., K.M.J.; Visualization: Y.L., J.J.; Supervision: J.G., J.J., K.M.J.; Project administration: J.J., K.M.J.; Funding acquisition: J.J., K.M.J.
Funding
This work was supported by National Institutes of Health grant GM062916 to K.M.J. and J.J. Deposited in PMC for release after 12 months.
References
- Bao X., Cai W., Deng H., Zhang W., Krencik R., Girton J. and Johansen K. M. (2008). The COOH-terminal domain of the JIL-1 histone H3S10 kinase interacts with histone H3 and is required for correct targeting to chromatin. J. Biol. Chem. 283, 32741-32750. 10.1074/jbc.M806227200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm A. K., Saunders A., Werner J. and Lis J. T. (2003). Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol. Cell. Biol. 23, 7628-7637. 10.1128/MCB.23.21.7628-7637.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W., Bao X., Deng H., Jin Y., Girton J., Johansen J. and Johansen K. M. (2008). RNA polymerase II-mediated transcription at active loci does not require histone H3S10 phosphorylation in Drosophila. Development 135, 2917-2925. 10.1242/dev.024927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W., Jin Y., Girton J., Johansen J. and Johansen K. M. (2010). Preparation of polytene chromosome squashes for antibody labeling. J. Vis. Exp. 36, pii:1748 10.3791/1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W., Wang C., Li Y., Yao C., Shen L., Liu S., Bao X., Schnable P. S., Girton J., Johansen J. et al. (2014). Genome-wide analysis of regulation of gene expression and H3K9me2 distribution by JIL-1 mediated histone H3S10 phosphorylation in Drosophila. Nucleic Acids Res. 42, 5456-5467. 10.1093/nar/gku173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H., Zhang W., Bao X., Martin J. N., Girton J., Johansen J. and Johansen K. M. (2005). The JIL-1 kinase regulates the structure of Drosophila polytene chromosomes. Chromosoma 114, 173-182. 10.1007/s00412-005-0006-8 [DOI] [PubMed] [Google Scholar]
- Deng H., Bao X., Zhang W., Girton J., Johansen J. and Johansen K. M. (2007). Reduced levels of Su(var)3-9 but not Su(var)2-5 (HP1) counteract the effects on chromatin structure and viability in loss-of-function mutants of the JIL-1 histone H3S10 kinase. Genetics 177, 79-87. 10.1534/genetics.107.075143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H., Bao X., Cai W., Blacketer M. J., Belmont A. S., Girton J., Johansen J. and Johansen K. M. (2008). Ectopic histone H3S10 phosphorylation causes chromatin structure remodeling in Drosophila. Development 135, 699-705. 10.1242/dev.015362 [DOI] [PubMed] [Google Scholar]
- Girton J., Wang C., Johansen J. and Johansen K. M. (2013). The effect of JIL-1 on position-effect variegation is proportional to the total amount of heterochromatin in the genome. Fly 7, 129-133. 10.4161/fly.24266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong W. J. and Golic K. G. (2004). Genomic deletions of the Drosophila melanogaster Hsp70 genes. Genetics 168, 1467-1476. 10.1534/genetics.104.030874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y., Landesman Y., Drees B., Bare J. W., Saumweber H., Paddy M. R., Sedat J. W., Smith D. E., Benton B. M. and Fisher P. (1988). Drosophila nuclear lamin precursor Dm0 is translated from either of two developmentally regulated mRNA species apparently encoded by a single gene. J. Cell Biol. 106, 585-596. 10.1083/jcb.106.3.585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivaldi M. S., Karam C. S. and Corces V. G. (2007). Phosphorylation of histone H3 at Ser10 facilitates RNA polymerase II release from promoter-proximal pausing in Drosophila. Genes Dev. 21, 2818-2831. 10.1101/gad.1604007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y. and Tulin A. V. (2009). Poly(ADP-ribosyl)ation of heterogeneous nuclear ribonucleoproteins modulates splicing. Nucleic Acids Res. 37, 3501-3513. 10.1093/nar/gkp218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y., Rath U., Girton J., Johansen K. M. and Johansen J. (2005). D-Hillarin, a novel W180-domain protein, affects cytokinesis through interaction with the septin family member Pnut. J. Neurobiol. 64, 157-169. 10.1002/neu.20131 [DOI] [PubMed] [Google Scholar]
