Abstract
Rhipicephalus microplus – the cattle tick – is the most significant ectoparasite in terms of economic impact on livestock as a vector of several pathogens. Efforts have been dedicated to the cattle tick control to diminish its deleterious effects, with focus on the discovery of vaccine candidates, such as BM86, located on the surface of the tick gut epithelial cells. Current research focuses upon the utilization of cDNA and genomic libraries, to screen for other vaccine candidates. The isolation of tick gut cells constitutes an important advantage in investigating the composition of surface proteins upon the tick gut cells membrane. This paper constitutes a novel and feasible method for the isolation of epithelial cells, from the tick gut contents of semi-engorged R. microplus. This protocol utilizes TCEP and EDTA to release the epithelial cells from the subepithelial support tissues and a discontinuous density centrifugation gradient to separate epithelial cells from other cell types. Cell surface proteins were biotinylated and isolated from the tick gut epithelial cells, using streptavidin-linked magnetic beads allowing for downstream applications in FACS or LC-MS/MS-analysis.
Keywords: Biochemistry, Issue 125, Cell surface proteins, biotinylation, ticks, microplus, gut epithelial cells
Introduction
Rhipicephalus microplus, the cattle tick, is the most significant ectoparasite in terms of economic impact on the cattle industry of tropical and sub-tropical regions as it vectors bovine tick fever (babesiosis), anaplasmosis and equine piroplasmosis1,2,3,4. Efforts have been dedicated to cattle tick control, to diminish the deleterious effect, however conventional methods such as the use of chemical acaricides have implicit drawbacks, such as the presence of chemical residues in milk and meat, and the increase in prevalence of chemically resistant ticks5,6,7. Consequently, the development of alternative methods of tick control have been studied, such as the use of natural resistance cattle, biological control (biopesticides) and vaccines4,5,6,7,8,9.
In the pursuit of proteins capable of being utilized as vaccine candidates, current research is focused upon the tick gut. The midgut wall is built from a single layer of epithelial cells resting on a thin basal lamina, with the outside of the basal lamina forming a network of muscle. Light and electron microscope observations indicate that the midgut consists of three types of cells: reserve (undifferentiated), secretory, and digestive. The number of cell types varies considerably depending upon the physiological phase. Secretory and digestive cells both originate from reserve cells18,19,20.
The construction of cDNA libraries to examine the composition of the tick gut has led to the identification of antigenic proteins, such as Bm86, as potential vaccine candidates2,3,4. The glycoprotein Bm86 is localized at the surface of tick gut cells and induces a protective immune response against the cattle tick (R. microplus) in vaccinated cattle. Anti-Bm86 IgGs produced by the immunized host are ingested by the tick, recognize this antigen on the surface of tick gut cells, and subsequently disturb tick gut tissue function and integrity. Vaccines based upon Bm86 antigens have shown effective control of R. microplus and Rhipicephalus annulatus, by reducing the number, weight and reproductive capacity of engorging females, resulting in a reduced larval infestation in subsequent tick generations4. However, Bm86 based vaccines are not effective against all tick stages and have demonstrated unsatisfactory efficacy against some geographical strains of R. microplus, consequently the beef and dairy industries have poorly adopted these vaccines2,4.
The ability to isolate epithelial cells from the tick gut is a significant innovation which would enable the progression of research to determine protein membrane composition including morphology and physiology under different environmental conditions. The method described here utilizes the chelating agent ethylenediaminetetraacetic acid (EDTA) and the reducing agent tris(2-carboxyethyl)phosphine (TCEP) to release the epithelium from its sub-epithelial support tissues10. The epithelium is recovered following mechanical disruption of the tissues by shaking, followed by discontinuous gradient centrifugation in Percoll. This paper describes a feasible and novel technique for the isolation of tick gut epithelial cells. Biotinylated cell surface proteins, isolated from the surface of these epithelial cells can subsequently analyzed in downstream applications such as FACS and/or LC-MS/MS-analysis.
