Abstract
An experimental design mimicking natural plant-microbe interactions is very important to delineate the complex plant-microbe signaling processes. Arabidopsis thaliana-Agrobacterium tumefaciensprovides an excellent model system to study bacterial pathogenesis and plant interactions. Previous studies of plant-Agrobacterium interactions have largely relied on plant cell suspension cultures, the artificial wounding of plants, or the artificial induction of microbial virulence factors or plant defenses by synthetic chemicals. However, these methods are distinct from the natural signaling in planta, where plants and microbes recognize and respond in spatial and temporal manners. This work presents a hydroponic cocultivation system where intact plants are supported by metal mesh screens and cocultivated with Agrobacterium. In this cocultivation system, no synthetic phytohormone or chemical that induces microbial virulence or plant defense is supplemented. The hydroponic cocultivation system closely resembles natural plant-microbe interactions and signaling homeostasis in planta. Plant roots can be separated from the medium containing Agrobacterium, and the signaling and responses of both the plant hosts and the interacting microbes can be investigated simultaneously and systematically. At any given timepoint/interval, plant tissues or bacteria can be harvested separately for various "omics" analyses, demonstrating the power and efficacy of this system. The hydroponic cocultivation system can be easily adapted to study: 1) the reciprocal signaling of diverse plant-microbe systems, 2) signaling between a plant host and multiple microbial species (i.e. microbial consortia or microbiomes), 3) how nutrients and chemicals are implicated in plant-microbe signaling, and 4) how microbes interact with plant hosts and contribute to plant tolerance to biotic or abiotic stresses.
Keywords: Molecular Biology, Issue 125, Molecular plant-microbe interaction, Agrobacterium tumefaciens, Arabidopsis thaliana, hydroponics, co-cultivation, plant stress response, pathogenicity, bacteria, microbiome
Introduction
Plant-associated microbes play important roles in biogeochemical cycling, bioremediation, mitigation of climate change, plant growth and health, and plant tolerance to biotic and abiotic stresses. Microorganisms interact with plants both directly through plant cell wall contact and indirectly via chemical secretion and signaling1,2,3. As sessile organisms, plants have developed direct and indirect mechanisms to resist infection by pathogens. Direct defenses include structural defenses and the expression of defense proteins, whereas indirect defenses include secondary plant metabolite production and the attraction of organisms antagonistic to invading pathogens4,5. Plant-derived root exudates, secretions, mucilages, mucigel, and lysates alter the physical-chemical properties of the rhizosphere to attract or repel microbes towards their hosts6. The chemical composition of root secretion is species-specific, thereby serving as a selective filter that allows certain microorganisms capable of recognizing such compounds to flourish in the rhizosphere6. Thus, compatible microbial species may be stimulated to activate and enhance their associations, either to the benefit or detriment of the plant host1.
Understanding plant-microbe interactions in the rhizosphere is key to enhancing plant productivity and ecosystem functioning, since a majority of the microbial and chemical exposure occurs at the root structure and soil-air interface2,6,7,8. However, the examination of subterranean plant-microbe interactions and reciprocal responses has been a challenge due to its intriguingly complex and dynamic nature and the lack of suitable experimental models with natural root structure and plant morphology under tightly controllable growth conditions. As one of the most heavily studied phytopathogens, Agrobacterium infects a wide range of plants with agricultural and horticultural importance, including cherry, apple, pear, grape, and rose9. Agrobacterium is an important model organism for understanding plant-pathogen interactions and is a powerful tool in plant transformation and plant engineering10,11,12,13,14.
Molecular plant-Agrobacterium interactions have been well studied for several decades, and the current understanding of Agrobacterium pathogenicity is extensive9,11,15,16. Agrobacterium pathogenicity is largely attributed to its evolved capabilities of perceiving plant-derived signals, resulting in the fine modulation of its virulence program and cell-to-cell communication, so-called quorum sensing17. The Agrobacterium virulence program is regulated by several signals available in the rhizosphere and involves two sets of 2-component systems, the ChvG/I system and the VirA/G system. Acidic conditions in the rhizosphere activate the transcription of chvG/I, virA/G, and several other genes involved in Agrobacterium pathogenicity, including virE0, virE1, virH1, virH2, and genes of the type VI secretion system (T6SS)18. Plant-derived phenolic compounds, including acetosyringone (4'-hydroxy-3',5'-dimethoxyacetophenone), activate the VirA/G 2-component system through phosphorylation signaling mechanisms19. VirA/G then activates the entire vir regulon, resulting in the transfer and integration of a ~20 kb bacterial DNA fragment called transfer DNA (T-DNA) from its tumor-inducing (Ti) plasmid into the plant nucleus16. T-DNA carries genes responsible for the synthesis of the plant hormones indole-3-acetic acid (IAA) (iaaM and iaaH) and cytokinin (ipt), and once expressed in plant cells, large amounts of these phytohormones are produced. This results in abnormal tissue proliferation and plant tumor development, known as crown gall disease, which is a chronic and resurgent problem for plants9,11,20. IAA also acts collectively with salicylic acid and gamma-amino butyric acid to repress Agrobacterium virulence or to reduce Agrobacterium quorum sensing (QS)17,21,22. To counter this repression, T-DNA also carries genes for opine biosynthesis, which activates Agrobacterium quorum sensing to promote Agrobacterium pathogenicity and also serves as a nutrient source for the pathogen22,23.
Despite an overall deep understanding of Agrobacterium-plant interactions and the resultant T-DNA transfer into the plant host, the complex signaling events at the initial stage of interaction are less well understood. This is partially due to the limitations of conventional approaches to investigate Agrobacterium-plant signaling. Plant cell suspension cultures and artificial site-specific wounding are commonly used to study molecular plant-microbe interactions24,26,27. However, cell suspensions lack typical plant morphology; in particular, plant suspension cells do not have root structures and root exudates, which are very important for activating microbial chemotaxis and virulence28,29. The maintenance of plant morphology and root structure has been addressed by artificially wounding plants, which facilitates site-specific infection, resulting in the detection of induced plant defense-related genes in directly infected plant tissue30,31. However, artificial wounding is significantly different from pathogen infection in nature, particularly as wounding leads to jasmonic acid (JA) accumulation, which systemically interferes with natural plant signaling and defense26. In addition, synthetic chemicals are typically used to artificially induce plant host responses or pathogen virulence. Although the supplementation of such chemical compounds reflective of concentrations in planta is possible, such supplementation does not account for the diffusion of root exudates gradually into the surrounding rhizosphere, which generates a chemotactic gradient sensed by microbes28,32. Given the limitations of conventional approaches to study plant-microbe interactions, the accuracy and depth of the data obtained might be impeded and restrictive, and the knowledge generated from the conventional approaches may not translate directly in planta. Many aspects of plant-Agrobacterium signaling are not yet fully understood, particularly at the early stage of interactions, when the disease symptoms have not yet developed.
