Abstract
Development of an effective, safe, and convenient method for gene delivery to the pancreas is a critical step toward gene therapy for pancreatic diseases. Therefore, we tested the possibility of applying the principle of hydrodynamic gene delivery for successful gene transfer to pancreas using rats as a model. The established procedure involves the insertion of a catheter into the superior mesenteric vein with temporary blood flow occlusion at the portal vein and hydrodynamic injection of DNA solution. We demonstrated that our procedure achieved efficient pancreas-specific gene expression that was 2,000-fold higher than that seen in the pancreas after the systemic hydrodynamic gene delivery. In addition, the level of gene expression achieved in the pancreas by the pancreas-specific gene delivery was comparable to the level in the liver achieved by a liver-specific hydrodynamic gene delivery. The optimal level of reporter gene expression in the pancreas requires an injection volume equivalent to 2.0% body weight with flow rate of 1 mL/s and plasmid DNA concentration at 5 μg/mL. With the exception of transient expansion of intercellular spaces and elevation of serum amylase levels, which recovered within 3 days, no permanent tissue damage was observed. These results suggest that pancreas-targeted hydrodynamic gene delivery is an effective and safe method for gene delivery to the pancreas and clinically applicable.
Keywords: gene therapy, hydrodynamic gene delivery, non-viral vectors, pancreas
Introduction
The pancreas lies transversely on the posterior abdominal wall and is surrounded by various ducts and vessels.1 The arteries feeding the pancreas arise from the gastroduodenal artery, superior mesenteric artery, and splenic artery, forming a complex arcade. The veins drain into the portal vein (PV), either directly or through the superior mesenteric or splenic vein. The pancreas has both endocrine and exocrine functions; the pancreatic islet consists of the acinar cells and serves an endocrine function, while the duct consists of columnar epithelial cells and serves an exocrine function.1 Pancreatic diseases include diabetes mellitus, pancreatitis, and pancreatic cancer, often resulting in dysfunction of pancreas. Among them, the incidence of pancreatic cancer is increasing, and it is now the fourth-most-common cause of cancer-related mortality in the US and Europe and has a poor prognosis, with a 5-year survival rate at 6%.2, 3 Various chemotherapies have been tried, but the results are far from satisfactory. Recently, the applicability of gene and cell therapy has been tested.4 To date, a few clinical trials have been performed using the modalities of gene addition, gene editing, ex vivo gene therapy, and oncolytic viral vector-mediated therapy (for recent review, see Singh et al.4). These studies showed feasibility of the gene therapy approach in preclinical studies; however, the use of these approaches has been limited in clinics due to safety concerns and lack of pancreas specificifity.4 Therefore, the development of a new method for pancreas-targeted gene delivery is an unmet need to advance medical treatment of pancreatic diseases.
Therefore, in this study, we explored the possibility of achieving pancreatic gene delivery using the principles of hydrodynamic gene delivery (HGD). HGD employs a physical force of dynamic pressure generated by a rapid injection of a large volume of fluid into a blood vessel to permeabilize the cell membrane and facilitate intracellular gene transfer.5, 6 The method of HGD was established 18 years ago and has been successfully employed for delivery of proteins, oligonucleotides, siRNA, viral vectors, and small chemicals to hepatocytes in rodents following a simple tail-vein injection.5, 6, 7, 8, 9 More recent work has extended the use of HGD to large animals to achieve organ-specific gene delivery to the liver, muscle, heart, and kidneys. Target-specific gene delivery has the advantage of avoiding delivery of genes to non-targeted cells and is commonly achieved by hydrodynamic injection into a major vein of the target organ where gene delivery is needed. The techniques of imaging-guided catheter insertion and surgical exposure of blood vessels for needle insertion have been developed to aid target-specific HGD.10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20 A significant advantage of this method is the achievement of therapeutic levels of gene expression in target organs of the transfected animals.19, 20 Consequently, HGD has become one of the most commonly used techniques for gene therapy and function studies in vivo.
Based on these successes, we investigated the possibility of pancreas-targeted HGD toward a safe and effective gene therapy for pancreatic diseases, including cancers, for which no effective therapy has been developed to date. The experiments were designed to assess the effects of hydrodynamic parameters on gene delivery efficiency and potential tissue damage. We demonstrate for the first time that efficient HGD to the pancreas can be achieved by hydrodynamic injection of plasmid DNA in saline (5 μg/mL) into superior mesenteric vein (SMV) with injection volume equivalent to 2.0% body weight (BW) and an injection rate at 1 mL/s. These results provide direct evidence supporting the safety and efficacy of pancreas-targeted HGD and its potential use in clinic for treatment of pancreatic diseases.
Results
Development of Pancreas-Targeted HGD
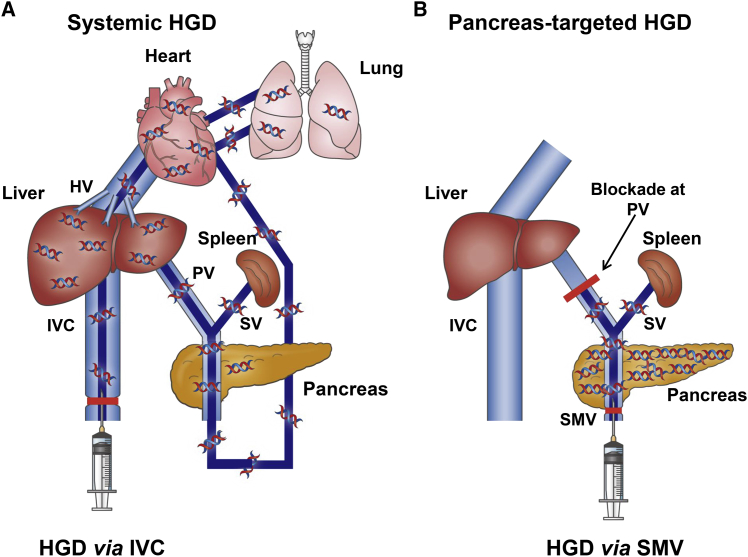
The experimental design for catheter insertion is shown in Figure 1. Systemic HGD was done in rats by inserting a catheter from the inferior vena cava (IVC), as shown in Figure 1A. HGD via IVC induces a retrograde flow of DNA solution into the liver. With time, the injected DNA solution joins the blood flow, enters the heart, passes through the mesenteric arteries into the SMV and PV, fills the liver, and then returns to the IVC. While this method showed an efficient gene delivery to the liver, no effective distribution of the solution was seen in the pancreas. For our deign of pancreatic gene delivery (Figure 1B), hydrodynamic injection is performed through the SMV with occlusion at the PV to maximize DNA distribution and hydrodynamic impacts on pancreas, and to prevent leakage of injected DNA solution into the liver.
