Abstract
l-Asparaginase is employed in leukaemic treatment and in processing starchy foods. The in silico analysis of Lactobacillus reuteri DSM 20016 reveals the presence of an l-asparaginase gene with theoretical pI value of 4.99. 3D structure prediction was carried out and one model was selected based on the validation scores of 86.293 for ERRAT, 92.10% for VERIFY 3D and Ramachandran plot. Multiple sequence alignment of the protein sequences of l-asparaginases I and II of Escherichia coli, Erwinia chrysanthemum and Homo sapiens shows their sequence similarity. The ORF LREU_RS09880 from L. reuteri DSM 20016 genome was cloned and expressed in E. coli. The recombinant protein was purified to homogeneity using Ni–NTA chromatography and showed higher substrate specificity for l-asparagine. Kinetic parameters like K m and V max of recombinant l-asparaginase were calculated as 0.3332 mM, 14.06 mM/min, respectively. Temperature and pH profile of recombinant l-asparaginase were analysed and maximum activity was found between 30 and 40 °C and at pH 6. The recombinant enzyme was thermally stable up to 24 h at 28 °C. Recombinant l-asparaginase has a recovery percentage of 92 and 10.5 fold purification. HPLC–MS–MS and SDS-PAGE analysis of the purified protein indicated a molecular weight of 35 kDa as a monomer.
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-017-0974-4) contains supplementary material, which is available to authorized users.
Keywords: 3D structure, Km, Lactobacillus reuteri DSM 20016, l-Asparaginase, HPLC–MS–MS, Ni–NTA Chromatography, Vmax
Introduction
l-Asparaginase (EC 3.5.1.1) metabolizes l-asparagine to l-aspartate and ammonia (Doria and Kumar 2016) thereby finds its wide application in pharmaceutical and food industry (Cachumba et al. 2016). l-asparaginase is used to treat patients suffering from Acute Lymphoblastic Leukaemia, Acute Myeloid Leukaemia and non-Hodgkin’s lymphoma. Intravenous administration of l-asparaginase depletes l-asparagine in the blood stream, thus making the leukaemic cells devoid of extracellular l-asparagine while the normal cells synthesize l-asparagine. l-asparaginases in commercial use are isolated from many sources including Escherichia coli and Erwinia caratovora. The commercial enzyme has the following side effects widely ranging from fever, chills, vomiting, weight loss, pancreatitis to hepatocellular dysfunction, reported death due to asparaginase-associated pancreatitis (Raja et al. 2014) and death due to steatosis, necrosis in the liver and renal failure (Bodmer et al. 2006). The commercially used l-asparaginases have proven side effects so there is a need to look for a newer source. In food industry, this enzyme finds a major application by reducing the formation of acrylamide, a potent carcinogen by hydrolysing the l-asparagine (Sanghvi et al. 2016). The l-asparagine rich foods produce acrylamide by-product during baking, frying, roasting, etc. This formation can be reduced by pre-treating the foods with l-asparaginase to reduce the risk of cancer.
Asparaginases from various sources like E. coli MTCC 739 (Vidya et al. 2011), Bacillus licheniformis (Sudhir et al. 2014; Mahajan et al. 2014), Streptomyces griseoluteus WS3/1 (Kumari et al. 2013), Pseudomonas fluorescens (Sinha et al. 2015), Pencillium sp (Soniyambi et al. 2011), Rhizomucor miehei (Huang et al. 2014) have been studied. The 3D structure of the l-asparaginase from Bacillus subtilis (Onishi et al. 2015), human asparaginase-like protein (Gunda et al. 2016) has been modelled. The l-asparaginase gene from the above sources has been cloned into E. coli system for overexpression. Diverse purification protocols have been employed like Ni-IDA (Huang et al. 2014), DEAE cellulose (Kumar and Manonmani 2013; Mahajan et al. 2014; Soniyambi et al. 2011), Ni–NTA chromatography (Sudhir et al. 2014; Vidya et al. 2011), Gel filtration using Sephadex (Kumari et al. 2013) Sepharose (Kishore et al. 2015). The molecular mass of the recombinant protein was characterized using SDS-PAGE (Sudhir et al. 2014; Vidya et al. 2011; Sinha et al. 2015; Kishore et al. 2015) and Gel filtration chromatography (Kumar and Manonmani 2013; Mahajan et al. 2014; Kumari et al. 2013). The enzyme characteristics like optimum temperature, pH, thermostability, effect of metal ions and kinetic parameters are studied. Anticancer activity of the enzyme against various cancer cell lines JURKAT E6-1, MCF 7, K562 (Mahajan et al. 2014) were also studied. The effect of the enzyme on acrylamide reduction by pre-treating the dough (Huang et al. 2014), potato and potato chips (Sanghvi et al. 2016; Onishi et al. 2015) have also been studied. Lactobacillus species are well established probiotics which are also known to harbour the asparaginase genes (Amer et al. 2013) that are yet to be characterized.
The present work is to study the properties of the l-asparaginase gene (LREU_RS09880) from the probiotic strain Lactobacillus reuteri DSM 20016 using bioinformatics tools, to clone the l-asparaginase gene, express it in E. coli system and purify using Ni–NTA chromatography. The characteristics and the kinetic parameters of the recombinant protein are studied.
Materials and methods
Bacterial strains
Lactobacillus reuteri DSM 20016 was procured from DSMZ, Germany, and maintained on MRS medium at 37 °C. E. coli cultures were grown at 37 °C with agitation in LB medium, supplemented with 50 µg/ml of Streptomycin for E.coli TOP 10 and Kanamycin for recombinant E.coli BL21(DE3).
