Abstract
From 2002 to 2003, a global pandemic of severe acute respiratory syndrome (SARS) spread to 5 continents and caused 8000 respiratory infections and 800 deaths. To ameliorate the effects of future outbreaks as well as to prepare for biodefense, a process for the production of a recombinant protein vaccine candidate is under development. Previously, we reported the 5 L scale expression and purification of a promising recombinant SARS vaccine candidate, RBD219-N1, the 218–amino acid residue receptor-binding domain (RBD) of SARS coronavirus expressed in yeast–Pichia pastoris X-33. When adjuvanted with aluminum hydroxide, this protein elicited high neutralizing antibody titers and high RBD-specific antibody titers. However, the yield of RBD219-N1 (60 mg RBD219-N1 per liter of fermentation supernatant; 60 mg/L FS) still required improvement to reach our target of >100 mg/L FS. In this study, we optimized the 10 L scale production process and increased the fermentation yield 6- to 7-fold to 400 mg/L FS with purification recovery >50%. A panel of characterization tests indicated that the process is reproducible and that the purified, tag-free RBD219-N1 protein has high purity and a well-defined structure and is therefore a suitable candidate for production under current Good Manufacturing Practice and future phase-1 clinical trials.
Keywords: protein purification, protein characterization, Pichia pastoris, hydrophobic interaction chromatography, circular dichroism
Abbreviations used: SARS, severe acute respiratory syndrome; CoV, coronavirus; S, spike; RBD, receptor-binding domain; DO, dissolved oxygen; CV, column volume; % CV, coefficient of variation
Introduction
Severe acute respiratory syndrome (SARS) is a respiratory disease caused by SARS coronavirus (SARS-CoV), a category C pathogen as defined by the U.S. National Institute of Allergy and Infectious Diseases of the U.S. National Institutes of Health. The original outbreak in Guangdong Province, China became a pandemic between 2002 and 2003 and caused approximately 800 deaths and more than 8000 infections. Although the overall mortality rate was about 10 percent, the mortality exceeded 50 percent among older adults.1 In preparation for future pandemics and in light of concerns about the use of SARS-CoV in acts of terrorism, scientists have spent intensive efforts to develop vaccines against SARS. Such vaccines would ideally be stockpiled for future use in the event of an outbreak.
In the past decade, several different antigens have been identified and developed as SARS vaccine candidates, including inactivated whole virus, the SARS S protein, and the receptor-binding domain (RBD) of the S protein. Initially, vaccines containing inactivated whole virus adjuvanted with Alhydrogel® were developed1; however, eosinophilic pathology was observed in mice, likely due to antibody-dependent immune enhancement (ADE).2, 3 Later, vaccines using the recombinant S protein of SARS-CoV were developed,1 but the full-length S-protein still seemed to induce ADE.4
As an alternative approach, the RBD of the S protein has been developed as a substitute for the full-length S protein.5, 6 Recombinant RBD formulated with Freund's adjuvant and with Sigma adjuvant system® (monophosphoryl-lipid A and trehalose dicorynomycolate adjuvant) has been shown to elicit neutralizing antibodies and highly protective immunity in vaccinated animals while significantly reducing or eliminating ADE and other harmful inflammatory and immune responses.7, 8, 9, 10, 11 However, these recombinant wild-type RBD proteins had only been expressed at small scale in Escherichia coli, insect cells (Sf9), and mammalian cells (293T, CHO-K1).5, 7, 8, 11, 12 In addition, these wild-type proteins were either tagged with a hexahistidine sequence and purified by immobilized metal affinity chromatography7, 8, 12 or tagged with an Fc fragment and purified by protein A chromatography.5, 11 Although tags simplify the purification process, they usually serve no purpose in the final product and may potentially cause unwanted immune responses and raise safety concerns. Thus, they are generally not considered desirable for use in human therapeutic proteins.
In our previous study,13 we had developed a production process to express and purify several tag-free recombinant RBD constructs in yeast and had identified one of the yeast-expressed RBDs, RBD219-N1 (residues 319-536), as a promising SARS vaccine candidate due to its ability to induce a stronger RBD-specific antibody response and a high level of neutralizing antibodies in immunized mice when adjuvanted with aluminum hydroxide. The designation N1 refers to the fact that the first amino acid (residue 318) was deleted to avoid glycosylation at the N-terminus.14 However, the default production process used for RBD219-N1 still had several areas that could be improved or optimized before translating to a suitable pilot manufacturing scale. These included improving the fermentation yield and purification recovery, as the fermentation yield using the default process was only 60 mg RBD219-N1/L of fermentation supernatant (FS) and the purification process had not been optimized. In this study, we optimized the upstream process to generate a fermentation yield of approximately 400 mg RBD219-N1/L FS. The purification process was also optimized to a final recovery exceeding 50 percent. This final optimized production process was validated in 3 identical production runs, and an array of characterization procedures for the process indicated high reproducibility and robustness. Further characterization of the purified RBD219-N1 demonstrated that the protein was highly pure with well-defined secondary and tertiary structures. The process has since been transferred to a pilot manufacturing plant and a 60 L scale manufacture has been performed under current Good Manufacturing Practice (cGMP). The cGMP grade–purified RBD219-N1 (drug substance) is to be used in support of future phase-1 trials.
Materials and Methods
Reselection of Clone and Seed Stock Generation
Cloning, expression, and clone selection to generate the RBD219-N1 seed stock were performed as previously described13 with some modifications. Briefly, the recombinant plasmid encoding RBD219-N1 was transformed into Pichia pastoris X-33 cells by electroporation. Transformed cells were streaked consecutively on YPD plates with 0.1 mg/mL Zeocin and 0.5 mg/mL Zeocin. A total of 80 colonies were chosen for 5-10 mL expression cultures, and the colony providing the highest expression level, as judged by SDS-PAGE, was used to generate the seed stock.14 The seed stock from the previous study13 and the optimized seed stock were then compared in the fermentation process described in the following as part of the fermentation optimization. The optimized seed stock was used in the final optimized production process.
Fermentation
To rapidly screen for the optimal conditions during the development phase, recombinant RBD219-N1 was expressed in 5-10 mL scale as described previously14 with some modifications, including induction at different temperatures (22-30°C) and pH (pH 4.7-7.19) and the addition of additives. Following the small-scale screening, 5-10 L fermentation runs were performed under different induction conditions, such as pH (6.0 and 6.5), temperature (24-30°C), carbon feeds (sorbitol co-feed), methanol flow rate, and fermentation media (basal salt media or low salt media15 [LS; 4.55 g/L potassium sulfate, 3.73 g/L magnesium sulfate heptahydrate, 1.03 g/L potassium hydroxide, 0.23 g/L calcium sulfate dehydrate, 10.9 mL/L phosphoric acid {85%}, and 40 g/L glycerol]). The default fermentation procedure was described in the previous study13 and used to generate a baseline to compare with the optimized fermentation procedure developed here. One milliliter of seed stock was inoculated into 500 mL buffered minimal glycerol medium, and the culture was incubated overnight at 30°C with constant shaking at 250 rpm until an OD600 of ∼10. Approximately 250 mL overnight culture was inoculated into 5 L sterile LS medium containing 3.5 mL/L PTM1 Trace Elements and 3.5 mL/L 0.02% D-Biotin. Fermentation was initiated and maintained at 30°C and pH 5.0. Gas and agitation were adjusted to maintain the dissolved oxygen concentration at 30%. On exhaustion of glycerol during the batch phase (dissolved oxygen spike), the pH was ramped up to 6.5 using 14% ammonium hydroxide, and the temperature was lowered to 25°C over 1 h. After the pH and temperature ramping, the methanol induction phase was initiated. Methanol was added from 1 mL/L/h to 11 mL/L/h over 6 h. After this methanol adaptation phase, the methanol feed was maintained at 11 mL/L/h for 18 h, elevated from 11 to 13 mL/L/h over 6 h and then maintained at 13 mL/L/h for 18 h. Finally, the methanol feed rate was further increased from 13 to 15 mL/L/h over 6 h and maintained at 15 mL/L/h until the end of fermentation (∼70 h of methanol induction). After fermentation, cells were removed from the culture by centrifugation at 7000 rpm for 30 min at 4°C using a Beckman Avanti J-26 XPI High-Speed Centrifuge equipped with a JLA 8.1000 rotor. After centrifugation, the wet cell weight was measured and the supernatant was used for further purification.
