Abstract
The use of microtechnology for the highly selective isolation and sensitive detection of circulating tumor cells has shown enormous promise. One challenge for this technology is that the small feature sizes – which are the key to this technology’s performance – can result in low sample throughput and susceptibility to clogging. Additionally, conventional molecular analysis of CTCs often requires cells to be taken off-chip for sample preparation and purification before analysis, leading to the loss of rare cells. To address these challenges, we have developed a microchip platform that combines fast, magnetic micropore based negative immunomagnetic selection (>10 mL/hr) with rapid on-chip in-situ RNA profiling (>100× faster than conventional RNA labeling). This integrated chip can isolate both rare circulating cells and cell clusters directly from whole blood and allow individual cells to be profiled for multiple RNA cancer biomarkers, achieving sample-to-answer in less than 1 hour for 10 mL of whole blood. To demonstrate the power of this approach, we applied our device to the circulating tumor cell based diagnosis of pancreatic cancer. We used a genetically engineered lineage-labeled mouse model of pancreatic cancer (KPCY) to validate the performance of our chip. We show that in a cohort of patient samples (N = 25) that this device can detect and perform in-situ RNA analysis on circulating tumor cells in patients with pancreatic cancer, even in those with extremely sparse CTCs (< 1 CTC / mL of whole blood).
Graphical abstract
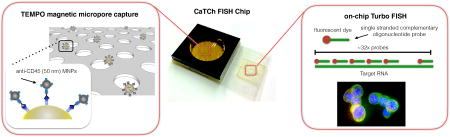
We have developed a microchip platform that combines fast, magnetic micropore based negative immunomagnetic selection (>10 mL/hr) with rapid on-chip in-situ RNA profiling (>100× faster than conventional RNA labeling).
Introduction
The detection and molecular profiling of circulating tumor cells (CTCs) have demonstrated enormous utility for the diagnosis and monitoring of cancer1,2. In particular, platforms that use micrometer-scale structures, where dimensions are designed to match those of CTCs, have been used with great success to selectively and sensitively sort3–6 and detect7–10 rare cells. However, there is an inherent mismatch between the throughput of microfluidic devices that can sort cells based on specific surface markers (Φ ≅ 1–10 mL/hr) and the large sample volume of blood (V > 10 mL) necessary for ultra-rare cell detection (< 5 cells/mL), resulting in long run-times (T > 1–10 hrs). Furthermore, conventional downstream molecular analysis of CTCs, such as single cell quantitative PCR11,12 or sequencing13, requires cells to be taken off-chip for sample preparation and purification before analysis, leading to the loss of target cells and the decay of molecular biomarkers14,15.
To address these challenges, we have developed a microchip-based platform to isolate and analyze rare cells directly from whole blood. The overall operation of our platform, which we have coined the Circulating Tumor Cell Fluorescence In-Situ Hybridization (CaTCh FISH) Chip, can be broken into three steps. First, rather than isolate CTCs based on any one of their heterogeneous properties4,16, we instead remove the large fraction of cells that are non-cancer cells. White blood cells (WBCs), which can be similarly sized to CTCs, are labeled with CD45 functionalized 50 nm magnetic nanoparticles and then isolated from the surrounding complex sample using a novel high throughput magnetic micropore filter. Downstream, a micropore size-based sorting structure is used to remove red blood cells (RBCs) and platelets based on their smaller size (< 8 µm) relative to CTCs (d > 8 µm). Single cell RNA analysis is performed on this micropore structure, which now contains a population of cells enriched for CTCs concentrated into a small field-of-view (12 mm2). To perform single cell RNA analysis, we use a newly developed rapid in situ hybridization (Turbo FISH)17(< 5 min hybridization) strategy, to both identify CTCs and profile their molecular state with single molecule sensitivity.
The CaTCh FISH combines several key features and innovations that differentiate it from previous work in the field of CTC isolation and analysis. CaTCh FISH combines the benefits of micro-scale, surface marker specific sorting with fast flow rates (>10 mL/hr), allowing extremely rare cells (1 CTC / mL) to be detected in large volume samples (>10 mL). On our chip, both CTCs and CTC cluster populations that are heterogenous in both size and surface marker expression can be isolated and profiled individually, without bias towards any assumed CTC surface markers (e.g. EpCAM expression). In comparison to prior CTC chips that use negative selection4, our chip differentiates itself in its high flow rates, its ability to capture both single cells and clusters, and its integrated on-chip single molecule RNA analysis. In comparison to previous work, wherein extremely high flow rates have been achieved using size-based sorting,51–53 our surface-marker specific isolation most differentiates itself in its ability to reduce co-purification and loss of circulating tumor cells. With these features, the CaTCh FISH chip offers a powerful new approach for both the discovery of circulating rare cell biomarkers and for rapid translation of these biomarkers into the clinic to improve patient care.
We chose to validate the clinical utility of CaTCh FISH by applying it to the diagnosis of pancreatic cancer. Pancreatic cancer is the third most common cause of cancer related death in the United States, with a five-year survival rate of approximately 8%18. Better tools to detect the disease early and to guide treatment more effectively, based on molecular biomarkers, are predicted to lead to significant improvements in these outcomes19. Because pancreatic tumor cells are localized in difficult to access parts of the body, molecular measurements currently rely on invasive procedures (i.e. biopsy), which limit their practical diagnostic use20. We have recently shown that circulating pancreatic cells can be detected in the blood at the onset of the disease cycle (pre-Stage 1) in both mice21 and humans22. For these reasons, there is both an enormous clinical need and potential for a non-invasive, highly accurate diagnostic test for pancreatic cancer based on the detection of CTCs in the blood. Here, we used a genetically engineered lineage-labeled mouse model of pancreatic cancer (KPCY)23 to validate the performance of our chip. Importantly for this study, the YFP expressed by every CTC in this model serves as a built-in positive control, facilitating the evaluation of selected RNA FISH targets in individual CTCs. In addition, we validated the clinical utility of our chip by profiling CTCs in blood samples from patients with advanced pancreatic cancer, achieving sample-to-answer in less than 1 hr.
Materials and Methods
CaTCh FISH Design
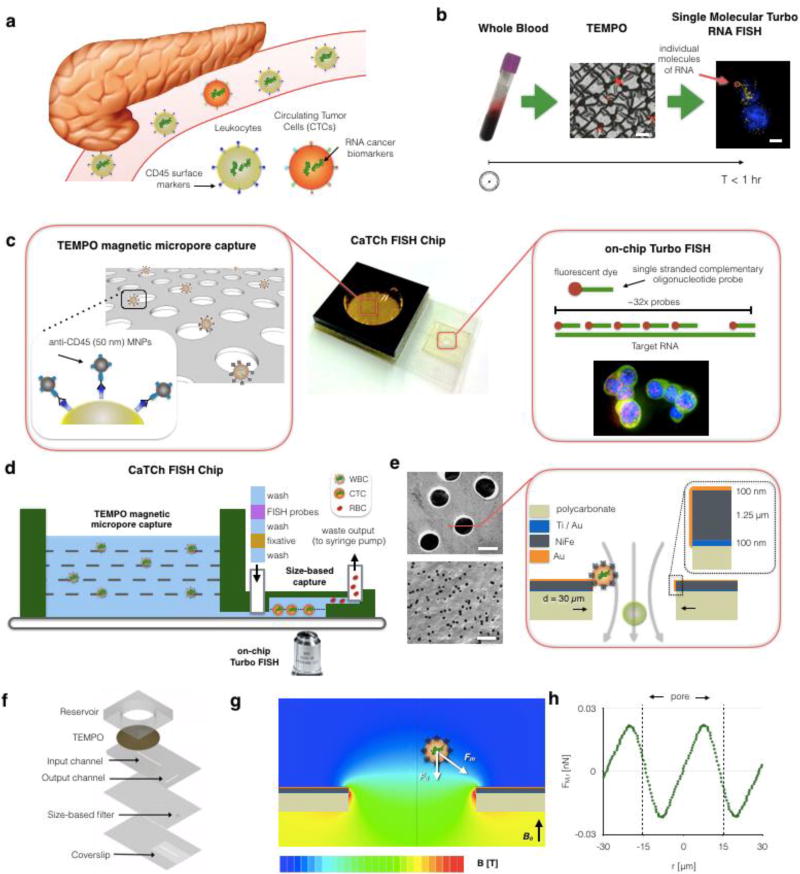
The extremely sparse and often heterogeneous4,24 CTCs that our chip aims to detect are suspended amongst 5×107 white blood cells and 5×1010 red blood cells in a 10 mL sample of whole blood (Fig. 1a). Our CaTCh FISH Chip isolates these rare cells and performs multiplexed, single molecular RNA analysis to identify and analyze each individual CTC (Fig. 1b)(Fig. S1) To accomplish these goals, we incorporated two key innovations onto an integrated microchip: (i) Track Etched Magnetic Micropore (TEMPO) sorting and (ii) microfluidic Turbo FISH.
