Abstract
To investigate the influence of Debaryomyces hansenii treatment on intestinal microorganisms in mice with antibiotics-induced diarrhea, mouse model of antibiotics-induced diarrhea was created by gavaging mice with mixed antibiotics (23.33 mL/kg/days) composed of gentamycin sulfate and cefradine for 5 days. Mice with the symptom of diarrhea were then treated with D. hansenii by intragastric administration. The control group mice were given with sterile water. After 4 day treatment, total DNA of intestinal microflora of treated and control mice was extracted, and their quantities were measured by sequencing the V4 region of 16S rDNA. The results showed that when compared to the control (sterile water), treatment with D. hansenii increased the operational taxonomic units (OTUs) of intestinal bacteria. The Chao index in diarrhea treated group was higher than diarrhea control group and was similar to healthy control group, while all differences had no significance (P > 0.05). D. hansenii treatment increased the Shannon index but not significantly (P > 0.05). Moreover, there was not significant impact on density and diversity of intestinal bacterial population at phylum and genus levels (P > 0.05). Interestingly, D. hansenii treatment recovered the population density of certain bacterium species, such as Bacteroidaceae (in family level) (P < 0.05). Our results indicate that D. hansenii has potency of adjusting the density and diversity of intestinal bacteria and recovering the population density of Bacteroidaceae in family level.
Keywords: Antibiotics-induced diarrhea, Debaryomyces hansenii, Bacterial diversity, V4 region sequencing of 16S rDNA
Introduction
Diarrhea is a heath condition with the symptom featured by over three times of watery stools daily. There are two types of diarrhea in clinic (Zong et al. 2006): the acute one, which usually takes 1 or 2 days to recover, and the chronic diarrhea which takes much longer (up to weeks). Usually, acute diarrhea is caused by microbial infection, such as bacteria, virus, and even fungi. However, chronic diarrhea is usually associated with disorders in bowel or intestine (Liu 2010), such as Crohn’s disease. In theory, when the symptom occurs, the volume of fluid in intestine is increased due to lack of absorption capability, or increased production by different causes such as infection. Interestingly, antibiotics application can also cause such symptoms as the bacteria population, diversity, and density are changed in certain parts of intestines when antibiotics are applied (Lu 2010). Particularly, instead of balancing various microorganisms, the antibiotics-sensitive strains are killed and drug-resistant strains survive and are even over-propagated, which result in the disruption of healthy microenvironment or microecosystem. In clinic, it is called dysbacteriosis, superinfection, or antibiotics-induced diarrhea (Wu and She 2007).
It is well known that the intestinal microorganisms are very important for maintaining the normal functions of intestine (Fu 2003; Zhang et al. 2010). Approximately 1014 microbes and about 1000 species of bacteria inhabit in human intestinal tract (Thompson et al. 2015; Li et al. 2014). Gut microbiota can be regarded as a large organ of human beings. It plays critical roles in nutrient digestibility, metabolism, and immunity (Niu et al. 2015). Meanwhile, it is frequently associated with many diseases, such as diabetes, obesity, cardiovascular disease, and acute respiratory distress syndrome (Li et al. 2014; Guo et al. 2015a, b). Antibiotics are known to save lives by working against bacterial infections. As such, antibiotics are commonly used for treating infectious diseases, especially for the ones caused by bacteria in developing countries (Li et al. 2001). Unfortunately, the antibiotics-induced diarrhea frequently occurs in the patients, particularly when it is misused. Misuse of antibiotics might be due to the misunderstanding on the effect of antibiotics that it can work against all infections. In fact, antibiotics only fight infections caused by bacteria, fungi, and certain parasites but have no effect on infections caused by viruses. Using antibiotics can cause antibiotic resistance when it is not needed or improperly used (Harold 1992).