Figure 1.
Schematic Presentation of Hydrodynamic Gene Delivery
(A) Schema of the standard hydrodynamic gene delivery (HGD) via the inferior vena cava (IVC). (B) Schema of pancreas-targeted HGD from the superior mesenteric vein (SMV) with a temporary vascular blockade at the portal vein (PV). HV, hepatic vein; SV, splenic vein.
Efficiency of Pancreatic HGD
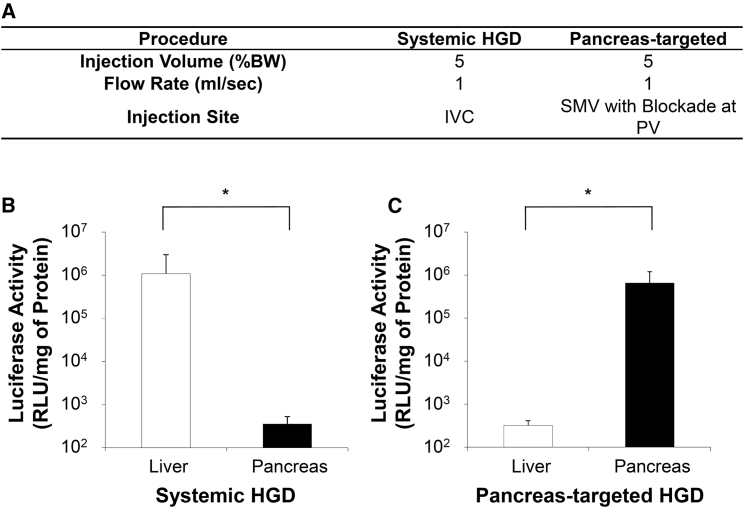
The efficiency of HGD via the IVC and pancreas-targeted HGD on gene expression in the pancreas was evaluated using a pCMV-Luc reporter plasmid. The injection parameters used are derived from previous studies21, 22, 23 and shown in Figure 2A. The injection volume and flow rate were set at 5.0% BW, which was 10 mL in 200 g rats, and 1 mL/s, respectively. The level of luciferase activity by HGD via the IVC showed 1.1 × 106 relative light units (RLU)/mg of protein in the liver, which was approximately 3,000-fold higher than the 3.6 × 102 RLU/mg of protein achieved in the pancreas (p < 0.05; Figure 2B). However, pancreas-targeted HGD achieved a level of 6.5 × 105 RLU/mg of protein in the pancreas, which was 2,000-fold higher than the 3.2 × 102 RLU/mg of protein seen in the liver (p < 0.05; Figure 2C). Other than pancreas and residual level in the liver, pancreas-targeted HGD did not result in detectable level of luciferase activity in other organs, including the brain, heart, lungs, spleen, and kidneys.
Figure 2.
Effect of Hydrodynamic Gene Delivery on Gene Expression in the Pancreas
(A) Summary of the procedures performed. (B) The level of luciferase activity in the liver and pancreas 4 hr after HGD via IVC. (C) The level of luciferase activity in the liver and pancreas 4 hr after pancreas-targeted HGD. The values represent mean ± SD (n = 10 for each value); *p < 0.05 (t test).
Effect of Hydrodynamic Injection Volume on Gene Delivery Efficiency
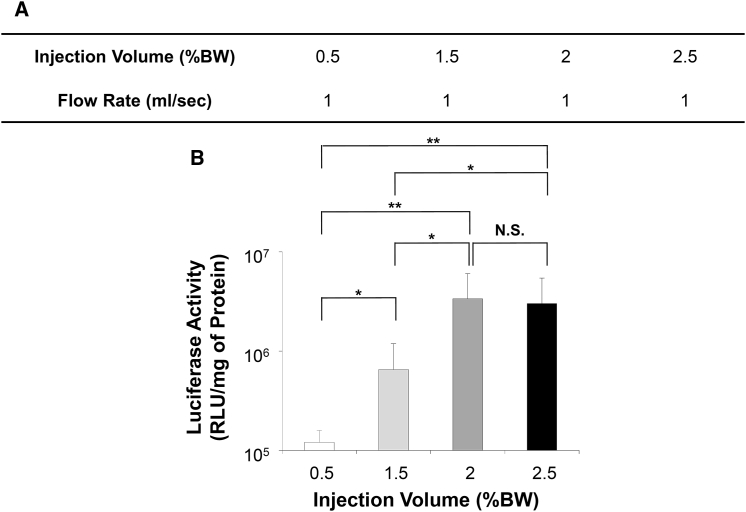
The impact of injection volume of DNA solution on gene delivery efficiency was examined using the same condition utilized in Figure 2. The flow rate (1 mL/s) and plasmid concentration (5 μg/mL) in saline were kept constant and the injection volume varied from 0.5% BW to 2.5% BW equivalent to 1–5 mL for a 200 g rat (Figure 3A). Figure 3B shows that the highest level of luciferase expression of 3.4 × 106 RLU/mg of protein was achieved with an injection volume of 2.0% BW, which was significantly higher than that of 1.1 × 105 RLU/mg and 6.5 × 105 RLU/mg of protein achieved by the injections of 0.5% BW (p < 0.01) and 1.5% BW (p < 0.05), respectively. In addition, luciferase gene expression appears higher than the level of 6.5 × 105 RLU/mg of protein achieved with a volume of 5.0% BW (p < 0.05; Figure 2C). The injection volume of 2.5% BW resulted in 3.0 × 106 RLU/mg of protein, similar to the level achieved with injection volume of 2.0% BW (no statistical significance [NS]). These results suggest that an injection volume of 2.0% BW containing 20 μg of plasmid DNA was optimal for pancreas-targeted HGD.
Figure 3.