Protocol
1. Dissection of the Gut Epithelium from R. microplus
Collect semi-engorged ticks from cattle on the day of experiment. Dissect ticks within 24 h after removal from the host.
Adhere a strip of duct tape to the bottom of the 92 mm x 16 mm Petri dish. Add a drop of super glue to the tape. Place the tick, ventral side down on the super glue, allow to dry for 2 min.
Pour 100 mL of phosphate buffered saline (PBS) into the petri dish, or until the tick is completely submerged.
Utilizing a size 11 scalpel, cut from the top of the eyes to the bottom festoons, on both sides of the tick.
Using sterile forceps, completely remove the scutum and alloscutum, to expose the internal organs.
Remove the fine white thread-like organs (trachea) and other membranes to prevent contamination.
Remove the gut using forceps, by pinching the upper region and pulling from the carcass. Remove any remaining gut tissues, ensuring that no other tissues have been dissected.
Store gut in ice-cold Hank's Balanced Salt Solution (HBSS) without calcium chloride and magnesium sulfate with proteinase-inhibitor cocktail (PIC). Snap freeze the guts in dry ice and store at -20 °C. Note: To assist with gut dissection and details of the tick internal organs refer to Chapter 3.1 of D. Sonenshine, "Biology of Ticks"19.
2. Epithelial Cell Dissociation
Pour the dissected gut onto a 70 µm cell strainer inside a 50 mL tube.
Flush the gut tissue with 50 mL ice cold HBSS with PIC until the solution runs clear, and the guts take on a white/clear appearance.
Re-suspend guts in 30 mL ice cold HBSS with PIC, mix gently and centrifuge at 500 x g at 4 °C for 10 min to pellet the gut. Remove the supernatant and repeat the wash process three times.
To dislodge epithelial gut cells, re-suspend the gut in 10 mL of cell culture medium Dulbecco's modified eagle medium (DMEM), 2% fetal calf serum (FCS), 0.5 mM ethylenediaminetetraacetic acid (EDTA), 1 mM Tris(2-carboxyethyl)phosphine (TCEP), PIC and incubate for 60 min at 37 °C under slow rotation using a roller at 6 rpm.
Filter the suspension through a 250 µm cell strainer, vortex the flow-through and filter through a 70 µm cell strainer collecting the remaining flow-through.
Centrifuge the suspension at 500 x g at 4 °C for 20 min to pellet the single cells.
3. Isolation of Single Epithelial Cells using a density centrifugation gradient
Prepare density centrifugation gradient (e.g., Percoll) by filtering through an AP15 pre-filter paper. Prepare 40% and a 20% Percoll in mqH20 (v/v) and cool at 4 °C for 1 h prior to layering the gradient.
Using a peristaltic pump set at the lowest speed, layer 3 mL of 40% density centrifugation gradient into a 16 mL ultracentrifuge tube, allowing it to settle on ice for 15 min. Speed of the pump should lead to a <1 mL per min flow rate.
Tilting the tube to a 45° angle, use the peristaltic pump to layer the 20% density centrifugation gradient on top of the 40% layer. Speed of the pump should lead to <1 mL per min flow rate. Allow the layers to settle on ice for 15 min.
Use the peristaltic pump at <1 mL per min flow rate to layer 3 mL of DMEM medium containing tick gut cells over the 20-40% density centrifugation gradient.
Program the centrifuge for maximum acceleration and minimum deceleration. Centrifuge at 600 x g for 10 min. Collect interphases between the DMEM:20% density centrifugation gradient, and the 20%: 40% density centrifugation gradients to isolate epithelial single cells. Store collected interphases at 4 °C for subsequent analyses.
4. Assessment of Cell Isolation
- Hemacytometer
- Clean the hemacytometer slide with alcohol.
- .Re-suspend the cells by gently pipetting the cells up and down. Pipette 100 µL of the cell suspension and place in a 1.5 mL microcentrifuge tube.