Probiotics confer a health benefit on the host, mostly by bacteria, such as Lactobacillus strain (Isolauri et al. 1991). Interestingly, recent investigations show that yeast belongs to probiotics and can be a potential treatment for diarrhea, since it can alleviate the symptom dramatically. The effective yeast strain was found to be Saccharomyces boulardii (Chen 2013; Czerucka et al. 2007). As shown in the reports, it might mediate or balance the intestinal microenvironment by either producing metabolites or digesting the substances which causes diarrhea. As such, S. boulardii, is widely applied for the treatment of antibiotics-associated diarrhea. In addition, D. hansenii, which was isolated from food or intestinal contents of experimental mice, was identified as a halotolerant yeast with a high biotechnological potential, particularly in food industry (Banjara et al. 2016). It has been demonstrated that D. hansenii adjusts the microecosystem balance and has curative effect on antibiotic-associated diarrhea (Banjara et al. 2016; Xiao et al. 2016; Guo et al. 2015a, b). The previous studies on D. hansenii mainly focus on its application in food industry and in suppressing some pathogenic microorganism such as Candida albicans and Candida tropicalis (Corbo et al. 2001; Corral et al. 2015; Banjara et al. 2016). The present study aimed to find out by examining the bacteria living in intestine how the treatment of antibiotic-associated diarrhea with substance derived from D. hansenii works. To determine the population and density of the bacteria, we applied metagenomic sequencing of intestinal microorganisms from D. hansenii-treated or untreated mice. Our results showed that antibiotics application in mice disrupted the intestinal microecosystem balance leading to dysbacteriosis and D. hansenii treatment partially restored the density and diversity of intestinal microbiota. Therefore, our study suggests that D. hansenii has the potential for treating antibiotics-induced diarrhea in patients.
Materials and methods
Reagents
Gentamycin sulphate and cefradine capsules were purchased from Yichang Humanwell Pharmaceutical Co., Ltd. Lysozyme, proteinase K, dNTP, Taq polymerase, acetate alcohol, acetone, TE buffer, Tris-saturated phenol–chloroform–isoamyl alcohol (25:24:1), Chloroform–isoamyl alcohol (24:1), and 10× Taq buffer were purchased from Beijing Dingguo Changsheng Biotechnology Co., Ltd. Solutions such as PBS, 10% SDS, 3 mol/L sodium acetate, 5 mol/L NaCl, CTAB/NaCl, Tris, 0.5 mol/L EDTA, and 70% ethanol were prepared in the lab.
Debaryomyces hansenii was isolated and identified in our lab as previously reported (Xiao et al. 2016). The D. hansenii strain was inoculated into 200 mL liquid Potato Sucrose medium in a 300-mL Erlenmeyer flask and shaken for 36 h at 28 °C. The cells were collected by centrifugation at 2000 rpm for 4 min whereupon they were washed 1–2 times using sterile stroke-physiological saline solution repeatedly. Then, the right amount of sterile stroke-physiological saline solution was added into above cells. After counting by hemocytometer, cells were diluted to 1010 cells/mL and stored at 4 °C for later use (Guo 2014).
Animals and procedures
Mice were purchased from Hunan Slaccas Jingda Laboratory Animal Company with license number SCXK(Xiang)2013-0004. All procedures involving animals were performed according to protocols approved by the Institutional Animal Care and Use Committee of Hunan University of Chinese Medicine. The experiment included equal number of male and female mice. Mice were randomly selected into three groups: healthy control group (qck), diarrhea treated group (qj), and diarrhea control group (qm). The qck was gavaged with 0.35 mL PBS. The qm and qj groups were treated with antibiotics mixture composed of gentamycin sulfate and cefradine at the concentration of 62.5 g/L (0.35 mL, 40,000 U/mL) by intragastric administration. After diarrhea was induced, D. hansenii solution (0.35 mL, 1010/mL) (Guo 2014) was administrated twice a day for 4 days in qj group. Both qck and qm groups were treated with PBS. The intestine contents were collected after mice were euthanized by cervical dislocations, and then were frozen immediately and stored at −20 °C.