In silico analysis of l-asparaginase
The protein sequence of l-asparaginase was retrieved from UniProt (UniProt entry: A5VMR3) and used for further in silico analysis of the protein. The physico-chemical parameters of the protein sequence were analysed using ExPASy-Protparam tool (http://web.expasy.org/protparam/). The 3D structure prediction was carried out using the tool Easy Modeller 4.0. The structures were validated using ERRAT (http://services.mbi.ucla.edu/ERRAT/), VERIFY 3D (http://services.mbi.ucla.edu/Verify_3D/) and Ramachandran plot (http://mordred.bioc.cam.ac.uk/~rapper/rampage.php). The sequence alignment was performed using PRALINE multiple sequence alignment (http://www.ibi.vu.nl/programs/pralinewww/) to know the sequence similarity between human (UniProt entry: Q7L266), E. coli (UniProt entry: P0A962, P00805), Erwinia chrysanthemi (UniProt entry: A0A0A3ZNT4, P06608) l-asparaginases I and II and the l-asparaginase of L. reuteri DSM 20016.
Cloning of LREU_RS09880 gene in E. coli
Genomic DNA was isolated from L. reuteri DSM 20016 (Cheng and Jiang 2006) and the l-asparaginase gene (new locus tag: LREU_RS09880 and old locus tag: Lreu_1900) was amplified using gene-specific forward (1900F-AGACCCATATGACGAAAAAATTATTACTCCTTTCAAC) and reverse (1900R-GGCCTCTCGAGATCTAAAGTTACTTCACCAGCAACAT) primers. The amplicon was cloned into pET28a(+) using NdeI and XhoI (sites were introduced in the primers) and transformed into E. coli TOP10 competent cells. The recombinant expression plasmid was subsequently transformed into BL21 (DE3) competent cells for expression studies.
Plate assay
Modified M-9 medium with composition Na2HPO4.2 H2O 6 g; KH2PO4 3 g; NaCl 0.5 g; l-asparagine 5 g; 1 M MgSO4·7H2O 2 ml; 0.1 M CaCl2.2H2O 1 ml; 20% glucose stock 10 ml; agar 20 g; 0.007% bromothymol blue (BTB) (Mahajan et al. 2014) was prepared. E. coli BL21(DE3)(pET28a(+):LREU_RS09880) was analysed for l-asparaginase activity with E. coli BL21(DE3) as a control in the above medium.
Overexpression of l-asparaginase
The E. coli BL21(DE3) expression strain harbouring LREU_RS09880 gene was inoculated in LB broth with kanamycin (50 µg/ml) and induced at OD600 0.6 with 0.05 mM IPTG for 4 h. Cells were harvested and suspended in 1% v/v of equilibration buffer (50 mM sodium phosphate, pH 8, with 0.3 M sodium chloride and 10 mM imidazole) and sonicated (XL-2000, QSonica, USA) for 10 min. The crude lysate was centrifuged at 10,000 rpm for 10 min at 4 °C and the supernatant was stored at 4 °C for further use.
Purification using Ni–NTA column chromatography
The protein was purified to homogeneity using HIS-Select® HF Nickel Affinity Gel column chromatography. Ni–NTA column was equilibrated using five column volumes of equilibration buffer and recombinant His-tagged protein was eluted using six column volumes of elution buffer (50 mM sodium phosphate, pH 8.0 with 0.3 M sodium chloride and 250 mM imidazole). The eluted fractions were analysed on 12% SDS-PAGE to ensure homogeneity. Purified protein was immediately subjected to dialysis. Dialysis membrane (Dialysis Membrane-110, HIMEDIA) was pretreated by boiling with 1 mM EDTA for 10 min, washed thrice with distilled water and boiled for 10 min in distilled water. Dialysis is performed for 1 h against freshly prepared and filter sterilized 50 mM Tris buffer pH 7.
Enzyme assay
Nesslerization assay (Imada et al. 1973) was carried out using the following protocol: 0.5 ml of recombinant l-asparaginase enzyme with 0.5 ml of 0.05 M Tris–HCl Buffer (pH 8.6), 0.5 ml of 0.2 M l-asparagine and incubated for 30 min at 37 °C. Enzyme activity was stopped by the addition of 0.5 ml Trichloro Acetic Acid (10% W/V) and centrifuged at 10,000 rpm for 5 min. 0.2 ml of Nessler’s reagent is added to 0.1 ml of supernatant and made up to 4 ml with distilled water, incubated for 20 min at 20 °C and the colour formation is measured at 450 nm (Synergy HTX, multimode reader, BioTeK, USA). All assays were performed in triplicates. The corresponding ammonia concentrations were obtained from the standard graph of ammonia for calculation of enzyme activity. One unit of l-asparaginase activity is defined as the amount of enzyme required to release of 1 µM of ammonia per hour at 37 °C and pH 8.6.
Temperature and pH profile
Temperature profile of the purified recombinant enzyme was analysed by subjecting the reaction mixture at various temperatures from 10 to 80 °C. The pH profile was analysed by varying pH from 2 to 9 using 50 mM Citrate Buffer for pH 2 and 3, 50 mM Sodium Acetate Buffer for pH 4, 5 and 6, 50 mM Tris HCl Buffer for pH 7, 8 and 9 (Rajnish et al. 2008). All assays were performed in triplicates.
