Abstract
Gliomas possess complex and heterogeneous vasculatures with abnormal hemodynamics. Despite considerable advances in diagnostic and therapeutic techniques for improving tumor management and patient care in recent years, the prognosis of malignant gliomas remains dismal. Perfusion-weighted magnetic resonance imaging techniques that could noninvasively provide superior information on vascular functionality have attracted much attention for evaluating brain tumors. However, nonconsensus imaging protocols and postprocessing analysis among different institutions impede their integration into standard-of-care imaging in clinic. And there have been very few studies providing a comprehensive evidence-based and systematic summary. This review first outlines the status of glioma theranostics and tumor-associated vascular pathology and then presents an overview of the principles of dynamic contrast-enhanced MRI (DCE-MRI) and dynamic susceptibility contrast-MRI (DSC-MRI), with emphasis on their recent clinical applications in gliomas including tumor grading, identification of molecular characteristics, differentiation of glioma from other brain tumors, treatment response assessment, and predicting prognosis. Current challenges and future perspectives are also highlighted.
1. Introduction
Gliomas are the most common primary brain tumors in adults with varying malignancy ranging from pilocytic astrocytoma to glioblastoma multiforme (GBM) [1]. Despite considerable advances in various diagnostic and therapeutic techniques in recent years, the prognosis of malignant gliomas remains dismal, with median survival less than 5 years for anaplastic glioma and approximately 14.5–16.6 months for GBM [2, 3]. Glioma-associated neovascularization with aberrant structure and functionality is a typical tumor hallmark participating in multiple biological behaviors such as tumor progression, invasiveness, and therapy resistance [4]. Visualization of tumor vasculatures is of great importance for improved glioma management.
Magnetic resonance imaging (MRI) is currently the prior choice for clinical applications in brain tumors [5]. Although conventional MRI sequences can provide exquisite anatomical information of tumors, they have the inability to quantitatively evaluate vascular physiology and capture tumor biology at molecular/cellular levels, which contribute to tumor grading [6], therapeutic assessment [7], and prognosis prediction [8]. Furthermore, nonenhancing regions representing peritumoral edema with infiltrative tumor cells are not visualized on conventional MRI, hindering the maximum safe surgical resection and therapy response assessment [9, 10]. Perfusion-weighted magnetic resonance imaging (PW-MRI) techniques, such as dynamic contrast-enhanced MRI (DCE-MRI) and dynamic susceptibility contrast-MRI (DSC-MRI), have demonstrated much potential as powerful imaging biomarkers for glioma management as they can provide information of vascular hemodynamics [11–13]. PW-MRI is now rapidly expanding its application spectrum by noninvasively exploring the relationship between imaging parameters and the molecular characteristics of gliomas [14] (Figure 1).
Figure 1.
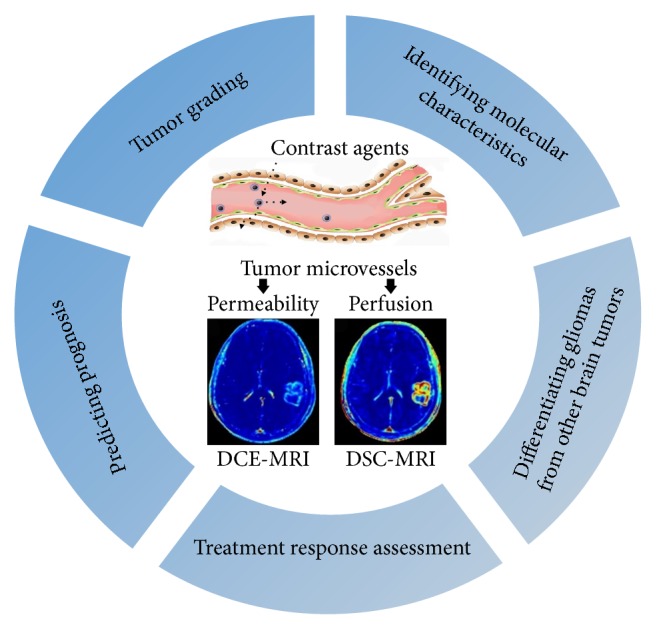
The versatile clinical applications of contrast-enhanced perfusion MRI techniques in gliomas.
Despite that numerous studies have explored PW-MRI for evaluating gliomas, there have been very few studies providing a comprehensive evidence-based and systematic summary. This review first outlines the status of glioma theranostics and tumor-associated vascular pathology and then presents an overview of the principles of DCE-MRI and DSC-MRI, with emphasis on their recent clinical applications in gliomas including tumor grading, identification of molecular characteristics, differentiation of glioma from other brain tumors, treatment response assessment, and predicting prognosis. Current challenges and future perspectives are also highlighted.
2. Glioma Vascular Pathology
Malignant gliomas possess exuberant neovascularization characterized by disorganized, irregular, and tortuous vessels with arteriovenous shunting [15, 16]. In low grade glioma (LGG), tumor vessels are mainly composed of normal endothelial cells (ECs), with cell-to-cell tight junction and relatively intact blood brain barrier (BBB) [17]. However, the vascular ultrastructure of high grade glioma (HGG) is characterized by large caliber and aberrant vascular walls, composed of abundant immature ECs with loose conjunction, fenestrated structure, and discontinuous membrane [15]. The garland-like formation of glomerular capillary loops, consisting of multilayered, actively mitotic ECs and perivascular cells, is the typical architecture of the abnormal microvascular proliferation in GBM [18]. Glioma-associated vessels exhibit prominent spatial heterogeneity. The marginal tumor area is rich of proliferative and invasive cells, with increased microvessel density (MVD) and active neovascularization. However, compressed and tortuous vascular networks with reduced vascular perfusion are observed in the lesion core, resulting in hypoxia, cell metabolic scarcity, and necrosis [19, 20]. The abnormal tumor vascular structure consequently causes abnormal vascular function with increased permeability and perfusion.
3. Principle of PW-MRI
Cerebral vascular hemodynamics can be assessed with PW-MRI, including DCE-MRI, DSC-MRI, and arterial spin-labeling (ASL) techniques. Using exogenous gadolinium-based contrast agents (GBCAs), PW-MRI can characterize tumor vascular perfusion and permeability with multiple parameters, by emphasizing either the T1 relaxivity properties of GBCAs through T1-weighted DCE-MRI or their susceptibility effects through T2/T2∗-weighted DSC-MRI [21]. ASL-MRI is a much less frequently used perfusion modality that involves magnetically labeled arterial blood water protons rather than GBCAs for perfusion characterization. As contrast-enhanced MRI is the most commonly used for brain tumors assessment in clinical setting, DCE-MRI and DSC-MRI will be discussed in detail below.
3.1. DCE-MRI
DCE-MRI is based on T1 relaxivity of GBCAs with fast imaging acquisition. Due to the BBB disruption and vascular hyperpermeability in gliomas, the GBCAs administered intravenously are easy to leak from intravascular compartment to extravascular extracellular space (EES), leading to an increase in T1 signal intensity induced by paramagnetic effect [22]. By consecutively acquiring a serial of T1 weighted images before, during, and after GBCAs administration, the dynamic T1 signal intensity can be measured and proportionally depict the concentration distribution of GBCAs between intravascular space and EES using mode-free (semiquantitative) and model-dependent (quantitative) parameters.
Model-free parameters are calculated based on signal intensity-acquisition time curve, reflecting an overall kinetics of GBCAs perfusion (Figure 2(a)). This approach is simple and straightforward without fitting complicated pharmacokinetic (PK) models. However, it often encounters limited temporal resolution and is weak in providing specific physiological information of tumor vasculatures (e.g., permeability and blood flow) [23]. Furthermore, measurement of these parameters is more susceptible to subjectivity, prone to errors due to experience and bias [24].
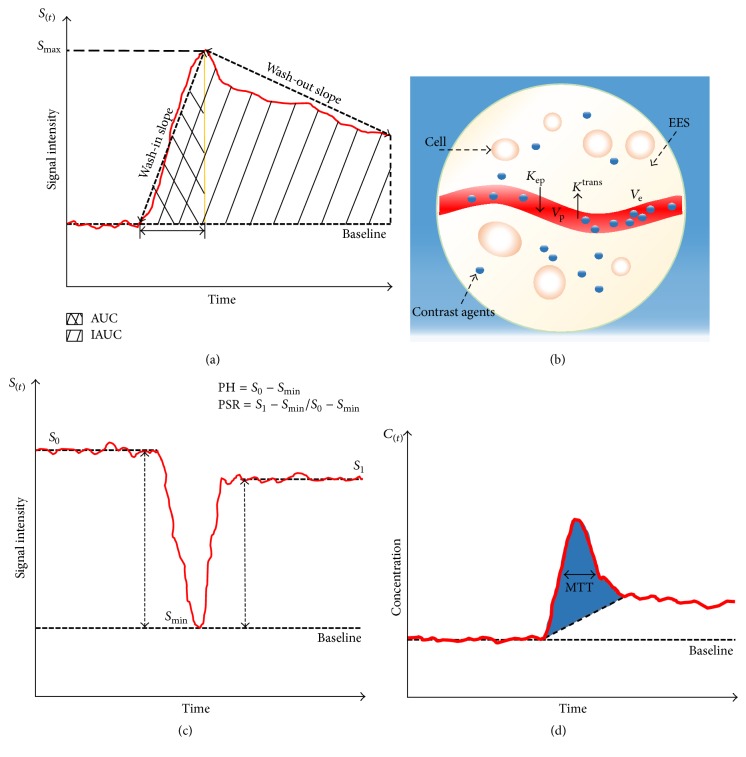
Figure 2.
An illustration of parameters derived from DCE-MRI and DSC-MRI. (a) Semiquantitative parameters from signal intensity curve in DCE-MRI. (b) Schematic diagram of ETK model from DCE-MRI. (c) Calculation of PSR and PH from DSC-MRI. (d) Contrast concentration-time course curve of DSC-MRI. CBV is proportional to determined area under contrast concentration-time course curve (blue shaded area), and CBF is easily calculated given the relationship of MTT and CBV.
Model-dependent parameters can be calculated by fitting various mathematical PK models. Common-used PK models for brain tumors include classic Tofts-Kermode (TK) model and extended TK (ETK) model [25]. Of them ETK model is the most commonly employed in clinical applications (Figure 2(b)). ETK model-derived parameters are summarized in Table 1. These quantitative parameters are physiologically interpretable and better characterize the hemodynamics of vasculatures with more accurate and less data noise compared with model-free parameters [24, 26].
Table 1.
Main perfusion parameters derived from DCE-MRI and DSC-MRI.
| Parameters | Full name | Definition and meaning |
|---|---|---|
| DCE-MRI | ||
| K trans | Volume transfer constant between blood plasma and EES | It describes the leakage rate of GBCAs from the blood plasma towards EES |
| V e | Extravascular extracellular volume fraction | Quantification of cellularity and necrosis in EES. Ve together with Ktrans characterize the microvascular permeability |
| V P | Blood plasma volume | Quantification of the volume of blood plasma |
| K ep | Transfer constant from EES into blood plasma | It is determined by the equation Kep = Ktrans/Ve |
| DSC-MRI | ||
| CBV | Cerebral blood volume | The blood volume in a given region of brain tissue (unit, mL/100 g). It is calculated by integrating the area under the CC-TCC |
| CBF | Cerebral blood flow | The blood volume passing through a given region of brain tissue per unit of time (unit, mL/min/100 g) |
| MTT | Mean transit time | The average time in which blood passes through a given region of brain tissue (unit, s). It is estimated from the CC-TCC as width of the curve at half maximum height |
| PH | Peak height | The maximal drop of signal intensity from precontrast baseline during the first-pass bolus phase of GBCAs. It is correlated with CBV and reflects total blood volume |
| PSR | Percentage of signal intensity recovery | It reflects capillary permeability indirectly, providing information like Ktrans |
| rCBV | Relative cerebral blood volume | Measurement of the relative lesion blood volume compared with that of contralateral white matter. It is proportional to the area under the CC-TCC, providing an estimate of MVD and angiogenesis |
| rCBF | Relative cerebral blood flow | Measurement of the relative lesion blood flow compared with that of contralateral white matter |
| K 2 | Leakage coefficient | Quantification of the degree of vascular permeability using algorithm method for leakage effect correction |
3.2. DSC-MRI
DSC-MRI is based on a dynamic series acquisition of T2/T2∗-weighted images. During the first pass of a bolus GBCAs injection through the vessels, the changes of T2/T2∗ signal intensity induced by the magnetic susceptibility effect are described [27]. Using tracer kinetic modeling and indicator dilution theory, hemodynamic measurements can be evaluated by several kinetic parameters derived from the signal intensity-time course curve (SI-TCC) and corresponding contrast concentration-time course curve (CC-TCC) (Figures 2(c)-2(d)) (Table 1).
Although promising in vascular perfusion evaluation, DSC-MRI has some limitations. The T2∗-weighted technique may generate strong susceptibility artifacts, rendering DSC-MRI insufficient for assessing infratentorial lesions [27]. More importantly, it is assumed that GBCAs remain in intravascular space with intact BBB in postprocessing PK models [28], which is frequently invalid in gliomas characterized by BBB disruption and vascular hyperpermeability. The GBCAs extravasation can produce a strong and competing T1 contrast effect, known as T1 shine-through effect [29], resulting in rCBV misestimate. To this end, several methods have been proposed to minimize T1 contamination such as preenhancement, focusing analysis on nonenhancing portions, gamma-variate fitting, and low flip angles [30, 31], and hence the extent of vascular permeability can be quantified by K2, a leakage coefficient determined by linear fitting of T2∗ signal intensity curve.
4. Applications of DCE-MRI and DSC-MRI in Gliomas
4.1. Tumor Grading
Accurate glioma grading is of great importance for clinical decision making and personalized management. Histopathologic biopsy is currently the gold standard for glioma grading in clinical practice. However, it encounters inherent sampling bias, invasive procedure, and interobserver variability. Moreover, biopsy specimen may not be representative of the tumor panorama characteristics due to the improper resection and intratumoral heterogeneity. It is crucial to establish an accurate diagnosis without biopsy if (1) the lesion is located at critical functional brain areas or require no surgical removal and (2) patients are in poor general condition. Conventional structural MRI techniques are insufficient for accurate glioma grading due to the relatively poor sensitivity and specificity of patterns and extent of contrast enhancement [32]. Up to 45% of nonenhancing gliomas are malignant and approximately 20% of enhancing oligodendrogliomas are benign [33, 34]. PW-MRI techniques enable qualitative and quantitative delineation of the entire tumor microvascular hemodynamics, helping in tumor grading and targeted biopsy (Table 2).
Table 2.