Effects of Injection Volume on Gene Delivery Efficiency
(A) Summary of the injection volumes examined. (B) The level of luciferase activity in the pancreas 4 hr after pancreas-targeted HGD. The values represent mean ± SD (n = 10 for each value); *p < 0.05, **p < 0.01, and NS, no statistical significance; one-way ANOVA followed by Bonferroni’s multiple comparison test.
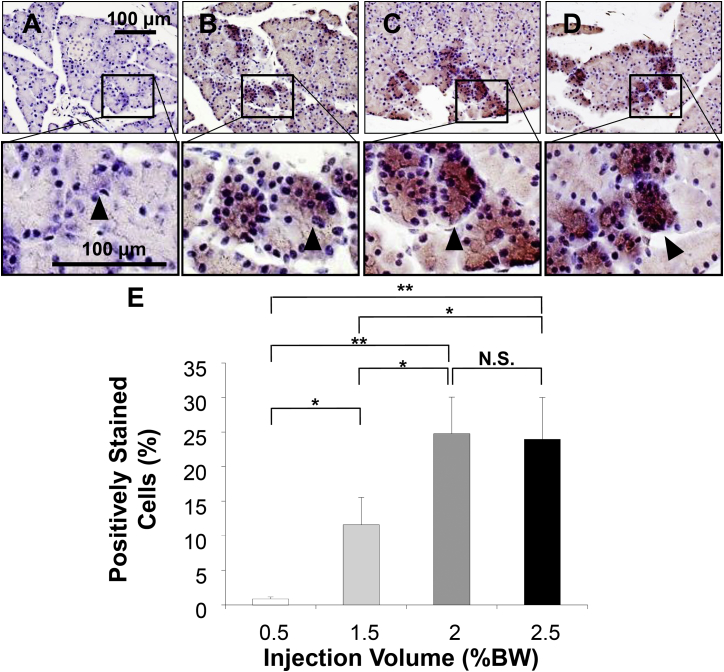
Immunohistochemical analyses were performed to confirm luciferase gene expression in pancreases of animals hydrodynamically transfected with different volumes of DNA solution. Results in Figure 4 show about 25% ± 5% of pancreatic cells stained positive in rats hydrodynamically transfected with the volume of 2.0% BW (Figure 4E). This level was significantly higher than the level achieved with injection volume of 0.5% BW (Figure 4A) (0.8% ± 1.6%, p < 0.01) and 1.5% BW solution (Figure 4B) (12% ± 4%, p < 0.05). No statistical difference was seen with an injection volume of 2.5% BW (Figure 4D) (24% ± 6%, NS). Other than a few cells stained positive in the intestine (Figure S1), no expression of luciferase reporter gene was observed in other organs assessed, including the brain, heart, lungs, spleen, and kidneys. These results confirm that a volume of 2.0% BW is optimal for pancreatic gene delivery under our experimental conditions. Luciferase-positive cells appear clustered in the pancreas, with higher density seen in one region, but not in others. The highest transfection efficiency obtained was 30% of cells in the areas stained positive with anti-luciferase antibodies, mostly acinar cells (Figures 4C and 4E).
Figure 4.
Histological Analyses of Luciferase Expression in the Pancreas
Immunohistochemical staining with anti-luciferase antibody was performed on pancreatic tissues collected 4 hr after HGD. Scale bar represents 100 μm. Injection volumes were (A) 0.5% BW, (B) 1.5% BW, (C) 2% BW, and (D) 2.5% BW. Black arrowhead indicates the positively stained cells. (E) Quantitative analysis of positively stained cells. Ten different pancreas sections from each of five rats in each group were immunohistochemically stained with anti-luciferase antibody, and a quantitative analysis was performed using the ImageJ software. The values represent mean ± SD (n = 50); *p < 0.05, **p < 0.01, and NS, no statistical significance; one-way ANOVA followed by Bonferroni’s multiple comparison test.
Impact of Hydrodynamic Injection on the Pancreas
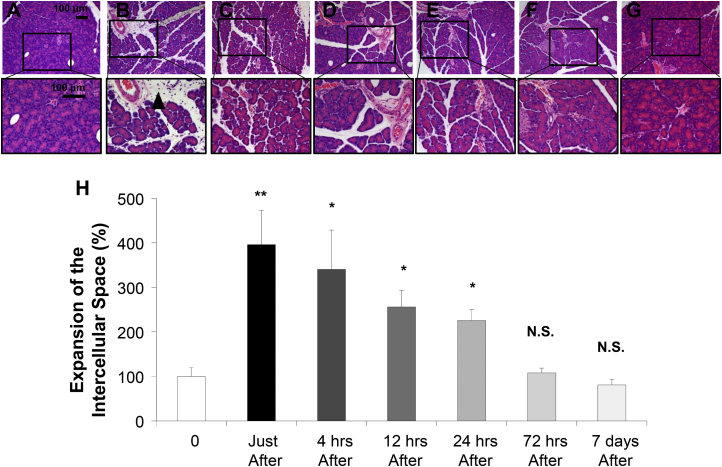
To examine the impact of pancreas-targeted HGD on pancreatic tissue, a histological analysis was performed on samples collected at different times before and after the procedure using an injection volume of 2.0% BW (Figure 5). H&E staining showed significant expansion of the intercellular space in the pancreas (Figure 5A) up to 396% of the original immediately after the procedure (p < 0.01) (Figure 5B). The expanded tissue returned gradually to its normal size: 340% in 4 hr (Figure 5C), 245% in 12 hr (Figure 5D), 226% in 24 hr (Figure 5E), and 108% in 72 hr (Figure 5F). No fibrosis or other tissue damage was noticed 7 days after the injection (Figure 5G). These results suggest that the impact of pancreas-targeted HGD on the pancreas is transient and reversible.
Figure 5.
Impact of Pancreas-Targeted HGD on Structure of Pancreas
H&E staining was performed on the pancreatic samples. (A) Before, (B) immediately following, (C) 4 hr, (D) 12 hr, (E) 24 hr, (F) 72 hr, and (G) 7 days after the pancreas-targeted HGD in rats. The scale bar represents 100 μm. Black arrowhead indicates the expansion of the intercellular space in the pancreas. (H) Quantitative measurement of intercellular space in the pancreas. Ten different pancreas sections from each of five rats per group were stained and images were captured. Quantitative analysis was performed using the ImageJ software (version 1.6.0_20, National Institutes of Health, USA). The values represent mean ± SD (n = 50); *p < 0.05, **p < 0.01, and NS, no statistical significance; one-way ANOVA followed by Bonferroni’s multiple comparison test.