Thermostability
To analyse the thermostability the recombinant l-asparaginase was incubated at 28 and 37 °C. Enzyme was aliquoted at different time intervals from 15, 30, 45 min, 1, 2, 4, 6, 8, 12 and 24 h (Kishore et al. 2015) and Nesslerization assay was carried out in triplicates.
Determination of kinetic parameters
Kinetic parameters K m and V max of the recombinant l-asparaginase were generated from Lineweaver–Burk plot (Kishore et al. 2015) by varying the substrate concentration from 1 to 150 mM in Tris buffer (pH 8.6) using GraphPad Prism 7.02.
Liquid chromatography–mass-spectrometry (LC–MS) and MS–MS
The purified recombinant protein sample (200 µg) was loaded and resolved on 12% SDS-PAGE. After Coomassie staining, the protein band was excised from the gel and subjected to trypsin digestion. The digested sample was analysed in Bruker impact HD 1819696.00315 (ESI-QTime of Flight detector). Mascot software was used to analyse the LC–MS and MS–MS data.
Results
In silico analysis of l-asparaginase of Lactobacillus reuteri DSM 20016
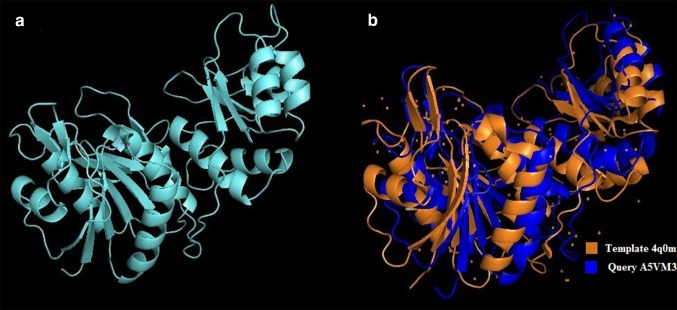
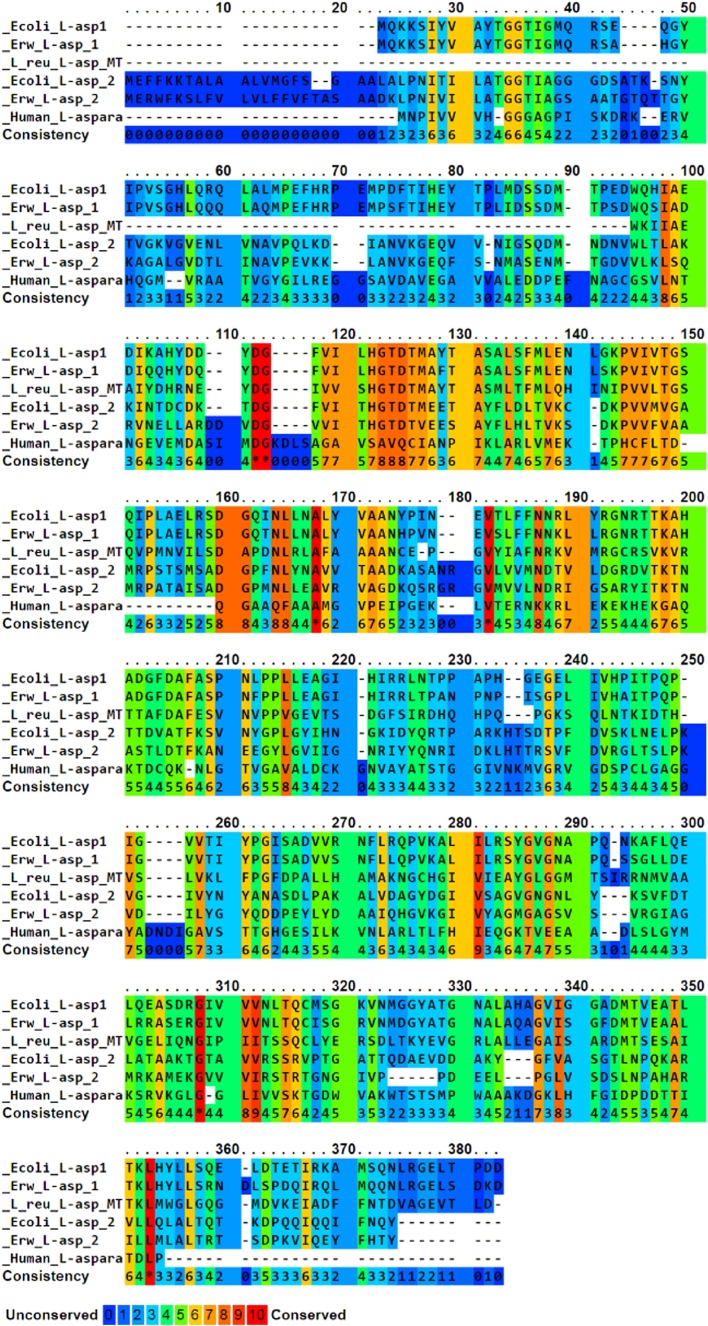
In silico analysis of the l-asparaginase gene from L. reuteri DSM 20016 showed that the protein sequence has 329 amino acids coding a protein for 35.7 kDa in the monomeric form. The theoretical pI of the protein is an important parameter of study as this value is important during protein purification and was found to be 4.99 for the l-asparaginase gene from L. reuteri DSM 20016. The instability index was computed to be 32.08 and this classifies the protein to be stable. The 3D structure of the l-asparaginase of L. reuteri DSM 20016 (A5VMR3) was predicted using Easy Modeller 4.0 with the crystal structure of l-asparaginase of Pyrococcus furiosus (sequence ID: 4Q0M_A) as template since the identity was 42% with the query sequence. Nine models were generated and validated using validation tools. The protein 3D structure (Fig. 1) with the highest validation score of 86.293 for ERRAT (Online Resource 1) and 92.10% for VERIFY 3D was chosen. The torsion angles phi and psi were analysed using Ramachandran Plot (Online Resource 2) to validate a stable conformation. The number of residues in the favoured region was 312 (95.4%), in allowed region 12 (3.7%) and 3 (0.9%) residues in the outlier region which approximate to 99% in the favoured region validating the model to be stable. The conserved sequences between the l-asparaginases of E. coli, E. chrysanthemi, human and L. reuteri DSM 20016 were studied using PRALINE multiple sequence alignment (Fig. 2).