To amend the limitations of conventional approaches, this work presents an inexpensive, tightly controllable, and flexible hydroponic cocultivation system that allows researchers to gain deeper insights into the complex signaling and response pathways at the initial stage of molecular plant-microbe interactions. Hydroponics has been widely used to study plant nutrients, root exudates, growth conditions, and the effects of metallic toxicity on plants33,34. There are several advantages of hydroponic models, including the small spatial requirements, the accessibility of various plant tissues, the tight control of nutrient/environmental conditions, and the pest/disease control. Hydroponic systems are also less limiting to plant growth in comparison to agar/phytoagar plating techniques, which typically restrict growth after 2-3 weeks. Importantly, the maintenance of whole-plant structures facilitates the natural root secretion necessary for microbial chemotaxis and virulence induction8,29. The system described here is simpler and less labor-intensive than the alternatives33,34. It uses fewer parts and does not require any tools other than standard scissors. It uses metal mesh (as opposed to nylon33) as a strong support for plant growth and a simple method of aeration under sterile conditions through shaking to support microbial growth. In addition, the system can use metal mesh of various sizes to support plant growth, which accommodates diverse plant species without restricting the width of their roots.
In the hydroponic cocultivation system presented here, plants are cultivated in a sterile hydroponic system where the plant roots secrete organic compounds supporting the growth of inoculated bacteria. In this cocultivation system, no artificial chemicals, such as plant hormones, defense elicitor, or virulence-inducing chemicals, are supplemented, which reflects the natural cell-signaling homeostasis during plant-microbe interactions. With this hydroponic cocultivation system, it was possible to simultaneously determine gene expression in Arabidopsis thaliana Col-0 root tissue upon infection by Agrobacterium, as well as the activation of Agrobacterium genes upon cocultivation with Arabidopsis. It was further demonstrated that this system is suitable to study Agrobacterium attachment to plant roots, as well as the plant root secretome profile, upon cocultivation (infection) with Agrobacterium (Figure 1).
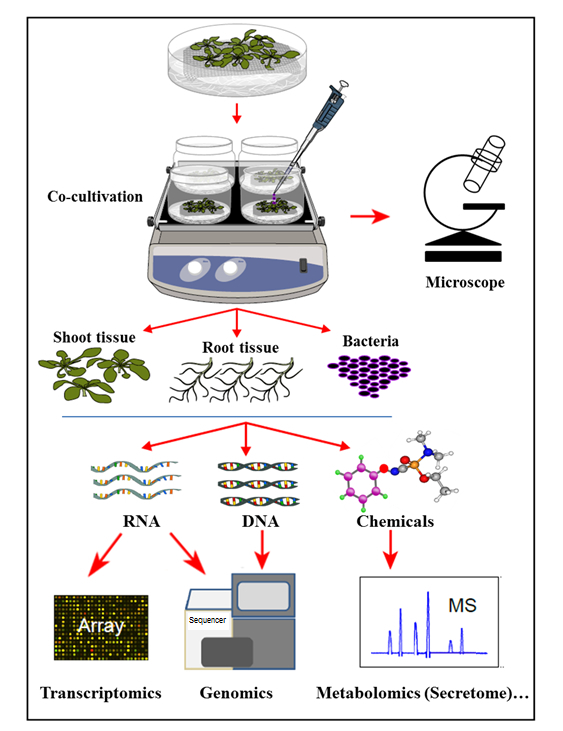
Figure 1: Overview of the Hydroponic Cocultivation System, with Sample Analyses. Plants are grown on top of the mesh (shoots above the mesh), with the roots immersed in hydroponic medium that is then inoculated with bacteria for coculture. Plant tissues and bacteria are then separated for simultaneous extractions and analyses. This figure has been modified from reference35.
Protocol
1. Experimental Planning
Determine the specific goals of the experiment.
- Read through the entire protocol below. Determine which sections are relevant to the specific goals and whether any modifications need to be made. See below for examples.
- Consider whether the experiments will use A. thaliana plants and A. tumefaciens bacterium, as below, or another plant-microbe combination. NOTE: While various plant species can be used, the thickness of the plant roots may require a different-sized mesh (step 3.3), and the growth parameters (steps 3.10-3.11, and 4.4) and hydroponic tank size and medium volume (step 4.2) may also require adjustment (Figure 2). Microbes can easily be inoculated as pure cultures, mixed species (consortia), or from environmental samples (microbiomes), but any changes can affect the protocol, particularly step 4.5.
- Consider the types of experiments that will be used to analyze the samples. NOTE: Fluorescence microscopy (step 5) requires organisms that are labeled with a fluorescent reporter construct.
Design appropriate controls. Include hydroponic tanks without seedlings that are inoculated with control bacteria and tanks with control seedlings that are not inoculated with bacteria.
Plan experiments with the appropriate number of seeds and hydroponic tanks. Consider controls, biological replicates (3 at minimum), and amounts of tissue required for all analyses.
Determine the appropriate length of cocultivation for the experimental goals (step 4.8).
Revise any steps below, as required.
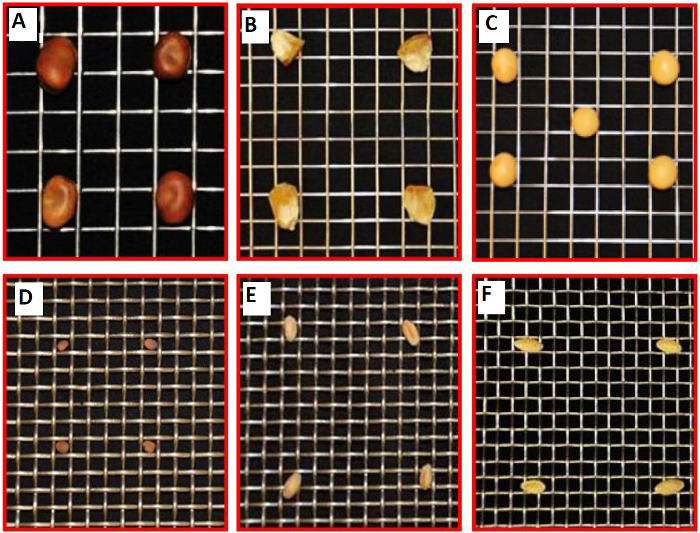
Figure 2. Examples of Other Plants that Could be Cultivated in the Hydroponic System, Supported by a Platform of Metal Mesh. Compatibility of a set of metal meshes for a variety of plant seeds and cultivation. (A) Stainless-steel type 304 weldmesh 3×3 mesh ×.047" dia wire for Vicia faba. (B) Stainless-steel type 304 weldmesh 4×4 mesh ×.035" dia wire for Zea mays. (C) Stainless-steel type 304 weldmesh 4×4 mesh ×.032" dia wire for Glycine max (soybean). (D) Stainless-steel type 304 weldmesh 6×6 mesh ×.047" dia wire for Raphanus sativus (winter radish). (E) Stainless-steel type 304 weldmesh 6×6 mesh ×.047" dia wire for Triticum spp. (F) Stainless-steel type 304 weldmesh 6×6 mesh ×.035" dia wire for Cucumis sativus. This figure has been modified from reference35. Please click here to view a larger version of this figure.