- Jin Y., Wang Y., Walker D. L., Dong H., Conley C., Johansen J. and Johansen K. M. (1999). JIL-1: a novel chromosomal tandem kinase implicated in transcriptional regulation in Drosophila. Mol. Cell 4, 129-135. 10.1016/S1097-2765(00)80195-1 [DOI] [PubMed] [Google Scholar]
- Jin Y., Wang Y., Johansen J. and Johansen K. M. (2000). JIL-1, a chromosomal kinase implicated in regulation of chromatin structure, associates with the MSL dosage compensation complex. J. Cell Biol. 149, 1005-1010. 10.1083/jcb.149.5.1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen K. M. and Johansen J. (2003). Studying nuclear organization in embryos using antibody tools. Drosophila Cytogenetics Protocols (ed. Henderson D.S.), pp. 215-234. Totowa, New Jersey: Humana Press. [DOI] [PubMed] [Google Scholar]
- Johansen K. M., Cai W., Deng H., Bao X., Zhang W., Girton J. and Johansen J. (2009). Methods for studying transcription and epigenetic chromatin modification in Drosophila polytene chromosome squash preparations using antibodies. Methods 48, 387-397. 10.1016/j.ymeth.2009.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotova E., Jarnik M. and Tulin A. V. (2010). Uncoupling of the transactivation and transrepression functions of PARP1 protein. Proc. Natl. Acad. Sci. USA 107, 6406-6411. 10.1073/pnas.0914152107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake C. M., Holsclaw J. K., Bellendir S. P., Sekelsky J. and Hawley R. S. (2013). The development of a monoclonal antibody recognizing the Drosophila melanogaster phosphorylated histone H2A variant (γ-H2AV). G3 (Bethesda) 3, 1539-1543. 10.1534/g3.113.006833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerach S., Zhang W., Deng H., Bao X., Girton J., Johansen J. and Johansen K. M. (2005). JIL-1 kinase, a member of the male-specific lethal (MSL) complex, is necessary for proper dosage compensation of eye pigmentation in Drosophila. Genesis 43, 213-215. 10.1002/gene.20172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerach S., Zhang W., Bao X., Deng H., Girton J., Johansen J. and Johansen K. M. (2006). Loss-of-function alleles of the JIL-1 kinase are strong suppressors of position effect variegation of the wm4 allele in Drosophila. Genetics 173, 2403-2406. 10.1534/genetics.106.059253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Cai W., Wang C., Deng H., Bao X., Zhang W., Girton J., Johansen J. and Johansen K. M. (2012). The Mof acetyltransferase is required for JIL-1 H3S10ph kinase stability in Drosophila males. Mol. Biol. Cell 23, 734 10.1091/mbc.E12-10-0757 [DOI] [Google Scholar]
- Li Y., Cai W., Wang C., Yao C., Bao X., Deng H., Girton J., Johansen J. and Johansen K. M. (2013). Domain requirements of the JIL-1 tandem kinase for histone H3 serine10 phosphorylation and chromatin remodeling in vivo. J. Biol. Chem. 288, 19441-19449. 10.1074/jbc.M113.464271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley D. L. and Zimm G. G. (1992). The Genome of Drosophila melanogaster. New York, NY: Academic Press. [Google Scholar]
- Madigan J. P., Chotkowski H. L. and Glaser R. L. (2002). DNA double-strand break-induced phosphorylation of Drosophila histone variant H2Av helps prevent radiation-induced apoptosis. Nucleic Acids Res. 30, 3698-3705. 10.1093/nar/gkf496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed M. F. and Hollfelder F. (2012). Efficient crosswise catalytic promiscuity among enzymes that catalyze phosphoryl transfer. Biochem. Biophys. Acta 1834, 417-424. 10.1016/j.bbapap.2012.07.015 [DOI] [PubMed] [Google Scholar]
- Nowak S. J., Pai C.-Y. and Corces V. G. (2003). Protein phosphatase 2A activity affects histone H3 phosphorylation and transcription in Drosophila melanogaster. Mol. Cell. Biol. 23, 6129-6138. 10.1128/MCB.23.17.6129-6138.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck S. C. (2006). Analysis of protein phosphorylation: methods and strategies for studying kinases and substrates. Plant J. 45, 512-522. 10.1111/j.1365-313X.2005.02613.x [DOI] [PubMed] [Google Scholar]
- Regnard C., Straub T., Mitterweger A., Dahlsveen I. K., Fabian V. and Becker P. B. (2011). Global analysis of the relationship between JIL-1 kinase and transcription. PLoS Genet. 7, e1001327 10.1371/journal.pgen.1001327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts D. B. (1998). Drosophila: A Practical Approach. Oxford, UK: IRL Press. [Google Scholar]
