Abstract
BACKGROUND: Although the contrast-enhancing (CE) lesion on T1-weighted MR images is widely used as a surrogate for glioblastoma (GBM), there are also non-enhancing regions of infiltrative tumor within the T2-weighted lesion, which elude radiologic detection. Because non-enhancing GBM (Enh−) challenges clinical patient management as latent disease, this study sought to characterize ex vivo metabolic profiles from Enh− and CE GBM (Enh+) samples, alongside histological and in vivo MR parameters, to assist in defining criteria for estimating total tumor burden. Methods: Fifty-six patients with newly diagnosed GBM received a multi-parametric pre-surgical MR examination. Targets for obtaining image-guided tissue samples were defined based on in vivo parameters that were suspicious for tumor. The actual location from where tissue samples were obtained was recorded, and half of each sample was analyzed for histopathology while the other half was scanned using HR-MAS spectroscopy. Results: The Enh+ and Enh− tumor samples demonstrated comparable mitotic activity, but also significant heterogeneity in microvascular morphology. Ex vivo spectroscopic parameters indicated similar levels of total choline and N-acetylaspartate between these contrast-based radiographic subtypes of GBM, and characteristic differences in the levels of myo-inositol, creatine/phosphocreatine, and phosphoethanolamine. Analysis of in vivo parameters at the sample locations were consistent with histological and ex vivo metabolic data. CONCLUSIONS: The similarity between ex vivo levels of choline and NAA, and between in vivo levels of choline, NAA and nADC in Enh+ and Enh− tumor, indicate that these parameters can be used in defining non-invasive metrics of total tumor burden for patients with GBM.
Introduction
Glioblastoma (GBM) is the most common form of infiltrating glioma in adults and exhibits the highest level of malignancy based upon histopathologic criteria developed by the World Health Organization [1]. Defining tumor margins for GBM is complicated by its potential for proliferation and invasion into surrounding parenchyma. Median survival with standard-of-care treatment is around 14 months [2] and has remained unchanged over recent years, despite vigorous research into novel therapeutic agents and whole genome mutational analyses. Given the relatively rapid course of the disease, patients are routinely examined every 2 months by magnetic resonance (MR) imaging to discern whether radiographic progression has occurred and to modify clinical intervention accordingly.
Routine MR examinations exploit the vascular pathology of GBM, which is characterized by immature and tortuous growth of tumor microvasculature. This leads to blood–brain barrier (BBB) permeability that can be interrogated via the leakage of T1-sensitive paramagnetic contrast agents. The most commonly utilized agent in clinical practice is gadolinium (Gd) that has been chelated with diethylene triamine pentaacetic acid (Gd-DTPA) to reduce toxicity to patients [3]. Extravasation of Gd-DTPA causes a local reduction in the T1 relaxation time and reveals regions of elevated signal where the BBB is compromised on T1-weighted MR images. Increased blood volume and vessel permeability within and beyond the contrast enhancing (CE) lesion can be evaluated using advanced techniques such as dynamic susceptibility contrast (DSC) and dynamic contrast enhanced (DCE) perfusion-weighted MR imaging.
The other type of MR data used in the routine evaluation of GBM is the T2-weighted fluid-attenuated inversion recovery (FLAIR) image. Regions that are hyperintense on FLAIR imaging and extend beyond the borders of the CE lesion are referred to as the T2-weighted (T2w) lesion. Although part of the T2w lesion contains vasogenic edema and other non-specific changes, there may also be substantial regions of infiltrative tumor [4]. The recent use of anti-angiogenic agents has added to the ambiguity in interpreting imaging data, because they act by diminishing vascular permeability and can cause a reduction in the size of both the CE and T2w lesions, while not necessarily changing the number of tumor cells present.
Other imaging modalities that are thought to be associated with tumor characteristics include diffusion-weighted imaging (DWI) and metabolic imaging using in vivo 1H MRSI. Although there have been reports of a linear correlation between tumor cellularity and restricted diffusivity, it is unclear whether this assumption applies to tumor in T2w as well as CE lesions. Numerous studies from our own as well as other groups have demonstrated the ability of 1H MRSI to highlight areas of tumor in the T2w lesion based upon a combination of increased levels of choline and reduced levels of NAA [5].
The purpose of this study was to obtain image-guided tumor samples of enhancing (Enh+) and non-enhancing (Enh−) GBM from CE and T2w regions, in order to characterize the similarities and differences between their histological and ex vivo spectroscopic profiles. By saving the locations where the actual tissue samples were obtained during surgery, it was also possible to compare the corresponding in vivo anatomic, 1H MRSI, DWI, and DSC imaging parameters between these radiographic subtypes of GBM.
Materials and Methods
Patient Population
The institutionally approved study comprised 56 treatment-naïve patients who underwent surgical resection and whose lesions were subsequently diagnosed as being GBM by a single pathologist according to standard histological criteria [1]. Informed consent was obtained from each patient for the removal of image-guided tissue samples during surgery.
Preoperative MR Exam
A multiparametric MR examination was conducted within 24 hours prior to surgery using either 1.5- or 3-T whole-body MR scanners (GE Healthcare Technologies, Milwaukee, WI, USA) with an 8-channel, phased-array headcoil for signal reception (MRI Devices, Knaresborough, United Kingdom). The data obtained included 3-D pre- and post-contrast T1-weighted inversion recovery spoiled-gradient echo (IRSPGR) images (TR/TE/TI = 8/3/400 ms, 1.5 mm slices, 15o flip angle, 256 × 256 × 124 matrix, 26 × 26 cm2 in-plane field of view) and T2-weighted anatomic FLAIR and fast spin echo (FSE) images (see Supplementary Material). In 45/56 cases, diffusion-weighted imaging was acquired in the axial plane with spin-echo echo-planar-imaging (TR/TE = 1000/108 ms; voxel size, 1.7 × 1.7 × 3 mm3; 6 gradient directions; 4 NEX; b = 1000 s/mm2) and, in 24/56 cases, lactate (Lac)-edited three-dimensional 1H MRSI at 3 T. The latter applied point-resolved spectroscopic selection (PRESS) for volume localization and very selective saturation (VSS) pulses for lipid (Lip) signal suppression (excited volume, ~80 × 80 × 40 mm3; overpress factor, 1.5; TR/TE = 1104/144 ms; field of view, 16 × 16 × 16 cm3; nominal voxel size, 1 × 1 × 1 cm3; flyback echo-planar readout gradient in the superior–inferior direction; 712 dwell points; sweep width, 988 Hz) [6]. DSC perfusion imaging was acquired in 45/56 cases as a series of T2*-weighted echo-planar images (TR/TE/flip angle = 1250–1500 ms/54–56 ms/30–35 degrees, 128 × 128 matrix, 26 × 26 cm field of view, slice thickness = 3–4 mm, 7–15 slices with 60–80 time points) using a 5 ml/s bolus injection of 0.1 mmol/kg body weight Gd-DTPA (Magnevist, gadopentetate dimeglumine).
