SUMMARY
Imbalances in secretory proteostasis induced by genetic, environmental or aging related insults are pathologically associated with etiologically diverse protein misfolding diseases. To protect the secretory proteome from these insults, organisms evolved stress-responsive signaling pathways that regulate the composition and activity of biologic pathways involved in secretory proteostasis maintenance. The most prominent of these is the endoplasmic reticulum (ER) Unfolded Protein Response (UPR), which functions to regulate ER proteostasis in response to ER stress. While the signaling mechanisms involved in UPR activation are well-defined, the impact of UPR activation on secretory proteostasis is only becoming clear. Here, we highlight recent reports defining how activation of select UPR signaling pathways influences proteostasis within the ER and downstream secretory environments. Furthermore, we describe recent evidence that highlights the therapeutic potential for targeting UPR signaling pathways to correct pathologic disruption in secretory proteostasis associated with diverse types of protein misfolding diseases.
Secretory proteostasis is regulated by the Unfolded Protein Response (UPR)
Nearly one-third of the human proteome is targeted to secretory environments consisting of the endoplasmic reticulum (ER), lysosome, plasma membrane or the extracellular space. These proteins are involved in many aspects of cellular and organismal function including lipid metabolism, protein degradation, intracellular signal transduction, and cell-cell signaling. Thus, maintaining the integrity of secretory proteins is essential to prevent pathologic disruption of these important functions.
Secretory proteome integrity (also referred to as secretory proteostasis) is primarily maintained by biologic pathways localized within the ER through a process termed ER quality control [1–3]. ER quality control functions to regulate the integrity of secreted proteins by partitioning non-native protein conformations between biologic pathways involved in ER protein folding or degradation (Fig. 1)[4, 5]. ER folding pathways (consisting of ATP-dependent chaperones, lectin chaperones, protein disulfide isomerases, and other folding factors) facilitate the folding of proteins into native, functional conformations, which are then packaged into vesicles for trafficking to downstream secretory environments. However, proteins unable to attain a natively folded conformation through iterative interactions with ER folding pathways are instead partitioned towards degradation, where they are removed from the ER and degraded through mechanisms including ER-associated degradation (ERAD) or autophagy [6, 7]. Through these mechanisms, ER quality control prevents secretion of non-native conformations that could induce toxic proteostasis imbalances in downstream secretory environments.
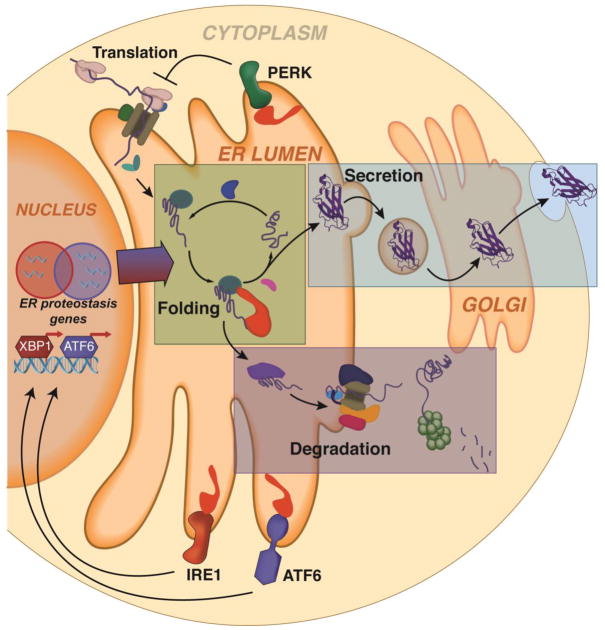
Figure 1. Protein Secretion Through the Secretory Pathway is Regulated by the Activity of ER Protein Folding, Trafficking and Degradation Pathways.
Proteins co-translationally entering into the ER in non-native conformations engage ER-localized chaperones (e.g., BiP, GRP94, CNX, CRT) and folding factors (e.g., PDIs, PPIases) that facilitate their folding into their proper three-dimensional conformation (green box). These proteins are then packaged into vesicles for trafficking to downstream secretory environments such as the extracellular space (blue box). However, proteins unable to fold in the ER are directed towards degradation pathways such as ER-associated degradation where they are retrotranslocated from the ER to the cytosol and degraded by the ubiquitin-proteasome pathway (purple box). This partitioning of proteins between ER protein folding/trafficking or degradation pathways is referred to as ER quality control and functions to limit secretion of non-native protein conformations to downstream secretory environments. The primary impact of activating each UPR signaling arm on ER quality control is also depicted. PERK-dependent translation attenuation decreases the import of newly-synthesized proteins entering the ER, reducing ER protein folding load (top). Alternatively, IRE1/XBP1s and ATF6 activation induces transcriptional remodeling of ER proteostasis pathways involved in protein import, folding, degradation and trafficking.
The two main determinant that influence ER quality control for an individual protein are the inherent energetic stability of the protein fold and the relative activities of ER quality control pathways involved in protein folding, trafficking or degradation [5]. The stability of the protein fold (defined by both kinetic and thermodynamic parameters) influences the intrinsic capacity for a polypeptide to attain a native, folded conformation. This control over the relative population of folded and non-folded conformations influences the partitioning for given polypeptides between ER protein folding/trafficking and degradation. For example, energetically destabilized proteins are generally targeted to degradation pathways because their low stability prevents them from attaining a folded conformation within the steady-state ER environment. However, stable proteins can efficiently fold in the ER environment, increasing their trafficking to downstream secretory environments. In contrast, the composition and relative activities of ER protein folding and degradation pathways (collectively ER proteostasis pathways) influence ER quality control for specific proteins through extrinsic alterations in their partitioning toward folding or degradation. Since the components of both these pathways engage non-native conformations, changing their relative activities and stoichiometries directly influences the relative flux of polypeptides through folding or degradation pathways. For example, increasing the activity of ER protein folding pathways by raising the levels of ER chaperones and folding factors can antagonize the targeting of non-native proteins to degradation pathways and facilitate the folding of these polypeptides through iterative chaperone cycles [5]. In contrast, increasing the activities of ER degradation pathways can decrease chaperone-assisted folding of non-native polypeptides by increasing their flux towards degradation. Hence, the capacity of ER protein folding and degradation pathways, which is dictated by the composition and activity of the individual pathways and the ER protein folding load, is a key determinant in matching ER quality control efficiency to the secretory demands of different cell types and tissues.
Many genetic, environmental, or aging-related insults induce imbalances in ER quality control that result in the ER accumulation of non-native protein conformations. This type of condition is termed ER stress. ER stress-induced alterations in ER quality control are a direct threat to secretory proteostasis as it challenges the ability for the ER to fold and traffic secretory proteins in properly folded, functional conformations. Furthermore, the aberrant secretion of non-native protein conformations can disrupt proteostasis and function in downstream secretory environments. In order to protect the secretory proteome from these types of insults, cells evolved a stress responsive signaling pathway called the the Unfolded Protein Response (UPR). The UPR is a tripartite signaling pathway activated downstream of three ER transmembrane sensor proteins: protein-kinase R-like endoplasmic reticulum kinase (PERK), inositol requiring enzyme 1 (IRE1), and activating transcription factor 6 (ATF6). These sensor proteins are all activated in response to ER stress through mechanisms well-described in other reviews [8, 9] and only briefly summarized in Fig. 2. A primary function of the UPR is to alleviate the ER accumulation of non-native proteins and reestablish ER quality control and secretory proteostasis in response to ER stress. The UPR also influences other aspects of cellular physiology including the regulation of ER function [10], mitochondria biology [11], lipid metabolism [12, 13], and apoptotic signaling [14]. However, in this review, we exclusively focus on the importance of UPR signaling in regulating ER quality control and secretory proteostasis, which is achieved through UPR-dependent reductions in ER protein folding load and enhancement of ER quality control capacity [5].
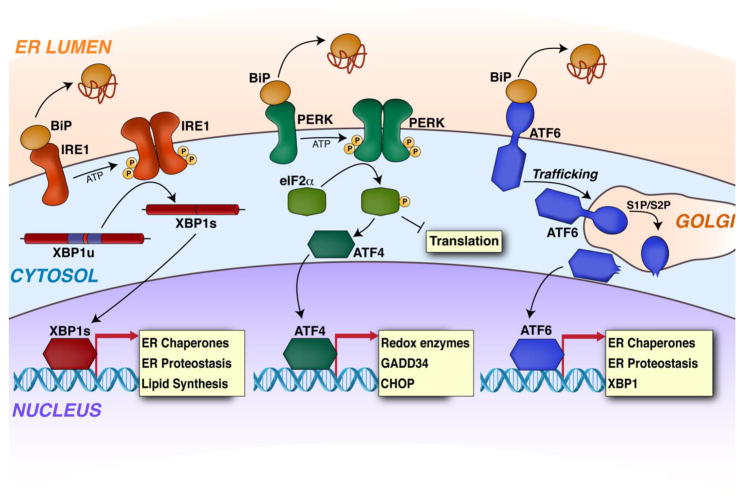
Figure 2. The Unfolded Protein Response.