Fig. 1.
Three-dimensional structure prediction of l-asparaginase from Lactobacillus reuteri DSM 20016 using homology modelling with Pyrococcus furiosus (PDB ID: 4Q0 M) sequence as a template. a 3D structure of Lactobacillus reuteri DSM 20016 l-asparaginase and b comparison of the 3D structure of the query Lactobacillus reuteri DSM 20016 l-asparaginase and template Pyrococcus furiosus
Fig. 2.
Multiple sequence alignment of l-asparaginases gene of Lactobacillus reuteri DSM 20016, E. coli, Erwinia chrysanthemi, Homo sapiens using PRALINE multiple sequence alignment tool; conserved regions are identified using the colour code provided
Cloning and expression of LREU_RS09880 gene in Escherichia coli
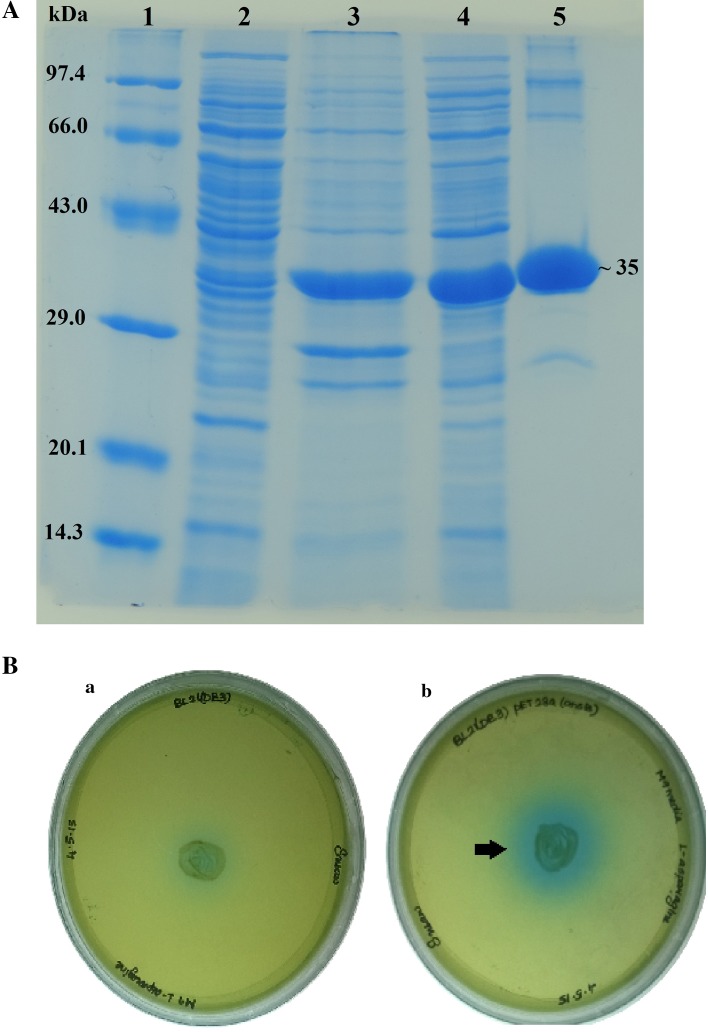
The ORF (LREU_RS09880) was 990 nucleotides in length and encodes a protein of 329 amino acids. The deduced protein sequence has a predicted monomer molecular weight of 35.7 kDa. The l-asparaginase gene from L. reuteri DSM 20016 was cloned in pET28a(+) vector using E. coli TOP 10. The recombinant plasmids were isolated and the insert sequence was confirmed using automated Sanger sequencing. The clone was subsequently transformed into BL21(DE3) competent cells. The protein was expressed and purified using HIS-Select® HF Nickel Affinity Gel column chromatography and the eluted fractions were analysed on SDS-PAGE. Dialysis was performed to remove the higher concentrations of imidazole and the integrity of the protein was checked by performing the enzyme assay and SDS-PAGE. The molecular weight of the purified protein corresponded to ~35 kDa (Fig. 3A). The results of the purification of l-asparaginase enzyme are given in Table 1.
Fig. 3.
A SDS PAGE analysis of the cell lysates. Lane 1: protein molecular weight marker medium range (GeNei™), lane 2: uninduced BL21(DE3)(pET28a(+):LREU_RS09880), lane 3: cell pellet after sonication of induced BL21(DE3)(pET28a(+):LREU_RS09880) with 0.05 mM IPTG for 4 h, lane 4: supernatant obtained after sonication of induced BL21(DE3)(pET28a(+):LREU_RS09880) with 0.05 mM IPTG for 4 h, lane 5: purified and dialysed recombinant l-asparaginase showing the molecular weight of ~35 kDa. B Detection of l-asparaginase production using modified M9 medium plate assay with BTB dye. a Plate inoculated with E. coli BL21(DE3). b Plate inoculated with E. coli BL21(DE3)(pET28a(+):LREU_RS09880)
Table 1.
