Abstract
Studies using Saccharomyces cerevisiae, the common baker’s or brewer’s yeast, have progressed over the past twenty years from knowing which sphingolipids are present in cells and a basic outline of how they are made to a complete or nearly complete directory of the genes that catalyze their anabolism and catabolism. In addition, cellular processes that depend upon sphingolipids have been identified including protein trafficking/exocytosis, endocytosis and actin cytoskeleton dynamics, membrane microdomains, calcium signaling, regulation of transcription and translation, cell cycle control, stress resistance, nutrient uptake and aging. These will be summarized here along with new data not previously reviewed. Advances in our knowledge of sphingolipids and their roles in yeast are impressive but molecular mechanisms remain elusive and are a primary challenge for further progress in understanding the specific functions of sphingolipids.
Keywords: Actin, aging, endocytosis, exocytosis, calcium signaling, heat stress, lipid raft, long-chain base, longevity, myriocin, phytosphingosine, Pkc1, Pkh1, Pkh2, protein trafficking, rapamycin, Sch9, Slm1, Slm2, signal transduction, sphingolipids, TORC1, TORC2, Ypk1, Ypk2
Introduction
Besides providing many satiating bakery and brewery products for human pleasures, Saccharomyces cerevisiae, has been a remarkably informative host for discovering how sphingolipids are made and degraded and the cellular functions they perform. Carter’s laboratory identified the long-chain bases (LCBs) dihydrosphingosine (DHS, sphinganine) and phytosphingosine (PHS) in the 1950s and early 1960s.1 In the 1970s and 1980s the Lester laboratory identified the three classes of inositol phosphate-containing complex sphingolipids in yeast and their mode of synthesis and in doing so devised excellent methods for their extraction and analysis.2, 3 These advances and others enabled yeast genes to be identified and manipulated in every imaginable way and have permitted in elucidation of most, perhaps all, genes for making and degrading sphingolipids in S. cerevisiae, the first organism for which this has been achieved (reviewed in ref.4–6). Many of these genes made possible the identification of homologs in organisms ranging from bacteria to plants to man. In this review I try to summarize our knowledge of sphingolipids in S. cerevisiae and suggest where future research is needed.
Sphingolipid Metabolism in S. cerevisiae
Sphingolipids are abundant in S. cerevisiae representing about 7% of the mass of the plasma membrane or 30% of phospholipids.7 S. cerevisiae sphingolipid metabolism including metabolites, enzymes and their cognate genes are diagramed in Fig. 1. Synthesis begins with condensation of serine and a fattyacyl-CoA, typically palmitoyl-CoA, in every organism that has been examined, and generates the short-lived intermediate 3-ketodihydrosphingosine, which is reduced to yield DHS, the first LCB in the pathway. DHS can either be N-acylated with a fatty acid to give ceramide or hydroxylated on C4 to give PHS which is N-acylated to yield phytoceramide. The C1 hydroxyl of phytoceramide is decorated with polar head groups by the three sequential reactions diagramed in Fig. 1 which yield three species of complex sphingolipids including inositol phosphoceramide (IPC), mannose inositol phosphoceramide (MIPC) and mannose-(inositol-P)2-ceramide M(IP)2C. The genes and enzymes for the steps in sphingolipid metabolism have been discussed thoroughly in previous reviews.2–5, 8–10
Fig. 1.
Pathways of sphingolipid metabolism in Saccharomyces cerevisiae. Metabolic intermediates and complex sphingolipids are shown in bold, genes are indicated by italics and enzyme names are in regular lettering. When grown aerobically the fatty acid in complex sphingolipids is often hydroxylated at C2 and sometimes at C3 (not shown), a reaction that requires Scs7 (not shown).107 Ceramides can be hydrolyzed by two ceramidases, Ydc1 and Ypc1 (not shown), to yield a fatty acid and an LCB.35, 36 Structures of the indicated compounds are presented in previous publications.4, 8 Adapted from ref.6
Sphingolipid synthesis begins in the endoplasmic reticulum and generates ceramides which are transported by both vesicular and nonvesicular transport to the Golgi apparatus for addition of the polar head groups.11 Most enzymes involved in sphingolipid synthesis have been localized to these two compartments although there are exceptions.12 Movement of sphingolipids between these compartments has been examined in detail although much remains to be elucidated (reviewed in refs.8, 9, 13, 14). Most complex sphingolipids are then transported to the plasma membrane but small amounts are found in other membrane compartments15, 16 including mitochondria.17 The function of sphingolipids in these other cellular compartments is not understood. About three-fourths of the mass of the sphingolipids in S. cerevisiae cells is M(IP)2C with the rest being equal parts of IPC and MIPC.18 It is not known how these ratios are determined, but they are probably important in ways that remain to be identified. The shear mass of sphingolipids and their negative charge are likely to affect processes dependent upon the plasma membrane. Many types of sphingolipids in mammals are localized to one or the other leaflet of the plasma membrane, but this has not been determined for yeast sphingolipids.
A unique and distinguishing feature of S. cerevisiae sphingolipids is a C26 fatty acid, although a small percentage of C22 and C24 fatty are also present.16, 19 These long-chain fatty acids are synthesized by an elongation system19–21 whose components have now been identified and this has led to a model for how the length of the fatty acid is determined.22 The functions of such a long-chain fatty in sphingolipids are not well defined, although it may have something to do with the fact that they span both leaflets of a membrane bilayer. They have been suggested to play roles in nuclear membrane pores.23 Others have argued that sphingolipids do not need them but that they are necessary to perform some function that remains to be identified.24 While it is true that cells making sphingolipids with shorter acyl chains can survive in laboratory situations, it is highly unlikely that such strains would survive in the wild, thus arguing that the C26 fatty acid is an essential component of yeast sphingolipids under natural conditions.
While S. cerevisiae only makes inositolphosphoceramides and no other type of sphingolipids such as glycosylceramides, many other fungi make both classes of sphingolipids3 as do plants.25, 26 Phosphoinositol-containing sphingolipids have not been identified in mammals. It probably is no accident that S. cerevisiae and other fungi have sphingolipids that are more like plants than mammals given that the natural habitat of S. cerevisiae appears to be forests, particularly oaks and other broadleaf trees.27
Like most other membrane components, sphingolipids are broken down as a normal part of membrane remodeling. The need for turnover is apparent in mammals where defects in enzymes that catalyze turnover lead to debilitating diseases termed sphingolipidoses.28 Such a need has been much less apparent in yeast but this notion is changing. One type of sphingolipidoses is Niemann Pick type C, a fatal neurodegenerative disorder caused by a defect in the human NPC1 gene. The yeast homolog of this gene, NCR1, has been suggested to play a role in recycling of sphingolipids.29 The yeast sequence similarity between a bacterial neutral sphingomyelinase and the Isc1 protein of S. cerevisiae lead to the demonstration that Isc1 has phospholipase C-type activity and cleaves the polar head group from yeast sphingolipids, much like mammalian sphingomyelinases cleave the polar head group from sphingomyelin.30, 31
Unraveling the physiological importance of sphingolipid turnover in S. cerevisiae has been challenging and much remains to be learned. At least in some strains backgrounds, Isc1 enzyme activity is necessary for growth on nonfermentable carbon sources, implying a role in mitochondrial respiration32, and several types of experiments support this mitochondrial connection as discussed in detail in a previous review.6 Another role for Isc1 is to regulate the concentration of IPC which is toxic above its physiological concentration.33, 34 In response to heat stress and perhaps other stress that perturb the plasma membrane, Isc1 appears to be activated to breakdown complex sphingolipids (we don’t know their location) by a pathway involving synthesis of phosphatidylinositol 4,5-bisphosphate (PIP2) as described below along.
A good deal of effort has gone into trying to determine the physiological functions of the other enzymes that degrade sphingolipids including the ceramidases Ypc135 and Ydc136, the LCB kinases Lcb4 and Lcb5, the LCB-phosphate phosphatases Lcb2 and Ysr3 and the LCB lyase Dpl1. Some may disagree, but I think it is fair to say that other than their basic catalytic function, we do not know the real physiological roles of these enzymes and their precursors and products in S. cerevisiae (reviewed in refs. 2–5, 8, 9) They must be important because most are conserved in fungi and other organisms.
Both natural and synthetic inhibitors of several enzymes in the sphingolipid biosynthesis pathway have been identified and have been very useful in a range of experiments (reviewed in refs.3, 37). These include myriocin, sphingofungins, lipoxamycin and viridiofungins that inhibit serine palmitoyltransferase (SPT), the first enzyme in sphingolipid biosynthesis (Fig. 1), australifungin and the fumonisins that inhibit ceramide synthase and aureobasidin and khafrefungin that inhibit IPC synthase.
Membrane-Associated Functions and Processes
A screen for mutants defective in endocytosis gave the first indication that sphingolipids are necessary for this process.38 The screen identified end8-1, later shown to be allelic with LCB1 and renamed lcb1-100.39 The lcb1-100 mutation causes serine palmitoyltransferase (Fig. 1) activity to be labile even in cells grown at a permissive temperature40 and sphingolipid synthesis becomes limiting for growth at the restrictive temperature of 37°C41, most likely because LCB levels drop quickly and limit sphingolipid synthesis.42 The lcb1-100 mutation has been extremely useful for studying sphingolipid metabolism and functions. Sphingolipids affect multiple aspects of endocytosis including the actin cytoskeleton and these will be described below in the section on signal transduction. Other experiments implicating sphingolipids in endocytosis used strains defective in SNC1 and/or SNC2 that encode v-SNARE proteins.43 Sphingolipids have been shown to play several roles in endocytosis of the uracil transporter Fur4 (reviewed in ref.9) as have the Slm proteins44 (see below).