DNA extraction
According to a previous report (Wu et al. 2012), 2.0 g of intestinal faces was weighed in a sterile environment and was homogenized in 5 mL of 0.1 mol/L phosphate buffer solution (PBS), followed by centrifugation at 200 g for 2 min. After wash twice with PBS, the whole supernatant were transferred into new tubes and centrifuged for 8 min at 10,000×g. The sediment were gathered, washed once with PBS, twice with acetone, and three times with PBS, and then resuspended in 4 mL TE buffer. 500 μL of spreadhead were added with 45 μL TE buffer, 5 μL proteinase K, and 20 μL lysozyme, and homogenized in 1.5-mL germ-free EP tubes. Samples were incubated at 37 °C for 30 min, and then mixed with 30 μL of 10% SDS, followed by incubation at 37 °C for 40 min with turning upside down once every 10 min. Afterward, 100 μL of 5 mol/L NaCl and 80 μL of CTAB/NaCl were added and mixed well. The mixture was reacted at 65 °C for 10 min. An equal volume of Tris-saturated phenol–chloroform–isoamyl alcohol (25:24:1) then were added to the sample, mixed well, and were centrifuged at 10,000×g for 3 min. The supernatant was transferred to fresh sterile tubes, mixed with an equal volume of chloroform–isoamyl alcohol (24:1), and centrifuged at 10,000×g for 3 min. The supernatant was transferred into fresh sterile tubes and mixed with an equal volume of chloroform–isoamyl alcohol (24:1) again. After centrifugation at 10,000×g for 3 min, the supernatant were transferred into new sterile tubes, added with 1/10 volume of 3 mol/L sodium acetate and double volume of absolute ethyl alcohol, and were precipitated at −20 °C overnight. Samples were centrifuged at 10,000×g for 3 min. The acquired sediment were washed with 70% ethanol, dried, and eventually dissolved in 50 μL TE buffer.
PCR amplification of 16S rDNA V4 region and sequencing
Universal primer 520F: (5-AYTGGGYDTAAAGNG-3) and 802R: (5-TACNVGGGTATCTAATCC-3) were synthesized by Shanghai Personal Biotechnology Co., Ltd. PCR mixture (25 μL) contained 2 μL 2.5 mmol/L dNTP Mixture, 5.0 μL 5× Q5 Reaction Buffer, 5.0 μL 5× Q5 high Enhancer, 1.0 μL 10 μmol/L 520F, 1.0 μL 10 μmol/L 802R, 2.0 μL 0.2 ng/μL template DNA, 0.25 μL 5 U/μL Q5 Polymerase, and 8.75 μL sterilized ddH2O. Cycling parameters were as follows: initial denaturation at 98 °C for 30 s; denaturation at 98 °C for 30 s, annealing at 50 °C for 30 s and extension at 72 °C for 30 s, repeated for 25 cycles; last cycle of final extension at 72 °C for 5 min. Then, PCR products were detected by 2% Agarose gel electrophoresis. Ultimately, the amplified V4 region of 16S rDNA was sequenced by Shanghai Personal Biotechnology Co., Ltd.
Data analyses
Three methods were applied for analyzing the variety of microorganisms by determining operational taxonomic unit (OTU), including community composition analysis (CCA), alpha diversity analysis (ADA), and principal components analysis (PCA). OTU was developed to measure the last node in phylogenetic or population genetics (Blaxter et al. 2005). Literally, the genetic DNA was broken into number of units and OTU according to the homogeneity. The analysis was conducted by clustering using software Qiime (Altschul et al. 2015; Edgar 2010). While CCA tests the genetic similarity at various levels, such as phylum, class, order, and so on, ADA is a method of measuring population density and variety by its Chao index and Shannon index, respectively. Higher Chao index indicates higher population density, and higher Shannon index means higher diversity (Pitta et al. 2014; Shannon 1997; Mahaffee and Kloepper 1997). PCA is a statistical method of interpreting variable variance. The first component occupies majority of total variance, and the second one has majority of variance of the rest variance. Different components are orthogonal to each other. It was conducted using package of R.
The significant differences among three groups were determined using one-way ANOVA. Differences among groups were considered significant when P values <0.05.
Results
Metagenome DNA sequence analysis
The genomic DNA collected in the experiment was actual mixtures from bacteria, yeast, and other sources. Therefore, DNA sequencing data were first filtered by Uchime (version 1.35.1, http://www.mothur.org/) to remove chimeric sequence, followed by analysis using Qiime (version 1.7.0, http://qiime.org/) to compare homogeneity. Our results showed that the procedure was effective to collect DNA information specifically from bacteria. In brief, the proportion of bacterial DNA information in total sample DNA information were 93, 88, and 92% for healthy control group, diarrhea treated group, and diarrhea control group, respectively (Table 1).
Table 1.
