Abstract
Although antimicrobial peptides (AMPs) have been used as feed additives, only a few studies have examined their use in ruminants. In this study, we evaluated the use of AMPs(recombinant swine defensin and a fly antibacterial peptide were mixed by 1:1) as a medicated feed additive for juvenile goats. Dietary treatments included control groups (group I: 300 g concentrate; group III: 600 g concentrate), and AMP-supplemented groups (group II: 300 g concentrate + 3.0 g AMPs; group IV: 600 g concentrate + 3.0 g AMPs). AMP-treated groups exhibited an increase in bacterial genera, including Fibrobacter, Anaerovibrio, and Succiniclasticum, and the ciliate genus Ophryoscolex; as well a reduction in bacterial genera, such as Selenomonas, Succinivibrio, and Treponema, and the ciliate genera Polyplastron, Entodinium, and Isotricha. The changes in Fibrobacter, Anaerovibrio, Ophryoscolex, Polyplastron, Entodinium, and Isotricha were related to the concentrate. AMP treatment led to increased body weight, average daily weight gain, enzymatic activity (pectinase, xylanase, and lipase), especially in the normal concentrate group, and influence on ruminal fermentation function. In addition, goats treated with AMPs had higher rumen microorganism diversity indices than the control groups. Our results demonstrate that AMPs can be utilized as feed additives for juvenile goats.
Introduction
The microbial environment in the rumen is quite complex and dynamic; this is due to several factors including type of diet1,2. The microbial community consists of bacteria (1010–1011 cells/mL), methanogenic archaea (107–109 cells/mL), ciliate protozoa (104–106 cells/mL), anaerobic fungi (103–106 cells/mL), and bacteriophages (109–1010 particles/mL) present3. A major function of the microbiome is to ferment plant materials that can be ingested by ruminant animals4–6. Rumen regulation is one of the most important methods for improving feed efficiency, ruminant health, and ruminant livestock production performance. Several antibiotic compounds, such as monensin, hainanmycin, and virginiamycin, have been used to improve ruminal fermentation and the efficiency of nutrient utilization7–9. However, the overuse of antibiotics has raised concerns regarding product safety and environmental health, therefore, the use of antibiotics as animal feed additives has been banned in the European Union (European Union, 2003).
Antimicrobial peptides (AMPs) are widespread in bacteria, animals, and plants and provide opportunities for novel research. In addition to antimicrobial properties10, previous studies have demonstrated antifungal11, antiviral12, anti-parasitic13, and antitumor activities14. AMP-induced immunoregulatory and antioxidant activities have been shown to be mediated by cationic charge, amphipathicity, amino acid composition, and structure15. AMPs have also been demonstrated to improve performance, nutrient retention, and intestinal morphology, and to reduce the incidence of diarrhoea in livestock animals16–19. Peng et al.20 demonstrated that dietary supplementation with crude rpBD2 (recombinant porcine β-defensin 2) has beneficial effects on growth and intestinal morphology of weaned piglets, reducing the incidence of post-weaning diarrhoea and the numbers of potential pathogens in the caecum. AMPs could therefore serve as potential alternatives to antibiotics in livestock production. However, there is insufficient information on the effects of AMPs on rumen digestion, as only a limited number of inconclusive studies have examined the use of AMPs as alternatives to feed antibiotics and growth promoters in ruminant nutrition. Previous studies in our laboratory have shown that adding AMPs (composed of recombinant swine defensin and a fly antibacterial peptide at a blending ratio of 50:50) in feed can improve growth and immunity of weaned piglets15. Based on our previous findings and the reported bactericidal effects of AMPs, we hypothesized that dietary AMP supplementation could affect rumen microbiology, and therefore ruminal fermentation. In the present study, we investigated the effects of AMPs on rumen fermentation function and rumen microbial community structure in Chuanzhong black goats.
Results
Growth performance
The mean initial body weights in groups I, II, III, and IV were 15.54 kg, 15.51 kg, 16.31 kg, and 16.70 kg, respectively. The weights increased to 18.96 kg, 19.93 kg, 21.60 kg, and 22.99 kg, respectively, following 60 days of experimental feeding (Table 1 and Fig. 1). The average daily gain (g) was significantly higher (P < 0.05) in the AMP-supplemented groups (II, IV) than in the control groups (I, III; Table 2).
Table 1.
Changes in goat body weight and average daily gain.
| Item | Time point(day)/Time range | Groups | P-Value | ||||||
|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | I VS II | III VS IV | I VS III | II VS IV | ||
| weight (kg) | 0d | 15.54 ± 0.21 | 15.51 ± 0.84 | 16.31 ± 1.96 | 16.70 ± 0.97 | 0.890 | 0.698 | 0.465 | 0.067 |
| 20d | 16.91 ± 0.18 | 17.49 ± 0.59 | 18.13 ± 0.49 | 19.23 ± 0.61 | 0.167 | 0.099 | 0.011a | 0.024a | |
| 60d | 18.96 ± 0.19 | 19.93 ± 0.18 | 21.60 ± 0.77 | 22.99 ± 0.72 | 0.003a | 0.094 | 0.005a | 0.004a | |
| average daily gain (g/d) | 0d-20d | 68.50 ± 7.45 | 90.63 ± 3.15 | 99.88 ± 5.81 | 126.26 ± 37.50 | 0.024a | 0.280 | 0.010a | 0.163 |
| 20d-60d | 51.38 ± 6.19 | 86.88 ± 8.26 | 61.06 ± 5.72 | 94.06 ± 19.04 | 0.002a | 0.033a | 0.197 | 0.639 | |
| 0d-60d | 57.08 ± 1.89 | 88.12 ± 6.25 | 74.00 ± 3.37 | 104.79 ± 13.55 | 0.002a | 0.009a | 0.007a | 0.168 | |
aMean significant difference (P < 0.05).
Figure 1.
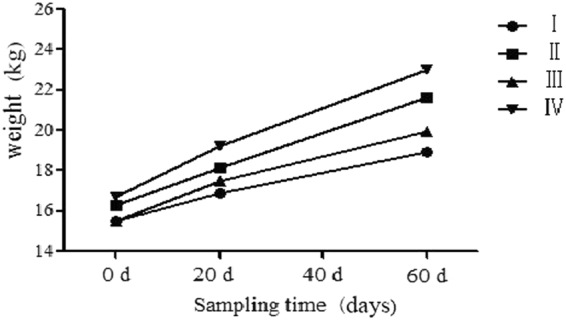
Weight changes in goats per group.
Table 2.
Changes in ruminal fermentation parameters in goat rumen fluid.