Purification steps for recombinant l-asparaginase from Lactobacillus reuteri DSM 20016 overexpressed in E. coli
| Step | Total protein (mg/ml) | Total activity (U) | Specific activity (U/mg) | Recovery (%) | Purification fold |
|---|---|---|---|---|---|
| Crude | 1360 | 78.24 | 0.06 | 100 | 1 |
| Ni–NTA | 114.4 | 72.08 | 0.63 | 92 | 10.5 |
Plate assay
A green colour is formed at a neutral pH and dark blue zone in the hydrolysed region. The enzymatic activity of the recombinant protein was analysed by plate assay in which the hydrolysis of the substrate l-asparagine by the enzyme l-asparaginase was confirmed by distinct blue zone formation (Fig. 3B) around the recombinant E. coli BL21(DE3)(pET28a:LREU_RS09880) with respect to that of absence in E. coli BL21(DE3).
Temperature and pH profile
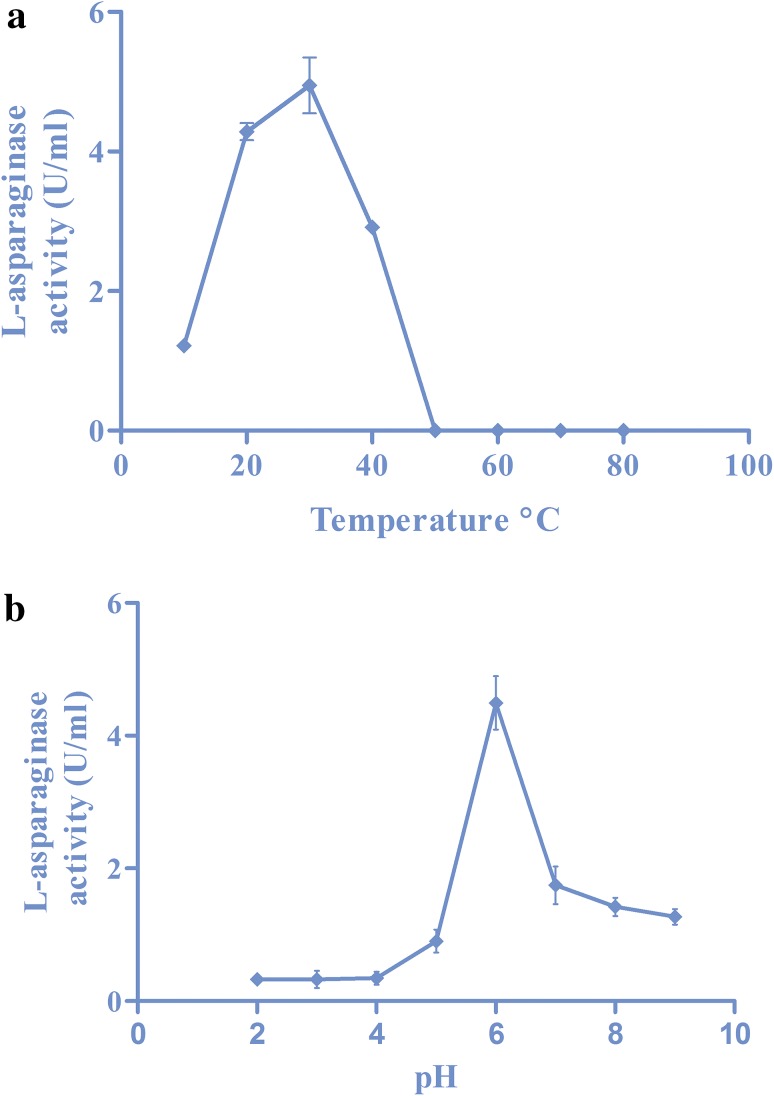
The purified recombinant l-asparaginase was subjected to various temperatures from 10 to 80 °C. The enzyme was active from 10 °C and the activity increased with corresponding increase in temperatures till 30 °C. There was a reduction in the activity after 40 °C and no activity thereafter (Fig. 4a). The effect of various pH on the recombinant enzyme was analysed and the purified enzyme showed maximum activity at pH 6 (Fig. 4b).
Fig. 4.
a The temperature profile of recombinant l-asparaginase enzyme. b pH profile of recombinant l-asparaginase enzyme
Thermostability
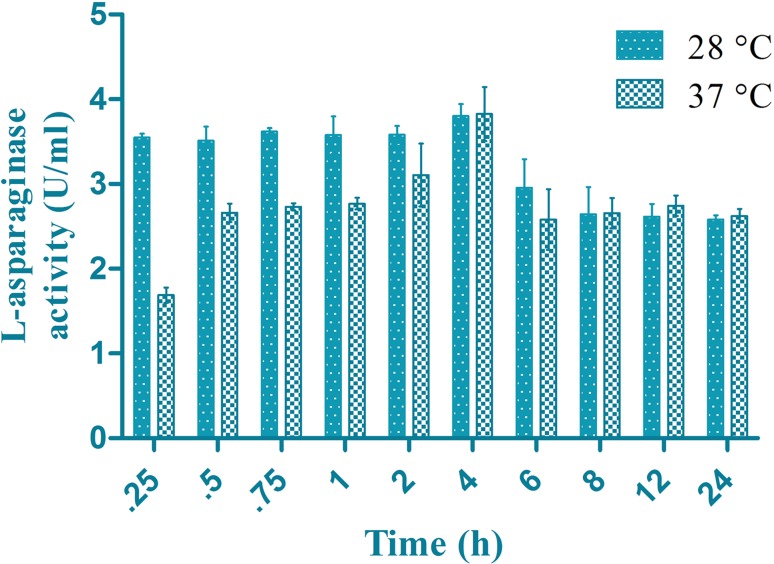
The enzyme retained maximum activity at 4 h at 28 °C and 37 °C (Fig. 5). The enzyme was found to be stable for longer hours displaying a good activity at 28 °C.