Statistics of DNA sequences identified in samples of different groups
| Samples | Effective sequences | High-quality sequences | Proportion (%) |
|---|---|---|---|
| qck | 136,835 | 126,865 | 92.63 |
| qj | 234,969 | 207,661 | 88.45 |
| qm | 135,978 | 123,132 | 92.23 |
| Total | 507,782 | 457,658 | 90.10 |
Effective sequences: index exactly matched sequence; high-quality sequences: the effective sequences after filtering and removing chimeric sequences
Effect of D. hansenii treatment on OTUs of intestinal microbiota
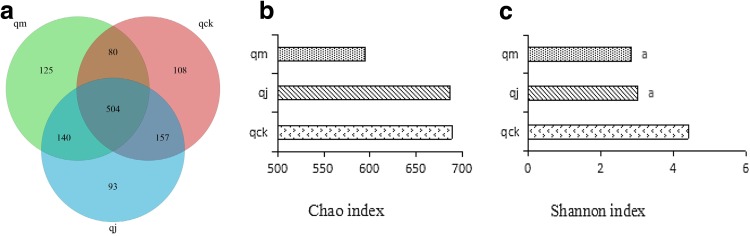
Based on sequence homogeneity, the sequences with over 97% similarity were aligned and grouped into individual OTUs. The alignment was conducted using Qiime. As shown in Fig. 1a, over 1207 OTUs were identified in total. 849, 849, and 894 OTUs were found from healthy control group, diarrhea control group, and diarrhea treated group, respectively. The results indicate that the D. hansenii treatment could stimulate growth or proliferation of intestinal bacteria. In addition, the numbers of unique OTUs identified from these three groups were 108, 125, and 93, respectively. The data suggest that the variety of bacteria was recovered upon the treatment of D. hansenii. The difference tested by ADA of Shannon index was shown to be significant with P value <0.01. However, the Chao index testing did not show any significance (Fig. 1b, c).
Fig. 1.
Comparison of the OTUs and ADA in three groups. a Venn diagram of OTUs based on the sequences with over 97% similarity under the similar level of clustering. b, c Alpha diversity analysis of Chao and Shannon indexes, respectively. Compared with the qck, a: P < 0.01
Impact of D. hansenii treatment on intestinal microbiota in mice
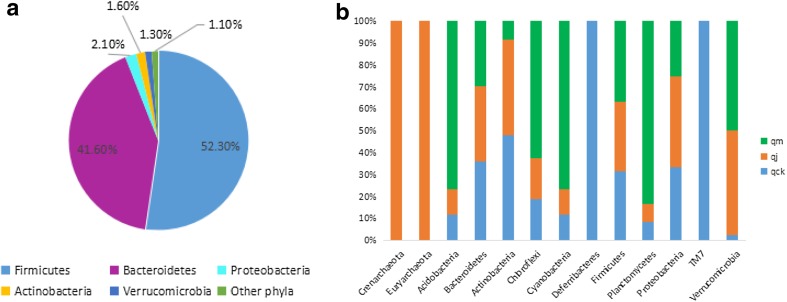
The microbiota from the mice guts was isolated. We found they basically belong to Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, and Verrucomicrobia at the phylum level. Interestingly, the bacteria from Firmicutes were the most abundant, followed by Bacteroidetes and Proteobacteria. The percentage of different phyla in bacteria community was approximately 1.1% for Crenarchaeota, Euryarchaeota, Acidobacteria, Chloroflexi, Cyanobacteria, Deferribacteres, Planctomycetes, and bacteria TM7 (Fig. 2a) in all three groups. In addition, totally 13 phyla were found. Acidobacteria, Chloroflexi, Cyanobacteria, Bacteroidetes, Actinobacteria, Planctomycetes, Firmicutes, Proteobacteria, and Verrucomicrobia were found in all three groups, whereas Crenarchaeota, Euryarchaeota, Deferribacteres, and bacteria TM7 were unique to diarrhea treated group and healthy control group, respectively. The relative abundance ratio of Acidobacteria, Actinobacteria, Chloroflexi, Cyanobacteria, and Planctomycetes in qck group is similar to that of qj group and is different from qm group. The relative percentage of Verrucomicrobia was similar between qm group and qj group, which was higher than that of qck. Qm group had the lowest relative density of Bacteroidetes but the highest relative density of Firmicutes. All differences were insignificant (P > 0.05) (Fig. 2b).