| Parameter | Groups | P-Value | |||||||
|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | I VS II | III VS IV | I VS III | II VS IV | ||
| pH | 0d | 6.89 ± 0.03 | 6.88 ± 0.03 | 6.88 ± 0.03 | 6.87 ± 0.04 | 0.215 | 0.893 | 0.504 | 0.714 |
| 20d | 6.87 ± 0.05 | 6.82 ± 0.04 | 6.78 ± 0.04 | 6.74 ± 0.01 | 0.48 | 0.054 | 0.006a | 0.119 | |
| 60d | 6.95 ± 0.01 | 6.92 ± 0.06 | 6.85 ± 0.27 | 6.79 ± 0.03 | 0.49 | 0.659 | 0.515 | 0.024 | |
| Ammonia (mg/100 mL) | 0d | 11.19 ± 0.21 | 11.19 ± 0.36 | 11.01 ± 0.24 | 11.06 ± 0.22 | 0.99 | 0.067 | 0.256 | 0.334 |
| 20d | 9.55 ± 0.29 | 8.86 ± 0.31 | 12.53 ± 0.42 | 11.95 ± 0.37 | 0.016a | 0.189 | 0.003a | 0.003a | |
| 60d | 10.32 ± 0.15 | 9.64 ± 0.33 | 17.79 ± 0.86 | 16.49 ± 0.51 | 0.008a | 0.108 | 0.001a | 0.001a | |
| T-VFA (mmol/L) | 0d | 75.59 ± 1.27 | 75.48 ± 0.89 | 75.28 ± 1.39 | 75.33 ± 1.18 | 0.801 | 0.916 | 0.591 | 0.734 |
| 20d | 69.22 ± 1.25 | 88.83 ± 2.13 | 86.97 ± 2.27 | 72.67 ± 2.23 | 0.001a | 0.001a | 0.001a | 0.002a | |
| 60d | 63.97 ± 1.52 | 68.83 ± 1.85 | 69.03 ± 2.20 | 62.57 ± 1.68 | 0.06 | 0.007a | 0.001a | 0.043a | |
| Acetate (mmol/l) | 0d | 51.70 ± 1.60 | 51.61 ± 0.91 | 51.46 ± 1.59 | 51.31 ± 1.52 | 0.849 | 0.76 | 0.73 | 0.719 |
| 20d | 46.21 ± 1.50 | 61.12 ± 1.99 | 61.52 ± 2.25 | 48.58 ± 1.75 | 0.004a | 0.001a | 0.001a | 0.002a | |
| 60d | 41.94 ± 1.36 | 45.51 ± 2.07 | 46.91 ± 1.78 | 40.14 ± 1.54 | 0.096 | 0.002a | 0.003a | 0.064 | |
| Propionate (mmol/l) | 0d | 15.30 ± 0.40 | 15.27 ± 0.20 | 15.22 ± 0.34 | 15.41 ± 0.41 | 0.909 | 0.354 | 0.671 | 0.573 |
| 20d | 14.38 ± 0.48 | 19.05 ± 0.85 | 15.26 ± 0.71 | 15.47 ± 0.88 | 0.003a | 0.52 | 0.003a | 0.02a | |
| 60d | 13.24 ± 0.47 | 14.43 ± 0.73 | 12.88 ± 0.58 | 13.46 ± 0.60 | 0.099 | 0.374 | 0.523 | 0.039a | |
| Butyrate (mmol/l) | 0d | 8.60 ± 0.15 | 8.59 ± 0.14 | 8.60 ± 0.19 | 8.61 ± 0.23 | 0.945 | 0.782 | 0.885 | 0.926 |
| 20d | 8.64 ± 0.44 | 8.67 ± 0.30 | 10.20 ± 0.58 | 8.61 ± 0.53 | 0.838 | 0.021a | 0.033a | 0.863 | |
| 60d | 8.79 ± 0.34 | 8.88 ± 0.52 | 9.24 ± 0.22 | 8.98 ± 0.17 | 0.507 | 0.124 | 0.108 | 0.711 | |
| Acetate + Butyrate to Propionate ratio | 0d | 3.95 ± 0.20 | 3.94 ± 0.09 | 3.95 ± 0.17 | 3.89 ± 0.19 | 0.906 | 0.195 | 0.973 | 0.634 |
| 20d | 3.82 ± 0.17 | 3.67 ± 0.24 | 4.71 ± 0.26 | 3.71 ± 0.24 | 0.512 | 0.002a | 0.002a | 0.887 | |
| 60d | 3.84 ± 0.21 | 3.78 ± 0.23 | 4.36 ± 0.15 | 3.65 ± 0.23 | 0.672 | 0.026a | 0.069 | 0.238 | |
| Urea nitrogen (mg/mL) | 0d | 1.78 ± 0.03 | 1.74 ± 0.08 | 1.79 ± 0.06 | 1.77 ± 0.04 | 0.229 | 0.432 | 0.924 | 0.409 |
| 20d | 1.96 ± 0.17 | 1.77 ± 0.22 | 3.07 ± 0.61 | 2.09 ± 0.38 | 0.215 | 0.037a | 0.05 | 0.304 | |
| 60d | 2.59 ± 0.28 | 2.55 ± 0.27 | 3.83 ± 0.66 | 2.63 ± 0.58 | 0.898 | 0.116 | 0.081 | 0.766 | |
| MCP (mg/mL) | 0d | 1.30 ± 0.07 | 1.31 ± 0.04 | 1.33 ± 0.05 | 1.33 ± 0.04 | 0.88 | 0.486 | 0.607 | 0.221 |
| 20d | 1.35 ± 0.03 | 1.37 ± 0.09 | 2.31 ± 0.48 | 2.39 ± 0.09 | 0.613 | 0.794 | 0.025a | 0.001a | |
| 60d | 1.33 ± 0.02 | 1.35 ± 0.04 | 3.04 ± 0.33 | 3.60 ± 0.26 | 0.67 | 0.003a | 0.002a | 0.001a | |
aMean significant difference (P < 0.05).
Ruminal fermentation function
The mean ruminal pH of samples from AMP-treated goats ranged from 6.74 to 6.92, which is within the normal physiological range. No significant difference in ruminal pH was observed between AMP-treated groups and control group (P > 0.05; Table 2).
Total volatile fatty acid (T-VFA) and acetate concentrations increased in goats fed AMPs with normal concentrate (significant difference on day 20), but decreased in goats fed double concentrate compared to the control groups (I, III; P < 0.05). The concentrations of ammonia (significant difference with normal concentrate) and urea nitrogen decreased in AMP-treated groups. The acetate + butyrate-to-propionate ratio decreased in AMP-treated groups; however, significant differences were only observed with double concentrate (P < 0.05). The concentrations of microbial protein (MCP, significant difference on day 60 with double concentrate) and propionate (significant difference on day 20 with normal concentrate) increased in AMP-treated groups.
In addition, all indicators (except ruminal pH and urea nitrogen) were elevated in AMP-treated groups compared with groups I and III; the concentrations of T-VFA, acetate, ammonia, and MCP were significantly increased (P < 0.05). Similarly, the concentrations of propionate, butyrate, and the acetate + butyrate-to-propionate ratio were significantly increased on day 20 (P < 0.05) with double concentrate.
Enzyme activity
Pectinase activity appeared to increase in the AMP-supplemented groups (Table 3), and was higher in AMP-supplemented goats than in the control groups (I, III; P < 0.05, except on day 60 with double concentrate). Changes in xylanase, lipase, and amylase activity were associated with concentrate. Xylanase increased with normal concentrate (P < 0.05) and decreased with double concentrate; lipase increased with normal concentrate (P < 0.05) but did not change with double concentrate; and amylase decreased with normal concentrate (P < 0.05) but did not change with double concentrate. No differences in β-glucosidase, carboxymethyl cellulase (CMCase), and protease activity could be detected between AMP-treated and control animals (P > 0.05).
Table 3.
Changes of the activity of enzymes in rumen fluid of goats.