Fig. 5.
Thermal stability of the recombinant enzyme at 28 and 37 °C in time durations from 15, 30, 45 min, 1, 2, 4, 6, 8, 12 and 24 h
Determination of kinetic parameters
Lineweaver–Burk plot of the recombinant l-asparaginase enzyme (Online Resource 3) was derived using enzyme kinetics in GraphPad Prism 7.02 software. The K m, V max values were 0.3332 mM and 14.06 mM/min, respectively.
Liquid chromatography–mass-spectrometry (LC–MS) and MS–MS analysis
The purified protein band was subjected to trypsin digestion and was analysed using LC–MS and MS–MS. The protein peaks were distinct stating the proper trypsin digestion of the protein (Fig. 6). The molecular weight was found to be 35,910 Da in agreement with the SDS PAGE results. The obtained mass results were analysed using Mascot database and the protein sequences were retrieved and assembled which confirmed the source organism L. reuteri DSM 20016.
Fig. 6.
Total ion chromatogram of LC-MS/MS of recombinant L-asparaginase. Protein was resolved on a 12% SDS PAGE excised and digested with trypsin. Protein was dissolved in 0.1% Formic acid and the chromatogram was obtained from the LC-MS instrument. The separation of digested protein as peaks in respective time (min), mass to charge ratio obtained for the peaks analysed using the MS instrument
Discussion
Substantiating the in silico analysis of the whole genome sequence of L. reuteri DSM 20016 revealed the presence of a 990-bp-long gene sequence coding for l-asparaginase. The putative LREU_RS09880 was cloned into E. coli expression system and purified to homogeneity using Ni–NTA chromatography. The overall GC content is about 42.72 which is similar to that of the chromosomes of Lactobacillus species (Morita and Toh 2008). A comparison table (Online Resource 4) highlights certain properties of l-asparaginase from other microbial sources. The comparison reveals that certain elemental properties of the recombinant l-asparaginase from L. reuteri DSM 20016 are similar to l-asparaginases from previous reports.
The in silico analysis of the gene sequence of l-asparaginase from L. reuteri DSM 20016 used in the prediction of physico-chemical parameters states that the protein is stable. The homology modelling of the l-asparaginase from L. reuteri DSM 20016 uses l-asparaginase of P. furiosus as a template similar to the cases of E. coli (ErAIII) l-asparaginase for modelling B. subtilis (BsAII) l-asparaginase (Onishi et al. 2011) and human asparaginase-like protein (Gunda et al. 2016). The structures were validated using Ramachandran plot, ERRAT and VERIFY 3D in the case of l-asparaginase from L. reuteri DSM 20016 as three validation tools are required to validate a structure. In multiple sequence alignment, certain amino acids were found to be conserved in all the l-asparaginases of aligned species. The amino acids Aspartic acid, Glycine, Alanine and Valine were highly conserved in l-asparaginases of E. coli, Erwinia, L. reuteri DSM 20016 and humans.
The Ni–NTA chromatography employed in this study results in 92% recovery with 10.5-fold purification. Sudhir et al. 2014 report a 52% recovery with 3.02 purification fold, whereas 40.9 and 19.1% in DEAE-Toyopearl and affinity chromatography, respectively (Yano et al. 2008), 49.29% in Anion exchange (DEAE cellulose) and 32.95% in Gel filtration (Sephadex G-150) as reported by Mahajan et al. 2014. The purification fold is not always corresponding to yield percentage as in the case of having a purification fold of 2.79 with the yield being 40.9% while using DEAE-Toyopearl and 4.55-fold purification with the yield of 19.1% (Yano et al. 2008). These results are resembling 18.09- and 30.17-fold purification with 49.28 and 32.95% yield, respectively, for Anion exchange (DEAE cellulose) and Gel filtration (Sephadex G-150) (Mahajan et al. 2014). Hence, Ni–NTA chromatography is the better choice for purification with higher yield.
The formation of blue colour around the inoculum is indicated as a reduction in pH in the Modified M9 medium. The usage of BTB over phenol red gives a vivid description of the zone formation also as reported in B. licheniformis and Aspergillus niger (Mahajan et al. 2013). The recombinant l-asparaginase maintained maximum activity within 30–40 °C and pH 6 which makes it appropriate to be used as a chemotherapy drug as it is active at human physiological temperature. These results are in agreement with l-asparaginases from other producers like B. licheniformis MTCC 429 (Sudhir et al. 2014), E. coli MTCC 739 (Vidya et al. 2011), Pencillium sp. (Soniyambi et al. 2011) and B. licheniformis (Mahajan et al. 2014). The K m value for other asparaginases were comparatively similar to other l-asparaginase producers like 0.67 mM for B. licheniformis MTCC 429, 3 mM for Erwinia aroideae, 2.5 mM for Corynebacterium glutamicum, 1 mM for Cylindrocarpon obtusisporum, 2.8 mM for Thermus thermophilus, 2.06 mM for B. subtilis BsAIIS40 Ma and 7.06 mM for B. subtilis BsAIID49 Ma (Sudhir et al. 2014). The recombinant enzyme showed maximum activity in 4 h at 28 and 37 °C. The activity was retained to a level of 3.5 IU till 4 h and maintained an activity of 2.5 IU till 24 h at 28 °C which makes the storage effortless. The enzyme shows increasing activity at 37 °C with a highest activity in 4 h and activity drops thereafter. These results are very similar to l-asparaginase from Pseudomonas fluorescens (Kishore et al. 2015) where the enzyme shows maximum activity at 4 h incubation period.