Fig. 2.
Phylum distribution of species. a Percentage of phyla founded in three groups. Other phyla include Crenarchaeota, Euryarchaeota, Acidobacteria, Chloroflexi, Cyanobacteria, Deferribacteres, Planctomycetes, and TM7. b Relative density ratio of bacteria between experimental groups (P > 0.05)
We further investigated the difference of bacterial population among three groups. We totally identified 30 families: nine reached the analytical levels (Table 2), but other 21 families only had trace amounts. For the relative density of Bacteroidaceae, the diarrhea treated group was significantly lower than other two groups (P < 0.01); furthermore, the diarrhea control group was significantly higher than that in healthy control group (P < 0.01). Moreover, the relative densities of Ruminococcaceae and Erysipelotrichaceae in the healthy control group were significantly lower than those of other two groups (P < 0.01), whereas the difference between the diarrhea control group and the diarrhea treated group was not significant (P > 0.05). Furthermore, D. hansenii treatment recovered the density of Porphyromonadaceae and even increased the density of Bacteroidales S24-7, albeit not significantly (P > 0.05) (Table 2).
Table 2.
Relative density of the gut bacteria at the family level
| Family | qck | qm | qj |
|---|---|---|---|
| Bacteroidaceae | 0.060 ± 0.024 | 0.224 ± 0.027a | 0.005 ± 0.005Ab |
| Porphyromonadaceae | 0.022 ± 0.009 | 0.096 ± 0.094 | 0.016 ± 0.012 |
| Bacteroidales S24-7 | 0.285 ± 0.043 | 0.202 ± 0.281 | 0.334 ± 0.130 |
| Lactobacillaceae | 0.086 ± 0.049 | 0.034 ± 0.053 | 0.033 ± 0.050 |
| Lachnospiraceae | 0.094 ± 0.024 | 0.234 ± 0.137 | 0.276 ± 0.132 |
| Ruminococcaceae | 0.090 ± 0.035 | 0.019 ± 0.005a | 0.018 ± 0.010a |
| Erysipelotrichaceae | 0.012 ± 0.004 | 0.177 ± 0.053a | 0.139 ± 0.028a |
| Enterobacteriaceae | 0.001 ± 0.001 | 0.001 ± 0.001 | 0.008 ± 0.012 |
| Verrucomicrobiaceae | 0.001 ± 0.001 | 0.019 ± 0.021 | 0.018 ± 0.012 |
Each value is the mean ± standard deviation. Data were analyzed using one-way ANOVA. In comparison with qck, a: P < 0.01, A: P < 0.05. In comparison with qm, b: P < 0.01
β-Diversity analysis
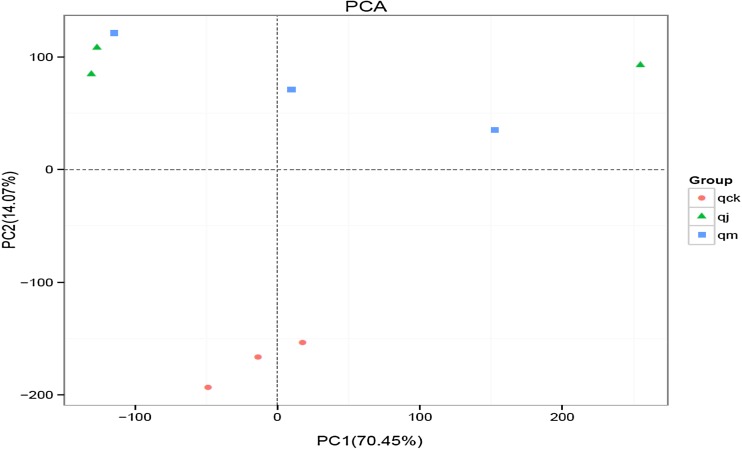
In addition, PCA was conducted to analyze the bacterial similarity at genus level based on the DNA sequence data. The results showed that qck group was well separated from that other two groups and that there was no significant difference between qj group and qm group. The percentage contributed to variation of PC1 and PC2 was 70.45 and 14.07%, respectively (Fig. 3).
Fig. 3.
