Abstract
Purpose
In recent years, fusion of multi-parametric MRI (mp-MRI) with transrectal ultrasound (TRUS) guided biopsy has enabled targeted prostate biopsy with improved cancer yield. Target identification is solely based on information from mp-MRI, which is subsequently transferred to the subject coordinates through an image registration approach. mp-MRI has shown to be highly sensitive to detect higher grade prostate cancer, but suffers from a high rate of false positives for lower grade cancer, leading to unnecessary biopsies. This paper utilizes a machine-learning framework to further improve the sensitivity of targeted biopsy through analyzing temporal ultrasound data backscattered from the prostate tissue.
Methods
Temporal ultrasound data was acquired during targeted fusion prostate biopsy from suspicious cancer foci identified in mp-MRI. Several spectral features, representing the signature of backscattered signal from the tissue, were extracted from the temporal ultrasound data. A supervised support vector machine classification model was trained to relate the features to the result of histopathology analysis of biopsy cores obtained from cancer foci. The model was used to predict the label of biopsy cores for mp-MRI identified targets in an independent group of subjects.
Results
Training of the classier was performed on data obtained from 35 biopsy cores. A five-fold cross-validation strategy was utilized to examine the consistency of the selected features from temporal ultrasound data, where we achieved the classification accuracy and area under receiver operating characteristic curve of 94% and 0.98, respectively. Subsequently, an independent group of 25 biopsy cores was used for validation of the model, in which mp-MRI had identified suspicious cancer foci. Using the trained model, we predicted the tissue pathology using temporal ultrasound data: 16 out of 17 benign cores, as well as all three higher grade cancer cores, were correctly identified.
Conclusion
The results show that temporal analysis of ultrasound data is potentially an effective approach to complement mp-MRI-TRUS-guided prostate cancer biopsy, specially to reduce the number of unnecessary biopsies and to reliably identify higher grade cancers.
Keywords: Temporal ultrasound data, cancer diagnosis, prostate cancer
1 Introduction
Prostate cancer (PCa) is the most frequently diagnosed malignancy and the third leading cause of cancer related death in North American men. According to the American and Canadian Cancer Societies, 262,000 new cases are diagnosed annually and 33,600 die of PCa in North America. These numbers will nearly double by 2025 when the baby boomer generation reaches the age of peak prevalence. If diagnosed early, PCa can be managed with a long-term disease free survival rate of above 95%1. Early stage PCa, which represents the majority of cases diagnosed today, has many therapy options, including surgery, radiation therapy, brachytherapy, thermal ablation, and active surveillance. Selection of the optimal therapy and therapeutic dosage are chiefly determined by diagnosis and staging. Definitive diagnosis of PCa requires core needle biopsy, typically guided by transrectal ultrasound (TRUS). Current biopsy regimens systematically sample the prostate, by collecting eight or more samples from predefined anatomical locations. PCa is seldom detectable in TRUS, hence the biopsy regimen is scaled to the prostate gland based on its size but otherwise not tailored to the individual. As a result, conventional systematic biopsy under TRUS guidance has rather poor sensitivity, with positive predictive values between 40–60%[5,23]. In order to enable patient-specific targeting, other modes of ultrasound imaging [18], such as radio frequency (RF) data analysis[3], elastography [13,17,19,20], and Doppler imaging[24,28] have been explored. These technologies, individually, have not entirely succeeded in accurate identification of PCa.
Alternative imaging modalities such as MRI have also been used for patient-specific targeting [11]. Guidelines for standardized reporting of PCa assessment based on multi-parametric MRI have been developed, involving simultaneous examination of T2-weighted, dynamic contrast-enhanced, T1-weighted, and diffusion weighted sequences [1]. It has been shown that MRI improves detection of higher grade PCa, by enabling targeting of the cancer foci predetermined in MRI during TRUS-guided biopsy [11]. While the sensitivity of MRI is about 80% in detecting larger sizes of high grade PCa, it cannot reliably identify smaller sizes of high grade cancer or differentiate between cancer grades [4,6].
Recently, we proposed “ultrasound time series” technology for tissue typing, comprising the analysis of a temporal sequence of Radio Frequency (RF) ultrasound frames from a stationary tissue location. This method uses a supervised machine learning framework to determine the correlation between features extracted from the temporal ultrasound data with the tissue label provided by histopathology. In addition to several ex vivo experiments [14, 15], we have demonstrated, in a series of in vivo experiments[7,16,27], that the technique can distinguish benign and cancerous tissue.
In this paper, we aim to demonstrate that the time series technology can be used to augment MRI-TRUS-guided targeted biopsy to reduce the high number of false positives of mp-MRI. In a study consisting of 60 biopsy cores obtained from 38 subjects, we develop a classification model to compare the labels assigned to the prostate tissue from temporal ultrasound data with those determined from mp-MRI, and use histopathological analysis of biopsy cores as the gold standard. Our results demonstrate that the technology is capable of accurately identifying higher grade cancer cases, and all but one of the benign cases.