Definition of Targets For Tissue Sampling
In vivo data from the pre-operative examination were transferred to a Unix workstation and in-house software were applied to derive estimates of diffusion, spectroscopic and perfusion parameters. Maps of the apparent diffusion coefficient (ADC) and fractional anisotropy (FA) were generated on a pixel-by-pixel basis [7]. The 1H MRSI data were processed as described previously [8] in order to quantify peak heights and areas of total choline (tCho), creatine (Cr/PCr), N-acetylaspartate (NAA), lactate (Lac) and lipid (Lip). Maps of the tCho-to-NAA index (CNI) were derived using an automated iterative linear regression-based algorithm that sequentially removes outlying values and represents the changes in tCho and NAA levels relative to the normal voxels from the same subject [9]. DSC perfusion data were processed to generate estimates of cerebral blood volume (CBV) by either fitting dynamic curves using a modified gamma-variate function with leakage correction [10], [11] or using a non-parametric procedure to estimate peak height (PH) and percent recovery (REC) [12].
Image-Guided Tissue Sampling
One to four tissue targets were identified, based upon observations from previous studies, as being within the CE or T2w region using (i) ADC values less than 1.5 times normal appearing white matter, (ii) CNI values greater than 2, or (iii) CBV values greater than 3. These targets were superimposed upon the T1-weighted post-contrast images as spherical regions of interest (ROIs) with a diameter of 5 mm, and transferred to the surgical navigation workstation (BrainLAB Inc., Munich, Germany).
While it was not always feasible to resect samples from the pre-defined tissue targets, the surgeon used the image guidance software to provide samples from the nearest accessible regions. The actual sample location was saved on the BrainLAB workstation as both screenshots and files of RAS coordinates relative to the pre-surgery images; all of the in vivo imaging was registered together along with the sample location. After excision, the tissue samples were immediately bisected: half was snap frozen in liquid nitrogen less than 1 min after removal and stored at −80°C for 1H HR-MAS spectroscopy; the other half was fixed in 10% zinc formalin, dehydrated by graded ethanols and embedded in Paraplast Plus wax (McCormick Scientific, St. Louis, MO, USA) using standardized techniques for pathological analysis.
Definition of InVivo Parameters
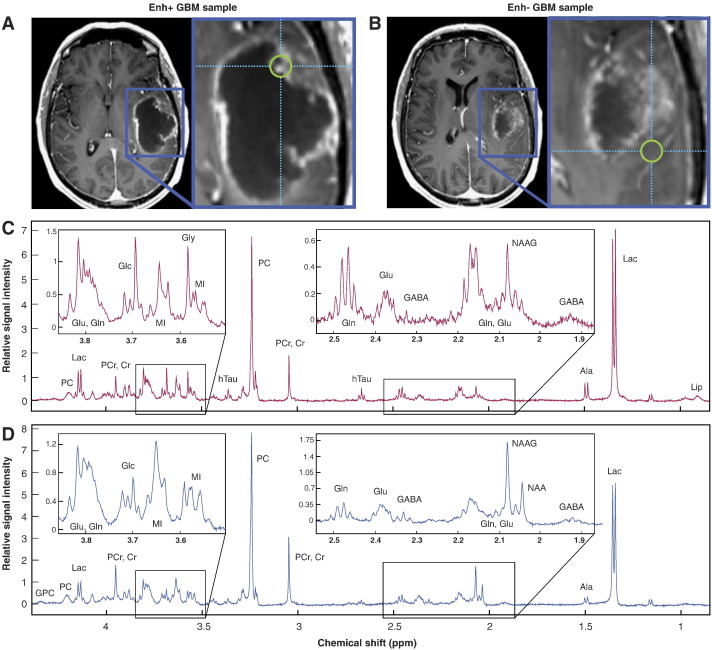
ROIs corresponding to the CE lesion and any area of necrosis (NEC) were manually segmented on the T1 post-contrast images. The T2w lesion was defined as the region of hyperintensity on the T2-weighted FLAIR image minus the CE lesion and NEC. A brain region of interest was defined using the brain extraction tool in FSL [13], and normal-appearing brain (NAB) was defined as the entire brain minus the lesion and NEC ROIs. Spherical regions with 5-mm-diameter and centered at the coordinates obtained by BrainLab from the actual site of tissue removal were defined and visually confirmed as corresponding to the screenshots acquired during surgery. Histologically-confirmed tumor with greater than 50% contrast-enhancement in the target ROIs was classified as enhancing GBM (Enh+), while tumor confined to the T2w lesion was considered non-enhancing GBM (Enh−). Figure 1 depicts regions of Enh+ (A) and Enh− (B) tumor from a single patient.
Figure 1.
Image-guided tissue from a patient with GBM designated as Enh+ (A) or Enh− (B) on the basis of post-contrast T1 IRSPGR images. Corresponding CPMG 1H HR-MAS spectra for Enh+ (C) and Enh− (D) tumor samples. NAAG, N-acetylaspartylglutamate.
In vivo parameters [14] from within sample ROIs were determined by histogram analysis of pixel intensities. For the anatomic and DWI parameters, normalized intensity values were obtained by dividing median intensities in the corresponding ROI by the mode of the intensity in the ROI from NAB. For the H-1 spectroscopic data, metabolite levels were obtained by sinc-interpolating these maps to create intensities at the location of the center of the sample ROI. Normalization factors for tCho, Cr and NAA were defined as median metabolite level values from the subset of ‘normal’ voxels that were defined from the CNI algorithm. Levels of Lac and Lip were normalized relative to the NAA in normal voxels. The CBV, PH and REC were obtained within sample ROIs as described previously [10], [12].
Histological Analysis of the Tumor
An experienced neuropathologist evaluated the slides prepared from tissue samples with hematoxylin and eosin (H&E) stains to determine the relative contribution of tumor cells to the overall cellularity. A score of ‘0’ denoted neuropil without tumor; ‘1’ indicated an infiltrating tumor margin containing detectable but not abundant numbers of tumor cells; ‘2’ denoted a more cellular infiltrated zone; and ‘3’ denoted highly cellular tumor with relatively few non-neoplastic cells. Only tissue samples with scores greater than zero were included in the analysis. Further H&E and immunohistochemical analyses were performed to determine overall cellularity, necrosis content, mitoses (MIB-1), hypoxia (CA9), axonal disruption (SMI-31), and microvascular morphology (Factor VIII) (see Supplementary Material) [14]. As described in the Supplementary Material, Factor VIII was used to grade microvasculature morphology as delicate (resembling normal cerebral vessels), simple microvascular hyperplasia (circumferential single cell hyperplasia with definitive lumen), or complex microvascular hyperplasia (circumferential multi-layered and glomeruloid-type vessels). IDH wild-type status relevant to establishing some level of molecular homogeneity among primary GBM was evaluated via the absence of 2-hydroxyglutarate (2HG) in HR-MAS spectra.