- Add 400 µL of 0.4% Trypan Blue. Gently mix by flicking the tube.
- Pipette 100 µL of the Trypan Blue-treated cell suspension to slowly fill both chambers of the hemocytometer.
- Place the hemocytometer under a light microscope, focusing upon the grid lines of the hemocytometer with a 10X objective.
- Using a hand-held tally counter, count the live unstained cells within a set of 16 squares. Within the same square, count the blue dead cells. Continue counting until four sets of 16 squares are counted.
- Calculate the total cells per mL by using the formula:
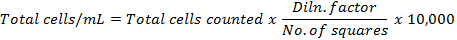
- Calculate the percentage cell viability by using the formula:
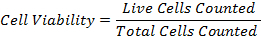
- Cell Isolate Visualization
- Dilute 1 µL of the isolated cells in 9 µL of HBSS in a 1.5 mL microcentrifuge tube. Flick the microcentrifuge tube gently to mix.
- Pipette 5 µL of isolated cells in HBSS into the middle of a glass slide. Apply three drops of mounting medium with 4', 6-diamidino-2-phenylindole (DAPI).
- Incubate the slide at room temperature for 5 min. Carefully place a cover slip over the preparation, avoiding air bubbles.
- Visualize cells stained with DAPI at excitation 360 nm and emission at 460 nm under a fluorescent microscope.
5. Cell Surface Protein Biotinylation
Biotinylate 100 µL of single cell epithelial cells using Biotin (Type A) Conjugation kit, at a molar ratio of 1:1 surface protein to conjugate, as per manufacturer's instructions
For cell lysis, add 100 µL of PBS, 1% Triton X-100, 10% glycerol, 100 µM oxidized glutathione and PIC to the biotinylated cells. Incubate on ice for 1 h with gentle mixing every 10 min.
Centrifuge the biotinylated cells at 20,000 x g at 4 °C for 20 min to pellet insoluble material. Collect the supernatant containing cytoplasmic, and biotinylated membrane proteins.
Determine the protein concentration using the Bradford assay.
6. Isolation of Biotinylated Surface Proteins
Add 50 µL of Streptavidin Magnetic Beads into a 1.5 mL microcentrifuge tube.
Place the tube into a magnetic stand, collecting the beads against the side of the tube. Remove and discard the supernatant.
Add 1000 µL of TBS, 0.1% Tween-20 to the tube. Mix gently and collect the beads with the magnetic stand. Remove and discard the supernatant.
Combine 40 µg of biotinylated cell surface proteins, diluted to 300 µL in 1x PBS with washed magnetic beads. Incubate for 2 h at room temperature with agitation.
Collect the beads with a magnetic stand, remove and discard the supernatant.
Add 300 µL of TBS, 0.1% Tween-20 to the tube, gently mixing to re-suspend the beads. Collect beads, remove and discard the supernatant. Repeat this wash step twice.
Add 100 µL of the 0.1 M glycine pH 2.0 to the magnetic beads, and incubate at room temperature for 5 min. Collect beads and remove supernatant containing eluted biotinylated surface proteins.
Visualize isolated surface proteins on a 4-20% Tris-MOPS SDS-PAGE gel.
7. Assessment of Biotinylated Surface Protein
- Dot Blot
- Cut a 7 cm x 3 cm strip of nitrocellulose membrane.
- Apply 10 µg total tick gut contents (from 1.8 above), and 10 µg of biotinylated surface protein to the membrane. Allow drying for 15 min at room temperature.
- Transfer to a container and submerge in 100 mL of blocking buffer. Incubate at room temperature with agitation for an hour. Discard blocking buffer.
- Wash nitrocellulose membrane in 100 µL PBS, 0.05% Tween-20 for 5 min with agitation. Discard wash buffer and repeat the washes for three times.