2 Methods and Materials
2.1 Data
This study was approved by the institutional ethics review board of the National Cancer Institute of the National Institutes of Health (NIH), MD. Subjects eligible for this study were consented and informed appropriately of the potential harms and benefits. We acquired ultrasound time series data from 60 biopsy cores of 38 subjects; 39 cores were non-cancerous, while there were 3, 7, 2, 7, and 2 biopsy cores with Gleason Scores (GSs) of 3+3, 3+4, 4+3, 4+4, and 4+5, respectively, based on pathology results. GS is the most common grading system to determine the level of abnormality, and describes the summation of cancer grades of two areas making up most of the cancer. Clinically, a GS of 4+3 and above is considered a strong indication of clinically significant cancer while a GS of 3+3 and below is an indicator of indolent cancer. Hence, for a tissue typing approach to be effective and most useful clinically, it should be able to reliably identify higher grades of cancer.
2.1.1 Multi-parametric MRI
Prior to the biopsy session, every subject had multi-parametric MR imaging using a 3.0 Tesla MRI scanner (Achieva, Philips, Healthcare, Best, The Netherlands) with a 6-channel cardiac surface coil (SENSE, Philips Healthcare) positioned over the pelvis, and an endorectal coil (BPX-30, Medrad, Pittsburgh, Pennsylvania). Tri-planar T2-weighted, axial dynamic contrast enhanced (DCE), and axial diffusion weighted imaging (DWI) with apparent diffusion coefficient mapping MRI sequences were acquired according to protocol. Details of these imaging sequences have been described previously [25, 26]. Suspicious lesions in MRI were identified and scored by two independent radiologists as low, moderate, and high.
2.1.2 Ultrasound
Temporal ultrasound RF data was acquired on a Philips iU22 US imaging platform during MRI-guided targeted TRUS biopsies using Philips UroNav MR/US fusion system (Fig. 1). Pre-procedure T2-weighted MR images, with delineated biopsy targets, were automatically fused with real-time TRUS images of the prostate of the same subject using UroNav [21]. In the beginning of the biopsy procedure, a series of electromagnetically (EM) tracked 2D TRUS images of the prostate were acquired from the base to the apex of the prostate gland. Next, a 3D US image volume was reconstructed from these images and semi-automatically registered to the MRI scan, in UroNav. See[12,29,30] for details of the registration approach. Following the registration of US and MR volumes, the target biopsy locations were transformed to the subject coordinate frame. During biopsy, the clinician navigated through the prostate volume to reach the desired target location; immediately prior to acquiring the biopsy core, the clinician held the TRUS transducer steady for 2–3 sec to collect RF time series data. Typically, 100 frames of RF time series data were acquired from each biopsy core. RF time series data was collected from at most two MR-identified targets per subject.
Fig. 1.
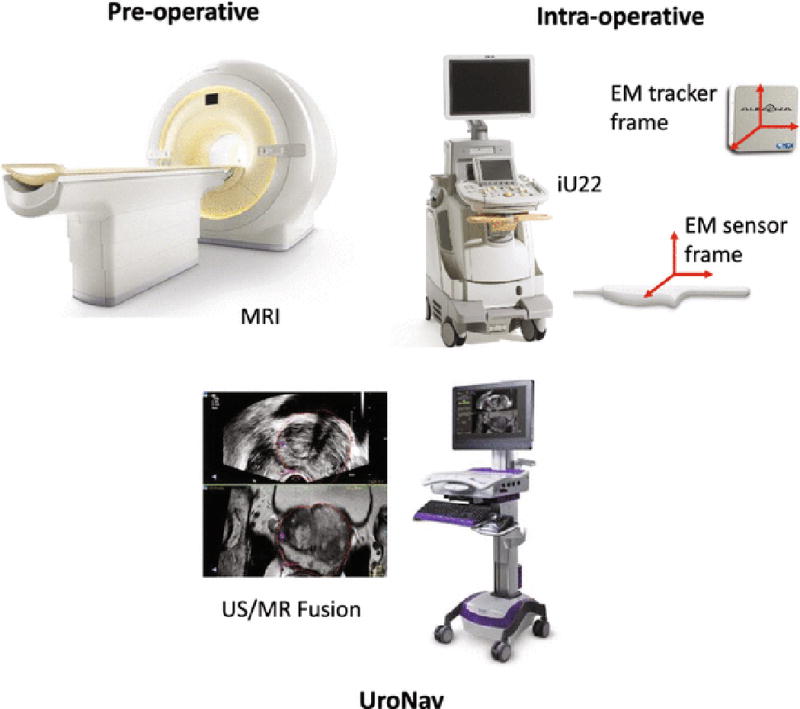
An overview of the MRI-TRUS targeted biopsy system using the Philips UroNav platform.
2.1.3 Ground Truth Labeling based on Histopathology
We pre-determined the cancer foci as targets based on the information from mp-MR. Then, biopsy targets were identified intra-operatively in ultrasound images by fusion of MR and TRUS using UroNav. For every identified target location, two biopsies were acquired from axial and sagittal planes, respectively, and histopathology is reported for each of them. For generating the ground truth labeling, we only considered histopathology reporting in the axial plane, even though for some cases, the histopathology reporting from axial and sagittal planes were not in agreement. Since RF time series data are collocated with biopsy cores, ground truth labeling is automatically registered to this data.