Examples of perfusion MRI for glioma grading.
| Study (year) (ref) | Group (n) | Average age (years) | Imaging modality (method or model; parameter analysis) | Indexes | Results | Limitations |
|---|---|---|---|---|---|---|
| Maia et al. (2005) [35] | Grade II (13) Grade III (7) |
36 | DSC-MRI (leakage effect uncorrected; ROI-based analysis) | rCBV | Positive correlation between rCBV and tumor grade and VEGF expression | Impact of leakage effect on rCBV accuracy; small sample size |
|
| ||||||
| Boxerman et al. (2006) [38] | Grade II (11) Grade III (9) Grade IV (23) |
52 | DSC-MRI (algorithm for leakage correction; ROI-based analysis) | rCBV | Significant correlation between tumor grade and corrected rCBV | rCBV threshold to discriminate tumor grade was not provided |
|
| ||||||
| Law et al. (2007) [41] | Grade II (31) Grade III (30) Grade IV (31) |
43 | DSC-MRI (γ-variate function for leakage correction; histogram analysis) | rCBV | Positive correlation between all parameters and tumor grade; more specific than rCBVmax using histogram analysis | Histogram was based on whole tumor ROI probably including normal brain tissues |
|
| ||||||
| Emblem et al. (2008) [42] | LGG (24) HGG (29) |
52 | DSC-MRI (γ-variate function for leakage correction; histogram analysis) | rCBV | Increased diagnosis accuracy and interobserver agreement were obtained using histogram method | Only the peak height of histogram distribution was measured |
|
| ||||||
| Server et al. (2011) [39] | Grade II (18) Grade III (14) Grade IV (47) |
57 | DSC-MRI (algorithm for leakage correction; ROI-based analysis) | rCBV rCBF K2 |
All parameters were correlated with tumor grade; the diagnostic power of rCBV was better than K2 | Influence of steroid treatment on correlation between K2 and tumor grade |
|
| ||||||
| Yoon et al. (2014) [57] | LGG (12) HGG (48) |
50 | DSC-MRI (γ-variate function for leakage correction; ROI-based analysis) | rCBV | Significant difference of rCBV between HGG and LGG | Subjectivity and neglect of the heterogeneity using ROI-based analysis |
|
| ||||||
| Aprile et al. (2015) [45] | HGG (31) LGG (18) |
55 | DSC-MRI (preload for leakage correction; ROI-based analysis) | PSR rCBV |
Both the two parameters were significantly different between LGG and HGG; PSR was better than rCBV for grading | The relative small sample number of grade III glioma |
|
| ||||||
| Smitha et al. (2015) [46] | HGG (25) LGG (39) |
38 | DSC-MRI (leakage effect uncorrected; ROI-based analysis) | rPSR rCBV rCBF |
Positive correlation between all parameters and tumor grade; the diagnosis performance of rPSR was better than rCBV and rCBF | Impact of leakage effect on rCBV accuracy |
|
| ||||||
| Choi et al. (2013) [47] | LGG (10) HGG (23) |
51 | DCE-MRI (ETK model; ROI-based analysis), DWI |
K
trans Ve Vp ADC |
Significant difference in Ktrans and Ve between LGG and HGG; no statistic difference in Vp and ADC between the two tumor groups | Small sample size |
|
| ||||||
| Zhao et al. (2015) [52] | LGG (9) HGG (15) |
46 | DCE-MRI (TK model; ROI-based analysis), DWI |
K
trans Ve IAUC ADC |
Significant difference of all parameters between LGG and HGG; Ktrans was the most valuable parameter | Small sample size; lack of correlation between histopathology and imaging biomarkers |
|
| ||||||
| Jung et al. (2014) [53] | Grade II (7) Grade III (8) Grade IV (13) |
49 | DCE-MRI (ETK model; histogram analysis) |
K
trans Ve Vp |
Positive correlation between all parameters and tumor grade | Small sample size of LGG; lack the percentile of parameters ranging from 0 to 50 |
|
| ||||||
| Li et al. (2015) [48] | Grade II (15) Grade III (8) Grade IV (9) |
42 | DCE-MRI (TK model; ROI-based analysis), SWI |
K
trans Ve ITSS |
All parameters could distinguish tumor grade except for grade III and grade IV | Small sample size; lack of voxel-to-voxel correlation between imaging features and pathological specimens |
|
| ||||||
| Nguyen et al. (2015) [54] | Grade II (9) Grade III (11) Grade IV (28) |
57 | DCE-MRI (ETK model, phase-derived AIF; ROI-based analysis), DSC-MRI (bookend method; ROI-based analysis) |
K
trans Vp rCBV |
Significant difference between all parameters and tumor grade; improved diagnostic power of parameters using phase-derived AIF method | Only 2 flip angles were used for estimation of the precontrast T1 map; sampling error of histopathological biopsy; some patients received steroids before imaging |
|
| ||||||
| Santarosa et al. (2016) [49] | Grade II (9) Grade III (4) Grade IV (13) |
55 | DCE-MRI (ETK model; histogram/ROI-based analysis), DSC-MRI (algorithm for leakage correction; histogram/ROI-based analysis) |
K
trans Vp rCBV |
Significant difference of all parameters between HGG and LGG; histogram analysis is better than ROI-based method | Small sample size |
Early studies demonstrated that increased rCBV was correlated with more active angiogenesis and aggressive tumor malignancy, being a potential imaging biomarker for preoperative tumor grading [35–37]. Considering the leakage effect, several studies introduced correction methods such as preload and algorithm to improve the rCBV accuracy [38, 39]. The corrected rCBV for tumor grading was more accurate than uncorrected rCBV. Nevertheless, rCBV obtained from region of interest- (ROI-) based method is inefficient for oligodendroglioma grading, which demonstrates elevated rCBV regardless of tumor grade [40]. rCBV from histogram analysis allows more objective and reliable evaluation for glioma grading than ROI-based methods. It could quantify the extent of tumor heterogeneity and discriminate oligodendroglioma from LGG [41–43].
Increased vascular permeability is another predominant characteristic of tumor vessels, playing an adjuvant role for glioma grading. PSR was found to be inversely correlated with vascular permeability [44]. Lower PSR reflects higher vascular leakiness, indicating higher tumor grade [44–46]. Similarly, significantly elevated Ktrans and Ve values reflect greater extent of BBB disruption and higher tumor grade [37, 47–51]. Gliomas are of vascular spatial heterogeneity. Zhao et al. [52] presented a comprehensive analysis of the grading efficacy of quantitative DCE-MRI parameters in different tumor areas. In the tumor parenchyma region, Ve showed the highest diagnostic power, and Ktrans the most specific, and Kep the most sensitive, respectively. While in the peritumoral region, only Ktrans could aid in tumor grading. Histogram analysis and phase-derived arterial input function (AIF) could improve the diagnostic accuracy of DCE-MRI perfusion parameters, allowing us to differentiate grade III from grade IV glioma [53–55]. The rCBV/permeability surface-area product (PS) ratio may also serve as a potential imaging biomarker for glioma grading [56]. It was the highest in grade II and the lowest in grade IV. Moreover, the rCBV/PS ratio was suggestive of different vasculature formation occurring at the microvasculature level, with high value to vessel cooption and low to sprouting angiogenesis. This finding helps investigators to better understand the pathologic basis of the two imaging parameters.
In spite of serving as potential imaging biomarkers for glioma grading, the perfusion parameters are overlapped to some extent among different tumor grades. The thresholds of perfusion indexes, specificity, and sensitivity from different institutions vary considerably, making the comparison difficult. This may be partly attributed to the difference in sample sizes, enrollment criteria, and especially imaging methods. Although there have been a variety of imaging strategies (e.g., bookend technique and phase-derived arterial input function) for improving the accuracy and reproducibility of indexes estimation, standardization and improvement of the imaging acquisition methodology are indispensable for further clinical applications.
4.2. Identification of Molecular Characteristics
Recent in-depth molecular/genetic investigations have led to a profound shift in glioma theranostics based on the substantial progress in genetic alteration profiles. The latest 2016 WHO classification for central nervous system (CNS) tumors integrates the molecular/genetic criteria into histological diagnostics [1]. It emphasizes the molecular classification for gliomas, such as isocitrate dehydrogenase (IDH) gene mutations, epidermal growth factor receptor (EGFR) status, methyl-guanine methyltransferase (MGMT) promoter methylation status, and chromosome 1p/19q codeletion. Preoperative identification of these molecular/genomic characteristics is greatly beneficial for precise diagnosis and personalized therapeutics, guiding treatment decision and improving outcome prediction. The current available method is surgical biopsy along with subsequent genomic and proteomic analysis. The procedure has inherent sampling error due to the tumor heterogeneity, inevitably resulting in erroneous determination. Furthermore, it is invasive, time consuming, and expensive. Imaging genomics bidirectionally links radiographic features to molecular/genomic expression patterns and creates specific imaging biomarkers for noninvasive genomic profiling [58, 59]. Recently, perfusion MRI modalities have attracted considerable attention to distinguish the genotypic profiles of gliomas (Table 3).
Table 3.
Examples of perfusion MRI for identifying molecular characterization.
| Study (year) (ref) | Group (n) | Average age (year) | Imaging modality (method or model; parameter analysis) | Indexes | Results | Limitations |
|---|---|---|---|---|---|---|
| Kickingereder et al. (2015) [60] | Grades II and III: IDH (+) (59) IDH (−) (14) |
49 | DSC-MRI (algorithm for leakage correction; histogram analysis) | rCBV | rCBV was significantly different between IDH mutation and wild-type tumors | Only including grades II and III tumors |
|
| ||||||
| Lee et al. (2015) [61] | HGG: IDH (+) (16) IDH (−) (36) |
50 | DSC-MRI (algorithm for leakage correction; histogram analysis) | nCBV | Significant difference of nCBV between IDH mutation and wildtype; higher heterogeneity in mutation tumor than the wild-type | Not including LGG. Not excluding influence of MGMT mutation |
|
| ||||||
| Tykocinski et al. (2012) [62] | GBM: EGFRvIII (+) (30) EGFRvIII (−) (102) |
61 | DSC-MRI (preload for leakage correction; ROI-based analysis) | rCBV | Strong correlation between rCBV and EGFRvIII status | Relative small sample size of EGFRvIII-positive tumors |
|
| ||||||
| Gupta et al. (2015) [63] | GBM: EGFRam (+) (44) EGFRam (−) (62) EGFRvIII (+) (18) EGFRvIII (−) (47) |
66 | DSC-MRI (γ-variate function for leakage correction; ROI-based analysis) | PSR rPH rCBV |
Higher rCBV and lower PSR were associated with EGFRam; higher rPH was related to EGFRvIII mutation | Pathologic sampling may not be consistent with ROI selection |
|
| ||||||
| Arevalo-Perez et al. (2015) [64] | GBM: EGFRvIII (+) (24) EGFRvIII (−) (58) |
66 | DCE-MRI (ETK model; histogram analysis) |
K
trans Vp |
Strong correlation between both parameters and EGFRvIII status; Vp was better than Ktrans for diagnosis | Not eliminating influence of other molecular mutations |
|
| ||||||
| Jung et al. (2013) [65] | GBM: MGMT (+) (16) MGMT (−) (9) |
52 | DSC-MRI (γ-variate function for leakage correction; ROI-based analysis) | nCBV | nCBV was higher in MGMT-negative tumors than in MGMT-positive tumors | Small sample size |
|
| ||||||
| Moon et al. (2012) [66] | HGG: MGMT (+) (11) MGMT (−) (13) |
51 | DSC-MRI (leakage effect uncorrected; ROI-based analysis) DTI | rCBV ADC |
No significant correlation between rCBV and MGMT | Small sample size; impact leakage effect of rCBV accuracy |
|
| ||||||
| Ahn et al. (2014) [67] | GBM: MGMT (+) (16) MGMT (−) (27) |
58 | DCE-MRI (TK model; ROI-based analysis); DTI |
K
trans Kep Ve ADC |
Only Ktrans was correlated with MGMT | Subjectivity of ROI-based method |
|
| ||||||
| Jenkinson et al. (2006) [68] | Grades II and III: Codeletion (+) (19) Codeletion (−) (18) |
44 | DSC-MRI (leakage effect uncorrected; ROI-based analysis) | rCBV | rCBV was associated with 1p/19q codeletion in oligodendroglioma | Subjectivity of ROI-based method; impact leakage effect of rCBV accuracy |
|
| ||||||
| Emblem et al. (2008) [42] | Grades II and III: Codeletion (+) (11) Codeletion (−) (11) |
52 | DSC-MRI (algorithm for leakage correction; histogram analysis) | rCBV | Histogram analysis of rCBV could differentiate 1p/19q genotype in astrocytic and oligodendroglial tumors | Small sample size; only the peak height of histogram distribution was assessed |
4.2.1. IDH Gene Mutation
IDH (IDH-1/IDH-2) enzymes catalyze isocitrate oxidative decarboxylation to form α-ketoglutarate (α-KG), protecting cells against oxidative damage [69, 70]. IDH gene mutations are present in approximately 50%–80% of grades II and III glioma and nearly all secondary GBM, with IDH-1 much more common than IDH-2 [71–74]. Mutated IDH-1 induces a neomorphic enzyme activity, leading to the overproduction of metabolite 2-hydroxyglutarate (2-HG) [75]. The accumulated 2-HG in excess can competitively inhibit the function of α-KG [76, 77]. Patients with IDH1 gene mutations experience more favorable prognosis than those with wild-type IDH1 gliomas. It has demonstrated that IDH1 mutations could serve as independent prognostic indicators [73, 78, 79]. Noninvasive detection of IDH gene mutations is of great benefit in glioma stratification management.
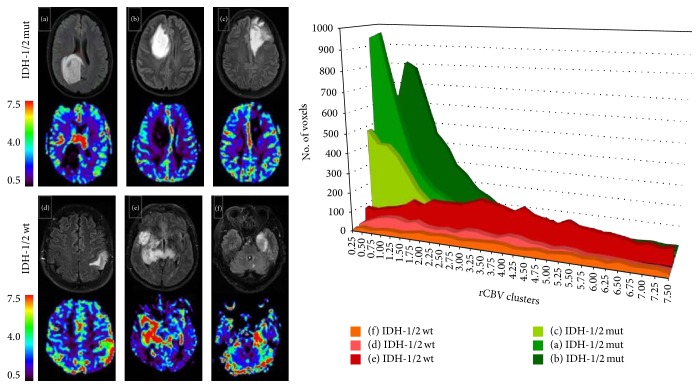
Water suppressed proton-magnetic resonance spectroscopy (1H-MRS) has been explored to noninvasively detect 2-HG in gliomas for identification of IDH-1 gene mutation [80, 81]. However, caution is currently warranted owing to the frequent false negative results and it remains to determine whether 2-HG levels could be qualified to serve as biomarkers for evaluating treatment response, tumor aggressiveness, and other malignant features [82]. Considering that IDH mutation status is associated with hypoxia induced factor-1α, a driving factor in hypoxia-dependent angiogenesis, perfusion MRI may predict this genetic alteration indirectly. Kickingereder et al. [60] found the potential of rCBV for predicting IDH mutation status in LGG and anaplastic glioma. The IDH mutant glioma clustered at decreased rCBV compared with the wild-type counterparts (Figure 3). A one-unit increase in rCBV corresponded to a 2/3 decrease in the odds for an IDH-1/2 mutation, verified successfully in 88% of patients. Similar findings were confirmed by using histogram/ROI-based analysis of normalized CBV (nCBV) mapping and ASL technique [61, 83, 84]. IDH-1 wild-type tumors demonstrated much higher blood perfusion regardless of histologic grade [84]. Lee et al. [61] demonstrated that the slopes between the 10th and 90th of cumulative nCBV histograms were the significant variables in differentiation of IDH-1 genetic status. The results suggested that IDH-1 wild-type glioma possessed more active angiogenesis and less heterogeneous microenvironment. rCBV could be a robust and noninvasive imaging biomarker for predicting IDH mutation status.
Figure 3.
DSC-MRI for identification of IDH mutation status in GBM. Six sets of representative FLAIR and corresponding rCBV images from IDH1/2 mutant and wild-type GBM. Histogram analysis demonstrates that IDH1/2 mutant tumors have substantially lower rCBV value than the wild-type. Reproduce with permission from Kickingereder et al. [60].
4.2.2. EGFR Mutation
EGFR is a transmembrane glycoprotein belonging to receptor tyrosine kinase (RTK) family [85]. Various mutations in EGFR occur in approximately 57% of GBM patients, accompanied with EGFR rearrangement/amplification [78]. EGFR variant III (EGFRvIII), characterized by exons 2–7 deletion in the extracellular domain, is the most common variant of EGFR present in 25%–35% of GBM patients [86]. Cross-talk between EGFR and EGFRvIII enables activating downstream signal pathways such as phosphoinositide 3-kinase, RTK, and phosphatase and tensin homolog, participating in tumor progression, angiogenesis, and treatment resistance [85, 87]. GBM carrying EGFRvIII mutation has a grim prognosis [88]. It has been recognized that EGFR was a potential target for immune-mediated therapy such as tyrosine kinase inhibitors [89], chimeric antigen receptor T-cell (CAR-T) [90], and EGFRvIII-targeted peptide vaccine [91]. Establishing robust imaging biomarkers is of great significance for predicting EGFR-defined subtypes of glioma, to help in clinical decision making.
Previous studies showed that higher contrast enhancement volume and enhancement/necrosis ratio on conventional MRI were associated with EGFR overexpression [92, 93]. Increased T2 intensity to enhancing volume ratio was more likely to reveal EGFRvIII mutation [94]. It indicated that tumor angiogenesis with abnormal perfusion and permeability may reflect the EGFR status. Tykocinski et al. [62] demonstrated that rCBV was remarkably higher in EGFRvIII-positive GBM compared with the negative. The rCBV threshold value of 4.34 acquired on 1.5 T system corresponded with 100% sensitivity and specificity. Gupta et al. [63] analyzed the correlation between EGFR amplification and preoperative DSC-MRI metrics including rCBV, PSR, and relative peak height (rPH). They found that GBM with EGFR amplification presented as higher median rCBV and lower PSR. Also, higher median rPH was associated with EGFRvIII mutation. Recently, Arevalo-Perez et al. [64] evaluated the ability of DCE-MRI for reflecting EGFRvIII expression in GBM patients. Significantly increased Ktrans and Vp mean/histogram values were observed in EGFRvIII-positive GBM, and the predictive power of Vp outperformed those of Ktrans.