Tangential Flow Filtration With Salt Concentration Adjustment
To purify RBD219-N1, 5-6 L of fermentation supernatant were first filtered through a 0.45 μm polyethylene sulfone (PES) filter and concentrated 3- to 4-fold to approximately 1.5 L with a 10 kDa Millipore Pellicon 2 Mini Cassette (0.1 m2 surface area). After the fermentation supernatant was concentrated, 400-450 g of ammonium sulfate were added to reach a target concentration of 2 M. The fermentation supernatant containing 2 M ammonium sulfate was then centrifuged at 13,000× g for 30 min in a Beckman J-26 XPI High-Speed Centrifuge equipped with a JLA 8.1000 rotor. After centrifugation, the supernatant was collected, filtered through a 0.22 μm filter, and loaded onto a Butyl Sepharose High Performance (Butyl HP) column.
Hydrophobic Interaction Chromatography–Butyl HP
Butyl HP (GE Healthcare) was packed in a Millipore Vantage A2 column with an internal diameter of 8.9 cm and bed height of 13 cm. The total column volume (CV) was 808 mL. The column was pre-equilibrated with 2 CVs of 20 mM Tris, pH 8.0, and 2 M ammonium sulfate. After the column was equilibrated, it was loaded with concentrated fermentation supernatant containing 2 M ammonium sulfate at 50 cm/h and then washed with 2 CVs of 20 mM Tris, 2 M ammonium sulfate, pH 8.0 to remove loosely bound contaminants. Another wash was performed using 10 CVs of 20 mM Tris, 0.7 M ammonium sulfate, pH 8.0. Finally, the target protein was eluted using 7 CVs of 20 mM Tris, pH 8.0. Approximately 2 L of the hydrophobic interaction chromatography (HIC) elution pool containing RBD219-N1 were collected.
Second Tangential Flow Filtration (Post-HIC TFF) Followed by Size Exclusion Chromatography
After the HIC step, the HIC elution pool was further concentrated 20-fold to reduce the volume to approximately 100 mL using a 10 kDa Sartorius Sartocon Slice 200 Hydrosart TFF Cassette (0.02 m2). After the HIC elution pool was concentrated, size exclusion chromatography (SEC)75 chromatography was performed. Superdex 75 prep grade resin (GE Healthcare) was packed in a GE Healthcare Index 70 column (internal diameter 7 cm × bed height 65 cm) with a column volume of 2501 mL. Two identical cycles (loading volume: approximately 50 mL each) were performed to process the concentrated HIC elution pool (post-HIC TFF). The column was first equilibrated with 20 mM Tris, 150 mM NaCl, pH 7.5, and the post-HIC TFF pool was loaded at 30 cm/h and eluted at the same flow rate over 1.5 CVs. The eluate from both SEC cycles was pooled for a total of 800 mL and filtered through a 0.22 μm PES filter. The final protein concentration of purified RBD219-N1 was determined using the absorbance at 280 nm, a theoretical molar extinction coefficient of 40,903 L mol−1 cm−1 and a molecular weight of 24.49 kDa.
Process Recovery/Purity by Quantitative SDS-PAGE and Host Cell Protein Content Assessment by Slot Blot
The concentration of RBD219-N1 in the fermentation supernatant and in the in-process samples was determined in triplicate by SDS-PAGE under reducing condition on 14% Tris glycine gels (Invitrogen) following the manufacturer's instructions. Initially, bovine serum albumin (BSA) with known concentrations was used as the standard for quantitative analysis by densitometry but was eventually replaced with purified RBD219-N1 after it became available. After electrophoresis, the SDS-PAGE gels were stained with Coomassie Blue and scanned using a GE ImageScanner II with the Image Quant software version 8.1. After the concentration of RBD219-N1 in each in-process sample had been quantified, various volumes of in-process samples which contained approximately 3 μg RBD219-N1 were loaded on 4%-20% Tris glycine gels to determine the process purity using densitometry, as described previously. In these experiments, we observed that equal amounts of BSA and RBD219-N1, as determined by spectrophotometry, react differently with Coomassie Blue. In fact, RBD219-N1 appears to bind Coomassie 1.33 times less than BSA. Thus, we have used this conversion factor when appropriate (Supplementary Table S1). Finally, host cell protein (HCP) content in the in-process samples was analyzed using a slot blot assay as described previously with minor procedure changes.16, 17 A log-log plot of HCP intensity versus known HCP concentration was graphed and the HCP concentration of the in-process samples was calculated using the linear regression from the plot.
Purity Assessment by HPLC–Reverse Phase
A Waters® HPLC system (Alliance® 2695 Separation Module) equipped with a Waters® Symmetry C4 Column (300 Å, 5 μm, 4.6 mm × 150 mm) and Waters® Symmetric C4 guard column (300 Å, 5 μm, 3.9 mm × 20 mm) was used to assess the purity of the RBD219-N1 preparation by reverse-phase chromatography in buffer A (0.1% TFA in HPLC grade water) and buffer B (0.1% TFA in acetonitrile). The column was first equilibrated with 30% buffer B at a flow rate of 1.0 mL/min and a temperature of 45°C. After the column was fully equilibrated, 50 μg purified RBD219-N1, in triplicate, was injected into the system and eluted with 30% buffer B for 4 min, followed by a gradient of 30%-90% buffer B over 15 min. After the gradient elution, the column was washed with 90% buffer B for 2.5 min to elute remaining proteins. Finally, the column was re-equilibrated with 30% buffer B for 8.5 min. The run was monitored using absorbance at 280 nm.