1H HR-MAS Spectroscopy
Tissue samples weighing between 1.11 and 30.42 mg (median, 10.25 mg) were evaluated. A 35-μL zirconia rotor (custom-designed by Varian, Palo Alto, CA, USA) was used with 3 μL of 99.9% atom-D deuterium oxide containing 0.75 wt% 3-(trimethylsilyl)propionic acid (Sigma-Aldrich, St. Louis, MO, USA) for chemical shift referencing. Data were acquired at 11.7 T at 1°C with a spin rate of 2250 Hz in a 4-mm gHX nanoprobe with a Varian INOVA 500-MHz multi-nuclear spectrometer.
A rotor-synchronized, one-dimensional, Carr–Purcell–Meiboom–Gill (CPMG) pulse sequence was run with TR/TE = 4 s/144 ms, 512 scans, 40,000 acquired points, 90° pulse and spectral width of 20 kHz for a total time of 35 min. A relatively long TE was used to maximally suppress the macromolecular background for purposes of metabolite fitting. The electronic reference to access in vivo concentrations (ERETIC) method was used to generate an artificial electronic signal that served as an external standard for the estimation of metabolite levels [15].
Pre-processing of the spectra was performed in the time domain using the Java-based Magnetic Resonance User Interface (jMRUI) [16]. The estimation of relative one-dimensional metabolite levels [14] was achieved with the semi-parametric algorithm, high-resolution quantum estimation (HR-QUEST), which fits a customized basis set of metabolites to the spectrum [17]. The basis set used in this study comprised spectra from 26 metabolite solutions commonly studied in the human brain (Sigma-Aldrich): NAA, free choline (Cho), phosphocholine (PC), glycerophosphocholine (GPC), ethanolamine (Eth), phosphoethanolamine (PE), creatine/phosphocreatine (Cr/PCr), myo-inositol (MI), scyllo-inositol (SI), glucose (Glc), glycine (Gly), total glutathione [tGSH: glutathione (GSH) + glutathione disulfide (GSSG)], glutamate (Glu), glutamine (Gln), 2HG, γ-aminobutyric acid (GABA), taurine (Tau), hypotaurine (hTau), threonine (Thr), acetate (Ace), Lac, alanine (Ala), betaine (Bet), aspartate (Asp), valine (Val) and succinate (Suc). Total choline (tCho) was defined as [PC + GPC + Cho] for comparison with the in vivo data. Only metabolite estimates with Cramer-Rao errors less than 12% were included in subsequent analyses. In order to estimate lipid (Lip) content, the area under the methyl Lip resonance, centered at approximately 0.9 ppm, was evaluated over the interval [0.936 0.850] ppm in MATLAB, and normalized by sample weight and ERETIC peak area. Figure 1 shows ex vivo spectra corresponding to regions of Enh+ (Figure 1, A and C) and Enh− (Figure 1, B and D) tumor.
Statistical Analysis
Tissue samples with tumor scores of 1–3 were evaluated according to a random mixed effects model in R v3.3.2 [18] that accounted for patients with multiple tissue samples. Odds ratios and P-values are reported for continuous imaging and histopathological parameters that were determined to predict enhancement status when accounting for repeated specimen sampling. Ordinal histopathology parameters were compared between Enh+ and Enh− tumor samples using a proportional odds logistic regression model that also accounted for repeated sampling [19]; mixed effects models of ordinal-valued outcomes were analyzed with Proc Genmod in SAS v9.2. Pairwise correlations were assessed for continuous and ordinal variables using the Pearson product–moment correlation test and Kendall tau rank correlation, respectively. Each correlation was run 50 times with a random tumor sample selected per patient upon every iteration; the mean and standard deviations for tau estimates are reported along with median P-values. Bonferroni correction was applied for multiple correlations among 116 derived parameters.
Results
Summary of Analyzed Tumor Tissue
As shown in Table 1, a total of 102 tumor samples (70 Enh+, 32 Enh−) were acquired from the 56 subjects that were evaluated.
Table 1.
Summary of Patients and Image-Guided Samples of GBM Classified According to Enhancement Status
| GBM Classification | Number of Patients | Number of Tissue Samples |
|
|---|---|---|---|
| Enh+ | Enh− | ||
| Enh+ | 34 | 56 | 0 |
| Enh− | 11 | 0 | 19 |
| Mixed | 11 | 14 | 13 |
| Total | 56 | 102 (70 | 32) |
Enh+, contrast-enhancing; Enh−, non-enhancing.
1H HR-MAS Spectroscopy
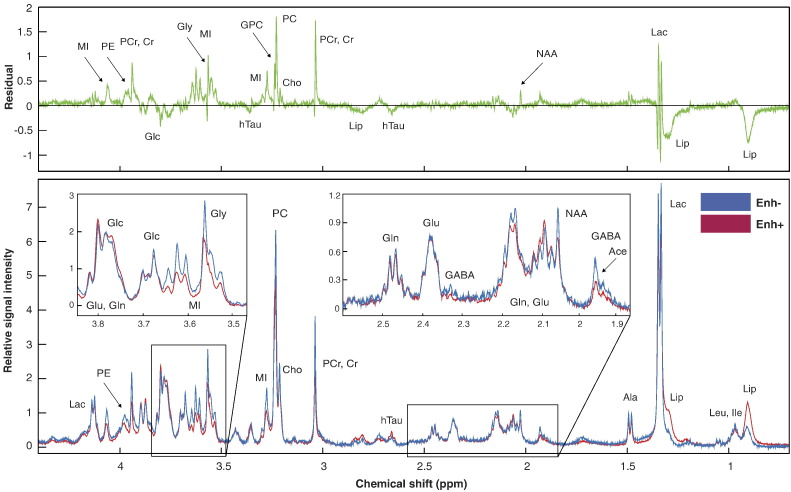
To provide a visual comparison between metabolite profiles of Enh+ and Enh− GBM samples, individual spectra were normalized by the sample weight and ERETIC peak area, then averaged together according to enhancement status. The composite spectra shown in Figure 2 for Enh+ (red) and Enh− (blue) GBM reveal the characteristic features of these distinct radiographic entities. These demonstrate that phosphocholine (PC), rather than glycero-phosphocholine (GPC), is the dominant choline species. Trace levels of N-acetylaspartate (NAA) and the presence of hypo-taurine (hTau) in these samples corroborated the pathological analysis of the tissue. The Enh− tissue samples displayed elevated levels of phosphoethanolamine (PE), creatine/phosphocreatine (Cr/PCr), glycine (Gly), myo-inositol (MI), and GPC. The Enh+ samples demonstrated higher methyl lipid (Lip).