Assessment of Safety of Pancreas-Targeted HGD
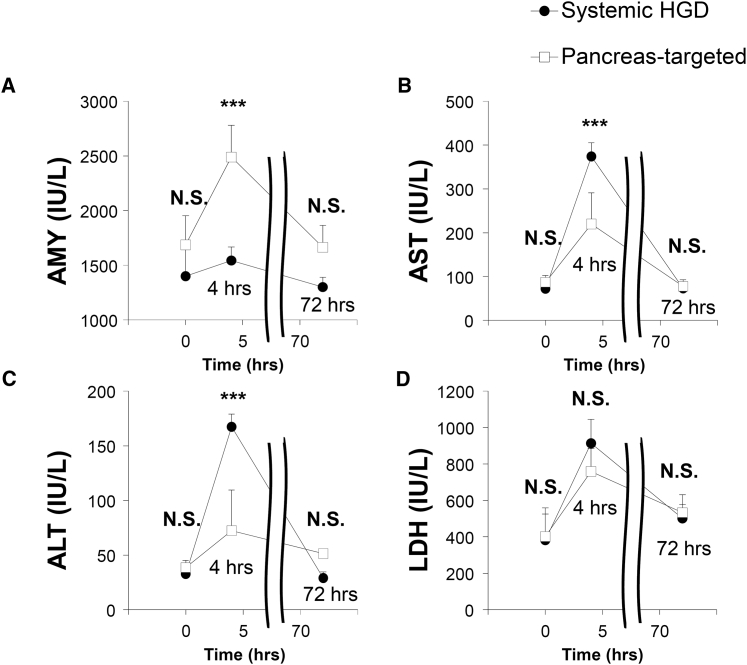
To assess potential tissue damage of the procedure, serum biochemical analyses were performed using blood samples collected before and at 4 and 72 hr after pancreas-targeted HGD with the injection volume of 2.0% BW. As shown in Figure 6, serum levels of amylase (AMY) in animals received the HGD via IVC or pancreas-targeted HGD showed a transient increase from 1,400 ± 270 IU/l and 1,700 ± 300 IU/l (before, NS) to 1,500 ± 290 IU/l and 2,500 ± 120 IU/l, respectively, at 4 hr, which is significantly higher in pancreas-targeted HGD (p < 0.001) than in HGD via IVC. The elevated levels returned to background levels of 1,300 ± 200 IU/l and 1,700 ± 90 IU/l, respectively, within 72 hr (NS) (Figure 6A). An additional examination was performed using serum AMY level as an indicator to see whether plasmid DNA is responsible for transient increases of cellular enzymes. Results shown in Figure S2 demonstrate that plasmid DNA does not play any role. It is the hydrodynamic pressure that not only facilitates intracellular gene transfer but also induces minor release of cellular content. In addition, serum levels of aspartate aminotransferase (AST) by systemic HGD via IVC and pancreas-targeted HGD groups increased from the background levels of 72 ± 16 IU/l and 87 ± 4 IU/l (before, NS) to 370 ± 71 IU/l and 220 ± 31 IU/l, respectively, at 4 hr, showing significantly higher levels in animals receiving HGD via IVC (p < 0.001). The elevated levels returned to background levels of 73 ± 15 and 77 ± 13 IU/l, respectively, within 72 hr (Figure 6B). A similar pattern was seen for alanine aminotransferase (ALT) in animals with HGD via IVC and pancreas-targeted HGD, showing a transient increase from 33 ± 6 IU/l and 39 ± 8 IU/l (NS) to 167 ± 37 IU/l and 72 ± 12 IU/l, respectively, at 4 hr (p < 0001), which returned to 29 ± 4 IU/l and 51 ± 6 IU/l (NS), respectively, within 72 hr (Figure 6C). No significant differences were seen in the levels of lactate dehydrogenase (LDH) between the groups (Figure 6D). The concentrations of other blood components including creatinine, albumin, blood urea nitrogen, sodium, chloride, and potassium, showed no change (data not shown).
Figure 6.
Impact of Pancreas-Targeted HGD on Serum Biochemistry
The standard serum biochemical analysis was performed with sera collected before (time = 0) and at 4 hr and 72 hr after the HGD via IVC or pancreas-targeted HGD. Concentrations of (A) amylase (AMY), (B) aspartate aminotransferase (AST), (C) alanine aminotransferase (ALT), and (D) lactate dehydrogenase (LDH). Black circles and white squares represent values of systemic (black circles) and pancreas-targeted HGD (white squares) groups, respectively. The values represent mean ± SD (n = 5). ***p < 0.001, and N.S., no statistical significance; two-way factor repeated-measure ANOVA followed by Bonferroni’s multiple comparison test.
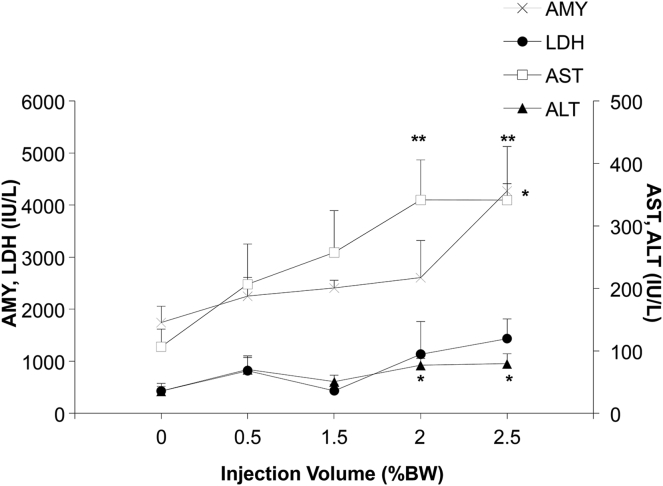
The relationship between the impacts of the procedure and injection volume was established 4 hr after pancreas-targeted HGD. The level of AMY with the injection volume of 2.5% BW was the highest, at 4,300 ± 850 IU/l at 4 hr, which was significantly higher than those with injection volumes of 0.5%, 1.5%, and 2.0% BW (2,300 ± 360 IU/l, 2,400 ± 150 IU/l, and 2,600 ± 710 IU/l, respectively) (p < 0.05). AST showed significantly higher levels with injection volume of 2.0% and 2.5% BW (340 ± 64 IU/l and 340 ± 26 IU/l, respectively), than those of 0.5% and 1.5% BW (p < 0.01). ALT showed a similar pattern to AST with significantly higher levels at 2.0% and 2.5% BW (77 ± 11 IU/l and 79 ± 16 IU/l) than 0.5% and 1.5% BW (p < 0.05). LDH showed no significant difference among the injection volumes tested (Figure 7). No procedure-related animal deaths were seen, with injection volumes up to 2.5% BW and animals were active and eating well. These results suggest that although a transient increase of intracellular marker enzymes was seen, the injection volume of 2.0% BW solution is well tolerated.