| Parameter | Groups | P-Value | |||||||
|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | I VS II | III VS IV | I VS III | II VS IV | ||
| CMCase (U/mL) | 0d | 74.39 ± 1.87 | 73.50 ± 1.50 | 73.13 ± 1.39 | 73.20 ± 1.38 | 0.54 | 0.79 | 0.453 | 0.824 |
| 20d | 85.89 ± 2.11 | 81.03 ± 2.51 | 89.38 ± 3.21 | 88.15 ± 1.45 | 0.15 | 0.626 | 0.099 | 0.016a | |
| 60d | 112.06 ± 3.33 | 109.07 ± 3.22 | 67.97 ± 2.10 | 71.97 ± 2.43 | 0.266 | 0.186 | 0.001a | 0.002a | |
| Xylanase (U/mL) | 0d | 10.03 ± 0.33 | 10.02 ± 0.41 | 10.24 ± 0.47 | 10.20 ± 0.35 | 0.975 | 0.701 | 0.424 | 0.216 |
| 20d | 14.56 ± 0.40 | 18.14 ± 1.27 | 15.74 ± 1.97 | 11.27 ± 1.15 | 0.016a | 0.019a | 0.246 | 0.013a | |
| 60d | 21.25 ± 0.64 | 34.57 ± 2.35 | 26.73 ± 4.34 | 19.94 ± 1.19 | 0.001a | 0.084 | 0.073 | 0.005a | |
| Pectinase (U /mL) | 0d | 45.51 ± 3.01 | 45.15 ± 2.14 | 45.14 ± 1.65 | 45.16 ± 1.71 | 0.728 | 0.684 | 0.829 | 0.992 |
| 20d | 37.42 ± 4.56 | 60.04 ± 1.87 | 44.23 ± 2.70 | 47.36 ± 1.34 | 0.002a | 0.033a | 0.013a | 0.001a | |
| 60d | 17.19 ± 2.57 | 26.69 ± 0.53 | 20.13 ± 2.49 | 21.01 ± 2.25 | 0.016a | 0.433 | 0.275 | 0.013a | |
| β-glucosidase (U/ mL) | 0d | 72.62 ± 3.31 | 72.52 ± 3.23 | 71.90 ± 2.48 | 72.05 ± 2.40 | 0.954 | 0.75 | 0.68 | 0.407 |
| 20d | 68.90 ± 4.03 | 62.40 ± 2.67 | 60.79 ± 2.69 | 66.18 ± 3.98 | 0.168 | 0.177 | 0.005a | 0.006a | |
| 60d | 59.89 ± 0.49 | 55.97 ± 2.79 | 50.82 ± 3.54 | 59.33 ± 3.61 | 0.13 | 0.051 | 0.016a | 0.004a | |
| Protease (µg /min.mL−1) | 0d | 3.25 ± 0.80 | 3.14 ± 0.35 | 3.15 ± 0.30 | 3.17 ± 0.28 | 0.8 | 0.571 | 0.827 | 0.58 |
| 20d | 3.28 ± 0.66 | 3.18 ± 0.26 | 2.74 ± 0.62 | 3.20 ± 0.35 | 0.775 | 0.095 | 0.244 | 0.957 | |
| 60d | 4.49 ± 0.43 | 4.42 ± 0.16 | 4.16 ± 0.32 | 4.72 ± 0.48 | 0.839 | 0.164 | 0.471 | 0.411 | |
| Amylase (U/dL) | 0d | 20.92 ± 0.78 | 20.89 ± 0.37 | 20.76 ± 1.17 | 20.78 ± 0.68 | 0.962 | 0.967 | 0.854 | 0.778 |
| 20d | 24.88 ± 0.33 | 21.17 ± 1.50 | 25.59 ± 0.83 | 25.92 ± 0.61 | 0.029a | 0.244 | 0.25 | 0.024a | |
| 60d | 27.62 ± 0.59 | 25.02 ± 0.58 | 26.26 ± 1.14 | 27.71 ± 1.04 | 0.006a | 0.163 | 0.145 | 0.033a | |
| Lipase (U/ L) | 0d | 19.24 ± 1.69 | 18.99 ± 1.09 | 19.92 ± 1.40 | 19.85 ± 1.38 | 0.862 | 0.861 | 0.316 | 0.529 |
| 20d | 18.81 ± 1.12 | 23.05 ± 1.36 | 18.25 ± 2.48 | 18.76 ± 0.69 | 0.037a | 0.634 | 0.742 | 0.007a | |
| 60d | 21.13 ± 2.32 | 30.50 ± 3.37 | 32.42 ± 4.18 | 33.38 ± 3.82 | 0.041a | 0.713 | 0.014a | 0.468 | |
aMean significant difference (P < 0.05).
In addition, β-glucosidase and CMCase (except on day 20) activities appeared to be significantly lower in group III compared to group I (P < 0.05); whereas pectinase (except on day 60) and lipase (except on day 20) activities appeared to be significantly higher in group III compared to group I (P < 0.05). No differences in xylanase, amylase, and protease activity could be detected between groups I and III (P > 0.05).
Rumen microorganisms
Bacterial community structure
Following the removal of low-quality reads from sequencing data, we obtained 1,786,781 total reads for bacteria, with an average of 49,632 reads per sample. The identified bacterial phyla and genera are detailed in Tables 4 and 5 and their respective community compositions are detailed in Supplementary Fig. S1A and B. Bacteroidetes was the dominant bacterial phylum in all goat rumen samples (expect in group III), accounting on average for 40.85% of the bacterial community. The next seven most abundant phyla were Firmicutes, Proteobacteria, Verrucomicrobia, Fibrobacteres, Tenericutes, Spirochaetes, and Cyanobacteria.
Table 4.
Influence of AMPs on proportion of different bacterial phyla.
| Bacterial phylum | Groups | P-value | |||||||
|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | I VS II | III VS IV | I VS III | II VS IV | ||
| Bacteroidetes | 0d | 36.00 ± 2.07 | 36.15 ± 5.15 | 36.20 ± 4.35 | 35.93 ± 3.71 | 0.964 | 0.638 | 0.939 | 0.828 |
| 20d | 40.87 ± 2.19 | 43.68 ± 3.53 | 34.30 ± 3.67 | 38.52 ± 2.95 | 0.483 | 0.287 | 0.026a | 0.105 | |
| 60d | 47.12 ± 1.10 | 52.77 ± 4.33 | 33.57 ± 2.66 | 35.94 ± 3.72 | 0.213 | 0.585 | 0.023a | 0.065 | |
| Firmicutes | 0d | 27.02 ± 5.86 | 28.08 ± 2.58 | 27.79 ± 4.10 | 27.57 ± 3.68 | 0.645 | 0.735 | 0.732 | 0.537 |
| 20d | 27.19 ± 1.77 | 29.65 ± 3.32 | 35.29 ± 1.53 | 31.91 ± 1.98 | 0.387 | 0.058 | 0.016a | 0.474 | |
| 60d | 18.05 ± 1.07 | 22.70 ± 1.70 | 33.72 ± 3.06 | 26.76 ± 3.56 | 0.051 | 0.199 | 0.022a | 0.296 | |
| Proteobacteria | 0d | 19.92 ± 6.46 | 19.69 ± 4.14 | 18.99 ± 1.55 | 18.65 ± 3.53 | 0.973 | 0.805 | 0.859 | 0.213 |
| 20d | 19.23 ± 2.88 | 7.73 ± 2.46 | 12.54 ± 2.55 | 7.20 ± 1.31 | 0.032a | 0.042a | 0.042a | 0.800 | |
| 60d | 19.99 ± 0.17 | 3.29 ± 0.46 | 14.00 ± 0.33 | 10.64 ± 0.51 | 0.001a | 0.001a | 0.001a | 0.006a | |
| Verrucomicrobia | 0d | 4.60 ± 1.73 | 5.06 ± 0.38 | 5.57 ± 1.23 | 4.58 ± 2.66 | 0.613 | 0.557 | 0.563 | 0.811 |
| 20d | 4.34 ± 0.34 | 4.45 ± 0.40 | 6.66 ± 2.20 | 7.89 ± 0.53 | 0.760 | 0.393 | 0.215 | 0.005a | |
| 60d | 2.69 ± 0.35 | 7.81 ± 2.43 | 4.82 ± 0.87 | 8.23 ± 2.02 | 0.086 | 0.043a | 0.034a | 0.863 | |
| Tenericutes | 0d | 1.75 ± 0.40 | 2.45 ± 1.23 | 2.51 ± 0.42 | 2.60 ± 1.60 | 0.524 | 0.936 | 0.194 | 0.562 |
| 20d | 1.83 ± 0.58 | 3.72 ± 0.92 | 1.94 ± 0.50 | 3.67 ± 1.33 | 0.094 | 0.203 | 0.825 | 0.970 | |
| 60d | 2.43 ± 0.44 | 4.56 ± 0.96 | 3.12 ± 0.52 | 5.39 ± 0.71 | 0.026a | 0.020a | 0.317 | 0.479 | |
| Spirochaetes | 0d | 0.95 ± 0.24 | 0.69 ± 0.32 | 0.90 ± 0.25 | 0.88 ± 0.31 | 0.499 | 0.900 | 0.696 | 0.428 |
| 20d | 1.25 ± 0.17 | 0.41 ± 0.08 | 3.02 ± 0.43 | 1.43 ± 0.40 | 0.007a | 0.001a | 0.023a | 0.058 | |
| 60d | 3.00 ± 0.71 | 1.35 ± 0.21 | 4.01 ± 0.32 | 2.73 ± 0.64 | 0.077 | 0.101 | 0.216 | 0.031a | |
| Cyanobacteria | 0d | 1.67 ± 0.72 | 1.24 ± 0.44 | 1.49 ± 0.41 | 1.48 ± 0.79 | 0.550 | 0.999 | 0.786 | 0.352 |
| 20d | 1.13 ± 0.19 | 2.48 ± 0.20 | 1.30 ± 0.07 | 1.60 ± 0.15 | 0.003a | 0.143 | 0.355 | 0.004a | |
| 60d | 0.60 ± 0.11 | 1.45 ± 0.35 | 0.77 ± 0.03 | 1.61 ± 0.55 | 0.056 | 0.130 | 0.076 | 0.769 | |
| Fibrobacteres | 0d | 5.24 ± 1.00 | 5.14 ± 0.97 | 4.68 ± 1.21 | 5.49 ± 1.10 | 0.938 | 0.611 | 0.696 | 0.778 |
| 20d | 3.93 ± 0.26 | 5.37 ± 0.18 | 3.01 ± 0.26 | 4.25 ± 0.18 | 0.002a | 0.002a | 0.025a | 0.006a | |
| 60d | 2.63 ± 0.40 | 4.36 ± 0.31 | 2.74 ± 0.21 | 4.39 ± 0.36 | 0.008a | 0.017a | 0.761 | 0.463 | |
amean significant difference (P < 0.05).