Integrity Assessment by Coomassie/Silver-Stained SDS-PAGE and Western Blot
After purification, various amounts of RBD219-N1 were loaded on 4%-20% Tris glycine gels, subsequently stained with Coomassie Blue or silver as described in Curti et al.,18 or transferred to a polyvinylidene difluoride membrane and analyzed by Western blot using the conformational monoclonal anti-RBD antibody 33G4, as described previously.9
Hydrophobicity Assessment by Extrinsic Fluorescence
Purified RBD219-N1 was buffer exchanged in the buffer systems at pH 4.0, 5.0, 6.5, 7.5, 8.5, and 9.5 (Supplementary Table S2). Extrinsic fluorescence of purified RBD219-N1 (0.023-1.5 mg/mL) at pH 4.0-9.5 was measured using Nile Red (Sigma-Aldrich) as a probe.19 BSA and lysozyme at pH 7.5 were used as controls. In all the samples, Nile Red was added to 1.25 μM from a 250 μM stock solution in dimethyl sulfoxide. The relative fluorescence intensity was measured using a Spectra Max M3 multimode plate reader (Molecular Devices). The samples were excited at 520 nm, and the emission spectra were detected from 550 to 700 nm. The surface hydrophobicity of RBD219-N1, BSA, and lysozyme was determined using the slope of relative fluorescence intensity of the emission peak versus protein concentration plot as described previously.20, 21
Hydrophobicity Assessment by Dynamic Light Scattering
Purified RBD219-N1 was concentrated to approximately 8-9 mg/mL and buffer exchanged in the buffer systems at pH 4.0, 5.0, 6.5, 7.5, 8.5, and 9.5 (Supplementary Table S2). The stock solutions at different pH were then filtered with either 0.2 μm filter (pH 4.0 and 5.0 stock solutions) or 0.02 μm filter (pH 6.5, 7.5, 8.5, and 9.5 solutions) followed by serial dilution to a concentration of approximately 1 mg/mL. For each of the serially diluted samples, 40 μL was loaded in duplicate into each well in a 384-well microtiter plate. The diffusion coefficient (D) was measured using DynaPro Plate Reader II (Wyatt Technology). The diffusion interaction parameter (kD) was calculated from linear regression of the measured diffusion coefficient and concentration using the following equation22:
where D is the measured extinction coefficient, D0 is the coefficient of the solute at infinite dilution, and kD is the diffusion interaction parameter.
Structural Assessment by Circular Dichroism
Purified RBD219-N1 was diluted with deionized water to a final concentration of 0.3-0.4 mg/mL and analyzed by circular dichroism (CD) on a Jasco-810 spectropolarimeter (Easton, MD) with a 6-position Peltier temperature controller. The CD spectra were obtained from 260 to 185 nm with the Jasco J-1500s spectrophotometer set at 100 nm/min and a response time of 1 s at 25°C. The obtained CD data were analyzed using CDPro software by comparing to the reference sets (SP43, SP37, SDP48, and SDP42) using 2 data-fitting programs (CONTIN and CDSSTR). The analyzed secondary structure was also compared with the secondary structure predicted by Phyre2 (Protein Homology/analogY Recognition Engine; http://www.sbg.bio.ic.ac.uk/phyre2/). In addition, the sample was heated from 25°C to 85°C for a denaturation profile analysis.
Structural Assessment by Thermal Shift Assay
Purified RBD219-N1 was diluted with deionized water to a final concentration of approximately 0.3 mg/mL. The protein thermal shift assay was then performed by heating the sample from 25°C to 95°C and monitoring the fluorescence intensity change using Protein Thermal Shift™ reagents, a ViiA™ 7 Real-Time PCR system and Protein Thermal Shift™ software version 2.0.
Results
Optimization of Production Process
Fermentation
During development, we performed a series of 5-10 mL cultures and several 5-10 L fermentation runs to evaluate induction parameters (Supplementary Fig. S1 and Table S3). We thus generated an optimized seed stock, as well as improved the 10 L scale fermentation process for the production of RBD219-N1. BSA was used as a heterologous reference standard for Quantitative SDS-PAGE (densitometry) in early development phase to help quantify the fermentation yield and evaluate expression levels, although different proteins may vary in their affinity for Coomassie Blue.23 Once pure RBD219-N1 became available, we switched to the more appropriate homologous standard. The key parameters using the default and optimized seed stocks and of the default and optimized fermentation processes are shown in Table 1 .
Table 1.
Comparison of the Fermentation Yields During Optimization
| Seed Stock | Fermentation Process | Final Wet Cell Weight (mg/mL) | Fermentation Yield by Densitometry Using BSA Standards (mg RBD219-N1/L FS) | Fermentation Yield by Densitometry Using RBD219-N1 Standards (mg RBD219-N1/L FS) |
|---|---|---|---|---|
| Default | Default | 401 | 45 | 60b |
| Default | Optimized | 460 | 70 | 93b |
| Optimized | Default | 359 | 72 | 95b |
| Optimized | Optimized | 432 ± 23a | 288 ± 8a | 409 ± 9a |
Two identical runs were performed during development phase; the value provided was the mean value ± standard deviation for these 2 runs.
Calculated using a conversion factor of 1.33 (Supplementary Table S1).
The 2 seed stocks were used to compare each fermentation optimization run. The final wet cell weight remained consistent (410 ± 50 mg/mL), suggesting that culture growth was robust during independent fermentation runs with 2 different seed stocks. However, the yield of RBD219-N1 in the fermentation supernatant varied significantly depending on which seed stock was used. (We note that the yield of RBD219-N1 also varied for each fermentation run when comparing the quantification by densitometry with BSA standards vs. quantification by densitometry with purified RBD219-N1 standard; to account for this discrepancy, a conversion factor of 1.33 was used as addressed in the Materials and Methods section.) Before optimization, the fermentation yield using the default process and default seed stock was 60 mg RBD219-N1/L of FS. After the optimization of the seed stock, the default fermentation process yielded 95 mg RBD219-N1/L FS, a 1.6-fold increase compared with the default seed stock. Using the default seed stock with the optimized fermentation process, the yield also increased 1.5-fold to 93 mg RBD219-N1/L FS. Finally, combining the optimized seed stock with the optimized fermentation process, the yield increased significantly to 409 mg RBD219-N1/L FS, a more than 6-fold increase compared with the original yield.
The optimized seed stock and the optimized fermentation process were therefore chosen to evaluate process reproducibility and process lockdown for eventual technology transfer into a cGMP facility.
Purification
The purification process was optimized to increase the binding of the target protein to a butyl HP hydrophobic interaction chromatography (HIC) column. In the default purification process reported in Chen et al.,13 1 part of the fermentation supernatant was diluted with 2 parts of 3 M ammonium sulfate to adjust the ammonium sulfate concentration to 2 M. We determined that less than 0.49 mg RBD219-N1 was bound per mL butyl HP resin using this approach. However, in the optimized purification process, the fermentation supernatant was concentrated 3- to 4-fold before the addition of ammonium sulfate, and protein binding which increased to more than 4.5 mg RBD219-N1/mL butyl HP resin, an approximately 9-fold improvement.
Reproducibility and Robustness of the Production Process
The optimized seed stock and the optimized fermentation and purification processes were validated in 3 identical production runs (Table 2 ). From a 10 L fermentation run, we obtained 5.7 L of FS, with, on average, 425 ± 1 mg RBD219-N1 per L FS (coefficient of variation [%CV] = 0.3 %). After purification, a total of 800 mL of purified RBD219-N1 was obtained from each run with a concentration of 1.62 ± 0.18 mg/mL (total: 1296 ± 144 mg). This corresponds to 226 ± 20 mg of RBD219-N1 per liter of FS with a %CV of 8.9 %. This low %CV suggests that the optimized process was robust and reproducible.