The LC–MS/MS and SDS PAGE analysis of the purified recombinant l-asparaginase states that the protein has a molecular weight of ~35 kDa as a monomer. These results are similar to l-asparaginase from Helicobacter pylori where the protein sample as a monomer was 37 kDa (Scotti et al. 2010), 36 kDa from B. subtilis (Yano et al. 2008) and 36.7 kDa in E. coli (Upadhyay et al. 2014).
Conclusion
The ans (LREU_RS09880) gene coding for l-asparaginase from L. reuteri DSM 20016 was cloned and overexpressed in E. coli expression system. The in silico analysis of the gene gives a vivid description of the theoretical pI, stability index, 3D structure and the sequence similarity between other asparaginases. The identity of the recombinant enzyme was confirmed by the activity in Nesslerisation enzyme assay. The recombinant enzyme was further characterized for temperature, pH, thermostability and enzyme kinetics. The molecular weight of the recombinant enzyme was determined using SDS PAGE and LC–MS/MS.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Authors thank the SRM-DBT Partnership Platform for Contemporary Research Services and Skill Development in Advanced Life Sciences Technologies (No. BT/PR12987/INF/22/205/2015) for providing the LC–MS/MS facility.
Author contributions
Study conception and design: Dr. KNR. Performed the experiments: SSA. Analysed the data: Dr. KNR, Dr. SI, SSA. Contributed reagents/materials/analysis tools: Dr. SI, KVL. Wrote the manuscript: SSA.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-017-0974-4) contains supplementary material, which is available to authorized users.
Contributor Information
Suresh Susan Aishwarya, Email: susanaishwarya@gmail.com.
Sellamuthu Iyappan, Email: iyappansbt@gmail.com.
Kamepali Vijaya Lakshmi, Email: kamepali.sriviji@gmail.com.
Kandathil Narayanan Rajnish, Phone: 044 - 27417817, Email: knrmku@gmail.com.
References
- Amer MN, Mansour NM, El-Diwany AI, Dawoud IE, Rashad FM. Isolation of probiotic Lactobacillus strains harbouring l-asparaginase and arginine deiminase genes from human infant feces for their potential application in cancer prevention. Ann Microbiol. 2013;63:1121–1129. doi: 10.1007/s13213-012-0569-6. [DOI] [Google Scholar]
- Bodmer M, Sulz M, Stadlmann S, Droll A, Terracciano L, Krahenbuhl S. Fatal liver failure in an adult patient with acute lymphoblastic leukemia following treatment with l-asparaginase. Digestion. 2006;74(1):28–32. doi: 10.1159/000095827. [DOI] [PubMed] [Google Scholar]
- Cachumba JJM, Antunes FAF, Peres GFD, Brumano LP, Santos JCD, Da Silva SS. Current applications and different approaches for microbial l-asparaginase production. Braz J Microbiol. 2016;47S:77–85. doi: 10.1016/j.bjm.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H-R, Jiang N. Extremely rapid extraction of DNA from bacteria and yeasts. Biotechnol Lett. 2006;28:55–59. doi: 10.1007/s10529-005-4688-z. [DOI] [PubMed] [Google Scholar]
- Costa IM, et al. Recombinant l-asparaginase 1 from Saccharomyces cerevisiae: an allosteric enzyme with antineoplastic activity. Sci Rep. 2016;6:36239. doi: 10.1038/srep36239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria K, Kumar DS. Isolation and screening of L-asparaginase free from glutaminase and urea from fungal sp. 3 Biotech. 2016;6:239. doi: 10.1007/s13205-016-0544-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Naggar NE, et al. Purification, characterization, cytotoxicity and anticancer activities of l-asparaginase, anti-colon cancer protein, from the newly isolated alkaliphilic Streptomyces fradiae NEAE-82. Sci Rep. 2016;6:32926. doi: 10.1038/srep32926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunda SK, Rudroju SC, Bandi S, Bodhankar R. Homology modelling of human asparaginase like protein and its molecular interaction studies with natural flavanoids. Int J Pharm Sci Res. 2016;7(1):287–289. [Google Scholar]
- Huang L, Liu Y, Sun Y, Yan Q, Jiang Z. Biochemical characterization of a novel l-asparaginase with low glutaminase activity from Rhizomucor miehei and its application in food safety and leukemia treatment. Appl Environ Microbiol. 2014;80(5):1561–1569. doi: 10.1128/AEM.03523-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imada A, Igarasi S, Nakahama K, Isono M. Asparaginase and glutaminase activities of micro-organisms. J Gen Microbiol. 1973;76:85–99. doi: 10.1099/00221287-76-1-85. [DOI] [PubMed] [Google Scholar]
- Kishore V, Nishita KP, Manonmani HK. Cloning, expression and characterization of l-asparaginase from Pseudomonas fluorescens for large scale production in E. coli BL21. 3 Biotech. 2015;5:975–981. doi: 10.1007/s13205-015-0300-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar NSM, Manonmani HK. Purification, characterization and kinetic properties of extracellular l-asparaginase produced by Cladosporium sp. World J Microbiol Biotechnol. 2013;29:577–587. doi: 10.1007/s11274-012-1213-0. [DOI] [PubMed] [Google Scholar]