Figure 7.
Impact of the Injection Volume on Serum Biochemistry
The serum biochemical analysis was performed on the sera collected 4 hr after pancreas-targeted HGD with injection volumes of 0.5% BW, 1.5% BW, 2% BW, and 2.5% BW. The values represent mean ± SD, (n = 5 for each group) of concentrations of AMY (×), LDH (black circle), AST (white square), and ALT (black triangle), *p < 0.05 and **p < 0.01; one-way ANOVA followed by Bonferroni’s multiple comparison test.
Discussion
The incidence of pancreatic cancer is increasing and is currently the fourth-most-common cause of cancer-related mortality in US and Europe with poor prognosis; therefore, the development of effective therapeutic methods is essential.2, 3 Although surgical procedures are known to be effective, complex anatomical structures surrounding the pancreas, leading to vascular invasion and metastasis, preclude this option in many patients. Various chemotherapies, including gemcitabine,24 FOLFIRINOX (fluorouracil plus leucovorin, irinotecan, and oxaliplatin),25 and albumin-bound paclitaxel26 have shown positive results but far from satisfactory with systemic side effects affecting the patient’s general condition and prognosis.27 The poor clinical outcomes and systemic side effects with conventional treatments are partially due to the lack of pancreas-specific delivery. Therefore, the development of new and innovative approaches with pancreas-specific therapy, including gene therapy, is important.4 Toward this end, only a few clinical trials of gene therapy have been performed, utilizing an adenoviral vector expressing IL-228 or TNF-α,29, 30 a retroviral vector expressing TNF-α,31, 32 or dsDNA expressing diphtheria toxin A.33 Basic research is ongoing to demonstrate the efficacy of tumor vaccines, viral-vector-mediated therapy, and gene editing34, 35 for achievement of antitumor effects or management of pain associated with cancer progression.36 To extend the applicability of gene therapy for pancreatic diseases, we focused our effort in this study on development of a pancreas-specific gene delivery utilizing the principles of HGD using rats as an animal model and luciferase marker gene. The results in Figures 2, 3, 4, 5, and 6 clearly demonstrate that the procedure developed is pancreas specific, efficient, and safe.
HGD has been developed as a safe and effective method for gene transfer to the liver of small animals by injecting naked DNA into the tail vein.5, 6, 9, 37, 38, 39 Further applicability of the procedure was evidenced by the development of catheter-based, organ-specific HGD,40 and its safety and efficiency have been reported in the liver,10, 11, 13, 14, 17, 19, 20, 22, 23, 41 kidney,42 and muscle18, 43 of large animals. As the pancreas-specific delivery method is a key to improving the treatment of pancreatic diseases, we developed a catheter-based pancreas-targeted gene-delivery system, demonstrating the safety and efficacy of the procedure in this study. The complex vascular structures surrounding the pancreas have been considered major concerns for pancreatic gene delivery in the past, compared to the systemic HGD. Our results suggest that such technical barriers can be overcome by gene injection to the SMV, a major vein linking to pancreatic veins to lead venous blood to the PV (Figure 1). Gene delivery efficiency of the procedure was significantly better than HGD via IVC (Figure 2), implicating that this pancreas-specific injection may be considered for administrating other therapeutic agents for better treatment of pancreatic diseases with fewer systemic side effects.
As we have previously reported regarding liver-specific gene delivery,19, 21 injection volume and flow rate are key factors for safety and efficacy. The optimal parameters determined for pancreatic gene delivery include an injection volume of 2.0% BW and a flow rate of 1 mL/s with plasmid DNA concentration of 5 μg/mL (Figures 3, 4, and 7). A transient increase of AMY concentration is likely due to the transient impact of HGD on the pancreatic tissue, supported by the evidence of a time-dependent change in intercellular space expansion (Figures 5 and 6) depending on injection volume (Figure 7), very similar to liver-specific HGD.20, 21 The mild and transient increase of AST and ALT concentrations is caused by temporary occlusion of the PV (Figure 6) for around 6–10 s.
Further studies are needed to assess the procedure and injection parameters optimized in rats in human-size animals. In addition, the procedure established needs to be evaluated in disease-carrying animals, as there might be changes in pancreas and surrounding tissue of animals with chronic pancreatitis and cancer.1, 2, 3, 4 For this purpose, the strategy we have reported in various animal species and tissues17, 18, 19, 44 using computer-controlled HGD21, 42 could be considered.
In summary, we report for the first time, the development of a method for pancreas-targeted HGD and demonstrate its safety and efficacy. We believe that pancreas-specific delivery of therapeutic genes could offer great advantages for treatment of pancreatic diseases.
Materials and Methods
Materials
The pCMV-Luc plasmid, containing firefly luciferase cDNA driven by a CMV promoter, was purified using a Plasmid Mega Kit (QIAGEN, Hilde, Germany). The purity of the plasmid preparation was verified on the basis of absorbency at 260 nm and 280 nm and 1% agarose gel electrophoresis. Luciferase assay kits were purchased from Wako Pure Chemical Industries (Chuo-ku, Osaka, Japan). Wistar rats (n = 50, female, 7–8 weeks of age, 200–250 g) were purchased from Japan SLC (Hamamatsu, Shizuoka, Japan) and fed a standard chow. Ten animals were used for each group of the experiment.