PCA of bacterial similarity at genus level based on the DNA sequence data. Each point in the figure represents a sample. Points with the same color belong to the same group. The closer the distance between two points, the smaller difference of the microbial community
Discussion
Meta 16 seq is a newly developed method for specifying or identifying origination of DNA from species. Basically, it amplifies the nine highly variable regions of 16S rDNA (Chen et al. 2015). Since those regions are conserved among species from bacteria to human being, it is a promising method for identifying the origins of different DNAs in a mixed population. The method includes PCR amplification and the subsequent high-throughput deep sequencing (Zhang et al. 2014). Therefore, it is an effective method for our current study to differentiate DNAs from a mix of bacteria, yeast, or even cells from mouse intestines. The current study aimed to understand the influence of D. hansenii on intestinal microbiota of mice with antibiotic-induced diarrhea at genus level by mainly using the high-throughput sequencing.
PCR-amplified sequences include many chimera sequences that could affect the accuracy of analysis. Therefore, the method of uchime in mothur package was used to remove chimera sequences according to the data filtering standard (Schloss et al. 2009; Edgar et al. 2011). In addition, the following sequences were also removed: the sequences containing fuzzy base (N base), the sequences with more than eight continuous identical base, the sequences longer than 150 bp, and the sequences more than one mismatch base. The average proportion of high-quality sequences was more than 90% after removing the chimera sequences, ensuring the accuracy and reliability of the following analysis.
Our recent study showed that mice had diarrhea after they were given with antibiotics by intragastric administration (Zeng et al. 2012). Our current study demonstrated that intestinal microbiota was changed after antibiotics-induced diarrhea and partially restored after the treatment with D. hansenii, which is consistent with the findings in the recent reports (Long et al. 2015; Guo 2014). In our study, the OTUs were applied for measuring the density and diversity of bacterial population. Our results showed that the diarrhea treated group had higher OTUs and the diarrhea control group had lower Chao index and Shannon index. These results indicated that the bacterial density and diversity were decreased upon antibiotics-induced diarrhea, but were restored by D. hansenii treatment. However, there was no significant curative effect (P > 0.05). Our findings suggest that the treatment may improve bacterial growth and proliferation.
Intestinal bacteria include mainly Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, and Verrucomicrobia. There are two subgroups within Firmicutes, Clostridium coccoides subgroup (cluster XIVa) and Clostridium leptum subgroup (cluster IV). These taxa are shown to be associated with obesity and intestinal diseases. Bacteroidetes actually play many roles in maintaining intestinal functions, such as carbohydrate fermentation, glucose metabolism, and bile acid and steroid metabolism. While many bacteria within Bacteroidaceae family are pathogenic, Porphyromonadaceae and S24-7 are beneficial to human beings. Interestingly, although Actinobacteria is not predominant in intestinal tract, Bifidobacterium is probiotic and plays an important role in human health, for example, reducing the incidence rate of diarrhea (Li 2009; Zhang et al. 2015; Saavedra et al. 1994). Our results showed that antibiotics-induced diarrhea reduced the relative densities of Bacteroidetes and Actinobacteria at the phylum level, which was partially restored by the treatment with D. hansenii (P > 0.05). At family level, the relative density of Bacteroidaceae was decreased (P < 0.05) and that of S24-7 was increased (P > 0.05). The relative density of Porphyromonadaceae was recovered (P > 0.05) upon the treatment with D. hansenii. The PCA results showed that the density and diversity of bacteria had no change after the treatment at genus level. Taken together, D. hansenii has the potential to recover intestinal bacteria, albeit non-significantly. Possible reasons are that the treating time was short and that individual differences existed.
In summary, antibiotics-induced diarrhea impacted the intestinal bacterial population density at the levels of phylum, family, and genus, whereas D. hansenii treatment readjusted them at family level, but not at both phylum and genus level. Our current study sheds new light on the mechanisms underlying intestinal microecosystem balancing, which will help developing therapeutic strategies for treating patients with antibiotics-associated diarrhea.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (No. 81573951).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Nenqun Xiao, Phone: (+86) 13974954942, Email: xiaonenqun@sohu.com.
Zhoujin Tan, Phone: (+86) 13974954942, Email: tanzhjin@sohu.com.