Figure 1. Overview of the CaTCh FISH platform.
a. Circulating tumor cells (CTCs) are shed from the primary tumor and circulate in the blood. Due to their scarcity relative to blood cells and their heterogeneous biomarker expression, CTCs are difficult to isolate and analyze. b. Schematic of the CaTCh FISH workflow, in which whole blood is isolated is isolated, CTCs are enriched through Track Etched Magnetic MicroPOre (TEMPO)-mediated negative selection, and then molecularly characterized with single molecule RNA FISH, all in under an hour. Scale bars are 60 µm and 8 µm respectively. c. Higher resolution schematic illustrating the key components of the CatTCh FISH platform: the TEMPO filter and Turbo RNA FISH. d. A cross-section of the CaTCh FISH chip. e. The TEMPO filter is fabricated by coating track etched polycarbonate with a film of magnetic material (Ni80Fe20), enabling millions of micro-scale magnetic traps to act in parallel for selective, ultra-fast isolation. f. The TEMPO and TurboFISH components are incorporated into a single monolithic chip using laser micro-machined laminate microfluidics. g. Finite element simulations of magnetic field are used to design the TEMPO, such that the magnetic force can be measured and compared to the competing drag force (h). The pore has a diameter of 30 µm.
Track Etched Magnetic Micropore (TEMPO) sorting
In the last decade, the sorting of magnetic nanoparticle (MNP) labeled cells in microfluidic devices has emerged as a powerful technique to isolate rare CTCs9. This sorting requires minimal sample purification and yet achieves a high signal-to-background contrast, due to the negligible magnetic background of biological samples. Moreover, microscale magnets can be fabricated to match the size of cells, enabling highly specific capture of MNP labeled cells using micromagnetic traps in microfluidic devices25,26. However, due to the limited throughput of microfluidics (Φ < 10 mL/hr), which sort cells based on specific surface markers, these technologies operate too slowly for large volume of samples (V > 10 mL) and are susceptible to clogging from unprocessed whole blood.
To address these challenges, we have devised a new approach to micromagnetic sorting that achieves significantly higher flow rates by using millions of micromagnetic traps operating in parallel (Fig. 1c). Our design rotates the conventional approach by 90° to form magnetic traps at the edges of pores instead of in microfluidic channels. The TEMPO filter consists of a polycarbonate membrane track etched with 30 µm diameter pores coated with a 1.25 µm thick electroplated soft magnetic film (permalloy, Ni80Fe20). The chip sits in a large uniform magnetic field |B| = 0.4 T oriented normal to the surface of the TEMPO, provided by a centimeter sized NdFeB magnet. This field magnetizes both MNP labeled cells and the soft magnetic material on the magnetic micropore. We have previously demonstrated this technology using 5 µm pores to isolate pathogens from complex media6, and adopt it here for the rapid depletion of leukocytes and isolation of CTCs.
The TEMPO geometry retains the benefits of conventional microscale sorting, but offers several improvements essential for the rapid sorting of CTCs from whole blood. Similar to the strong field gradients created by lithographically defined micromagnets in conventional microfluidic systems, strong, highly localized microscale field gradients are formed at the edge of each micropore to selectively trap magnetically labeled cells. Analogous to the role of microfluidic channels in conventional devices, the microscale-sized pores force the cells into close proximity of the magnetic traps where they are selectively trapped. However, unlike conventional microfluidic devices, because the flow rate is distributed over millions of microscale traps in parallel, high flow rates can be achieved. Additionally, because the occlusion of any single pore does not significantly change the overall behavior of the device, TEMPO is robust against unprocessed clinical samples that would clog conventional microfluidic devices. By utilizing track-etching to fabricate TEMPO, our device can be fabricated with microscale features over areas much larger than economically feasible using conventional microfabrication A > 1 cm2 at a cost (< 5 cents / cm2) many times less than conventional microfabrication27.
Microfluidic Turbo FISH
Most current rare cell diagnostic tests profile either proteins, using immunofluorescence or enzyme-linked immunosorbent assay (ELISA), or nucleic acids, using RT-PCR. Protein-based diagnostics use antibodies, which require long development times at high costs.58 In contrast, nucleic acid detection is highly sensitive and highly specific and allows easier development of new assays. Detecting nucleic acids by RT-PCR, however, requires a thermal cycler and does not easily allow for on-chip single-cell analysis. A complementary approach for nucleic acid detection is direct labeling via RNA fluorescence in situ hybridization (RNA FISH).58,42 Conventionally, RNA FISH has suffered from three main drawbacks preventing its use as a clinical diagnostic: sensitivity, long assay times (6–12 hours), and many complex steps requiring laboratory training. We overcome the time requirement by developing a rapid hybridization protocol that utilizes alcohol based fixatives and high concentrations of oligonucleotide probe sets, achieving a < 5 minute assay17,28. To overcome sensitivity limitations, we use a variant of RNA FISH that involves hybridization of 20–50 short, fluorescently-labeled oligonucleotide probes to the target RNA (Fig. 1c). We design these oligonucleotides to bind to different segments of the target RNA via Watson-Crick base pairing, and the combined fluorescence from the fluorophores at the single RNA leads to a fluorescent spot of intensity much higher than that of the background. Each probe was designed to maximize binding to the target regions and to minimize off-site binding using an algorithm developed by the Raj lab (University of Pennsylvania). The entire Turbo FISH assay, including imaging, is carried out on our CaTCh FISH chip, enabling automated use for clinical diagnostics.
We designed these RNA probes by first obtaining sequences of targeted mRNA from the UCSC genome browser. Next, the targeted sequences were added as an input to the customized code developed by the Raj lab, which generates ~32 oligo RNA FISH probes that are optimized through cross-matching, probe elimination, and oligonucleotide placement memoization 45. Next, the information was sent to Biosearch Technologies for synthesis. For fluorophore conjugation, 32 oligos were first pooled and dried using SpeedVac (Thermo). Next, the oligo pellet was resuspended in 10 µl of 0.1M sodium bicarbonate. A volume of 0.2M sodium bicarbonate equal to the volume of dye was added. Then, the dye was added and incubated at room temperature for 4 hours. Ethanol precipitation was performed to remove excess dye. High-performance liquid chromatography (HPLC) was performed to purify dye conjugated RNA probes.