4.2.3. MGMT Methylation Status
MGMT is a ubiquitous DNA repair enzyme in glioma cells. The MGMT promoter methylation could induce epigenetic silencing of this gene and consequently result in DNA damage and cell death [95]. MGMT methylation has been reported in 30%–60% of GBM and 50%–84% of anaplastic glioma [96–98]. These patients have more favorable prognosis and prolonged survival [99], better response to temozolomide chemotherapy [100], and increased occurrence of pseudoprogression [101]. Currently, the most universal analytic techniques for MGMT testing include methylation-specific sequencing and methylation-specific reverse-transcription polymerase chain reaction (RT-PCR) [88]. They require invasive procedures and are often subjected to insufficient biopsy sampling due to the intratumoral heterogeneity [102]. Noninvasive detection of MGMT promoter methylation status with preoperative imaging is greatly meaningful.
Some conventional imaging features (such as enhancement pattern, tumor margin characteristic and T2/FLAIR signal intensity) appear to be associated with MGMT promoter methylation status but have some discrepancies among institutions [66, 103, 104]. Part of the explanation may be the nonspecificity of the anatomic imaging features. Several studies have demonstrated perfusion parameters as noninvasive radiophenotypic surrogates for predicting MGMT methylation in GBM. The GBM with MGMT methylation have lower nCBV, with 73.3% sensitivity and 85.7% specificity for discrimination [65]. Ahn et al. [67] evaluated the efficacy of conventional imaging features, quantitative parameters from diffusion tensor imaging (ADC, fractional anisotropy), and DCE-MRI (Ktrans, Kep, and Ve) for predicting MGMT methylation status in GBM. They found that only Ktrans was associated with this genetic alteration. Interestingly, GBM with MGMT methylation showed significantly higher Ktrans, indicating that MGMT methylation may be involved in glioma-associated angiogenesis characterized by high endothelial permeability vasculatures. Although promising, very few studies reported the relationship between MGMT status and PW-MRI parameters. Relevant studies need to be extended to large-sample trials and great efforts are essential to provide a deeper insight into the underlying mechanism of the correlation between imaging features and MGMT status.
4.2.4. Chromosome 1p/19q Codeletion
The unbalanced translocation between chromosome arm 1p and 19q results in loss of heterozygosity (LOH) [105, 106]. The 1p/19q codeletion is a typical characteristic in 40%–90% of oligodendroglioma [107]. Oligodendrogliomas harboring 1p/19q codeletion are associated with higher sensitivity to chemoradiotherapy and prolonged survival than those with intact 1p/19q alleles, irrespective of the tumor grade [108, 109]. Noninvasive identification of this genetic profile is of prominent benefit for prognosis prediction and improved treatment strategies.
Jenkinson et al. [110] reported that rCBV was associated with 1p/19q genotype of oligodendroglioma using ROI-based analysis. Higher rCBV was suggestive of intact 1p/19q alleles and shorter PFS and OS following vincristine chemotherapy but not predictive of chemosensitivity, indicating that rCBV seemed merely a prognostic biomarker in oligodendroglioma with different 1p/19q genotypes. The histogram analysis of rCBV maps could identify low grade oligodendroglial tumor without 1p/19q LOH with high interobserver agreement, with 100% sensitivity and 91% specificity [42]. Combined use of multiparameters from different imaging techniques may improve the discriminative performance in preoperative genetic profiling. High rCBV is associated with angiogenesis and increased mitotic activity. In a recent study by Chawla et al. [111], rCBVmax was used for guiding the selection of optimal 1H-MRS voxels. The incorporation of rCBVmax and metabolite ratios provided improved diagnostic accuracy in distinguishing 1p/19q genotypic profile of oligodendroglioma.
Above-mentioned studies demonstrate that PW-MRI parameters hold great potential implications for reflecting glioma-associated molecular characteristics. However, given the intrinsic limitations of PW-MRI imaging technique, the physiologic description or significance of perfusion parameters is intricate at molecular level and is difficult to recapitulate a certain molecule/gene characterization. For example, EGFR amplification and mutation can result in the overexpression of various downstream effector molecules such as VEGF, interleukin-18, and angiopoietin-like 4 to make synergic effect on tumor neovascularization, consequently altering the vascular structure and function [112–114]. Therefore, perfusion parameters are the comprehensive embodiment of multiple molecule characteristics of glioma indeed. Multimodal and multiparametric imaging based on radiomics and imaging genomics could be a foreground strategy to narrow the gap between imaging features and gene status. Large-scale prospective studies are warranted before being translated into clinical routine.
4.3. Differentiation of Gliomas from Other Brain Tumors
The therapeutics and prognosis of different CNS tumors are of extreme disparity. Preoperative differentiation of gliomas from other brain tumors is important for preoperative staging, intraoperative management, and postoperative treatment. Conventional MRI cannot provide pathophysiological information for identifying glioma, solitary brain metastasis (MET), and primary central nervous system lymphoma (PCNSL), due to their similar imaging performance such as space-occupying and enhancing patterns [115]. Perfusion MRI techniques can delineate the characteristics of tumor vascularity and quantifying vascular perfusion and permeability. They have shown satisfactory efficacy to differentiate glioma from other intracranial tumors (Table 4).
Table 4.
Differential diagnosis in glioma, metastasis, and PCNSL.
| Study (year) (ref) | Tumor type (n) | Average age (year) | Imaging modality (method or model; parameter analysis) | Indexes | Results | Limitations |
|---|---|---|---|---|---|---|
| Law et al. (2002) [116] | HGG (24) MET (12) |
52 | DSC-MRI (leakage effect uncorrected; ROI-based analysis) | rCBV | rCBV in peritumoral region was significantly different between HGG and MET | The peritumoral region was not defined clearly; the threshold value was not provided |
|
| ||||||
| Cha et al. (2007) [117] | GBM (27) MET (16) |
52 | DSC-MRI (alteration of TE and flip angle for leakage correction; ROI-based analysis) | PSR PH |
Significant difference of all parameters between GBM and MET; PSR was the most powerful with 100% specificity | Small sample size; some cases were not confirmed by histopathology |
|
| ||||||
| Mangla et al. (2011) [118] | GBM (22) MET (22) PCNSL (22) |
54 | DSC-MRI (preload for leakage correction; ROI-based analysis) | rCBV PSR |
PSR was better than rCBV for differentiation | Small sample size; impact of steroid treatment on parameter evaluation |
|
| ||||||
| Toh et al. (2013) [119] | GBM (20) PCNSL (15) |
60 | DSC-MRI (algorithm for leakage correction; ROI-based analysis) | rCBV K2 |
Uncorrected rCBV is much better for differentiating | Lack of direct correlation between parameters and histopathologic features |
|
| ||||||
| Xing et al. (2014) [120] | HGG (26) PCNSL (12) |
51 | DSC-MRI (leakage effect uncorrected; ROI-based analysis) | rCBV PSR |
The combination of rCBV with PSR might help in more accurate differentiation | Impact of leakage effect on parameter measurements |
|
| ||||||
| Kickingereder et al. (2014) [121] | GBM (60) PCNSL (11) |
N/A | DCE-MRI (TK model; ROI-based analysis) |
K
trans Ve Kep |
K trans and Kep could identify the two tumors. Ktrans was the optimum parameter | Relative small sample size of PCNSL |
|
| ||||||
| Kickingereder et al. (2014) [122] | GBM (28) PCNSL (19) |
66 | DSC-MRI (preload for leakage correction; ROI-based analysis), DWI, SWI | rCBV ADC ITSS |
Multiparametric MRI allowed differentiation of GBM from PCNSL | Small sample size |
|
| ||||||
| Zhao et al. (2015) [52] | LGG (9) HGG (15) MET (5) |
46 | DCE-MRI (TK model; ROI-based analysis) |
K
trans Ve IAUC |
All parameters were significantly different between LGG, HGG, and MET. IAUC had the most diagnostic power | Small sample size; subjectivity of ROI selection |
|
| ||||||
| Jung et al. (2016) [123] | GBM (26) MET (32) |
N/A | DCE-MRI (ETK model, ROI-based analysis) |
K
trans Vp AUC Washout log slope |
Semiquantitative parameters could differentiate between GBM and hypovascular metastasis | Subjectivity of ROI selection |
4.3.1. Solitary Brain Metastasis
GBM and metastatic brain tumor are the two most common malignant intracranial tumors representing similar imaging appearances and enhancing patterns on conventional MRI [124], whereas therapeutic decisions and prognosis are substantially different. Accurate differentiation of the two distinct entities is of great importance for clinical management. The morphology and functional status of tumor vasculature differ greatly between the two types of tumors. GBM is characterized by increased perfusion and heterogeneous BBB disruption with microvascular morphology and permeability varying from relatively normal to increased [125]. The tumor margin represents vasogenic edema with infiltrative tumor cells along with perivascular spaces [126]. In contrast, the absence of BBB components in brain metastasis often results in relatively low perfusion and uniformly increased capillary permeability throughout the tumor, causing pure vasogenic edema without infiltrative tumor cells or abundant angiogenesis [126, 127]. DCE-MRI and DSC-MRI can provide physiological information which is unavailable on conventional MRI to settle the diagnostic dilemma.
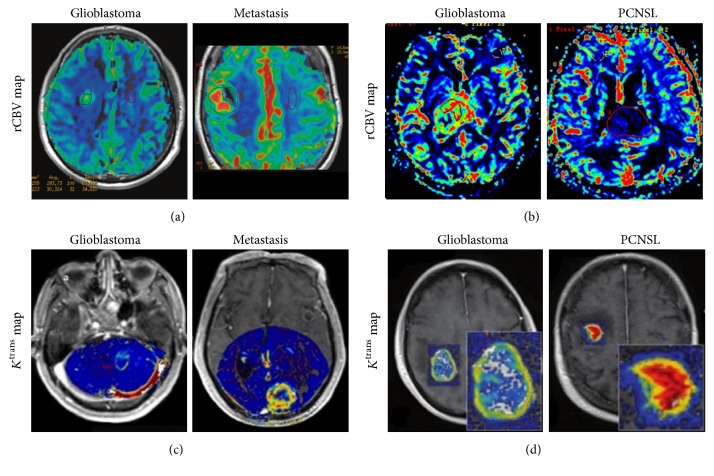
DSC-MRI could differentiate subtle differences of vascular perfusion. Higher rCBVmean in the peritumoral region and higher PSR were present in HGG compared with metastasis (Figure 4(a)) [116–118, 128]. Similarly, DCE-MRI can also identify the two malignancies. Although there is no difference for Ktrans and Vp between GBM and melanoma metastasis, hypovascular metastasis could be differentiated from GBM using logarithmic slope of the wash-out phase and AUC [123]. Zhao et al. [52] found that Ve and IAUC in the tumor parenchyma and Ktrans in peritumoral area could discriminate HGG from solitary metastasis (Figure 4(a)). All the parameters in LGG, HGG, and metastasis were lower, intermediate, and higher, respectively.
Figure 4.
DSC-MRI (a) and DCE-MRI (b) for differentiation of GBM, PCNSL, and metastasis. rCBV maps demonstrate different characteristic features in the three distinct entities, with significantly higher rCBV value of GBM compared with metastasis and PCNSL. The Ktrans value of GBM is significantly lower than metastasis and PCNSL. Reproduce with permission from Mangla et al. [118], Xing et al. [120], Zhao et al. [52], and Kickingereder et al. [121].
Although PW-MRI provides valuable information for antidiastole between gliomas and solitary brain metastases, it is undeniable that the threshold of indexes for diagnosis varies among the studies because of different origin of metastases except for various imaging acquisitions. More importantly, DCE-MRI is weak in differentiating GBM and highly vascular brain metastasis such as melanoma metastasis on account of their similar vascular function. DWI-derived ADC value could be an alternative and complementary imaging biomarker to differentiate the two tumor entities [129].
4.3.2. Primary Central Nervous System Lymphoma (PCNSL)
PCNSL is a rare neoplasm constituting up to 6% of intracranial malignant tumors [130]. The diffusely infiltrative pattern of PCNSL resembles the infiltrative behavior of gliomas [131]. PCNSL is also known to have greatly destroyed vessel architecture and lack abundant neovascularization, thus demonstrating relatively low blood perfusion and increased vascular permeability [132]. The medical staging, surgical planning, and therapeutic decisions between PCNSL and HGG are completely different. Despite having some characteristics on conventional MRI, differentiation of the imaging appearances of PCNSL from those of HGG is difficult or even impractical [133, 134]. Preoperative differentiation of HGG from PCNSL using advanced imaging techniques is of great clinical significance. PW-MRI has gained an important clinical role for differentiation of GBM from PCNSL.
Higher rCBV and lower PSR were suggestive of GBM (Figure 4(b)) [120, 135–138]. Despite the consistent results, the cut-off values of rCBV and PSR were considerably variable among different studies [118–120]. It seems to indicate that these indexes not only reflect the pathophysiologic features but also are influenced by different imaging protocols and acquisitions. rCBV with leakage correction is regarded to own improved accuracy. However, Toh et al. [119] found that uncorrected rCBV seemed to have better diagnostic performance than corrected rCBV in differentiating PCNSL from GBM. This may be partly explained by the greater restoration of CBV in PCNSL because of its higher vascular permeability, leading to decreased CBV differences between the two tumors. Similar results were observed by Nakajima et al. [137]. Thus, it is more rational and reliable to evaluate the vascular permeability for differentiation. As expected, PCNSL demonstrated significantly higher Ktrans, K2, and Kep than GBM due to their severe vascular leakage [52, 119, 137], and Ktrans had far superior diagnostic performance than K2 [119, 121] (Figure 4(b)). Furthermore, integration of advanced MRI techniques has been explored to improve the diagnostic performance by various studies [122, 139, 140]. Kickingereder et al. [122] demonstrated that combined evaluation of mean ADC, mean rCBV, and presence of intratumoral susceptibility signals (ITSS) improved the probability for differentiating PCNSL from atypical GBM. The integrated multiparametric assessment correctly predicted histologic results in 95% of PCNSL and 96% of atypical GBM. However, one recent study showed that relative Vp from DCE-MRI did not outperform ADC alone, or in combination for diagnostic accuracy [141]. Despite the fact that more prospective studies are necessary to confirm these findings, PW-MRI may be helpful to support presumed diagnosis of GBM marked by higher blood perfusion and decreased permeability.
4.4. Treatment Response Assessment
The current standard of care for GBM is concomitant and adjuvant chemoradiotherapy following maximum safe surgical resection. The treatment options are influenced by various factors and need to be timely adjusted at different stages of care. Accurate treatment response assessment is greatly important to clinical decision making and personalized medicine. Macdonald Criteria is based on treatment response assessment via evaluation of the contrast-enhancing areas on MRI [142]. This criterion has critical limitation as it only focuses on the contrast-enhanced component of the tumor. With the recognition of the importance of nonenhancing region when monitoring therapeutic response, the nonenhancing region of the tumor is taken into account in updated guidelines for Response Assessment in Neuro-Oncology (RANO) [143, 144]. Yet, the morphologic features underlying complicated treatment response (such as pseudoprogression, pseudoresponse, and radiation reaction), tumor progression, recurrent lesion, and detection of nonenhancing region with conventional MRI are insufficient to fully evaluate therapy response [145]. Perfusion MRI techniques offering vascular functional information have demonstrated their powerful capacity to help characterize these treatment-related imaging changes.
4.4.1. Pseudoprogression
Approximately up to 50% of glioma patients treated with chemoradiotherapy can develop transient new areas of increasing contrast enhancement or edema, termed pseudoprogression (PsP), which is easily confounded with true progressive disease (PD) [146]. PsP is typically recognized at the follow-up MRI examinations obtained within the first 3 months after chemoradiotherapy. It is characterized by increased capillary permeability with edema and reduced overall vessel perfusion, considered to be induced by chemoradiotherapy-related vascular damage/inflammation [147]. This reaction is often clinically asymptomatic and can resolve spontaneously. PsP has been found to be associated with increased survival, possibly because of more active inflammatory response and increased probability of MGMT promoter methylation in this population [147, 148]. Failure to accurately identify PsP would lead to needless surgical intervention, premature termination of an effective treatment, or redundant chemotherapeutics [149]. DCE-MRI and DSC-MRI have been widely proposed to differentiate PsP from PD (Table 5).
Table 5.