Table 5.
Influence of AMPs on proportion of different bacterial genus.
| Bacterial genus | Groups | P-value | |||||||
|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | I VS II | III VS IV | I VS III | II VS IV | ||
| Undefined genera | 0d | 39.16 ± 2.73 | 36.66 ± 1.86 | 38.09 ± 2.38 | 38.93 ± 2.07 | 0.365 | 0.723 | 0.158 | 0.069 |
| 20d | 40.27 ± 2.71 | 39.96 ± 2.76 | 36.58 ± 3.12 | 38.41 ± 1.82 | 0.864 | 0.280 | 0.100 | 0.378 | |
| 60d | 35.57 ± 1.26 | 39.34 ± 1.26 | 35.83 ± 1.46 | 42.70 ± 1.26 | 0.081 | 0.047 | 0.384 | 0.126 | |
| Prevotella | 0d | 22.20 ± 1.25 | 22.71 ± 1.55 | 20.71 ± 1.53 | 22.73 ± 2.41 | 0.782 | 0.466 | 0.059 | 0.973 |
| 20d | 25.54 ± 2.66 | 28.71 ± 4.78 | 31.58 ± 3.90 | 33.89 ± 3.63 | 0.467 | 0.538 | 0.048a | 0.349 | |
| 60d | 27.67 ± 2.54 | 32.97 ± 6.85 | 35.82 ± 2.67 | 35.60 ± 2.95 | 0.393 | 0.890 | 0.029a | 0.567 | |
| [Paraprevotellaceae]CF231 | 0d | 7.36 ± 0.52 | 7.71 ± 1.52 | 7.63 ± 2.09 | 7.14 ± 2.95 | 0.696 | 0.874 | 0.870 | 0.606 |
| 20d | 6.03 ± 1.08 | 5.71 ± 0.81 | 3.60 ± 1.31 | 3.26 ± 0.60 | 0.763 | 0.687 | 0.151 | 0.037a | |
| 60d | 8.79 ± 1.03 | 4.72 ± 0.39 | 4.99 ± 0.21 | 3.06 ± 0.84 | 0.119 | 0.043a | 0.111 | 0.130 | |
| Butyrivibrio | 0d | 6.51 ± 0.89 | 6.50 ± 2.03 | 6.99 ± 0.52 | 6.71 ± 1.13 | 0.998 | 0.719 | 0.414 | 0.876 |
| 20d | 6.31 ± 0.86 | 6.52 ± 0.45 | 7.03 ± 0.73 | 6.68 ± 0.29 | 0.555 | 0.302 | 0.331 | 0.685 | |
| 60d | 6.15 ± 0.07 | 6.23 ± 0.17 | 6.54 ± 0.48 | 6.71 ± 0.22 | 0.624 | 0.530 | 0.323 | 0.009a | |
| Succinivibrio | 0d | 8.23 ± 0.40 | 7.98 ± 0.54 | 7.81 ± 0.35 | 8.37 ± 0.82 | 0.653 | 0.413 | 0.069 | 0.661 |
| 20d | 7.56 ± 0.69 | 1.00 ± 0.13 | 4.85 ± 0.45 | 1.20 ± 0.38 | 0.003a | 0.001a | 0.003a | 0.375 | |
| 60d | 3.99 ± 0.52 | 1.33 ± 0.24 | 2.28 ± 0.58 | 2.11 ± 0.94 | 0.026a | 0.714 | 0.040a | 0.371 | |
| Fibrobacter | 0d | 4.60 ± 0.32 | 4.79 ± 0.68 | 4.73 ± 0.62 | 4.61 ± 0.87 | 0.697 | 0.885 | 0.814 | 0.805 |
| 20d | 3.60 ± 0.32 | 5.20 ± 0.14 | 2.95 ± 0.25 | 3.22 ± 0.15 | 0.005a | 0.299 | 0.008a | 0.004a | |
| 60d | 2.63 ± 0.40 | 3.69 ± 0.11 | 3.07 ± 1.36 | 3.32 ± 0.45 | 0.046a | 0.785 | 0.678 | 0.286 | |
| Selenomonas | 0d 0d | 3.39 ± 0.44 | 3.21 ± 1.09 | 3.27 ± 0.49 | 3.19 ± 1.37 | 0.851 | 0.903 | 0.813 | 0.988 |
| 20d | 2.95 ± 0.16 | 1.75 ± 0.45 | 2.75 ± 0.65 | 2.99 ± 0.20 | 0.042a | 0.625 | 0.659 | 0.046a | |
| 60d | 1.53 ± 0.23 | 0.57 ± 0.16 | 0.74 ± 0.22 | 0.32 ± 0.11 | 0.025a | 0.026a | 0.008a | 0.095 | |
| Anaerovibrio | 0d | 1.92 ± 0.48 | 2.07 ± 0.24 | 2.16 ± 0.33 | 1.96 ± 0.19 | 0.747 | 0.159 | 0.366 | 0.689 |
| 20d | 1.48 ± 0.46 | 3.35 ± 0.26 | 1.69 ± 0.36 | 1.10 ± 0.25 | 0.046a | 0.209 | 0.468 | 0.012 | |
| 60d | 1.23 ± 0.27 | 2.65 ± 0.12 | 2.22 ± 0.17 | 2.04 ± 0.20 | 0.009a | 0.183 | 0.049a | 0.073 | |
| Succiniclasticum | 0d | 1.45 ± 0.58 | 1.57 ± 0.50 | 1.55 ± 0.51 | 1.48 ± 0.36 | 0.753 | 0.867 | 0.878 | 0.542 |
| 20d | 1.12 ± 0.09 | 1.80 ± 0.09 | 0.85 ± 0.26 | 1.97 ± 0.14 | 0.012a | 0.004a | 0.289 | 0.168 | |
| 60d | 0.04 ± 0.01 | 1.48 ± 0.36 | 0.07 ± 0.01 | 0.53 ± 0.08 | 0.019a | 0.016a | 0.477 | 0.029a | |
| Treponema | 0d | 0.98 ± 0.07 | 1.15 ± 0.58 | 1.05 ± 0.35 | 1.21 ± 0.36 | 0.701 | 0.287 | 0.797 | 0.883 |
| 20d | 1.22 ± 0.15 | 0.38 ± 0.11 | 2.35 ± 0.37 | 1.67 ± 0.24 | 0.011a | 0.179 | 0.012a | 0.023 | |
| 60d | 2.95 ± 0.70 | 1.45 ± 0.15 | 3.98 ± 0.86 | 1.73 ± 0.48 | 0.031a | 0.043a | 0.530a | 0.396 | |
amean significant difference (P <0.05).
At the phylum level, Proteobacteria appeared to significantly decrease (P < 0.05; Table 4) and Fibrobacteres appeared to significantly increase (P < 0.05; Table 4) in the AMP-supplemented groups compared with the control groups (I and III). In addition, Bacteroidetes and Proteobacteria appeared to significantly decrease (P < 0.05) and Firmicutes appeared to significantly increase (P < 0.05) in group III compared with group I.