HGD to Rat Pancreas
All animal experiments were approved by and conducted in full compliance with the regulations of the Institutional Animal Care and Use Committee at Niigata University, Niigata, Japan. Systemic HGD via IVC in rats was performed as previously described.21 In brief, a midline skin incision was made on the rats under general anesthesia using isoflurane and 2,2,2-tribromoethanol (concentration, 0.016 g/mL in 0.9% saline; dose, 1.25 mL/100 g bodyweight). An injection catheter (SURFLO 22 gauge, Terumo, Shibuya-ku, Tokyo, Japan) was inserted into the IVC and its tip was placed right below the junction of the hepatic vein. Saline-containing plasmid DNA (pCMV-Luc, 5 μg/mL) was hydrodynamically injected into the liver via the catheter with temporary blood flow occlusion at the infra-hepatic IVC. For the pancreas-targeted HGD, the PV in the hilus and the SMV were dissected out and isolated. And the catheter (SURFLO 22 gauge, Terumo, Shibuya-ku, Tokyo, Japan) was inserted into the SMV with temporary occluding blood flow at the PV by vessel loops followed by hydrodynamic injection of pCMV-Luc (5 μg/mL) at a flow rate of 1 mL/s. The doses of plasmid DNA were 5 μg in 1 mL (0.5% BW), 15 μg in 3 mL (1.5%BW), 20 μg in 4 mL (2.0% BW), and 25 μg in 5 mL, (2.5% BW), and the time for PV occlusions including the preparation time were around 6, 8, 9, and 10 s for injection of 1, 3, 4, and 5 mL, respectively. The abdominal median incision was sutured after the procedure. For this surgical procedure, female rats were used in order to be consistent with our previous studies,21, 22, 23 and there is no effect of hormonal status on this gene expression study.
Luciferase Assay
Rats were euthanized 4 hr after the injection of pCMV-Luc plasmid DNA, and tissue samples were collected from liver, pancreas, and other organs as previously described21, 22, 23 and kept at −80°C until use. Lysis buffer (2 mL) (0.1 M Tris-HCL, 2 mM EDTA, and 0.1% Triton X-100 [pH 7.8]) was added to each sample (∼200 mg wet tissue), and the tissue samples were homogenized for 30 s with the tissue homogenizer (ULTRA-TURRAX T25 digital, IKA, Staufen, Germany) at maximum speed. The tissue homogenates were centrifuged in a microcentrifuge for 10 min at 13,000 × g at 4°C. The protein concentration of the supernatant was determined using a protein assay kit (Bio-Rad, Hercules, CA, USA) based on Coomassie blue assay strategy. Supernatant (10 μL) was mixed with luciferase assay reagent (100 μL), and the luciferase activity was measured in a luminometer (Luminescencer Octa AB-2270, ATTO, Bunkyo-ku, Tokyo, Japan) for 10 s. The luciferase activity is presented as relative light units per mg of protein.
Histological Analysis
Tissue samples for immunohistochemical staining were collected 4 hr after the HGD of pCMV-Luc plasmid upon euthanasia and fixed in 10% formalin upon tissue collection before embedding in paraffin. A total of 10 sections (10 μm) were cut from each of the 5 rats randomly chosen from a group of 10 animals in the group, and standard immunohistochemistry was performed using goat anti-Luciferase polyclonal antibody (G7451, 1:100 dilution; Promega, Madison, WI, USA), Vecstain Elite ABC Goat IgG kit (PK-6105; Vector Laboratories, Burlingame, CA, USA), and DAB chromogen tablet (Muto Pure Chemicals, Bunkyo-ku, Tokyo, Japan). Images were captured from each tissue section randomly, and a quantitative analysis of positively stained cells was performed using ImageJ software (version 1.6.0_20, National Institutes of Health, USA) as previously described.45
Assessment of Tissue Damage
Blood samples were collected from each rat before (time = 0) and at 4 hr, and 72 hr after HGD from tail vein or IVC. The serum biochemical analysis was performed by BML Inc. (Shibuya-ku, Tokyo, Japan). Tissue samples for H&E staining were collected before, immediately following, and at 4 hr, 12 hr, 24 hr, 72 hr, and 7 days after HGD. Pancreas sections from each of five randomly selected rats of seven groups were stained, and images were captured. Quantitative analysis was performed using ImageJ software (version 1.6.0_20, National Institutes of Health, USA).45
Statistical Analyses
The data of luciferase assays, histological analyses, and biochemical analyses were statistically evaluated by analyses of variance followed by Bonferroni’s multiple comparison test and the t test.
Author Contributions
K.O., Y.K., H.A., T.Y., N.S., T.N., and G.Z. conducted the experiments; K.K. and S.T. designed the experiments and wrote the paper; A.S., S.A., K.H., S.I., J.K., and K.K. analyzed data; M.T., Y.A., D.L., K.K., and S.T. interpreted data.
Conflicts of Interest
The authors declare that they have no conflict of interest.
Acknowledgments
The authors would like to thank Takao Tsuchida in the Division of Gastroenterology and Hepatology at the Niigata University for his excellent assistance in histological analyses. The authors would also like to thank Nobuyoshi Fujisawa, Yoshitaka Maeda, Toshikuni Sasaoka, and all staff members at the Division of Laboratory Animal Resources in Niigata University. They also thank Enago and Dr. Megan Morgan for the critical reading of the manuscript and English language editing. The research in the authors’ laboratories has been supported in part by Grant-in-Aid for Scientific Research from the Japanese Society for the Promotion of Sciences (22890064, 23790595, 26860354, and 17K09408 to K.K.; 24592084 to M.T.; and 16K19333 to T.Y.), a Takara Bio Award from JSGT to K.K., and a Research grant from Pancreas Research Foundation of Japan to K.K.
Footnotes
Supplemental Information includes Supplemental Materials and Methods and two figures and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2017.08.009.