- Incubate in 1/5000 Streptavidin- horseradish peroxidase (HRP)in 100 µL PBS, 0.05% Tween-20 for 2 h with agitation at room temperature, and discard any remaining solution.
- Wash nitrocellulose membrane in 100 µL of PBS, 0.05% Tween-20 for 5 min with agitation. Discard wash buffer and repeat the wash three times.
- For detection, dissolve 1 tablet of 4-chloro-1-napthol in 10 mL of ice cold methanol. Add 4 mL of methanol stock to 20 mL of TBS. Add 10 µL of fresh 30% hydrogen peroxide and immediately apply to nitrocellulose membrane.
- Incubate with agitation at room temperature until substrate produces an insoluble blue end product. This may take between 2-15 min.
- Discard detection solution and wash the membrane three times in 100 µL mqH2O.
- ELISA Assay
- Dilute 200 ng of each sample with 400 µL of 100 mM Carbonate coating buffer and coat 2 lanes of the first row (A#) of the ELISA plate (flat bottom wells)
- Add 100 µL of 100 mM carbonate coating buffer to all other corresponding wells in rows (B-H).
- Prepare serial dilutions of each sample by pipetting 100 µL from each well in row A, and transferring into row B. Mix gently by pipetting and avoid producing bubbles. Repeat the dilutions down each row, discarding the final 100 µL from each well in row H.
- Cover the plate with parafilm and incubate at 4 °C overnight.
- Wash the plate with 200 µL PBS, 0.05% Tween-20 per well three times.
- Add 200 µL of blocking buffer to the coated wells, cover with Parafilm and incubate at 4 °C overnight.
- Dilute 1/15,000 Streptavidin-HRP in blocking buffer and add 100 µL to each well of the plate. Cover with parafilm and incubate at 4 °C overnight.
- Wash the plate in 200 µL PBS, 0.05% Tween-20 per well five times.
- To detect, add 100 µL of TMB reagent per well. After sufficient color development, usually between 10-15 min, add 100 µL of 1 M phosphoric acid to stop the reaction.
- Read the absorbance of each well at λ = 450nm.
Representative Results
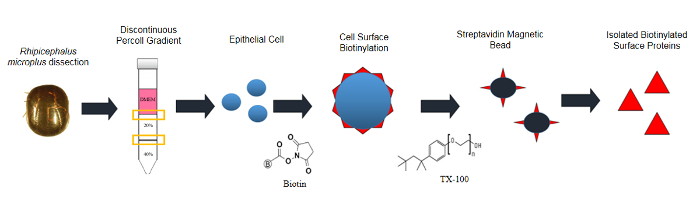
Epithelial cells were isolated from the gut tissues of R. microplus as per the schematic presented in Figure 1.Representative fluorescence microscopy imagery of tick gut epithelial cells prepared using this protocol are shown in Figure 2Aand 2B.As the cell isolation is conducted upon semi-engorged R. microplus, cells appear as singular, spherical, smooth surface morphology and a consistent size throughout the sample. Differences in the size and type of gut cell populations, are more evident once the tick proceeds to become fully-engorged adults.