Sphingolipids along with sterols (cholesterol in mammals and ergosterol in fungi and plants) are critical for formation of microdomains within membranes that have been referred to as lipid rafts and are typically isolated by treating a yeast extract with detergent at low temperature to form what are referred to as detergent-insoluble complexes or detergent-resistant membranes.45, 46 In S. cerevisiae rafts are vital for sorting and delivering membrane-bound proteins to their proper cellular address and they are also necessary for fusion of cells during mating. 45, 47, 48 Such proteins include Pma1, which exports protons across the plasma membrane to maintain intracellular pH and also to generate a proton gradient necessary for cells to take up nutrients from their surroundings.49–54 Other membrane proteins requiring rafts/sphingolipids include Gas1, a Beta-1,3-glucanosyltransferase and Nce249, Fus2, Fig 1, Sho1, Ste1 and Prm155, Fur442, 56, and Can1, an arginine transporter.57 The general amino acid permease Gap1 depends upon sphingolipids for its transport to the plasma membrane in an active conformation capable of amino acid transport and that can resist degradation.58
Roles for sphingolipids in exocytosis based upon suppressor mutant analyses were reviewed previously.4, 9 In wild-type cells the C26-fatty acid in sphingolipids is required for raft association and stable surface transport of newly made Pma1 to the plasma membrane.59 Recently, a large-scale visual screen for genes that play roles in the sorting of proteins in the trans-Golgi network for delivery by the exocytosis pathway to the cell surface identified genes in sphingolipid metabolism (SUR2, SUR4, and YPC1).48 It is not clear why the Sur2, Sur3 and Ypc1 proteins are required for protein sorting. Ypc1 is particularly interesting because it degrades phytoceramides.35
In mammals, sphingosine-1-phosphate is involved in intracellular calcium signaling through an unknown mechanism.60 Likewise, sphingolipids may regulate calcium fluxes and signaling pathways in yeast, but the mechanisms are unknown (reviewed in ref.2) and see also ref.61) Cells defective in CSG1 or CSG2 (Fig. 1) are sensitive to 100 mM calcium and this phenotype has played a key role in identifying genes in sphingolipid metabolism.33, 34, 62, 63 Why mutants defective in CSG1 or CSG2 are sensitive to calcium is unclear, but it indicates a connection of some type between sphingolipids and calcium metabolism or signal transduction and these potential connections are presented below in the section on Signal Transduction Pathways.
Nutrient transport is affected by LCBs as first suggested by the extreme sensitivity of auxotrophic strains to LCBs in the culture medium.64 PHS, but not other LCBs, was found to block uptake of tryptophan, leucine, histidine and uracil.65 It is not entirely clear how LCBs are regulating nutrient transport, except in the case of uracil where PHS has been shown to be important for heat-induced, ubiquitin-mediated breakdown of Fur4, the uracil transporter.40 Other data also support the idea that heat-induced LCBs promote ubiquitination of proteins66 and that the immunosuppressant drug, FYT720, a synthetic sphingolipid-like molecule, acts in ways very similar to PHS to inhibit yeast growth.29 As described below in Signal Transduction Pathways, LCBs maybe regulating nutrient transporters via activation of the Pkh1/2 kinases (Fig. 2) or their downstream kinases such as Ypk1 or Sch9, which play roles in nutrient sensing.67–69
Fig. 2.
S. cerevisiae signal transduction pathways regulated by LCBs. LCBs transiently increase during a heat stress and are hypothesized to activate Pkh1 and Pkh2. As discussed in the text, Pkh2 is probably more dependent upon LCBs than is Pkh1 although this has not been defined for each cellular response. Kinase assays with purified proteins suggest that LCBs can directly trigger a small increase in Ypk1, Ypk2 and Sch9 activity (indicated by a dotted line).97. Pkh1/2 phosphorylate Ypk1, Ypk2, Sch9 and Pkc1 in their activation loop (PDK1 site) but the proteins are not enzymatically active. To become active they also need to be phosphorylated in a hydrophobic region (PDK2 site) and in a turn motif in their C-terminus. Phosphorylation of these sites in Ypk2 is mediated by TORC2124 and for Sch9 phosphorylation is mediated by TORC1.94 Ypk1/2 and Pkc1 are shown working in parallel pathways to control cell wall integrity, but data also support an alternative pathway in which Ypk1/2 work upstream of Pkc1.90, 91 Adapted from ref.6
Interactions of some sort or cross-talk between sphingolipids and both ergosterol and glycerophospholipids has been observed. For example, defects in ergosterol synthesis are suppressed by mutations in SUR470 and a decrease in ergosterol content is compensated by an increase in sphingolipids.71 Other data show that defects in ergosterol synthesis affect hydroxylation of yeast sphingolipids.72 Yeast cells also have mechanisms for maintaining asymmetry in the distribution of sphingolipids and glycerophospholipids in the two leaflets of the plasma membrane so that a change in one class of lipids is compensated by a change in another class.73
While the complex yeast sphingolipids, IPCs, MIPCs and M(IP)2Cs, are abundant, progress in identifying unique functions for them has been slow. The antifungal action of the plant defensin DmAMP1, a peptide defensin produced by Dahlia merckii, requires M(IP)2C, which serves as a high affinity receptor.74 DmAMP1 is postulated to bind or interact with M(IP)2C-containing lipid rafts making the plasma membrane more permeable thereby disrupting essential cellular processes.75 Syringomycin E is an antifungal cyclic lipodepsinonapeptide that interacts with the plasma membrane and inhibits growth of S. cerevisiae cells by forming ion channels. Yeast mutants defective in ipt1, fen1 or sur4, scs7 and sur2 are drug resistant, showing that M(IP)2C with a C26-fatty acid and PHS but not DHS is essential for the antifungal action of syringomycin E.76–78 While interesting, these results do not represent normal functions of the complex sphingolipids beyond their role as structural components of the plasma membrane and essential elements of lipid rafts. One would imagine that they interact in physiologically essential ways with plasma membrane proteins, but evidence is lacking and will require advances in three dimensional structure of membrane proteins bound to natural lipids.
Sphingolipids play roles in exocytosis of glycosylphosphatidylinositol-anchored proteins (reviewed in ref.79). For example, transport of Gas1p from the endoplasmic reticulum to the Golgi apparatus requires ceramide or a related sphingolipid for transport.39, 80, 81 Furthermore, the diacylglycerol moiety in glycosylphosphatidylinositol anchors is often replaced by ceramide.79 Sphingolipids and the Slm protein (see below) are required for exocytosis of arginine transporter Can182 and references therein). Sphingolipids have also been shown to play roles in generating a functional V1 component of the vacuolar ATPase.83
Signal Transduction Pathways that Require Sphingolipids
One of the most exciting and extensively explored functions of sphingolipids is the regulation of signal transduction pathways and ceramides and sphingosine-1-phosphate in mammals are the most well documented signaling species.84–86 There is no firm evidence that ceramides and long-chain base phosphates play similar roles in yeast. Instead, LCBs appear to regulate signaling pathways. The one piece of evidence that would firmly establish LCBs as regulators of signaling pathways is to show that they bind to specific proteins such as the protein kinases discussed below that are implicated to be regulated by LCBs (Fig. 2).
The first clue that LCBs might be intracellular signaling molecules or second messengers was the observation that they rapidly but transiently increase following heat stress typically executed in the laboratory by shifting cells from 25°C to 37°C or 39°C87, 88 and reviewed in detail in ref.4, 9, 10
Insight into LCB signaling pathways was first found during a screen to identify genes whose overexpression bypassed growth inhibition by myriocin89, an inhibitor of serine palmitoyltransferase (Fig. 1). The YPK1 gene bypassed the myriocin block. The Ypk1 protein kinase has a role in cell wall maintenance and actin cytoskeleton dynamics90, 91, endocytosis92 and translation during nitrogen starvation and nutrient sensing.67 Ypk1 and its paralog Ypk2 are structural and functional homologs of mammalian serum and glucocorticoid-inducible kinase (SGK).93 Ypk1 is phosphorylated and activated by Pkh193 and multiple copies of PKH1 were found to also bypass the myriocin block.89 One explanation for these results is that a sphingolipid acts upstream of and activates the Pkh1/Ypk1-Ypk2 the signaling pathway. Myriocin-treated cells lacked a phosphorylated and presumably active form of Ypk1 but this form reappeared in vivo following PHS-treatment of cells. The original and still accepted view of these experiments is that PHS activates Pkh1 or its homolog Pkh2, which then phosphorylate and activate Ypk1 and probably Ypk2. Many laboratories (reviewed in ref.9, 10) have contributed data supporting this hypothesis and expanded it to include activation of other kinases including Sch994 and Pkc1 which are activated by the LCB-Pkh1/2 pathway (Fig. 2) and by the Target of Rapamycin (TOR) pathways as indicated in Fig. 2.