References
- Altschul SF, Gish W, Miller W, Myers EW, David J. Basic local alignment search tool. J Mol Biol. 2015;3:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Banjara N, Nickerson KW, Suhr MJ, Heather EH. Killer toxin from several food-derived Debaryomyces hansenii strains effective against pathogenic Candida yeasts. Int J Food Microbiol. 2016;222:23–29. doi: 10.1016/j.ijfoodmicro.2016.01.016. [DOI] [PubMed] [Google Scholar]
- Blaxter M, Mann J, Chapman T, Thomas F, Whitton C, Floyd R, Abebe E. Defining operational taxonomic units using DNA barcode data. Philos Trans R Soc Lond B. 2005;360(1462):1935–1943. doi: 10.1098/rstb.2005.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JB. Effect of Saccharomyces boulardii on prevention of antibiotic-associated diarrhea in children. Chin Foreign Med Res. 2013;11(30):15–16. [Google Scholar]
- Chen ZB, Li B, Wang DK, Yu L, Xu SG, Ren Z, Jin S, Zhang YF, Peng SJ. Diversity of endophytic bacteria in walnut analyzed by Illumina MiSeq high-throughput sequencing technology. Jiangsu J Agric Sci. 2015;31(5):1129–1133. [Google Scholar]
- Corbo MR, Lanciotti R, Albenzio M. Occurrence and characterization of yeasts isolated from milks and dairy products of Apulia region. Int J Food Microbiol. 2001;69:147–152. doi: 10.1016/S0168-1605(01)00585-2. [DOI] [PubMed] [Google Scholar]
- Corral S, Salvador A, Belloch C, Flores M. Improvement the aroma of reduced fat and salt fermented sausages by Debaromyces hansenii inoculation. Food Control. 2015;47:526–535. doi: 10.1016/j.foodcont.2014.08.001. [DOI] [Google Scholar]
- Czerucka D, Piche T, Rampa P. Review article: yeast as probiotics—Saccharomyces boulardii. Aliment Pharmacol Ther. 2007;26:767–778. doi: 10.1111/j.1365-2036.2007.03442.x. [DOI] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. Uchime improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu DR (2003) Impact of lactic acid bacteria on human health. Xinjiang Animal Husbandry (zl):41–43
- Guo KX (2014) The effects of ultra-micro powder Qiweibaizhusan collaborative with yeast on dysbacteriotic diarrheal mice. Dissertation, Hunan University of Chinese Medicine
- Guo HL, Shao YY, Menghe BLG, Zhang HP. Research on the relation between gastrointestinal microbiota and disease. Microbiol China. 2015;42(2):400–410. [Google Scholar]
- Guo KX, Tan ZJ, Xie MZ, She Y, Wang XH. The synergic effect of ultro-micro powder qiweibaizhusan combined with yeast on dysbacteriotic diarrhea mice. Chin J Appl Environ Biol. 2015;21(1):61–67. [Google Scholar]
- Harold C. The crisis in antibiotic resistance. Science. 1992;257(5073):1064–1073. doi: 10.1126/science.257.5073.1064. [DOI] [PubMed] [Google Scholar]
- Isolauri E, Juntunen M, Rautanen T, Sillanaukee P, Koivula T. A human Lactobacillus strain (Lactobacillus casei sp strain GG) promotes recovery from acute diarrhea in children. Pediatrics. 1991;88(1):90–97. [PubMed] [Google Scholar]
- Li M (2009) Co-variation analysis of human gut microbial structure and host global metabolism. Dissertation, Shanghai Jiao Tong University
- Li GQ, Du RL, Liu GL, Dou LC, Xiao CY. Investigation on application status of antibacterial agents. Clin Med. 2001;21(11):47–48. [Google Scholar]
- Li Y, Liu XY, Ma MM, Qi ZJ, Zhang XQ, Li Z, Cao GH, Li J, Zhu WW, Wang XZ. Chagnges in intestinal microflora in rats with acute respiratory distress syndrome. World J Gastroenterol. 2014;20(19):5849–5858. doi: 10.3748/wjg.v20.i19.5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WZ. Diagnosis and management of chronic diarrhea. Chin J Gastroenterol. 2010;15(5):257–260. [Google Scholar]