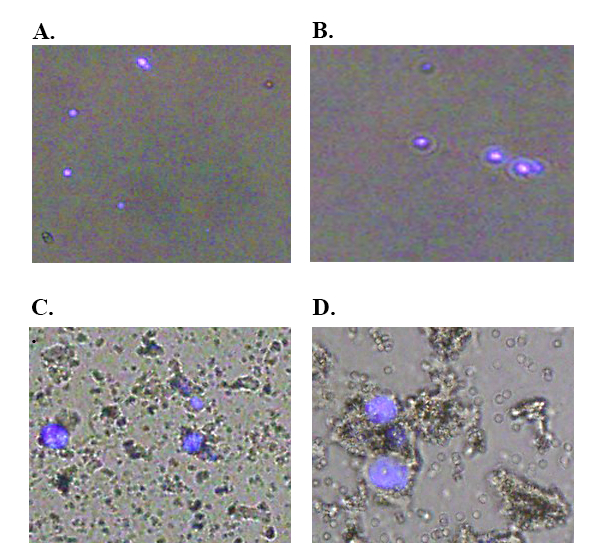
Fluorescent staining of the nucleus of isolated epithelial cells with DAPI assists to visualize the cells. Poor isolations are visualized to contain incomplete dissociation of epithelial cells with varying cell populations identified through the varying cell sizes and morphologies (Figure 2C and 2D)
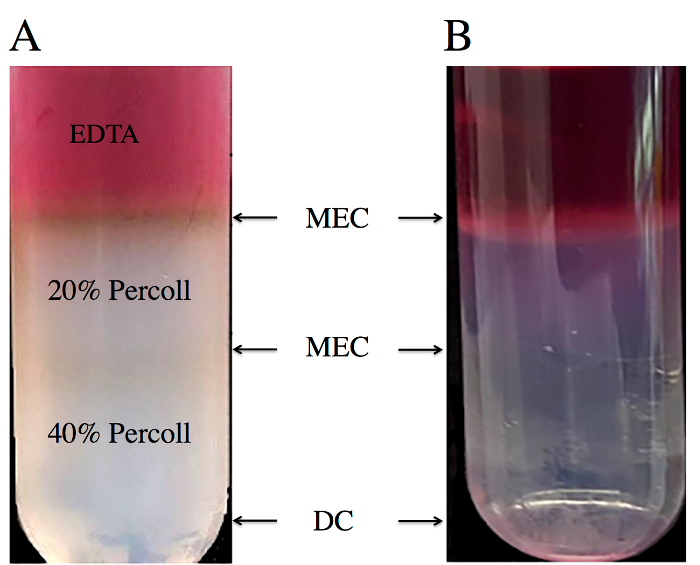
Utilizing this protocol, approximately 1.2 x 107 cell per/mL from 50 tick gut dissections, with 75-80% viability were successfully isolated from R. microplus tick gut. Cross-contamination from host proteins (Figure 3A) can be minimized by adequately rinsing tick guts until they have a white appearance that result in a white/clear Percoll gradient(Figure 3B).
Surface-bound proteins were isolated through biotinylation of surface-bound proteins, destruction of cellular membranes and the purification of biotinylated surface proteins with magnetic streptavidin beads. A total of 20-24 µg of purified biotinylated surface proteins can be isolated for down-stream applications from an initial dissection of 50x tick guts utilizing this protocol. Comparison of proteins by SDS-PAGE (Figure 4A), Silver stain (Figure 4B), Dot blot (Figure 5) and ELISA (Figure 6) indicates that the described methodology successfully purified biotinylated surface proteins from epithelial cells isolated from R. microplus tick gut.
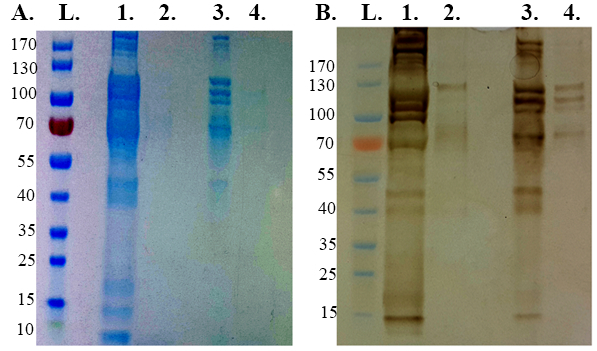
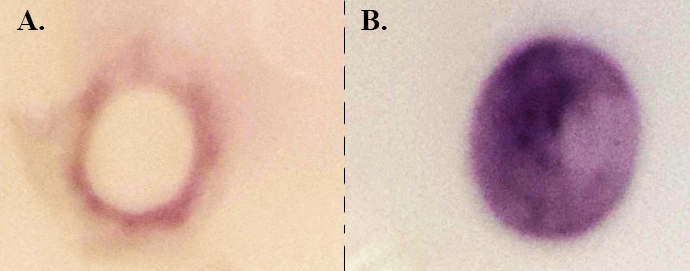
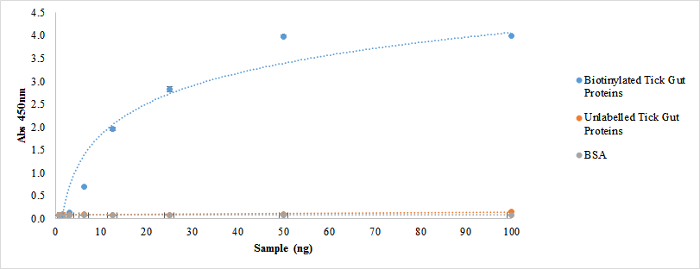
Figure 1 : Schematic representation to isolate biotinylated proteins present in the surface of the R. microplus midgut cells. Please click here to view a larger version of this figure.