Figure 2.
Mean CPMG 1H HR-MAS spectra for image-guided Enh+ (red, n = 70) and Enh− (blue, n = 32) GBM samples. The residual of Enh− minus Enh+ spectra is shown in green.
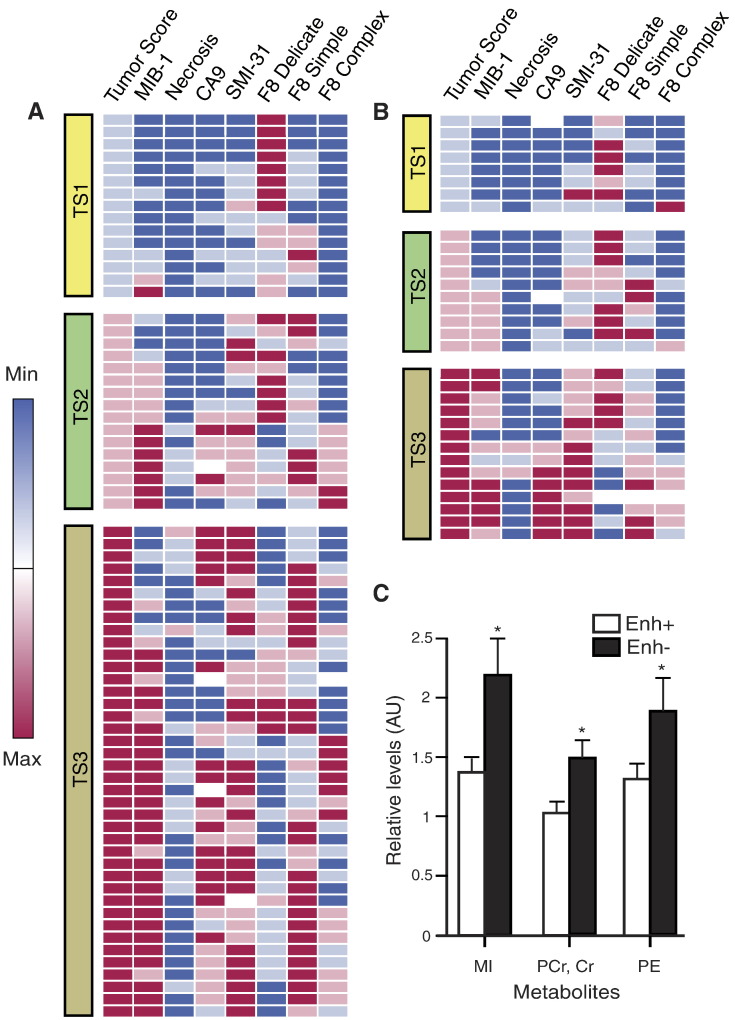
Results from the random mixed effects model were consistent with the visual assessments and are shown in Table 2. Lactate (Lac) was not considered in the analysis because of its lack of reliability in the ex vivo setting [20]. The Enh− GBM had significantly elevated MI (P = .003), Cr/PCr (P = .009), and PE (P = .05) compared to Enh+ samples, with a similar trend for Gly. Enh+ tumor samples showed a trend toward elevated methyl Lip (P = .07), but there was no difference observed in tCho levels (Table 2).
Table 2.
Regional Summary of Continuous Imaging and Histopathological Parameters
| MR/histology Parameter | Mean ± SE (n) |
Odds Ratio (95% CI) | P-value | |
|---|---|---|---|---|
| Enh+ | Enh− | |||
| HR-MASa | ||||
| MI | 1.37 ± 0.12 (53) | 2.19 ± 0.35 (23) | 0.57 (0.39–0.83) | 0.003 |
| Cr/PCr | 1.03 ± 0.10 (52) | 1.48 ± 0.15 (21) | 0.44 (0.24–0.82) | 0.009 |
| PE | 1.32 ± 0.12 (53) | 1.88 ± 0.28 (24) | 0.51 (0.28–0.94) | 0.05 |
| Lip | 8.51 ± 1.70 (70) | 4.67 ± 0.53 (32) | 1.19 (0.99–1.43) | 0.07 |
| Gly | 2.21 ± 0.34 (48) | 4.34 ± 1.38 (16) | 0.86 (0.73–1.00) | 0.06 |
| GPC | 0.58 ± 0.07 (46) | 0.81 ± 0.13 (21) | 0.44 (0.18–1.07) | 0.07 |
| Anatomic | ||||
| nT1c | 1.52 ± 0.01 (68) | 0.91 ± 0.01 (29) | 140 (23–880) | 1 × 10−7 |
| nFL | 1.69 ± 0.01 (67) | 1.61 ± 0.01 (29) | NA | NS |
| nFSE | 2.17 ± 0.01 (63) | 1.96 ± 0.03 (29) | NA | NS |
| Perfusion | ||||
| nCBV | 2.91 ± 0.48 (28) | 1.39 ± 0.11 (20) | 3.51 (1.46–8.41) | 0.005 |
| nPH | 2.23 ± 0.33 (28) | 1.23 ± 0.09 (20) | 4.31 (1.30–14.30) | 0.02 |
| REC (%) | 75.94 ± 2.46 (28) | 84.27 ± 2.27 (20) | 0.93 (0.87–1.00) | 0.04 |
| Diffusion | ||||
| Median nADC | 1.54 ± 0.01 (53) | 1.44 ± 0.01 (28) | NA | NS |
| Median nFA | 0.73 ± 0.01 (51) | 0.97 ± 0.01 (25) | 0.14 (0.02–0.55) | 0.01 |
| 1H MRSI | ||||
| nCr/PCr | 0.51 ± 0.03 (26) | 0.93 ± 0.10 (11) | 0.22 (0.04–1.26) | 0.09 |
| nLac | 0.52 ± 0.07 (26) | 0.30 ± 0.06 (11) | 17.65 (0.84–370) | 0.07 |
| nLip | 0.86 ± 0.19 (26) | 0.59 ± 0.14 (11) | NA | NS |
| CNI | 6.75 ± 0.90 (26) | 6.69 ± 0.84 (11) | NA | NS |
| Histopathology | ||||
| MIB-1 (%) | 14.86 ± 0.33 (68) | 12.88 ± 0.23 (30) | NA | NS |
| Cellularityb (cells) | 286 ± 2 (67) | 287 ± 6 (30) | NA | NS |
The estimated difference in effect between Enh+ and Enh− (reference) GBM is summarized by the odds ratio/P-value from a mixed effects model.