Device Fabrication
The TEMPO and size-based filter are integrated into a laser-cut laminate sheet microfluidic chip29. The size-based filter consists of a d = 3 µm track etched polycarbonate filter (Whatman, Nuclepore). The enriched CTCs are delivered to this filter through a 100 µm thick fluid channel, which sits directly on top of a #1 glass coverslip. To fabricate the TEMPO, we coated a polycarbonate track-etched film (Whatman, Nuclepore) with an evaporated metal layer of Ti/Au (100 nm) followed by an electroplated layer of Ni80Fe20 (1.25 µm) and finally with an electroplated passivation layer of Au (Transene) (Fig. 1e). To electroplate Ni80Fe20 we used nickel foil (1 mm thick, 99.5%, Alfa Aesar) and NiFe electroplating solution, containing 200 g/l NiSO4-6H2O, 8 g/l FeSO4-7H2O, 5 g/l NiCl2-6H2O, 25 g/l H3BO3, and 3 g/l saccharin (pH=2.5–3). The overall microfluidic chip is constructed from laser cut laminate sheets of mylar and acrylic (Fig. 1f). Whole blood is introduced into the device in an acrylic reservoir, fabricated above the TEMPO filter. The device is connected to a syringe pump using blunt syringe tips (McMaster Carr). The device is pretreated with Pluronic F-127 (Sigma–Aldrich) to minimize nonspecific retention of cells to the channel walls or to the TEMPO. Because the device does not require accurately controlled flow rates, it is well suited for mobile use where flow can be driven by inexpensive pumps or capillary action30,31.
Finite Element Modeling of TEMPO
We use finite element field simulations (Comsol) to aid the design and characterization of the magnetic micropores. The simulated field strength B is plotted on the cross-section of the magnetic filter (Fig. 1g). The field strength drops rapidly in distance from the filter creating gradients that lead to strong forces. The micropore was modeled as an axially symmetric pore with boundary conditions of zero field at large distances. The magnetophoretic force Fm on a magnetic nanoparticle labeled cell as it passes through a micropore is calculated by combining the finite element simulation from Fig. 1h with a simplified model for the cell 6,32. The model assumes that magnetic particles are fully magnetized by the applied field (|B| ≈ 0.4 T). The force Fm is calculated by combining this magnetic moment m with the simulated magnetic field B. The magnetic moment of the cell is proportional to the number of MNPs n and the moment mp of the particle (|m| = n * |mp|). For the 50 nm microbeads (Miltenyi) that we use, |mp| = 106 Bohr magnetons33. We assume the total number of particles per cell to be n = 104 particles, a conservative estimate, based on the 105 CD45 receptors per leukocyte34. The Stoke’s drag of a trapped cell is Fd = 6πηrv, where η = 0.8 mPa*s is the viscosity of water and r is the radius of the cell. The average flow velocity can be calculated vavg =Φ/(ρApA) where ρ = 1–5×105 pores*cm−2 is the pore density (Whatman), Ap is the cross-sectional area of an individual pore, and A = 10.2 cm2 is the cross-sectional area of the membrane. Moreover, the drag force Fd is minimized at the edges of the pore due to the no slip boundary condition.35
The capture of a cell on a TEMPO occurs in two sequential steps, which must both occur for capture. First, as a cell approaches a micropore, the magnetic force Fm must be great enough to pull the cell to the edge of the pore along the polar axis ȓ before the cell passes through the trap along the cylindrical axis ẑ. Second, once the cell reaches the edge of the trap, the magnetic trapping force Fm must oppose the drag force Fd, such that the cell will remain in the trap. Using this model, once trapped the magnetic trapping force on a 10 µm cell is >100× the drag force, and thus it will remain trapped at flow rates Φ >> 100 mL / hr. We conclude that the flow rate limit of the device does not arise from the competition of the magnetic and drag force for trapped cells, but instead comes from the fraction of cells that are successfully translated by the magnetic force to the trap at the pore’s edge. Based on this insight, we hypothesized that for this chip the capture rate could be improved by placing multiple pores in series to give each cell multiple chances to get trapped.
Characterization of enrichment of CTCs using an in vitro model
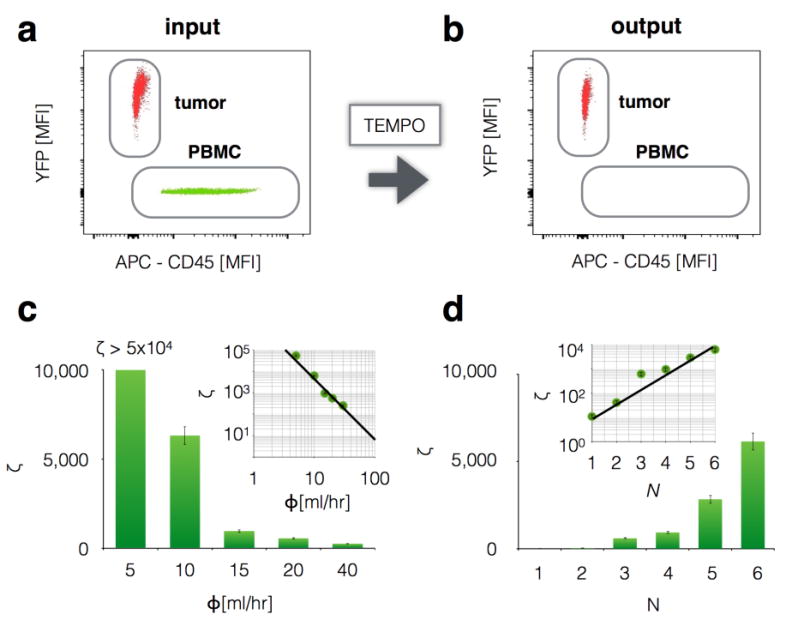
The ability of TEMPO to isolate magnetically targeted cells was first tested using an in vitro model. We sorted peripheral blood mononuclear cells (PBMCs) from YFP+ pancreatic cancer cells (PD7591). These two types of cells were mixed at a ratio of 100:1 (PBMC:PD7591) and PBMCs were stained (APC) and labeled with anti-CD45 magnetic microbeads. The input (Fig. 2a) and output (Fig. 2b) were measured using flow cytometry (BD FACSVerse flow cytometer, LSR II Flow Cytometer; BD Biosciences), and the performance of the TEMPO was quantified. In these devices we do not include a size-based filter, such that the performance of the TEMPO can be independently evaluated. To characterize the device’s capability to purify CTCs from white blood cells using magnetic sorting, we calculated the device’s ability to deplete nucleated cells ζ = (C1p/C1m)/(C0p/C0m) measured as a function of flow rate Φ, where C0p and C1p are the concentration of non-targeted cells before and after sorting respectively, and C0m and C1m are the concentration of targeted cells before and after sorting respectively. The depletion rate ζ using our micro-magnetic sorting strategy is >1,000× greater than only a macroscopic magnet.6 The purity of the output of the TEMPO is a function of the number of CTCs in a given sample and can be calculated by multiplying ζ by the number of CTCs to the number of leukocytes in a given sample.
Figure 2. Characterization of TEMPO.
Magnetic nanoparticle labeled cultured tumor cells positive for YFP were separated from mouse leukocytes using TEMPO, and quantified by flow cytometry a. before and b. after filtration. c. Very high depletion of leukocytes was achieved ζ >104 at flow rates Φ> 10 mL h−1 using N = 6 filters in series. Inset: depletion ζ depended on flow rate Φ as a power law. d. Depletion at 10 mL/hr could be improved exponentially in N by placing N filters in series. Thus, flow rate can be further increased beyond 10 mL/hr and depletion conserved, by continuing to add filters in series.
We made a series of measurements to determine the scaling relationship of the TEMPO’s performance ζ to flow rate Φ, the area of the device A, and the number of filters n. These scaling relationships allow the TEMPO to be tailored specifically to isolating rare CTCs directly from whole blood samples. We found that as flow rate increased, depletion decreased, dropping as a power law(ζ ∝ Φ−3.13) (Fig. 2c). However, depletion of leukocytes at high flow rates can be recovered by increasing the filter’s area A, which allows the flow rate to be scaled linearly with A while keeping ζ constant, Φ ∝ A.6 Additionally, depletion can be further enhanced by stacking multiple TEMPO membranes in series. By increasing from n = 1 to n = 6 for an A = 10.2 cm2 at Φ = 10 ml/hr, depletion was improved >500× (Fig. 2d). As the TEMPO membranes are vertically stacked, the subset of the cells missed by the previous membrane can be captured on the next membrane, which leads to this exponential increase in the depletion of leukocytes. To maximize depletion, we used n = 6 membranes for all the experiments. We challenged a device that included only a TEMPO with healthy donor blood, and profiled both the leukocytes in the input and those that passed-through the TEMPO for CD45 expression using flow cytometry (Flow Cytometry Core, University of Pennsylvania). Moreover, we evaluated the effect of leukocyte’s heterogeneity in CD45 expression on the performance of the device. We found that the rare leukocytes that pass through the TEMPO do not have a significantly different CD45 expression than those that are trapped. (Fig. S2) Performance could be further increased by labeling white blood cells using a cocktail of surface markers.