2.1.4 Data Categorization
We used two data sets (referred to as data sets A and B, hereafter) to build and verify a model for cancer detection. To build the model, the aim is to use homogeneous tissue regions. From all available data we, therefore, selected biopsy cores with at least tumor-in-core-length ≥ 8 mm for a core length of 20 mm (or 4 mm for a core length of 10 mm) or benign cases for training our model. 35 cores fulfilled these conditions, which we labeled as data set A, presented in Fig. 2. Data set B contained 25 biopsy cores with any tumor-in-core-length and non-cancerous pathology including fibromuscular, chronic inflammation, atrophy, and Prostatic Intraepithelial Neoplasia (PIN) as presented in Fig. 3.
Fig. 2.
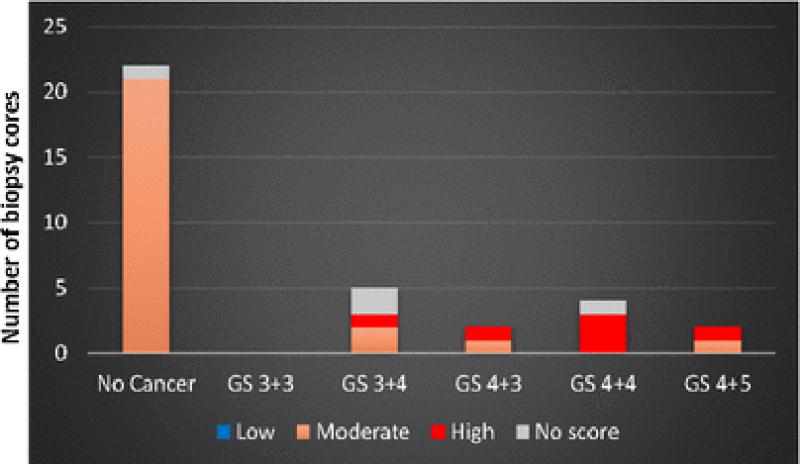
Gleason score distribution and MRI grades of the 35 biopsy cores of data set A used for training a model to characterize cancerous versus non-cancerous cores.
Fig. 3.
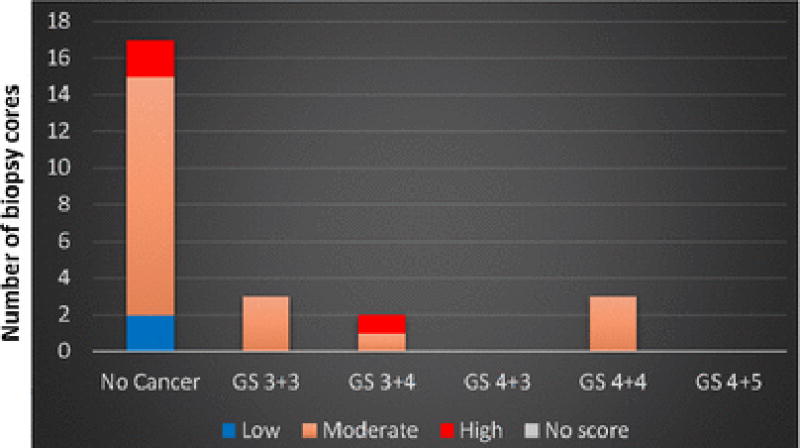
Gleason score distribution and MRI grades of the 25 biopsy cores of data set B used for testing the generated model.
2.2 Regions of Interest Selection
We extracted ultrasound time series features from regions of interest (ROIs) of each biopsy core. To account for possible hand motion during data collection, hence misalignment of ultrasound and pathology data, we opted to analyze about half of the typical biopsy core length (0.5×20 = 10 mm). As a result, we selected an area of 2 mm×10 mm in lateral and axial directions along the needle trajectory and centered on the target in the TRUS image. We then divided the selected 2 mm×10 mm area into 20 ROIs; the size of each ROI was 1 mm×1 mm. The number of samples in the RF data within each ROI varied between 90–480 due to scan conversion and the depth of biopsy cores, while the length of RF time series was 100 time points.
2.3 Feature Extraction
We analyzed the spectrum information of the temporal ultrasound data in order to determine the characteristics of the non-cancerous and cancerous cores. Figure 4 shows typical spectrograms where the x-axis is the frequency content of the temporal data and the y-axis is time. The colormap represents the log-compressed distribution of spectral components with red (blue) referring to the higher (lower) power in the signal, in the spectral band marked by the values of the x-axis. In general, non-cancerous cores present with two trends: they either show high power across the entire spectrum, or high power in a narrow frequency range between 0–2 Hz. On the other hand, cancerous cores present a spectrum that is clearly different. To characterize these phenomena and differentiate the core types based on the spectrum range they cover, we used the short time Fourier transform (STFT) and the Fast Fourier Transform (FFT) of the temporal ultrasound data, averaged for each ROI. STFT helps reserve localized information in time. For instance, since the tissue could be moving during the temporal ultrasound acquisition period, the characteristics of cancerous tissue may only exist part of the time in the temporal ultrasound data, and can only be picked up by STFT.
Fig. 4.
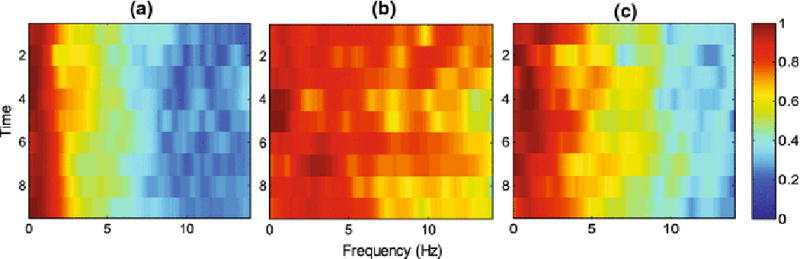
Typical spectrograms for two trends of non-cancerous cores (a and b), and cancerous cores (c).