Figure 2: Fluorescence microscope visualization (100x) of the R. microplus midgut epithelial cells stained with DAPI. The software was used to conduct the fluorescence overlay. A&B) Represent epithelial cells successfully isolated from the R. microplus midgut. C/D) Epithelial cells isolated from the R. microplus gut contaminated with larger tick tissues.
Figure 3: Percoll gradient containing DMEM: 20% Percoll:40% Percoll. The epithelial cell layers were formed between the DMEM:20% Percoll and the 20:40% Percoll. (A) Tick gut dissection without adequate wash step. (B) Tick gut dissection with adequate wash step. Please click here to view a larger version of this figure.
Figure 4: Electrophoretic separations of biotinylated surface proteins run on 4-20% Tris-MOPS gel, at 140 V for 55 min with 10 µg of sample utilized. (L) PageRuler Prestained protein ladder (1) Unlabelled R. microplus whole gut sample (2) Biotinylated proteins extracted from R. microplus whole gut as per step 6. (3) Proteins from epithelial cells extracted from unlabelled R. microplus gut (4) Biotinylated proteins extracted from R. microplus epithelial cells as per step 6. (A) SDS-PAGE stained by Comassie blue (B) Silver stain.
Figure 5: Dot blot analysis. Streptavidin-HRP conjugated diluted 1/5,000. (A) A total of 10 µg of raw tick gut protein extract (B) Biotinylated surface proteins extracted from purified epithelial cells (10 µg).
Figure 6: Assessment of Biotinylated surface proteins viability utilizing ELISA. ELISA of surface proteins was developed by Strep-HRP conjugated antibody diluted 1/15,000 against different concentrations of biotinylated tick gut proteins (from 0.7 ng to 100 ng). Unlabeled tick gut proteins and bovine serum albumin (BSA) were used as negative control of the ELISA. Please click here to view a larger version of this figure.
Discussion
Cattle tick infestations constitute a major problem for the cattle industry in tropical and subtropical regions of the world, with the most common method of control reliant upon the use of acaricides1,4. Bm86 was previously identified within the tick gut epithelial surface as a protective antigen against R. microplus infestation10, with limited success as a vaccine strategy due to Bm86 geographic sequence variation and the requirement for regular boosting4.
Previous publications focusing upon epithelial isolation methodologies were principally focused upon vertebrates or insects species9,11,12,13,14. For example, early fractionation attempts to isolate midgut utilizing mammalian techniques, found that the same methodology could not be applied to insects due to the different cell structure and organizations12. Furthermore, mammalian techniques rely upon the use of either dipase or collagenase, to enzymatically digest the cell-cell junction proteins9,13. Dipase and collagenase considerably affect cell surface bound proteins, and therefore techniques dependent on their use are not suitable for cell-surface protein studies15. Insect midgut cell isolations currently focus upon two techniques. The first utilizes mechanical disruption of midgut through ultrasound followed by separation over a continuous linear sucrose gradient8,11,12,14. This mechanical technique produces a samples almost clear of contaminants, however produces a low yield of microvillarmembranes14. The second technique relies upon Tris disruption of membranes to release microvillar membranes8.