Differentiation of pseudoprogression from true progression.
| Study (year) (ref) | Group (n) | Average age (year) | Imaging modality (method or model; parameter analysis) | Indexes | Threshold (Sp%, Sn%) | limitations |
|---|---|---|---|---|---|---|
| Mangla et al. (2010) [150] | PsP (7) TP (12) |
61 | DSC-MRI (algorithm for leakage effect correction; ROI-based analysis) | rCBV | Percentage change in rCBV for discrimination of PsP and TP (85.7%, 76.9%) | Retrospective; different treatment management; small sample size |
|
| ||||||
| Martínez-Martínez and Martínez-Bosch (2014) [151] | PsP (17) TP (7) |
48 | DSR-MRI (leakage effect uncorrected; ROI-based analysis) | rCBV rPSR rPH |
rPH = 1.37 (82.2%, 88%) rCBV = 0.9 (100%, 100%) rPSR = 99% (70.6%, 100%) |
Retrospective; small sample size; impact of corticoid therapy on parameter evaluation; lack of histological confirmation |
|
| ||||||
| Prager et al. (2015) [152] | PsP (8) TP (43) |
55 | DSC-MRI (γ-variate function for leakage correction; ROI-based analysis) | rCBVlesion rCBVROI |
rCBVlesion = 1.07 (75%, 100%) rCBVROI = 1.74 (75%, 92.9%) |
Retrospective; small sample size of PsP; MGMT in some patients may affect the perfusion parameters |
|
| ||||||
| Baek et al. (2012) [153] | PsP (37) TP (42) |
49 | DSC-MRI (γ-variate function for leakage correction; histogram analysis) | nCBV | Percent change of skewness: 1.27% (79.2%, 85.7%) Percent change of kurtosis: 14% (73.0%, 61.9%) |
Different therapies in patients; results were obtained from only one observer |
|
| ||||||
| Tsien et al. (2010) [154] | PsP (13) TP (14) |
52 | DSC-MRI (leakage effect uncorrected, parametric response map) | rCBV rCBF |
Not provided; patients with progressive had reduced rCBV | Leakage effect may underestimate rCBV value |
|
| ||||||
| Gahramanov et al. (2013) [155] | PsP (9) TP (10) |
N/A | DSC-MRI (ferumoxytol for leakage correction) | rCBV | rCBV = 1.5 (Sp%, Sn% not provided) | Lack of histopathologic confirmation; small sample size |
|
| ||||||
| Suh et al. (2013) [156] | PsP (36) TP (43) |
50 | DCE-MRI (nonmodel fitting; histogram analysis) | AUCR mAUCRH |
mAUCRH = 0.31 (82.9%, 90.1%) AUC50 = 0.19 (83.1%, 87.2%) |
Lack of correlation between imaging measurements and specimen histology |
|
| ||||||
| Yun et al. (2015) [157] | PsP (16) TP (17) |
55 | DCE-MRI (ETK model; histogram analysis) |
K
trans Ve Vp |
K
trans = 0.347 (94%, 59%) Ve = 0.570 (56%, 88%) No significant difference of Vp between PsP and TP group |
Small relative sample size; lack of histological confirmation |
PD demonstrated higher rCBV and lower PSR, while PsP exhibited decreased rCBV and rPH [145, 150–152, 158, 159] (Figure 5(a)). Considering the significant tumor heterogeneity and series changes of chemoradiation-induced vascular architectures, rCBVmean from ROI-based method is subjective and insufficient for delineating the exhaustive tumor characteristics. Percent changes of skewness and kurtosis on nCBV histograms were effective in predicting early treatment response, and the histographic pattern of nCBV demonstrated the best independent predictive efficacy [153]. Tsien et al. [154] developed parametric response map (PRM), a voxel-wise analytic approach, for quantifying treatment-associated hemodynamic alterations in HGG. Paradoxically, they found that decreased PRMrCBV at week 3 after chemoradiotherapy was associated with true progression. One possible explanation is that the parameters obtained at different time points only reflect the vascular characteristics of a specific stage. The decreased rCBV may actually be attributed to the higher BBB permeability at tumor progressive stage and the nonuse of leakage correction. Ferumoxytol is a nanosized blood pool agent requiring no contrast agent leakage correction. rCBVmean using ferumoxytol has been found to be superior to that of gadoteridol for differentiation of PsP from tumor progression [155, 160].
Figure 5.
Discrimination of PsP from PD using DSC-MRI and DCE-MRI. (a) Contrast-enhanced T1WI of GBM treated with temozolomide demonstrates increased contrast enhancement suspicious for both PsP (top row) and PD (bottom row). Corresponding rCBV maps show low perfusion in PsP and high perfusion in PD; (b) Ktrans maps demonstrate decreased Ktrans value in PsP (top row) compared with PD (bottom row). Reproduce with permission from Shin et al. [158] and Thomas et al. [161].
DSC-MRI has intrinsic sensitivity to susceptibility artifact, commonly caused by posttreatment hemorrhage and calcification [11]. Therefore, DCE-MRI has advantages over DSC-MRI for differentiating PsP from PD. Variations of Ktrans, Ve, and Vp are effective diagnostic indicators [157, 161] (Figure 5(b)). However, these quantitative parameters are inevitably affected by various methodological factors, such as parameter coupling, AIF measurement, and model fitting instability [162]. Semiquantitative parameters, while not physiologic, can be easily obtained and have also been investigated for treatment response assessment. The maximum slope of initial enhancement and final area under the time-signal intensity curve ratio (AUCR) could differentiate PsP from early tumor progression in GBM patients [156, 163]. Suh et al. [156] showed that the mean AUCR at a higher curve (mAUCRH) and the 50th cumulative AUCR histogram parameter (AUCR50) were the best and the most specific independent predictor of PsP, respectively.
While a number of studies have employed PW-MRI to discriminate PsP from PD in GBM, cut-off values of parameters with specificity and sensitivity across institutions are somewhat different even not comparable because of small sample size, as well as lack of standardization of imaging protocols and accordant inclusive criterion of individuals. Accuracy and reproducibility of perfusion parameters are inevitably affected by technical aspects (e.g., leakage correction, types of GBCAs, and PK model fitting) and parameter analysis (e.g., ROI-based/histogram analysis and parametric response map). The inclusion of patients who have already received corticoid therapy may bias the results of parameters evaluation. In addition, the initial and end timing for imaging monitoring, types, and doses of drug are inconsistent. Therefore, more well-controlled studies and coregistration of PW-MRI with corresponding histological mapping are urgently needed for reconfirmation of these results.
4.4.2. Pseudoresponse
Antiangiogenic therapies (such as bevacizumab and cediranib) could induce early decrease in contrast enhancement and edema on conventional MRI due to the restored BBB integrity and reduced endothelial permeability, resulting in prolonged progress-free survival (PFS) but modest benefit of overall survival (OS) [164, 165]. This phenomenon is termed as pseudoresponse. The explanation may be attributed to transient vascular normalization [166, 167], rather than true improvement in tumor status. Rebound enhancement and edema appeared when a “drug holiday” is encountered, arising from the reversal of vascular normalization. And pseudoresponse could occur when restarting antiangiogenic therapy [168]. Conventional MRI fails to prognosticate and stratify OS of patients treated with antiangiogenic therapy. Although the degree of decreased contrast enhancement to these therapies after one day of treatment is associated well with survival, progressive enhancement is predictive of shorter OS. However, patients with improved enhancement corresponding to those with stable enhancement have no survival benefits because of pseudoresponse [167, 169]. It is of great importance to stratify early therapy response and predict treatment success after antiangiogenic therapy initiation.
PW-MRI may help differentiate true response from PD by predicting OS. A multicenter trial investigated the efficacy of standardized rCBV (sRCBV) and mean tumor rCBV normalized to white matter (nRCBV) for predicting OS in recurrent GBM after treatment initiation [170]. The nRCBV at week 2 and sRCBV at week 16 significantly decreased in patients surviving at least one year (OS-1). Increased rCBV values indicated significantly shorter OS, being a good prognostic marker for OS-1. Similarly, reduced Ktrans and Ve could be predictive of pharmacodynamic effect as early as one day following antiangiogenic treatment initiation [171–173]. Sorensen et al. [167] described vascular normalization index incorporating Ktrans, CBV, and circulating collagen IV. The index was a potential early candidate predictor for PFS and OS. Similarly, by comparing the baseline and 1-day posttreatment value of DSC-MRI indexes using leakage correction method, Emblem et al. [174] showed a novel vascular normalization parameter combining CBV and apparent transfer constant (Ka) to predict PFS and OS in GBM patients after anti-VEGF treatment.
Due to the diverse imaging protocols applied, the use of standardized parameters (sRCBV) and model-free parameters (IAUC) could be alternative to reduce variability and improve accuracy and reproducibility when comparing results from multiple institutions or using different acquisition strategies.
4.4.3. Nonenhancing Regions of Tumor
The current standard response assessment of glioma is lined with the RANO criteria, especially including the abnormal hyperintensity of T2/FLAIR in nonenhancing regions [143]. However, vasogenic edema and gliosis in nonenhancing regions always confound the changes related to antiangiogenic treatment, which may mislead the response assessment. Differentiation of vasogenic edema from infiltrative tumors is of great significance. In a study by Artzi et al. [175] the nonenhancing hyperintense area on FLAIR was classified into vasogenic edema and infiltrative tumor area based on multiple MRI parameters. The former was characterized by decreased rCBV, rCBF and increased FLAIR values, and the latter increased perfusion. All perfusion parameters were correlated with PFS after bevacizumab therapy. Subsequently, they segmented GBM into three components using DSC-MRI and DCE-MRI [176], which include enhancing permeable area, the nonenhancing hypoperfusion area representing vasogenic edema, and the nonenhancing hyperperfusion area representing infiltrative tumor. Alternatively, DSC-MRI data with FL temporal principal component analysis in GBM could help discriminate peritumoral regions infiltrated with tumor cells from surrounding normal tissues [177]. Higher rCBV in nonenhancing tumor region was also suggestive of shorter OS and served as an independent prognostic marker [178]. Recently, Akbari et al. [179] reported multiparametric imaging pattern analysis including rCBV to delineate surrounding infiltrative tumor margin. The visually imperceptible imaging patterns on conventional MRI were revealed. They could delineate the extent of infiltrative tumor and predict the location of tumor recurrence. Integrating perfusion MRI and conventional MRI could hence improve the therapeutic response assessment and pave the way for personalized treatment strategies.
4.4.4. Tumor Recurrence and Late Radiation Necrosis
Radiation-induced brain injuries are mainly classified into three stages based on the occurrence time: acute (during radiation), subacute (within 3 months after radiation), and late (months to years after radiation). The acute and early subacute injuries are mainly caused by vasodilation, BBB disruption, and edema, usually present as relatively unchanged MR appearance [147]. The late radiation necrosis (RN) frequently occurred in GBM patients within 3 to 12 months after radiotherapy [180]. Due to the fibrinous necrosis triggered by ischemia, vasodilation and endothelial damage, late RN can present as brain edema, new lesions, or progressive contrast enhancement on conventional MRI, which is indistinguishable from that of recurrent tumor lesions [181]. Accurate differentiation of tumor recurrence from treatment-related changes is clinically important for follow-up patient management strategies. Perfusion MRI has shown great capability to differentiate the two entities (Table 6).
Table 6.
Discrimination of recurrent glioma from radiation necrosis.
| Study (year) (ref) | Group (n) | Average age (year) | Imaging modality (method or model; parameter analysis) | Indexes | Threshold (Sp%, Sn%) | Limitations |
|---|---|---|---|---|---|---|
| Barajas et al. (2009) [182] | RN (17) rGB (40) |
54 | DSC-MRI (alteration of TE and flip angle for leakage correction, ROI-based analysis) | rCBV rPH rPSR |
rPH = 1.38 (81.38%, 89.32%) rPSR = 87.3% (76.19%, 78.26%) rCBV = 1.75 (71.58%, 78.92%) |
Impact of partial volume averaging effect on parameter evaluation |
|
| ||||||
| Hu et al. (2009) [183] | rHGG (24) RN (16) |
47 | DSC-MRI (baseline subtraction method for leakage correction; ROI-based analysis) | rCBV | rCBV = 0.71 (100%, 91.7%) | Various tumor types; inconsistent radiation dose and different therapies |
|
| ||||||
| Bisdas et al. (2011) [184] | rHGG (12) RN (6) |
N/A | DCE-MRI (TK model; ROI-based analysis) |
K
trans Ve Vp IAUC |
K
trans = 0.19 (83%, 100%) IAUC = 15.35 (71%, 71%) No significant difference of Ve and Vp between RN and rHGG |
Small sample size; lack of histopathologic confirmation in some cases |
|
| ||||||
| Shin et al. (2014) [158] | Recurrent glioma (19) RN (4) |
55 | DCE-MRI (TK model; ROI-based analysis), DSC-MRI (preload for leakage corrected; ROI-based analysis) | rKtrans rIAUC rCBV |
rCBV = 2.33 (70%, 72.2%) rKtrans = 2.1 (80%, 61.1%) rIAUC = 2.29 (70%, 66.6%) |
Relative small sample size; ROI-based method was not comprehensive |
|
| ||||||
| Larsen et al. (2013) [185] | Recurrent glioma (11) RN (3) |
56 | DCE-MRI (deconvolution technique) | CBV | CBV = 2.0 ml/100 g (100%, 100%) | Small sample size; sample bias in histological analysis; various tumor types |
|
| ||||||
| Masch et al. (2016) [186] | Recurrent glioma (16) RN (8) |
51 | DSC-MRI (preload for leakage correction; ROI-based analysis) | rCBV | Not provided; elevated rCBV in recurrent lesion compared with RN | Various tumor types; lack of histological confirmation in some cases |
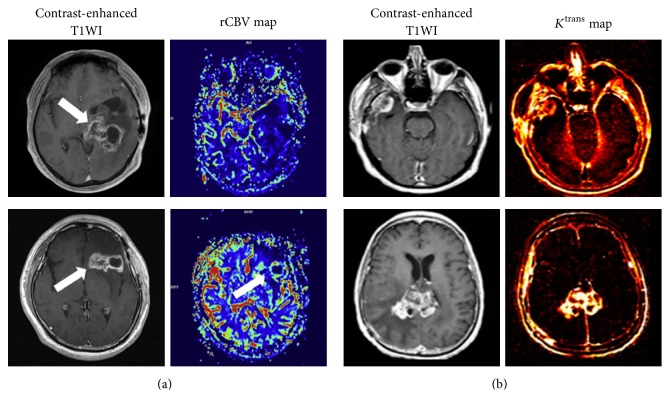
Several studies demonstrated that recurrent glioma owned higher rCBV and lower PSR compared with radiation injury [152, 182, 183]. However, there is an overlap of DSC-MRI parameters between RN and recurrent tumor. It presents as variable cut-off values among institutions, leading to inconsistent sensitivity and specificity [182, 183]. As the vascular permeability in recurrent HGG differs from RN, Bisdas et al. [184] showed increased Ktrans and IAUC indicating recurrent lesions and decreased values for radiation injury (Figure 6(a)), whereas Ve and Kep held no differentiating value. Shin et al. [158] compared the utility of DCE-MRI and DSC-MRI. It showed no significant difference for differentiating performance between these two imaging modalities using single index. However, when combination of relative Ktrans (rKtrans) and relative IAUC (rIAUC) was used, DCE-MRI seemed to outweigh DSC-MRI. CBV measured by DCE-MRI using deconvolution technique could offer equivalent or improved evaluation compared to fluorodeoxyglucose-positron emission tomography (FDG-PET) for differentiation [185]. The CBV threshold of 2.0 ml/100 g enabled the detection of regressing lesions with 100% sensitivity and 100% specificity. In addition, combined assessment using diffusion tensor imaging (ADC, mean parallel eigenvalues, and mean perpendicular eigenvalues) and DSC-MRI (rCBV) characteristics showed improved differentiation, particularly in the lesions with increased rCBV and decreased ADC values [186] (Figure 6(b)).
Figure 6.
Discrimination of RN from recurrent GBM using DCE-MRI (a) and DSC-MRI (b). Contrast-enhanced T1WI demonstrates similar contrast enhancement in recurrent glioblastoma (top row) and RN (bottom row). Corresponding rCBV and Ktrans maps show significant difference between these two entities, with higher Ktrans and rCBV for recurrent tumor (top row) but low for RN (bottom row). Reproduce with permission from Bisdas et al. [184] and Masch et al. [186].
Nonstandardized imaging acquisition renders a wide range of sensitivity and specificity using PW-MRI. Some other factors such as different inclusive criteria, tumor grades and radiation timing, and dose may also disturb the diagnostic accuracy of perfusion parameters. Moreover, histopathological validation lacks in published studies. Further work on significant improvement of imaging method and correlation between the imaging and histologic features is warranted to draw a definite conclusion.