Arbitrary units without correction for T2-dependence.
Average number of cells per 200X field.
There were significant correlations between a number of metabolites for either all samples or the Enh+ samples. In particular, levels of PE were shown to correspond to levels of tCho (P = .0001), glutamate (Glu, P = .002), threonine (Thr, P = .002), and alanine (Ala, P = .00002), which were intercorrelated (see Supplementary Table S1). For the same groups of samples, levels of MI were linked to those of tCho (P = .009) (Table S1). No significant correlations were found among the Enh− GBM after correction for multiple comparisons.
InVivo Imaging
As expected, the normalized T1 post-contrast (nT1c) intensities were much higher in the Enh+ ROIs corresponding to the regions where enhancing GBM was sampled intra-operatively (P = 1 × 10−7) (Table 2). The normalized FLAIR (nFL) and FSE values showed no significant difference in intensities. Perfusion-derived parameters demonstrated clear distinctions between Enh+ versus Enh− tissue, with the measures of perfusion peak height (nPH, P = .02) and cerebral blood volume (nCBV, P = .005) being elevated in Enh+ relative to Enh− GBM, while the percent recovery (REC, P = .04) was found to be reduced in Enh+ GBM, indicating greater leakage (Table 2). Although the nADC values showed no difference, the median normalized values of fractional anisotropy (nFA) were lower in Enh+ (P = .01) relative to Enh− targets (Table 2).
Lactate-edited 1H MRSI data were obtained for a smaller subset of patients because it was not always possible to add the additional sequence in the clinical setting and because the selected volume did not always cover the tissue sample location. The Enh− targets showed trends toward higher levels of normalized Cr (P = .09) and lower levels of normalized Lac (P = .07) compared to Enh+ targets, but with no significant difference in levels of Cho, NAA or Lip (Table 2). CNI values were elevated in both Enh+ and Enh− samples (Table 2). There was an inverse correlation between nADC and both tCho (P = .01) and CNI (P = .007) for all samples (Table S1).
Histopathology
Tumor scores were similar between Enh+ and Enh− GBM, with only ~12% more Enh+ samples classified as representing maximal tumor content (Figure 3; Table 3). Mean overall cell densities were almost identical at 286 (Enh+) versus 287 (Enh−) cells/200x field (Table 2). There was no significant difference in the MIB-1 indices for Enh+ and Enh− samples (14.66% versus 12.88% - see Figure 3 and Table 2). Delicate microvasculature predominated in 45% versus 24% of Enh− and Enh+ samples, respectively, and was completely absent in almost a third of the Enh+ samples (OR, P = .42, 0.08) (Figure 3; Table 3). Simple microvasculature overwhelmingly characterized 41% of Enh+ samples compared to 19% of Enh− samples (OR, P = 2.18, 0.07) (Figure 3; Table 3), while Enh+ samples had relatively higher scores for complex microvasculature (Figure 3; Table 3). Hypoxia (CA-9) and necrosis scores were not significantly different with regard to enhancement status, and axonal disruption (SMI-31) was comparable across all tumor samples.
Figure 3.
Histopathological parameters are depicted in heatmaps for Enh+ (A) and Enh− (B) GBM samples with deterministic hierarchical clustering applied to samples within each tumor score (TS, 1–3) group. Mean levels of ex vivo metabolites showing statistically significant differences (*) on the basis of enhancement status; error bars represent SE (C).
Table 3.
Analysis of Regional Histopathological Features With Categorical Scoring
|
Categorical Features | ||||||||
|---|---|---|---|---|---|---|---|---|
| Histology Parameter | Tissue Region | Number of Samples | Frequency Distribution of Categorical Scores (%) |
Random Effect Model Analysis |
||||
| 0 | 1 | 2 | 3 | Odds Ratio (95% CI) | P-value | |||
| Tumor score | Enh− | 32 | 0 | 25.0 | 31.3 | 43.8 | 1.47 (0.67–3.23) | NS |
| Enh+ | 70 | 0 | 21.4 | 22.9 | 55.7 | |||
| Necrosis | Enh− | 32 | 90.6 | 3.1 | 6.3 | NA | NA | NA |
| Enh+ | 70 | 75.7 | 21.4 | 2.9 | NA | |||
| Hypoxia | Enh− | 30 | 60.0 | 13.3 | 6.7 | 20.0 | NA | NA |
| Enh+ | 67 | 37.3 | 14.9 | 20.9 | 26.9 | |||
| AD | Enh− | 32 | 15.6 | 28.1 | 28.1 | 28.1 | 1.53 (0.65–3.56) | NS |
| Enh+ | 69 | 11.6 | 21.7 | 29.0 | 37.7 | |||
| Delicate vasculature | Enh− | 31 | 12.9 | 25.8 | 16.1 | 45.2 | 0.42 (0.16–1.10) | 0.08 |
| Enh+ | 70 | 27.1 | 30.0 | 18.6 | 24.3 | |||
| Simple MVH | Enh− | 31 | 19.4 | 38.7 | 22.6 | 19.4 | 2.18 (0.92–5.15) | 0.07 |
| Enh+ | 70 | 11.4 | 30.0 | 17.1 | 41.4 | |||
| Complex MVH | Enh− | 31 | 71.0 | 9.7 | 16.1 | 3.2 | 2.14 (0.69–6.65) | 0.18 |
| Enh+ | 69 | 53.6 | 14.5 | 20.3 | 11.6 | |||
The estimated difference in effect between Enh+ and Enh− (reference) GBM is summarized by the odds ratio/P-value from a random effects model. AD, axonal disruption; MVH, microvascular hyperplasia; NA, not applicable owing to insufficient non-zero scores; NS, not significant.
There were several instances where the histological scores and values showed similar trends. Although both Enh+ and Enh− samples independently showed a strong correlation between tumor score and SMI31 (P = .002, 0.004), the majority of correlations related to the enhancing tissue samples (Table S1). Among Enh+ GBM, tumor score was inversely correlated with the presence of delicate microvasculature (P = .01) and positively correlated with CA9 expression (P = .02), while CA9 correlated with necrosis (P = .009) and had an inverse relationship with delicate vasculature (P = .005). Microvascular hyperplasia in Enh+ samples was weakly related to MIB-1 (P = .05). Both Enh+ and Enh− samples showed a relationship between MIB-1 and average cellularity (P = .002, 0.009).