Illustration showing the primary signaling mechanisms activated downstream of the ER stress sensing proteins IRE1, PERK, and ATF6. ER stress induces PERK autophosphorylation and dimerization (middle). This leads to activation of a PERK cytosolic kinase domain that phosphorylates the α subunit of eukaryotic initiation factor 2 (eIF2α). Phosphorylated eIF2α induces transient translation attenuation and activation of effector stress-responsive transcription factors including the activating transcription factor 4 (ATF4). IRE1 activation also proceeds through a mechanism involving autophosphorylation and oligomerization (left). This leads to activation of a cytosolic IRE1 endoribonuclease domain, which primarily functions in the selective splicing of XBP1 mRNA. Spliced XBP1 mRNA (XBP1s) encodes the active bZIP XBP1s transcription factor, which is the primary effector of IRE1 signaling. Finally, ATF6 activation is initiated through a mechanism involving ER stress induced trafficking to the Golgi where the protein is proteolytically processed by site-1 and site-2 proteases (right). Proteolytic cleavage of ATF6 releases an active, N-terminal bZIP transcription factor, herein referred to as ATF6. While the image shown in this figure represent the basic mechanisms associated with UPR signaling, it is becoming increasing clear that many other factors can influence UPR signaling through direct interactions with components of these pathways. These types of interactions are well described elsewhere [95].
One mechanism by which the UPR influences ER quality control is by regulating ER protein folding load. ER protein folding load is primarily regulated by the transient attenuation of new protein synthesis induced downstream of PERK-dependent eIF2α phosphorylation (Fig. 2). This decreases the population of newly-synthesized proteins entering into the ER, freeing components of ER protein folding and degradation pathways to engage misfolded proteins that accumulate during ER stress. This capacity to regulate protein load provides a mechanism to match ER proteostasis capacity to the population of non-native proteins, preventing the potentially toxic overload of ER protein folding, trafficking, and degradation pathways during conditions of stress. For instance, PERK-dependent translation attenuation is important to regulate the high levels of insulin produced in pancreatic β-cells [15]. IRE1 can also regulate ER protein folding load through a promiscuous mRNA degradation mechanism referred to as regulated IRE1-dependent decay of mRNA (RIDD) [16] – an alternative function of the IRE1 endoribonuclease domain to the more commonly described XBP1 splicing. RIDD reduces the translation/import of secretory proteins entering into the ER environment, thus decreasing ER protein folding load in response to prolonged ER stress.
Secondly, the UPR influences ER quality control capacity through the transcriptional remodeling of ER proteostasis pathways. This is primarily achieved through the activity of the UPR-associated transcription factors XBP1s (activated downstream of IRE1) and ATF6 (the cleaved product of full-length ATF6) (Fig. 2). These two bZIP transcription factors homo- or hetero-dimerize to induce overlapping, but distinct sets of genes involved in ER quality control. XBP1s induces expression of genes involved in all aspects of ER function including import, N-glycosylation, ATP-dependent chaperoning, disulfide isomerization, ERAD, and vesicular trafficking [17–19]. The global impact of IRE1/XBP1s activation on the composition of ER proteostasis pathways is consistent with this pathway being the most conserved signaling arm of the UPR found in all eukaryotes from yeast to humans. In contrast, ATF6 induces expression of a smaller set of ER factors predominantly involved in core ER quality control pathways such as protein folding and degradation [17, 19, 20]. XBP1 is also a transcriptional target of ATF6, reflecting integration between these two transcription factors in the regulation of ER quality control pathways [17, 21, 22]. This is further evident as XBP1s and ATF6 can heterodimerize to synergistically induce expression of ER proteostasis factors predominantly involved in ERAD [17, 22]. This heterodimerization, along with the transcriptional cross-regulation, provides a mechanism for IRE1/XBP1s and ATF6 signaling to integrate and sensitively adapt ER quality control to diverse types of ER stresses.
While the signaling mechanisms and transcriptional profiles associated with IRE1/XBP1s and ATF6 activation are well-established, the functional implications of activating these UPR-associated transcriptional programs on ER quality control and the regulation of secretory proteostasis are only beginning to become clear. Here, we highlight recent results describing the direct and indirect mechanisms by which IRE1/XBP1s and/or ATF6 activation can influence ER quality control and secretory proteostasis for destabilized, disease-relevant proteins. Furthermore, we describe recent advances in the development of new therapeutic strategies to correct pathologic imbalances in secretory proteostasis by targeting the ATF6 arm of the UPR.
Disruptions in ER quality control are a threat to secretory proteostasis
Despite the general effectiveness of ER quality control, imbalances in secretory proteostasis are associated with the onset and pathogenesis of etiologically diverse human diseases (Table 1) [4, 23]. The extracellular aggregation of destabilized, aggregation-prone variants of proteins such as transthyretin (TTR) or immunoglobulin light chain (LC) are implicated in the pathogenesis of systemic amyloid diseases such as TTR amyloidosis and Light Chain Amyloidosis (AL), respectively [10, 24]. Similarly, intracellular aggregation of destabilized variants of secretory proteins such as rhodopsin or α-1-antitrypsin (A1AT) induce toxicity associated with retinal degeneration and A1AT deficiency [10]. The pathologic aggregation of these secretory proteins can be attributed, at least in part, to a failure of ER quality control to direct these destabilized variants to degradation pathways. This leads to increased concentrations of these proteins in secretory environments such as the extracellular space, which facilitates their concentration-dependent aggregation into toxic oligomers, aggregates and amyloid fibrils [10, 24]. In contrast, premature degradation of destabilized secretory proteins such as β-glucocerebrosidase, α-galactosidase, and γ-aminobutyric acid, Type A (GABAA) receptor is implicated in the pathogenesis of loss-of-function protein misfolding disorders including Gaucher disease, Fabry disease, and idiopathic epilepsy, respectively [25–28]. In these cases, premature protein degradation precludes proper folding and subsequent trafficking of these proteins to downstream functional environments such as the lysosome or plasma membrane. This reduced trafficking leads to pathology by decreasing the activity of these proteins in downstream secretory environments. Inhibition of ER degradation for some of these destabilized proteins can facilitate their proper folding and subsequent trafficking to increase downstream biologic activity [29–32], suggesting that many of these destabilized variants are capable of folding into functional conformations. This finding further highlights that their premature partitioning to degradation, and not necessarily their inability to fold, can be a key factor in dictating the pathogenesis of loss-of-function protein misfolding diseases.
Table 1.
Protein folding diseases impacted by UPR-dependent modulation of secretory proteostasis
| Protein | Disease | Pathophysiology | Correction by targeting secretory proteostasis | Ref | |
|---|---|---|---|---|---|
| Pathway targeted | Outcome | ||||
| Gain-of-toxicity (aggregation) diseases | |||||
| TTR | Senile systemic amyloidosis (SSA), familial amyloid polyneuropathy (FAP) | Extracellular aggregation and amyloid formation of WT (age-related) or destabilized mutant variants of TTR | ATF6 activation | Reduced secretion, increased degradation, co-secretion with ERdj3 co-chaperone | [17, 38, 43, 76] |
| Light Chain (LC) | LC Amyloidosis (AL) | Extracellular aggregation, amyloid formation of destabilized LC sequences | ATF6 activation XBP1s activation |
Reduced secretion, increased ER chaperone interactions Reduced secretion, increased proteosomal and lysosmal degradation | [39, 76] |
| Rhodopsin | Retinal degeneration | Cellular toxicity in retinal cells due to intracellular aggregate formation, developmental defects | ATF6 activation; IRE1 activation | Reduced trafficking and intracellular aggregation through increased targeting to ERAD or lysosomal degradation | [40, 41] |
| α1-antitrypsin (A1AT) | A1AD-associated emphyse ma, liver cirrhosis | A1AT deficiency due to intracellular aggregate formation of destabilized variants | ATF6 activation | Decreased intracellular aggregates, increased ERAD or autophagy | [42] |
| Loss-of-function diseases | |||||
| β-glucocerebrosidase (GC) | Gaucher Disease | Pre-mature degradation of destabilized variants and loss of lysosomal enzyme function, abnormal accumulation of glucocerebroside | ERAD inhibition (modulation of ER Ca(II) homeostasis); ER stress (IRE1 activation) | Enhancement of folding of mutant GC variants, trafficking, and lysosomal activity | [29, 32, 92] |
| α-galactosidase | Fabry Disease | Pre-mature degradation of destabilized variants and loss of lysosomal enzyme function, abnormal accumulation of globotriaosylceramide | |||
| γ-aminobutyric acid, Type A (GABAA) receptor | Idiopathic epilepsy | Excessive degradation of destabilized variants, reduced trafficking to plasma membrane, deficiency in excitatory-inhibitory balance of neurons | Inhibition of VCP (component of ERAD); Inhibition of ER Ca(II) receptors | Prevents degradation and enhances the trafficking of destabilized mutant α1-subunit containing GABAA receptors to plasma membrane, rescues receptor function | [31, 93, 94] |
Environmental insults that disrupt ER quality control (e.g., ER stress) can also influence proteostasis in downstream secretory environments by increasing secretion of proteins in non-native, non-functional conformations. For example, ER stress increases secretion of destabilized TTR variants in non-native conformations that accumulate in the extracellular space as soluble oligomers commonly associated with proteotoxicity [33]. Similarly, ER stress increases the trafficking of destabilized variants of the GPI-anchored prion protein or the integral membrane protein ABCA1 to the plasma membrane in misfolded, non-functional conformations [34–36]. These results indicate that ER stress challenges ER quality control to regulate the conformational integrity of secreted proteins, which can disrupt secretory proteostasis and function. Interestingly, the ability to respond to ER stress through mechanisms such as the UPR declines with age [37]. Therefore, aging could further exacerbate imbalances in ER quality control, and secretory proteostasis, providing a potential explanation for the contributions of aging in the onset and pathogenesis of many protein aggregation and loss-of-function protein misfolding diseases [4, 10, 23, 24].