The diagram shown in Fig. 2 suggests that Pkh1/2 and the kinases and cellular processes downstream of them are equally regulated by LCBs, but current data suggest that this is probably an oversimplification. First, initial analysis in vitro showed that purified Pkh2 was more strongly activated by LCBs than was Pkh1.95 Second, as discussed below, the Slm2 protein is more responsive to sphingolipids (likely to be LCBs) than is Slm1, consistent with finding that slm1Δ cells are more sensitive to growth inhibition by myriocin than are slm2Δ cells.44 In support of this latter result, a recent large-scale screen found both heterozygous and homozygous diploid slm1Δ cells to be extremely sensitive to myriocin while slm2Δ cells were not sensitive at all.96 Likewise, homozygous diploid pkh1Δ cells were 1000-fold more sensitive to myriocin than pkh2Δ cells, consistent with LCBs having a stronger affect on Pkh2 than on Pkh1. Additionally, this screen found both heterozygous and homozygous diploid ypk1Δ cells to be extremely sensitive to myriocin while ypk2Δ cells were not sensitive, implying that LCBs regulate Ypk2 but not Ypk1 activity. These myriocin data only apply to a process or processes that are necessary for growth: non-essential processes may depend upon LCBs to regulate Pkh1 and Ypk1, as suggested by the finding that in vitro the activity of Pkh1 and both Ypk1 and Ypk2 is stimulated by LCBs.97 As mentioned above, demonstrating which proteins bind LCBs would help to clarify their physiological roles.
Pkh1/2 are primarily found on eisosomes98, newly described very large structures, estimated to contain about 2000 copies each of the Pil1 and Lsp1 proteins, which bind to the cytoplasmic face of the plasma membrane.99 Pil1 and Lsp1 are highly conserved but seem to be found only in fungi. Eisosomes play roles in endocytosis of lipids and some proteins and localize at cites of endocytosis where they may physically interact with the actin cytoskeleton based on confocal fluorescent microscopy and genetic interaction studies 99. Pil98, 100 and Lsp1 (Dickson, et al. unpublished data) are highly phosphorylated by Pkh1/2 and phosphorylation plays roles in assembly and disassembly of eisosomes. Eisosomes are also thought to be involved in sensing changes in the plasma membrane caused by stresses such as heat and then signaling the cell to adjust the lipid and protein composition of the membrane by endocytosis and exocytosis in order for cells to be more stress tolerant.98, 100 Earlier data95 had indicated that Pil1 and Lsp1 indeed play roles in resisting heat stress and they may do so in several ways including serving as binding sites for Pkh1/2 to enable them to properly control downstream kinases with known roles in heat and other stress responses (Fig. 2). It is not known if some fraction of Pkh1 or Pkh2 also resides in other cellular locations or whether they cycle on and off of eisosomes. Clearly there is much to learn about eisosomes and the role they play in regulating cellular processes.
Previous results from genetic suppression experiments34, 101 suggested that sphingolipids interacted in some manner with phosphoinositides, probably phosphatidylinositol-5, 4-bisphosphate (PIP2), and with the TOR pathways. These novel interactions along with the calcineurin signaling pathway have now been shown by genetic and biochemical assays to control phosphorylation and dephosphorylation of the Slm1 and Slm2 proteins during heat stress thereby modulating actin polarization, endocytosis and sphingolipid metabolism.44, 82, 102
The Slm proteins have overlapping functions and at least one is required for viability.103 They also have a PH domain enabling binding to PIP2, known to arise transiently during stresses such as during heat shock on the inner leaflet of the plasma membrane.103, 104 Slm binding to the membrane is further strengthened by interactions with the Avo2 and Bit61 subunits of the Target Of Rapamycin Complex 2 ( TORC2)103, 105 and these protein-protein interactions promote phosphorylation of Slm1 and Slm2 by TORC2.103 The phosphorylated Slm proteins then mediate downstream effects of PIP2 and TORC2 that control roles of the actin cytoskeleton essential for growth, cell wall integrity and receptor-mediated endocytosis (Fig. 3). A recent study suggests that TORC2 is located on eisosomes, if true, then eisosomes may play a role in Slm protein function.106
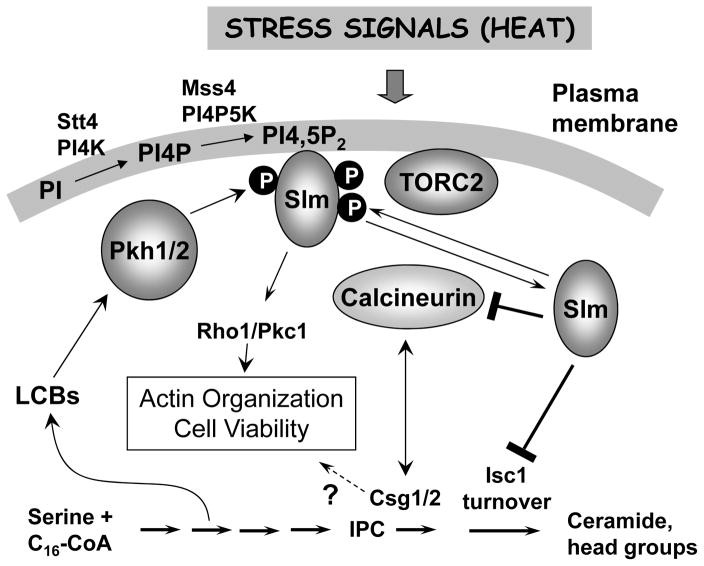
Fig. 3.
Synthesis and turnover of complex sphingolipids along with actin dynamics are regulated by PI4,5P2 and the Slm1 and Slm2 proteins. PI4,5P2, synthesized by Stt4 and Mss4 on the plasma membrane, in cooperation with the TORC2 and the Pkh1/2 protein kinases are proposed to activate Slm1 and Slm2. The Slm proteins then impair turnover of complex sphingolipids, particularly IPC, by inhibiting Isc1 and they also regulate the calcium/calmodulin-regulated protein phosphatase calcineurin which dephosphorylates and inactivates the Slm proteins and also interacts with Csg1/2 in an unknown manner to regulate conversion of IPC to MIPC by the Csg1/2 enzymes. Regulation of the Rho1/Pkc1 pathway by Slm1/2 works independently from regulation of sphingolipid metabolism. Genetic interactions suggest that IPC plays a role in actin organization, but the mechanism is unknown. Pkh1/2 are probably attached to eisosomes98 (not shown) as depicted in Fig. 2. Adapted from ref.6
Data supporting a role for sphingolipids in regulating the Slm proteins and how they in turn regulate sphingolipid synthesis and turnover were recently reviewed in ref 6 and are summarized diagrammatically in Fig. 3. The model depicts events during unstressed growth and during heat stress: whether similar events are active during other stresses is unknown.44, 82, 102 Central to this model are the Slm proteins functioning downstream of the PIP2, TORC2 and LCB-Pkh1/2 signaling pathways. Heat stress causes a transient increase in PIP2 thereby recruiting the Slm proteins to the plasma membrane where they are phosphorylated by TORC2 and also by Pkh1/2, which are activated also by the transient burst of LCBs induced by heat. Activated Slm proteins control movement of the actin cytoskeleton via the Rho1/Pkc1 pathway. Unexpectedly, the Slm proteins also control sphingolipid metabolism by down-regulating Isc1 activity, thus slowing breakdown of complex sphingolipids, especially the very abundant IPC-C (IPC with one hydroxyl group on the C26 fatty acid.107 The Slm proteins also down-regulate calcineurin phosphatase activity, which interacts with Csg2, in an unknown manner, to regulate conversion of IPC-C to MIPC. Hence, during heat stress the Slm proteins are thought to interface between phosphoinositides and sphingolipids and orchestrate changes in membrane lipid composition to promote survival.
Once cells adjust to a heat stress they down-regulate activated Slm proteins by calcineurin-mediated dephosphorylation in what appears to be a negative feedback loop (Fig. 3). It is also likely that IPC, possibly a specific pool of IPC-C, regulates actin organization and viability.102, 108 Identifying this pool of IPC-C could provide important clues for understanding how such regulation occurs.
Recognition that the LCB-Pkh1/2, PIP2 and TORC2 pathways use the Slm proteins and calcineurin to regulate sphingolipid metabolism provides a framework for deciphering the molecular basis for maintaining sphingolipid levels that are optimal for growth in the absence of stress and for surviving a heat stress. Many questions remain unanswered. Do Pkh1/2 directly phosphorylate Slm1 and Slm2 or does one of the protein kinases regulated by Pkh1/2 (Fig. 2)? Available evidence suggests that Pkh1/2 do not phosphorylate the Slm proteins nor do Ypk1/2, but the data do not exclude these possibilities and more work is needed.82 How do the Slm proteins regulate Isc1 activity? Does Isc1 play a direct role in regulating the actin cytoskeleton and how do Csg1\2 interact with calcineurin?
Slm2 seems more dependent upon sphingolipids for function than does Slm1. First, haploid slm1Δ cells are more sensitive to growth inhibition by myriocin than slm2Δ cells. Second, heat-induced and sphingolipid-dependent endocytosis of the uracil transporter requires Slm2 but not Slm1.44 Lastly, a recent large-scale screen of deletion mutants found that slm1Δ heterozygous and homozygous diploid cells were extremely sensitive to myriocin compared to all other gene deletion mutants including slm2Δ cells.96
Longevity and cellular aging
Long before it was realized that LAG1 (Longevity-Assurance Gene109) encodes a ceramide synthase110, 111, it had been found to affect replicative lifespan, measured by determining how many times a cell can bud. Deletion of LAG1 increases replicative lifespan by 50%109, but the mechanism remains unknown.