Discussion
This study characterized the metabolomic profiles of tissue samples obtained from enhancing and non-enhancing regions in newly diagnosed GBM that were confirmed to contain tumor by histological analysis. Ex vivo 1H HR-MAS metabolite levels were similar for tCho and NAA, but differed for a number of other metabolites that can be evaluated through in vivo methods utilizing short echo times. Analysis of the associated histopathology and multi-parametric MR examinations helped to contextualize and corroborate these findings. Given the heterogeneity of anatomic images with regard to contrast extravasation, having non-invasive parameters that can highlight non-enhancing tumor is of interest for planning surgical resection and for the ongoing assessment of total tumor burden.
The heterogeneity in ex vivo metabolite expression between Enh+ and Enh− samples may reflect biological differences in the tumor cells and their microenvironment. While Enh+ samples displayed a trend toward elevated methyl Lip that has commonly been linked with necrosis, the chemical profile of Enh− counterparts more closely resembled the increases in MI, Cr/PCr and Gly that were previously observed in samples from de novo grade III glioma, which lack necrosis and have limited microvascular proliferation [21]. Although failing to reach statistical significance, the trend toward higher levels of GPC in Enh− GBM is also consistent with the metabolic profile displayed by grade III glioma. Elevation in the glia-associated metabolite MI, which has been extensively employed for classifying infiltrating disease with respect to pathological grade, has also been reported for regions of tumor that are less malignant or contain gliosis based upon standard histological criteria [22], [23].
From a neurochemical perspective, the metabolomic profile exhibited by Enh− samples appears to confirm the heterogeneous nature of this disease, and suggests further investigation into the underlying significance of metabolite differences. The heightened levels of PCr/Cr observed in Enh− samples are likely due to their greater bioenergetic potential, as PCr contributes a reservoir of phosphate for rapid ATP generation [24]. Trending levels of Gly, a non-essential amino acid and inhibitory neurotransmitter that is commonly found in high-grade glioma [25], are suggestive of malignant pathology, though no explicit relationship has been established. Owing to the in vivo overlap between Gly's singlet and the M-spin resonance of MI centered at ~3.52 ppm, measurements of these two metabolites may be difficult to distinguish without application of techniques such as 2D J-resolved spectroscopy and spectral editing [26].
In assessing the pathobiology of GBM, consideration must also be given to the unique lipid metabolism that has been demonstrated in these lesions. Excess lipid has historically been associated with a poor prognosis for patients [27], and may relate to an aberrant oncogenic signaling EGFR/PI3K/Akt pathway that promotes lipid metabolism via the master transcriptional factor SREBP-1 [28]. Previous HR-MAS investigations suggested that such lipid elevations are cytoplasmic in origin and indicative of necrosis, which is a pathological hallmark exclusive to GBM within the WHO criteria [29], [30], [31]. While necrosis was not statistically differentiated in this study, it was noted as >2.5x more common in Enh+ samples. One of the above studies additionally revealed that the presence of cholesteryl esters and triglycerides in cell extracts from GBM was correlated with the extent of vascular proliferation using both 1H and 13C HR-MAS techniques [31]. Given the apparent relevance of lipid fractions to pathology, it is worth noting the elevation of PE in Enh− GBM, which suggests that there are differences in phospholipid metabolism associated with the Kennedy pathway [32].
Within Enh+ GBM, PE also showed multiple exceptionally strong correlations with other metabolites, including tCho and threonine. Because the cytidine diphophospate (CDP)-ethanolamine (Eth) pathway represents half of the Kennedy pathway, there are plausible relations of PE to choline species. The CDP-Eth pathway is additionally known to regulate diacylglycerol, a key activator of serine–threonine kinases, thus providing support for the association with threonine [33].
While limited in coverage and employment, the acquisition of in vivo 1H MRSI data proved valuable for corroborating HR-MAS findings. Both in vivo and ex vivo spectroscopy demonstrated levels of tCho that were not statistically different between Enh+ and Enh− tumor, but nevertheless abnormally elevated relative to NAA, as demonstrated by the in vivo CNI values. Previous investigations have shown that the PC resonance forms the dominant contribution to the in vivo tCho peak in GBM, whether as primary tumor or secondary tumor that has transitioned from grade II or III glioma [21], [34]. Because ex vivo Lac levels are considered unreliable given the ongoing metabolism of tissue following extraction, it was instructive to observe increased in vivo Lac levels in Enh+ GBM. Such increases may be attributed to heightened anaerobic respiration and/or interpolation of data with nearby necrosis on account of the coarse spatial resolution of MRSI.
Among the metabolites that distinguished Enh− samples, Cr/PCr and MI + Gly offer the greatest promise for in vivo characterization of differences between contrast-based radiographic subtypes of GBM. While the ex vivo elevation of Cr/PCr levels in Enh− relative to Enh+ samples was seen as a trend for in vivo 1H MRSI data, the long echo time utilized in the current study prevented the detection of MI + Gly. Because of the T2-shortening effects from MI's complex coupling, either a point-resolved spectroscopy (PRESS) sequence with a TE ≤ 35 ms or some Carr-Purcell-Meiboom-Gill (CPMG)-inspired sequence would be required for future studies [35].
The differences in vascular physiology assessed by in vivo perfusion-weighted imaging in this study were found to accurately reflect the microvascular heterogeneity of GBM represented by Factor VIII immunostaining. Elevations in nPH and nCBV in Enh+ regions were consistent with the microvascular hyperplasia and gross hypervascularity that are associated with angiogenesis, and similar to the results obtained by Essock-Burns et al. [36]. Reduced REC within the enhancing tumor is similarly indicative of immature, perforated vasculature whose permeability provides greater access to the extravascular, extracellular space. Although Enh+ regions displayed aberrant perfusion, the in vivo parameters observed in Enh− GBM remained within the range that is considered normal [37], and therefore present a challenge for defining total tumor burden.
Analysis of the DWI data indicated that nADC values in tumor were similar irrespective of enhancement status, with median values near 1.5 reflecting moderately restricted diffusion. This finding is supported by the fact that overall cell density and tumor cellularity were comparable between Enh+ and Enh− samples. The in vivo nADC was also found to inversely correlate with measures of tCho and CNI, which inform on tumor cellularity via choline species involved in membrane turnover. The higher values of nFA exhibited by Enh− samples demonstrated that non-enhancing components of the tumor may retain greater integrity of the surrounding tissue compared to that in the CE lesion, despite no clear differences in axonal disruption measured via SMI-31. Regardless of the potentially favorable structural status of Enh− samples, histopathologic analysis of MIB-1 mitotic indices indicated that there were similar levels of proliferation as in Enh+ samples. This suggests that Enh− tumor may bear a high degree of malignancy, even in the absence of patent contrast extravasation.