Impacting ER quality control of disease-associated protein through UPR activation
Imbalances in ER quality control threaten the integrity and function of proteins localized to downstream secretory environments. UPR signaling can adjust ER quality control capacity through remodeling of ER proteostasis pathways, mainly IRE1/XBP1s and/or ATF6 activation. Addressing the functional importance of IRE1/XBP1s and/or ATF6 activation on ER quality control has long been challenging because of the difficulty in monitoring ER function in the presence of ER stresses commonly used to activate these pathways (e.g., thapsigargin, DTT or tunicamycin). However, chemical biologic strategies have recently been developed for the ER stress-independent activation of IRE1/XBP1s and/or ATF6 to physiologic levels. These tools have revealed the unique contributions of IRE1/XBP1s and/or ATF6 activation to remodeling of the ER proteostasis environment and to the regulation of ER quality control and secretory proteostasis (Box 1).
Box 1. Chemical Biologic approaches to activate UPR signaling pathways independent of ER stress.
Chemical biologic approaches to activate the individual UPR signaling pathways IRE1/XBP1s, PERK, and ATF6 in the absence of ER stress have facilitated elucidation of unique functional contributions of each arm on ER function and secretory proteostasis. Initially, activation of the IRE1/XBP1s branch was achieved by a chemical genetic strategy targeting the kinase active site of IRE1. Mutation of a gate-keeper residue in the ATP-binding pocket of IRE1 renders the enzyme sensitized to the small molecule ATP mimetic 1NM-PP1, bypassing the need for IRE1 phosphorylation to activate its endoribonuclease function [81, 82]. The discovery that kinase inhibitors can induce IRE1 conformational changes that activate its RNAse activity led to subsequent identification of more potent pharmacologic IRE1 activators [83, 84]. Nonetheless, potential off-target effects limit the general utility of these compounds, which for instance can also target PERK [84]. Selective activation of the PERK pathway was achieved by taking advantage of another aspect of the activation mechanism: dimerization of the kinase required for trans-autophosphorylation. Fusion of the cytosolic PERK kinase domain to two modified FK506 binding domains (Fv2E) allows induced dimerization through addition of the small molecule ligand AP20187 [85]. This ligand-induced dimerization recapitulates PERK autosphosphorylation and downstream pathway activation through eIF2α phosphorylation, although it is difficult to accurately recapitulate the transient nature of PERK-dependent eIF2α phosphorylation using this system due to the difficulty in reversing AP20187-induced dimerization. However, stress-independent Fv2E-PERK activation has defined the protective role of the PERK pathway on ER function, as well as its anti-proliferative, pro-apoptotic function [85, 86]. The chemically induced dimerization approach has also been applied to IRE1 [87]. Strategies to activate the ATF6 pathway in the absence of ER stress have centered on exogenous posttranslational control of the ATF6 bZIP transcription factor domain. ATF6 fusion to a mutant estrogen receptor domain (MER) allows ligand-dependent regulation of this construct through addition of tamoxifen [68, 69]. Stress-independent activation of ATF6-MER in transgenic mice demonstrated ATF6 mediated protection of heart damage from ischemia/reperfusion injury [68]. In an alternative approach, the active ATF6 transcription factor is fused to a destabilized mutant dihydrofolate reductase (DHFR) protein. Destabilization of the DHFR-ATF6 causes constitutive degradation, while stabilization of the entire fusion protein and subsequent ATF6 activity can be achieved through addition of the small molecule ligand trimethoprim (TMP) [17]. Importantly, titration of TMP allows dose-dependent induction of ATF6 transcriptional targets to near-endogenous levels. Similar approaches have also been applied to regulate XBP1 activity [88]. DHFR-ATF6 expression was combined with doxycycline-mediated induction of XBP1s in a single mammalian cell line to allow orthogonal, small molecule-dependent activation of the IRE1/XBP1s and/or ATF6 pathways [17]. This cell line facilitated the characterization of transcriptional reprogramming of ER proteostasis pathways by the two UPR arms separately and synergistically [17]. Furthermore, these tools have defined the distinct roles for IRE1/XBP1s or ATF6 in regulating ER secretory proteostasis for a number of proteins that aggregate in association with diverse protein aggregation diseases [17, 33, 39, 43].
Using these chemical biologic approaches, it is becoming clear that activation of IRE1/XBP1s and/or ATF6 have distinct roles in regulating ER quality control for disease-associated proteins. This has been best demonstrated by evaluating how UPR signaling pathways influence the partitioning of destabilized, disease-associated proteins between folding/trafficking or degradation pathways within the ER. For example, stress-independent activation of ATF6, but not XBP1s, selectively reduces secretion of destabilized, aggregation prone variants of TTR (Table 1) [17, 33]. This reduced secretion corresponds with increased targeting of these variants to degradation through mechanisms such as ERAD and autophagy. Thus, ATF6 activation alters ER quality control for destabilized TTR variants by increasing the partitioning of these proteins for degradation. The extent of ATF6-dependent reductions in TTR secretion is a function of protein destabilization afforded by a specific mutation (defined by the in vitro-derived thermodynamic and kinetic stability of each mutant) [38]. This result, which can be predicted from mathematical modeling of ER quality control [5], indicates that ATF6-dependent remodeling of ER proteostasis pathways increases ER quality control stringency for TTR, requiring higher TTR stability to promote efficient folding and trafficking.
While only stress-independent activation of ATF6 reduces TTR secretion, activation of either XBP1s or ATF6 reduces secretion of a destabilized, amyloidogenic variant of immunoglobulin light chain (ALLC) associated with light chain amyloidosis (Table 1) [39]. However, the mechanisms by which XBP1s or ATF6 activation reduce ALLC secretion are distinct. XBP1 activation reduces ALLC secretion through increased targeting to ER degradation. In contrast, ATF6 activation reduces ALLC secretion through increased interactions with ER chaperones. The differential mechanisms by which IRE1/XBP1s or ATF6 activation influences ALLC secretion highlights the unique remodeling of ER quality control pathways afforded by independent activation of these two UPR-associated transcription factors [17]. Interestingly, the co-activation of both XBP1s and ATF6 in a single cell does not show cooperative reduction in ALLC secretion, but an intermediate level of ALLC degradation. This indicates that the altered partitioning of ALLC between ER folding and degradation pathways afforded by XBP1 or ATF6 activation are competing mechanisms to reduce ALLC secretion. This observation further highlights the unique functional impact of activating these UPR-associated transcription factors on ALLC ER quality control.
IRE1/XBP1s or ATF6 activation can also influence the stability of destabilized secretory proteins that undergo pathologic intracellular aggregation in the context of human disease. For example, stress-independent activation of either IRE1 or ATF6 reduces intracellular levels of destabilized, aggregation-prone variants of rhodopsin through increased partitioning to ERAD and lysosomal degradation [40, 41]. Similarly, ATF6 activation reduces intracellular levels of the destabilized, aggregation-prone A1AT Z-variant by increasing partitioning to ERAD (Table 1) [42]. Also, these results highlight that activation of IRE1/XBP1s or ATF6 can differentially influence ER quality control of structurally-diverse, disease-associated proteins.