The isolation of rare cells from whole blood
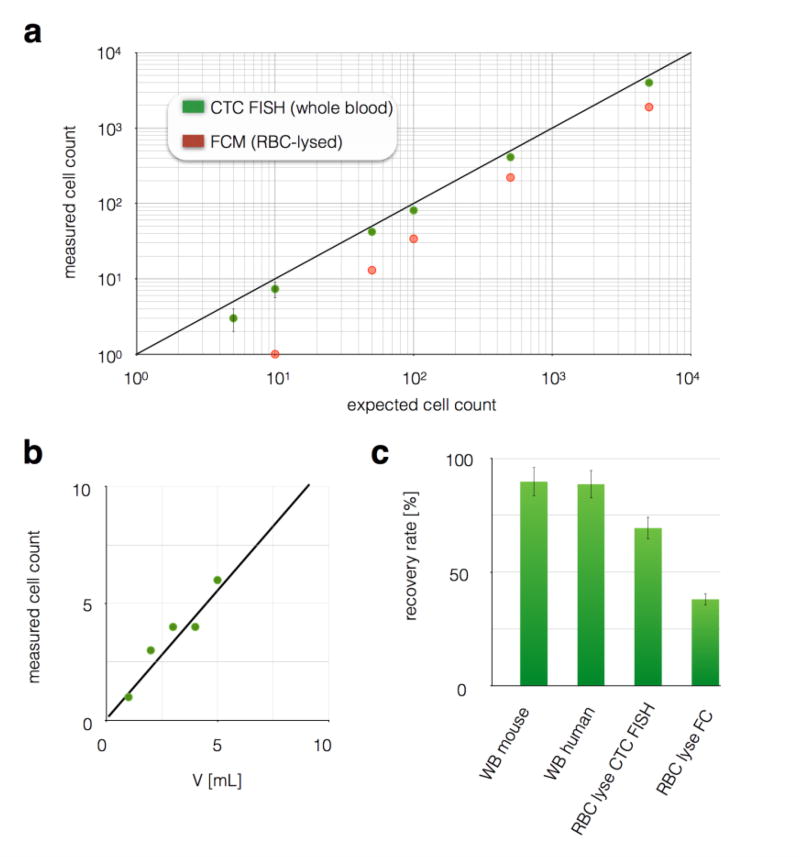
To test the sensitivity of our chip to rare cells in a complex background, we used healthy whole blood spiked with a known number of PD7591 cultured cancer cells. Cells were counted on the CaTCh FISH chip’s size-based filter using fluorescence microscopy (Leica, DMi8). A CaTCh FISH chip was used that contained n = 6 TEMPO membranes. The enumeration of these spiked cells using our chip showed excellent agreement with expected cell numbers (R2 = 0.99) and large dynamic range (1 to >104 cells) (Fig. 3a). Additionally, we showed that the limit of detection (LOD) could be increased by increasing the volume of blood input into our device. (Fig. 3b) We used whole blood with a concentration of 1 cancer cell / mL and ran samples with volumes from 1 to 5 mL. As expected, we observed a linear increase in the number of cells counted as a function of volume. In comparison to CaTCh FISH, flow cytometry resulted in an LOD of > 50 cells / mL whole blood, due to additional processing related loss and background autofluorescence (Fig. 3c). And, in comparison to competing CTC isolation technologies1–5,7,9 that report LOD > 1 CTC, because our device can rapidly (> 10 ml/hr) and selectively isolate rate cells from large volumes, it can detect ultra rare cells (< 1 CTC / mL) from large volume sample (V > 30 mL). At increasingly large sample volumes, a limiting factor for the CaTCh FISH’s performance becomes the saturation of the TEMPO membranes with WBCs. Because each TEMPO membrane contained ~5×106 pores, and in this study we used n = 6 membranes per device, and each pore could trap 3 WBCs, each chip could process >10 mL of blood (at 5×106 WBCs/ml) without becoming saturated. Because of CaTCh FISH’s design, more membranes or greater membrane area could be added to further increase this capacity. Experimentally, we showed that we were able to isolate CTCs from 10 mL of human whole blood and we did not observe any degradation of performance compared to smaller sample volumes.
Figure 3. Characterization of rare cell recovery on CaTCh FISH.
a. To validate CaTCh FISH’s ability to detect rare cells in whole blood, various numbers of cultured cells were spiked into whole blood and then measured using either CaTCh FISH or conventional flow cytometry. CaTCh FISH measured the cells directly in whole blood (WB), whereas RBCs were lysed prior to flow cytometry. CaTCh FISH achieved LOD ~ 1 cell / mL blood. Conventional cytometry had an LOD = 100 cell/mL whole blood. b. LOD of CaTCh FISH can be further increased by measuring larger volume samples. The graph shows a sample with 1 CTC/mL measured at volumes ranging from 1 to 5 mL. c. We evaluated CaTCh FISH’s insensitivity to background by comparing the recovery rate of spiked cells in whole mouse blood, whole human blood, RBC lysed mouse blood, and RBC lysed mouse blood measured on a flow cytometer (FC).
To verify that no tumor cells could pass through our size-based filter, and to confirm that the removal of leukocytes occurred on the TEMPO and not the size based filter, we performed the following experiments. We challenged our device with a sample containing a known number of leukocytes (37 million cells) and collected the flow-through of this device to measure any leukocytes that passed through the size-based filter. At Φ = 10 mL/hr we found 0 leukocytes had passed through the size based filter. To verify that the leukocytes were in fact isolated on the TEMPO, we performed the same experiment with the size-based filter removed. We found that the TEMPO captured 99.99983% of the leukocytes. Thus, out of 37 million cells, only 52 were missed by the TEMPO, which in the CaTCh FISH system would be trapped on the size-based filter, where they are distinguished from CTCs based on Turbo FISH. We performed the above experiments using both mouse pancreatic cancer cell lines (7–12 µm) and human pancreatic cancer cell lines (9–15 µm) and we did not observe any cancer cells that were able to pass through the size-based filter.
Characterization of background insensitivity
Our device’s sensitivity to background was tested by enumerating tumor cells (PD7591) across various backgrounds. We compared measurements in buffer with measurements in both mouse and human whole blood. Our chip successfully enumerated >90% of cells in 1 mL of mouse whole blood and in 1 mL of human whole blood. Additionally, we showed that by lysing and washing away RBCs prior to running the sample through our chip, there was an additional loss (70% recovery rate). Even with RBC-lysed samples, our chip still showed 2× improved recovery compared to flow cytometry (38% recovery rate) (Fig. 3c). Our chip’s recovery rate of 90% matches or exceeds that of other microfluidic techniques reported.3–5,7,9 For the current clinical gold standard, CellSearch, the recovery rate for EpCAM positive cells is 69%36 but can be much less if there are a large fraction of cells that are EpCAM negative.4,7
Design and fabrication of optofluidics for closed format ultra-rapid RNA FISH
Downstream of TEMPO, the population of trapped cells enriched for CTCs was analyzed using Turbo RNA FISH. To perform FISH, our device isolated CTCs downstream of the magnetic micropore device using a track-etched polycarbonate size-based filter (3 µm). This size-based filter serves two functions: (i) the pore size d = 3 µm was small enough to capture CTCs, but large enough to let RBCs pass. This size-based filter achieves a high capture rate (100%) of CTCs at fast flow rates (Φ > 10 ml/hr) over small areas (12 mm2) and is robust to unprocessed samples; and (ii) the size-based filter is built directly on top of a glass coverslip, enabling high resolution on-chip imaging. Because of the small field-of-view (12 mm2) afforded by this filter, the captured cells can be rapidly imaged with a 100× objective, with an average time less than 1 hour.