Besides providing evidence of comparable proliferation and cellularity between Enh+ and Enh− samples from GBM, the histopathology also showed internal relationships among several parameters. Both types of samples displayed the expected correlations between tumor score and axonal disruption, as well as between proliferation and average cellularity. For Enh+ samples, the association of hypoxia (as estimated by CA-9 scores) with regional necrosis provided validation of these respective assays. As might be anticipated, delicate vasculature, which is considered normal from a histological standpoint, was found to be inversely correlated with hypoxia for the Enh+ samples.
Conclusion
The results of this study provide extensive characterization of CE and non-enhancing GBM from image-guided tumor samples. While these contrast-based subtypes of disease demonstrated comparable mitotic activity and cellularity, their microvascular morphology provided a basis for histological differentiation. Given the similarity in ex vivo tCho and NAA levels measured via HR-MAS spectroscopy, there is strong evidence to support the use of CNI as a metric for defining total tumor burden in vivo. The ADC values were also found to be similar across Enh− and Enh+ GBM, and may be evaluated in conjunction with CNI as advanced criteria for detection of non-enhancing disease. Differences in ex vivo levels of Cr/PCr, as well as in the factor VIII scores and in vivo DSC parameters, were consistent with the expectation that Enh− tumor has more normal-appearing vasculature. Despite the proven utility of DSC perfusion in highlighting tumor, the vascular profile of Enh− GBM was shown to be such that it eludes detection in vivo.
Acknowledgments
Acknowledgements
We would like to acknowledge support from the Brain Tumor Research Center at the University of California, San Francisco (UCSF) in collecting tissue. Grant funding was provided by NIH PO1 CA118816, The Cancer Center Support Grant (P30 CA82103), and the German Research Foundation (DFG, MA 7292/1-1).
Footnotes
Conflicts of Interest: None.
Funding: NIH PO1 CA118816, The Cancer Center Support Grant (P30 CA82103), German Research Foundation (DFG, MA 7292/1-1).
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.tranon.2017.08.009.
Appendix A. Supplementary data
Supplementary material. Immunohistochemistry methods, T2-weighted imaging methods, and correlations among imaging and histopathologic parameters.
References
- 1.Lois DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 2.Johnson DR, O'Neill BP. Glioblastoma survival in the United States before and during the temozolomide era. J Neurooncol. 2012;107:359–364. doi: 10.1007/s11060-011-0749-4. [DOI] [PubMed] [Google Scholar]
- 3.Sherry AD, Caravan P, Lenkinski R. Primer on gadolinium chemistry. J Magn Reson Imaging. 2009;30(6):1240–1248. doi: 10.1002/jmri.21966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu S, Ahn D, Johnson G, Cha S. Peritumoral diffusion tensor imaging of high-grade gliomas and metastatic brain tumors. Am J Neuroradiol. 2003;24:937–941. [PMC free article] [PubMed] [Google Scholar]
- 5.McKnight TR, von dem Bussche MH, Vigneron DB, Lu Y, Berger MS, McDermott MW, Dillon WP, Graves EE, Pirzkall A, Nelson SJ. Histopathological validation of a three-dimensional magnetic resonance spectroscopy index as a predictor of tumor presence. J Neurosurg. 2002;97(4):794–802. doi: 10.3171/jns.2002.97.4.0794. [DOI] [PubMed] [Google Scholar]
- 6.Park I, Chen AP, Zierhut M, Ozturk-Isik E, Vigneron DB, Nelson SJ. Implementation of 3T lactate-edited 3D 1H MR spectroscopic imaging with flyback echo-planar readout for glioma patients. Ann Biomed Eng. 2011;39:193–204. doi: 10.1007/s10439-010-0128-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basser PJ, Pierpaoli C. Microstructural and physiological features of tissues elucidated by quantitative diffusion-tensor MRI. J Magn Reson B. 1996;111:209–219. doi: 10.1006/jmrb.1996.0086. [DOI] [PubMed] [Google Scholar]
- 8.Li Y, Osorio JA, Ozturk-Isik E, Chen AP, Xu D, Crane JC, Cha S, Chang S, Berger MS, Vigneron DB, Nelson SJ. Considerations in applying 3D PRESS H1 brain MRSI with and eight-channel phased-array coil at 3T. Magn Reson Imaging. 2006;24(10):1295–1302. doi: 10.1016/j.mri.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 9.McKnight TR, Noworolski SM, Vigneron DB, Nelson SJ. An automated technique for the quantitative assessment of 3D-MRSI data from patients with glioma. J Magn Reson Imaging. 2001;13:167–177. doi: 10.1002/1522-2586(200102)13:2<167::aid-jmri1026>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 10.Lee MC, Cha S, Chang SM, Nelson SJ. Dynamic susceptibility contrast perfusion imaging of radiation effects in normal-appearing brain tissue: changes in the first-pass and recirculation phases. J Magn Reson Imaging. 2005;21(6):683–693. doi: 10.1002/jmri.20298. [DOI] [PubMed] [Google Scholar]
- 11.Boxerman JL, Schmainda KM, Weisskoff RM. Relative cerebral blood volume maps corrected for contrast agent extravasation significantly correlate with glioma tumor grade, whereas uncorrected maps do not. Am J Neuroradiol. 2006;27(4):859–867. [PMC free article] [PubMed] [Google Scholar]
- 12.Lupo JM, Cha S, Chang SM, Nelson SJ. Dynamic susceptibility-weighted perfusion imaging of high-grade gliomas: characterization of spatial heterogeneity. Am J Neuroradiol. 2005;26(6):1446–1454. [PMC free article] [PubMed] [Google Scholar]
- 13.Smith SM. Fast robust automated brain extraction. Human Brain Mapping. 2002;17(3):143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Autry A, Phillips J, Maleschlijski S, Roy R, Molinaro AM, Chang SM, Cha S, Lupo JM, Nelson SJ. Mendeley Data. 2017. Data for: Characterization of metabolic, diffusion, and perfusion properties in GBM: contrast-enhancing versus non-enhancing tumor. [dataset] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Albers MJ, Butler TN, Rahwa I, Bao N, Keshari KR, Swanson MG, Kurhanewicz J. Evaluation of the ERETIC method as an improved quantitative reference for 1H HR-MAS spectroscopy of prostate tissue. Magn Reson Med. 2009;61:525–532. doi: 10.1002/mrm.21808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stefan D, Cesare FD, Andrasescu A, Popa E, Lazariev A, Vescovo E, Strbak O, Williams S, Starcuk Z, Cabanas M. Quantitation of magnetic resonance spectroscopy signals: the jMRUI software package. Meas Sci Technol. 2009;20:104035. [9 pp.] [Google Scholar]