At the genus level, Prevotella dominated the assignable sequences; on average it accounted for 31.35% of total bacteria. Prevotella was followed in average relative abundance by Butyrivibrio (6.52%), [Paraprevotellaceae]CF231 (5.02%), Fibrobacter (3.75%), Succinivibrio (3.04%), and Anaerovibrio (1.93%).
Fibrobacter and Anaerovibrio appeared to increase in the AMP-supplemented groups although a significant increase was only apparent with normal concentrate. Succiniclasticum appeared to increase (Table 5), whereas Succinivibrio, Selenomonas, and Treponema appeared to decrease in the AMP-treated groups (Table 5) compared with the control groups (I and III). In addition, Prevotella, Anaerovibrio (except on day 20), and Treponema appeared to significantly increase (P < 0.05); whereas Succinivibrio, Selenomonas (except on day 20), and Fibrobacter (except on day 60) appeared to significantly decrease (P < 0.05) in group III compared with group I. No differences in [Paraprevotellaceae]CF231, Butyrivibrio, and Succiniclasticum were observed between groups I and III (P > 0.05).
The Chao1, ACE, Simpson, and Shannon diversity index values of each sample (at the bacterial and ciliate genus level) are shown in Tables 6 and 7, all indices were elevated in the AMP-supplemented groups, especially on day 60. Moreover, all indices were reduced in group III, although these decreases were not statistically significant. These results indicate that AMP supplementation may enhance microbial diversity in the rumen whereas increasing concentrate may reduce it.
Table 6.
Diversity estimation based on sequence analysis of 16 S rRNA gene libraries of the goat rumen.
| erParamet | Bacterial | ||||
|---|---|---|---|---|---|
| I | II | III | IV | ||
| OUT | 0d | 1221 ± 101 | 1202 ± 144 | 1205 ± 153 | 1239 ± 105 |
| 20d | 1211 ± 171.52 | 1192 ± 168.82 | 948 ± 172 | 1058 ± 88 | |
| 60d | 953 ± 90 | 1290 ± 111 | 746 ± 117 A | 944 ± 105B | |
| Chao1 | 0d | 934 ± 103 | 948 ± 58 | 929 ± 54 | 917 ± 89 |
| 20d | 911 ± 167 | 914 ± 158 | 676 ± 136 | 754 ± 61 | |
| 60d | 713 ± 121 | 988 ± 103 | 559 ± 111 A | 725 ± 126B | |
| ACE | 0d | 1012 ± 118.40 | 1023 ± 60.99 | 1016 ± 152.75 | 1018 ± 64.38 |
| 20d | 1024.85 ± 167.14 | 1029.74 ± 146.54 | 750.04 ± 156.02 | 814.05 ± 113.19 | |
| 60d | 793.03 ± 106.73 | 1093.57 ± 106.45 | 614.01 ± 106.97 A | 796.81 ± 120.62B | |
| Simpson | 0d | 0.950 ± 0.049 | 0.949 ± 0.022 | 0.951 ± 0.019 | 0.956 ± 0.036 |
| 20d | 0.952 ± 0.050 | 0.947 ± 0.025 | 0.957 ± 0.018 | 0.964 ± 0.025 | |
| 60d | 0.950 ± 0.044 | 0.975 ± 0.015 | 0.939 ± 0.040 | 0.969 ± 0.014 | |
| Shannon | 0d | 6.560 ± 0.729 | 6.606 ± 0.516 | 6.532 ± 0.415 | 6.599 ± 0.208 |
| 20d | 6.650 ± 1.244 | 6.573 ± 0.687 | 6.217 ± 0.449 | 6.562 ± 0.486 | |
| 60d | 6.228 ± 1.116 | 7.290 ± 0.335 | 5.755 ± 0.849 | 6.663 ± 0.791 | |
bThe operational taxonomic units (OTUs) were defined with 3% dissimilarity. The diversity indices (Chao1, ACE, Shannon and Simpson) were calculated. A,BValues with different superscripts in the same row differ significantly (P < 0.05).
Table 7.
Diversity estimation based on sequence analysis of 18 S rRNA gene libraries of the goat rumen.
| Parameter | Ciliate | ||||
|---|---|---|---|---|---|
| I | II | III | IV | ||
| OUT | 0d | 116 ± 19 | 121 ± 8 | 119 ± 21 | 124 ± 8 |
| 20d | 123 ± 23 | 130 ± 18 | 103 ± 15 | 110 ± 15 | |
| 60d | 118 ± 19 | 141 ± 22 | 108 ± 14 | 122 ± 7 | |
| Chao1 | 0d | 91 ± 13 | 95 ± 18 | 89 ± 10 | 96 ± 12 |
| 20d | 98 ± 23 | 95 ± 18 | 76 ± 14 | 85 ± 19 | |
| 60d | 98 ± 24 | 116 ± 14 | 87 ± 14 | 95 ± 3 | |
| ACE | 0d | 103 ± 24 | 104 ± 17 | 105 ± 10 | 101 ± 21 |
| 20d | 106.42 ± 27.99 | 104.06 ± 20.98 | 83.82 ± 12.15 | 99.17 ± 106.42 | |
| 60d | 107.07 ± 24.21 | 128.27 ± 21.05 | 95.50 ± 17.63 | 108.40 ± 1.72 | |
| Simpson | 0d | 0.764 ± 0.073 | 0.747 ± 0.046 | 0.758 ± 0.028 | 0.765 ± 0.012 |
| 20d | 0.766 ± 0.142 | 0.720 ± 0.128 | 0.728 ± 0.091 | 0.769 ± 0.082 | |
| 60d | 0.784 ± 0.055 | 0.769 ± 0.071 | 0.741 ± 0.070 | 0.811 ± 0.050 | |
| Shannon | 0d | 2.987 ± 0.133 | 3.019 ± 0.233 | 3.029 ± 0.058 | 2.991 ± 0.126 |
| 20d | 3.014 ± 0.666 | 2.819 ± 0.664 | 2.707 ± 0.593 | 2.918 ± 0.572 | |
| 60d | 3.081 ± 0.563 | 3.074 ± 0.431 | 2.780 ± 0.311 | 3.146 ± 0.230 | |
bThe operational taxonomic units (OTUs) were defined with 3% dissimilarity. The diversity indices (Chao1, ACE, Shannon and Simpson) were calculated.
Ciliate community structure
A total of 631,179 quality protozoa sequences were obtained from the 36 samples, with an average of 17,532 reads per rumen sample. Although all animal groups were fed the same diet, there was a high level of variation between individuals in terms of ciliate community composition at the genus level and their respective community compositions are detailed in Fig. S2. The only characteristic in common was the dominant role of Polyplastron and Ophryoscolex (Table 8).
Table 8.
Influence of diet and AMPs on proportion of ciliates genera.