Despite this capacity for IRE1/XBP1s or ATF6 activation to influence ER quality control for destabilized proteins, the secretion of wild-type or stable non-amyloidogenic variants of TTR, LC, rhodopsin or A1AT is not significantly reduced by stress-independent IRE1/XBP1s and/or ATF6 activation [17, 33, 39–42]. Similarly, secretion of endogenous secretory proteome does not appear to be significantly impacted by activation of these UPR-associated transcription factors [17]. While it cannot be excluded that activating these signaling pathways influences secretion of some lower expressed secretory proteins, these results indicate that remodeling of ER proteostasis pathways induced by UPR activation evolved to selectively increase ER quality control stringency for proteins whose folding is challenged by genetic or environmental insults.
IRE1/XBP1s or ATF6 activation could also attenuate the premature degradation of destabilized proteins implicated in loss-of-function protein misfolding diseases, potentially increasing their folding and trafficking to downstream functional environments. Experimental evidence suggests that this approach could increase secretion and downstream function of certain destabilized proteins. For example, ER stress increases the trafficking and function of a destabilized β-glucocerebrosidase mutants associated with Gaucher disease through a process requiring IRE1 (Table 1) [32], suggesting that stress-independent activation of IRE1/XBP1 could similarly increase the folding and trafficking of these destabilized mutants. Moving forward, it will be interesting to see whether chemical biologic activation of IRE1/XBP1 or ATF6 can attenuate premature degradation and promote proper folding and trafficking of destabilized proteins associated with loss-of-function protein misfolding diseases.
The above results highlight the potential for IRE1/XBP1s or ATF6 activation to differentially influence ER quality control of destabilized, misfolding-prone proteins associated with diverse protein misfolding diseases. However, many questions still remain to understand the molecular mechanism by which UPR-dependent remodeling of ER quality control pathways influences the partitioning of substrates between protein folding or degradation. For example, what are the ER quality control pathways responsible for the reduced secretion of specific proteins afforded by IRE1/XBP1s or ATF6 activation? What are the structural features of ER substrates that make them selectively sensitive to ER quality control remodeling induced by either XBP1s or ATF6 activation? Can combined IRE1/XBP1s and ATF6 activation synergistically influence ER quality control for certain substrates? Does activation of XBP1s and/or ATF6 influence the conformation or ‘quality’ of proteins secreted during ER stress? Addressing these questions will improve our understanding of the functional role for UPR activation in regulating ER quality control, and reveal both pathologic consequences of how altered UPR signaling could influence protein misfolding disease pathology and new therapeutic opportunities to intervene.
Influencing downstream secretory proteostasis environments through UPR activation
The UPR provides a powerful mechanism to protect the downstream secretory environment during conditions of ER stress. The reduced secretion of destabilized, aggregation-prone proteins afforded by IRE1/XBP1s and/or ATF6-dependent remodeling of ER proteostasis pathways will decrease the concentrations of these proteins in downstream secretory environments. These reduced protein levels will attenuate their concentration-dependent aggregation into toxic oligomers and aggregates consequently protecting the downstream secretory environment. Consistent with this, ATF6-dependent reduced secretion of destabilized TTR or LC variants decreases their extracellular accumulation as soluble oligomers commonly associated with disease toxicity [17, 38, 39]. Similarly, increased degradation of destabilized rhodopsin or A1AT variants afforded by IRE1/XBP1s or ATF6 activation, decreases intracellular aggregation populations of the disease-associated proteins [40–42]. These results highlight that UPR-dependent remodeling of ER quality control indirectly regulates downstream secretory environments by controlling the trafficking and subsequent concentrations of destabilized, aggregation-prone proteins in secretory environments.
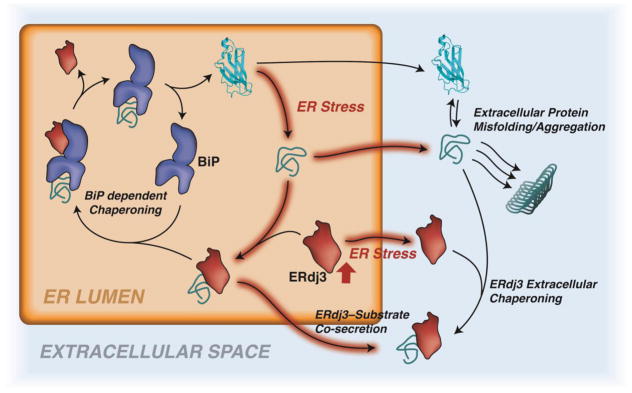
The UPR also directly protects downstream secretory environments during ER stress through the regulated secretion of the ER HSP40 co-chaperone ERdj3/DNAJB11 (Fig. 3). ERdj3 is a tetrameric HSP40 co-chaperone that coordinates ER and secretory proteostasis in response to ER stress [43–45]. ER stress induces ERdj3 secretion both in vitro and in vivo [43], and this increased secretion can be recapitulated by stress-independent ATF6 activation, indicating that this process is regulated by the UPR [43]. Increased ERdj3 secretion can promote extracellular proteostasis through two distinct mechanisms. First, secreted ERdj3 can bind non-native protein conformations in the extracellular space and attenuate their misfolding and/or aggregation into toxic conformations – two functional hallmarks of other extracellular chaperones [46]. Second, ERdj3 can be co-secreted with destabilized, aggregation-prone proteins in a process regulated by the availability of the ER HSP70 chaperone BiP (Fig. 3). When BiP is available, ERdj3 functions as a canonical ER HSP40 co-chaperone, delivering misfolded substrates to BiP and stimulating ATP-dependent BiP chaperoning activity [47]. However, when free BiP is limiting, as can occur during ER stress, ERdj3 cannot deliver misfolded proteins to BiP and instead is co-secreted with misfolded substrates. ERdj3-substrate co-secretion functions to remove misfolded, aggregation-prone proteins from the ER when chaperoning pathways are overwhelmed. This provides a mechanism to preemptively protect downstream secretory environments from toxic protein conformations that can escape ER quality control during ER stress by secreting these proteins in complex with a chaperone. Once secreted, ERdj3-substrate complexes are likely targeted for endocytic degradation, which is a mechanism used by other extracellular chaperones to remove non-native protein conformations from extracellular environments [46]; however, this remains to be established for ERdj3.
Figure 3. UPR-dependent Regulation of ERdj3 Secretion.
Illustration showing the mechanisms by which secreted ERdj3 protects the extracellular space from misfolded, aggregation prone proteins. In the ER, ERdj3 binds to misfolded protein conformations that can accumulate during ER stress. When BiP is available, ERdj3 delivers the misfolded substrate to BiP for ATP-dependent chaperoning (regular arrows). However, when BiP is limiting (due to ER stress, red-highlighted arrows), ERdj3 can be secreted when bound to non-native protein conformations, preemptively protecting the extracellular proteostasis environment from toxic aggregation-prone proteins. Alternatively, ERdj3 can be secreted in the absence of a bound protein during ER stress. This unbound ERdj3 can bind to non-native protein conformations in the extracellular space and prevent their misfolding and/or aggregation into toxic protein conformations and aggregates.
Interestingly, the UPR-dependent increase in ERdj3 secretion corresponds with reduced secretion of the prominent extracellular chaperone clusterin [48]. Clusterin is the most abundant extracellular chaperone and functions to regulate proteostasis [46]. In response to ER stress, clusterin secretion is reduced through a poorly defined mechanism involving clusterin retrotranslocation from the ER to the cytosol [48]. The involvement of the UPR in regulating clusterin secretion and the functional importance of this reduced secretion on secretory proteostasis is currently unknown. However, it is intriguing that ER stress-dependent increases in ERdj3 secretion correspond with reduced clusterin secretion, as it suggests that changing the properties of extracellular chaperones trafficking through the secretory pathway may be important for regulating secretory proteostasis during ER stress.
The capacity to regulate secretory proteostasis through mechanisms such as ERdj3 co-secretion allows cells to protect downstream secretory environments from non-native protein conformations that can escape ER quality control and be secreted during ER stress. As new approaches are established to monitor protein conformation and trafficking through the secretory pathway, it will be interesting to see if other similar mechanisms exist to regulate the secretion of other protein classes (e.g., plasma membrane proteins) during conditions of ER stress. In addition, defining how ERdj3 co-secretion integrates with UPR-dependent remodeling of ER quality control pathways will provide important insights into the global role for UPR signaling in regulating secretory proteostasis during ER stress. Finally, it will be interesting to determine if UPR-dependent regulation of secretory chaperones such as ERdj3 have non-proteostasis roles in regulating organismal physiology. For example, secreted ERdj3 is involved in multiple signaling pathways during development [49, 50]. This suggests that UPR-dependent regulation of ERdj3 secretion could function as an extracellular signal to coordinate organismal physiology during ER stress through mechanisms such as the non-cell autonomous UPR signaling observed in C. elegans [51]. However, a potential role for UPR-dependent increases in secreted ERdj3 in this type of non-autonomous cell signaling remains to be defined.