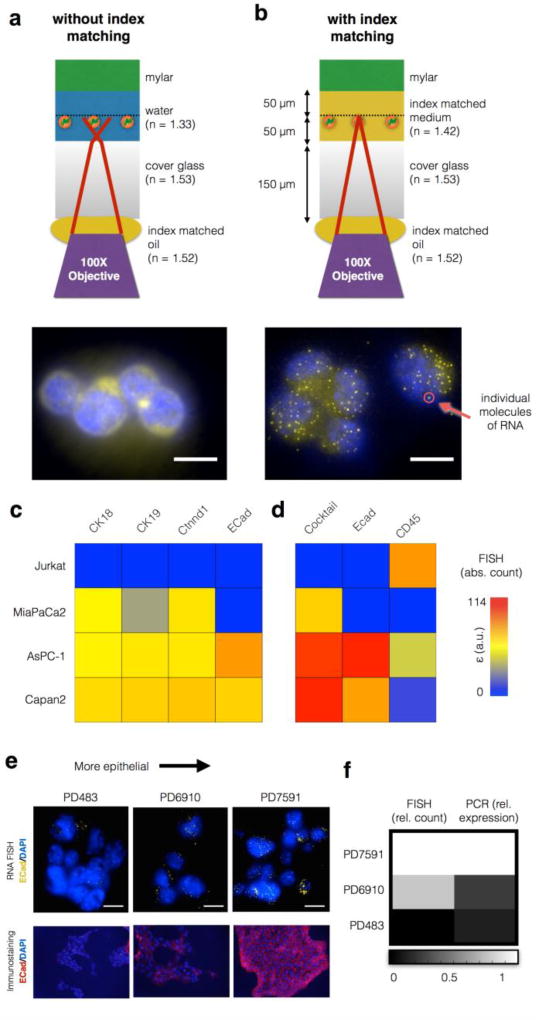
The incorporation of on-chip high resolution imaging into our platform led to a technological problem, which we solved using an optofluidic approach. To resolve multiply labeled single molecules of RNA, a 100× oil immersion, large numerical aperture (NA) objective is needed. Because an oil immersion lens is used, the introduction of 50 µm of water (n = 1.33) between the cells and the glass cover slip (n = 1.53) introduces image distortion, which destroys resolution (Fig. 4a). The cells are trapped on our track-etched filter, which lies above a 50 µm tall microfluidic channel that delivers cells to the filter. To overcome this challenge, we replaced the water in the channel prior to imaging with a low viscosity high index fluid (n = 1.42, AntiFade, Life Technologies) which is partially index matched to the glass. Index matching restored high resolution imaging (Fig. 4b). Other materials can achieve better index matching, but their high viscosity can lead to excessive drag forces on trapped cells, increasing cell loss.
Figure 4. Single Molecule In Situ RNA analysis on the CaTCh FISH chip.
With a 100× objective, individual mll1 RNA molecules were impossible to resolve without index matching (a), but became easily resolved with index matching (b). Scale bar:10 µm. c. RNA FISH on the CaTCh FISH chip was used to enumerate the number of RNA in individual cells for a variety of cell types, represented in a heat map. d. By using a cocktail of RNA markers (CK18, CK19, and Ctnnd1) and CD45 as a negative marker, all of the cell types can be identified. Additionally, ECad can be used to analyze the EMT state of the cell. e. Comparison of RNA FISH and immunofluorescence for ECad in PD483 (mesenchymal), PD6910 (intermediate), and PD7591 (epithelial) cultured cells. Scale bar is 10 µm. f. Comparison of Turbo FISH and quantitative PCR results for the cell lines in (e).
To characterize the sensitivity and specificity of our Turbo FISH method, we compared the number of CD45 RNA measured per Jurkat cells, using both conventional single molecule FISH and our Turbo FISH protocol (Fig. S3) No significant difference was detected.(P > 0.25) Additionally, we measured the rate of falsely positive and falsely negative single molecules of RNA by labeling the same RNA target (BABAM) using two differently colored probes, and we use co-localization to identify the true positives. (Fig. S4) For this measurement, as has been done previously,48 we partitioned the probe set (32 oligos) to the even and odd numbered oligonucleotides and coupled each subset with a different fluorophore (evens: Cy3, odds: Alexa 594). We hybridized the two probe sets and imaged each color and found that 76% of our punctates co-registered, consistent with previously published results using conventional single molecule RNA FISH.48 Because in this experiment, due to the odds- and evens- splitting of the probe set, only half as many oligo probes are used as in the rest of our studies, reducing the signal to background. Thus, we expect this performance metric to be exceeded in our clinical studies. The punctates were quantified using automated software, as described previously,45 which is open source and available for free use online.
To perform on-chip FISH, we ran the cold methanol (stored in −20°C) through the chip to fix the cells for 2 mins. After fixation, we washed the cells with 200 µl of wash buffer (10% formamide and 2× SCC) at 20 ml/hr. Next, we added 1 µl of each RNA FISH probe (stock concentrations ranging from 1288 ng/µL to 5800 ng/µL) into a hybridization buffer in 1:50 ratio. We added 50 µl of the mixed solution and stopped the flow and placed the chip on a 37°C hotplate for 5 mins. After hybridization, we washed the cells with 1 ml of wash buffer at 37°C. For imaging, wash buffer was replaced with the SlowFade Diamond Antifade mountant (Thermo Fisher Scientific). We imaged the size-based filter area of the chip on a Nikon Ti-E inverted fluorescence microscope using 100× Plan-Apo objective (numerical aperture of 1.40) and a cooled CCD camera (Andor iKon 934). Using four different fluorescence channels (DAPI, Alexa 594, Cy3, and Atto 647N), we acquired three-dimensional stacks of images. After imaging acquisition, the image stacks were projected into a single image using maximum projection, using ImageJ software.
RNA biomarker selection and validation for pancreatic cancer
To determine which RNA biomarkers to target with our RNA FISH, we searched for markers using published RNA sequencing databases that were highly expressed in CTCs and at low or undetectable levels expressed by leukocytes37. We found that no one marker was positive for all CTCs, and so instead we defined a cocktail of three markers (CK18, CK19, and Ctnnd1) that are positive in combination for all pancreatic CTCs. For each cell, we also separately measured E-cadherin (ECad) to profile the epithelial-to-mesenchymal transition (EMT), and CD45 as a negative marker to identify leukocytes. As CTCs of the KPCY mouse model we used for this study express YFP, we were able to use it as a built-in endogenously expressed positive control. We designed sets of oligonucleotides FISH probes for each of these targets, where each set was designed to bind to different segments of the target RNA via Watson-Crick base pairing. Two sets of FISH probes were created for each of the targets, one set for human and one set for mouse experiments. To validate these markers, we tested them using in vitro model systems, including the following human pancreatic cancer cell lines: MiaPaCa2, AsPC-1, and Capan2, as well as Jurkat cells, which were used as a convenient model for background leukocytes. The three human pancreatic cancer cell lines are representative of expected pancreatic cancer cell heterogeneity, covering the spectrum of epithelial to mesenchymal cells. As expected, no single marker was able to identify every one of these cell types (Fig. 4c). However, by using the cocktail of positive markers (CK18, CK19, and Ctnnd1), as well as CD45 as a negative marker, all three pancreatic cancer cell lines could be distinguished from the Jurkat cells (Fig. 4d, Fig. S5).