| Protozoal genus | Groups | P-value | |||||||
|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | I VS II | III VS IV | I VS III | II VS IV | ||
| Polyplastron | 0d | 40.07 ± 4.64 | 41.23 ± 4.37 | 40.21 ± 4.06 | 42.57 ± 2.07 | 0.785 | 0.299 | 0.977 | 0.716 |
| 20d | 45.37 ± 0.64 | 33.37 ± 4.71 | 51.44 ± 7.60 | 49.09 ± 7.80 | 0.031a | 0.783 | 0.313 | 0.031a | |
| 60d | 56.78 ± 4.55 | 41.28 ± 1.70 | 65.59 ± 2.93 | 63.67 ± 2.74 | 0.013a | 0.599 | 0.074 | 0.003a | |
| Diploplastron | 0d | 7.39 ± 1.41 | 6.80 ± 1.45 | 7.46 ± 1.27 | 6.98 ± 0.30 | 0.719 | 0.589 | 0.959 | 0.846 |
| 20d | 6.17 ± 1.04 | 6.41 ± 0.32 | 2.60 ± 0.91 | 2.83 ± 1.71 | 0.642 | 0.884 | 0.086 | 0.051 | |
| 60d | 3.31 ± 0.54 | 3.36 ± 0.37 | 1.51 ± 0.27 | 1.81 ± 0.75 | 0.881 | 0.581 | 0.016a | 0.131 | |
| Entodinium | 0d | 4.43 ± 1.05 | 4.12 ± 0.78 | 3.67 ± 0.66 | 4.07 ± 0.21 | 0.776 | 0.502 | 0.207 | 0.924 |
| 20d | 2.65 ± 0.50 | 0.46 ± 0.16 | 0.94 ± 0.18 | 1.01 ± 0.49 | 0.022a | 0.726 | 0.037a | 0.105 | |
| 60d | 1.38 ± 0.12 | 0.60 ± 0.13 | 1.50 ± 0.21 | 1.04 ± 0.29 | 0.002a | 0.163 | 0.497 | 0.165 | |
| Ophryoscolex | 0d | 10.86 ± 1.43 | 11.30 ± 2.98 | 9.90 ± 4.22 | 10.31 ± 1.72 | 0.866 | 0.886 | 0.795 | 0.741 |
| 20d | 14.99 ± 7.23 | 45.07 ± 4.14 | 24.64 ± 2.60 | 33.19 ± 4.77 | 0.006a | 0.058 | 0.113 | 0.014a | |
| 60d | 27.98 ± 3.44 | 52.09 ± 2.13 | 29.09 ± 2.56 | 31.52 ± 2.07 | 0.001a | 0.450 | 0.641 | 0.006a | |
| Enoploplastron | 0d | 0 | 0 | 0 | 0 | — | — | — | — |
| 20d | 0 | 0 | 0 | 0 | — | — | — | — | |
| 60d | 5.79 ± 1.40 | 0.16 ± 0.14 | 0 | 0 | 0.023a | — | — | — | |
| Dasytricha | 0d | 0.99 ± 0.25 | 0.79 ± 0.47 | 1.02 ± 0.29 | 0.81 ± 0.19 | 0.626 | 0.476 | 0.914 | 0.936 |
| 20d | 0.32 ± 0.40 | 0.74 ± 0.32 | 0.99 ± 0.42 | 0.78 ± 0.54 | 0.397 | 0.201 | 0.261 | 0.800 | |
| 60d | 0 | 0.50 ± 0.42 | 0.89 ± 0.20 | 0.93 ± 0.13 | 0.126 | 0.829 | 0.014a | 0.309 | |
| Isotricha | 0d | 36.09 ± 3.74 | 37.20 ± 3.16 | 38.02 ± 4.44 | 36.70 ± 2.92 | 0.538 | 0.785 | 0.427 | 0.876 |
| 20d | 29.87 ± 5.49 | 13.95 ± 1.36 | 18.80 ± 5.11 | 12.89 ± 4.01 | 0.042a | 0.362 | 0.284 | 0.759 | |
| 60d | 4.21 ± 0.90 | 2.01 ± 0.46 | 1.42 ± 0.31 | 1.04 ± 0.40 | 0.038a | 0.357 | 0.057 | 0.007a | |
amean significant difference (P < 0.05).
Compared with the control groups (I and III), Ophryoscolex appeared to increase in the AMP-supplemented groups (Table 8), although a significant increase was only apparent with normal concentrate. Polyplastron, Entodinium, and Isotricha appeared to decrease in the AMP-supplemented groups, although a significant decrease was only observed with normal concentrate. No differences in Diploplastron and Dasytricha were detected between AMP-treated goats and control animals (P > 0.05). Moreover, no differences in Polyplastron, Ophryoscolex, and Isotricha were evident between groups I and III (P > 0.05).
Discussion
Microbial community composition in ruminants has previously been linked with animal production traits21,22. In the present study, we found that Bacteroidetes was the dominant phylum in all samples (except group III), followed by Firmicutes, Proteobacteria, and Verrucomicrobia. This structure is similar to the rumen bacterial community of sheep inferred from multiplex 454 Titanium pyrosequencing23. At the genus level, Prevotella, known as an abundant member of the rumen microbiome24–26, was the most abundant genus detected, followed by Butyrivibrio, [Paraprevotellaceae]CF231, Fibrobacter, Succinivibrio, and Anaerovibrio. Many of these genera include organisms that are important cellulose and hemicellulose-degraders; this indicates a rumen bacterial community highly oriented towards fibre degradation. Polyplastron and Ophryoscolex were the most abundant ciliate genera in this study; the protozoal community composition is similar to that of the A type (dominated by Polyplastron, Ostracodinium, Dasytricha, and Entodinium)27. However, many studies have identified Entodinium as the predominant protozoal group in ruminants28–31. This discrepancy may be due to diet. In this study, forage grass was the main fodder and xylanase and glucanase activities of Polyplastron and Ophryoscolex are much higher than those of Entodinium 27. In addition, high-throughput sequencing technology could also affect the true composition of rumen ciliates. Kittelmann et al.32 reported that smaller-celled genera, such as Entodinium, Charonina, and Diplodinium, tended to be underrepresented, while larger-celled genera, such as Metadinium, Epidinium, Eudiplodinium, Ostracodinium, and Polyplastron, tended to be overrepresented using the pyrosequencing approach.
Antimicrobial peptides possess broad-spectrum antimicrobial activity and have been used as a new type of feed additive in animal husbandry. A number of recent studies have suggested that dietary supplementation containing an antimicrobial peptide, such as lactoferricin and the lactoferrampin fusion peptide, potato protein, cecropin AD, or antimicrobial peptide P5, reduced the total numbers of aerobes while simultaneously enhancing the total amount of anaerobes and beneficial lactobacilli, thus improving growth performance in weanling pigs33–36. In this study, we have shown that dietary supplementation with AMPs improved growth of juvenile goats under two types of concentrate conditions. These results suggest that AMPs can be used to promote growth performance in goats. This is consistent with the finding of Yoon et al.36 who observed an improvement in the average daily gain and feed efficiency of weanling pigs fed diets supplemented with antimicrobial peptide-A3. Similarly, Jin et al.35,37 observed an improvement in the average daily gain(ADG) of weanling pigs fed diets supplemented with antimicrobial peptides from Solanum tuberosum. Antimicrobial peptides beneficially affect host animals by improving their intestinal balance and creating gut microecological conditions38–40. In this study, we found that Proteobacteria were significantly decreased in the AMP-supplemented groups, while Fibrobacteres were significantly increased. This may be due to the fact that Fibrobacteres are anaerobic bacteria41, whereas Proteobacteria consist of aerobic bacteria that are mostly pathogenic42; the antibacterial peptide could have inhibited the pathogenic bacteria while enhancing the total amount of anaerobes17. Dietary supplementation with AMPs has the potential to increase bacterial genera, such as Fibrobacter, Anaerovibrio, Succiniclasticum, and the ciliate genus Ophryoscolex, while reducing bacterial genera, including Selenomonas, Succinivibrio, and Treponema, and ciliate genera such as Polyplastron, Entodinium, and Isotricha. However, changes in Fibrobacter, Anaerovibrio, Ophryoscolex, Polyplastron, Entodinium, and Isotricha were related to the amount of concentrate that no significant different in the double concentrate group. Of these, Fibrobacter 43,44, Treponema 45, Ophryoscolex 46, and Polyplastron 47 are cellulose-degrading microbes and Succiniclasticum 48, Entodinium, and Isotricha 47 are starch-degrading microbes. Selenomonas and Succinivibrio degrade both starch and cellulose and Anaerovibrio 49 are fat-degrading bacteria. Therefore, we hypothesize that the increase in the relative abundance of Fibrobacter and Ophryoscolex in the normal concentrate group was due to an increase in xylanase and pectinase activities. Similarly, the decrease in the relative abundance of Isotricha and Entodinium was caused by a decrease in amylase activity in the normal concentrate group; whereas the increase in the relative abundance of Anaerovibrio was due to an increase in lipase activity in the same group.