Other non-canonical secretion mechanisms are also influenced by ER stress (Box 2). Both the rapid ER stress induced export (RESET) and secretory autophagy pathways can direct proteins from the ER to the plasma membrane in response to ER stress in a process that ultimately leads to their degradation. While these pathways are well-described elsewhere [52–55], it is important to note that they provide additional mechanisms to degrade non-native or potentially damaged proteins during ER stress.
Box 2. ER stress-dependent modulation of secretory proteostasis through unconventional secretion pathways.
Recently, non-canonical secretory mechanisms have been identified by which ER stress can influence the secretion of destabilized proteins. In the initial phases of ER stress prior to UPR activation, several GPI anchored proteins, such as destabilized variants of prion protein PrP, are rapidly trafficked to the plasma membrane in non-native conformations through a process called rapid ER stress induced export (RESET)[34]. At the plasma membrane, these proteins are redirected to the lysosome for degradation. Interestingly, RESET, like ERdj3 co-secretion, is regulated by the activity of an ER chaperone, in this case calnexin. In the absence of ER stress, destabilized GPI-anchored proteins are retained within the ER and maintained in a soluble conformation through interactions with calnexin. In response to ER stress, these proteins are released from calnexin and trafficked from the ER in a process involving the export receptor Tmp21. Through this RESET mechanism, cells can prevent the potentially toxic aggregation of GPI-anchored proteins in the ER and secretory environments. Consistent with this, genetic impairment of this RESET pathway leads to intracellular aggregation of destabilized prion proteins and neurodegeneration in mouse models of prion disease, highlighting the importance of this mechanism in regulating secretory proteostasis [34, 89]. In a second non-canonical secretion pathway, destabilized ΔF508 CFTR was observed to traffic more efficiently to the plasma membrane under ER stress conditions, albeit in its immature core-glycosylated form [90]. This trafficking bypasses the Golgi and instead depends on direct interactions between CFTR and Golgi reassembly stacking proteins (GRASPs) that relocalize to the ER and facilitate the direct export to the plasma membrane [90]. A stress-mediated Golgi-independent transport mechanism has also been described for a misfolded pendrin variant, but here the export depends on interactions with the co-chaperone DNAJC14 [91]. Interestingly, the unconventional secretion of both pendrin and CFTR under ER stress conditions has been suggested to depend on IRE1 kinase activity, although further studies are necessary to define a specific role for IRE1 in this mechanism of unconventional protein secretion [90, 91].
Therapeutic targeting of UPR signaling pathways to ameliorate pathologic imbalances in secretory proteostasis
The potential to influence secretory proteostasis through UPR activation has led to speculation that targeting IRE1/XBP1, PERK, or ATF6 signaling pathways could offer a unique therapeutic opportunity to correct pathologic imbalances in secretory proteostasis in human disease. Genetic strategies to increase XBP1s using adeno-associated virus (AAV) have been shown to attenuate neurodegenerative phenotypes associated with diverse neurodegenerative diseases [56]. In addition, numerous small molecules are available to activate (or inhibit) IRE1/XBP1s and PERK signaling, which have significant potential to therapeutically modulate ER proteostasis in the context of etiologically-diverse human disease. Multiple excellent reviews have focused on the use of AAV and small molecule strategies to target IRE1/XBP1s or PERK in human disease [56–61]. Therefore, we focus on recent human genetic and pharmacologic evidence that highlights the therapeutic potential for targeting ATF6 to regulate secretory proteostasis in human disease.
ATF6 activation provides a unique opportunity to influence ER quality control of multiple destabilized proteins that are pathologically associated with human disease (Table 1), indicating that ATF6 is an attractive therapeutic target to intervene in these disorders. However, it remains to be known whether there are pathologic consequences of altered ATF6 activity on organismal physiology. Recent human genetic evidence has begun to answer this question. Numerous mutations in ATF6α have now been identified in humans presenting with the developmental eye disorder achromatopsia, where alterations in ATF6 activity lead to impaired retinal development [62–64]. Interestingly, these mutations differentially influence ATF6 activity. Certain mutations impair ER stress induced ATF6 activation, indicating a decreased capacity to signal through this UPR pathway [65]. However, other mutations lead to constitutive ATF6 activation [65]. Despite presenting with achromatopsia, patients harboring these mutations do not appear to suffer from severe systemic or neurological phenotypes, indicating that constitutive activation or inhibition of ATF6 activity does not globally influence organismal physiology. This is consistent with results observed in ATF6α−/− mice, which show no overt developmental phenotype, although they are more sensitive to ER stress [22, 66]. While it is possible that other cellular mechanisms compensate to regulate ATF6 signaling in these patients (e.g., constitutive ATF6 activity could be attenuated through nonsense mediated mRNA decay [65]), these results highlight the therapeutic potential for targeting ATF6 to develop strategies to correct pathologic imbalances in secretory proteostasis associated with disease.
One potential strategy to selectively activate the ATF6 arm of the UPR is to use the same AAV approach successfully employed for XBP1s in neurodegenerative diseases [56]. The delivery of the active ATF6 transcription factor using AAV has been shown to rescue cardiac defects in ATF6-deficient mice, indicating that this approach can be employed to deliver active ATF6 to specific tissues [67]. However, a potential challenge in implementing this approach for ATF6 is that high, non-physiologic levels of ATF6 activity, which can result from exogenous expression of the active, ATF6 N-terminal transcription factor domain, can lead to detrimental consequences such as global UPR activation and reductions in cell viability [17]. Thus, when implementing AAV approaches, it will be important to tightly control ATF6 activity to achieve protective remodeling of ER proteostasis pathways without negative consequences. One potential strategy to address this issue could be to combine AAV with ligand-regulated approaches to control ATF6 activity such a DHFR-ATF6 or MER-ATF6 where the protein levels and transcriptional activity can be dose-dependently and temporally controlled through the addition of small molecules (see Box 1) [17, 68, 69].
Unlike genetic strategies, pharmacologic approaches to activate ATF6 have the advantage of targeting the endogenous transcriptional pathway, circumventing the need for exogenous ATF6 overexpression, and are therefore more likely to achieve near-physiological levels of ATF6 pathway activity. However, pharmacologic targeting of ATF6 signaling is complicated. Although the ATF6 structure remains to be established, no enzyme active sites or allosteric binding pockets, which could be targeted by small molecules, are predicted for this protein. Furthermore, while it is clear that ATF6 activation involves ER stress induced trafficking to the Golgi and subsequent processing by Site 1 and Site 2 proteases (Fig. 2), the molecular mechanism(s) responsible for ATF6 activation remain to be firmly established. ER stress-dependent increases in ATF6 trafficking have been proposed to involve diverse signaling events, including dissociation of the ER HSP70 chaperone BiP, reduction in ATF6 N-linked glycosylation, dissociation of ATF6 oligomers, and alterations in inter and intra-molecular disulfides within the ATF6 luminal domain [70–72]. The lack of a clear mechanism challenges the ability to target specific upstream regulators of ATF6 signaling to promote its trafficking to the Golgi for activation. Finally, it is challenging to develop approaches to selectively activate ATF6 independent of other UPR signaling arms, all of which are activated by similar types of ER stress. However, these challenges have been largely overcome using phenotypic screening to identify small molecules that selectively activate ATF6 transcriptional activity. The first such compound, BiX, was identified using a high-throughput screen (HTS) monitoring expression of the ATF6-regulated chaperone BiP [73]. BiX induces BiP through an ATF6-dependent mechanism, and while it does not induce other arms of the UPR, BiX also does not significantly induce other ATF6 target genes [73], likely limiting its ability to regulate secretory proteostasis in the context of protein misfolding diseases. However, the addition of BiX has been shown to attenuate ER stress induced neurodegeneration in both cell culture and mouse models, suggesting this compound has potential for preventing pathologic ER stress [73–75].