- Long CX, Liu YJ, Deng TH, Yin KK, Tan ZJ. The influence of sucrose on the effective of qiwei baizhu powder combined with yeast in treatment of dysbacteriotic diarrhea in mice. Chin J Microecol. 2015;27(11):1241–1249. [Google Scholar]
- Lu LL. Probiotics in the prevention of antibiotics-associated diarrhea: a meta-analysis of randomized control trails. J Clin Pediatr. 2010;28(11):1083–1085. [Google Scholar]
- Mahaffee WF, Kloepper JW. Temporal changes in the bacterial communities of soil, rhizosphere, and endorhiza associated with field-grown cucumber (Cucumis sativus L.) Microb Ecol. 1997;34(3):210–223. doi: 10.1007/s002489900050. [DOI] [PubMed] [Google Scholar]
- Niu Q, Li P, Hao S, Zhang Y, Kim SW, Li H, Ma X, Gao S, He L, Wu W, Huang X, Hua J, Zhou B, Huang R. Dynamic distribution of the gut microbiota and the relationship with apparent crude fiber digestibility and growth stages in pigs. Sci Rep. 2015;5:1–7. doi: 10.1038/srep09938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitta DW, Parmar N, Patel AK, Indugu N, Kumar S, Prajapathi KB, Patel AB, Reddy B, Joshi C. Bacterial diversity dynamics associated with different diets and different primer pairs in the rumen of kankrej cattle. PLoS ONE. 2014;9(11):e111710. doi: 10.1371/journal.pone.0111710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra JM, Bauman NA, Oung I, Perman JA, Yolken RH. Feeding of Bifidobacterium bifidum and Streptococcus thermophilus to infants in hospital for prevention of diarrhea and shedding of rotavirus. Lancet. 1994;334(8929):1046–1049. doi: 10.1016/S0140-6736(94)91708-6. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75(23):7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon CE. The mathematical theory of communication. MD Comput. 1997;14(4):306–317. [PubMed] [Google Scholar]
- Thompson JA, Oliveira RA, Djukovic A, Ubeda C, Xavier KB. Manipulation of the quorum sensing signal Al-2 effects the antibiotic-treated gut microbiota. Cell Rep. 2015;10:1861–1871. doi: 10.1016/j.celrep.2015.02.049. [DOI] [PubMed] [Google Scholar]
- Wu D, She KX. Antibiotic-associated diarrhea in critical patients. Chin J Nosocomiol. 2007;17(5):587–588. [Google Scholar]
- Wu H, Zhou SN, Guo C, Tan ZJ, Cai GX, Zeng A, Zhang HL. A metagenome DNA extracting method of intestinal flora in mice for molecular diversity analysis based on PCR technology. Chin J Microecol. 2012;24(7):648–651. [Google Scholar]
- Xiao XY, Liu YJ, Deng YL, Guo KX, Tan ZJ. Isolation and identification of a yeast strain from intestine of mice. J Hunan Agric Univ (Nat Sci) 2016;42(4):419–423. [Google Scholar]
- Zeng A, Zhang HL, Tan ZJ, Cai Y, Cai GX, Zhou SN. The construction of mice diarrhea model due to dysbacteriosis and curative effect of ultra-micro qiweibaizhusan. Microbiology. 2012;39(9):1341–1348. [Google Scholar]
- Zhang Q, Luo Y, Yang EH, Wu Q, Tao J, Zhong Y. Research progress in isolation and identification methods of Bifidobacterium and its application. Anim Husb Feed Sci. 2010;31(3):10–12. [Google Scholar]
- Zhang QL, Peng XX, Wu H, Peng MJ, Cao R, Wang H, Xiao NQ, Tan ZJ. Molecular diversity of intestinal microbiota in dysbacteria diarrheal mice associated with ultra-micro qiweibaizhudan. Int J Curr Microbiol Appl Sci. 2014;3(2):198–207. [Google Scholar]
- Zhang Q, Zhou ZK, Ren XC. Comparision of intestinal microbiota in high-throughput sequencing. J Good Saf Qual. 2015;6(5):1776–1782. [Google Scholar]
- Zong Y, Zhao HY, Liang XM, Zhang ST, Lv Z. Intestinal flora changes in patients with acute or chronic diarrhea. J Clin Intern Med. 2006;23(2):89–90. [Google Scholar]