Sch9 (Fig. 2) plays roles in both replicative and chronological lifespan, measured by how long cells survive in stationary phase when division has ceased. Deletion of SCH9 increases chronological lifespan by 300%112 and increases both the mean and maximal replicative lifespan.113 No one has yet determined if LCBs play a role in lifespan by regulating Pkh1/2 which in turn phosphorylate Sch9 on threonine 570 in its activation domain.94, 97 To be active, Sch9 must also be phosphorylated by TORC1 at several serine and threonine residues located in its C-terminus.94
Isc1, which cleaves polar head groups from complex sphingolipids to generate ceramides, has been shown to play a role in oxidative stress resistance (hydrogen peroxide) and chronological lifespan.114 For lifespan measurements, cells were grown to log phase or early stationary phase in YP medium containing glycerol as the carbon source and then transferred to water, a severe form of calorie restriction. Both log and stationary phase isc1Δ cells died extremely rapidly compared to ISC1 cells, indicating that viability under these experimental conditions depends on Isc1 function. The oxidative stress theory of aging argues that cells die because of oxidative stress and several but not all measures of oxidative stress were elevated in isc1Δ cells. Apoptosis is induced by oxidative stress and during aging115–118 and isc1Δ cells showed classic signs of apoptosis including DNA fragmentation and activation of Yca1 metacaspase activity. The rapid loss of viability in calorie-restricted stationary phase isc1Δ cells was completely prevented by deletion of YCA1, arguing that Isc1 is involved in a pathway that regulates Yca1 and apoptosis.
To begin to elucidate the role of Isc1 in oxidative stress resistance and chronological lifespan, a global analysis of mRNAs was performed using microarray technology.114 Expression of 72 genes was found to increase and sorting these into biological processes showed that six were involved in iron uptake. The authors reasoned that induction of these genes during iron abundance could cause iron overloading and enhance oxidative stress. They found iron increased in both log and stationary phase isc1Δ cells and data from several types of experiments supported the notion that such accumulation contributed to but was not entirely responsible for increased oxidative stress and cell death. At this time it is not clear how Isc1 protects cells against oxidative stress and apoptosis. A basic question is whether the ceramide generated by Isc1 activity is necessary for stress protection or whether accumulation of one or more complex sphingolipids such as IPC1-C in isc1Δ cells causes cell death. Others have argued that the increased level of M(IP2)C found in isc1Δ cells30, 114 is the cause of cell death. It would be interesting and novel if ceramide production by Isc1 protected against oxidative stress, since ceramide plays an opposite role in mammals during stresses and promotes cell death.84, 119, 120 Other roles for Isc1 were recently reviewed and readers are encouraged to examine previous work on this interesting enzyme.6, 9
Finally, recent studies showed that overexpression of the Ydc1 ceramidase (Fig. 1) shortened chronological lifespan, but this shortening probably results from fragmentation of mitochondria and vacuoles and increased apoptosis and it is not clear how these studies relate to normal physiological functions of Ydc1 and sphingolipids.121 Analysis of ceramides in Ydc1 overproducing cells showed that both dihydroceramides and phytoceramides were reduced, suggesting that the enzyme does not specifically hydrolyze dihydroceramides in vivo. These studies are supportative of a role for ceramide as an inducer of apoptosis in yeast similar to what transpires in mammals.
Regulation of sphingolipid biosynthesis
I think it is safe to say that we do not really understand how de novo sphingolipid synthesis is regulated in any organism. Readers should consult previous reviews describing what we do know about the regulation of sphingolipid biosynthesis including regulation of gene transcription as only the latest studies will be discussed herein.4–6, 8–10
Recently it was shown that TORC2 controls ceramide synthase activity.122 The TOR protein kinases sense nutrients and stresses and coordinate metabolism both temporarily and spatially to regulate cell growth.123 All eukaryotes that have been examined contain two TOR protein complexes, TORC1, which is inhibited by rapamycin, a bacterial macrocyclic lactone, and TORC2, which is rapamycin-insensitive. A genetic screen had implied a link between sphingolipids, the TOR proteins and calcium homeostasis in yeast. This screen used the sensitivity of a csg2 mutant to 100 mM Ca++ to identify temperature-sensitive mutations that bypassed the calcium-sensitivity.62 Bypass mutations occurred in several genes including TOR2 and AVO3/TSC11 that encode components of TORC2.34 TORC2 specifically controls the organization of the actin cytoskeleton in yeast and mammals.123
A critical advance in understanding the connection between TOR signaling and sphingolipids relied upon the isolation of a temperature-sensitive allele of AVO3 (avo3-30) that diminished growth even at 30°C. Three hours after shifting avo3-30 cells from 25°C to 30°C the major yeast ceramide species containing PHS and a C26-fatty acid was 5-fold lower in concentration compared to wild-type cells and minor ceramide species having shorter fatty acid chains were reduced about 10-fold.122 Even when grown at 25°C, the concentration of the major ceramides was reduced 2-fold in avo3-30 cells. These results implied a reduction in ceramide synthase activity in avo3-30 cells and this prediction was confirmed by measuring enzyme activity in cells grown at 30°C.
How might TORC2 influence ceramide synthase activity? The Ypk2 protein kinase was known to be activated by TORC2124 and mutant ypk2 and avo3-30 cells showed similar defects in cell wall integrity and actin polarization, suggesting that Ypk2 working downstream of TORC2 and Pkh1/2 (Fig. 2) might regulate ceramide synthase activity. This possibility was supported by data showing that a constitutively active allele of YPK2 reversed all avo3-30 phenotypes including the ceramide deficiency.122 These results argue that Ypk2 acts downstream of TORC2 to activate ceramide synthase activity. The molecular details of how Ypk2 governs ceramide synthase activity remains to be determined. Further analysis implicated calcineurin as a regulator of ceramide levels. Deletion of the CNB1 gene, which encodes the calcineurin regulatory subunit B, restored ceramide levels in avo3-30 cells and promoted growth at 30°C, suggesting that calcineurin down-regulates ceramide synthase activity. Perhaps calcineurin dephosphorylates ceramide synthase and one wonders if the Slm proteins (Fig. 3) are somehow involved is such regulation. Surprisingly, deletion of CNB1 did not restore actin polarization to avo3-30 cells, implying that it is controlled by TORC2 in a distinctly different manner from ceramide synthesis activity.
A reduction in ceramide synthase activity predicts an increase in DHS and PHS based upon previous work (reviewed in ref.6) and avo3-30 cells did indeed have elevated DHS and PHS levels.122 These elevated LCBs could act in a feed-forward manner to activate ceramide synthase activity and promote synthesis of complex sphingolipids. This may occur because DHS and PHS activate Pkh1 and Pkh2 which then phosphorylate Ypk2 in its activation domain while TORC2 phosphorylate residues in the C-terminus (Fig. 2). To be active, Ypk2 must be phosphorylated in both domains. The possibility of similar types of regulation of ceramide synthase in mammals has been discussed.125
These studies form a basis for understanding how yeast cells coordinate ceramide and sphingolipid synthesis with nutrient availability and cell growth and how synthesis is reduced during stress when cells need to shift from growth to survival mode. But there is likely to be a lot more to this story than is apparent now and there are likely to be many more layers of regulation including regulation of Isc1 as depicted in Fig. 3.
Summary and future developments
Since sphingolipids are abundant in the plasma membrane and also present in smaller amounts in other cellular membranes and compartments it is not surprising that they have been found to play roles in many processes occurring in or on membranes (Figs. 2 and 3 and see also Fig. 3 in ref.9). Most of what we know about sphingolipid involvement in these processes has come from directed, small-scale experiments involving one or a few proteins or cellular processes as readouts and such studies will continue to be required to elucidate molecular mechanisms. Larger-scale, genome or proteome-wide experiments have started to identify new roles for sphingolipids and proteins that require sphingolipids for function.12, 48, 96, 126–130 More large-scale experiments that interrogate all yeast proteins need to be devised to determine if there are general rules for why proteins depend upon sphingolipids for function or whether such functional dependence is unique for each protein. The ability to analyze and quantify (“profile”) nearly all species of sphingolipids in a single sample by mass spectrometry is a great advance in methodology that will likely become the standard way to measure sphingolipids in yeast16, 24, 131, 132 much as it has become the standard way to analyze sphingolipids in mammals.131, 133 One of the most challenging problems will be to advance our understanding of the roles that sphingolipids play in cells from phenomenology and associations to molecular mechanisms. This will require techniques to measure sphingolipid binding to proteins either in vivo or in vitro and to demonstrate that binding promotes or inhibits a cellular process. For example, do LCBs directly interact with Pkh1/2 to stimulate their activity or do they act indirectly such as by binding to eisosomes which then activate Pkh1/2?
Acknowledgments
Work in the author’s laboratory was supported by research grant from the National Institutes of Health (AG024377) and by core facilities supported by Grant P20-RR020171 from the National Center for Research Resources, a component of the National Institutes of Health.