Another HTS screen using a luciferase reporter of ATF6 activation identified a set of structurally-diverse, non-toxic small molecule ER proteostasis regulators that activate the ATF6 arm of the UPR [76]. Using a combination of transcriptional and proteomic profiling, these compounds were shown to preferentially induce the entire set of genes regulated by ATF6 relative to genes induced by the IRE1/XBP1s or PERK arms of the UPR. In addition, these compounds did not induce expression of stress-responsive genes regulated by other signaling pathways (e.g., the heat shock response or oxidative stress response). Thus, unlike BiX, these compounds induce global ATF6-dependent remodeling of secretory proteostasis, suggesting that these compounds should phenocopy the capacity for genetic ATF6 activation to correct ER quality control defects for destabilized proteins. Consistent with this, these ER proteostasis regulators reduce secretion and extracellular aggregation of destabilized, aggregation-prone variants of TTR or LC from physiologically-relevant human disease models without impacting on secretion of wild-type TTR, non-amyloidogenic light chains or immunoglobulins, or the endogenous secretory proteome [76]. These results mirror what has been observed during stress-independent, genetic ATF6 activation [17, 38, 39]. This highlights the therapeutic potential of these compounds to influence secretory proteostasis in disease without globally influencing secretion of proteins localized throughout the secretory pathway.
Apart from ATF6 activation, HTS screening has also identified small molecules, called Ceapins, which inhibit ER stress induced ATF6 activation [77, 78]. These compounds stabilize ATF6 oligomers in the ER, desensitizing the full-length protein to ER stress-induced trafficking to the Golgi and subsequent proteolytic activation. While the impact of ATF6 inhibition on secretory proteostasis has not yet been determined, the identification of these ATF6 inhibitors offers a unique opportunity to define the involvement of ATF6 activity in human disease and potentially develop new therapeutic approaches to influence secretory proteostasis through selective inhibition of the ATF6 signaling pathway.
The recent identification of pharmacologic approaches to target ATF6 activity, combined with the human genetic evidence that constitutive ATF6 activation (or inhibition) does not induce severe phenotypes in humans, provides a strong foundation for the further development of small molecule ATF6 modulators as a therapeutic strategy to intervene in human disease. ATF6 activation has significant potential to correct pathologic imbalances in diverse human protein misfolding diseases including TTR amyloid diseases, AL, retinal degeneration and A1AT deficiency [17, 38–40, 42] (Table 1). Furthermore, ATF6 activation has the potential to influence other pathophysiologic states such as cardiac function in ischemic reperfusion injury [67, 79] and pancreatic beta cell death in diabetes [80]. These ATF6-regulating compounds provide a critical resource to define the therapeutic potential for targeting this UPR signaling pathway in cellular and organismal models of these and many other human diseases associated with imbalances in secretory proteostasis. However, key questions related to the therapeutic potential for targeting ATF6 remain. What are the underlying molecular mechanisms by which small molecules activate (or inhibit) ATF6? Do these compounds have sufficient bioavailability and bioactivity for further drug development? What other diseases are potentially amenable to therapeutic, small molecule-dependent ATF6 activation? Are there specific subsets of compounds that are best suited to ameliorate tissue-specific defects in secretory proteostasis associated with distinct diseases? What are the potential consequences of chronic, small molecule-dependent ATF6 activation? As these, and other, small molecule regulators of ATF6 signaling continue to be used by the research community to address these questions, we predict that the therapeutic potential for targeting ATF6 will become increasingly apparent.
Concluding Remarks
We have only begun to define the functional mechanisms by which UPR activation regulates secretory proteostasis in the context of health and disease. It is now becoming clear that the UPR has a key role in regulating secretory proteostasis through diverse mechanisms such as ER quality control pathway remodeling and ERdj3 secretion. In addition, the establishment and implementation of chemical biologic, genetic, and small molecule approaches to activate select UPR signaling pathways has highlighted the therapeutic potential for arm-selective UPR activation to correct pathologic imbalances in secretory proteostasis associated with many protein misfolding diseases. As these and new approaches are applied to diverse experimental systems, new mechanisms by which the UPR regulates secretory proteostasis will be defined, providing additional information for how these UPR signaling pathways integrate to protect the secretory proteome during stress. Furthermore, future studies will yield therapeutic opportunities to influence secretory proteostasis that can be harnessed to intervene in etiologically-diverse classes of human disease (see Outstanding Questions).
Outstanding Questions.
What are the ER quality control pathways responsible for the reduced secretion of destabilized proteins afforded by IRE1/XBP1s or ATF6 activation?
What are the structural features of ER substrates that make them sensitive to ER quality control remodeling induced by XBP1s or ATF6 activation?
Does activation of XBP1s or ATF6 influence the conformation and/or function of proteins secreted during ER stress?
How do UPR-dependent and UPR-independent mechanisms integrate to regulate secretory proteostasis in response to ER stress?
Are there non-proteostasis functions of UPR-regulated secreted chaperones important for dictating organismal physiology during ER stress?
What are the underlying molecular mechanisms by which small molecule ATF6 modulators influence ATF6 activity?
What other diseases are amenable to therapeutic, small molecule-dependent ATF6 activation?
Are there specific subsets of ATF6 activating compounds that are best suited to ameliorate tissue-specific defects in secretory proteostasis associated with distinct diseases?
What are the potential consequences of chronic, small molecule-dependent ATF6 activation on organismal physiology?
Trends Box.
Activation of the IRE1/XBP1s or ATF6 Unfolded Protein Response (UPR) signaling pathways differentially influence endoplasmic reticulum (ER) quality control decisions for destabilized, disease-associated proteins.
XBP1s- or ATF6-dependent reductions in the secretion of aggregation-prone proteins indirectly protects secretory proteostasis by lowering concentrations of these proteins available for toxic aggregation.
The UPR integrates with other ER stress-responsive pathways to directly influence secretory proteostasis through the regulated secretion of extracellular chaperones.
Human ATF6 mutations indicate that chronic ATF6 activation does not globally disrupt organismal physiology.
Small molecules that target ATF6 activity have significant promise for ameliorate pathologic defects in secretory proteostasis associated with etiologically-diverse human diseases.
Acknowledgments
We apologize to all of our colleagues whose work we were unable to cite due to space limitations. We thank Evan Powers, Bibiana Rius, Julia Dendle, Isabelle Noxon (TSRI) and Joseph Genereux (UC-Riverside) for their critical reading of the manuscript and help with figure preparation. This work is supported by the NIH (DK102635, DK107604, and NS092829 to RLW) and the Leukemia & Lymphoma Society Career Development Program (to LP).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Guerriero CJ, Brodsky JL. The delicate balance between secreted protein folding and endoplasmic reticulum-associated degradation in human physiology. Physiol Rev. 2012;92(2):537–76. doi: 10.1152/physrev.00027.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braakman I, Bulleid NJ. Protein folding and modification in the mammalian endoplasmic reticulum. Annu Rev Biochem. 2011;80:71–99. doi: 10.1146/annurev-biochem-062209-093836. [DOI] [PubMed] [Google Scholar]
- 3.McCaffrey K, Braakman I. Protein quality control at the endoplasmic reticulum. Essays Biochem. 2016;60(2):227–235. doi: 10.1042/EBC20160003. [DOI] [PubMed] [Google Scholar]
- 4.Powers ET, et al. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem. 2009;78:959–91. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- 5.Wiseman RL, et al. An adaptable standard for protein export from the endoplasmic reticulum. Cell. 2007;131(4):809–21. doi: 10.1016/j.cell.2007.10.025. [DOI] [PubMed] [Google Scholar]
- 6.Smith MH, Ploegh HL, Weissman JS. Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science. 2011;334(6059):1086–90. doi: 10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brodsky JL. Cleaning up: ER-associated degradation to the rescue. Cell. 2012;151(6):1163–7. doi: 10.1016/j.cell.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334(6059):1081–6. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 9.Korennykh A, Walter P. Structural basis of the unfolded protein response. Annu Rev Cell Dev Biol. 2012;28:251–77. doi: 10.1146/annurev-cellbio-101011-155826. [DOI] [PubMed] [Google Scholar]