4.5. Predicting Prognosis
Initial patient stratification is clinically important for optimized and individualized therapeutic regimens. Multiple efforts are ongoing for survival prediction in glioma patients. Glioma is characterized by abnormal vasculature with active angiogenesis. Perfusion MRI techniques providing physiologic information have been widely investigated for noninvasive prognosis prediction in glioma patients.
rCBV has demonstrated predictive value for gliomas regardless of treatment [150, 178, 187–189]. Elevated rCBV in untreated glioma was associated with OS [178]. It is because that tumor angiogenesis induces increased CBV, resulting in aggressive tumor growth. High rCBV (>1.75) indicated more rapid and earlier progression [188]. Increased rCBV could predict the malignant transformation of LGGs as early as 12 months in advance compared to apparent contrast enhancement on T1-weighted imaging [187]. In addition, rCBV of the nonenhancing region in GBM was associated with OS and PFS and could provide unique prognostic information independent of the morphologic, genomic, and clinical features [190]. DCE-MRI parameters (Ktrans and Vp) also appear to be prognostic markers [191, 192]. Very recently, Kim et al. [193] evaluated the prognostic value of T2 high signal intensity lesions without enhancement in GBM using DCE-MRI. They found that the percentile of Ktrans, Ve, and Vp could identify early disease progression. The 99th percentile of Ktrans holds potential as a candidate prognostic imaging biomarker. Combination of Ktrans and rCBV seems to be more powerful than single parameter for survival prediction of newly diagnosed GBM patients [194]. Burth et al. [195] found that clinical parameters (age, sex, resection extent, and Karnofsky performance scale) outperformed MRI parameters (Ktrans, rCBV, and ADC) for predicting prognosis of GBM patients. It suggests that physiologic MRI parameters may be auxiliary indexes for patient prognostication but offer additional values to clinical data for improved prognosis prediction.
5. Current Challenges
Contrast-enhanced PW-MRI techniques are becoming increasingly common approaches for clinical applications in gliomas. They facilitate better understanding of a variety of hemodynamic pathologies and the underlying mechanisms of tumor neovascularization. However, there are still some unresolved issues when implementing PW-MRI in contemporary radiology practice. We noted that perfusion parameters are inevitably influenced by various hemodynamic factors, types of GBCAs, and total acquisition time. For example, Ktrans is determined by blood flow and PS. In high leakage condition or low-molecular-weight contrast agents administrated, Ktrans depends almost entirely on blood flow. Increased acquisition time with low temporal resolution can also disturb the accuracy of Ktrans, resulting in underestimate. Furthermore, glioma vasculatures describe the anatomical and functional abnormalities within tumors in spatiality. The frequently used hotspot analysis pays too little attention to the tumor heterogeneity, which cannot realistically delineate the tumor panorama. Heterogeneity analysis such as histogram and texture methods can provide more detailed information and benefit over simple “average value” measurement [196, 197]. In addition, several studies focused on optimizing the postprocessing analysis [198–200]. Standardization of rCBV combined with leakage correction may be better than normalized rCBV for eliminating the subjective ROI selection, helping to reduce variability of quantitative comparison across studies [200].
Perfusion parameters are affected by a complex interaction of factors. In multicenter clinical trials, even minor differences of benchmarked standards may result in significant changes in perfusion parameters. These variables include (1) MR scanners (e.g., field strength, gradient system and manufacturer, and pulse sequences); (2) imaging acquisition protocols (e.g., acquisition parameters, spatial and temporal resolution, and coverage); (3) GBCAs administration (e.g., preload, dynamic bolus, injection dose and rate, and timing); and (4) postprocessing methods (e.g., modeling selection, leakage correction, AIF determination, and ROI/histogram/voxel-wise analysis). These integrated factors across institutions hinder the accuracy and reproducibility of results and thus impede further development of these two powerful imaging modalities into routine clinical setting. A recent joint meeting provided consensus recommendations for a standardized Brain Tumor Imaging Protocol (BTIP) for multicenter studies in GBM [201]. The Clinical Practice Committee of the American Society of Functional Neuroradiology (ASFNR) proposed recommendations for DSC-MRI acquisition protocols and validation of imaging biomarkers [31]. And, the Quantitative Imaging Biomarkers Alliance of the Radiological Society of North American (QIBA of RSNA) established an updated technical guideline for DCE-MRI data acquisition and analysis, in which Ktrans and IAUC were recommended as standard endpoints [202, 203]. These proposed recommendations will significantly reduce variability and allow interpretation of imaging results and also provide benchmarks for comparison to further improvements and innovations.
6. Conclusion and Perspectives
Despite some clinical limitations and unsolved issues, the current evidence available demonstrated the tremendous foreground of PW-MRI for improving glioma management. Imaging protocols standardization is urgently demanded for accelerating the translation of PW-MRI into routine clinical applications. For DSC-MRI, sustained and focused efforts on exploiting novel imaging sequences, contrast agents, and better algorithm to maximally eliminate T1 and T2∗-dominant extravasation effects, reduce susceptibility artifacts, and enhance imaging signal-noise ratio will better augment parameters accuracy and repeatability in glioma settings. For DCE-MRI, modeling more exquisite PK models based on real transvascular transport process and calculating more physiologic indicators will comprehensively recapitulate the tumor vascular microenvironment and elaborate a certain specific tumor biology process. With the rapid development of imaging genomics and the latest 2016 WHO classification criteria for CNS tumors, ongoing research is needed to illuminate and define the molecular mechanism or genotype underlying the variation of perfusion parameters. Establishing the correlation between glioma genetic characteristics and PW-MRI features will provide deep insight into tumor angiogenesis processes and vascular heterogeneity, significantly improving our understanding of tumor biology and finally allowing more precise diagnosis and individual therapeutics. Meanwhile, multimodal and parametric imaging strategies incorporating anatomy, permeability, perfusion, and other characterizations of tumor biology like cellularity from DWI and metabolism from MRS consist of big data archive to delineate cancer landscape. This will tremendously push forward the development of glioma management and theranostics.
Acknowledgments
This work was supported by National Natural Science Foundation of China (nos. 81271626 and 81511660), Chongqing Science and Technology R&D Base Construction (International Cooperation) Project (cstc2014gjhz110002), and Clinical Scientific Research Fund of Daping Hospital, Institute of Surgery Research, Third Military Medical University (2014YLC03).
Abbreviations
- GBM:
Glioblastoma multiforme
- PW-MRI:
Perfusion-weighted magnetic resonance imaging
- DCE-MRI:
Dynamic contrast-enhanced MRI
- DSC-MRI:
Dynamic susceptibility contrast-MRI
- GBM:
Glioblastoma multiforme
- FLAIR:
Fluid attenuated inversion recovery
- VEGF:
Vascular endothelial growth factor
- LGG:
Low grade glioma
- BBB:
Blood brain barrier
- HGG:
High grade glioma
- MVD:
Microvessel density
- ECs:
Endothelial cells
- ASL:
Arterial spin-labeling
- GBCAs:
Gadolinium-based contrast agents
- EES:
Extravascular extracellular space
- IAUC:
Initial area under the concentration-time curve
- AUC:
Area under the concentration-time curve
- Smax:
Peak signal intensity
- TK:
Tofts-Kermode
- ETK:
Extended Tofts-Kermode
- Ktrans:
The volume transfer constant between blood plasma and EES
- Ve:
The volume of EES per unit volume of tissue
- Kep:
Rate constant between EES and blood plasma
- Vp:
Fractional plasma volume per unit of tissue volume
- SE-EPI:
Spin echo-echo planar imaging
- GRE-EPI:
Gradient echo-echo planar imaging
- SI-TCC:
Signal intensity-time course curve
- CC-TCC:
Contrast concentration-time course curve
- CBV:
Cerebral blood volume
- CBF:
Cerebral blood flow
- PH:
Peak height
- MTT:
Mean transit time
- PSR:
Percentage of signal intensity recovery
- rCBV:
Relative CBV
- ROI:
Region of interest
- PS:
Permeability surface-area product
- CNS:
Central nervous system
- IDH:
Isocitrate dehydrogenase
- EGFR:
Epidermal growth factor receptor
- EGFRam:
EGFR amplification
- MGMT:
Methyl-guanine methyltransferase
- α-KG:
α-Ketoglutarate
- 2-HG:
2-Hydroxyglutarate
- 1H -MRS:
Water suppressed proton-magnetic resonance spectroscopy
- nCBV:
Normalized CBV
- ADC:
Apparent diffusion coefficient
- EGFRvIII:
EGFR variant III
- CAR-T:
Chimeric antigen receptor T-cell
- rPH:
Relative peak height
- RT-PCR:
Reverse-transcription polymerase chain reaction
- LOH:
Loss of heterozygosity
- MET:
Metastasis
- PCNSL:
Primary central nervous system lymphoma
- ITSS:
Intratumoral susceptibility signals
- RANO:
Response Assessment in Neuro-Oncology
- PRM:
Parametric response map
- AUCR:
The initial and final area under the time-signal intensity curves ratio
- mAUCRH:
The mean AUCR at a higher curve
- AUCR50:
The 50th cumulative AUCR histogram parameter
- nRCBV:
rCBV normalized to white matter
- sRCBV:
Standardized rCBV
- PsP:
Pseudoprogression
- PD:
Progressive disease
- RN:
Radiation necrosis
- Sp%:
Percentage of specificity
- Sn%:
Percentage of sensitivity
- Ka:
Apparent transfer constant
- PFS:
Progress-free survival
- OS:
Overall survival
- rCBF:
Relative cerebral blood flow
- BTIP:
Brain Tumor Imaging Protocol
- ASFNR:
American Society of Functional Neuroradiology
- QIBA of RSNA:
Quantitative Imaging Biomarkers Alliance of the Radiological Society of North American.
Conflicts of Interest
The authors declared that they have no conflicts of interest.
Authors' Contributions
Junfeng Zhang and Heng Liu contributed equally to this work.
References
- 1.Louis D. N., Perry A., Reifenberger G., et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathologica. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 2.Wen P. Y., Reardon D. A. Neuro-oncology in 2015: progress in glioma diagnosis, classification and treatment. Nature Reviews Neurology. 2016;12(2):69–70. doi: 10.1038/nrneurol.2015.242. [DOI] [PubMed] [Google Scholar]
- 3.Weller M., van den Bent M., Hopkins K., et al. EANO guideline for the diagnosis and treatment of anaplastic gliomas and glioblastoma. The Lancet Oncology. 2014;15(9):e395–e403. doi: 10.1016/S1470-2045(14)70011-7. [DOI] [PubMed] [Google Scholar]
- 4.Hardee M. E., Zagzag D. Mechanisms of glioma-associated neovascularization. American Journal of Pathology. 2012;181(4):1126–1141. doi: 10.1016/j.ajpath.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kimura M., da Cruz L. C. Multiparametric MR imaging in the assessment of brain tumors. Magnetic Resonance Imaging Clinics of North America. 2016;24(1):87–122. doi: 10.1016/j.mric.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 6.Jain R., Gutierrez J., Narang J., et al. In vivo correlation of tumor blood volume and permeability with histologic and molecular angiogenic markers in gliomas. American Journal of Neuroradiology. 2011;32(2):388–394. doi: 10.3174/ajnr.A2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang W., Huang Y., An Y., Kim B. Y. S. Remodeling tumor vasculature to enhance delivery of intermediate-sized nanoparticles. ACS Nano. 2015;9(9):8689–8696. doi: 10.1021/acsnano.5b02028. [DOI] [PubMed] [Google Scholar]
- 8.Sorensen A. G., Emblem K. E., Polaskova P., et al. Increased survival of glioblastoma patients who respond to antiangiogenic therapy with elevated blood perfusion. Cancer Research. 2012;72(2):402–407. doi: 10.1158/0008-5472.CAN-11-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lemée J.-M., Clavreul A., Menei P. Intratumoral heterogeneity in glioblastoma: don't forget the peritumoral brain zone. Neuro-Oncology. 2015;17(10):1322–1332. doi: 10.1093/neuonc/nov119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellingson B. M., Wen P. Y., van den Bent M. J., Cloughesy T. F. Pros and cons of current brain tumor imaging. Neuro-oncology. 2014;16:vii2–vii11. doi: 10.1093/neuonc/nou224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jain R. Measurements of tumor vascular leakiness using DCE in brain tumors: clinical applications. NMR in Biomedicine. 2013;26(8):1042–1049. doi: 10.1002/nbm.2994. [DOI] [PubMed] [Google Scholar]
- 12.O'Connor J. P. B., Jackson A., Parker G. J. M., Roberts C., Jayson G. C. Dynamic contrast-enhanced MRI in clinical trials of antivascular therapies. Nature Reviews Clinical Oncology. 2012;9(3):167–177. doi: 10.1038/nrclinonc.2012.2. [DOI] [PubMed] [Google Scholar]
- 13.Barajas R. F. R., Cha S. Benefits of dynamic susceptibility-weighted contrast-enhanced perfusion MRI for glioma diagnosis and therapy. CNS oncology. 2014;3(6):407–419. doi: 10.2217/cns.14.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung C., Metser U., Ménard C. Advances in magnetic resonance imaging and positron emission tomography imaging for grading and molecular characterization of glioma. Seminars in Radiation Oncology. 2015;25(3):164–171. doi: 10.1016/j.semradonc.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Jain R. K., Di Tomaso E., Duda D. G., Loeffler J. S., Sorensen A. G., Batchelor T. T. Angiogenesis in brain tumours. Nature Reviews Neuroscience. 2007;8(8):610–622. doi: 10.1038/nrn2175. [DOI] [PubMed] [Google Scholar]
- 16.Soda Y., Myskiw C., Rommel A., Verma I. M. Mechanisms of neovascularization and resistance to anti-angiogenic therapies in glioblastoma multiforme. Journal of Molecular Medicine. 2013;91(4):439–448. doi: 10.1007/s00109-013-1019-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolburg H., Noell S., Fallier-Becker P., MacK A. F., Wolburg-Buchholz K. The disturbed blood-brain barrier in human glioblastoma. Molecular Aspects of Medicine. 2012;33(5-6):579–589. doi: 10.1016/j.mam.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Onishi M., Ichikawa T., Kurozumi K., Date I. Angiogenesis and invasion in glioma. Brain Tumor Pathology. 2011;28(1):13–24. doi: 10.1007/s10014-010-0007-z. [DOI] [PubMed] [Google Scholar]
- 19.Padera T. P., Stoll B. R., Tooredman J. B., Capen D., Di Tomaso E., Jain R. K. Cancer cells compress intratumour vessels. Nature. 2004;427, article 695 doi: 10.1038/427695a. [DOI] [PubMed] [Google Scholar]
- 20.Vartanian A., Singh S. K., Agnihotri S., et al. GBM's multifaceted landscape: highlighting regional and microenvironmental heterogeneity. Neuro-Oncology. 2014;16(9):1167–1175. doi: 10.1093/neuonc/nou035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peet A. C., Arvanitis T. N., Leach M. O., Waldman A. D. Functional imaging in adult and paediatric brain tumours. Nature Reviews Clinical Oncology. 2012;9(12):700–711. doi: 10.1038/nrclinonc.2012.187. [DOI] [PubMed] [Google Scholar]
- 22.Hylton N. Dynamic contrast-enhanced magnetic resonance imaging as an imaging biomarker. Journal of Clinical Oncology. 2006;24(20):3293–3298. doi: 10.1200/JCO.2006.06.8080. [DOI] [PubMed] [Google Scholar]
- 23.O'Connor J. P., Tofts P. S., Miles K. A., Parkes L. M., Thompson G., Jackson A. Dynamic contrast-enhanced imaging techniques: CT and MRI. The British Journal of Radiology. 2011;84(2):S112–S120. doi: 10.1259/bjr/55166688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khalifa F., Soliman A., El-Baz A., et al. Models and methods for analyzing DCE-MRI: a review. Medical Physics. 2014;41(12) doi: 10.1118/1.4898202.124301 [DOI] [PubMed] [Google Scholar]
- 25.Bergamino M., Bonzano L., Levrero F., Mancardi G. L., Roccatagliata L. A review of technical aspects of T1-weighted dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) in human brain tumors. Physica Medica. 2014;30(6):635–643. doi: 10.1016/j.ejmp.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 26.Sourbron S. P., Buckley D. L. Classic models for dynamic contrast-enhanced MRI. NMR in Biomedicine. 2013;26(8):1004–1027. doi: 10.1002/nbm.2940. [DOI] [PubMed] [Google Scholar]