References
- 1.Carter HE, Hendrickson HS. Biochemistry of the sphingolipids. XV. Structure of phytosphingosine and dehydrophytospingosine. Biochemistry. 1963;2:389–93. doi: 10.1021/bi00902a036. [DOI] [PubMed] [Google Scholar]
- 2.Dickson RC, Lester RL. Metabolism and selected functions of sphingolipids in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 1999;1438(3):305–21. doi: 10.1016/s1388-1981(99)00068-2. [DOI] [PubMed] [Google Scholar]
- 3.Dickson RC, Lester RL. Yeast sphingolipids. Biochim Biophys Acta. 1999;1426(2):347–357. doi: 10.1016/s0304-4165(98)00135-4. [DOI] [PubMed] [Google Scholar]
- 4.Dickson RC, Lester RL. Sphingolipid functions in Saccharomyces cerevisiae. Biochim Biophys Acta. 2002;1583(1):13–25. doi: 10.1016/s1388-1981(02)00210-x. [DOI] [PubMed] [Google Scholar]
- 5.Sims KJ, Spassieva SD, Voit EO, Obeid LM. Yeast sphingolipid metabolism: clues and connections. Biochem Cell Biol. 2004;82(1):45–61. doi: 10.1139/o03-086. [DOI] [PubMed] [Google Scholar]
- 6.Dickson RC. Thematic review series: sphingolipids. New insights into sphingolipid metabolism and function in budding yeast. J Lipid Res. 2008;49(5):909–21. doi: 10.1194/jlr.R800003-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patton JL, Lester RL. The phosphoinositol sphingolipids of Saccharomyces cerevisiae are highly localized in the plasma membrane. J Bacteriol. 1991;173(10):3101–3108. doi: 10.1128/jb.173.10.3101-3108.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Funato K, Vallee B, Riezman H. Biosynthesis and trafficking of sphingolipids in the yeast Saccharomyces cerevisiae. Biochemistry. 2002;41(51):15105–15114. doi: 10.1021/bi026616d. [DOI] [PubMed] [Google Scholar]
- 9.Dickson RC, Sumanasekera C, Lester RL. Functions and metabolism of sphingolipids in Saccharomyces cerevisiae. Prog Lipid Res. 2006;45(6):447–65. doi: 10.1016/j.plipres.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Cowart LA, Obeid LM. Yeast sphingolipids: recent developments in understanding biosynthesis, regulation, and function. Biochim Biophys Acta. 2007;1771(3):421–31. doi: 10.1016/j.bbalip.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Funato K, Riezman H. Vesicular and nonvesicular transport of ceramide from ER to the Golgi apparatus in yeast. J Cell Biol. 2001;155(6):949–59. doi: 10.1083/jcb.200105033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Natter K, Leitner P, Faschinger A, Wolinski H, McCraith S, Fields S, et al. The Spatial Organization of Lipid Synthesis in the Yeast Saccharomyces cerevisiae Derived from Large Scale Green Fluorescent Protein Tagging and High Resolution Microscopy. Mol Cell Proteomics. 2005;4(5):662–672. doi: 10.1074/mcp.M400123-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Hechtberger P, Daum G. Intracellular transport of inositol-containing sphingolipids in the yeast, Saccharomyces cerevisiae. FEBS Lett. 1995;367(2):201–204. doi: 10.1016/0014-5793(95)00567-s. [DOI] [PubMed] [Google Scholar]
- 14.Futerman AH, Riezman H. The ins and outs of sphingolipid synthesis. Trends Cell Biol. 2005;15(6):312–8. doi: 10.1016/j.tcb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 15.Hechtberger P, Zinser E, Saf R, Hummel K, Paltauf F, Daum G. Characterization, quantification and subcellular localization of inositol-containing sphingolipids of the yeast, Saccharomyces cerevisiae. Eur J Biochem. 1994;225(2):641–649. doi: 10.1111/j.1432-1033.1994.00641.x. [DOI] [PubMed] [Google Scholar]
- 16.Schneiter R, Brugger B, Sandhoff R, Zellnig G, Leber A, Lampl M, et al. Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J Cell Biol. 1999;146(4):741–54. doi: 10.1083/jcb.146.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kitagaki H, Cowart LA, Matmati N, Vaena de Avalos S, Novgorodov SA, Zeidan YH, et al. Isc1 regulates sphingolipid metabolism in yeast mitochondria. Biochim Biophys Acta. 2007;1768(11):2849–61. doi: 10.1016/j.bbamem.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith SW, Lester RL. Inositol phosphorylceramide, a novel substance and a chief member of a major group of yeast sphingolipids containing single inositol phosphate. Journal of Biological Chemistry. 1974;249:3395–3405. [PubMed] [Google Scholar]
- 19.Oh CS, Toke DA, Mandala S, Martin CE. ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J Biol Chem. 1997;272(28):17376–84. doi: 10.1074/jbc.272.28.17376. [DOI] [PubMed] [Google Scholar]
- 20.Kohlwein SD, Eder S, Oh CS, Martin CE, Gable K, Bacikova D, et al. Tsc13p is required for fatty acid elongation and localizes to a novel structure at the nuclear-vacuolar interface in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21(1):109–25. doi: 10.1128/MCB.21.1.109-125.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rossler H, Rieck C, Delong T, Hoja U, Schweizer E. Functional differentiation and selective inactivation of multiple Saccharomyces cerevisiae genes involved in very-long-chain fatty acid synthesis. Mol Genet Genomics. 2003;269(2):290–8. doi: 10.1007/s00438-003-0836-0. [DOI] [PubMed] [Google Scholar]
- 22.Denic V, Weissman JS. A molecular caliper mechanism for determining very long-chain fatty acid length. Cell. 2007;130(4):663–77. doi: 10.1016/j.cell.2007.06.031. [DOI] [PubMed] [Google Scholar]
- 23.Schneiter R, Hitomi M, Ivessa AS, Fasch EV, Kohlwein SD, Tartakoff AM. A yeast acetyl coenzyme a carboxylase mutant links very-long-chain fatty acid synthesis to the structure and function of the nuclear membrane-pore complex. Molecular and Cellular Biology. 1996;16(12):7161–7172. doi: 10.1128/mcb.16.12.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cerantola V, Vionnet C, Aebischer OF, Jenny T, Knudsen J, Conzelmann A. Yeast sphingolipids do not need to contain very long chain fatty acids. Biochem J. 2007;401(1):205–16. doi: 10.1042/BJ20061128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lynch V, Dunn T. An introduction to plant sphingolipids and a review of recent advances in understanding their metabolism and function. New Phytologist. 2004;16(3):677–702. doi: 10.1111/j.1469-8137.2004.00992.x. [DOI] [PubMed] [Google Scholar]
- 26.Markham JE, Li J, Cahoon EB, Jaworski JG. Separation and identification of major plant sphingolipid classes from leaves. J Biol Chem. 2006;281(32):22684–94. doi: 10.1074/jbc.M604050200. [DOI] [PubMed] [Google Scholar]
- 27.Replansky T, Koufopanou V, Greig D, Bell G. Saccharomyces sensu stricto as a model system for evolution and ecology. Trends Ecol Evol. 2008;23(9):494–501. doi: 10.1016/j.tree.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 28.Raas-Rothschild A, Pankova-Kholmyansky I, Kacher Y, Futerman AH. Glycosphingolipidoses: beyond the enzymatic defect. Glycoconj J. 2004;21(6):295–304. doi: 10.1023/B:GLYC.0000046272.38480.ef. [DOI] [PubMed] [Google Scholar]
- 29.Welsch CA, Roth LW, Goetschy JF, Movva NR. Genetic, biochemical, and transcriptional responses of Saccharomyces cerevisiae to the novel immunomodulator FTY720 largely mimic those of the natural sphingolipid phytosphingosine. J Biol Chem. 2004;279(35):36720–31. doi: 10.1074/jbc.M406179200. [DOI] [PubMed] [Google Scholar]
- 30.Sawai H, Okamoto Y, Luberto C, Mao C, Bielawska A, Domae N, et al. Identification of ISC1 (YER019w) as inositol phosphosphingolipid phospholipase C in Saccharomyces cerevisiae. J Biol Chem. 2000;275(50):39793–8. doi: 10.1074/jbc.M007721200. [DOI] [PubMed] [Google Scholar]
- 31.Wells GB, Dickson RC, Lester RL. Isolation and composition of inositolphosphorylceramide-type sphingolipids of hyphal forms of Candida albicans. Journal of Bacteriology. 1996;178(21):6223–6226. doi: 10.1128/jb.178.21.6223-6226.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaena de Avalos S, Su X, Zhang M, Okamoto Y, Dowhan W, Hannun YA. The phosphatidylglycerol/cardiolipin biosynthetic pathway is required for the activation of inositol phosphosphingolipid phospholipase C, Isc1p, during growth of Saccharomyces cerevisiae. J Biol Chem. 2005;280(8):7170–7. doi: 10.1074/jbc.M411058200. [DOI] [PubMed] [Google Scholar]
- 33.Zhao C, Beeler T, Dunn T. Suppressors of the Ca(2+)-sensitive yeast mutant (csg2) identify genes involved in sphingolipid biosynthesis. Cloning and characterization of SCS1, a gene required for serine palmitoyltransferase activity. J Biol Chem. 1994;269(34):21480–8. [PubMed] [Google Scholar]
- 34.Beeler T, Bacikova D, Gable K, Hopkins L, Johnson C, Slife H, et al. The Saccharomyces cerevisiae TSC10/YBR265w gene encoding 3-ketosphinganine reductase is identified in a screen for temperature-sensitive suppressors of the Ca2+-sensitive csg2 Delta mutant. J Biol Chem. 1998;273(46):30688–94. doi: 10.1074/jbc.273.46.30688. [DOI] [PubMed] [Google Scholar]