Next, we validated that the CaTCh FISH chip could quantify the RNA expression within each individual cell. The molecular analysis of CTCs offers a window into the molecular mechanism of metastasis. It is widely believed that a phenotypic change known as epithelial-mesenchymal transition (EMT) – where cells lose their epithelial characteristics and become mesenchymal – is important for invasion and bloodstream entry38,39. To validate that our chip could profile the EMT transition, three different pancreatic mouse cell lines with varying degrees of expression of the epithelial marker, ECad, were tested, including a mesenchymal cell line PD483, an epithelial cell line PD7591, and an intermediate cell line PD6910. The quantity of ECad RNA measured by RNA FISH was greater for cells that were more epithelial, and agreed with immunostaining (Fig. 4e). Additionally, the quantity of ECad mRNA measured per cell on the CaTCh FISH was compared with quantitative PCR, and the Ecad expression increased as the cells were more epithelial in both measurements (Fig. 4f).
Results
In vivo testing in KPCY mice
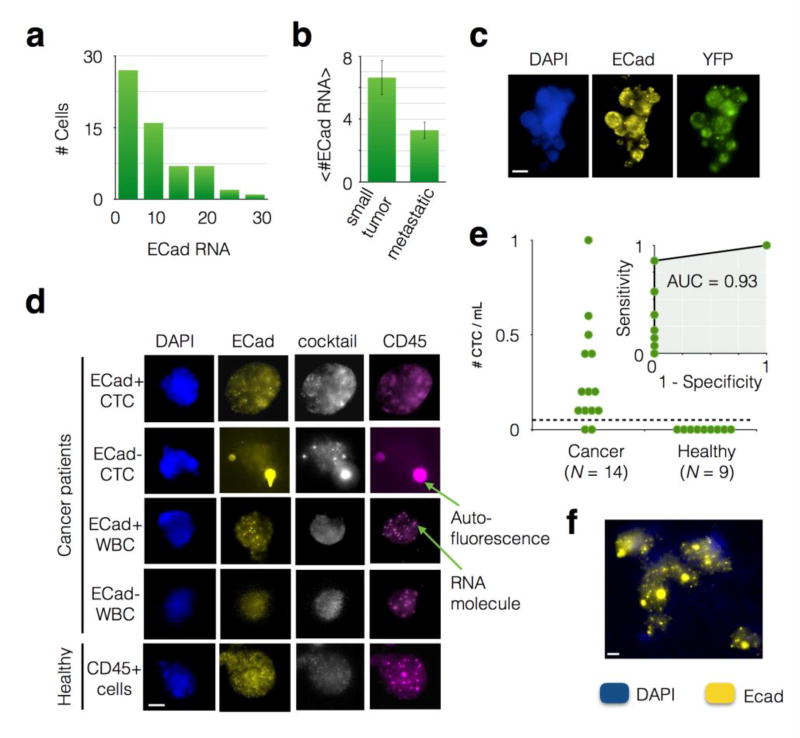
To validate that we could isolate and analyze CTCs in an in vivo system that closely recapitulates human pancreatic cancer, we began by isolating CTCs from whole blood obtained from tumor-bearing KPCY mice. Each CTC in the KPCY mouse expresses YFP, enabling us to readily identify CTCs independent of the markers that we have chosen. We isolated CTCs from whole mouse blood (N = 5 KPCY mice; mean 30 cells per mouse) and quantified the number of ECad RNA punctae in each CTC (Fig. 5a). All experimental protocols were approved and animal care and use was in accordance with the guidelines specified by the Institutional Animal Care and Use Committee (IACUC) of the University of Pennsylvania. We isolated CTCs from the whole blood volume of mouse (~ 1 ml), where on average < 50 WBCs were co-purified with CTCs, at Φ = 10 mL/hr and n = 6 TEMPO membranes. We found that CTCs captured from mice with low disease burden had significantly more ECad punctae per cell than CTCs from mice that were metastatic (P < 0.05) (Fig. 5b). In addition to individual CTCs, we also observed clusters of cells, as were observed in a closely-related pancreatic cancer model24, the largest containing 13 cells (Fig. 5c). Within these clusters we observed heterogeneity of ECad expression, with some cells expressing as few as zero copies of ECad RNA and some as many as 18.
Figure 5. Application of CaTCh FISH Chip to murine and patient whole blood samples.
a. A histogram of the number of ECad RNA per CTC for blood taken from an individual KPCY mouse, showing the heterogeneity of ECad expression amongst CTCs. All cells in this histogram were cocktail+ (CK18, CK19, and Ctnnd1) and CD45-, identifying them as CTCs. b. The average number of ECad punctae per cell <#ECad RNA> for mice with small tumors versus mice with advanced lesions. A statistically significant difference P = 0.01 was found. Error bars represent standard error. c. An image of a circulating tumor cell cluster recovered from a KPCY mouse. Scale bar is 10 µm. d. Cells captured from advanced pancreatic cancer patients and healthy donors, including both ECad+ and ECad- CTCs and WBCs. e. The number of CTCs captured in patients with advanded pancreatic cancer versus those from healthy controls. The dashed line represents a threshold that can be defined to identify patients that have cancer. Inset: A Receiver Operator Characteristic (ROC) curve for the CatCH FISH, achieving an Area Under the Curve (AUC = 0.93). f. A CTC cluster recovered from a patient with pancreatic cancer. Scale bar is 4 µm.
Detection of CTCs in clinical samples
To validate the use of our CaTCh FISH Chip on clinical samples, we measured blood samples obtained from patients with advanced pancreatic cancer. We processed samples from N = 14 patient samples, all with metastatic or locally advanced disease (10 patients with one time point and 2 patients with two time points) and N = 9 healthy controls. Patient demographics are included in Fig.S6. Peripheral whole blood was obtained in EDTA tubes from pancreatic ductal adenocarcinoma (PDAC) patients with advanced pancreatic cancer and from healthy age- and gender-matched controls at the University of Pennsylvania Health System. All patients and healthy donors provided written informed consent for blood donation on approved institutional protocols. For each patient with pancreatic cancer, we also obtained CA19-9 values (units/ml). Of these patient samples, 11 out of 12 were positive for CA19-9. We confirmed that each clinical sample had a small number of CTCs (<1 cell/mL) by sending aliquots to Epic Sciences (N = 5). Consistent with prior reports47, CTC counts for pancreatic cancer patients are low or undetectable relative to other cancers of epithelial origin such as breast or prostate cancer. Epic Sciences identifies CTCs using a high throughput optical imaging system that inspects 3 million nucleated cells per glass slide (0.5 mL of whole blood), and defines CTCs as being Cytokeratin (CK) + and CD45-.
By measuring large volumes of sample V > 10 mL, the CaTCh FISH chip was able to identify extremely rare CTCs (<1 cell/ml). On our chip we defined CTCs as having an RNA FISH signal that was positive for our cocktail (CK18, CK19, and Ctnnd1) and negative for CD45. The cocktail (CK18, CK19, and Ctnnd1) that we used to identify circulating tumor cells was highly expressed by CTCs in the clinical samples (Fig. 5d). Using ECad to evaluate the EMT state of the CTCs, we were able to detect both epithelial CTCs (cocktail+, ECad+) and mesenchymal CTCs (cocktail+, ECad-). The CD45 FISH probes were useful as a negative selection marker to exclude the few leukocytes that made it past the TEMPO (leukocytes were defined as CD45+ and cocktail -). Interestingly, a subset of leukocytes expressed a small number of ECad RNA (CD45+, cocktail -, and ECad+), as has been seen previously observed.40 On CaTCh FISH, an average of 2.8 CTCs were detected per 10 mL of whole blood in the N = 14 patient samples with advanced disease that we measured.(Fig. 5e) We additionally ran N = 9 negative controls, consisting of 10 mL of whole blood obtained from healthy patients (Zen Bio), and did not find a single false positive in any of the samples. The extremely high specificity of CaTCh FISH can be attributed to the specificity of TURBO FISH, the use of multiple biomarkers to identify CTCs (CD45-, and either CK18+, CK19+, or Ctnnd1+), and the highly selective depletion of leukocytes by TEMPO. To quantify the performance of the device, a receiver operator characteristic curve was generated and the CaTCh FISH was found to have an AUC = 0.93 for the classification of patients with pancreatic cancer (N = 14) versus healthy controls (N = 9).(Fig. 5e - inset) In addition to single CTCs, we also identified cell clusters, containing as many as five cells from a metastatic patient sample (Fig. 5f). Within these clusters we observed heterogeneity of ECad expression, with some cells expressing as few as three copies of ECad RNA and some as many as thirteen. The bright circular patterns that are present in some of our images are hypothesized to be platelets adhered to the CTCs. This autofluorescence signal was easily ignored in analysis due to its presence in all fluorescence channels41.