2. Plant Seed Surface Sterilization
Vortex (at high speed) approximately 200 seeds of A. thaliana with 500 µL of deionized water in a microcentrifuge tube. For plant species with larger seeds, use a large tube to facilitate the surface sterilization.
Centrifuge this at approximately 9,000 x g for 30 s in a table-top microcentrifuge. Remove the water (supernatant) using a pipette.
Add 300 µL of 2% sodium hypochlorite to the seeds and vortex. Leave at room temperature for 1 min. For plant species with larger seeds, use larger volumes to cover the seeds.
Centrifuge at approximately 9,000 x g for 30 s in a table-top microcentrifuge. Remove the sodium hypochlorite solution using a pipette.
Add 500 µL of sterile, deionized water to the seeds and vortex. Centrifuge at approximately 9,000 x g for 30 s and remove the water using a sterile pipette.
Repeat step 2.5 an additional 4 times.
Add 500 µL of 70% ethanol to the seeds and vortex. Leave at room temperature for 1 min.
Centrifuge at approximately 9,000 x g for 30 s in a table-top microcentrifuge. Remove the 70% ethanol using a pipette.
Repeat step 2.5 an additional 5 times.
Resuspend the sterilized seeds in 500 µL of sterile, ultrapure water.
3. Seed Germination and Semi-solid Cultivation of Plants
Prepare semi-solid Murashige and Skoog (MS) medium: 2.165 g/L MS basal salts; 10 g/L sucrose; 0.25 g/L MES; and 59 mL/L B5 vitamin mix, pH 5.75, with 4 g/L phytoagar.
Autoclave the MS medium. Pour 25 mL of it into each sterile deep Petri dish (100 x 25 mm2).
For each Petri dish, cut out a 90 x 90 mm square of stainless-steel mesh. NOTE: The recommended mesh for A. thaliana has a grade of 304 (standard grade), mesh count (number of holes per linear inch) of 40 x 40, and a wire diameter of 0.01" (0.0254 cm). A larger mesh square is required for larger hydropinic tanks or higher platforms.
Bend the corners of each square mesh just enough so that the mesh fits inside the Petri plate. Bend the corners at a 90° angle to the bulk of the mesh to allow the mesh to be propped up by the corners, leaving enough space below for root development (Figure 3a).
Put the cut mesh squares in a beaker, cover with aluminum foil, and sterilize by autoclaving using a 30 min dry cycle.
Once the medium has solidified in the Petri dishes and the mesh is sterile, place each sterilized mesh square on top of the semi-solid medium, with the bent corners facing down.
Push the mesh so that the corners penetrate the medium and the bulk of the mesh touches the top surface of the medium (Figure 3b).
Use a 200 µL transfer pipette to draw up individual surface-sterilized A. thaliana seeds (seeds will stay on the tip of the pipette due to vacuum force) and gently transfer each seed on top of the mesh. Place the seeds so that they are spaced appropriately for experimental needs (e.g., 4 6 seeds/plate).
Seal the Petri plates with lids, placing porous surgical tape around the edges.
Stratify the seeds by placing the entire Petri dish at 4 °C for 2 d in the dark (e.g., wrap in tinfoil to simulate darkness.)
Cultivate the seeds in the Petri dish at 22-24 °C with a 16 h photoperiod for 10-14 days (Figure 3c).
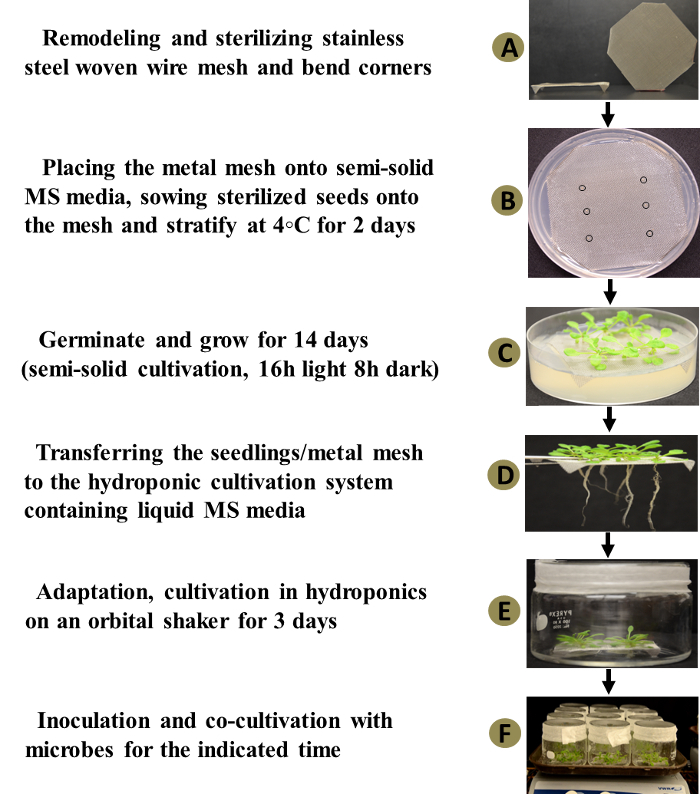
Figure 3: Hydroponic Plant-microbe Cocultivation System. The left panel represents a flow chart outlining the six key steps in assembling and operating the hydroponic co-cultivation system. The right panel demonstrates the actual experimental materials, equipment, and operational procedures for the hydroponic cocultivation system when studying Arabidopsis-Agrobacterium interactions. The inoculation step and final step for plant or bacterial sampling are now shown. This figure has been modified from reference35. Please click here to view a larger version of this figure.
4. Hydroponic Cocultivation System
Prepare liquid Murashige and Skoog (MS) medium: 2.165 g/L MS basal salts, 10 g/L sucrose, 0.25 g/L MES, and 59 mL/L B5 vitamin mix, pH 5.75.
Autoclave the medium. Pour 18 mL of it into each sterile cylindrical glass or clear plastic tank (crystallizing dishes, 100 x 80 mm2) with sterile lids. NOTE: Presterilize the tanks and lids by wrapping them in aluminum foil and autoclaving.
In a sterilized flow hood, use sterile forceps to gently transfer each mesh square with 10-14 day-old seedlings from the semi-solid medium to a hydroponic cylindrical tank (Figure 3d). Seal the lids onto the tanks using porous surgical tape.
- Cultivate the seedlings on the mesh in the tank for 72 h at 22-24 °C, with a 16 h photoperiod and shaking at 50 rpm for aeration (Figure 3e). NOTE: This allows plants to adapt to the hydroponic environment prior to inoculation (cocultivation) with microorganisms. This waiting period will also allow accidental microbial contamination to be apparent prior to the inoculation.
- Begin the next step the day prior to the end of the incubation period.