- 10.Ryno LM, Wiseman RL, Kelly JW. Targeting unfolded protein response signaling pathways to ameliorate protein misfolding diseases. Curr Opin Chem Biol. 2013;17(3):346–52. doi: 10.1016/j.cbpa.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rainbolt TK, Saunders JM, Wiseman RL. Stress-responsive regulation of mitochondria through the ER unfolded protein response. Trends Endocrinol Metab. 2014;25(10):528–37. doi: 10.1016/j.tem.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 12.Wang S, Kaufman RJ. How does protein misfolding in the endoplasmic reticulum affect lipid metabolism in the liver? Curr Opin Lipidol. 2014;25(2):125–32. doi: 10.1097/MOL.0000000000000056. [DOI] [PubMed] [Google Scholar]
- 13.Lee AH, Glimcher LH. Intersection of the unfolded protein response and hepatic lipid metabolism. Cell Mol Life Sci. 2009;66(17):2835–50. doi: 10.1007/s00018-009-0049-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hiramatsu N, et al. Multiple Mechanisms of Unfolded Protein Response-Induced Cell Death. Am J Pathol. 2015;185(7):1800–8. doi: 10.1016/j.ajpath.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harding HP, et al. Diabetes mellitus and exocrine pancreatic dysfunction in perk−/− mice reveals a role for translational control in secretory cell survival. Mol Cell. 2001;7(6):1153–63. doi: 10.1016/s1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- 16.Maurel M, et al. Getting RIDD of RNA: IRE1 in cell fate regulation. Trends Biochem Sci. 2014;39(5):245–54. doi: 10.1016/j.tibs.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 17.Shoulders MD, et al. Stress-independent activation of XBP1s and/or ATF6 reveals three functionally diverse ER proteostasis environments. Cell Rep. 2013;3(4):1279–92. doi: 10.1016/j.celrep.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23(21):7448–59. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamamoto K, et al. Differential contributions of ATF6 and XBP1 to the activation of endoplasmic reticulum stress-responsive cis-acting elements ERSE, UPRE and ERSE-II. J Biochem. 2004;136(3):343–50. doi: 10.1093/jb/mvh122. [DOI] [PubMed] [Google Scholar]
- 20.Adachi Y, et al. ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct Funct. 2008;33(1):75–89. doi: 10.1247/csf.07044. [DOI] [PubMed] [Google Scholar]
- 21.Lee K, et al. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002;16(4):452–66. doi: 10.1101/gad.964702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamamoto K, et al. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007;13(3):365–76. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 23.Kaushik S, Cuervo AM. Proteostasis and aging. Nat Med. 2015;21(12):1406–15. doi: 10.1038/nm.4001. [DOI] [PubMed] [Google Scholar]
- 24.Eisele YS, et al. Targeting protein aggregation for the treatment of degenerative diseases. Nat Rev Drug Discov. 2015;14(11):759–80. doi: 10.1038/nrd4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valastyan JS, Lindquist S. Mechanisms of protein-folding diseases at a glance. Dis Model Mech. 2014;7(1):9–14. doi: 10.1242/dmm.013474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang YJ, Di XJ, Mu TW. Using pharmacological chaperones to restore proteostasis. Pharmacol Res. 2014;83:3–9. doi: 10.1016/j.phrs.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steinlein OK. Ion channel mutations in neuronal diseases: a genetics perspective. Chem Rev. 2012;112(12):6334–52. doi: 10.1021/cr300044d. [DOI] [PubMed] [Google Scholar]
- 28.Lindquist SL, Kelly JW. Chemical and biological approaches for adapting proteostasis to ameliorate protein misfolding and aggregation diseases: progress and prognosis. Cold Spring Harb Perspect Biol. 2011;3(12) doi: 10.1101/cshperspect.a004507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang F, Segatori L. Remodeling the proteostasis network to rescue glucocerebrosidase variants by inhibiting ER-associated degradation and enhancing ER folding. PLoS One. 2013;8(4):e61418. doi: 10.1371/journal.pone.0061418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang F, et al. Inhibition of endoplasmic reticulum-associated degradation rescues native folding in loss of function protein misfolding diseases. J Biol Chem. 2011;286(50):43454–64. doi: 10.1074/jbc.M111.274332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han DY, et al. Combining valosin-containing protein (VCP) inhibition and suberanilohydroxamic acid (SAHA) treatment additively enhances the folding, trafficking, and function of epilepsy-associated gamma-aminobutyric acid, type A (GABAA) receptors. J Biol Chem. 2015;290(1):325–37. doi: 10.1074/jbc.M114.580324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mu TW, et al. Chemical and biological approaches synergize to ameliorate protein-folding diseases. Cell. 2008;134(5):769–81. doi: 10.1016/j.cell.2008.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen JJ, et al. Endoplasmic Reticulum Proteostasis Influences the Oligomeric State of an Amyloidogenic Protein Secreted from Mammalian Cells. Cell Chem Biol. 2016;23(10):1282–1293. doi: 10.1016/j.chembiol.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Satpute-Krishnan P, et al. ER stress-induced clearance of misfolded GPI-anchored proteins via the secretory pathway. Cell. 2014;158(3):522–33. doi: 10.1016/j.cell.2014.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanaka AR, et al. The ABCA1 Q597R mutant undergoes trafficking from the ER upon ER stress. Biochem Biophys Res Commun. 2008;369(4):1174–8. doi: 10.1016/j.bbrc.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 36.Nunziante M, et al. Proteasomal dysfunction and endoplasmic reticulum stress enhance trafficking of prion protein aggregates through the secretory pathway and increase accumulation of pathologic prion protein. J Biol Chem. 2011;286(39):33942–53. doi: 10.1074/jbc.M111.272617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor RC. Aging and the UPR(ER) Brain Res. 2016;1648(Pt B):588–593. doi: 10.1016/j.brainres.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 38.Chen JJ, et al. ATF6 activation reduces the secretion and extracellular aggregation of destabilized variants of an amyloidogenic protein. Chem Biol. 2014;21(11):1564–74. doi: 10.1016/j.chembiol.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cooley CB, et al. Unfolded protein response activation reduces secretion and extracellular aggregation of amyloidogenic immunoglobulin light chain. Proc Natl Acad Sci U S A. 2014;111(36):13046–51. doi: 10.1073/pnas.1406050111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiang WC, et al. Selective activation of ATF6 and PERK endoplasmic reticulum stress signaling pathways prevent mutant rhodopsin accumulation. Invest Ophthalmol Vis Sci. 2012;53(11):7159–66. doi: 10.1167/iovs.12-10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiang WC, Messah C, Lin JH. IRE1 directs proteasomal and lysosomal degradation of misfolded rhodopsin. Mol Biol Cell. 2012;23(5):758–70. doi: 10.1091/mbc.E11-08-0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith SE, et al. Activating transcription factor 6 limits intracellular accumulation of mutant alpha(1)-antitrypsin Z and mitochondrial damage in hepatoma cells. J Biol Chem. 2011;286(48):41563–77. doi: 10.1074/jbc.M111.280073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Genereux JC, et al. Unfolded protein response-induced ERdj3 secretion links ER stress to extracellular proteostasis. EMBO J. 2015;34(1):4–19. doi: 10.15252/embj.201488896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Genereux JC, Wiseman RL. Regulating extracellular proteostasis capacity through the unfolded protein response. Prion. 2015;9(1):10–21. doi: 10.1080/19336896.2015.1011887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen KC, et al. The Endoplasmic Reticulum HSP40 Co-Chaperone ERdj3/DNAJB11 Assembles and Functions as a Tetramer. EMBO J. 2017 doi: 10.15252/embj.201695616. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wyatt AR, et al. Extracellular chaperones and proteostasis. Annu Rev Biochem. 2013;82:295–322. doi: 10.1146/annurev-biochem-072711-163904. [DOI] [PubMed] [Google Scholar]
- 47.Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11(8):579–92. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nizard P, et al. Stress-induced retrotranslocation of clusterin/ApoJ into the cytosol. Traffic. 2007;8(5):554–65. doi: 10.1111/j.1600-0854.2007.00549.x. [DOI] [PubMed] [Google Scholar]
- 49.Wong YH, et al. Protogenin defines a transition stage during embryonic neurogenesis and prevents precocious neuronal differentiation. J Neurosci. 2010;30(12):4428–39. doi: 10.1523/JNEUROSCI.0473-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee JY, et al. Maintenance of Stem Cell Niche Integrity by a Novel Activator of Integrin Signaling. PLoS Genet. 2016;12(5):e1006043. doi: 10.1371/journal.pgen.1006043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taylor RC, Dillin A. XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell. 2013;153(7):1435–47. doi: 10.1016/j.cell.2013.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ponpuak M, et al. Secretory autophagy. Curr Opin Cell Biol. 2015;35:106–16. doi: 10.1016/j.ceb.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Noack J, Molinari M. Protein trafficking: RESETting proteostasis. Nat Chem Biol. 2014;10(11):881–2. doi: 10.1038/nchembio.1652. [DOI] [PubMed] [Google Scholar]