Discussion
CaTCh FISH chip offers rapid, ultra-sensitive detection and quantitative single cell molecular analysis of rare circulating cells. Due to the CaTCh FISH chip’s ability to isolate targeted cells regardless of their heterogeneous physical or molecular properties, and its ability to profile a broad-range of RNA biomarkers within these individual cells, the platform can be adopted to study a wide variety of cell types, such as endothelial cells, immune cells, and stem cells. Due to the highly parallel nature of this device, it can rapidly process samples (sample to answer in less than 1 hour for a 10 mL blood sample) and it is insensitive to clogging from unprocessed whole blood. In comparison, other negative depletion technologies have achieved flow rates of 0.12 – 2 mL/hr. 3, 55–57 In comparison to previous work, where surface marker specific isolation has been achieved at flow rates as high as 10 mL/hr,53,54 our device’s throughput scales with the device area rather than device width for conventional devices, which allows scaling to greater than 100 mL/hr on centimeter-scale devices.
Beyond the performance demonstrated in this paper, the CaTCH FISH platform offers several key benefits which will enable further advancements: i. The throughput demonstrated in this paper can be further improved due to favorable scaling features. By either increasing the area of the TEMPO (Φ∝A) or vertically stacking more TEMPO filters (ζ ∝eN), the device can be optimized for the rapid enrichment of rare cells in a wide range of samples and sample volumes. Moreover, newly developed master-replica fabrication strategies can be used to fabricate magnetic micropore devices with improved throughput.49 ii. The use of Turbo RNA FISH also provides an opportunity to expand this platform. While we used five sets of RNA FISH probes to inspect each cell in this work, this can be expanded to much larger numbers of both RNA and DNA biomarkers using advanced FISH techniques to gain a more detailed picture of the cell42,43,44 iii. Imaging using a conventional microscope, as is done in this paper, requires acquisition of several fields of view, stitched together in software. Alternatively, synthetic aperture imaging offers an opportunity to image our entire field-of-view in a single shot,43, 50 thus enabling high resolution, wide field-of-view microscopy to be easily incorporated into a clinical diagnostic. Overall, based on CaTCh FISH’s favorable properties, and its opportunities for continued improvement, this new approach to rare cell sorting and analysis offers an enormous opportunity as a rare-cell diagnostic.
Supplementary Material
Acknowledgments
Issadore was supported by an American Cancer Society - CEOs Against Cancer - CA Division Research Scholar Grant (RSG-15-227-01-CSM), a grant from The Hartwell Foundation, and NIH R21 5R21CA182336. Arjun Raj was supported by NIH: R33-EB-019767 and U01- HL129998 and NSF MCB-1350601. Stanger was supported by NIH: UC4-DK104196, R01CA182869-01A1, R01-DK-083355 and DOD: W81XWH-15-1-0457. Carpenter was supported by NIH: R01 CA207643-01, P30 CA016520-33 and The Pancreatic Cancer Action Network.
Footnotes
Statement of Contributions: JK helped conceive and perform all experiments in this study, as well as prepare the manuscript and figures. N.B., S.Y., T.B, C.R., J.R., M.O., helped conceive and perform experiments on murine and clinical testing of our device. B.S., A.R., and E.C. helped conceive all experiments in this study, as well as prepare the manuscripts and figures. DI conceived and oversaw all aspects of this study, and prepared the manuscript. All authors reviewed the manuscript.
References
- 1.Parkinson DR, et al. Considerations in the development of circulating tumor cell technology for clinical use. J Transl Med. 2012;10:138. doi: 10.1186/1479-5876-10-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Franken B, et al. Circulating tumor cells, disease recurrence and survival in newly diagnosed breast cancer. Breast Cancer Res. 2012;14:R133. doi: 10.1186/bcr3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagrath S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ozkumur E, et al. Inertial focusing for tumor antigen--dependent and--independent sorting of rare circulating tumor cells. Science translational medicine. 2013;5:179ra47–179ra47. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheng W, Ogunwobi OO, Chen T, Zhang J, George TJ, Liu C, Fan ZH. Capture, release and culture of circulating tumor cells from pancreatic cancer patients using an enhanced mixing chip. Lab on a Chip. 2014;14:89–98. doi: 10.1039/c3lc51017d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muluneh M, Shang W, Issadore D. Track-Etched Magnetic Micropores for Immunomagnetic Isolation of Pathogens. Advanced healthcare materials. 2014;3:1078–1085. doi: 10.1002/adhm.201300502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Issadore D, et al. Ultrasensitive clinical enumeration of rare cells ex vivo using a micro-hall detector. Science translational medicine. 2012;4:141ra92–141ra92. doi: 10.1126/scitranslmed.3003747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee H, Sun E, Ham D, Weissleder R. Chip--NMR biosensor for detection and molecular analysis of cells. Nature medicine. 2008;14:869–874. doi: 10.1038/nm.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J, Li J, Sun Y. Microfluidic approaches for cancer cell detection, characterization, and separation. Lab on a Chip. 2012;12:1753–1767. doi: 10.1039/c2lc21273k. [DOI] [PubMed] [Google Scholar]
- 10.Castro CM, et al. Miniaturized nuclear magnetic resonance platform for detection and profiling of circulating tumor cells. Lab on a Chip. 2014;14:14–23. doi: 10.1039/c3lc50621e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gasch C, et al. Heterogeneity of epidermal growth factor receptor status and mutations of KRAS/PIK3CA in circulating tumor cells of patients with colorectal cancer. Clinical chemistry. 2013;59:252–260. doi: 10.1373/clinchem.2012.188557. [DOI] [PubMed] [Google Scholar]
- 12.Alix-Panabieres C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clinical chemistry. 2013;59:110–118. doi: 10.1373/clinchem.2012.194258. [DOI] [PubMed] [Google Scholar]
- 13.Heitzer E, et al. Complex tumor genomes inferred from single circulating tumor cells by array-CGH and next-generation sequencing. Cancer research. 2013;73:2965–2975. doi: 10.1158/0008-5472.CAN-12-4140. [DOI] [PubMed] [Google Scholar]
- 14.Issadore D, Westervelt RM. Point-of-care Diagnostics on a Chip. Springer Science & Business Media; 2013. [Google Scholar]
- 15.Ko J, Carpenter E, Issadore D. Detection and isolation of circulating exosomes and microvesicles for cancer monitoring and diagnostics using micro-/nano-based devices. Analyst. 2015 doi: 10.1039/c5an01610j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Powell AA, et al. Single cell profiling of circulating tumor cells: transcriptional heterogeneity and diversity from breast cancer cell lines. PloS one. 2012;7:e33788. doi: 10.1371/journal.pone.0033788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaffer SM, Wu MT, Levesque MJ, Raj A. Turbo FISH: a method for rapid single molecule RNA FISH. PloS one. 2013;8:e75120. doi: 10.1371/journal.pone.0075120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA: A cancer journal for clinicians. 2015 doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 19.Ghatnekar O, et al. Modelling the benefits of early diagnosis of pancreatic cancer using a biomarker signature. International Journal of Cancer. 2013;133:2392–2397. doi: 10.1002/ijc.28256. [DOI] [PubMed] [Google Scholar]