Supplemental Information
References
- 1.Agur A.M.R., Dalley A.F., II . Thirteenth Edition. Lippincott Williams and Wilkins; 2013. Grant’s Atlas of Anatomy. [Google Scholar]
- 2.Ferlay J., Steliarova-Foucher E., Lortet-Tieulent J., Rosso S., Coebergh J.W., Comber H., Forman D., Bray F. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur. J. Cancer. 2013;49:1374–1403. doi: 10.1016/j.ejca.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R., Ma J., Zou Z., Jemal A. Cancer statistics, 2014. CA Cancer J. Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 4.Singh H.M., Ungerechts G., Tsimberidou A.M. Gene and cell therapy for pancreatic cancer. Expert Opin. Biol. Ther. 2015;15:505–516. doi: 10.1517/14712598.2015.1001734. [DOI] [PubMed] [Google Scholar]
- 5.Liu F., Song Y., Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6:1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- 6.Zhang G., Budker V., Wolff J.A. High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum. Gene Ther. 1999;10:1735–1737. doi: 10.1089/10430349950017734. [DOI] [PubMed] [Google Scholar]
- 7.Suda T., Liu D. Hydrodynamic gene delivery: its principles and applications. Mol. Ther. 2007;15:2063–2069. doi: 10.1038/sj.mt.6300314. [DOI] [PubMed] [Google Scholar]
- 8.Herweijer H., Wolff J.A. Gene therapy progress and prospects: hydrodynamic gene delivery. Gene Ther. 2007;14:99–107. doi: 10.1038/sj.gt.3302891. [DOI] [PubMed] [Google Scholar]
- 9.Kamimura K., Suda T., Zhang G., Liu D. Advances in gene delivery systems. Pharmaceut. Med. 2011;25:293–306. doi: 10.2165/11594020-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshino H., Hashizume K., Kobayashi E. Naked plasmid DNA transfer to the porcine liver using rapid injection with large volume. Gene Ther. 2006;13:1696–1702. doi: 10.1038/sj.gt.3302833. [DOI] [PubMed] [Google Scholar]
- 11.Aliño S.F., Herrero M.J., Noguera I., Dasí F., Sánchez M. Pig liver gene therapy by noninvasive interventionist catheterism. Gene Ther. 2007;14:334–343. doi: 10.1038/sj.gt.3302873. [DOI] [PubMed] [Google Scholar]
- 12.Aliño S.F., José Herrero M., Bodi V., Noguera I., Mainar L., Dasí F., Sempere A., Sánchez M., Díaz A., Sabater L., Lledó S. Naked DNA delivery to whole pig cardiac tissue by coronary sinus retrograde injection employing non-invasive catheterization. J. Gene Med. 2010;12:920–926. doi: 10.1002/jgm.1510. [DOI] [PubMed] [Google Scholar]
- 13.Eastman S.J., Baskin K.M., Hodges B.L., Chu Q., Gates A., Dreusicke R., Anderson S., Scheule R.K. Development of catheter-based procedures for transducing the isolated rabbit liver with plasmid DNA. Hum. Gene Ther. 2002;13:2065–2077. doi: 10.1089/10430340260395910. [DOI] [PubMed] [Google Scholar]
- 14.Fabre J.W., Grehan A., Whitehorne M., Sawyer G.J., Dong X., Salehi S., Eckley L., Zhang X., Seddon M., Shah A.M. Hydrodynamic gene delivery to the pig liver via an isolated segment of the inferior vena cava. Gene Ther. 2008;15:452–462. doi: 10.1038/sj.gt.3303079. [DOI] [PubMed] [Google Scholar]
- 15.Khorsandi S.E., Bachellier P., Weber J.C., Greget M., Jaeck D., Zacharoulis D., Rountas C., Helmy S., Helmy A., Al-Waracky M. Minimally invasive and selective hydrodynamic gene therapy of liver segments in the pig and human. Cancer Gene Ther. 2008;15:225–230. doi: 10.1038/sj.cgt.7701119. [DOI] [PubMed] [Google Scholar]
- 16.Carreño O., Sendra L., Montalvá E., Miguel A., Orbis F., Herrero M.J., Noguera I., Aliño S.F., Lopez-Andujar R. A surgical model for isolating the pig liver in vivo for gene therapy. Eur. Surg. Res. 2013;51:47–57. doi: 10.1159/000351339. [DOI] [PubMed] [Google Scholar]
- 17.Kamimura K., Suda T., Xu W., Zhang G., Liu D. Image-guided, lobe-specific hydrodynamic gene delivery to swine liver. Mol. Ther. 2009;17:491–499. doi: 10.1038/mt.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamimura K., Zhang G., Liu D. Image-guided, intravascular hydrodynamic gene delivery to skeletal muscle in pigs. Mol. Ther. 2010;18:93–100. doi: 10.1038/mt.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamimura K., Suda T., Zhang G., Aoyagi Y., Liu D. Parameters affecting image-guided, hydrodynamic gene delivery to swine liver. Mol. Ther. Nucleic Acids. 2013;2:e128. doi: 10.1038/mtna.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamimura K., Kanefuji T., Yokoo T., Abe H., Suda T., Kobayashi Y., Zhang G., Aoyagi Y., Liu D. Safety assessment of liver-targeted hydrodynamic gene delivery in dogs. PLoS ONE. 2014;9:e107203. doi: 10.1371/journal.pone.0107203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yokoo T., Kamimura K., Suda T., Kanefuji T., Oda M., Zhang G., Liu D., Aoyagi Y. Novel electric power-driven hydrodynamic injection system for gene delivery: safety and efficacy of human factor IX delivery in rats. Gene Ther. 2013;20:816–823. doi: 10.1038/gt.2013.2. [DOI] [PubMed] [Google Scholar]
- 22.Abe H., Kamimura K., Kobayashi Y., Ohtsuka M., Miura H., Ohashi R., Yokoo T., Kanefuji T., Suda T., Tsuchida M. Effective prevention of liver fibrosis by liver-targeted hydrodynamic gene delivery of matrix metalloproteinase-13 in a rat liver fibrosis model. Mol. Ther. Nucleic Acids. 2016;5:e276. doi: 10.1038/mtna.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi Y., Kamimura K., Abe H., Yokoo T., Ogawa K., Shinagawa-Kobayashi Y., Goto R., Inoue R., Ohtsuka M., Miura H. Effects of fibrotic tissue on liver-targeted hydrodynamic gene delivery. Mol. Ther. Nucleic Acids. 2016;5:e359. doi: 10.1038/mtna.2016.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burris H.A., 3rd, Moore M.J., Andersen J., Green M.R., Rothenberg M.L., Modiano M.R., Cripps M.C., Portenoy R.K., Storniolo A.M., Tarassoff P. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J. Clin. Oncol. 1997;15:2403–2413. doi: 10.1200/JCO.1997.15.6.2403. [DOI] [PubMed] [Google Scholar]