Grow A. tumefaciens in AB medium (0.5% w/v yeast extract, 0.5% w/v tryptone, 0.5% w/v sucrose, 50 mM MgSO4, pH 7.0) at 28 °C with shaking overnight until the culture reaches a final optical density of 1.0 at 600 nm (OD600), as measured using a spectrophotometer, with uninoculated AB medium as a blank. NOTE: An OD600 of 1.0 is equivalent to approximately 109 cells/mL.
Wash the A. tumefaciens three times in an equal volume of 0.85% NaCl. Resuspend in an equal volume of sterile double-distilled water.
- Prior to the inoculation, examine the tank with seedlings carefully to ensure that there is no contamination; the medium in the uncontaminated tanks should be clear and transparent.
- Discard any tank with cloudy liquid (contamination) and number the rest of the tanks. Sample a small amount (20 µL) of hydroponic medium from each numbered tank and spot it on a petri dish filled with Luria broth (LB). Incubate the dish at 28 °C.
- Immediately, add 50 µL of the A. tumefaciens suspension (approximately 5 x 107 cells, estimated from the OD600) into each numbered hydroponic tank. Cocultivate the A. thaliana seedlings and A. tumefaciens at 22-24 °C with a 16 h photoperiod and shaking at 50 rpm (Figure 3f).
- Examine the spotted LB plate from step 4.7.1. after 12 and 28 h incubation to verify the sterility of the tanks. If there is any contamination, do not use the corresponding numbered tank (inoculated with bacteria) for further downstream assays.
If desired, monitor the growth of A. tumefaciens at regular intervals by removing a sample of medium from the tank and measuring the OD600 using a spectrophotometer with medium from an un-inoculated control as a blank (Figure 4). NOTE: Plant disease symptoms should be apparent after 7 d (Figure 5).
Separate A. thaliana roots from the bacterial suspension by lifting the mesh plate; the timing of this step depends on downstream processes. See steps 5-8 for details.
If using roots for subsequent analyses, quickly rinse the roots in double-distilled water. If using leaf or other tissue, cut off the tissue. For RNA analyses, proceed immediately to step 7.1.
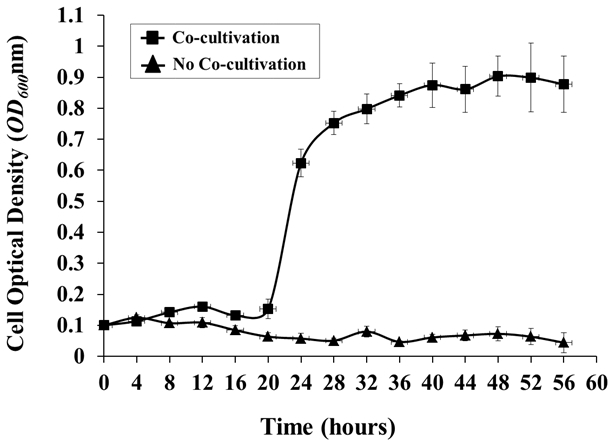
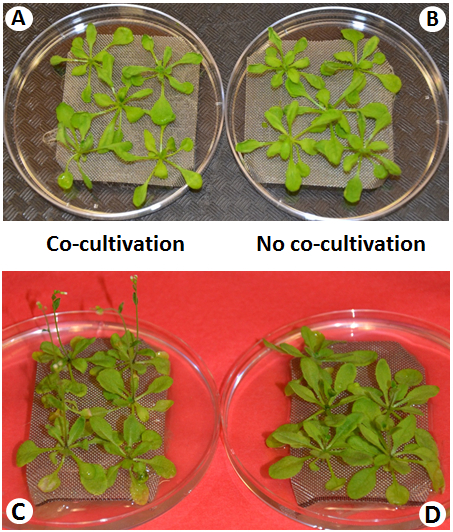
Figure 4: Agrobacterium Growth in the Hydroponic Cocultivation System. Agrobacterium growth in the presence or absence of a plant host (Arabidopsis) was monitored every 4 h. Agrobacterium cells were grown in AB medium O/N, washed 3x with 0.85% NaCl, and inoculated into the hydroponic system, with or without Arabidopsis co-cultivation, from an initial OD600 of around 0.1. The OD600 values are the means of three biological replicates, with standard deviations (OD600 of 1.0 = 1 x 109 cells/mL).
Figure 5: Representative Plant Phenotypes and Observable Disease symptoms during Cocultivation. Within 4 d after inoculation, no disease symptoms are visible (A) as compared to noninoculated plants (B). Following 7 d of Cocultivation (infection), disease symptoms are observed in infected plants (C), while noninoculated plants remain healthy (D).
5. Fluorescence Microscopy
Use A. tumefaciens harboring a reporter construct for an autofluorescent protein for the cocultivation setup in step 4.5. NOTE: A modified pJP2 plasmid expressing pCherry, a derivative of dsRed36, is used here.
Following an appropriate co-cultivation (e.g., 48 h) in step 4.8, separate individual secondary roots (side branches of the main root) using sterile scissors.
Rinse the roots in double-distilled water to remove loosely bound material. Submerge each root in 30 µL of water on a microscope slide and cover with a glass coverslip.
Seal the edges of the coverslip with nail polish to prevent the dehydration of the roots.
Visualize the attachment of A. tumefaciens to A. thaliana roots by fluorescence microscopy. NOTE: Here, a confocal microscope with excitation from a helium-neon (He-Ne) 543/594 nm laser is used to visualize pCherry red fluorescence at 590-630 nm under an inverted 63X water lens objective with a numerical aperture of 1.4 (Figure 6).
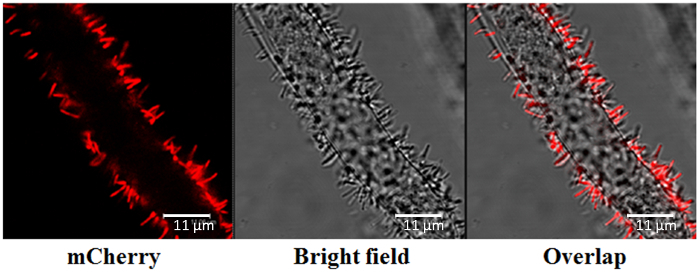
Figure 6: Agrobacterium Root Attachment, Determined by Confocal Microscopy. The pCherry red fluorescence-labelled Agrobacterium was visualized at 590-630 nm, with excitation from a helium-neon (He-Ne) 543/594 nm laser. Visualization was conducted under an inverted 63X water lens objective with a numerical aperture of 1.4. Scale bars = 11 µm. Please click here to view a larger version of this figure.
6. Bacterial Transcript Analyses
Following an appropriate co-cultivation (e.g., 8 h) in step 4.8, separate the roots from the hydroponic medium as in step 4.9. Transfer 1.5 mL of the hydroponic medium to a 1.5 mL microcentrifuge tube and centrifuge at 12,000 x g for 2 min to pellet the cells.
Remove the supernatant. Transfer another 1.5 mL of the hydroponic medium to the same 1.5 mL microcentrifuge tube and centrifuge at 12,000 g for 2 min to pellet the cells.
Remove the supernatant. NOTE: The protocol can be paused here by freezing the samples at -80 °C until use.