- 35.Mao C, Xu R, Bielawska A, Obeid LM. Cloning of an alkaline ceramidase from Saccharomyces cerevisiae. An enzyme with reverse (CoA-independent) ceramide synthase activity. J Biol Chem. 2000;275(10):6876–84. doi: 10.1074/jbc.275.10.6876. [DOI] [PubMed] [Google Scholar]
- 36.Mao C, Xu R, Bielawska A, Szulc ZM, Obeid LM. Cloning and characterization of a Saccharomyces cerevisiae alkaline ceramidase with specificity for dihydroceramide. J Biol Chem. 2000;275(40):31369–78. doi: 10.1074/jbc.M003683200. [DOI] [PubMed] [Google Scholar]
- 37.Pruett ST, Bushnev A, Hagedorn K, Adiga M, Haynes CA, Sullards MC, et al. Biodiversity of sphingoid bases (“sphingosines”) and related amino alcohols. J Lipid Res. 2008;49(8):1621–39. doi: 10.1194/jlr.R800012-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munn AL, Riezman H. Endocytosis is required for the growth of vacuolar H(+)-ATPase-defective yeast: identification of six new END genes. J Cell Biol. 1994;127(2):373–86. doi: 10.1083/jcb.127.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sutterlin C, Doering TL, Schimmoller F, Schroder S, Riezman H. Specific requirements for the ER to Golgi transport of GPI-anchored proteins in yeast. Journal of Cell Science. 1997;110(Pt 21):2703–2714. doi: 10.1242/jcs.110.21.2703. [DOI] [PubMed] [Google Scholar]
- 40.Chung N, Jenkins G, Hannun YA, Heitman J, Obeid LM. Sphingolipids signal heat stress-induced ubiquitin-dependent proteolysis. J Biol Chem. 2000;275(23):17229–32. doi: 10.1074/jbc.C000229200. [DOI] [PubMed] [Google Scholar]
- 41.Zanolari B, Friant S, Funato K, Sutterlin C, Stevenson BJ, Riezman H. Sphingoid base synthesis requirement for endocytosis in Saccharomyces cerevisiae. EMBO J. 2000;19(12):2824–2833. doi: 10.1093/emboj/19.12.2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hearn JD, Lester RL, Dickson RC. The uracil transporter Fur4p associates with lipid rafts. J Biol Chem. 2003;278(6):3679–3686. doi: 10.1074/jbc.M209170200. [DOI] [PubMed] [Google Scholar]
- 43.Grote E, Vlacich G, Pypaert M, Novick PJ. A snc1 endocytosis mutant: phenotypic analysis and suppression by overproduction of dihydrosphingosine phosphate lyase. Mol Biol Cell. 2000;11(12):4051–65. doi: 10.1091/mbc.11.12.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bultynck G, Heath VL, Majeed AP, Galan JM, Haguenauer-Tsapis R, Cyert MS. Slm1 and slm2 are novel substrates of the calcineurin phosphatase required for heat stress-induced endocytosis of the yeast uracil permease. Mol Cell Biol. 2006;26(12):4729–45. doi: 10.1128/MCB.01973-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bagnat M, Simons K. Lipid rafts in protein sorting and cell polarity in budding yeast Saccharomyces cerevisiae. Biol Chem. 2002;383(10):1475–80. doi: 10.1515/BC.2002.169. [DOI] [PubMed] [Google Scholar]
- 46.Simons K, Vaz WL. Model systems, lipid rafts, and cell membranes. Annu Rev Biophys Biomol Struct. 2004;33:269–95. doi: 10.1146/annurev.biophys.32.110601.141803. [DOI] [PubMed] [Google Scholar]
- 47.Lisman Q, Pomorski T, Vogelzangs C, Urli-Stam D, de Cocq van Delwijnen W, Holthuis JC. Protein sorting in the late Golgi of Saccharomyces cerevisiae does not require mannosylated sphingolipids. J Biol Chem. 2004;279(2):1020–9. doi: 10.1074/jbc.M306119200. [DOI] [PubMed] [Google Scholar]
- 48.Proszynski TJ, Klemm RW, Gravert M, Hsu PP, Gloor Y, Wagner J, et al. A genome-wide visual screen reveals a role for sphingolipids and ergosterol in cell surface delivery in yeast. Proc Natl Acad Sci U S A. 2005;102(50):17981–6. doi: 10.1073/pnas.0509107102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bagnat M, Keranen S, Shevchenko A, Simons K. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc Natl Acad Sci U S A. 2000;97(7):3254–9. doi: 10.1073/pnas.060034697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Q, Chang A. Sphingoid base synthesis is required for oligomerization and cell surface stability of the yeast plasma membrane ATPase, Pma1. Proc Natl Acad Sci U S A. 2002;99(20):12853–8. doi: 10.1073/pnas.202115499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee MC, Hamamoto S, Schekman R. Ceramide biosynthesis is required for the formation of the oligomeric H+-ATPase Pma1p in the yeast endoplasmic reticulum. J Biol Chem. 2002;277(25):22395–401. doi: 10.1074/jbc.M200450200. [DOI] [PubMed] [Google Scholar]
- 52.Eisenkolb M, Zenzmaier C, Leitner E, Schneiter R. A specific structural requirement for ergosterol in long-chain fatty acid synthesis mutants important for maintaining raft domains in yeast. Mol Biol Cell. 2002;13(12):4414–28. doi: 10.1091/mbc.E02-02-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gaigg B, Timischl B, Corbino L, Schneiter R. Synthesis of sphingolipids with very long chain fatty acids but not ergosterol is required for routing of newly synthesized plasma membrane ATPase to the cell surface of yeast. J Biol Chem. 2005;280(23):22515–22. doi: 10.1074/jbc.M413472200. [DOI] [PubMed] [Google Scholar]
- 54.Toulmay A, Schneiter R. Lipid-dependent surface transport of the proton pumping ATPase: a model to study plasma membrane biogenesis in yeast. Biochimie. 2007;89(2):249–54. doi: 10.1016/j.biochi.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 55.Bagnat M, Simons K. Cell surface polarization during yeast mating. Proc Natl Acad Sci U S A. 2002;99(22):14183–8. doi: 10.1073/pnas.172517799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dupre S, Haguenauer-Tsapis R. Raft partitioning of the yeast uracil permease during trafficking along the endocytic pathway. Traffic. 2003;4(2):83–96. doi: 10.1034/j.1600-0854.2003.40204.x. [DOI] [PubMed] [Google Scholar]
- 57.Opekarova M, Malinska K, Novakova L, Tanner W. Differential effect of phosphatidylethanolamine depletion on raft proteins: further evidence for diversity of rafts in Saccharomyces cerevisiae. Biochim Biophys Acta. 2005;1711(1):87–95. doi: 10.1016/j.bbamem.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 58.Lauwers E, Grossmann G, Andre B. Evidence for coupled biogenesis of yeast Gap1 permease and sphingolipids: essential role in transport activity and normal control by ubiquitination. Mol Biol Cell. 2007;18(8):3068–80. doi: 10.1091/mbc.E07-03-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gaigg B, Toulmay A, Schneiter R. Very long-chain fatty acid-containing lipids rather than sphingolipids per se are required for raft association and stable surface transport of newly synthesized plasma membrane ATPase in yeast. J Biol Chem. 2006;281(45):34135–45. doi: 10.1074/jbc.M603791200. [DOI] [PubMed] [Google Scholar]
- 60.Olivera A, Zhang H, Carlson RO, Mattie ME, Schmidt RR, Spiegel S. Stereospecificity of sphingosine-induced intracellular calcium mobilization and cellular proliferation. Journal of Biological Chemistry. 1994;269(27):17924–17930. [PubMed] [Google Scholar]
- 61.Birchwood CJ, Saba JD, Dickson RC, Cunningham KW. Calcium influx and signaling in yeast stimulated by intracellular sphingosine 1-phosphate accumulation. J Biol Chem. 2001;276(15):11712–8. doi: 10.1074/jbc.M010221200. [DOI] [PubMed] [Google Scholar]
- 62.Beeler T, Gable K, Zhao C, Dunn T. A novel protein, CSG2p, is required for Ca2+ regulation in Saccharomyces cerevisiae. J Biol Chem. 1994;269(10):7279–84. [PubMed] [Google Scholar]
- 63.Lisman Q, Urli-Stam D, Holthuis JC. HOR7, a multicopy suppressor of the Ca2+-induced growth defect in sphingolipid mannosyltransferase-deficient yeast. J Biol Chem. 2004;279(35):36390–6. doi: 10.1074/jbc.M406197200. [DOI] [PubMed] [Google Scholar]
- 64.Skrzypek MS, Nagiec MM, Lester RL, Dickson RC. Inhibition of amino acid transport by sphingoid long chain bases in Saccharomyces cerevisiae. J Biol Chem. 1998;273(5):2829–34. doi: 10.1074/jbc.273.5.2829. [DOI] [PubMed] [Google Scholar]
- 65.Chung N, Mao C, Heitman J, Hannun YA, Obeid LM. Phytosphingosine as a specific inhibitor of growth and nutrient import in Saccharomyces cerevisiae. J Biol Chem. 2001;276(38):35614–35621. doi: 10.1074/jbc.M105653200. [DOI] [PubMed] [Google Scholar]
- 66.Friant S, Meier KD, Riezman H. Increased ubiquitin-dependent degradation can replace the essential requirement for heat shock protein induction. EMBO J. 2003;22(15):3783–91. doi: 10.1093/emboj/cdg375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gelperin D, Horton L, DeChant A, Hensold J, Lemmon SK. Loss of ypk1 function causes rapamycin sensitivity, inhibition of translation initiation and synthetic lethality in 14–3-3-deficient yeast. Genetics. 2002;161(4):1453–1464. doi: 10.1093/genetics/161.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kraakman L, Lemaire K, Ma P, Teunissen AW, Donaton MC, Van Dijck P, et al. A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol Microbiol. 1999;32(5):1002–12. doi: 10.1046/j.1365-2958.1999.01413.x. [DOI] [PubMed] [Google Scholar]