To generate the features based on STFT, we used two window-lengths of the time series to characterize the spectrum at two different resolutions: 1) We divided the time series signal to six non-overlapping time windows of length 16; 2) we also computed the STFT features based on the analysis of the time series data with time windows of length 32, where there is 24 sample overlap between neighboring windows. We computed the average amplitude of log-compressed, normalized, zero-mean spectrum in frequency bands of 0–2 Hz, 2–4 Hz, 4–6 Hz, 6–8 Hz, and above 8 Hz. Moreover, we used the values of STFT spectrum at different frequencies as individual features.
To compute the features based on the Fourier Transform of the entire time series data, we divided the spectrum to 10 equally spaced frequency bands. In order to identify the spectrum bands with higher power than other bands, we computed the relative ratio of average spectrum between different frequency bands.
2.4 Classification
A Support Vector Machine (SVM) was used to generate a model for characterization of cancer using the ultrasound time series features extracted from biopsy targets. SVM has been demonstrated previously to differentiate various tissue types using temporal ultrasound data in in vivo and ex vivo studies, consistently and with high accuracies [7,8]. To build an SVM model, data is mapped to a higher dimension using a kernel function enabling separation of its two classes with maximum margin2. We used an RBF kernel function which only needs one parameter for initialization [7,8]. Tuning the SVM kernel and parameters provided us the flexibility to: i) choose from various linear and nonlinear transformation of input data and different decision boundaries, and ii) prevent overfitting of the classifier during training. In addition to the binary outputs of the classifier, we computed the estimated likelihoods of the ROIs to be cancerous. The details of this implementation is presented in[10,22].
2.5 Cross-validation
The machine learning approach proposed involved training an SVM model using cancerous and non-cancerous ROIs of a set of biopsy cores from data set A. To generate such model, we heuristically searched for the subset of features that led to the highest classification accuracy on this data.
In order to determine the consistency of selected features in the training data set A, we randomly divided this data set, 500 times, to 80% training and 20% cross-validation data. We computed the average classification result on the cross-validation data of all trials. We refer to this evaluation as five-fold cross-validation analysis.
Finally, we validated the classification model developed using data set A, with the independent data set B and reported its accuracy.
3 Results
Using heuristic search to identify time series features that result in highest classification accuracy for data set A, yielded seven features of STFT and FFT. The best two features are: F1) the summation of the amplitude between 0–2 Hz of the STFT spectrum, and F2) the ratio of the summation of the amplitude between 2–4 Hz over the summation of amplitude between 4–6 Hz of the STFT spectrum. The normalized histogram of these features (F1, F2) are presented in Fig. 5 for 13 cancerous and 22 benign cores of data set A. Table 1 shows the classification results and the area under Receiver Operating Characteristics curve (AUC) for five-fold classification analysis.
Fig. 5.
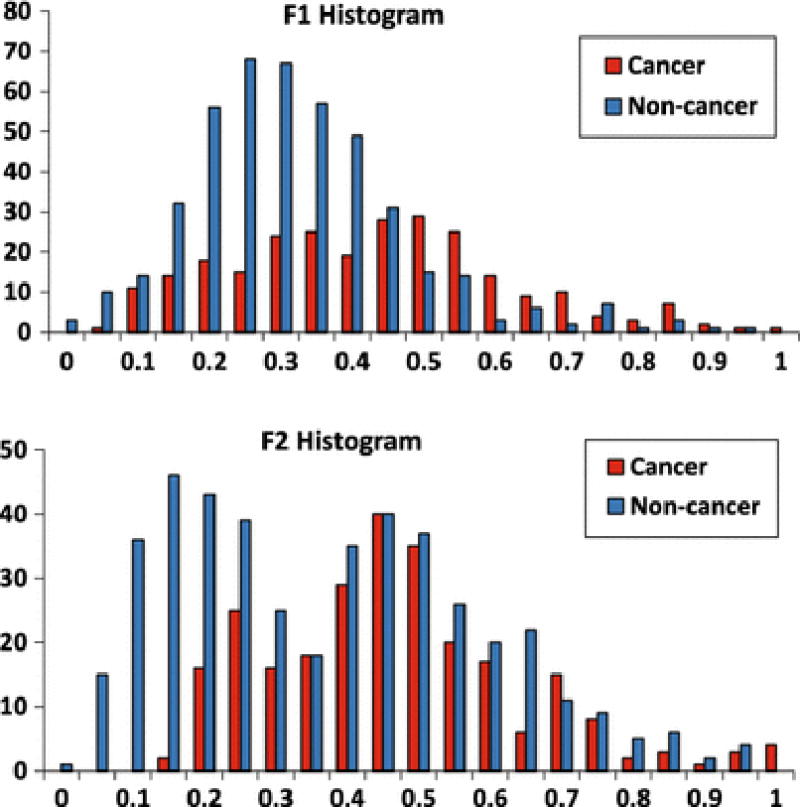
Histograms of two best features in pooled analysis. Each feature range has been normalized between 0 and 1.