Extract the RNA from the A. tumefaciens cells using standard methods37 or a suitable commercial RNA isolation and purification kit with DNase treatment to avoid DNA contamination. NOTE: The protocol can be paused here by freezing aliquots of RNA at -80 °C until use.
- Use the isolated RNA for a variety of analyses using standard methods, including the following.
- Use a commercial microarray according to the manufacturer's protocol to detect gene sets that are differentially expressed in cocultivated versus control culture.
- Use quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) to detect the expression of specific genes or to validate microarray data38.
7. Plant Transcript Analyses
Following an appropriate cocultivation (e.g., 8 h) in step 4.8, separate the roots from the hydroponic medium, as in step 4.9, or separate other tissues as needed. Immediately place approximately 150 mg of roots or other plant tissue into liquid nitrogen. NOTE: The protocol can be paused here by freezing the samples at -80 °C until use.
Grind the frozen samples to a powder in liquid nitrogen using a mortar and pestle.
Extract RNA from the powder using standard methods39 or a suitable commercial RNA isolation and purification kit with DNase treatment to avoid DNA contamination. NOTE: The protocol can be paused here by freezing aliquots of RNA at -80 °C until use.
- Use the isolated RNA for a variety of analyses using standard methods, including the following.
- Use a commercial microarray according to the manufacturer's protocol to detect gene sets that are differentially expressed in co-cultivated versus control plants.
- Use qRT-PCR to detect differential expression of specific genes or to validate microarray data38.
8. Secretome Profiling
Following an appropriate co-cultivation (e.g., 72 h) in step 4.8, separate the roots from the hydroponic medium as in step 4.9. Sterilize the 18 mL of hydroponic medium by passing it through a 0.2 µm pore filter into 50 mL conical tubes.
Freeze the samples at -80 °C O/N.
Loosen the caps on the 50 mL conical tubes and place them in a freeze-drying machine for 36 h. Proceed to store or process the samples (as described below) for HPLC analysis and compound detection. NOTE: The protocol can be paused here.
Resuspend each sample in 5 mL of double-distilled water in a sealable test tube.
Add 5 mL of ethyl acetate and mix by inverting the tube several times.
Allow the phases to separate at room temperature for 5 min.
Transfer the top (organic phase) to a new container by pipetting. If required, pool multiple organic fractions.
Dry under a gentle stream of nitrogen gas (~45 min).
Re-suspend the sample in an appropriate solution for HPLC (e.g., 100% methanol).
Perform HPLC according to standard methods40.
Detect compounds by appropriate means. NOTE: Electrospray Ionization Time-of-Flight Mass Spectrometry (ESI-TOF-MS)40 is used here.
Representative Results
Growth in the hydroponic co-cultivation system
The growth curve of A. tumefaciens C58 demonstrated a significant lag phase in the initial 16 h of cocultivation, followed by a very stable growth when co-cultivated with A. thaliana Col-0, up to a maximum OD600 of about 0.9 starting at 48 h postinoculation. By contrast, essentially no bacterial growth was observed in the control culture without A. thaliana (Figure 4). These results suggest that, without additional nutrients supplied artificially, plant root-secreted chemical compounds are able to support Agrobacterium growth in the hydroponics cocultivation system. The significant lag phase during the initial 16 h of cocultivation may be attributed to the rather complex host perception and gradual root secretion upon cocultivation with the pathogen.
Cocultivated A. thaliana plants showed no disease symptoms during the first 3 days, relative to control plants grown in the absence of A. tumefaciens (Figure 5a, 5b). However, after 5 days of cocultivation, differences gradually became visible and the mock-treated control plants were greener and healthier. After 7 days of cocultivation with Agrobacterium, plant tissue showed chlorosis and necrosis, as well as early flowering (Figure 5c, 5d). Early flowering in Arabidopsis is a general consequence of infection by a range of phylogenetically disparate pathogens41,42.
Microscopic visualization of Agrobacterium attachment to plant roots
After 48 h of cocultivation, A. tumefaciens labelled with pCherry red fluorescence was localized to the plant root structure with a typical rod-like or filamentous shape, approximately 0.5 µm in width and 3-5 µm in length (Figure 6). Most of the bacterial cells were vertically attached to the surface of the root cell wall, in accordance with previous findings that Agrobacterium bind to the cell wall in a polar fashion32.
These results suggest that the hydroponic cocultivation system can be used to study bacterial dynamics in planta, such as attachment to the root structure and the infection process, in real-time. In fact, in the absence of other microbes, this cocultivation system provides a noise-free platform for the direct observation of bacterium-plant attachment in planta.
Activation of Agrobacterium virulence transcripts
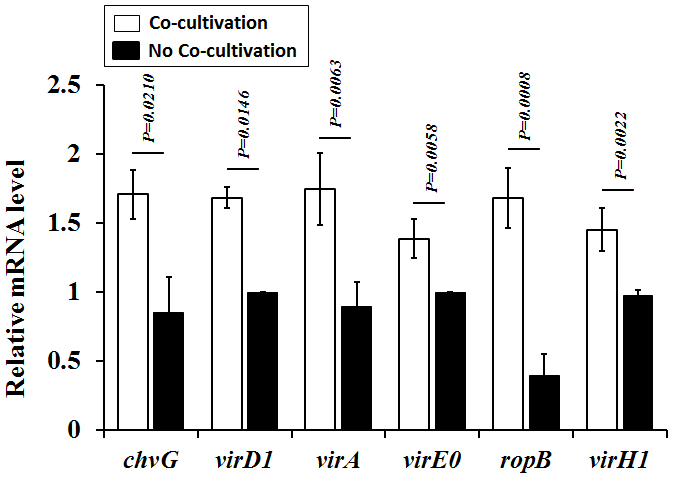
After 8 h of cocultivation, A. tumefaciens cells were harvested for RNA isolation, and the transcription levels of the representative virulence genes were evaluated by qRT-PCR (Figure 7). The expression of the virulence genes ropB, virA, virD1, virH1, virE0, and chvG were significantly upregulated during co-cultivation with the plant host, as compared to the control without A. thaliana. Specifically, the abundance of mRNA increased by 100% for chvG, by 70% for virD1, by 94% for virA, by 40% for virE0, by 330% for ropB, and by 48% for virH1.
These results suggest that, without supplementation with synthetic virulence-inducing chemicals, plants secreting chemicals in the hydroponic cocultivation system are able to activate Agrobacterium virulence programming. The induction of Agrobacterium virulence genes is indicative of the Agrobacteria response to plant-derived signals in the hydroponic cocultivation system. It is very interesting that Agrobacterium ropB and chvG are induced upon cocultivation with Arabidopsis, as previous studies have indicated that these genes are induced by acidic conditions18, but not by acetosyringone, a well-known virulence-inducing compound19. This result suggests that unidentified compounds from Arabidopsis roots are capable of inducing ropB and chvG expression. However, such virulence-inducing chemicals must be further identified.