Table 2.
Comparison and Overview for 3 Identical Production Runs of RBD219-N1
| Run# | Volume of FS (L) | Concentration of Unpurified RBD219-N1 in FS (mg RBD219-N1/L FS) | Final Concentration of Purified RBD219-N1 (mg RBD219-N1/mL) Determined by A280 | Total Yield (mg) (V = 800 mL) | Final Process Yield of Purified RBD (mg RBD219-N1/L FS) |
|---|---|---|---|---|---|
| Run 1 | 5.8 | 424 | 1.80 | 1440 | 250 |
| Run 2 | 5.5 | 427 | 1.38 | 1104 | 201 |
| Run 3 | 5.9 | 424 | 1.68 | 1344 | 228 |
| Mean | 5.7 | 425 | 1.62 | 1296 | 226 |
| SD | 0.2 | 1 | 0.18 | 141 | 20 |
| %CV | 3.0% | 0.3% | 10.9% | 10.9% | 8.9% |
The final volume of the purified protein for each run was 800 mL.
Analytical and Biochemical Evaluation of Protein Integrity and Purity of RBD219-N1
During purification, in-process samples were used to characterize protein integrity and analyze the process recovery (Table 3a ), as well as monitor sample purity (Fig. 1 ) by SDS-PAGE with Coomassie Blue staining. The HCP content was also analyzed using slot blot, a more sensitive method (Table 3b). The low purity of the target protein (67% under nonreducing conditions or 53% under reducing conditions) in the FS as determined by densitometry using purified RBD219-N1 (Fig. 1c), was likely due to heavy contamination with host cell impurities and hyperglycosylated RBD (Figs. 1a and 1b, lane 1). To purify the target protein, the FS was first filtered through a 0.22 μm PES filter and then concentrated 3- to 4-fold by tangential flow filtration (TFF; Figs. 1a and 1b, lane 2). At this first TFF step, the high recovery of 98% suggested that the loss of target protein was insignificant and that the purity of the concentrated target protein in the FS was unchanged. After the first TFF step, ammonium sulfate was added to the concentrated FS to adjust the salt concentration to 2 M. This concentrated FS was centrifuged, and the supernatant was loaded to a hydrophobic interaction chromatography Butyl HP column (HIC). In the HIC step, recovery was 67%, the purity of the RBD219-N1 in the HIC elution pool increased to 86% under nonreducing conditions and 84% under reducing conditions (Figs. 1a and 1b, lane 3), and 92% of HCPs were also removed. The eluted pool was further concentrated to reduce the loading volume by 20-fold using a second TFF step (post-HIC TFF). During this post-HIC TFF step (Figs. 1a and 1b, lane 4), the recovery dropped from 67.0% to 60.3% without a change in purity. Finally, the Post-HIC TFF pool was loaded onto a SEC75 column (SEC) to remove hyperglycosylated proteins (Figs. 1a and 1b, lane 5). After the SEC run, the overall recovery was determined to be 52%, and the purity of the final product was approximately 99% under nonreducing and reducing conditions; the slot blot also indicated that 99.9988% of HCPs were removed, and the final purified product contained approximately 0.5 μg HCP/mg RBD219-N1. The high purity of the final product and the high overall recovery of the process suggest that this production process is robust.
Table 3.
Analysis of the Process Recovery and Host Cell Protein Content Among 3 Identical Production Runs
| (a) | Overall Process Recovery |
Average Step Loss | ||||
|---|---|---|---|---|---|---|
| Run 1 | Run 2 | Run 3 | Mean | %CV | ||
| FS | 100% | 100% | 100% | 100% | – | – |
| TFF | 96% | 103% | 94% | 98 ± 4% | 4.0% | 2% |
| HIC | 68% | 63% | 70% | 67 ± 3% | 4.4% | 31% |
| Post-HIC TFF | 55% | 64% | 62% | 60 ± 4% | 6.4% | 7% |
| SEC | 54% | 48% | 54% | 52 ± 3% | 5.4% | 8% |
| (b) | HCP Content (mg) |
Percentage of HCP Remaining | Percentage of HCP Removed | ||||
|---|---|---|---|---|---|---|---|
| Run 1 | Run 2 | Run 3 | Mean | %CV | |||
| FS | 53,910 | 47,809 | 71,089 | 57,603 ± 9856 | 17% | 100% | – |
| TFF | 24,832 | 44,636 | 74,870 | 48,113 ± 20,567 | 43% | 83.5% | 16.5% |
| HIC | 5011 | 5832 | 3210 | 4684 ± 1095 | 23% | 8.1% | 91.9% |
| Post-HIC TFF | 6386 | 4421 | 2297 | 4368 ± 1670 | 38% | 7.6% | 92.4% |
| SEC | 0.696 | 0.648 | 0.720 | 0.688 ± 0.030 | 4% | 0.0012% | 99.9988% |
(a) The recovery was determined by SDS-PAGE followed by densitometry with purified RBD219-N1 standards. The yield of RBD219-N1 from fermentation supernatant was used as a baseline (i.e., 100%). (b) The HCP content was determined by slot blot. The Mean HCP content was used as a baseline to calculate the percentage of HCP remaining after each purification stage.
Figure 1.
Purity assessment for the production process. SDS-PAGE with Coomassie Blue staining under (a) nonreduced condition and (b) reduced condition. M, SeeBlue plus 2 protein marker; lane 1, original fermentation supernatant (FS); lane 2, concentrated FS after tangential flow filtration (TFF); lane 3, butyl HP elution pool (HIC); lane 4, concentrated HIC pool (post-HIC TFF); and lane 5, the SEC pool/final purified RBD219-N1 (SEC) and (c) quantified purity at different purification steps.
Purity of RBD219-N1 by HPLC–Reverse Phase
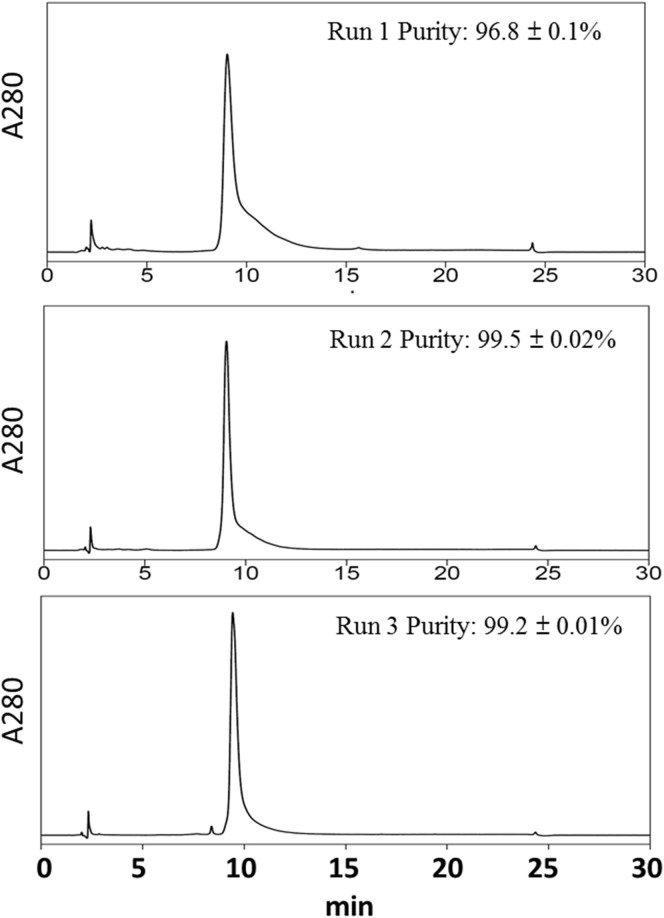
The purity of the final RBD219-N1 product from 3 identical runs was further analyzed by HPLC–reverse phase. The results for all 3 lots were consistent, and chromatograms with the analyzed purity results are shown in Figure 2 . Based on the results, the purity for these 3 lots of purified RBD219-N1 was approximately 99%, consistent with the purity determined by SDS-PAGE with Coomassie Blue staining (Fig. 1c).