- 69.Roosen J, Engelen K, Marchal K, Mathys J, Griffioen G, Cameroni E, et al. PKA and Sch9 control a molecular switch important for the proper adaptation to nutrient availability. Molecular Microbiology. 2005;55(3):862–880. doi: 10.1111/j.1365-2958.2004.04429.x. [DOI] [PubMed] [Google Scholar]
- 70.Valachovic M, Wilcox LI, Sturley SL, Bard M. A mutation in sphingolipid synthesis suppresses defects in yeast ergosterol metabolism. Lipids. 2004;39(8):747–52. doi: 10.1007/s11745-004-1291-6. [DOI] [PubMed] [Google Scholar]
- 71.Valachovic M, Bareither BM, Shah Alam Bhuiyan M, Eckstein J, Barbuch R, Balderes D, et al. Cumulative mutations affecting sterol biosynthesis in the yeast Saccharomyces cerevisiae result in synthetic lethality that is suppressed by alterations in sphingolipid profiles. Genetics. 2006;173(4):1893–908. doi: 10.1534/genetics.105.053025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Swain E, Baudry K, Stukey J, McDonough V, Germann M, Nickels JT., Jr Sterol-dependent Regulation of Sphingolipid Metabolism in Saccharomyces cerevisiae. J Biol Chem. 2002;277(29):26177–84. doi: 10.1074/jbc.M204115200. [DOI] [PubMed] [Google Scholar]
- 73.Kihara A, Igarashi Y. Cross talk between sphingolipids and glycerophospholipids in the establishment of plasma membrane asymmetry. Mol Biol Cell. 2004;15(11):4949–59. doi: 10.1091/mbc.E04-06-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thevissen K, Francois IE, Takemoto JY, Ferket KK, Meert EM, Cammue BP. DmAMP1, an antifungal plant defensin from dahlia (Dahlia merckii), interacts with sphingolipids from Saccharomyces cerevisiae. FEMS Microbiol Lett. 2003;226(1):169–73. doi: 10.1016/S0378-1097(03)00590-1. [DOI] [PubMed] [Google Scholar]
- 75.Thevissen K, Cammue BP, Lemaire K, Winderickx J, Dickson RC, Lester RL, et al. A gene encoding a sphingolipid biosynthesis enzyme determines the sensitivity of Saccharomyces cerevisiae to an antifungal plant defensin from dahlia (Dahlia merckii) Proc Natl Acad Sci U S A. 2000;97(17):9531–6. doi: 10.1073/pnas.160077797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stock SD, Hama H, Radding JA, Young DA, Takemoto JY. Syringomycin E inhibition of Saccharomyces cerevisiae: requirement for biosynthesis of sphingolipids with very-long-chain fatty acids and mannose- and phosphoinositol-containing head groups. Antimicrob Agents Chemother. 2000;44(5):1174–1180. doi: 10.1128/aac.44.5.1174-1180.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hama H, Youngb DA, Raddingb JA, Mab D, Tangb J, Stocka SD, et al. Requirement of sphingolipid alpha-hydroxylation for fungicidal action of syringomycin E. FEBS Lett. 2000;478(1–2):26–8. doi: 10.1016/s0014-5793(00)01821-4. [DOI] [PubMed] [Google Scholar]
- 78.Grilley MM, Stock SD, Dickson RC, Lester RL, Takemoto JY. Syringomycin action gene SYR2 is essential for sphingolipid 4-hydroxylation in Saccharomyces cerevisiae. J Biol Chem. 1998;273(18):11062–11068. doi: 10.1074/jbc.273.18.11062. [DOI] [PubMed] [Google Scholar]
- 79.Bosson R, Conzelmann A. Multiple functions of inositolphosphorylceramides in the formation and intracellular transport of glycosylphosphatidylinositol-anchored proteins in yeast. Biochem Soc Symp. 2007;(74):199–209. doi: 10.1042/BSS0740199. [DOI] [PubMed] [Google Scholar]
- 80.Horvath A, Sutterlin C, Manning-Krieg U, Movva NR, Riezman H. Ceramide synthesis enhances transport of GPI-anchored proteins to the Golgi apparatus in yeast. Embo J. 1994;13(16):3687–95. doi: 10.1002/j.1460-2075.1994.tb06678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Skrzypek M, Lester RL, Dickson RC. Suppressor gene analysis reveals an essential role for sphingolipids in transport of glycosylphosphatidylinositol-anchored proteins in Saccharomyces cerevisiae. J Bacteriol. 1997;179(5):1513–20. doi: 10.1128/jb.179.5.1513-1520.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Daquinag A, Fadri M, Jung SY, Qin J, Kunz J. The yeast PH domain proteins Slm1 and Slm2 are targets of sphingolipid signaling during the response to heat stress. Mol Cell Biol. 2007;27(2):633–50. doi: 10.1128/MCB.00461-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chung JH, Lester RL, Dickson RC. Sphingolipid requirement for generation of a functional v1 component of the vacuolar ATPase. J Biol Chem. 2003;278(31):28872–81. doi: 10.1074/jbc.M300943200. [DOI] [PubMed] [Google Scholar]
- 84.Carpinteiro A, Dumitru C, Schenck M, Gulbins E. Ceramide-induced cell death in malignant cells. Cancer Lett. 2008;264(1):1–10. doi: 10.1016/j.canlet.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 85.Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9(2):139–50. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- 86.Hait NC, Oskeritzian CA, Paugh SW, Milstien S, Spiegel S. Sphingosine kinases, sphingosine 1-phosphate, apoptosis and diseases. Biochim Biophys Acta. 2006;1758(12):2016–26. doi: 10.1016/j.bbamem.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 87.Dickson RC, Nagiec EE, Skrzypek M, Tillman P, Wells GB, Lester RL. Sphingolipids are potential heat stress signals in Saccharomyces. Journal of Biological Chemistry. 1997;272:30196–30200. doi: 10.1074/jbc.272.48.30196. [DOI] [PubMed] [Google Scholar]
- 88.Jenkins GM, Richards A, Wahl T, Mao C, Obeid L, Hannun Y. Involvement of yeast sphingolipids in the heat stress response of Saccharomyces cerevisiae. J Biol Chem. 1997;272(51):32566–32572. doi: 10.1074/jbc.272.51.32566. [DOI] [PubMed] [Google Scholar]
- 89.Sun Y, Taniguchi R, Tanoue D, Yamaji T, Takematsu H, Mori K, et al. Sli2 (Ypk1), a homologue of mammalian protein kinase SGK, is a downstream kinase in the sphingolipid-mediated signaling pathway of yeast. Mol Cell Biol. 2000;20(12):4411–4419. doi: 10.1128/mcb.20.12.4411-4419.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schmelzle T, Helliwell SB, Hall MN. Yeast Protein Kinases and the RHO1 Exchange Factor TUS1 Are Novel Components of the Cell Integrity Pathway in Yeast. Mol Cell Biol. 2002;22(5):1329–1339. doi: 10.1128/mcb.22.5.1329-1339.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Roelants FM, Torrance PD, Bezman N, Thorner J. Pkh1 and Pkh2 differentially phosphorylate and activate Ypk1 and Ykr2 and define protein kinase modules required for maintenance of cell wall integrity. Mol Biol Cell. 2002;13(9):3005–3028. doi: 10.1091/mbc.E02-04-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.deHart AK, Schnell JD, Allen DA, Hicke L. The conserved Pkh-Ypk kinase cascade is required for endocytosis in yeast. J Cell Biol. 2002;156(2):241–248. doi: 10.1083/jcb.200107135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Casamayor A, Torrance PD, Kobayashi T, Thorner J, Alessi DR. Functional counterparts of mammalian protein kinases PDK1 and SGK in budding yeast. Current Biology. 1999;9(4):186–197. doi: 10.1016/s0960-9822(99)80088-8. [DOI] [PubMed] [Google Scholar]
- 94.Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, et al. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell. 2007;26(5):663–74. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 95.Zhang X, Lester RL, Dickson RC. Pil1p and Lsp1p negatively regulate the 3-phosphoinositide-dependent protein kinase-like kinase Pkh1p and downstream signaling pathways Pkc1p and Ypk1p. J Biol Chem. 2004;279(21):22030–22038. doi: 10.1074/jbc.M400299200. [DOI] [PubMed] [Google Scholar]
- 96.Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320(5874):362–5. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu K, Zhang X, Lester RL, Dickson RC. The sphingoid long chain base phytosphingosine activates AGC-type protein kinases in Saccharomyces cerevisiae including Ypk1, Ypk2, and Sch9. J Biol Chem. 2005;280(24):22679–22987. doi: 10.1074/jbc.M502972200. [DOI] [PubMed] [Google Scholar]
- 98.Walther TC, Aguilar PS, Frohlich F, Chu F, Moreira K, Burlingame AL, et al. Pkh-kinases control eisosome assembly and organization. Embo J. 2007;26(24):4946–55. doi: 10.1038/sj.emboj.7601933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Walther TC, Brickner JH, Aguilar PS, Bernales S, Pantoja C, Walter P. Eisosomes mark static sites of endocytosis. Nature. 2006;439(7079):998–1003. doi: 10.1038/nature04472. [DOI] [PubMed] [Google Scholar]