- 17.Ratiney H, Albers MJ, Rabeson H, Kurhanewicz J. Semi-parametric time-domain quantification of HR-MAS data from prostate tissue. NMR Biomed. 2010;23:1–13. doi: 10.1002/nbm.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2013. R: A language and environment for statistical computing. [Google Scholar]
- 19.Barajas RF, Phillips JJ, Parvateneni R, Molinaro A, Essock-Burns E, Bourne G, Parsa AT, Aghi MK, McDermott MW, Berger MS. Regional variation in histopathologic features of tumor specimens from treatment-naïve glioblastoma correlates with anatomic and physiologic MR Imaging. Neuro Oncol. 2012;14(7):942–954. doi: 10.1093/neuonc/nos128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Opstad KS, Bell BA, Griffiths JR, Howe FA. An assessment of the effects of sample ischaemia and spinning time on the metabolic profile of brain tumour biopsy specimens as determined by high-resolution magic angle spinning 1H NMR. NMR Biomed. 2008;21(10):1138–1147. doi: 10.1002/nbm.1296. [DOI] [PubMed] [Google Scholar]
- 21.Elkhaled A, Llewellyn J, Constantin A, Yoshihara HA, Phillips JJ, Molinaro AM, Chang SM, Nelson SJ. Characterization of metabolites in infiltrating gliomas using ex vivo 1H high-resolution magic angle spinning spectroscopy. NMR Biomed. 2014;27(5):578–593. doi: 10.1002/nbm.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Candiota AP, Majos C, Julia-Sape M, Cabanas M, Acebes JJ, Moreno-Torres A, Griffiths JR, Arus C. Non-invasive grading of astrocytic tumours from the relative contents of myo-inositol and glycine measured by in vivo MRS. JBR-BTR. 2011;94(6):319–329. doi: 10.5334/jbr-btr.698. [DOI] [PubMed] [Google Scholar]
- 23.Srinivasan R, Phillips JJ, Vandenberg SR, Polley MY, Bourne G, Au A, Pirzkall A, Cha S, Chang SM, Nelson SJ. Ex vivo MR spectroscopic measure differentiates tumor from treatment effects in GBM. Neuro-Oncol. 2010;12:1152–1161. doi: 10.1093/neuonc/noq075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Govindaraju V, Young K, Maudsley AA. Proton NMR chemical shifts and coupling constants for brain metabolites. NMR Biomed. 2000;13:129–153. doi: 10.1002/1099-1492(200005)13:3<129::aid-nbm619>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 25.Righi V, Andronesi OC, Mintzopoulos D, Black MB, Tzika AA. High-resolution magic angle spinning magnetic resonance spectroscopy detects glycine as a biomarker in brain tumors. Int J Oncol. 2010;36(2):301–306. doi: 10.3892/ijo_00000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schulte RF, Boesiger P. ProFit: two-dimensional prior-knowledge fitting of J-resolved spectra. NMR Biomed. 2006;19:255–263. doi: 10.1002/nbm.1026. [DOI] [PubMed] [Google Scholar]
- 27.Yan Li, Lupo JM, Parvataneni R, Lamborn KR, Cha S, Chang SM, Nelson SJ. Survival analysis in patients with newly diagnosed glioblastoma using pre-and post radiotherapy MR spectroscopic imaging. Neuro Oncol. 2013;15(5):607–617. doi: 10.1093/neuonc/nos334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo D, Bell EH, Chakravarti A. Lipid metabolism emerges as a promising target for malignant glioma therapy. CNS Oncol. 2013;2(3):289–299. doi: 10.2217/cns.13.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng LL, Anthony DC, Comite AR, Black PM, Tzika AA, Gonzolez RG. Quantification of microheterogeneity in glioblastoma multiforme with ex vivo high-resolution magic-angle spinning (HRMAS) proton magnetic resonance spectroscopy. Neuro Oncol. 2000;2(2):87–95. doi: 10.1093/neuonc/2.2.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Opstad KS, Bell BA, Griffiths JR, Howe FA. An investigation of human brain tumour lipids by high-resolution magic angle spinning 1H MRS and histological analysis. NMR Biomed. 2008;21(7):677–685. doi: 10.1002/nbm.1239. [DOI] [PubMed] [Google Scholar]
- 31.Tugnoli V, Tosi MR, Tinti A, Trinchero A, Bottura G, Fini G. Characterization of lipids from human brain tissues by multinuclear magnetic resonance spectroscopy. Biopolymers. 2001;62(6):297–306. doi: 10.1002/bip.10005. [DOI] [PubMed] [Google Scholar]
- 32.Esmaeili M, Hamans BC, Navis AC, van Horssen R, Bathen TF, Gribbestad IS, Leenders WP, Heerschap A. IDH1 R132H mutation generates a distinct phospholipid metabolite profile in glioma. Cancer Res. 2014;74:4898. doi: 10.1158/0008-5472.CAN-14-0008. [DOI] [PubMed] [Google Scholar]
- 33.Selathurai A, Kowalski GM, Burch ML, Sepulveda P, Risis S, Lee-Young RS, Lamon S, Meikle PJ, Genders AJ, McGee SL. The CDP-ethanolamine pathway regulates skeletal muscle diacylglycerol content and mitochondrial biogenesis without altering insulin sensitivity. Cell Metab. 2015;21:718–730. doi: 10.1016/j.cmet.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 34.Glunde K, Bhujwalla ZM, Ronen SM. Choline metabolism in malignant transformation. Nat Rev Cancer. 2011;11(12):835–848. doi: 10.1038/nrc3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hancu I. Which pulse sequence is optimal for myo-inositol detection at 3T? NMR Biomed. 2009;22(4):426–435. doi: 10.1002/nbm.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Essock-Burns E, Phillips JJ, Molinaro AM, Lupo JM, Cha S, Chang SM, Nelson SJ. Comparison of DSC-MRI post-processing techniques in predicting microvascular histopathology in patients newly diagnosed with GBM. J Magn Reson Imaging. 2013;38(2):388–400. doi: 10.1002/jmri.23982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Law M, Young RJ, Babb JS, Peccerelli N, Chheang S, Gruber ML, Miller DC, Golfinos JG, Zagzag D, Johnson G. Gliomas: predicting time to progression or survival with cerebral blood volume measurements at dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging. Radiology. 2008;247:490–498. doi: 10.1148/radiol.2472070898. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material. Immunohistochemistry methods, T2-weighted imaging methods, and correlations among imaging and histopathologic parameters.