- 20.Khoja L, et al. A pilot study to explore circulating tumour cells in pancreatic cancer as a novel biomarker. British journal of cancer. 2012;106:508–516. doi: 10.1038/bjc.2011.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhim AD, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148:349–361. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhim AD, et al. Detection of circulating pancreas epithelial cells in patients with pancreatic cystic lesions. Gastroenterology. 2014;146:647–651. doi: 10.1053/j.gastro.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rhim AD, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148:349–361. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maddipati R, Stanger BZ. Pancreatic cancer metastases harbor evidence of polyclonality. Cancer discovery. 2015;5:1086–1097. doi: 10.1158/2159-8290.CD-15-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pamme N, Manz A. On-chip free-flow magnetophoresis: continuous flow separation of magnetic particles and agglomerates. Analytical Chemistry. 2004;76:7250–7256. doi: 10.1021/ac049183o. [DOI] [PubMed] [Google Scholar]
- 26.Adams JD, Kim U, Soh HT. Multitarget magnetic activated cell sorter. Proceedings of the National Academy of Sciences. 2008;105:18165–18170. doi: 10.1073/pnas.0809795105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Earhart CM, Wilson RJ, White RL, Pourmand N, Wang SX. Microfabricated magnetic sifter for high-throughput and high-gradient magnetic separation. Journal of magnetism and magnetic materials. 2009;321:1436–1439. doi: 10.1016/j.jmmm.2009.02.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaffer SM, et al. Multiplexed detection of viral infections using rapid in situ RNA analysis on a chip. Lab on a Chip. 2015;15:3170–3182. doi: 10.1039/c5lc00459d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weigl BH, Bardell R, Schulte T, Battrell F, Hayenga J. Design and rapid prototyping of thin-film laminate-based microfluidic devices. Biomedical Microdevices. 2001;3:267–274. [Google Scholar]
- 30.Chumo B, Muluneh M, Issadore D. Laser micromachined hybrid open/paper microfluidic chips. Biomicrofluidics. 2013;7:064109. doi: 10.1063/1.4840575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dimov IK, et al. Stand-alone self-powered integrated microfluidic blood analysis system (SIMBAS) Lab on a Chip. 2011;11:845–850. doi: 10.1039/c0lc00403k. [DOI] [PubMed] [Google Scholar]
- 32.Shevkoplyas SS, Siegel AC, Westervelt RM, Prentiss MG, Whitesides GM. The force acting on a superparamagnetic bead due to an applied magnetic field. Lab on a chip. 2007;7:1294–1302. doi: 10.1039/b705045c. [DOI] [PubMed] [Google Scholar]
- 33.Tchikov V, Fritsch J, Kabelitz D, Schutze S. Immunomagnetic isolation of subcellular compartments. Methods in Microbiology. 2010;37:21–33. [Google Scholar]
- 34.Sharma D, et al. Effective flow cytometric phenotyping of cells using minimal amounts of antibody. Biotechniques. 2012;53:57. doi: 10.2144/0000113854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hasimoto H. On the flow of a viscous fluid past a thin screen at small Reynolds numbers. Journal of the Physical Society of Japan. 1958;13:633–639. [Google Scholar]
- 36.Hillig T, Horn P, Nygaard AB, Haugaard AS, Nejlund S, Brandslund I, Sölétormos G. In vitro detection of circulating tumor cells compared by the CytoTrack and CellSearch methods. Tumor Biology. 2015;36:4597–4601. doi: 10.1007/s13277-015-3105-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu M. RNA sequencing of pancreatic circulating tumour cells implicates WNT signalling in metastasis. Nature. 2012;487:510–513. doi: 10.1038/nature11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thiery JP, Acloque H, Huang RYJ, Nieto MA. Epithelial-mesenchymal transitions in development and disease. cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 39.Ye X, Weinberg RA. Epithelial--Mesenchymal Plasticity: A Central Regulator of Cancer Progression. Trends in cell biology. 2015;25:675–686. doi: 10.1016/j.tcb.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Corn PGAS, Douglas B, Ruckdeschel Emily S, Douglas Donna, Baylin Stephen B, Herman James G. E-cadherin expression is silenced by 5′ CpG island methylation in acute leukemia. Clinical Cancer Research. 2000;6:4243–4248. [PubMed] [Google Scholar]
- 41.Chang C-L. Circulating tumor cell detection using a parallel flow micro-aperture chip system. Lab on a Chip. 2015;15:1677–1688. doi: 10.1039/c5lc00100e. [DOI] [PubMed] [Google Scholar]
- 42.Levesque MJ, Raj A. Single-chromosome transcriptional profiling reveals chromosomal gene expression regulation. Nature methods. 2013;10:246–248. doi: 10.1038/nmeth.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee CS, Park JB, Lightspeed Genomics, Inc. Illumination apparatus optimized for synthetic aperture optics imaging using minimum selective excitation patterns. 12/728,140. U.S. Patent Application. 2011
- 44.Schreiber sFS, et al. Successful growth and characterization of mouse pancreatic ductal cells: functional properties of the Ki-RASG12V oncogene. Gastroenterology. 2004;127:250–260. doi: 10.1053/j.gastro.2004.03.058. [DOI] [PubMed] [Google Scholar]
- 45.Raj A, Van DB, Patrick, Rifkin SA, Van O, Alexander, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nature methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park J-W. PhD Thesis, Georgia Tech. 2003. Core lamination technology for micromachined power inductive components. [Google Scholar]
- 47.Allard W, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clinical Cancer Research. 2004;10.20:6897–6904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 48.Cabili MN, et al. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biology. 2015;16.1:1. doi: 10.1186/s13059-015-0586-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ko J, et al. Magnetic Nickel iron Electroformed Trap (MagNET): a master/replica fabrication strategy for ultra-high throughput (>100 mL h− 1) immunomagnetic sorting. Lab on a Chip. 2016 doi: 10.1039/C6LC00487C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen KH, et al. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348.6233:aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hou HW, et al. Isolation and retrieval of circulating tumor cells using centrifugal forces. Scientific Reports. 2013;3:1259. doi: 10.1038/srep01259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khoo BL, et al. Clinical validation of an ultra high-throughput spiral microfluidics for the detection and enrichment of viable circulating tumor cells. PloS one. 2014;9.7:e99409. doi: 10.1371/journal.pone.0099409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murlidhar V, Rivera-Báez L, Nagrath S. Affinity Versus Label-Free Isolation of Circulating Tumor Cells: Who Wins? Small. 2016 doi: 10.1002/smll.201601394. [DOI] [PubMed] [Google Scholar]
- 54.Murlidhar V, et al. A Radial Flow Microfluidic Device for Ultra- High- Throughput Affinity- Based Isolation of Circulating Tumor Cells. Small. 2014;10.23:4895–4904. doi: 10.1002/smll.201400719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hyun KA, Lee TY, Jung H., II Negative enrichment of circulating tumor cells using a geometrically activated surface interaction chip. Anal. Chem. 2013;85:4439–4445. doi: 10.1021/ac3037766. [DOI] [PubMed] [Google Scholar]
- 56.Diéguez L, Winter MA, Pocock KJ, Bremmell KE, Thierry B. Efficient microfluidic negative enrichment of circulating tumor cells in blood using roughened PDMS. Analyst. 2015;140:3565–72. doi: 10.1039/c4an01768d. [DOI] [PubMed] [Google Scholar]
- 57.Bu J. Dual-patterned immunofiltration (DIF) device for the rapid efficient negative selection of heterogeneous circulating tumor cells. Lab Chip. 2016;351:781–791. doi: 10.1039/c6lc01179a. [DOI] [PubMed] [Google Scholar]
- 58.Ross JS. Rluorescence In Situ Hybridization Is the Preferred Approach over Immunohistochemistry for Determining HER2 Status. Clinical Chemistry. 2011;57:7. doi: 10.1373/clinchem.2010.160762. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.