Table 1.
Classification results for five-fold cross validation analysis.
| Evaluation | Classification accuracy | Sensitivity | Specificity | AUC |
|---|---|---|---|---|
| Five-fold cross-validation analysis | 94±1% | 93±1% | 96±1% | 0.98 |
Fig. 6 shows the result of validation of the generated model on data set B. Among 17 non-cancerous cores, 16 cases are predicted as non-cancerous and one case identified as cancerous with a threshold of 40% of the model output. In other words, if at least 40% of the ROIs of a core (8 out of 20 ROIs) are predicted by the model as cancerous, we declare the core as cancerous. Moreover, all cases of higher grade cancer with GS of 4+4, irrespective of tumor size, are identified correctly. The only cancerous cases where the model fails to correctly identify are: 1) two cases with less than 1 mm cancerous tissue of GS of 3+3, and 2) one case with GS of 3+4 with core length of 6 mm.
Fig. 6.
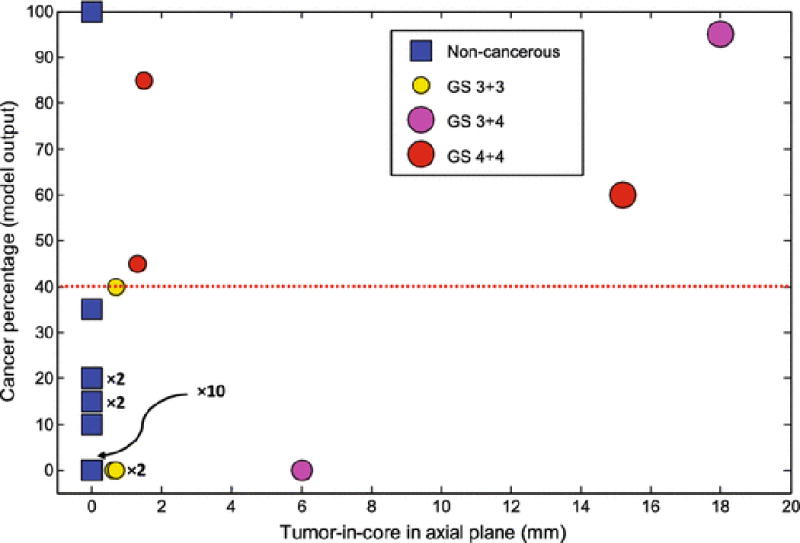
Cancer percentage of the model output versus tumor-in-core-length for validation data. Cases with GS of 3+3, 3+4, and 4+4, and benign are in different colors of yellow, magenta, red, and blue. Note that there are cases that are placed at the same location of the graph, ×m indicate m cases that place at the same location.
Fig. 7 and Table 2 present a different perspective of validation on data set B, where we compare the output of the model with the grade assigned to each suspicious cancer foci in MRI. Results show that for MRI grade of moderate, which encapsulates the majority of subjects, our approach can improve the sensitivity of MRI for detecting higher grade cancer cases. Moreover, except one benign case with MRI grade of low, the model correctly predicts the label of biopsy cores for cases with MRI grades of low and high.
Fig. 7.
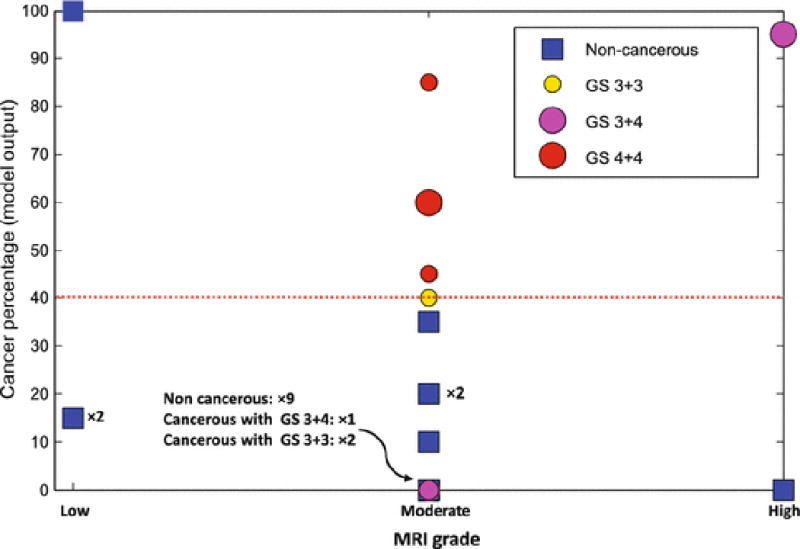
Cancer percentage of the model output versus MRI grade for validation data. The size of circles for cancerous cases indicates tumor-in-core-length; the larger circles have higher tumor lengths. Cases with GS of 3+3, 3+4, and 4+4, and benign are in different colors of yellow, magenta, red, and blue. Note that there are cases that are placed at the same location of the graph. ×m indicate m cases that place at the same location.
Table 2.