Figure 7. Activation of Agrobacterium Virulence in the Hydroponic Cocultivation System. The mRNA levels of six Agrobacterium virulence factors were quantitatively measured by qRT-PCR. The abundance of virulence factor transcripts was normalized to that of the 16S rRNA transcripts. The data represent the means ±standard deviation (error bars) of 3 biological replicates, with P-values obtained by student's t-test.
Expression of plant transcripts
qRT-PCR analysis was performed for five genes that were previously shown to be differentially expressed in A. thaliana (unpublished data). As expected, qRT-PCR confirmed that AGP30 (AT2G33790) and NAS1 (AT5G04950) were downregulated (by 97 & 89%, respectively) whereas HSP21 (AT4G27670), PGIP1 (AT5G06860), and ELIP1 (AT3G22840) were upregulated (by 3,800, 540, and 510%, respectively) in each of the 3 replicates versus the controls (Figure 8).
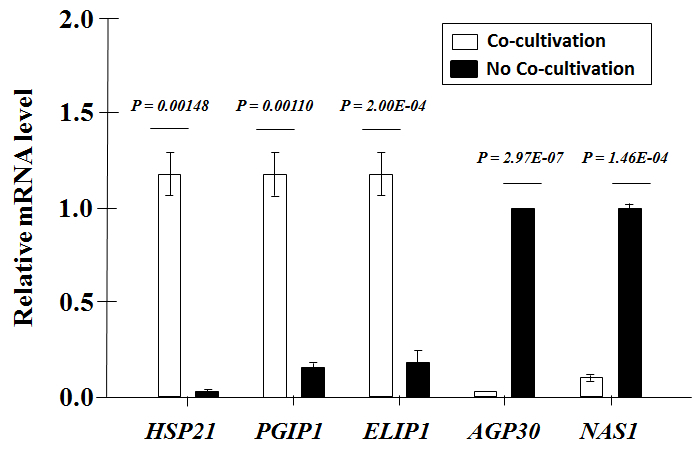
Figure 8: qRT-PCR of A. thaliana Root Transcripts. The abundance of each mRNA transcript was normalized to that of AtACTINII transcripts. The data are the averages of 3 biological replicates ±the standard deviation (error bars), with P-values obtained by student's t-test.
Alterations to host secretion profiles
Root-derived compounds have been shown to largely differ between plant species6,8. Root exudates include molecular signals that elicit various signaling responses during plant-microbe interactions1,43. However, progressive differences in host secretion profiles over time following microbe interaction are far less understood, except that root-secreted compounds are imperative to the attraction of beneficial bacteria29. Following 72 h of cocultivation with A. tumefaciens C58, differences in A. thaliana Col-0 secretome profiles were established using positive ion mode LC-MS analysis and comparisons to non-inoculated A. thaliana Col-0 (i.e. in the absence of Agrobacterium). A clear shift in secretome was observed, with new compounds appearing as a result of Agrobacterium inoculation (Figure 9). Altogether, 35 compounds were comparatively enhanced in the secretome of Agrobacterium-inoculated plants, while 76 were enhanced in the secretome of noninoculated plants (Figure 9). The latter likely represent compounds that A. tumefaciens C58 would encounter prior to infection. Although these compounds were not identified, and the differential secretion of specific root-derived compounds was not determined within the context of this study, the secretome analysis demonstrated detectable differences in host secretion profiles using this hydroponic cocultivation system.
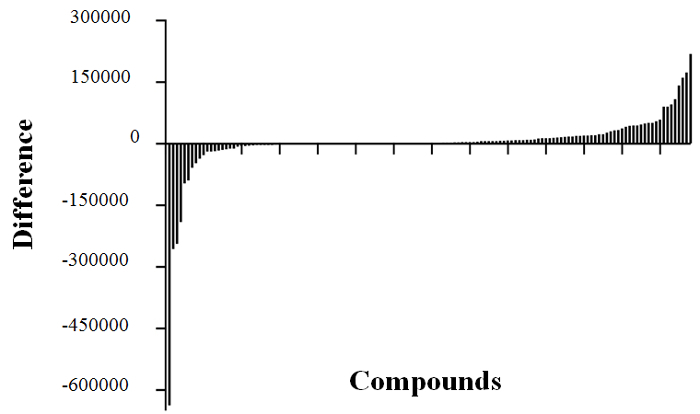
Figure 9: A. thaliana Col-0 Root Secretome Analysis. Analysis of A. thaliana Col-0 secretome profiles was completed in triplicate for mock-inoculated and inoculated samples. Peak areas for the 138 unique mass signals (compounds) found in the secretion profiles in each treatment group were averaged, and the average values were used to calculate the difference in abundance of each compound (mock-inoculated minus inoculated) in order to visualize differences in detectable compounds. Compounds with large negative difference values were more abundant in the secretome of inoculated plants, while those with large positive differences were more abundant in the secretome of mock-inoculated plants.
Discussion
Given the gradual nature of root secretion, the concentration of virulence-inducing chemicals produced in planta and their effects on dynamic plant-microbe interactions occur in spatial and temporal gradients. In this hydroponic co-cultivation system, no synthetic phytohormone or chemical that induces microbial virulence or plant defenses is supplemented. In contrast, using conventional approaches, the addition of synthetic chemicals, such as acetosyringone, creates a sudden, artificial spike in concentrations. Therefore, this hydroponic cocultivation system more closely mimics the natural environment, where microbes and host plants recognize and respond in spatial and temporal manners.
Here, it was possible to demonstrate that Agrobacterium virulence genes were activated upon co-cultivation with Arabidopsis. This suggests that natural plant-derived chemicals induce Agrobacterium virulence. Future effort is needed to identify these chemicals and to verify whether or not the virulence-inducing compound acetosyringone is among them. In addition, substantial changes in Arabidopsis root gene expression and secretome profiles were observed upon cocultivation with Agrobacterium, demonstrating the potential of the hydroponic cocultivation system to study plant host responses and secretion profiles during plant-microbe interactions. In fact, using the hydroponic cocultivation technique, gene expression and secretome profiles of plant hosts can be studied at any stage leading up to and following microbial interaction, which will help to elucidate how these transcriptional or secretome changes facilitate or deter subsequent plant-microbe interactions.
In addition to studying Arabidopsis-Agrobacterium signaling, the hydroponic cocultivation system can be adapted to study other important plant-microbe systems. It can be used to elucidate plant responses to a wide variety of pathogenic or beneficial microorganisms, acting individually or collectively, and to study microbial attachment and responses to different plant hosts in relatively "natural" conditions. The maintenance of typical seedling morphology is important for clarifying molecular plant-microbe interactions. In the case of other bacteria, fungi, or herbivory pathogens, additional nutrients may be supplemented to support microbial growth during the cocultivation.