Figure 2.
Purity assessment of purified RBD219-N1 by HPLC–reverse phase. The mean purity for the 3 identical runs was 98.5% with a %CV of 1.2%.
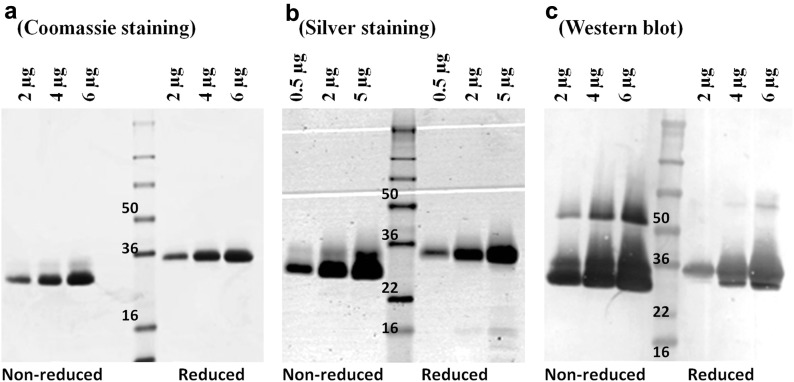
Integrity of RBD219-N1 by SDS-PAGE
The purified recombinant RBD219-N1 was characterized by SDS-PAGE to study its integrity (Fig. 3 ). When 2 μg of purified RBD219-N1 were loaded onto an SDS-PAGE gel and stained with Coomassie Blue (Fig. 3a), only a single band was observed at approximately 29 kDa under nonreducing conditions and 33 kDa under reducing conditions. When the load was increased to 4 and 6 μg, a faint additional band at approximately 32 kDa was observed under nonreducing conditions but not under reducing conditions. On the silver-stained SDS-PAGE gel, additional low molecular weight bands were observed under reducing conditions when 5 μg of purified RBD219-N1 was loaded (Fig. 3b), but no aggregation or dimer formation was observed. Using Western blotting with the RBD-specific antibody 33G4 (Fig. 3c), an RBD219-N1 dimer was observed under nonreducing conditions when 2 μg of RBD219-N1 were loaded (and could also be observed under reducing conditions when loading more than 4 μg).
Figure 3.
Integrity assessment for purified RBD by SDS-PAGE under reducing and nonreducing conditions followed by (a) Coomassie Blue staining: 2-6 μg of purified RBDs were loaded, (b) Silver staining: 0.5-5 μg of purified RBDs were loaded, or (c) Western blot: transferred to polyvinylidene difluoride membrane and probed with an RBD-specific antibody (33G4); 2-6 μg of purified RBDs were loaded.
Hydrophobicity Assessment Using Extrinsic Fluorescence and Dynamic Light Scattering
Extrinsic fluorescence of RBD219-N1 probed with Nile Red was used to determine its surface hydrophobicity S0 (Table 4a and Supplementary Fig. S3). Typically, the blue shift in emission peak wavelength λmax indicates an increase binding of probe to the protein, thus an increase in hydrophobicity. The emission peak λmax of RBD219-N1 at pH 7.5 was measured at 626 nm suggested RBD219-N1 was less hydrophobic than BSA (λmax = 620 nm) and more hydrophobic than lysozyme (λmax = 650 nm), which was consistent with the results on the calculated surface hydrophobicity (S0;BSA > S0;RBD219-N1> S0;Lysozyme). The effect of pH on the surface hydrophobicity of RBD219-N1 was also studied. The results showed that the hydrophobicity remained constant at pH 5.0-9.5 (λmax = 627 ± 3 and S0 = 52 ± 7 RFU·mL/mg) and increased at pH 4.0 (λmax = 618 nm and S0 = 76.8 RFU·mL/mg) suggesting possible denaturation at pH 4.0.
Table 4.
(a) Emission Peak Wavelength and Surface Hydrophobicity of RBD219-N1 at Different pH Values. BSA and Lysozyme at pH 7.5 Were Used as Controls. The Emission Peak of Nile Red Remains at Approximately 654 nm Between pH 4.0 and 9.5. (b) Diffusion Interaction Parameter of RBD219-N1 at Different pH Values Measured Using Dynamic Light Scattering
| Variable | (a) Extrinsic Fluorescence |
(b) Dynamic Light Scattering |
|
|---|---|---|---|
| Emission Peak Wavelength λmax (nm) | Surface Hydrophobicity S0 (RFU·mL/mg) | Diffusion Interaction Parameter, kD (mL/G) | |
| RBD219-N1 pH 4.0 | 618 | 76.8 | Aggregated |
| RBD219-N1 pH 5.0 | 624 | 45.6 | Aggregated |
| RBD219-N1 pH 6.5 | 628 | 50.6 | −16.0 |
| RBD219-N1 pH 7.5 | 626 | 57.4 | −15.9 |
| RBD219-N1 pH 8.5 | 630 | 51.4 | −9.6 |
| RBD219-N1 pH 9.5 | 628 | 58.8 | −5.6 |
| BSA, pH 7.5 | 620 | 139.9 | – |
| Lysozyme, pH 7.5 | 650 | 3.2 | – |
Dynamic light scattering (DLS) was used to further characterize the diffusion interaction parameter (kD) of purified RBD219-N1 at different pH (Table 4b). During the sample preparation, it was observed that the stock samples at pH 4.0 and pH 5.0 could not be filtered through 0.02 μm filter indicating protein aggregation and DLS results confirmed protein aggregation at pH 4.0 and 5.0. On the other hand, the samples at pH 6.5-9.5 remained monodisperse; however, the negative kD was obtained (−16.0 mL/g to −5.6 mL/g), which suggested protein-protein attraction and predisposition to form oligomers.