Model performance for classification of testing cores for different MRI grades.
| MRI grade | Non-cancerous | GS 3+3 | GS 3+4 | GS 4+4 | Predicted correctly |
|---|---|---|---|---|---|
| Low | 3 | 0 | 0 | 0 | 2 out of 3 |
| Moderate | 13 | 3 | 1 | 3 | 17 out of 20 |
| High | 1 | 0 | 1 | 0 | 2 out of 2 |
In this study, we have set the following parameters for characterization of PCa: i) SVM penalty factor = 11, ii) RBF kernel parameter = 2, iii) ROI size = 1 mm×1 mm, and iv) threshold of model output for cancerous core detection = 40%.
4 Discussion and Conclusion
In this paper, we presented an approach to augment mp-MRI-TRUS-guided biopsy with the analysis of temporal ultrasound data to improve cancer yield. In a study consisting of 60 biopsy cores, we developed a classification model to predict the pathology label of each core solely based on ultrasound time series data.
Using the pre-procedure acquired mp-MR images, suspicious prostate regions were delineated. Then, we identified the MRI identified targets in ultrasound images using the UroNav system. We built an SVM model for characterization of biopsy cores using a set of features determined from time series data of 35 biopsy cores. The model was validated over an independent set of 25 biopsy cores. As presented in Fig. 6 and Fig. 7, the results indicate that for the cases with MRI grade of moderate, the model can improve the sensitivity of mp-MRI. Most importantly, all higher grade cancer cases with GS of 4+4 were successfully identified. The model failed to detect two cases with GS of 3+3 and tumor length of less than 1 mm, and one case with GS of 3+4. All non-cancerous cases with moderate MRI grade were identified correctly. Moreover, for the three non-cancerous cases with low MRI grade, the model predicted the label of two cases correctly.
We emphasize that this study has focused on distinguishing cancerous from non-cancerous biopsy cores, where all cases had identified MR-positive lesions. The data set is also moderate suspicious dominant, where the cancer detection rate was only approximately 67% (per subject) or 40% (per suspicious cancer foci) by radiologists within our subject population. Our results strongly support the promise of ultrasound time series technology to complement mp-MRI for MRI-TRUS guided targeted prostate biopsy.
In this paper, we built a model for characterization of cancer using data set A by tuning the classifier penalty factor, and RBF kernel parameter. Through a comprehensive search in the parameter space, we determined those parameters that provided us with the best classification results in our training data set. In our previous study, we have shown that classification results achieved using random forest is similar to those obtained with SVM [27].
A challenge with the time series technology has been to explain the physical phenomenon underlying its interaction with tissue, and the interpretation of the derived features from this temporal signal. The interpretation is especially difficult in prostate tissue which is highly heterogeneous, and includes fundamentally different micro-structures within cancerous and non-cancerous regions. Even though the exact physical model for the time series interaction with tissue is not immediately available, we have previously studied the response of tissue to the time series signals [14]. We have shown that it is highly dependent on the density and size of scatterers in the tissue as well as the speed of sound. We have also shown that tissue temperature increase, induced by acoustic radiation during time series acquisition, plays a role in the physical phenomenon governing the tissue typing properties of this technology [2]. We are further exploring the understanding of this interaction through simulation and laboratory experiments. This paper is the first study where we show features that distinguish cancerous cores well, correlate directly with the response of the tissue to time series (Fig. 4).
To characterize PCa using temporal ultrasound data, RF time series should be collected without motion. Although in the current study, we removed the cases with considerable motion between RF frames, in the future, we are planning to: i) increase the frame rate and decrease the acquisition time to reduce the effect of hand motion, breathing, and perfusion, and ii) apply a deformable registration to minimize the impact of in-plane registration. Other future work will focus on increasing the size of the data set by acquiring temporal ultrasound data not only from the target biopsy locations identified by mp-MRI, but also from the standard sextant biopsy locations.
Currently, ultrasound time series data is acquired prior to the start of the biopsy procedure from registered mp-MRI targets. Naturally, there is a time lap between the acquisition of time series and obtaining the biopsy core sample. We expect that this process can lead to possible misalignment between the tissue location where the time series data was obtained from, and the tissue sample of the biopsy core. To minimize the effect of possible misalignment, we chose ROIs from a length of 10 mm along the needle trajectory centered on the biopsy target. The typical length of a biopsy core is 18 mm. Modifications in the ultrasound hardware are underway to enable the acquisition of time series data in real time during the biopsy procedure. The chosen ROI size (1 mm×1 mm) might impact the classification accuracy for the biopsy cores with smaller tumor size. As shown in Fig. 6, the model failed to detect two biopsy cores with tumor-in-core-length < 1 mm. However, the effect of misalignment on the classification results for biopsy cores with larger tumor size does not appear to be significant.
One should note that the registration accuracy between MRI and TRUS could affect our analyses that compare MRI grades with the model output. It is possible that the actual biopsies have been obtained from different tissue locations than what were identified as suspicious based on mp-MRI. In [9], we provided an accuracy assessment of TRUS-MRI registration as it is used in the UroNav guidance interface. In that study, based on data obtained from 10 patients, the mean registration error was determined as 2.4 mm. However, such error does not affect the results obtained from temporal ultrasound data analysis; this analysis is independent of MRI, and is verified directly using the histopathological information of the tissue samples obtained within the ultrasound image coordinates.
Acknowledgments
The authors would like to acknowledge the help of Jochen Kruecker and Pingkun Yan for assisting with the experiments. This work was supported in part by the Natural Sciences and Engineering Research Council of Canada (NSERC), and the Canadian Institutes of Health Research (CIHR).