However, there are a few parameters to consider for cocultivating with different plant-microbe systems. Depending on seed size and root morphology, metal mesh with larger holes and a bigger growth container may be required for larger seeds and seedlings to ensure that root structures fully develop below the mesh surface and that shoot tissues proliferate above. The setup may also need to be altered to include a higher platform. This could be accomplished by cutting a larger square of mesh and bending it further from the corners, to raise the mesh surface higher from the bottom of the container, or by cutting the mesh in the shape of a Greek cross and bending each flap at the indentations. Of course, there are limitations to every system, and the one presented here will not be useful for larger plants that are grown for longer periods (e.g., mature maize). Furthermore, some organisms may require more aeration than is provided by shaking when using this system.
Admittedly, it is very difficult or even impossible to design a system in a laboratory that exactly and perfectly replicates the dynamic plant-microbe interactions that occur in nature. Though the hydroponic cocultivation more closely represents natural conditions in the rhizosphere, at least conceptually, with intact root structures and plant morphology, there are no soil or soil microorganisms in the system. However, this system can be adapted to meet different needs, such as to study microbiomes in a controllable environment and to learn how plants or microbes perceive and respond to different biotic or abiotic stimuli. This hydroponic cocultivation system may also facilitate an understanding of microbiomes and their functions in hydroponics or ecosystems. This is very important for disease management and plant health for the hydroponic greenhouse industry or for the aquatic ecology of plants and microbes.
In addition, this hydroponic cocultivation system can be adapted to study bioremediation and to elucidate the relevant metabolic processes and regulatory pathways in microbes or plants. Moreover, this hydroponic cocultivation system facilitates investigation on the effects of pre-existing (allelochemicals) and/or applied (synthetic) nutrients and chemical signals/compounds on microbes or plant hosts44. The availability of indirectly affecting plant tissues (i.e. leaf, shoot) in the hydroponic cocultivation system provides the opportunity to explore other plant-defensive processes, such as plant priming, and how these signals are transduced.
The most critical part of this co-cultivation system is to prevent contamination. In particular, the transfer of plants from the semi-solid medium into the tank must be done very carefully to prevent the contamination of the hydropic medium. It is therefore necessary for this experimental step to be carried out under sterile conditions in a bio safety cabinet, and experimenters must wait 2-3 days after the transfer before any microorganisms are inoculated for the experiment.
In summary, the hydroponic cocultivation system ameliorates various limitations of conventional approaches by maintaining typical plant morphology and the integrity of plant root structures throughout the experimental process. In addition, the system avoids the supplementation of synthetic chemicals, such as plant hormones or defense/virulence-inducing chemicals. Thus, it more closely mimics the natural environment, where microbes and host plants recognize and respond in spatial and temporal manners. This cocultivation system can be adapted to study different plant-microbe systems in more "natural" and manageable conditions, including with endophytic or epiphytic microbes. Various aspects of interactions in planta, such as pathogenesis, symbiosis, or plant growth promotion, can be examined. The hydroponic cocultivation system can offer new insights into the physiological, biochemical, and molecular responses of both plants and microbes. At any given timepoint/interval or growth stage, bacteria or plants growing in the hydroponic cocultivation system can be sampled separately for various "omics" analyses, including plant and microbial transcriptomics, plant and microbial metabolomics, plant and microbial secretome, and bioremediation. Moreover, this hydroponic co-cultivation system can be adapted to study the interactions between plant hosts and microbiomes in a controllable environment. In summary, the advantages of the hydroponic cocultivation exemplify a superior system for uncovering molecular plant-microbe interaction and signaling.
Disclosures
The authors have nothing to disclose.
Acknowledgments
We would like to thank Brian Weselowski and Alexander W. Eastman for their help and useful discussion. We would also like to thank Drs. Eugene W. Nester, Lingrui Zhang, Haitao Shen, Yuhai Cui, and Greg Thorn for their help, useful discussions, and critical reading of the manuscript. This research was funded by Agriculture and Agri-Food Canada, Growing Forward-AgriFlex (RBPI number 2555) and Growing Forward II project number 1670, conducted by the authors as a part of their duties. This study was also partially funded by the Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant RGPIN-2015-06052 awarded to Z-C Yuan.
References
- Lambers H, Mougel C, Jaillard B, Hinsinger P. Plant-microbe-soil interactions in the rhizosphere: An evolutionary perspective. Plant Soil. 2009;321(1):83–115. [Google Scholar]
- Philippot L, Raaijmakers JM, Lemanceau P, vander Putten WH. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013;11(11):789–799. doi: 10.1038/nrmicro3109. [DOI] [PubMed] [Google Scholar]
- Somers E, Vanderleyden J, Srinivasan M. Rhizosphere bacterial signalling: A Love Parade beneath our feet. Crit. Rev. Microbiol. 2004;30(4):205–240. doi: 10.1080/10408410490468786. [DOI] [PubMed] [Google Scholar]
- Barah P, Winge P, Kusnierczyk A, Tran DH, Bones AM. Molecular signatures in Arabidopsis thaliana in response to insect attack and bacterial infection. PLoS ONE. 2013;8(3) doi: 10.1371/journal.pone.0058987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhou J-M. Plant immunity triggered by microbial molecular signatures. Mol. Plant. 2010;3(5):783–793. doi: 10.1093/mp/ssq035. [DOI] [PubMed] [Google Scholar]
- Paterson E, Gebbing T, Abel C, Sim A, Telfer G. Rhizodeposition shapes rhizosphere microbial community structure in organic soil. New Phytol. 2006;173(3):600–610. doi: 10.1111/j.1469-8137.2006.01931.x. [DOI] [PubMed] [Google Scholar]
- Hartmann A, Schmid M, van Tuinen D, Berg G. Plant-driven selection of microbes. Plant Soil. 2008;321(1-2):235–257. [Google Scholar]
- Micallef SA, Shiaris MP, Colon-Carmona A. Influence of Arabidopsis thaliana accessions on rhizobacterial communities and natural variation in root exudates. J. Exp. Bot. 2009;60(6):1729–1742. doi: 10.1093/jxb/erp053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr T, Otten L. Crown gall of grape: Biology and disease management. Annu. Rev. Phytopathol. 2001;37:53–80. doi: 10.1146/annurev.phyto.37.1.53. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Binns AN, Costantino P. The Agrobacterium oncogenes. In: Spaink HP, Kondorosi A, Hooykaas PJJ, editors. The Rhizobiaceae. Cham, Switzerland: Springer International Publishing AG; 1998. [Google Scholar]