- 27.Shiroishi M. S., Castellazzi G., Boxerman J. L., et al. Principles of T2∗-weighted dynamic susceptibility contrast MRI technique in brain tumor imaging. Journal of Magnetic Resonance Imaging. 2015;41(2):296–313. doi: 10.1002/jmri.24648. [DOI] [PubMed] [Google Scholar]
- 28.Bjornerud A., Sorensen A. G., Mouridsen K., Emblem K. E. T1- and T2∗-dominant extravasation correction in DSC-MRI: part I—theoretical considerations and implications for assessment of tumor hemodynamic properties. Journal of Cerebral Blood Flow and Metabolism. 2011;31(10):2041–2053. doi: 10.1038/jcbfm.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson G., Mills S. J., Coope D. J., O'Connor J. P. B., Jackson A. Imaging biomarkers of angiogenesis and the microvascular environment in cerebral tumours. British Journal of Radiology. 2011;84(2):S127–S144. doi: 10.1259/bjr/66316279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boxerman J. L., Paulson E. S., Prah M. A., Schmainda K. M. The effect of pulse sequence parameters and contrast agent dose on percentage signal recovery in DSC-MRI: implications for clinical applications. American Journal of Neuroradiology. 2013;34(7):1364–1369. doi: 10.3174/ajnr.a3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Welker K., Boxerman J., Kalnin A., Kaufmann T., Shiroishi M., Wintermark M. ASFNR recommendations for clinical performance of MR dynamic susceptibility contrast perfusion imaging of the brain. American Journal of Neuroradiology. 2015;36(6):E41–E51. doi: 10.3174/ajnr.A4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smits M. Imaging of oligodendroglioma. The British Journal of Radiology. 2016;89(1060) doi: 10.1259/bjr.20150857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scott J. N., Brasher P. M. A., Sevick R. J., Rewcastle N. B., Forsyth P. A. How often are nonenhancing supratentorial gliomas malignant? A population study. Neurology. 2002;59(6):947–949. doi: 10.1212/WNL.59.6.947. [DOI] [PubMed] [Google Scholar]
- 34.Whittle I. R. The dilemma of low grade glioma. Journal of Neurology, Neurosurgery & Psychiatry. 2004;75, supplement 2:ii31–ii36. doi: 10.1136/jnnp.2004.040501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maia A. C. M., Jr., Malheiros S. M. F., Da Rocha A. J., et al. MR cerebral blood volume maps correlated with vascular endothelial growth factor expression and tumor grade in nonenhancing gliomas. American Journal of Neuroradiology. 2005;26(4):777–783. [PMC free article] [PubMed] [Google Scholar]
- 36.Law M., Yang S., Wang H., et al. Glioma grading: sensitivity, specificity, and predictive values of perfusion MR imaging and proton MR spectroscopic imaging compared with conventional MR imaging. American Journal of Neuroradiology. 2003;24(10):1989–1998. [PMC free article] [PubMed] [Google Scholar]
- 37.Law M., Yang S., Babb J. S., et al. Comparison of cerebral blood volume and vascular permeability from dynamic susceptibility contrast-enhanced perfusion MR imaging with glioma grade. American Journal of Neuroradiology. 2004;25(5):746–755. [PMC free article] [PubMed] [Google Scholar]
- 38.Boxerman J. L., Schmainda K. M., Weisskoff R. M. Relative cerebral blood volume maps corrected for contrast agent extravasation significantly correlate with glioma tumor grade, whereas uncorrected maps do not. American Journal of Neuroradiology. 2006;27(4):859–867. [PMC free article] [PubMed] [Google Scholar]
- 39.Server A., Graff B. A., Orheim T. E. D., et al. Measurements of diagnostic examination performance and correlation analysis using microvascular leakage, cerebral blood volume, and blood flow derived from 3T dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging in glial tumor grading. Neuroradiology. 2011;53(6):435–447. doi: 10.1007/s00234-010-0770-x. [DOI] [PubMed] [Google Scholar]
- 40.Cha S. Update on brain tumor imaging: from anatomy to physiology. American Journal of Neuroradiology. 2006;27(3):475–487. [PMC free article] [PubMed] [Google Scholar]
- 41.Law M., Young R., Babb J., Pollack E., Johnson G. Histogram analysis versus region of interest analysis of dynamic susceptibility contrast perfusion MR imaging data in the grading of cerebral gliomas. American Journal of Neuroradiology. 2007;28(4):761–766. [PMC free article] [PubMed] [Google Scholar]
- 42.Emblem K. E., Scheie D., Due-Tonnessen P., et al. Histogram analysis of MR imaging-derived cerebral blood volume maps: combined glioma grading and identification of low-grade oligodendroglial subtypes. American Journal of Neuroradiology. 2008;29(9):1664–1670. doi: 10.3174/ajnr.a1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emblem K. E., Nedregaard B., Nome T., et al. Glioma grading by using histogram analysis of blood volume heterogeneity from MR-derived cerebral blood volume maps. Radiology. 2008;247(3):808–817. doi: 10.1148/radiol.2473070571. [DOI] [PubMed] [Google Scholar]
- 44.Lupo J. M., Cha S., Chang S. M., Nelson S. J. Dynamic susceptibility-weighted perfusion imaging of high-grade gliomas: characterization of spatial heterogeneity. American Journal of Neuroradiology. 2005;26(6):1446–1454. [PMC free article] [PubMed] [Google Scholar]
- 45.Aprile I., Giovannelli G., Fiaschini P., Muti M., Kouleridou A., Caputo N. High- and low-grade glioma differentiation: the role of percentage signal recovery evaluation in MR dynamic susceptibility contrast imaging. Radiologia Medica. 2015;120(10):967–974. doi: 10.1007/s11547-015-0511-7. [DOI] [PubMed] [Google Scholar]
- 46.Smitha K. A., Gupta A. K., Jayasree R. S. Relative percentage signal intensity recovery of perfusion metrics-an efficient tool for differentiating grades of glioma. British Journal of Radiology. 2015;88(1052) doi: 10.1259/bjr.20140784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi H. S., Kim A. H., Ahn S. S., Shin N.-Y., Kim J., Lee S.-K. Glioma grading capability: comparisons among parameters from dynamic contrast-enhanced MRI and ADC value on DWI. Korean Journal of Radiology. 2013;14(3):487–492. doi: 10.3348/kjr.2013.14.3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X., Zhu Y., Kang H., et al. Glioma grading by microvascular permeability parameters derived from dynamic contrast-enhanced MRI and intratumoral susceptibility signal on susceptibility weighted imaging. Cancer Imaging. 2015;15, article 4 doi: 10.1186/s40644-015-0039-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Santarosa C., Castellano A., Conte G. M., et al. Dynamic contrast-enhanced and dynamic susceptibility contrast perfusion MR imaging for glioma grading: preliminary comparison of vessel compartment and permeability parameters using hotspot and histogram analysis. European Journal of Radiology. 2016;85(6):1147–1156. doi: 10.1016/j.ejrad.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 50.Patankar T. F., Haroon H. A., Mills S. J., et al. Is volume transfer coefficient (Ktrans) related to histologic grade in human gliomas? American Journal of Neuroradiology. 2005;26(10):2455–2465. [PMC free article] [PubMed] [Google Scholar]
- 51.Jia Z., Geng D., Xie T., Zhang J., Liu Y. Quantitative analysis of neovascular permeability in glioma by dynamic contrast-enhanced MR imaging. Journal of Clinical Neuroscience. 2012;19(6):820–823. doi: 10.1016/j.jocn.2011.08.030. [DOI] [PubMed] [Google Scholar]
- 52.Zhao J., Yang Z.-Y., Luo B.-N., Yang J.-Y., Chu J.-P. Quantitative evaluation of diffusion and dynamic contrast-enhanced MR in tumor parenchyma and Peritumoral area for distinction of brain tumors. PLoS ONE. 2015;10(9) doi: 10.1371/journal.pone.0138573.e0138573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jung S. C., Yeom J. A., Kim J.-H., et al. Glioma: application of histogram analysis of pharmacokinetic parameters from T1-weighted dynamic contrast-enhanced MR imaging to tumor grading. American Journal of Neuroradiology. 2014;35(6):1103–1110. doi: 10.3174/ajnr.a3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nguyen T. B., Cron G. O., Perdrizet K., et al. Comparison of the diagnostic accuracy of DSC- and dynamic contrast-enhanced MRI in the preoperative grading of astrocytomas. American Journal of Neuroradiology. 2015;36(11):2017–2022. doi: 10.3174/ajnr.A4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nguyen T. B., Cron G. O., Mercier J. F., et al. Diagnostic accuracy of dynamic contrast-enhanced MR imaging using a phase-derived vascular input function in the preoperative grading of gliomas. American Journal of Neuroradiology. 2012;33(8):1539–1545. doi: 10.3174/ajnr.A3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jain R., Griffith B., Alotaibi F., et al. Glioma angiogenesis and perfusion imaging: understanding the relationship between tumor blood volume and leakiness with increasing glioma grade. American Journal of Neuroradiology. 2015;36(11):2030–2035. doi: 10.3174/ajnr.a4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoon J. H., Kim J.-H., Kang W. J., et al. Grading of cerebral glioma with multiparametric MR imaging and 18F-FDG-PET: concordance and accuracy. European Radiology. 2014;24(2):380–389. doi: 10.1007/s00330-013-3019-3. [DOI] [PubMed] [Google Scholar]
- 58.Pope W. B. Genomics of brain tumor imaging. Neuroimaging Clinics of North America. 2015;25(1):105–119. doi: 10.1016/j.nic.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 59.Itakura H., Achrol A. S., Mitchell L. A., et al. Magnetic resonance image features identify glioblastoma phenotypic subtypes with distinct molecular pathway activities. Science Translational Medicine. 2015;7(303):p. 303ra138. doi: 10.1126/scitranslmed.aaa7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kickingereder P., Sahm F., Radbruch A., et al. IDH mutation status is associated with a distinct hypoxia/angiogenesis transcriptome signature which is non-invasively predictable with rCBV imaging in human glioma. Scientific Reports. 2015;5 doi: 10.1038/srep16238.16238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee S., Choi S. H., Ryoo I., et al. Evaluation of the microenvironmental heterogeneity in high-grade gliomas with IDH1/2 gene mutation using histogram analysis of diffusion-weighted imaging and dynamic-susceptibility contrast perfusion imaging. Journal of Neuro-Oncology. 2015;121(1):141–150. doi: 10.1007/s11060-014-1614-z. [DOI] [PubMed] [Google Scholar]
- 62.Tykocinski E. S., Grant R. A., Kapoor G. S., et al. Use of magnetic perfusion-weighted imaging to determine epidermal growth factor receptor variant III expression in glioblastoma. Neuro-Oncology. 2012;14(5):613–623. doi: 10.1093/neuonc/nos073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gupta A., Young R. J., Shah A. D., et al. Pretreatment dynamic susceptibility contrast MRI perfusion in glioblastoma: prediction of EGFR gene amplification. Clinical Neuroradiology. 2015;25(2):143–150. doi: 10.1007/s00062-014-0289-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arevalo-Perez J., Thomas A. A., Kaley T., et al. T1-weighted dynamic contrast-enhanced MRI as a noninvasive biomarker of epidermal growth factor receptor VIII status. American Journal of Neuroradiology. 2015;36(12):2256–2261. doi: 10.3174/ajnr.A4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jung S. C., Choi S. H., Yeom J. A., et al. Cerebral blood volume analysis in glioblastomas using dynamic susceptibility contrast-enhanced perfusion MRI: A Comparison of Manual and Semiautomatic Segmentation Methods. PLoS ONE. 2013;8(8, article e69323) doi: 10.1371/journal.pone.0069323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moon W.-J., Choi J. W., Roh H. G., Lim S. D., Koh Y.-C. Imaging parameters of high grade gliomas in relation to the MGMT promoter methylation status: the CT, diffusion tensor imaging, and perfusion MR imaging. Neuroradiology. 2012;54(6):555–563. doi: 10.1007/s00234-011-0947-y. [DOI] [PubMed] [Google Scholar]
- 67.Ahn S. S., Shin N.-Y., Chang J. H., et al. Prediction of methylguanine methyltransferase promoter methylation in glioblastoma using dynamic contrast-enhanced magnetic resonance and diffusion tensor imaging. Journal of Neurosurgery. 2014;121(2):367–373. doi: 10.3171/2014.5.jns132279. [DOI] [PubMed] [Google Scholar]
- 68.Jenkinson M. D., Du Plessis D. G., Smith T. S., Joyce K. A., Warnke P. C., Walker C. Histological growth patterns and genotype in oligodendroglial tumours: correlation with MRI features. Brain. 2006;129(7):1884–1891. doi: 10.1093/brain/awl108. [DOI] [PubMed] [Google Scholar]
- 69.Lee S. M., Koh H.-J., Park D.-C., Song B. J., Huh T.-L., Park J.-W. Cytosolic NADP+-dependent isocitrate dehydrogenase status modulates oxidative damage to cells. Free Radical Biology and Medicine. 2002;32(11):1185–1196. doi: 10.1016/S0891-5849(02)00815-8. [DOI] [PubMed] [Google Scholar]
- 70.Kim S. Y., Lee S. M., Tak J. K., Choi K. S., Kwon T. K., Park J.-W. Regulation of singlet oxygen-induced apoptosis by cytosolic NADP+-dependent isocitrate dehydrogenase. Molecular and Cellular Biochemistry. 2007;302(1-2):27–34. doi: 10.1007/s11010-007-9421-x. [DOI] [PubMed] [Google Scholar]
- 71.Parsons D. W., Jones S., Zhang X., et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Goodenberger M. L., Jenkins R. B. Genetics of adult glioma. Cancer Genetics. 2012;205(12):613–621. doi: 10.1016/j.cancergen.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 73.Yan H., Parsons D. W., Jin G., et al. IDH1 and IDH2 mutations in gliomas. New England Journal of Medicine. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lai A., Kharbanda S., Pope W. B., et al. Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin. Journal of Clinical Oncology. 2011;29(34):4482–4490. doi: 10.1200/JCO.2010.33.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dang L., White D. W., Gross S., et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Turcan S., Rohle D., Goenka A., et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oermann E. K., Wu J., Guan K. L., Xiong Y. Alterations of metabolic genes and metabolites in cancer. Seminars in Cell & Developmental Biology. 2012;23(4):370–380. doi: 10.1016/j.semcdb.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brennan C. W., Verhaak R. G., McKenna A., et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.SongTao Q., Lei Y., Si G., et al. IDH mutations predict longer survival and response to temozolomide in secondary glioblastoma. Cancer Science. 2012;103(2):269–273. doi: 10.1111/j.1349-7006.2011.02134.x. [DOI] [PubMed] [Google Scholar]
- 80.Andronesi O. C., Kim G. S., Gerstner E., et al. Detection of 2-hydroxyglutarate in IDH-mutated glioma patients by in vivo spectral-editing and 2D correlation magnetic resonance spectroscopy. Science Translational Medicine. 2012;4(116) doi: 10.1126/scitranslmed.3002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pope W. B., Prins R. M., Thomas M. A., et al. Non-invasive detection of 2-hydroxyglutarate and other metabolites in IDH1 mutant glioma patients using magnetic resonance spectroscopy. Journal of Neuro-Oncology. 2012;107(1):197–205. doi: 10.1007/s11060-011-0737-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bertolino N., Marchionni C., Ghielmetti F., et al. Accuracy of 2-hydroxyglutarate quantification by short-echo proton-MRS at 3 T: a phantom study. Physica Medica. 2014;30(6):702–707. doi: 10.1016/j.ejmp.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 83.Yamashita K., Hiwatashi A., Togao O., et al. MR imaging-based analysis of glioblastoma multiforme: estimation of IDH1 mutation status. American Journal of Neuroradiology. 2016;37(1):58–65. doi: 10.3174/ajnr.a4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tan W., Xiong J., Huang W., Wu J., Zhan S., Geng D. Noninvasively detecting Isocitrate dehydrogenase 1 gene status in astrocytoma by dynamic susceptibility contrast MRI. Journal of Magnetic Resonance Imaging. 2017;45(2):492–499. doi: 10.1002/jmri.25358. [DOI] [PubMed] [Google Scholar]