- 25.Conroy T., Desseigne F., Ychou M., Bouché O., Guimbaud R., Bécouarn Y., Adenis A., Raoul J.L., Gourgou-Bourgade S., de la Fouchardière C., Groupe Tumeurs Digestives of Unicancer. PRODIGE Intergroup FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 2011;364:1817–1825. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- 26.Von Hoff D.D., Ervin T., Arena F.P., Chiorean E.G., Infante J., Moore M., Seay T., Tjulandin S.A., Ma W.W., Saleh M.N. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N. Engl. J. Med. 2013;369:1691–1703. doi: 10.1056/NEJMoa1304369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fogel E.L., Shahda S., Sandrasegaran K., DeWitt J., Easler J.J., Agarwal D.M., Eagleson M., Zyromski N.J., House M.G., Ellsworth S. A multidisciplinary approach to pancreas cancer in 2016: A Review. Am. J. Gastroenterol. 2017;112:537–554. doi: 10.1038/ajg.2016.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilly F.N., Beaujard A., Bienvenu J., Trillet Lenoir V., Glehen O., Thouvenot D., Malcus C., Favrot M., Dumontet C., Lombard-Bohas C. Gene therapy with Adv-IL-2 in unresectable digestive cancer: phase I-II study, intermediate report. Hepatogastroenterology. 1999;46(Suppl 1):1268–1273. [PubMed] [Google Scholar]
- 29.Hecht J.R., Farrell J.J., Senzer N., Nemunaitis J., Rosemurgy A., Chung T., Hanna N., Chang K.J., Javle M., Posner M. EUS or percutaneously guided intratumoral TNFerade biologic with 5-fluorouracil and radiotherapy for first-line treatment of locally advanced pancreatic cancer: a phase I/II study. Gastrointest. Endosc. 2012;75:332–338. doi: 10.1016/j.gie.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herman J.M., Wild A.T., Wang H., Tran P.T., Chang K.J., Taylor G.E., Donehower R.C., Pawlik T.M., Ziegler M.A., Cai H. Randomized phase III multi-institutional study of TNFerade biologic with fluorouracil and radiotherapy for locally advanced pancreatic cancer: final results. J. Clin. Oncol. 2013;31:886–894. doi: 10.1200/JCO.2012.44.7516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gordon E.M., Cornelio G.H., Lorenzo C.C., 3rd, Levy J.P., Reed R.A., Liu L., Hall F.L. First clinical experience using a ‘pathotropic’ injectable retroviral vector (Rexin-G) as intervention for stage IV pancreatic cancer. Int. J. Oncol. 2004;24:177–185. [PubMed] [Google Scholar]
- 32.Chawla S.P., Chua V.S., Fernandez L., Quon D., Blackwelder W.C., Gordon E.M., Hall F.L. Advanced phase I/II studies of targeted gene delivery in vivo: intravenous Rexin-G for gemcitabine-resistant metastatic pancreatic cancer. Mol. Ther. 2010;18:435–441. doi: 10.1038/mt.2009.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanna N., Ohana P., Konikoff F.M., Leichtmann G., Hubert A., Appelbaum L., Kopelman Y., Czerniak A., Hochberg A. Phase 1/2a, dose-escalation, safety, pharmacokinetic and preliminary efficacy study of intratumoral administration of BC-819 in patients with unresectable pancreatic cancer. Cancer Gene Ther. 2012;19:374–381. doi: 10.1038/cgt.2012.10. [DOI] [PubMed] [Google Scholar]
- 34.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aiuti A., Biasco L., Scaramuzza S., Ferrua F., Cicalese M.P., Baricordi C., Dionisio F., Calabria A., Giannelli S., Castiello M.C. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science. 2013;341:1233151. doi: 10.1126/science.1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Westlund K.N. Gene therapy for pancreatitis pain. Gene Ther. 2009;16:483–492. doi: 10.1038/gt.2009.27. [DOI] [PubMed] [Google Scholar]
- 37.Kamimura K., Liu D. Physical approaches for nucleic acid delivery to liver. AAPS J. 2008;10:589–595. doi: 10.1208/s12248-008-9067-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suda T., Gao X., Stolz D.B., Liu D. Structural impact of hydrodynamic injection on mouse liver. Gene Ther. 2007;14:129–137. doi: 10.1038/sj.gt.3302865. [DOI] [PubMed] [Google Scholar]
- 39.Zhang G., Gao X., Song Y.K., Vollmer R., Stolz D.B., Gasiorowski J.Z., Dean D.A., Liu D. Hydroporation as the mechanism of hydrodynamic delivery. Gene Ther. 2004;11:675–682. doi: 10.1038/sj.gt.3302210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kamimura K., Yokoo T., Abe H., Kobayashi Y., Ogawa K., Shinagawa Y., Inoue R., Terai S. Image-guided hydrodynamic gene delivery: current status and future directions. Pharmaceutics. 2015;7:213–223. doi: 10.3390/pharmaceutics7030213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yokoo T., Kamimura K., Abe H., Kobayashi Y., Kanefuji T., Ogawa K., Goto R., Oda M., Suda T., Terai S. Liver-targeted hydrodynamic gene therapy: recent advances in the technique. World J. Gastroenterol. 2016;22:8862–8868. doi: 10.3748/wjg.v22.i40.8862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suda T., Suda K., Liu D. Computer-assisted hydrodynamic gene delivery. Mol. Ther. 2008;16:1098–1104. doi: 10.1038/mt.2008.66. [DOI] [PubMed] [Google Scholar]
- 43.Hagstrom J.E., Hegge J., Zhang G., Noble M., Budker V., Lewis D.L., Herweijer H., Wolff J.A. A facile nonviral method for delivering genes and siRNAs to skeletal muscle of mammalian limbs. Mol. Ther. 2004;10:386–398. doi: 10.1016/j.ymthe.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 44.Yokoo T., Kanefuji T., Suda T., Kamimura K., Liu D., Terai S. Site-specific impact of a regional hydrodynamic injection: Computed tomography study during hydrodynamic injection targeting the swine liver. Pharmaceutics. 2015;7:334–343. doi: 10.3390/pharmaceutics7030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vrekoussis T., Chaniotis V., Navrozoglou I., Dousias V., Pavlakis K., Stathopoulos E.N., Zoras O. Image analysis of breast cancer immunohistochemistry-stained sections using ImageJ: an RGB-based model. Anticancer Res. 2009;29:4995–4998. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.