The technique outlined within this paper allows for the isolation of epithelial cells, biotinylation of surface proteins and their isolation16, permitting further studies through downstream applications such as mass spectrometry or FACS. The method described here utilizes the chelating agent EDTA and the reducing agent TCEP. EDTA functions to sequester calcium ions, inhibiting cadherins, breaking cadherin-mediated cell-cell junctions whereas TCEP reduces the disulfide bonds that confers viscosity of the glycan-rich mucus-like peritrophic matrix that separates the gut lumen from the epithelium. The epithelium is recovered following mechanical disruption of the tissues by shaking, followed by discontinuous gradient centrifugation in the density centrifugation gradient. Epithelial cells are isolated between the DMEM:20% density centrifugation gradient, and 20%:40% density centrifugation gradient layers.
Utilization of a higher temperature during epithelial cell dissociation will cause large tissues to dissociate from the gut, leading to failure to isolate epithelial cells. Ensuring that epithelial cell dissociation is conducted promptly post tick dissection; the utilization of ice-cold buffers and the avoidance of high centrifugation speeds is critical in retaining live viable cells. Despite this, the dislodgment of larger cells from the basal lamina can provide some contamination. Furthermore, the midgut epithelium of the tick alters dramatically dependent on the time from the last blood meal18,19,20.
As such, this protocol has been designed and accessed on R. microplus ticks starting digestion, and the techniques prescribed may have varying resulting cell populations dependent on when the tick's last blood meal was taken. The protocol has been designed to collect varying epithelial cell populations, however as the ticks utilized were semi-engorged the cells visualized (Figure 1) are of a uniform nature.
In conclusion, the methods utilized in this study successfully isolated epithelial cells from the whole gut of R. microplus. Proteins from the surface of R. microplus epithelial cells were obtained for further analysis and studies. Finally, the protocol developed has demonstrated the potential yield of epithelial cells from the tick gut, and the efficiency of biotinylated surface proteins of the cell. The protocol developed can be utilized in any tick species of economic significance in an effort to investigate tick:host interactions by studying the membrane protein composition of the gut.
Disclosures
The authors have nothing to disclose.
Acknowledgments
The authors wish to thank the Biosecurity Tick Colony (Queensland Department of Agriculture & Fisheries, Australia) for the provision of Rhipicephalus microplus ticks utilized for this study, and Lucas Karbanowicz for assistance with video filming.
References
- Rodriguez-Valle M, et al. Efficacy of Rhipicephalus (Boophilus) microplus Bm86 against Hyalomma dromedarii and Amblyomma cajennense tick infestations in camels and cattle. Vaccine. 2012;30:3453–3458. doi: 10.1016/j.vaccine.2012.03.020. [DOI] [PubMed] [Google Scholar]
- De Rose R, et al. Bm86 antigen induces a protective immune-response against Boophilus microplus following DNA and protein vaccination in sheep. Vet. Immunol. Immunopathol. 1999;71:151–160. doi: 10.1016/s0165-2427(99)00038-0. [DOI] [PubMed] [Google Scholar]
- García-García JC, et al. Sequence variations in the Boophilus microplus Bm86 locus and implications for immunoprotection in cattle vaccinated with this antigen. Exp. Appl. Acarol. 1999;23:883–895. doi: 10.1023/a:1006270615158. [DOI] [PubMed] [Google Scholar]
- Abbas RZ, Zaman MA, Colwell DD, Gilleard J, Iqbal Z. Acaricide resistance in cattle ticks and approaches to its management: The state of play. Vet. Parasitol. 2014;203:6–20. doi: 10.1016/j.vetpar.2014.03.006. [DOI] [PubMed] [Google Scholar]
- Kearney S. Acaricide (chemical) resistance in cattle ticks. 2013. AgNote No. K58. http://www.nt.gov.au/d/Content/File/p/Anim_Dis/845.pdf.