- Kumari PVK, Sankar GG, Prabhakar T, Lakshmi SS. Purification and characterization of l-asparaginase from Streptomyces griseoluteus WS3/1. Int J Pharm Sci Rev Res. 2013;23(2):198–202. [Google Scholar]
- Mahajan RV, Saran S, Saxena RK, Srivastava AK. A rapid, efficient and sensitive plate assay for detection and screening of l-asparaginase-producing microorganisms. FEMS Microbiol Lett. 2013;341:122–126. doi: 10.1111/1574-6968.12100. [DOI] [PubMed] [Google Scholar]
- Mahajan RV, Kumar V, Rajendran V, Saran S, Ghosh PC, et al. Purification and characterization of a novel and robust l-asparaginase having low glutaminase activity from Bacillus licheniformis: in vitro evaluation of anti-cancerous properties. PLoS One. 2014;9(6):e99037. doi: 10.1371/journal.pone.0099037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meena B, et al. l-Asparaginase from Streptomyces griseus NIOT-VKMA29 optimization of process variables using factorial designs and molecular characterization of l-asparaginase gene. Sci Rep. 2015;5:12404. doi: 10.1038/srep12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita H, Toh H. Comparative genome analysis of Lactobacillus reuteri and Lactobacillus fermentum reveal a genomic island for reuterin and cobalamin production. DNA Res. 2008;12:151–161. doi: 10.1093/dnares/dsn009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi Y, Yano S, Thongsanit J, Takagi K, Yoshimune K, Wakayama M. Expression in Escherichia coli of a gene encoding type II l-asparaginase from Bacillus subtilis and characterization of its unique properties. Ann Microbiol. 2011;61:517–524. doi: 10.1007/s13213-010-0167-4. [DOI] [Google Scholar]
- Onishi Y, Prihanto AA, Yano S, Takagi K, Umekawa M, Wakayama M. Effective treatment for suppression of acrylamide formation in fried potato chips using l-asparaginase from Bacillus subtilis. 3 Biotech. 2015;5:783–789. doi: 10.1007/s13205-015-0278-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja RA, Schmiegelow K, Albertson BA, Prunsild K, et al. Asparaginase-associated pancreatitis in children with acute lymphoblastic leukemia in the NOPHO ALL2008 protocol. Br J Haematol. 2014;165:126–133. doi: 10.1111/bjh.12733. [DOI] [PubMed] [Google Scholar]
- Rajnish KN, Choudhary GMK, Gunasekaran P. Functional characterization of a putative endoglucanase gene in the genome of Zymomonas mobilis. Biotechnol Lett. 2008;30:1461–1467. doi: 10.1007/s10529-008-9716-3. [DOI] [PubMed] [Google Scholar]
- Sanghvi G, Bhimani K, Vaishnav D, Oza T, Dave G, Kunjadia P, Sheth N. Mitigation of acrylamide by l-asparaginase from Bacillus subtilis KDPS1 and analysis of degradation products by HPLC and HPTLC. SpringerPlus. 2016;5:533. doi: 10.1186/s40064-016-2159-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti C, Sommi P, Pasquetto MV, Cappelletti D, Stivala S, et al. Cell-cycle inhibition by helicobacter pylori l-asparaginase. PLoS One. 2010;5(11):e13892. doi: 10.1371/journal.pone.0013892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha RK, Singh HR, Jha SK. Production, purification and kinetic characterization of l-asparaginase from Pseudomonas fluorescens. Int J Pharm Pharm Sci. 2015;7(1):135–138. [Google Scholar]
- Soniyambi AR, Lalitha S, Praveesh BV, Priyadarshini V. Isolation, production and anti-tumor activity of l-asparaginase of Pencillium sp. Int J Microbiol Res. 2011;2(1):38–42. [Google Scholar]
- Sudhir AP, Dave BR, Prajapati AS, Panchal K, Patel D, Subramanian RB. Characterization of a recombinant glutaminase-free l-asparaginase (ansA3) enzyme with high catalytic activity from Bacillus licheniformis. Appl Biochem Biotechnol. 2014;174:2504–2515. doi: 10.1007/s12010-014-1200-z. [DOI] [PubMed] [Google Scholar]
- Upadhyay AK, Singh A, Mukherjee KJ, Panda AK. Refolding and purification of recombinant l-asparaginase from inclusion bodies of E. coli into active tetrameric protein. Front Micobiol. 2014;486:1–10. doi: 10.3389/fmicb.2014.00486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidya J, Vasudevan UM, Soccol CR, Pandey A. Cloning, functional expression and characterization of l-asparaginase II from E. coli MTCC739. Food Technol Biotechnol. 2011;49(3):286–290. [Google Scholar]
- Yano S, Minato R, Thongsanit J, Tachiki T, Wakayama M. Overexpression of type I l-asparaginase of Bacillus subtilis in Escherichia coli, rapid purification and characterization of recombinant type I l-asparaginase. Ann Microbiol. 2008;58(4):711–716. doi: 10.1007/BF03175579. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.