- Gheysen G, Angenon G, Van Montagu M. Agrobacterium-mediated plant transformation: a scientifically intriguing story with significant applications. In: Lindsey K, editor. Transgenic Plant Research. Amsterdam, Netherlands: Harwood Academic; 1998. [Google Scholar]
- Valvekens D, Von Montagu MV, Van Lijsebettens M. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc. Natl. Acad. Sci. USA. 1988;85(15):5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Sussman MR. T-DNA as an insertional mutagen in Arabidopsis. Plant Cell. 1999;11(12):2283. doi: 10.1105/tpc.11.12.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilton MD, et al. Stable incorporation of plasmid DNA into higher plant cells: The molecular basis of crown gall tumorigenesis. Cell. 1977;11(2):263–271. doi: 10.1016/0092-8674(77)90043-5. [DOI] [PubMed] [Google Scholar]
- Gelvin SB. Agrobacterium in the genomics age. Plant Physiol. 2009;150(4):1665–1676. doi: 10.1104/pp.109.139873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramoni S, Nathoo N, Klimov E, Yuan Z-C. Agrobacterium tumefaciens responses to plant-derived signaling molecules. Front. Plant Sci. 2014;5:322. doi: 10.3389/fpls.2014.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Liu P, Saenkham P, Kerr K, Nester EW. Transcriptome profiling and functional analysis of Agrobacterium tumefaciens reveals a general conserved response to acidic conditions (pH 5.5) and a complex acid-mediated signaling involved in Agrobacterium-plant interactions. J. Bacteriol. 2008;190(2):494–507. doi: 10.1128/JB.01387-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachel SE, Messens E, Van Montagu M, Zambryski P. Identification of the signal molecules produced by wounded plant cells that activate T-DNA transfer in Agrobacterium tumefaciens. Nature. 1985;318(6047):624–629. [Google Scholar]
- Memelink J, de Pater BS, Hoge JHC, Schilperoort RA. T-DNA hormone biosynthetic genes: Phytohormones and gene expression in plants. Dev. Genet. 1987;8(5-6):321–337. [Google Scholar]
- Yuan Z-C, et al. The plant signal salicylic acid shuts down expression of the vir regulon and activates quormone-quenching genes in Agrobacterium. Proc. Natl. Acad. Sci. USA. 2007;104(28):11790–11795. doi: 10.1073/pnas.0704866104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z-C, Haudecoeur E, Faure D, Kerr KF, Nester EW. Comparative transcriptome analysis of Agrobacterium tumefaciens in response to plant signal salicylic acid, indole-3-acetic acid and γ-amino butyric acid reveals signalling cross-talk and Agrobacterium-plant co-evolution. Cell. Microbiol. 2008;10(11):2339–2354. doi: 10.1111/j.1462-5822.2008.01215.x. [DOI] [PubMed] [Google Scholar]
- Li PL, Farrand SK. The replicator of the nopaline-type Ti plasmid pTiC58 is a member of the repABC family and is influenced by the TraR-dependent quorum-sensing regulatory system. J. Bacteriol. 2000;182(1):179–188. doi: 10.1128/jb.182.1.179-188.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson MM, Huang J, Knopp JA. Hypersensitivity of suspension-cultured tobacco cells to pathogenic bacteria. Phytopathology. 1985;75(11):1270–1274. [Google Scholar]
- Veena , Jiang H, Doerge RW, Gelvin SB. Transfer of T-DNA and Vir proteins to plant cells by Agrobacterium tumefaciens induces expression of host genes involved in mediating transformation and suppresses host defense gene expression. Plant J. 2003;35(2):219–236. doi: 10.1046/j.1365-313x.2003.01796.x. [DOI] [PubMed] [Google Scholar]
- León J, Rojo E, Sanchez-Serrano JJ. Wound signalling in plants. J. Exp. Bot. 2001;52(354):1–9. doi: 10.1093/jexbot/52.354.1. [DOI] [PubMed] [Google Scholar]
- Ditt RF, Kerr KF, de Figueiredo P, Delrow J, Comai L, Nester EW. The Arabidopsis thaliana transcriptome in response to Agrobacterium tumefaciens. Mol. Plant Microbe In. 2006;19(6):665–681. doi: 10.1094/MPMI-19-0665. [DOI] [PubMed] [Google Scholar]
- Mandimba G, Heulin T, Bally R, Guckert A, Balandreau J. Chemotaxis of free-living nitrogen-fixing bacteria towards maize mucilage. Plant Soil. 1986;90(1-3):129–139. [Google Scholar]
- Rudrappa T, Czymmek KJ, Pare PW, Bais HP. Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol. 2008;148(3):1547–1556. doi: 10.1104/pp.108.127613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CW, et al. Agrobacterium tumefaciens promotes tumor induction by modulating pathogen defense in Arabidopsis thaliana. Plant Cell. 2009;21(9):2948–2962. doi: 10.1105/tpc.108.064576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE. Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell. 2000;12(5):707–720. doi: 10.1105/tpc.12.5.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthysse AG. Attachment of Agrobacterium to plant surfaces. Front. Plant Sci. 2014;5:252. doi: 10.3389/fpls.2014.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alatorre-Cobos F, et al. An improved, low-cost, hydroponic system for growing Arabidopsis and other plant species under aseptic conditions. BMC Plant Biol. 2014;14(1):69. doi: 10.1186/1471-2229-14-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn SJ, et al. Protocol: Optimising hydroponic growth systems for nutritional and physiological analysis of Arabidopsis thaliana and other plants. Plant Methods. 2013;9(1):4. doi: 10.1186/1746-4811-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathoo N. Identification of putative plant defense genes using a novel hydroponic co-cultivation technique for studying plant-pathogen interaction. University of Western Ontario; 2015. [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004;22(12):1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Wise AA, Liu H, Binns AN. Nucleic acid extraction from Agrobacterium strains. Methods Mol. Bio. 2006;343:67–76. doi: 10.1385/1-59745-130-4:67. [DOI] [PubMed] [Google Scholar]
- Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006;1(3):1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- Salter MG, Conlon HE. Extraction of plant RNA. Methods Mol. Bio. 2007;362:309–314. doi: 10.1007/978-1-59745-257-1_21. [DOI] [PubMed] [Google Scholar]
- Bao Y, Wang S, Yang X, Li T, Xia Y, Meng X. Metabolomic study of the intervention effects of Shuihonghuazi Formula, a Traditional Chinese Medicinal formulae, on hepatocellular carcinoma (HCC) rats using performance HPLC/ESI-TOF-MS. J. Ethnopharmacol. 2017;198:468–478. doi: 10.1016/j.jep.2017.01.029. [DOI] [PubMed] [Google Scholar]
- Korves TM, Bergelson J. A developmental response to pathogen infection in Arabidopsis. Plant Physiol. 2003;133(1):339–347. doi: 10.1104/pp.103.027094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons R, Rusu A, Stiller J, Powell J, Manners JM, Kazan K. Investigating the association between flowering time and defense in the Arabidopsis thaliana-Fusarium oxysporum interaction. PLoS ONE. 2015;10(6):e0127699. doi: 10.1371/journal.pone.0127699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badri DV, Weir TL, vander Lelie D, Vivanco JM. Rhizosphere chemical dialogues: Plant-microbe interactions. Curr. Opin. Biotechnol. 2009;20(6):642–650. doi: 10.1016/j.copbio.2009.09.014. [DOI] [PubMed] [Google Scholar]
- Baerson SR, et al. Detoxification and transcriptome response in Arabidopsis seedlings exposed to the allelochemical benzoxazolin-2(3H)-one. J. Biol. Chem. 2005;280(23):21867–21881. doi: 10.1074/jbc.M500694200. [DOI] [PubMed] [Google Scholar]