Footnotes
Canadian cancer society: http://www.cancer.ca/, and American cancer society: http://www.cancer.org
A Library for SVMs: http://www.csie.ntu.edu.tw/~cjlin/libsvm/
Conflict of Interest The authors declare that they have no conflict of interest.
Ethical Approval All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
References
- 1.Barentsz J, Richenberg J, Clements R, Choyke P, Verma S, Villeirs G, Rouviere O, Logager V, Futterer J. ESUR prostate MR guidelines. European Radiology. 2012;22(4):746–757. doi: 10.1007/s00330-011-2377-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daoud M, Mousavi P, Imani F, Rohling R, Abolmaesumi P. Tissue classification using ultrasound-induced variations in acoustic backscattering features. IEEE Transactions on Biomedical Engineering. 2013;60(2):310–320. doi: 10.1109/TBME.2012.2224111. [DOI] [PubMed] [Google Scholar]
- 3.Feleppa E, Porter C, Ketterling J, Dasgupta S, Ramachandran S, Sparks D. Recent advances in ultrasonic tissue-type imaging of the prostate. Acoustical Imaging. 2007;28:331–339. [Google Scholar]
- 4.Futterer J, Heijmink S, Scheenen T, Veltman J, Huisman H, Vos P, Hulsbergen-Van C, Witjes J, Krabbe P, Heerschap A, Barentsz J. Prostate cancer localization with dynamic contrast-enhanced MR imaging and proton MR spectroscopic imaging. Radiology. 2006;241(2):449–458. doi: 10.1148/radiol.2412051866. [DOI] [PubMed] [Google Scholar]
- 5.Goossen T, Wijkstra H. Transrectal ultrasound imaging and prostate cancer. Archivio italiano di urologia andrologia. 2003;75(1):68–74. [PubMed] [Google Scholar]
- 6.Hricak H, Choyke P, Eberhardt S, Leibel S, Scardino P. Imaging prostate cancer: a multidisciplinary perspective. Radiology. 2007;243(1):28–53. doi: 10.1148/radiol.2431030580. [DOI] [PubMed] [Google Scholar]
- 7.Imani F, Abolmaesumi P, Gibson E, Khojasteh A, Gaed M, Moussa M, Gomez J, Romagnoli C, Siemens D, Leveridge M, Chang S, Fenster A, Ward A, Mousavi P. Ultrasound-based characterization of prostate cancer: an in vivo clinical feasibility study. Medical Image Computing and Computer-Assisted Intervention. 2013:279–286. doi: 10.1007/978-3-642-40763-5_35. [DOI] [PubMed] [Google Scholar]
- 8.Imani F, Abolmaesumi P, Wu M, Lasso A, Burdette E, Ghoshal G, Heffter T, Williams E, Neubauer P, Fichtinger G, Mousavi P. Ultrasound-guided characterization of interstitial ablated tissue using RF time series: feasibility study. IEEE Transactions on Biomedical Engineering. 2013;60(6):1608–18. doi: 10.1109/TBME.2013.2240300. [DOI] [PubMed] [Google Scholar]
- 9.Krucker J, Xu S, Glossop N, Guion P, Choyke P, Ocak I, Singh AK, Wood BJ. Fusion of realtime transrectal ultrasound with pre-acquired MRI for multi-modality prostate imaging. SPIE. 2007:650,912–1. [Google Scholar]
- 10.Lin H, Lin C, Weng R. A note on platt’s probabilistic outputs for support vector machines. Technical report, Department of Computer Science, National Taiwan University. 2003;68:267–276. [Google Scholar]
- 11.Margel D, Yap S, Lawrentschuk N, Klotz L, Haider M, Hersey K, Finelli A, Zlotta A, Trachtenberg J, Fleshner N. Impact of multiparametric endorectal coil prostate magnetic resonance imaging on disease reclassification among active surveillance candidates: a prospective cohort study. The Journal of Urology. 2012;187(4):1247–1252. doi: 10.1016/j.juro.2011.11.112. [DOI] [PubMed] [Google Scholar]
- 12.Marks L, Young S, Natarajan S. MRI-ultrasound fusion for guidance of targeted prostate biopsy. Current Opinion in Urology. 2013;23(1):43–50. doi: 10.1097/MOU.0b013e32835ad3ee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyagawa T, Tsutsumi M, Matsumura T, Kawazoe N, Ishikawa S, Shimokama T, Miyanaga N, Akaza H. Real-time elastography for the diagnosis of prostate cancer: evaluation of elastographic moving images. Japanese Journal of Clinical Oncology. 2009;39(6):394–398. doi: 10.1093/jjco/hyp026. [DOI] [PubMed] [Google Scholar]
- 14.Moradi M, Abolmaesumi P, Mousavi P. Tissue typing using ultrasound RF time series: Experiments with animal tissue samples. Journal of Medical Physics. 2010;37(8):4401–4413. doi: 10.1118/1.3457710. [DOI] [PubMed] [Google Scholar]
- 15.Moradi M, Abolmaesumi P, Siemens DR, Sauerbrei EE, Boag A, Mousavi P. Augmenting detection of prostate cancer in transrectal ultrasound images using SVM and RF time series. IEEE Transactions on Biomedical Engineering. 2009;56(9):2214–2223. doi: 10.1109/TBME.2008.2009766. [DOI] [PubMed] [Google Scholar]