Structure Assessment Using Circular Dichroism and Thermal Shift
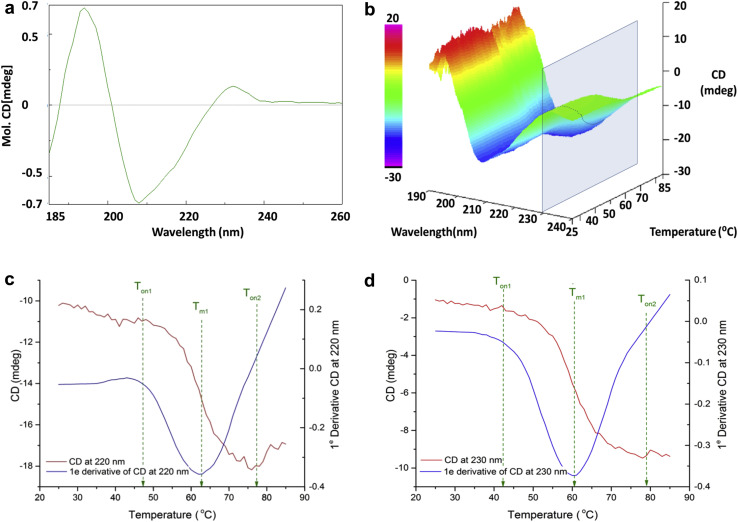
CD was performed to investigate the secondary structure of RBD219-N1. Based on the analysis of the far-UV CD region (Fig. 4a ), the yeast-expressed RBD219-N1 was rich in beta sheets (42% beta sheet, ∼5% alpha helix, and 20% turn/loop), consistent with the structure predicted by the online protein structure prediction tool Phyre2 (50% beta sheet and 7% helix). The thermal stability of the secondary structure was assessed by heating the sample from 25°C to 85°C (Figs. 4b-4d). The best signal-to-noise data were obtained at 220 nm (Fig. 4c) and 230 nm (Fig. 4d). The onset of structural changes was observed at approximately 43°C (Ton1) and based on the first derivative (blue curves; Figs. 4c and 4d), and the average melting temperature (Tm1) was estimated to be 61°C. Interestingly, a second CD change was observed (Ton2) at approximately 78°C at all wavelengths, indicating another structure-change event. Overall, the CD signal decreased further at all wavelengths as thermal stress increased (Fig. 4b and Supplementary Fig. S2), which may indicate transformation occurred within the secondary structure of RBD219-N1 rather than a complete denaturation at higher temperature.
Figure 4.
(a) Circular dichroism results for RBD219-N1 at 25°C, (b) CD profile for RBD219-N1 at different temperatures, (c) CD denaturation profile and the first CD derivative of RBD219-N1 at 220 nm, and (d) CD denaturation profile and the first CD derivative of RBD219-N1 at 230 nm.
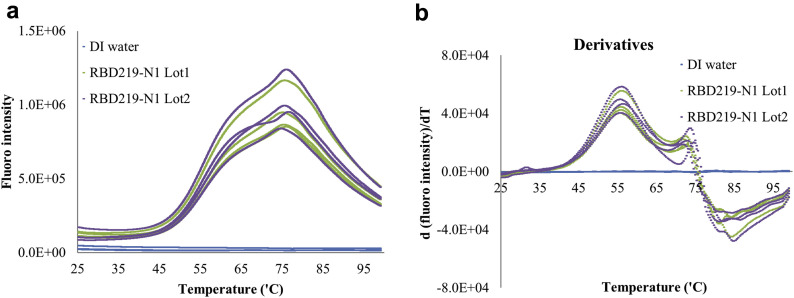
The thermal stability of the tertiary structure of the protein was tested by an extrinsic fluorescence-based thermal shift (Fig. 5a ). Consistent results from 2 different lots of RBD219-N1 are indicative of the reproducibility of the process. The denaturation profile (Fig. 5a) by thermal shift assay indicated loss of tertiary structure started at approximately 45°C, and the average melting temperature was estimated at approximately 57°C based on the first derivative (Fig. 5b). Similar to the secondary structure analysis by CD, a second structural change was observed at 73°C. The second structure-change event at higher temperature in both CD and thermal shift assay indicated there could be a second stage in the denaturation of RBD219-N1 monomers or a structure change in trace amounts of RBD219-N1 dimers. Overall, the CD and the thermal shift results indicated that the protein has well defined, with characteristic secondary and tertiary structures that can be used for the purpose of comparing and analyzing different batches.
Figure 5.
(a) Thermal shift assay results and (b) the derivatives of the intensity for purified RBD219-N1 lots. DI water was used as negative control.
Discussion
In preclinical testing, the recombinant RBD of the SARS-CoV S protein adjuvanted with Freund's adjuvant and with Sigma adjuvant system® has been proven as an effective and safe vaccine candidate against SARS-CoV infection.7, 8, 9, 10, 11, 24 Evidence for efficacy and low immunopathology include the induction of high titers of neutralizing antibodies, the prevention of infection against viral challenge, and the minimization or ablation of ADE.7, 8, 9, 10, 11 However, all the previously reported studies used recombinant RBDs containing heterologous amino acids (either tagged with a hexahistidine sequence or fused to the Fc domain), mainly for ease of purification purposes.5, 7, 8, 11, 12 The addition of hexahistidine or other tags in therapeutic proteins though is usually not favored for eventual advanced clinical development by the U.S. Food and Drug Administration and should be avoided when feasible. Thus, we had engineered a tag-free RBD expressed in yeast (RBD219-N1) that when adjuvanted with aluminum hydroxide induced strong RBD-specific antibody responses and a high level of neutralizing antibodies in immunized mice.13
To optimize the production process and improve yield and purity, improvements to clone selection and seed stock preparation, fermentation, and purification have been performed in this work. To optimize the seed stock, we first reselected the seed stock using increased antibiotic concentrations. The default fermentation process using this optimized seed stock showed a 1.6-fold increase compared with using the default seed stock. Likely, the increased antibiotic concentrations helped select for an increased copy number of the RBD219-N1 expressing plasmid, a process that had previously been shown to improve the expression levels of other recombinant proteins in yeast.25, 26, 27
To optimize the production process, a series of 5-10 mL scale cultures was used to screen for the best induction temperature, pH, and the possible need for additives (e.g., detergents to disrupt aggregates). Following this, optimizations at the 5-10 L fermentation scale were performed to evaluate other parameters, such as different carbon feeds, methanol flow rate, and fermentation media. Through a change of the fermentation medium to LS15 and minor adjustments to the induction pH (to 6.5) and the induction temperature (to 25°C), the overall fermentation yield was improved more than 6-fold.
With respect to the purification process, as the binding of the HIC column was low (<0.49 mg RBD219-N1/mL Butyl HP) in the default process,13 we aimed to optimize binding of the target protein to the HIC column. To this extent, several parameters were investigated including resin type, salt choice for the binding buffer, and concentration of the fermentation supernatant loaded onto the column. During the optimization, Capto Butyl resin (particle size 75 μm) and Butyl HP resin (particle size 34 μm) were compared, and Butyl HP was chosen on account of its better resolution and better ability to remove impurities. Because the RBD219-N1 in the default process was the last protein eluted from the HIC column using an ammonium sulfate gradient, 2 environmentally more friendly salts with lower ionic strength, sodium sulfate and sodium chloride, were investigated. However, neither salt provided the same high binding affinity and resolution achieved with ammonium sulfate. Finally, the concentration of the sample before application to the column was optimized. Rather than diluting the fermentation supernatant with buffer containing a high concentration of ammonium sulfate as described previously,13 we first concentrated the fermentation supernatant and then dissolved solid ammonium sulfate directly into the sample. This process increased the binding more than 9-fold, which reduced the required resin volume. However, to accommodate the high back pressure due to the smaller resin particle size and the higher viscosity of the more concentrated loading material, the flow rate had to be decreased from 100 to 50 cm/h with the bed height maintained at 11 ± 2 cm. Finally, to meet the industry standard for technology transfer, gradient elution for the HIC process was converted to stepwise elution. Although SEC columns are not ideal for large-scale processes, we decided to retain this step because it efficiently removed the remaining impurities. Intensive efforts were spent on the investigation of alternative purification schemes, including different capture, intermediate, or polishing steps (ion exchange, multimodel, etc.), but the recovery was low and the final product lacked sufficient purity.