- 54.Couve A, Hetz C. RESETing ER proteostasis: selective stress pathway hidden in the secretory route. EMBO J. 2014;33(21):2444–6. doi: 10.15252/embj.201489845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rabouille C. Pathways of Unconventional Protein Secretion. Trends Cell Biol. 2017;27(3):230–240. doi: 10.1016/j.tcb.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 56.Valenzuela V, et al. Gene therapy to target ER stress in brain diseases. Brain Res. 2016;1648(Pt B):561–570. doi: 10.1016/j.brainres.2016.04.064. [DOI] [PubMed] [Google Scholar]
- 57.Maly DJ, Papa FR. Druggable sensors of the unfolded protein response. Nat Chem Biol. 2014;10(11):892–901. doi: 10.1038/nchembio.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schneider K, Bertolotti A. Surviving protein quality control catastrophes--from cells to organisms. J Cell Sci. 2015;128(21):3861–9. doi: 10.1242/jcs.173047. [DOI] [PubMed] [Google Scholar]
- 59.Halliday M, Mallucci GR. Review: Modulating the unfolded protein response to prevent neurodegeneration and enhance memory. Neuropathol Appl Neurobiol. 2015;41(4):414–27. doi: 10.1111/nan.12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang D, Niwa M, Koong AC. Targeting the IRE1alpha-XBP1 branch of the unfolded protein response in human diseases. Semin Cancer Biol. 2015;33:48–56. doi: 10.1016/j.semcancer.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hetz C, Chevet E, Harding HP. Targeting the unfolded protein response in disease. Nat Rev Drug Discov. 2013;12(9):703–19. doi: 10.1038/nrd3976. [DOI] [PubMed] [Google Scholar]
- 62.Kohl S, et al. Mutations in the unfolded protein response regulator ATF6 cause the cone dysfunction disorder achromatopsia. Nat Genet. 2015;47(7):757–65. doi: 10.1038/ng.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu M, et al. ATF6 Is Mutated in Early Onset Photoreceptor Degeneration With Macular Involvement. Invest Ophthalmol Vis Sci. 2015;56(6):3889–95. doi: 10.1167/iovs.15-16778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ansar M, et al. Mutation of ATF6 causes autosomal recessive achromatopsia. Hum Genet. 2015;134(9):941–50. doi: 10.1007/s00439-015-1571-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chiang WC, et al. Achromatopsia mutations target sequential steps of ATF6 activation. Proc Natl Acad Sci U S A. 2017;114(2):400–405. doi: 10.1073/pnas.1606387114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu J, et al. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13(3):351–64. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 67.Jin JK, et al. ATF6 Decreases Myocardial Ischemia/Reperfusion Damage and Links ER Stress and Oxidative Stress Signaling Pathways in the Heart. Circ Res. 2016 doi: 10.1161/CIRCRESAHA.116.310266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martindale JJ, et al. Circulation Research. American Heart Association, Inc; 2006. Endoplasmic Reticulum Stress Gene Induction and Protection From Ischemia/Reperfusion Injury in the Hearts of Transgenic Mice With a Tamoxifen-Regulated Form of ATF6; pp. 1186–1193. [DOI] [PubMed] [Google Scholar]
- 69.Thuerauf DJ, et al. J Biol Chem. American Society for Biochemistry and Molecular Biology; 2007. Effects of the isoform-specific characteristics of ATF6 alpha and ATF6 beta on endoplasmic reticulum stress response gene expression and cell viability; pp. 22865–22878. [DOI] [PubMed] [Google Scholar]
- 70.Hong M, et al. Underglycosylation of ATF6 as a novel sensing mechanism for activation of the unfolded protein response. J Biol Chem. 2004;279(12):11354–63. doi: 10.1074/jbc.M309804200. [DOI] [PubMed] [Google Scholar]
- 71.Higa A, et al. Endoplasmic reticulum stress-activated transcription factor ATF6alpha requires the disulfide isomerase PDIA5 to modulate chemoresistance. Mol Cell Biol. 2014;34(10):1839–49. doi: 10.1128/MCB.01484-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shen J, et al. Stable binding of ATF6 to BiP in the endoplasmic reticulum stress response. Mol Cell Biol. 2005;25(3):921–32. doi: 10.1128/MCB.25.3.921-932.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kudo T, et al. A molecular chaperone inducer protects neurons from ER stress. Cell Death Differ. 2008;15(2):364–75. doi: 10.1038/sj.cdd.4402276. [DOI] [PubMed] [Google Scholar]
- 74.Inokuchi Y, et al. Effect of an inducer of BiP, a molecular chaperone, on endoplasmic reticulum (ER) stress-induced retinal cell death. Invest Ophthalmol Vis Sci. 2009;50(1):334–44. doi: 10.1167/iovs.08-2123. [DOI] [PubMed] [Google Scholar]
- 75.Oida Y, et al. Induction of BiP, an ER-resident protein, prevents the neuronal death induced by transient forebrain ischemia in gerbil. Brain Res. 2008;1208:217–24. doi: 10.1016/j.brainres.2008.02.068. [DOI] [PubMed] [Google Scholar]
- 76.Plate L, et al. Small molecule proteostasis regulators that reprogram the ER to reduce extracellular protein aggregation. Elife. 2016:5. doi: 10.7554/eLife.15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gallagher CM, Walter P. Ceapins inhibit ATF6alpha signaling by selectively preventing transport of ATF6alpha to the Golgi apparatus during ER stress. Elife. 2016:5. doi: 10.7554/eLife.11880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gallagher CM, et al. Ceapins are a new class of unfolded protein response inhibitors, selectively targeting the ATF6alpha branch. Elife. 2016:5. doi: 10.7554/eLife.11878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Glembotski CC. Roles for ATF6 and the sarco/endoplasmic reticulum protein quality control system in the heart. J Mol Cell Cardiol. 2014;71:11–5. doi: 10.1016/j.yjmcc.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sharma RB, et al. Insulin demand regulates beta cell number via the unfolded protein response. J Clin Invest. 2015;125(10):3831–46. doi: 10.1172/JCI79264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Papa FR, et al. Science. American Association for the Advancement of Science; 2003. Bypassing a Kinase Activity with an ATP-Competitive Drug; pp. 1533–1537. [DOI] [PubMed] [Google Scholar]
- 82.Lin JH, et al. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318(5852):944–9. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Korennykh AV, et al. The unfolded protein response signals through high-order assembly of Ire1. Nature. 2009:687–693. doi: 10.1038/nature07661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mendez AS, et al. Elife. eLife Sciences Publications Limited; 2015. Endoplasmic reticulum stress-independent activation of unfolded protein response kinases by a small molecule ATP-mimic; p. e05434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lu PD, et al. EMBO J. EMBO Press; 2004. Cytoprotection by pre - emptive conditional phosphorylation of translation initiation factor 2; pp. 169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lin JH, et al. PLoS ONE. Public Library of Science; 2009. Divergent Effects of PERK and IRE1 Signaling on Cell Viability; p. e4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Back SH, et al. J Biol Chem. American Society for Biochemistry and Molecular Biology; 2006. Cytoplasmic IRE1α-mediated XBP1 mRNA Splicing in the Absence of Nuclear Processing and Endoplasmic Reticulum Stress; pp. 18691–18706. [DOI] [PubMed] [Google Scholar]
- 88.Shoulders MD, et al. Broadly applicable methodology for the rapid and dosable small molecule-mediated regulation of transcription factors in human cells. J Am Chem Soc. 2013;135(22):8129–32. doi: 10.1021/ja402756p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Puig B, et al. Secretory pathway retention of mutant prion protein induces p38-MAPK activation and lethal disease in mice. Sci Rep. 2016;6:24970. doi: 10.1038/srep24970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gee HY, et al. Rescue of DeltaF508-CFTR trafficking via a GRASP-dependent unconventional secretion pathway. Cell. 2011;146(5):746–60. doi: 10.1016/j.cell.2011.07.021. [DOI] [PubMed] [Google Scholar]
- 91.Jung J, et al. The HSP70 co-chaperone DNAJC14 targets misfolded pendrin for unconventional protein secretion. Nat Commun. 2016;7:11386. doi: 10.1038/ncomms11386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ong DST, et al. Endoplasmic reticulum Ca2+ increases enhance mutant glucocerebrosidase proteostasis. - PubMed - NCBI. Nat Chem Biol. 2010:424–432. doi: 10.1038/nchembio.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Di X-J, et al. J Biol Chem. American Society for Biochemistry and Molecular Biology; 2016. Grp94 Protein Delivers γ-Aminobutyric Acid Type A (GABA A) Receptors to Hrd1 Protein-mediated Endoplasmic Reticulum-associated Degradation; pp. 9526–9539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Han D-Y, et al. ACS Chem Biol. American Chemical Society; 2015. L-type Calcium Channel Blockers Enhance Trafficking and Function of Epilepsy-associated α1(D219N) Subunits of GABA AReceptors; pp. 2135–2148. [DOI] [PubMed] [Google Scholar]
- 95.Woehlbier U, Hetz C. Modulating stress responses by the UPRosome: a matter of life and death. Trends Biochem Sci. 2011;36(6):329–37. doi: 10.1016/j.tibs.2011.03.001. [DOI] [PubMed] [Google Scholar]