Moreover, the fermentation products of Fibrobacter, Anaerovibrio, Treponema, Selenomonas, O phryoscolex, Polyplastron, and Isotricha are acetate, propionate, and succinate; the fermentation product of Succinivibrio is succinate; and the fermentation products of Butyrivibrio are acetate and butyrate. Therefore, an increase in the relative abundance of Fibrobacter, Anaerovibrio, Ophryoscolex in the normal concentrate group may have caused an increase in acetate; whereas a decrease in the relative abundance of Treponema, Selenomonas, Polyplastron, and Isotricha in the double concentrate group may have led to the decrease in acetate. Lack of any variation to the relative abundance of Butyrivibrio prevented a change in butyrate. Acetate, propionate, and butyrate are the main components in VFAs, accounting for 95% of the total volatile matter content50. A change of the acetate + butyrate-to-propionate ratio is related to rumen fermentation mode. Thus, changes in acetate can cause alterations to T-VFA content (increase with normal concentrate and decrease with double concentrate) and the acetate + butyrate-to-propionate ratio (significant decrease with double concentrate). These results indicate that the effects of AMPs on rumen fermentation function and rumen microorganisms in goats were related to the amount of concentrate. It is possible that increased dosage causes similar changes in the double concentrate groups. However, additional studies will be needed to thoroughly elucidate these changes. The alpha diversity indices were elevated in the AMP-supplemented groups in this study, especially on day 60; indicating that AMP supplementation could increase microbial diversity in the rumen.
Previous studies51,52 have demonstrated the importance of concentrate supplementation in goat growth and productivity. In this study, we found that the ADG increased with increasing concentrate amount. This result is consistent with the findings of Salim et al.53 who reported that feeding grazing goats with concentrate supplement may optimize growth performance. The main reason for this may be changes in the rumen bacterial composition of ruminants driven by the amount of dietary concentrate54. In the present study, Firmicutes replaced Bacteroidetes as the dominant phylum in group (III) and Proteobacteria were significantly fewer compared to group I. This is in good agreement with data reported by Liu et al.48 who reported that Firmicutes increased with a high concentrate diet. Similarly, Wetzels et al.55 observed that Proteobacteria decreased and Firmicutes increased with increasing concentrate doses because of the ability of many Firmicutes to easily degrade fermentable carbohydrates. In terms of bacterial and ciliate genera, Prevotella increased with increasing concentrate amount, as reported also by Khafipour et al.56 and Metzler-Zebeli et al.57. Prevotella is one of the most abundant genera in the rumen of goats because these bacteria possess highly diverse functions, in particular following a high-grain feeding regime. Anaerovibrio (on day 60) and Treponema increased significantly, whereas Succinivibrio, Fibrobacter (on day 20), Selenomonas (on day 60), and Diploplastron (on day 60) decreased significantly with increasing concentrate dosage. Therefore, the higher relative abundance of Prevotella and Anaerovibrio was due to augmented pectinase and lipase activities, which led to a further increase in T-VFA, ammonia, acetate, and MCP, and ultimately to enhanced goat growth performance.
Moreover, all indices were lower in group III compared to group I, indicating that bacterial diversity depended on dietary concentrate dosage. Similarly, Lillis et al.58 reported that bacterial diversity was affected to a greater degree by a 90:10 than a 50:50 concentrate:forage ratio.
In summary, this study demonstrates that dietary supplementation with AMPs has beneficial effects on the growth performance, ruminal fermentation function, enzymatic activity, and rumen morphology of juvenile goats; and that these effects are related to concentrate amount. Therefore, AMPs could potentially be used as feed additives for juvenile goats on commercial farms. The detailed mechanism(s) by which AMPs promote growth of juvenile goats and improve their rumen microbial community structure require further clarification.
Materials and Methods
Ethics statement
All experimental procedures and animal care performed in the present study were approved according to the recommendations of the Guide of the Sichuan Agricultural University Animal Care and Use Committee (Sichuan Agricultural University, Sichuan, China) under permit NO. DKYB20100805, and all efforts were made to minimize suffering. Field studies did not involve endangered or protected species. Chuanzhong black goats were housed at the experimental farm of the Animal Nutrition Institute of Sichuan Agricultural University.
Materials
Antimicrobial peptides used were provided by Rota BioEngineering Co., Ltd. (Sichuan, China). AMPs were composed of recombinant swine defensin PBD-mI(DHYICAKKGGTCNFSPCPLFNRIEGTCYSGKAKCCIR) and a fly antibacterial peptide LUC-n(ATCDLLSGTGVKHSACAAHCLLRGNRGGYCNGRAICVCRN) at a blending ratio of 1:115.
Animal handling
Twenty-four, approximately four-month old, non-castrated Chuanzhong black goats, of average weight (16.17 ± 0.72 kg), were acclimated for 7 days prior to the experiment. All goats were caged and randomly allotted to four dietary treatment groups: I-normal concentrate group (300 g concentrate [per head per day]), II-normal concentrate and antimicrobial peptide group (300 g concentrate + 3.0 g AMPs), III-double concentrate group (600 g concentrate), and IV-double concentrate and antimicrobial peptide group (600 g concentrate + 3.0 g AMPs).
The diet included concentrate (Table 9) and forage (fresh grass). The groups were composed of three replicate pens with 2 goats each, animals were maintained in a house with free access to water, and fed twice daily (at 09:00 and 18:00); the animals maintained their normal herd behaviour.
Table 9.
Composition and nutrient levels of the concentrate (DM basis).
| Ingredients | Content(%) | Nutrient levels | Content(%) |
|---|---|---|---|
| Corn grain | 51 | DE/(MJ/kg) | 13.34 |
| Wheat bran | 23 | DM | 84.27 |
| Rapeseed meal | 10 | CP | 16.66 |
| Rapeseed cake | 10 | CF | 4.17 |
| Fish meal | 3 | NDF | 13.72 |
| NaCl | 1 | ADF | 6.91 |
| Premix1) | 2 | ||
| Total | 100 |
1)Premix provides the following per kg of the diet:Fe(as ferrous sulfate) 30 mg,Cu (as copper sulfate) 10 mg, Zn (as zinc sulfate) 50 mg,Mn (as manganese sulfate) 60 mg,VA 2 937 IU,VD 343 IU,VE 30 IU.
Sampling and DNA extraction
Rumen fluid samples were collected using a stomach tube on days 0, 20 and 60, prior to morning feeding; the first part of the rumen fluid was discarded to prevent saliva interference. Three goats were selected from each group for sampling(one goats per pen). Rumen pH was measured immediately after collection using a portable pH meter (Model PHB-4, Shanghai Leica Scientific Instrument Co., Ltd., Shanghai, China). Solid feed particles were removed from the rumen fluid by filtration through 4 layers of cheesecloth. Samples were stored at −80 °C for later analysis. Microbial genomic DNA was extracted from rumen samples using a stool DNA kit (OMEGA Bio-Tek, Norcross, GA, USA), according to the manufacturer’s instructions.
Ruminal fermentation function and enzyme activity analysis
Samples were prepared for VFA analysis and chromatography according to Luo et al.59. The concentration of NH3-N was analysed using visible-light spectrophotometry (Scientific BioMate 3 s, Thermo). NH4Cl standards were prepared according to Broderick and Kang60. Microbial protein (MCP) in the rumen was analyzed by trichloroacetic acid protein precipitation61. The activities of CMCase, xylanase, pectinase and β-glucosidase were measured using commercially available ELISA kits (R&D Systems, Minneapolis, MN, USA). Protease activity was measured as follows: a reaction mixture containing 1 mL casein and 4 mL protease enzyme was incubated for 4 h at 38 °C; at this point, the reaction was stopped by adding 10% trichloroacetic acid. The sample was then centrifuged at 3500 × g for 15 min. Next, 1 mL of supernatant was removed and mixed with 5 mL 0.4 M Na2CO3 and 1 mL Folin-Ciocalteu’s phenol solution and incubated on the laboratory bench for 15 min. The hydrolysed protein was measured using visual-light spectrophotometry at 680 nm. Concentration and activity of lipase and amylase were measured using commercially available kits (NanJing JianCheng Bioengineering Institute, Nanjing, China).