- 85.Fan Q.-W., Cheng C., Gustafson W. C., et al. EGFR phosphorylates tumor-derived EGFRvIII driving STAT3/5 and progression in glioblastoma. Cancer Cell. 2013;24(4):438–449. doi: 10.1016/j.ccr.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Emlet D. R., Gupta P., Holgado-Madruga M., et al. Targeting a glioblastoma cancer stem-cell population defined by EGF receptor variant III. Cancer Research. 2014;74(4):1238–1249. doi: 10.1158/0008-5472.CAN-13-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zadeh G., Bhat K. P. L., Aldape K. EGFR and EGFRvIII in glioblastoma: partners in crime. Cancer Cell. 2013;24(4):403–404. doi: 10.1016/j.ccr.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 88.Siegal T. Clinical impact of molecular biomarkers in gliomas. Journal of Clinical Neuroscience. 2015;22(3):437–444. doi: 10.1016/j.jocn.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 89.Reardon D. A., Wen P. Y., Mellinghoff I. K. Targeted molecular therapies against epidermal growth factor receptor: past experiences and challenges. Neuro-Oncology. 2014;16:viii7–viii13. doi: 10.1093/neuonc/nou232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Johnson L. A., Scholler J., Ohkuri T., et al. Rational development and characterization of humanized anti-EGFR variant III chimeric antigen receptor T cells for glioblastoma. Science Translational Medicine. 2015;7(275):p. 275ra22. doi: 10.1126/scitranslmed.aaa4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Swartz A. M., Batich K. A., Fecci P. E., Sampson J. H. Peptide vaccines for the treatment of glioblastoma. Journal of Neuro-Oncology. 2015;123(3):433–440. doi: 10.1007/s11060-014-1676-y. [DOI] [PubMed] [Google Scholar]
- 92.Diehn M., Nardini C., Wang D. S., et al. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(13):5213–5218. doi: 10.1073/pnas.0801279105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gutman D. A., Cooper L. A. D., Hwang S. N., et al. MR imaging predictors of molecular profile and survival: multi-institutional study of the TCGA glioblastoma data set. Radiology. 2013;267(2):560–569. doi: 10.1148/radiol.13120118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Aghi M., Gaviani P., Henson J. W., Batchelor T. T., Louis D. N., Barker F. G., II Magnetic resonance imaging characteristics predict epidermal growth factor receptor amplification status in glioblastoma. Clinical Cancer Research. 2005;11(24):8600–8605. doi: 10.1158/1078-0432.CCR-05-0713. [DOI] [PubMed] [Google Scholar]
- 95.Riemenschneider M. J., Hegi M. E., Reifenberger G. MGMT promoter methylation in malignant gliomas. Targeted Oncology. 2010;5(3):161–165. doi: 10.1007/s11523-010-0153-6. [DOI] [PubMed] [Google Scholar]
- 96.Weller M., Stupp R., Reifenberger G., et al. MGMT promoter methylation in malignant gliomas: ready for personalized medicine? Nature Reviews Neurology. 2010;6(1):39–51. doi: 10.1038/nrneurol.2009.197. [DOI] [PubMed] [Google Scholar]
- 97.Cankovic M., Nikiforova M. N., Snuderl M., et al. The role of MGMT testing in clinical practice: a report of the association for molecular pathology. Journal of Molecular Diagnostics. 2013;15(5):539–555. doi: 10.1016/j.jmoldx.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 98.Ochs K., Kaina B. Apoptosis induced by DNA damage O6-methylguanine is Bcl-2 and caspase-9/3 regulated and Fas/caspase-8 independent. Cancer Research. 2000;60(20):5815–5824. [PubMed] [Google Scholar]
- 99.Weller M., Tabatabai G., Kästner B., et al. MGMT promoter methylation is a strong prognostic biomarker for benefit from dose-intensified temozolomide rechallenge in progressive glioblastoma: the DIRECTOR trial. Clinical Cancer Research. 2015;21(9):2057–2064. doi: 10.1158/1078-0432.ccr-14-2737. [DOI] [PubMed] [Google Scholar]
- 100.Rapkins R. W., Wang F., Nguyen H. T. N., et al. The MGMT promoter SNP rs16906252 is a risk factor for MGMT methylation in glioblastoma and is predictive of response to temozolomide. Neuro-Oncology. 2015;17(12):1589–1598. doi: 10.1093/neuonc/nov064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li H., Li J., Cheng G., Zhang J., Li X. IDH mutation and MGMT promoter methylation are associated with the pseudoprogression and improved prognosis of glioblastoma multiforme patients who have undergone concurrent and adjuvant temozolomide-based chemoradiotherapy. Clinical Neurology and Neurosurgery. 2016;151:31–36. doi: 10.1016/j.clineuro.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 102.Della Puppa A., Persano L., Masi G., et al. MGMT expression and promoter methylation status may depend on the site of surgical sample collection within glioblastoma: a possible pitfall in stratification of patients? Journal of Neuro-Oncology. 2012;106(1):33–41. doi: 10.1007/s11060-011-0639-9. [DOI] [PubMed] [Google Scholar]
- 103.Drabycz S., Roldán G., de Robles P., et al. An analysis of image texture, tumor location, and MGMT promoter methylation in glioblastoma using magnetic resonance imaging. NeuroImage. 2010;49(2):1398–1405. doi: 10.1016/j.neuroimage.2009.09.049. [DOI] [PubMed] [Google Scholar]
- 104.Eoli M., Menghi F., Bruzzone M. G., et al. Methylation of O6-methylguanine DNA methytransferase and loss of heterozygosity on 19q and/or 17p are overlapping features of secondary glioblastomas with prolonged survival. Clinical Cancer Research. 2007;13(9):2606–2613. doi: 10.1158/1078-0432.CCR-06-2184. [DOI] [PubMed] [Google Scholar]
- 105.Boots-Sprenger S. H. E., Sijben A., Rijntjes J., et al. Significance of complete 1p/19q co-deletion, IDH1 mutation and MGMT promoter methylation in gliomas: use with caution. Modern Pathology. 2013;26(7):922–929. doi: 10.1038/modpathol.2012.166. [DOI] [PubMed] [Google Scholar]
- 106.Weller M., Stupp R., Hegi M. E., et al. Personalized care in neuro-oncology coming of age: why we need MGMT and 1p/19q testing for malignant glioma patients in clinical practice. Neuro-Oncology. 2012;14(4):iv100–iv108. doi: 10.1093/neuonc/nos206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gladson C. L., Prayson R. A., Liu W. M. The pathobiology of glioma tumors. Annual Review of Pathology: Mechanisms of Disease. 2010;5(1):33–50. doi: 10.1146/annurev-pathol-121808-102109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cairncross G., Wang M., Shaw E., et al. Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: long-term results of RTOG 9402. Journal of Clinical Oncology. 2013;31(3):337–343. doi: 10.1200/jco.2012.43.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.van den Bent M. J., Brandes A. A., Taphoorn M. J. B., et al. Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long-term follow-up of EORTC brain tumor group study 26951. Journal of Clinical Oncology. 2013;31(3):344–350. doi: 10.1200/jco.2012.43.2229. [DOI] [PubMed] [Google Scholar]
- 110.Jenkinson M. D., Smith T. S., Joyce K. A., et al. Cerebral blood volume, genotype and chemosensitivity in oligodendroglial tumours. Neuroradiology. 2006;48(10):703–713. doi: 10.1007/s00234-006-0122-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chawla S., Krejza J., Vossough A., et al. Differentiation between oligodendroglioma genotypes using dynamic susceptibility contrast perfusion-weighted imaging and proton MR spectroscopy. American Journal of Neuroradiology. 2013;34(8):1542–1549. doi: 10.3174/ajnr.A3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ali M. M., Janic B., Babajani-Feremi A., et al. Changes in vascular permeability and expression of different angiogenic factors following anti-angiogenic treatment in rat glioma. PLoS ONE. 2010;5(1, article e8727) doi: 10.1371/journal.pone.0008727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Katanasaka Y., Kodera Y., Kitamura Y., Morimoto T., Tamura T., Koizumi F. Epidermal growth factor receptor variant type III markedly accelerates angiogenesis and tumor growth via inducing c-myc mediated angiopoietin-like 4 expression in malignant glioma. Molecular Cancer. 2013;12, article 31:241–249. doi: 10.1186/1476-4598-12-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Inda M.-D., Bonavia R., Mukasa A., et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes and Development. 2010;24(16):1731–1745. doi: 10.1101/gad.1890510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Omuro A. M., Leite C. C., Mokhtari K., Delattre J.-Y. Pitfalls in the diagnosis of brain tumours. Lancet Neurology. 2006;5(11):937–948. doi: 10.1016/S1474-4422(06)70597-X. [DOI] [PubMed] [Google Scholar]
- 116.Law M., Cha S., Knopp E. A., Johnson G., Arnett J., Litt A. W. High-grade gliomas and solitary metastases: differentiation by using perfusion and proton spectroscopic MR imaging. Radiology. 2002;222(3):715–721. doi: 10.1148/radiol.2223010558. [DOI] [PubMed] [Google Scholar]
- 117.Cha S., Lupo J. M., Chen M.-H., et al. Differentiation of glioblastoma multiforme and single brain metastasis by peak height and percentage of signal intensity recovery derived from dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging. American Journal of Neuroradiology. 2007;28(6):1078–1084. doi: 10.3174/ajnr.A0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mangla R., Kolar B., Zhu T., Zhong J., Almast J., Ekholm S. Percentage signal recovery derived from MR dynamic susceptibility contrast imaging is useful to differentiate common enhancing malignant lesions of the brain. American Journal of Neuroradiology. 2011;32(6):1004–1010. doi: 10.3174/ajnr.A2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Toh C. H., Wei K.-C., Chang C.-N., Ng S.-H., Wong H.-F. Differentiation of primary central nervous system lymphomas and glioblastomas: comparisons of diagnostic performance of dynamic susceptibility contrast-enhanced perfusion mr imaging without and with contrast-leakage correction. American Journal of Neuroradiology. 2013;34(6):1145–1149. doi: 10.3174/ajnr.a3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Xing Z., You R. X., Li J., Liu Y., Cao D. R. Differentiation of primary central nervous system lymphomas from high-grade gliomas by rCBV and percentage of signal intensity recovery derived from dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging. Clinical Neuroradiology. 2014;24(4):329–336. doi: 10.1007/s00062-013-0255-5. [DOI] [PubMed] [Google Scholar]
- 121.Kickingereder P., Sahm F., Wiestler B., et al. Evaluation of microvascular permeability with dynamic contrast-enhanced MRI for the differentiation of primary CNS lymphoma and glioblastoma: radiologic-pathologic correlation. American Journal of Neuroradiology. 2014;35(8):1503–1508. doi: 10.3174/ajnr.a3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kickingereder P., Wiestler B., Sahm F., et al. Primary central nervous system lymphoma and atypical glioblastoma: multiparametric differentiation by using diffusion-, perfusion-, and susceptibility-weighted MR imaging. Radiology. 2014;272(3):843–850. doi: 10.1148/radiol.14132740. [DOI] [PubMed] [Google Scholar]
- 123.Jung B. C., Arevalo-Perez J., Lyo J. K., et al. Comparison of glioblastomas and brain metastases using dynamic contrast-enhanced perfusion MRI. Journal of Neuroimaging. 2016;26(2):240–246. doi: 10.1111/jon.12281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Campos S., Davey P., Hird A., et al. Brain metastasis from an unknown primary, or primary brain tumour? A diagnostic dilemma. Current Oncology. 2009;16(1):62–66. doi: 10.3747/co.v16i1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nduom E. K., Yang C., Merrill M. J., Zhuang Z., Lonser R. R. Characterization of the blood-brain barrier of metastatic and primary malignant neoplasms. Journal of Neurosurgery. 2013;119(2):427–433. doi: 10.3171/2013.3.JNS122226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Klekner Á., Hutóczki G., Virga J., et al. Expression pattern of invasion-related molecules in the peritumoral brain. Clinical Neurology and Neurosurgery. 2015;139:138–143. doi: 10.1016/j.clineuro.2015.09.017. [DOI] [PubMed] [Google Scholar]
- 127.Fidler I. J. The biology of brain metastasis: challenges for therapy. The Cancer Journal. 2015;21(4):284–293. doi: 10.1097/ppo.0000000000000126. [DOI] [PubMed] [Google Scholar]
- 128.Rollin N., Guyotat J., Streichenberger N., Honnorat J., Tran Minh V.-A., Cotton F. Clinical relevance of diffusion and perfusion magnetic resonance imaging in assessing intra-axial brain tumors. Neuroradiology. 2006;48(3):150–159. doi: 10.1007/s00234-005-0030-7. [DOI] [PubMed] [Google Scholar]
- 129.Han C., Huang S., Guo J., Zhuang X., Han H. Use of a high b-value for diffusion weighted imaging of peritumoral regions to differentiate high-grade gliomas and solitary metastases. Journal of Magnetic Resonance Imaging. 2015;42(1):80–86. doi: 10.1002/jmri.24747. [DOI] [PubMed] [Google Scholar]
- 130.Ricard D., Idbaih A., Ducray F., Lahutte M., Hoang-Xuan K., Delattre J.-Y. Primary brain tumours in adults. The Lancet. 2012;379(9830):1984–1996. doi: 10.1016/S0140-6736(11)61346-9. [DOI] [PubMed] [Google Scholar]
- 131.Louis D. N., Ohgaki H., Wiestler O. D., et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathologica. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Takeuchi H., Matsuda K., Kitai R., Sato K., Kubota T. Angiogenesis in primary central nervous system lymphoma (PCNSL) Journal of Neuro-Oncology. 2007;84(2):141–145. doi: 10.1007/s11060-007-9363-x. [DOI] [PubMed] [Google Scholar]
- 133.Coulon A., Lafitte F., Hoang-Xuan K., et al. Radiographic findings in 37 cases of primary CNS lymphoma in immunocompetent patients. European Radiology. 2002;12(2):329–340. doi: 10.1007/s003300101037. [DOI] [PubMed] [Google Scholar]
- 134.Nabavizadeh S. A., Vossough A., Hajmomenian M., Assadsangabi R., Mohan S. Neuroimaging in central nervous system lymphoma. Hematology/Oncology Clinics of North America. 2016;30(4):799–821. doi: 10.1016/j.hoc.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 135.Blasel S., Jurcoane A., Bähr O., Weise L., Harter P. N., Hattingen E. MR perfusion in and around the contrast-enhancement of primary CNS lymphomas. Journal of Neuro-Oncology. 2013;114(1):127–134. doi: 10.1007/s11060-013-1161-z. [DOI] [PubMed] [Google Scholar]
- 136.Hartmann M., Heiland S., Harting I., et al. Distinguishing of primary cerebral lymphoma from high-grade glioma with perfusion-weighted magnetic resonance imaging. Neuroscience Letters. 2003;338(2):119–122. doi: 10.1016/S0304-3940(02)01367-8. [DOI] [PubMed] [Google Scholar]
- 137.Nakajima S., Okada T., Yamamoto A., et al. Differentiation between primary central nervous system lymphoma and glioblastoma: a comparative study of parameters derived from dynamic susceptibility contrast-enhanced perfusion-weighted MRI. Clinical Radiology. 2015;70(12):1393–1399. doi: 10.1016/j.crad.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 138.Liao W., Liu Y., Wang X., et al. Differentiation of primary central nervous system lymphoma and high-grade glioma with dynamic susceptibility contrast-enhanced perfusion magnetic resonance imaging. Acta Radiologica. 2009;50(2):217–225. doi: 10.1080/02841850802616752. [DOI] [PubMed] [Google Scholar]
- 139.Calli C., Kitis O., Yunten N., Yurtseven T., Islekel S., Akalin T. Perfusion and diffusion MR imaging in enhancing malignant cerebral tumors. European Journal of Radiology. 2006;58(3):394–403. doi: 10.1016/j.ejrad.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 140.Wang S., Kim S., Chawla S., et al. Differentiation between glioblastomas, solitary brain metastases, and primary cerebral lymphomas using diffusion tensor and dynamic susceptibility contrast-enhanced MR imaging. American Journal of Neuroradiology. 2011;32(3):507–514. doi: 10.3174/ajnr.A2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lin X., Lee M., Buck O., et al. Diagnostic accuracy of T1-weighted dynamic contrast-enhanced-MRI and DWI-ADC for differentiation of glioblastoma and primary CNS lymphoma. American Journal of Neuroradiology. 2017;38(3):485–491. doi: 10.3174/ajnr.a5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Macdonald D. R., Cascino T. L., Schold S. C., Jr., Cairncross J. G. Response criteria for phase II studies of supratentorial malignant glioma. Journal of Clinical Oncology. 1990;8(7):1277–1280. doi: 10.1200/JCO.1990.8.7.1277. [DOI] [PubMed] [Google Scholar]
- 143.Wen P. Y., Macdonald D. R., Reardon D. A., et al. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. Journal of Clinical Oncology. 2010;28(11):1963–1972. doi: 10.1200/jco.2009.26.3541. [DOI] [PubMed] [Google Scholar]
- 144.Sorensen A. G., Batchelor T. T., Wen P. Y., Zhang W. T., Jain R. K. Response criteria for glioma. Nature Clinical Practice Oncology. 2008;5(11):634–644. doi: 10.1038/ncponc1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shiroishi M. S., Boxerman J. L., Pope W. B. Physiologic MRI for assessment of response to therapy and prognosis in glioblastoma. Neuro-Oncology. 2016;18(4):467–478. doi: 10.1093/neuonc/nov179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Taal W., Brandsma D., De Bruin H. G., et al. Incidence of early pseudo-progression in a cohort of malignant glioma patients treated with chemoirradiation with temozolomide. Cancer. 2008;113(2):405–410. doi: 10.1002/cncr.23562. [DOI] [PubMed] [Google Scholar]
- 147.Brandsma D., Stalpers L., Taal W., Sminia P., van den Bent M. J. Clinical features, mechanisms, and management of pseudoprogression in malignant gliomas. The Lancet Oncology. 2008;9(5):453–461. doi: 10.1016/S1470-2045(08)70125-6. [DOI] [PubMed] [Google Scholar]