During quantification of the protein by SDS-PAGE with Coomassie Blue, BSA standards were replaced with purified RBD219-N1 standards. Although BSA has served as a common standard for several commercially available quantitative assays (e.g., the BCA assay) due to its high purity and easy availability, using a homologous standard is often preferred on account of protein-specific differential uptake of Coomassie Blue dye.23 Depending on the protein, Coomassie Blue reacts differently with each protein, which may cause intensity difference, even for the same protein concentration. In our case, RBD219-N1 only stained 75 ± 5% as efficient as BSA, leading to an underestimation of the protein concentration. The optimized quantitative densitometry assay was used to calculate the process recovery and the process yield. Typically, overall production yield of a recombinant protein relies on fermentation yield and purification recovery. Depending on the gene sequence and the size of the expressed protein, fermentation yield of a recombinant protein successfully expressed in yeast can vary dramatically ranging from 20 mg/L to 14.8 g/L,26, 28, 29 and biophysical properties of the expressed protein determine the purification process and the recovery. For a 10- 20 L scale production of subunit vaccine candidates expressed in yeast, the fermentation yield can range from 150 mg/L to 1 g/L with recovery of 31% to 55% and target protein purity of 97% to 99%.18, 30, 31 The optimized production process for RBD219-N1 has consistently and reproducibly provided high yields during fermentation (∼400 mg RBD219-N1/L FS), with an overall process recovery more than 50% (final purification yield of 226 mg RBD/L FS) and protein purity of ∼ 99%, which suggests the production process is comparable to other subunit vaccine production processes at similar scale.
Other assays to characterize the purified protein included HPLC–reverse phase and SDS-PAGE with silver staining, a staining reagent that is 100 times more sensitive than Coomassie Blue,32 as well as Western blotting, an immunological method able to detect trace amounts of aggregation or degradation. Using silver-stained SDS-PAGE, trace amounts of low molecular weight degradation products or host cell proteins were observed, whereas dimers were observed on Western blot, likely stemming from intermolecular disulfide bonds between the remaining free cysteine of RBD219-N1. Although only trace amounts of nontarget proteins were observed, highly sensitive assays allow increased ability to monitor the integrity and purity of the product that will ensure the level of degradation and aggregation does not increase and cause stability issues. Surface hydrophobicity of RBD219-N1 was also studied using extrinsic fluorescence of Nile Red. It is noted that surface hydrophobicity of RBD219-N1 remained unchanged at pH 5.0-9.5; however, an increase of surface hydrophobicity was observed at pH 4.0, which suggested possible denaturation, thus exposure of hydrophobic sites. Furthermore, DLS experiments on samples at pH 4.0 and 5.0 also indicated protein aggregation. Negative diffusion interaction parameter measured by DLS showed the propensity of oligomer formation or aggregation for all the pH tested, indicating extra attention should be taken for its long-term stability. Collectively, the results obtained from extrinsic fluorescence and DLS experiments suggested RBD219-N1 was most likely stable at pH 6.5-9.5 but instable at pH 4.0 and 5.0, which indicates any formulation at lower pH (>pH 5.0) should be avoided.
In the previous study,13 we confirmed the yeast-expressed RBD219-N1 was able to bind to its receptor-angiotensin-converting enzyme 2 (ACE2) and 4 different conformation-specific anti-RBD antibodies, which indirectly suggested it was properly folded and had preserved its functionality and antigenicity; in the present study, circular dichroism and thermal shift assay concordantly directly confirmed that the protein had retained defined secondary and tertiary conformation.
In conclusion, the production process for the recombinant protein–based SARS vaccine antigen, RBD219-N1, has been significantly improved. The optimized production process is highly reproducible and delivers high yields and high recovery. Characterization of the tag-free recombinant RBD219-N1 purified from the production process has shown that the protein is highly pure with defined structure. Currently, a series of stability studies including real-time long-term stability, accelerated stability, and freeze-thaw stability testing are underway to further characterize the protein. In the meantime, the technology of this production process has been transferred to a pilot manufacturer, and a batch of purified RBD219-N1 has been manufactured under cGMP to support downstream clinical trials.
Acknowledgments
The authors would like to express their high gratitude to Ms. Diane Niño for coordinating this study. The authors also acknowledge Drs. Lanying Du and Shibo Jiang for providing the anti-RBD monoclonal antibody 33G4 for western blot. This study was supported by a grant from the National Institutes of Health (R01AI098775).
Footnotes
Conflicts of interest: Several of the authors and investigators are patent holders on vaccines against SARS (patent number WO2015080973A1) and are in part supported by grants to develop these vaccines.
Current address for Chag: Department of Immunology, University of California at San Francisco, San Francisco, CA, 94143.
Current address for Rezende: Department of Pharmacology, Baylor College of Medicine, Houston, TX, 77030.
This article contains supplementary material available from the authors by request or via the Internet at http://dx.doi.org/10.1016/j.xphs.2017.04.037.