- Sambrook J. and Russell D. W. (2001). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Swaminathan J., Baxter E. M. and Corces V. G. (2005). The role of histone H2Av variant replacement and histone H4 acetylation in the establishment of Drosophila heterochromatin. Genes Dev. 19, 844-858. 10.1101/gad.1259105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. J., Kotova E., Andrake M., Adolf-Bryfolgle J., Glaser R., Regnard C. and Tulin A. V. (2014). Kinase-mediated changes in nucleosome conformation trigger chromatin decondensation via poly(ADP-rybosyl)ation. Mol. Cell 53, 831-842. 10.1016/j.molcel.2014.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulin A., Stewart D. and Spradling A. C. (2002). The Drosophila heterochromatic gene encoding poly(APD-ribose) polymerase (PARP) is required to modulate chromatin structure during development. Genes Dev. 16, 2108-2119. 10.1101/gad.1003902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Daal A. and Elgin S. C. R. (1992). A histone H2A variant, H2AvD, is essential in Drosophila melanogaster. Mol. Biol. Cell 3, 593-602. 10.1091/mbc.3.6.593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang W., Jin Y., Johansen J. and Johansen K. M. (2001). The JIL-1 tandem kinase mediates histone H3 phosphorylation and is required for maintenance of chromatin structure in Drosophila. Cell 105, 433-443. 10.1016/S0092-8674(01)00325-7 [DOI] [PubMed] [Google Scholar]
- Wang C., Girton J., Johansen J. and Johansen K. M. (2011a). A balance between euchromatic (JIL-1) and heterochromatic (SU(VAR)2-5 and SU(VAR)3-9) factors regulates position-effect variegation in Drosophila. Genetics 188, 745-748. 10.1534/genetics.111.129353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Cai W., Li Y., Deng H., Bao X., Girton J., Johansen J. and Johansen K. M. (2011b). The epigenetic H3S10 phosphorylation mark is required for counteracting heterochromatic spreading and gene silencing in Drosophila melanogaster. J. Cell Sci. 124, 4309-4317. 10.1242/jcs.092585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Cai W., Li Y., Girton J., Johansen J. and Johansen K. M. (2012). H3S10 phosphorylation by the JIL-1 kinase regulates H3K9 dimethylation and gene expression at the white locus in Drosophila . Fly 6, 1-5. 10.4161/fly.20029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Yao C., Li Y., Cai W., Bao X., Girton J., Johansen J. and Johansen K. M. (2013). Evidence against a role for the JIL-1 kinase in H3S28 phosphorylation and 14-3-3 recruitment to active genes in Drosophila. PLoS ONE 8, e62484 10.1371/journal.pone.0062484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Li Y., Cai W., Bao X., Girton J., Johansen J. and Johansen K. M. (2014). Histone H3S10 phosphorylation by the JIL-1 kinase in pericentric heterochromatin and on the fourth chromosome creates a composite H3S10phK9me2 epigenetic mark. Chromosoma 123, 273-280. 10.1007/s00412-014-0450-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks J. R., Hardin S. E., Shen J., Lee J. M. and Greenleaf A. L. (1993). Locus-specific variation in phoshorylation state of RNA polymerase II in vivo: correlations with gene activity and transcript processing. Genes Dev. 7, 2329-2344. 10.1101/gad.7.12a.2329 [DOI] [PubMed] [Google Scholar]
- Xue L., Wang W.-H., Iliuk A., Hu L., Galan J. A., Yu S., Hans M., Geahlen R. L. and Tao W. A. (2012). Sensitive kinase assay linked with phosphoproteomics for identifying direct kinase substrates. Proc. Natl. Acad. Sci. USA 109, 5615-5620. 10.1073/pnas.1119418109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Jin Y., Ji Y., Girton J., Johansen J. and Johansen K. M. (2003). Genetic and phenotypic analysis of alleles of the Drosophila chromosomal JIL-1 kinase reveals a functional requirement at multiple developmental stages. Genetics 165, 1341-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Deng H., Bao X., Lerach S., Girton J., Johansen J. and Johansen K. M. (2006). The JIL-1 histone H3S10 kinase regulates dimethyl H3K9 modifications and heterochromatic spreading in Drosophila. Development 133, 229-235. 10.1242/dev.02199 [DOI] [PubMed] [Google Scholar]