- 148.Gahramanov S., Varallyay C., Tyson R. M. A., et al. Diagnosis of pseudoprogression using MRI perfusion in patients with glioblastoma multiforme may predict improved survival. CNS oncology. 2014;3(6):389–400. doi: 10.2217/cns.14.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Khan M. N., Sharma A. M., Pitz M., et al. High-grade glioma management and response assessment—recent advances and current challenges. Current Oncology. 2016;23(4):e383–e391. doi: 10.3747/co.23.3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mangla R., Singh G., Ziegelitz D., et al. Changes in relative cerebral blood volume 1 month after radiation-temozolomide therapy can help predict overall survival in patients with glioblastoma. Radiology. 2010;256(2):575–584. doi: 10.1148/radiol.10091440. [DOI] [PubMed] [Google Scholar]
- 151.Martínez-Martínez A., Martínez-Bosch J. Perfusion magnetic resonance imaging for high grade astrocytomas: can cerebral blood volume, peak height, and percentage of signal intensity recovery distinguish between progression and pseudoprogression? Radiologia. 2014;56(1):35–43. doi: 10.1016/j.rx.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 152.Prager A. J., Martinez N., Beal K., Omuro A., Zhang Z., Young R. J. Diffusion and perfusion MRI to differentiate treatment-related changes including pseudoprogression from recurrent tumors in high-grade gliomas with histopathologic evidence. American Journal of Neuroradiology. 2015;36(5):877–885. doi: 10.3174/ajnr.a4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Baek H. J., Kim H. S., Kim N., Choi Y. J., Kim Y. J. Percent change of perfusion skewness and kurtosis: a potential imaging biomarker for early treatment response in patients with newly diagnosed glioblastomas. Radiology. 2012;264(3):834–843. doi: 10.1148/radiol.12112120. [DOI] [PubMed] [Google Scholar]
- 154.Tsien C., Galbán C. J., Chenevert T. L., et al. Parametric response map as an imaging biomarker to distinguish progression from pseudoprogression in high-grade glioma. Journal of Clinical Oncology. 2010;28(13):2293–2299. doi: 10.1200/JCO.2009.25.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gahramanov S., Muldoon L. L., Varallyay C. G., et al. Pseudoprogression of glioblastoma after chemo- and radiation therapy: diagnosis by using dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging with ferumoxytol versus gadoteridol and correlation with survival. Radiology. 2013;266(3):842–852. doi: 10.1148/radiol.12111472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Suh C. H., Kim H. S., Choi Y. J., Kim N., Kim S. J. Prediction of pseudoprogression in patients with glioblastomas using the initial and final area under the curves ratio derived from dynamic contrast-enhanced T1-weighted perfusion MR imaging. American Journal of Neuroradiology. 2013;34(12):2278–2286. doi: 10.3174/ajnr.A3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yun T. J., Park C.-K., Kim T. M., et al. Glioblastoma treated with concurrent radiation therapy and temozolomide chemotherapy: differentiation of true progression from pseudoprogression with quantitative dynamic contrast-enhanced MR Imaging. Radiology. 2015;274(3):830–840. doi: 10.1148/radiol.14132632. [DOI] [PubMed] [Google Scholar]
- 158.Shin K. E., Ahn K. J., Choi H. S., et al. DCE and DSC MR perfusion imaging in the differentiation of recurrent tumour from treatment-related changes in patients with glioma. Clinical Radiology. 2014;69(6):e264–e272. doi: 10.1016/j.crad.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 159.Young R. J., Gupta A., Shah A. D., et al. MRI perfusion in determining pseudoprogression in patients with glioblastoma. Clinical Imaging. 2013;37(1):41–49. doi: 10.1016/j.clinimag.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gahramanov S., Raslan A. M., Muldoon L. L., et al. Potential for differentiation of pseudoprogression from true tumor progression with dynamic susceptibility-weighted contrast-enhanced magnetic resonance imaging using ferumoxytol vs. gadoteridol: a pilot study. International Journal of Radiation Oncology Biology Physics. 2011;79(2):514–523. doi: 10.1016/j.ijrobp.2009.10.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Thomas A. A., Arevalo-Perez J., Kaley T., et al. Dynamic contrast enhanced T1 MRI perfusion differentiates pseudoprogression from recurrent glioblastoma. Journal of Neuro-Oncology. 2015;125(1):183–190. doi: 10.1007/s11060-015-1893-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cheng H.-L. M. Improved correlation to quantitative DCE-MRI pharmacokinetic parameters using a modified initial area under the uptake curve (mIAUC) approach. Journal of Magnetic Resonance Imaging. 2009;30(4):864–872. doi: 10.1002/jmri.21916. [DOI] [PubMed] [Google Scholar]
- 163.Narang J., Jain R., Arbab A. S., et al. Differentiating treatment-induced necrosis from recurrent/progressive brain tumor using nonmodel-based semiquantitative indices derived from dynamic contrast-enhanced T1-weighted MR perfusion. Neuro-Oncology. 2011;13(9):1037–1046. doi: 10.1093/neuonc/nor075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Carmeliet P., Jain R. K. Principles and mechanisms of vessel normalization for cancer and other angiogenic diseases. Nature Reviews Drug Discovery. 2011;10(6):417–427. doi: 10.1038/nrd3455. [DOI] [PubMed] [Google Scholar]
- 165.Batchelor T. T., Sorensen A. G., di Tomaso E., et al. AZD2171, a pan-VEGF receptor tyrosine kinase inhibitor, normalizes tumor vasculature and alleviates edema in glioblastoma patients. Cancer Cell. 2007;11(1):83–95. doi: 10.1016/j.ccr.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gerstner E. R., Duda D. G., di Tomaso E., et al. VEGF inhibitors in the treatment of cerebral edema in patients with brain cancer. Nature Reviews Clinical Oncology. 2009;6(4):229–236. doi: 10.1038/nrclinonc.2009.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sorensen A. G., Batchelor T. T., Zhang W.-T., et al. A ‘vascular normalization index’ as potential mechanistic biomarker to predict survival after a single dose of cediranib in recurrent glioblastoma patients. Cancer Research. 2009;69(13):5296–5300. doi: 10.1158/0008-5472.can-09-0814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Clarke J. L., Chang S. Pseudoprogression and pseudoresponse: challenges in brain tumor imaging. Current Neurology and Neuroscience Reports. 2009;9(3):241–246. doi: 10.1007/s11910-009-0035-4. [DOI] [PubMed] [Google Scholar]
- 169.Boxerman J. L., Zhang Z., Safriel Y., et al. Early post-bevacizumab progression on contrast-enhanced MRI as a prognostic marker for overall survival in recurrent glioblastoma: results from the ACRIN 6677/RTOG 0625 Central Reader Study. Neuro-Oncology. 2013;15(7):945–954. doi: 10.1093/neuonc/not049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Schmainda K. M., Prah M., Connelly J., et al. Dynamic-susceptibility contrast agent MRI measures of relative cerebral blood volume predict response to bevacizumab in recurrent high-grade glioma. Neuro-Oncology. 2014;16(6):880–888. doi: 10.1093/neuonc/not216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.O’Neill A. F., Qin L., Wen P. Y., de Groot J. F., Van den Abbeele A. D., Yap J. T. Demonstration of DCE-MRI as an early pharmacodynamic biomarker of response to VEGF Trap in glioblastoma. Journal of Neuro-Oncology. 2016;130(3):495–503. doi: 10.1007/s11060-016-2243-5. [DOI] [PubMed] [Google Scholar]
- 172.Lockhart A. C., Rothenberg M. L., Dupont J., et al. Phase I study of intravenous vascular endothelial growth factor trap, aflibercept, in patients with advanced solid tumors. Journal of Clinical Oncology. 2010;28(2):207–214. doi: 10.1200/JCO.2009.22.9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Piludu F., Marzi S., Pace A., et al. Early biomarkers from dynamic contrast-enhanced magnetic resonance imaging to predict the response to antiangiogenic therapy in high-grade gliomas. Neuroradiology. 2015;57(12):1269–1280. doi: 10.1007/s00234-015-1582-9. [DOI] [PubMed] [Google Scholar]
- 174.Emblem K. E., Bjornerud A., Mouridsen K., et al. T1-and T2∗-dominant extravasation correction in DSC-MRI: Part II—predicting patient outcome after a single dose of cediranib in recurrent glioblastoma patients. Journal of Cerebral Blood Flow and Metabolism. 2011;31(10):2054–2064. doi: 10.1038/jcbfm.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Artzi M., Bokstein F., Blumenthal D. T., et al. Differentiation between vasogenic-edema versus tumor-infiltrative area in patients with glioblastoma during bevacizumab therapy: a longitudinal MRI study. European Journal of Radiology. 2014;83(7):1250–1256. doi: 10.1016/j.ejrad.2014.03.026. [DOI] [PubMed] [Google Scholar]
- 176.Artzi M., Blumenthal D. T., Bokstein F., et al. Classification of tumor area using combined DCE and DSC MRI in patients with glioblastoma. Journal of Neuro-Oncology. 2015;121(2):349–357. doi: 10.1007/s11060-014-1639-3. [DOI] [PubMed] [Google Scholar]
- 177.Akbari H., Macyszyn L., Da X., et al. Pattern analysis of dynamic susceptibility contrast-enhanced MR imaging demonstrates peritumoral tissue heterogeneity. Radiology. 2014;273(2):502–510. doi: 10.1148/radiol.14132458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Jain R., Poisson L., Narang J., et al. Genomic mapping and survival prediction in glioblastoma: molecular subclassification strengthened by hemodynamic imaging biomarkers. Radiology. 2013;267(1):212–220. doi: 10.1148/radiol.12120846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Akbari H., Macyszyn L., Da X., et al. Imaging surrogates of infiltration obtained via multiparametric imaging pattern analysis predict subsequent location of recurrence of glioblastoma. Neurosurgery. 2016;78(4):572–580. doi: 10.1227/NEU.0000000000001202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Yang I., Aghi M. K. New advances that enable identification of glioblastoma recurrence. Nature Reviews Clinical Oncology. 2009;6(11):648–657. doi: 10.1038/nrclinonc.2009.150. [DOI] [PubMed] [Google Scholar]
- 181.Fajardo L. F. The pathology of ionizing radiation as defined by morphologic patterns. Acta Oncologica. 2005;44(1):13–22. doi: 10.1080/02841860510007440. [DOI] [PubMed] [Google Scholar]
- 182.Barajas R. F., Jr., Chang J. S., Segal M. R., et al. Differentiation of recurrent glioblastoma multiforme from radiation necrosis after external beam radiation therapy with dynamic susceptibility- weighted contrast-enhanced perfusion MR imaging. Radiology. 2009;253(2):486–496. doi: 10.1148/radiol.2532090007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hu L. S., Baxter L. C., Smith K. A., et al. Relative cerebral blood volume values to differentiate high-grade glioma recurrence from posttreatment radiation effect: direct correlation between image-guided tissue histopathology and localized dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging measurements. American Journal of Neuroradiology. 2009;30(3):552–558. doi: 10.3174/ajnr.a1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Bisdas S., Naegele T., Ritz R., et al. Distinguishing recurrent high-grade gliomas from radiation injury: a pilot study using dynamic contrast-enhanced MR imaging. Academic Radiology. 2011;18(5):575–583. doi: 10.1016/j.acra.2011.01.018. [DOI] [PubMed] [Google Scholar]
- 185.Larsen V. A., Simonsen H. J., Law I., Larsson H. B. W., Hansen A. E. Evaluation of dynamic contrast-enhanced T1-weighted perfusion MRI in the differentiation of tumor recurrence from radiation necrosis. Neuroradiology. 2013;55(3):361–369. doi: 10.1007/s00234-012-1127-4. [DOI] [PubMed] [Google Scholar]
- 186.Masch W. R., Wang P. I., Chenevert T. L., et al. Comparison of diffusion tensor imaging and magnetic resonance perfusion imaging in differentiating recurrent brain neoplasm from radiation necrosis. Academic Radiology. 2016;23(5):569–576. doi: 10.1016/j.acra.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Danchaivijitr N., Waldman A. D., Tozer D. J., et al. Low-grade gliomas: do changes in rCBV measurements at longitudinal perfusion-weighted MR imaging predict malignant transformation? Radiology. 2008;247(1):170–178. doi: 10.1148/radiol.2471062089. [DOI] [PubMed] [Google Scholar]
- 188.Law M., Young R. J., Babb J. S., et al. Gliomas: predicting time to progression or survival with cerebral blood volume measurements at dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging. Radiology. 2008;247(2):490–498. doi: 10.1148/radiol.2472070898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Kickingereder P., Wiestler B., Burth S., et al. Relative cerebral blood volume is a potential predictive imaging biomarker of bevacizumab efficacy in recurrent glioblastoma. Neuro-Oncology. 2015;17(8):1139–1147. doi: 10.1093/neuonc/nov028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Jain R., Poisson L. M., Gutman D., et al. Outcome prediction in patients with glioblastoma by using imaging, clinical, and genomic biomarkers: focus on the nonenhancing component of the tumor. Radiology. 2014;272(2):484–493. doi: 10.1148/radiol.14131691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Nguyen T. B., Cron G. O., Mercier J. F., et al. Preoperative prognostic value of dynamic contrast-Enhanced MRI-Derived contrast transfer coefficient and plasma volume in patients with cerebral gliomas. American Journal of Neuroradiology. 2015;36(1):63–69. doi: 10.3174/ajnr.A4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kickingereder P., Wiestler B., Graf M., et al. Evaluation of dynamic contrast-enhanced MRI derived microvascular permeability in recurrent glioblastoma treated with bevacizumab. Journal of Neuro-Oncology. 2015;121(2):373–380. doi: 10.1007/s11060-014-1644-6. [DOI] [PubMed] [Google Scholar]
- 193.Kim R., Choi S. H., Yun T. J., et al. Prognosis prediction of non-enhancing T2 high signal intensity lesions in glioblastoma patients after standard treatment: application of dynamic contrast-enhanced MR imaging. European Radiology. 2017;27(3):1176–1185. doi: 10.1007/s00330-016-4464-6. [DOI] [PubMed] [Google Scholar]
- 194.Bonekamp D., Deike K., Wiestler B., et al. Association of overall survival in patients with newly diagnosed glioblastoma with contrast-enhanced perfusion MRI: comparison of intraindividually matched T1—and T2 (∗) -based bolus techniques. Journal of Magnetic Resonance Imaging. 2015;42(1):87–96. doi: 10.1002/jmri.24756. [DOI] [PubMed] [Google Scholar]
- 195.Burth S., Kickingereder P., Eidel O., et al. Clinical parameters outweigh diffusion- and perfusion-derived MRI parameters in predicting survival in newly diagnosed glioblastoma. Neuro-Oncology. 2016;18(12):1673–1679. doi: 10.1093/neuonc/now122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.O'Connor J. P. B., Rose C. J., Waterton J. C., Carano R. A. D., Parker G. J. M., Jackson A. Imaging intratumor heterogeneity: role in therapy response, resistance, and clinical outcome. Clinical Cancer Research. 2015;21(2):249–257. doi: 10.1158/1078-0432.ccr-14-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Just N. Improving tumour heterogeneity MRI assessment with histograms. British Journal of Cancer. 2014;111(12):2205–2213. doi: 10.1038/bjc.2014.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Paulson E. S., Schmainda K. M. Comparison of dynamic susceptibility-weighted contrast-enhanced MR methods: recommendations for measuring relative cerebral blood volume in brain tumors. Radiology. 2008;249(2):601–613. doi: 10.1148/radiol.2492071659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Hu L. S., Kelm Z., Korfiatis P., et al. Impact of software modeling on the accuracy of perfusion MRI in glioma. American Journal of Neuroradiology. 2015;36(12):2242–2249. doi: 10.3174/ajnr.A4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Prah M. A., Stufflebeam S. M., Paulson E. S., et al. Repeatability of standardized and normalized relative CBV in patients with newly diagnosed glioblastoma. American Journal of Neuroradiology. 2015;36(9):1654–1661. doi: 10.3174/ajnr.A4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Ellingson B. M., Bendszus M., Boxerman J., et al. Consensus recommendations for a standardized Brain Tumor Imaging Protocol in clinical trials. Neuro-Oncology. 2015;17(9):1188–1198. doi: 10.1093/neuonc/nov095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Sullivan D. C., Obuchowski N. A., Kessler L. G., et al. Metrology standards for quantitative imaging biomarkers. Radiology. 2015;277(3):813–825. doi: 10.1148/radiol.2015142202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Quantitative Imaging Biomarkers Alliance, March 2012, http://qibawiki.rsna.org/index.php/QIBA_DCE-MRI_Technical_Committee_Call_Summaries.