Appendix. Supplementary Data
References
- 1.Du L., He Y., Zhou Y., Liu S., Zheng B.-J., Jiang S. The spike protein of SARS-CoV—a target for vaccine and therapeutic development. Nat Reviews Microbiol. 2009;7(3):226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perlman S., Dandekar A.A. Immunopathogenesis of coronavirus infections: implications for SARS. Nat Rev Immunol. 2005;5(12):917–927. doi: 10.1038/nri1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolles M., Deming D., Long K. A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J Virol. 2011;85(23):12201–12215. doi: 10.1128/JVI.06048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaume M., Yip M.S., Kam Y.W. SARS CoV subunit vaccine: antibody-mediated neutralisation and enhancement. Hong Kong Med J. 2012;18 Suppl 2:31–36. [PubMed] [Google Scholar]
- 5.Wong S.K., Li W., Moore M.J., Choe H., Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J Biol Chem. 2004;279(5):3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang S., Bottazzi M.E., Du L. Roadmap to developing a recombinant coronavirus S protein receptor-binding domain vaccine for severe acute respiratory syndrome. Expert Rev Vaccin. 2012;11(12):1405–1413. doi: 10.1586/erv.12.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du L., Zhao G., Li L. Antigenicity and immunogenicity of SARS-CoV S protein receptor-binding domain stably expressed in CHO cells. Biochem Biophys Res Commun. 2009;384(4):486–490. doi: 10.1016/j.bbrc.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du L., Zhao G., Chan C.C. Recombinant receptor-binding domain of SARS-CoV spike protein expressed in mammalian, insect and E. coli cells elicits potent neutralizing antibody and protective immunity. Virology. 2009;393(1):144–150. doi: 10.1016/j.virol.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He Y., Lu H., Siddiqui P., Zhou Y., Jiang S. Receptor-binding domain of severe acute respiratory syndrome coronavirus spike protein contains multiple conformation-dependent epitopes that induce highly potent neutralizing antibodies. J Immunol. 2005;174(8):4908–4915. doi: 10.4049/jimmunol.174.8.4908. [DOI] [PubMed] [Google Scholar]
- 10.He Y., Li J., Li W., Lustigman S., Farzan M., Jiang S. Cross-neutralization of human and palm civet severe acute respiratory syndrome coronaviruses by antibodies targeting the receptor-binding domain of spike protein. J Immunol. 2006;176(10):6085–6092. doi: 10.4049/jimmunol.176.10.6085. [DOI] [PubMed] [Google Scholar]
- 11.Du L., Zhao G., He Y. Receptor-binding domain of SARS-CoV spike protein induces long-term protective immunity in an animal model. Vaccine. 2007;25(15):2832–2838. doi: 10.1016/j.vaccine.2006.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du L., Zhao G., Chan C.C. A 219-mer CHO-expressing receptor-binding domain of SARS-CoV S protein induces potent immune responses and protective immunity. Viral Immunol. 2010;23(2):211–219. doi: 10.1089/vim.2009.0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen W.-H., Du L., Chag S.M. Yeast-expressed recombinant protein of the receptor-binding domain in SARS-CoV spike protein with deglycosylated forms as a SARS vaccine candidate. Hum Vaccin Immunother. 2014;10(3):648–658. doi: 10.4161/hv.27464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goud G.N., Zhan B., Ghosh K. Cloning, yeast expression, isolation, and vaccine testing of recombinant Ancylostoma-secreted protein (ASP)-1 and ASP-2 from Ancylostoma ceylanicum. J Infect Dis. 2004;189(5):919–929. doi: 10.1086/381901. [DOI] [PubMed] [Google Scholar]
- 15.Lin H., Kim T., Xiong F., Yang X. Enhancing the production of Fc fusion protein in fed-batch fermentation of Pichia pastoris by design of experiments. Biotechnol Prog. 2007;23(3):621–625. doi: 10.1021/bp0603199. [DOI] [PubMed] [Google Scholar]
- 16.Zhu D., Saul A.J., Miles A.P. A quantitative slot blot assay for host cell protein impurities in recombinant proteins expressed in E. coli. J Immunol Methods. 2005;306(1-2):40–50. doi: 10.1016/j.jim.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 17.Plieskatt J., Rezende W., Olsen C. Advances in vaccines against neglected tropical diseases. Hum Vaccin Immunother. 2012;8(6):765–776. doi: 10.4161/hv.19726. [DOI] [PubMed] [Google Scholar]
- 18.Curti E., Seid C.A., Hudspeth E. Optimization and revision of the production process of the Necator americanus glutathione S-transferase 1 (Na-GST-1), the lead hookworm vaccine recombinant protein candidate. Hum Vaccin Immunother. 2014;10(7):1914–1925. doi: 10.4161/hv.28872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sackett D.L., Wolff J. Nile red as a polarity-sensitive fluorescent probe of hydrophobic protein surfaces. Anal Biochem. 1987;167(2):228–234. doi: 10.1016/0003-2697(87)90157-6. [DOI] [PubMed] [Google Scholar]
- 20.Haskard C.A., Li-Chan E.C.Y. Hydrophobicity of bovine serum albumin and ovalbumin determined using uncharged (PRODAN) and anionic (ANS-) fluorescent probes. J Agric Food Chem. 1998;46(7):2671–2677. [Google Scholar]
- 21.Kato A., Nakai S. Hydrophobicity determined by a fluorescence probe method and its correlation with surface properties of proteins. Biochim Biophys Acta. 1980;624(1):13–20. doi: 10.1016/0005-2795(80)90220-2. [DOI] [PubMed] [Google Scholar]
- 22.Saluja A., Fesinmeyer R.M., Hogan S., Brems D.N., Gokarn Y.R. Diffusion and sedimentation interaction parameters for measuring the second virial coefficient and their utility as predictors of protein aggregation. Biophys J. 2010;99(8):2657–2665. doi: 10.1016/j.bpj.2010.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tal M., Silberstein A., Nusser E. Why does Coomassie Brilliant Blue R interact differently with different proteins? A partial answer. J Biol Chem. 1985;260(18):9976–9980. [PubMed] [Google Scholar]
- 24.Cao Z., Liu L., Du L. Potent and persistent antibody responses against the receptor-binding domain of SARS-CoV spike protein in recovered patients. Virol J. 2010;7(1):1–6. doi: 10.1186/1743-422X-7-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vedvick T., Buckholz R.G., Engel M. High-level secretion of biologically active aprotinin from the yeast Pichia pastoris. J Ind Microbiol. 1991;7(3):197–201. doi: 10.1007/BF01575883. [DOI] [PubMed] [Google Scholar]
- 26.Werten M.W., van den Bosch T.J., Wind R.D., Mooibroek H., de Wolf F.A. High-yield secretion of recombinant gelatins by Pichia pastoris. Yeast. 1999;15(11):1087–1096. doi: 10.1002/(SICI)1097-0061(199908)15:11<1087::AID-YEA436>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 27.Punt P.J., van Biezen N., Conesa A., Albers A., Mangnus J., van den Hondel C. Filamentous fungi as cell factories for heterologous protein production. Trends Biotechnol. 2002;20(5):200–206. doi: 10.1016/s0167-7799(02)01933-9. [DOI] [PubMed] [Google Scholar]
- 28.Cereghino G.P.L., Cereghino J.L., Ilgen C., Cregg J.M. Production of recombinant proteins in fermenter cultures of the yeast Pichia pastoris. Curr Opin Biotechnol. 2002;13(4):329–332. doi: 10.1016/s0958-1669(02)00330-0. [DOI] [PubMed] [Google Scholar]
- 29.Athmaram T.N., Singh A.K., Saraswat S. A simple Pichia pastoris fermentation and downstream processing strategy for making recombinant pandemic Swine Origin Influenza A virus Hemagglutinin protein. J Ind Microbiol Biotechnol. 2013;40(2):245–255. doi: 10.1007/s10295-012-1220-z. [DOI] [PubMed] [Google Scholar]
- 30.Goud G.N., Deumic V., Gupta R. Expression, purification, and molecular analysis of the Necator americanus glutathione S-transferase 1 (Na-GST-1): a production process developed for a lead candidate recombinant hookworm vaccine antigen. Protein Expr Purif. 2012;83(2):145–151. doi: 10.1016/j.pep.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 31.Curti E., Kwityn C., Zhan B. Expression at a 20L scale and purification of the extracellular domain of the Schistosoma mansoni TSP-2 recombinant protein: a vaccine candidate for human intestinal schistosomiasis. Hum Vaccin Immunother. 2012;9(11):2342–2350. doi: 10.4161/hv.25787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Switzer R.C., 3rd, Merril C.R., Shifrin S. A highly sensitive silver stain for detecting proteins and peptides in polyacrylamide gels. Anal Biochem. 1979;98(1):231–237. doi: 10.1016/0003-2697(79)90732-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.