- 100.Luo G, Gruhler A, Liu Y, Jensen ON, Dickson RC. The sphingolipid long-chain base-Pkh1/2-Ypk1/2 signaling pathway regulates eisosome assembly and turnover. J Biol Chem. 2008;283(16):10433–44. doi: 10.1074/jbc.M709972200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kobayashi T, Takematsu H, Yamaji T, Hiramoto S, Kozutsumi Y. Disturbance of sphingolipid biosynthesis abrogates the signaling of Mss4, phosphatidylinositol-4-phosphate 5-kinase, in yeast. J Biol Chem. 2005;280(18):18087–94. doi: 10.1074/jbc.M414138200. [DOI] [PubMed] [Google Scholar]
- 102.Tabuchi M, Audhya A, Parsons AB, Boone C, Emr SD. The phosphatidylinositol 4,5-biphosphate and TORC2 binding proteins Slm1 and Slm2 function in sphingolipid regulation. Mol Cell Biol. 2006;26(15):5861–75. doi: 10.1128/MCB.02403-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Audhya A, Loewith R, Parsons AB, Gao L, Tabuchi M, Zhou H, et al. Genome-wide lethality screen identifies new PI4,5P2 effectors that regulate the actin cytoskeleton. Embo J. 2004;23(19):3747–57. doi: 10.1038/sj.emboj.7600384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yu JW, Mendrola JM, Audhya A, Singh S, Keleti D, DeWald DB, et al. Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol Cell. 2004;13(5):677–88. doi: 10.1016/s1097-2765(04)00083-8. [DOI] [PubMed] [Google Scholar]
- 105.Fadri M, Daquinag A, Wang S, Xue T, Kunz J. The pleckstrin homology domain proteins Slm1 and Slm2 are required for actin cytoskeleton organization in yeast and bind phosphatidylinositol-4,5-bisphosphate and TORC2. Mol Biol Cell. 2005;16(4):1883–900. doi: 10.1091/mbc.E04-07-0564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sturgill TW, Cohen A, Diefenbacher M, Trautwein M, Martin DE, Hall MN. TOR1 and TOR2 have distinct locations in live cells. Eukaryot Cell. 2008;7(10):1819–30. doi: 10.1128/EC.00088-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dunn TM, Haak D, Monaghan E, Beeler TJ. Synthesis of monohydroxylated inositolphosphorylceramide (IPC-C) in Saccharomyces cerevisiae requires Scs7p, a protein with both a cytochrome b5-like domain and a hydroxylase/desaturase domain. Yeast. 1998;14(4):311–21. doi: 10.1002/(SICI)1097-0061(19980315)14:4<311::AID-YEA220>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 108.Brace JL, Lester RL, Dickson RC, Rudin CM. SVF1 regulates cell survival by affecting sphingolipid metabolism in Saccharomyces cerevisiae. Genetics. 2007;175(1):65–76. doi: 10.1534/genetics.106.064527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.D’Mello NP, Childress AM, Franklin DS, Kale SP, Pinswasdi C, Jazwinski SM. Cloning and characterization of LAG1, a longevity-assurance gene in yeast. J Biol Chem. 1994;269(22):15451–9. [PubMed] [Google Scholar]
- 110.Guillas I, Kirchman PA, Chuard R, Pfefferli M, Jiang JC, Jazwinski SM, et al. C26-CoA-dependent ceramide synthesis of Saccharomyces cerevisiae is operated by Lag1p and Lac1p. Embo J. 2001;20(11):2655–65. doi: 10.1093/emboj/20.11.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schorling S, Vallee B, Barz WP, Riezman H, Oesterhelt D. Lag1p and Lac1p Are Essential for the Acyl-CoA-dependent Ceramide Synthase Reaction in Saccharomyces cerevisae. Mol Biol Cell. 2001;12(11):3417–27. doi: 10.1091/mbc.12.11.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD. Regulation of longevity and stress resistance by Sch9 in yeast. Science. 2001;292(5515):288–290. doi: 10.1126/science.1059497. [DOI] [PubMed] [Google Scholar]
- 113.Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, Dang N, et al. Regulation of Yeast Replicative Life Span by TOR and Sch9 in Response to Nutrients. Science. 2005;310(5751):1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 114.Almeida T, Marques M, Mojzita D, Amorim MA, Silva RD, Almeida B, et al. Isc1p Plays a Key Role in Hydrogen Peroxide Resistance and Chronological Lifespan through Modulation of Iron Levels and Apoptosis. Mol Biol Cell. 2008;19(3):865–876. doi: 10.1091/mbc.E07-06-0604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Madeo F, Frohlich E, Ligr M, Grey M, Sigrist SJ, Wolf DH, et al. Oxygen stress: a regulator of apoptosis in yeast. J Cell Biol. 1999;145(4):757–67. doi: 10.1083/jcb.145.4.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Madeo F, Herker E, Maldener C, Wissing S, Lachelt S, Herlan M, et al. A caspase-related protease regulates apoptosis in yeast. Mol Cell. 2002;9(4):911–7. doi: 10.1016/s1097-2765(02)00501-4. [DOI] [PubMed] [Google Scholar]
- 117.Fabrizio P, Battistella L, Vardavas R, Gattazzo C, Liou LL, Diaspro A, et al. Superoxide is a mediator of an altruistic aging program in Saccharomyces cerevisiae. J Cell Biol. 2004;166(7):1055–67. doi: 10.1083/jcb.200404002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Herker E, Jungwirth H, Lehmann KA, Maldener C, Frohlich KU, Wissing S, et al. Chronological aging leads to apoptosis in yeast. J Cell Biol. 2004;164(4):501–7. doi: 10.1083/jcb.200310014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ogretmen B, Hannun YA. Biologically active sphingolipids in cancer pathogenesis and treatment. Nat Rev Cancer. 2004;4(8):604–16. doi: 10.1038/nrc1411. [DOI] [PubMed] [Google Scholar]
- 120.Taha TA, Mullen TD, Obeid LM. A house divided: ceramide, sphingosine, and sphingosine-1-phosphate in programmed cell death. Biochim Biophys Acta. 2006;1758(12):2027–36. doi: 10.1016/j.bbamem.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Aerts AM, Zabrocki P, Francois IE, Carmona-Gutierrez D, Govaert G, Mao C, et al. Ydc1p ceramidase triggers organelle fragmentation, apoptosis and accelerated ageing in yeast. Cell Mol Life Sci. 2008;65(12):1933–42. doi: 10.1007/s00018-008-8129-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Aronova S, Wedaman K, Aronov PA, Fontes K, Ramos K, Hammock BD, et al. Regulation of Ceramide Biosynthesis by TOR Complex 2. Cell Metab. 2008;7(2):148–58. doi: 10.1016/j.cmet.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124(3):471–84. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 124.Kamada Y, Fujioka Y, Suzuki NN, Inagaki F, Wullschleger S, Loewith R, et al. Tor2 directly phosphorylates the AGC kinase Ypk2 to regulate actin polarization. Mol Cell Biol. 2005;25(16):7239–48. doi: 10.1128/MCB.25.16.7239-7248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dickson RC. More Chores for TOR: De Novo Ceramide Synthesis. Cell Metab. 2008;7(2):99–100. doi: 10.1016/j.cmet.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 126.Bammert GF, Fostel JM. Genome-wide expression patterns in Saccharomyces cerevisiae: Comparison of drug treatments and genetic alterations affecting biosynthesis of ergosterol. Antimicrob Agents Chemother. 2000;44(5):1255–1265. doi: 10.1128/aac.44.5.1255-1265.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kwast KE, Lai LC, Menda N, James DT, 3rd, Aref S, Burke PV. Genomic analyses of anaerobically induced genes in Saccharomyces cerevisiae: functional roles of Rox1 and other factors in mediating the anoxic response. J Bacteriol. 2002;184(1):250–65. doi: 10.1128/JB.184.1.250-265.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cowart LA, Okamoto Y, Pinto FR, Gandy JL, Almeida JS, Hannun YA. Roles for sphingolipid biosynthesis in mediation of specific programs of the heat stress response determined through gene expression profiling. J Biol Chem. 2003;278(32):30328–38. doi: 10.1074/jbc.M300656200. [DOI] [PubMed] [Google Scholar]
- 129.Baetz K, McHardy L, Gable K, Tarling T, Reberioux D, Bryan J, et al. Yeast genome-wide drug-induced haploinsufficiency screen to determine drug mode of action. Proc Natl Acad Sci U S A. 2004;101(13):4525–30. doi: 10.1073/pnas.0307122101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Laun P, Ramachandran L, Jarolim S, Herker E, Liang P, Wang J, et al. A comparison of the aging and apoptotic transcriptome of Saccharomyces cerevisiae. FEMS Yeast Res. 2005;5(12):1261–72. doi: 10.1016/j.femsyr.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 131.Bielawski J, Szulc ZM, Hannun YA, Bielawska A. Simultaneous quantitative analysis of bioactive sphingolipids by high-performance liquid chromatography-tandem mass spectrometry. Methods. 2006;39(2):82–91. doi: 10.1016/j.ymeth.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 132.Guan XL, Wenk MR. Mass spectrometry-based profiling of phospholipids and sphingolipids in extracts from Saccharomyces cerevisiae. Yeast. 2006;23(6):465–77. doi: 10.1002/yea.1362. [DOI] [PubMed] [Google Scholar]
- 133.Sullards MC, Allegood JC, Kelly S, Wang E, Haynes CA, Park H, et al. Structure-specific, quantitative methods for analysis of sphingolipids by liquid chromatography-tandem mass spectrometry: “inside-out” sphingolipidomics. Methods Enzymol. 2007;432:83–115. doi: 10.1016/S0076-6879(07)32004-1. [DOI] [PubMed] [Google Scholar]