- 16.Moradi M, Mahdavi S, Nir G, Jones E, Goldenberg S, Salcudean S. Ultrasound RF time series for tissue typing: first in vivo clinical results. SPIE 8670, Medical Imaging. 2013:86,701I1–8. [Google Scholar]
- 17.Moradi M, Mahdavi S, Nir G, Mohareri O, Koupparis A, Gagnon L, Casey R, Ischia J, Jones E, Goldenberg S, Salcudean S. Multiparametric 3D in vivo ultrasound vibroelastography imaging of prostate cancer: Preliminary results. Medical Physics. 2014;41(7):073,505-1–073,505-12. doi: 10.1118/1.4884226. [DOI] [PubMed] [Google Scholar]
- 18.Moradi M, Mousavi P, Abolmaesumi P. Computer-aided diagnosis of prostate cancer with emphasis on ultrasound-based approaches: a review. Ultrasound in Medicine and Biology. 2007;33(7):1010–1028. doi: 10.1016/j.ultrasmedbio.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 19.Ophir J, Garra B, Kallel F, Konofagou E, Krouskop T, Righetti R, Varghese T. Elastographic imaging. Ultrasound in Medicine and Biology. 2000;26:S23–S29. doi: 10.1016/s0301-5629(00)00156-3. [DOI] [PubMed] [Google Scholar]
- 20.Pallwein L, Mitterberger M, Struve P, Pinggera G, Horninger W, Bartsch G, Aigner F, Lorenz A, Pedross F, Frauscher F. Real-time elastography for detecting prostate cancer: preliminary experience. BJU International. 2007;100(1):42–6. doi: 10.1111/j.1464-410X.2007.06851.x. [DOI] [PubMed] [Google Scholar]
- 21.Pinto P, Chung P, Rastinehad A. Ultrasound fusion guided prostate biopsy improves cancer detection following transrectal ultrasound biopsy and correlates with multiparametric magnetic resonance. The Journal of Urology. 2011;186(4):1281–1285. doi: 10.1016/j.juro.2011.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Platt J. Probabilistic outputs for support vector machines and comparisons to regularized likelihood methods. Advances in Large Margin Classifiers. 1999;10(3):61–74. [Google Scholar]
- 23.Rapiti E, Schaffar R, Iselin C, Miralbell R, Pelte M, Weber D, Zanetti R, Neyroud-Caspar I, Bouchardy C. Importance and determinants of Gleason score undergrading on biopsy sample of prostate cancer in a population-based study. BMC Urology. 2013;13:13–19. doi: 10.1186/1471-2490-13-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheipers U, Ermert H, Sommerfeld H, Garcia-Schurmann M, Senge T, Philippou S. Ultrasonic multifeature tissue characterization for prostate diagnostics. Ultrasound in Medicine and Biology. 2003;29(8):1137–49. doi: 10.1016/s0301-5629(03)00062-0. [DOI] [PubMed] [Google Scholar]
- 25.Turkbey B, Pinto P, Mani H, Bernardo M, Pang Y, McKinney Y, Khurana K, Ravizzini G, Albert P, Merino M, Choyke P. Prostate cancer: value of multiparametric MR imaging at 3 T for detection histopathologic correlation. Radiology. 2010;255(1):89–99. doi: 10.1148/radiol.09090475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turkbey B, Shah V, Pang Y, Bernardo M, Xu S, Kruecker J, Locklin J, Baccala AJ, Rastinehad A, Merino M, Shih J, Wood B, Pinto P, Choyke P. Is apparent diffusion coefficient associated with clinical risk scores for prostate cancers that are visible on 3-T mr images? Radiology. 2011;258(2):488–495. doi: 10.1148/radiol.10100667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uniyal N, Imani F, Tahmasebi A, Agarwal H, Bharat S, Yan P, Kruecker J, Kwak JT, Xu S, Wood B, Pinto P, Turkbey B, Choyke P, Abolmaesumi P, Mousavi P, Moradi M. Ultrasound-based prediction of prostate cancer in MRI-guided biopsy. Medical Image Computing and Computer-Assisted Intervention Workshop on Clinical Image-based Procedures. 2014 [Google Scholar]
- 28.Xie S, Li H, Du J, Xia J, Guo Y, Xin M, Li F. Influence of serum prostate-specific antigen (PSA) level, prostate volume, and PSA density on prostate cancer detection with contrast-enhanced sonography using contrast-tuned imaging technology. Journal of Ultrasound in Medicine. 2013;32(5):741–748. doi: 10.7863/ultra.32.5.741. [DOI] [PubMed] [Google Scholar]
- 29.Xu S, Kruecker J, Guion P, Glossop N, Neeman Z, Choyke P, Singh AK, Wood B. Closed-loop control in fused MR-TRUS image-guided prostate biopsy. Medical Image Computing and Computer-Assisted Intervention. 2007:128–135. doi: 10.1007/978-3-540-75757-3_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu S, Kruecker J, Turkbey B, Glossop N, Singh AK, Choyke P, Pinto P, Wood B. Real-time MRI-TRUS fusion for guidance of targeted prostate biopsies. Computer Aided Surgery. 2008;13(5):255–264. doi: 10.1080/10929080802364645. [DOI] [PMC free article] [PubMed] [Google Scholar]