- Foil LD, et al. Factors that influence the prevalence of acaricide resistance and tick-borne diseases. Vet. Parasitol. 2004;125:163–181. doi: 10.1016/j.vetpar.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Rodriguez M, et al. High level expression of the B. microplus Bm86 antigen in the yeast Pichia pastoris forming highly immunogenic particles for cattle. J Biotechnol. 1994;33:135–146. doi: 10.1016/0168-1656(94)90106-6. [DOI] [PubMed] [Google Scholar]
- Rodriguez M, et al. Effect of vaccination with a recombinant Bm86 antigen preparation on natural infestations of Boophilus microplus in grazing dairy and beef pure and cross-bred cattle in Brazil. Vaccine. 1995;13(18):1804–1808. doi: 10.1016/0264-410x(95)00119-l. [DOI] [PubMed] [Google Scholar]
- Lew-Tabor AE, Rodriguez Valle M. A review of reverse vaccinology approaches for the development of vaccines against ticks and tick borne diseases. Ticks Tick Borne Dis. 2016;7:573–585. doi: 10.1016/j.ttbdis.2015.12.012. [DOI] [PubMed] [Google Scholar]
- Capella AN, Terra WR, Ribeiro AF, Ferreira C. Cytoskeleton removal and characterization of the microvillar membranes isolated from two midgut regions of Spodoptera frugiperda (Lepidoptera) Insect Biochem. Mol. Biol. 1997;27:793–801. [Google Scholar]
- Cioffi M, Wolfersberger MG. Isolation of separate apical, lateral and basal plasma membrane from cells of an insect epithelium. A procedure based on tissue organization and ultrastructure. Tissue Cell. 1983;15:781–803. doi: 10.1016/0040-8166(83)90050-2. [DOI] [PubMed] [Google Scholar]
- Koefoed BM. A simple mechanical method to isolate the basal lamina of insect midgut epithelial cells. Tissue Cell. 1985;17:763–768. doi: 10.1016/0040-8166(85)90009-6. [DOI] [PubMed] [Google Scholar]
- Roche JK. Isolation of a purified epithelial cell population from human colon. Methods Mol. Med. 2001;50:15–20. doi: 10.1385/1-59259-084-5:15. [DOI] [PubMed] [Google Scholar]
- Terra WR, Costa RH, Ferreira C. Plasma membranes from insect midgut cells. An. Acad. Bras. Ciênc. 2006;78:255–269. doi: 10.1590/s0001-37652006000200007. [DOI] [PubMed] [Google Scholar]
- Vargas AE, Markoski MM, Cañedo AD, Helena F, Nardi NB. Identification, isolation and culture of intestinal epithelial stem cells from murine intestine. Stem Cells. 2012;879:479–490. [Google Scholar]
- Autengruber A, Gereke M, Hansen G, Hennig C, Bruder D. Impact of enzymatic tissue disintegration on the level of surface molecule expression and immune cell function. Eur. J. Microbiol. Immunol. 2012;2:112–120. doi: 10.1556/EuJMI.2.2012.2.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karhemo PR, et al. An optimized isolation of biotinylated cell surface proteins reveals novel players in cancer metastasis. J. Proteomics. 2012;77:87–100. doi: 10.1016/j.jprot.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obenchain FR, Galun R. Current Themes in Tropical Science Volume 1. Pergamon Press; 1982. Physiology of Ticks; pp. 201–205. [Google Scholar]
- Sonenshine D, Roe R. "Biology of Ticks". Two. Vol. 1. Oxford University Press; 2014. Chapter 3.1. [Google Scholar]
- Raikhel AS, Balashov YS. "An Atlas of Ixodid Tick Ultrastructure" (English Translation) Entomology Society of America Entomological Society of America; 1983. [Google Scholar]