Rumen microbial community analysis
The V4 regions of bacterial 16 S rRNA genes and ciliate protozoal 18 S rRNA genes were amplified. Bacterial sequences were amplified using primers 520 F 5′-GCACCTAAYTGGGYDTAAAGNG-3′ and 802 R 5′- TACNVGGGTATCTAATCC-3′; ciliate sequences were amplified using primers V547F 5′-CCAGCASCYGCGGTAATTCC-3′ and V4R 5′-ACTTTCGTTCTTGATYRA-3′. The bacterial amplification mixture consisted of 1 μL (10 μM) of each primer, 1 μL template DNA, 5 μL 5 × reaction buffer, 5 μL 5 × high GC buffer, 0.5 μL 10 mM dNTPs, 0.25 μL Q5 high-fidelity DNA polymerase and 11.25 μL ddH2O. The ciliate PCR was carried out in triplicate using 25 μL mixtures containing 1 μL (10 μM) of each primer, 2 μL template DNA, 5 μL 5 × Q5 reaction buffer, 5 μL 5 × Q5 GC high enhancer, 2 μL 2.5 mM dNTPs, and 0.25 μL (5 U/μL) Q5 polymerase. Amplification was performed as follows: initial denaturation at 98 °C for 5 min; 27 cycles of denaturation at 98 °C for 30 s, annealing at 50 °C for 30 s, and elongation at 72 °C for 30 s; plus a final 5-min extension step at 72 °C. PCR products were excised from 2% agarose gels and purified with a QIAquick Gel extraction kit (Qiagen, Venlo, The Netherlands). The remaining DNA was stored at −20 °C until it was used for sequencing. High quality DNA, was sent to Shanghai Paisennuo Biological Technology Co. Ltd for sequencing using an Illumina MiSeqPE250 (Illumina, San Diego, CA, USA).
Data analysis
Sequence reads were processed and analysed using QIIME pipeline software (version 1.8.0). Chimeric sequences were removed to generate high quality sequences. High-quality sequences were divided and aligned into Operational Taxonomic Units (OTUs) with 97% sequence similarity using the QIIME pipeline software. The highest abundance sequences were compared with template regions in the Greengenes database (Release 13.8, http://greengenes.secondgenome.com/) (bacterial) and NCBI (http://www.ncbi.nlm.nih.gov) database (Ciliate protozoal), and were used to acquire taxonomic information for each OTU and species composition. Alpha diversity indices (including the Simpson index and Shannon index) were obtained using QIIME pipeline software. R software was used to analyze microfloral population structures. The results of these various analyses are expressed as means ± standard error of the mean (SEM). Statistical comparisons were made using paired sample t test via a commercially available statistical software package (SPSS 19.0, Business Machines Corporation, Armonk, NY, USA). Differences among treatments were regarded as significant at P < 0.05.
Electronic supplementary material
Acknowledgements
This research was financially supported by grants from the Chang-jiang Scholars and Innovative Research Team for Universities (Grant No. IRT0848).
Author Contributions
Conceived and designed the experiments: Junliang Deng, Qi Liu, Zhihua Ren, Liuhong Shen. Performed the experiments: Qi Liu, Shuhua Yao, Yun Chen. Analyzed the data: Qi Liu, Shuhua Yao. Contributed reagents/materials/analysis tools: Qi Liu, Yanyi Yang, Shuang Gao, Hengmin Cui, Xiaoping Ma, Yanchun Hu, Shumin Yu. Wrote the paper: Qi Liu, Shuhua Yao.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Qi Liu and Shuhua Yao contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-12394-4.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wang, M. Z., Li-Huai, Y. U., Wang, H. R., Zhang, J. & Zhan-Jun, L. U. Effects of Dietary Structure on Protozoal Profile and Grazing Rate in Rumen of Goats. Chin J Anim Sci. (2009).
- 2.Franzolin R, Wright ADG. Microorganisms in the rumen and reticulum of buffalo (Bubalus bubalis) fed two different feeding systems. Bmc Research Notes. 2016;9(1):1–5. doi: 10.1186/s13104-016-2046-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adg W, Klieve AV. Does the complexity of the rumen microbial ecology preclude methane mitigation. Anim Feed Sci Techn. 2011;s166–167(7):248–253. [Google Scholar]
- 4.Yue ZB, Jin W, Liu XM, Yu HQ. Comparison of rumen microorganism and digester sludge dominated anaerobic digestion processes for aquatic plants. Renew Energ. 2012;46(5):255–258. doi: 10.1016/j.renene.2012.03.030. [DOI] [Google Scholar]
- 5.Madoni P. Estimates of ciliated protozoa biomass in activated sludge and biofilm. Bioresource Technol. 1994;48(3):245–249. doi: 10.1016/0960-8524(94)90153-8. [DOI] [Google Scholar]
- 6.Ladd JN, Walker DJ. Fermentation of lactic acid by the rumen microorganism, Peptostreptococcus elsdenii. Ann NY Acad Sci. 1965;119(3):1038. doi: 10.1111/j.1749-6632.1965.tb47460.x. [DOI] [PubMed] [Google Scholar]
- 7.Wang ZB, et al. Effects of hainanmycin or monensin supplementation on ruminal protein metabolism and populations of proteolytic bacteria in Holstein heifers. Anim Feed Sci Techn. 2015;201:99–103. doi: 10.1016/j.anifeedsci.2015.01.001. [DOI] [Google Scholar]
- 8.Candanosa E, Villagodoy A, Castillo DA, Mendoza GD. Effects of Monensin, Virginiamycin and Sodium Bicarbonate on Ruminal Fermentation and Acid-Base Status in Sheep. J Anim Vet Adv. 2012;7(2):184–189. [Google Scholar]
- 9.Bergen WG, Bates DB. Ionophores: their effect on production efficiency and mode of action. J Anim Sci. 1984;58(6):1465. doi: 10.2527/jas1984.5861465x. [DOI] [PubMed] [Google Scholar]
- 10.Saido-Sakanaka H, Ishibashi J, Momotani E, Amano F, Yamakawa M. In vitro and in vivo activity of antimicrobial peptides synthesized based on the insect defensin. Peptides. 2004;25(1):19–27. doi: 10.1016/j.peptides.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 11.Hwang B, Hwang JS, Lee J, Lee DG. The antimicrobial peptide, psacotheasin induces reactive oxygen species and triggers apoptosis in Candida albicans. Biochemical & Biophysical Research Communications. 2011;405(2):267–271. doi: 10.1016/j.bbrc.2011.01.026. [DOI] [PubMed] [Google Scholar]
- 12.Matanic VCA, Castilla V. Antiviral activity of antimicrobial cationic peptides against Junin virus and herpes simplex virus. Int J Antimicrob Ag. 2004;23(4):382–389. doi: 10.1016/j.ijantimicag.2003.07.022. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka T, et al. Parasiticidal activity of Haemaphysalis longicornis longicin P4 peptide against Toxoplasma gondii. Peptides. 2011;34(1):242–50. doi: 10.1016/j.peptides.2011.07.027. [DOI] [PubMed] [Google Scholar]
- 14.Maletzki C, et al. Host defense peptides for treatment of colorectal carcinoma – a comparative in vitro and in vivo analysis. Oncotarget. 2014;5(12):4467. doi: 10.18632/oncotarget.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ren ZH, et al. Effects of antibacterial peptide on cellular immunity in weaned piglets. J Anim Sci. 2015;93(1):127. doi: 10.2527/jas.2014-7933. [DOI] [PubMed] [Google Scholar]
- 16.Wu S, et al. Effects of the antimicrobial peptide cecropin AD on performance and intestinal health in weaned piglets challenged with Escherichia coli. Peptides. 2012;35(2):225. doi: 10.1016/j.peptides.2012.03.030. [DOI] [PubMed] [Google Scholar]
- 17.Yoon JH, et al. Effects of dietary supplementation of antimicrobial peptide-A3 on growth performance, nutrient digestibility, intestinal and fecal microflora and intestinal morphology in weanling pigs. Anim Feed Sci & Tech. 2012;177(177):98–107. doi: 10.1016/j.anifeedsci.2012.06.009. [DOI] [Google Scholar]
- 18.Lee SH, et al. Effects of dietary supplementation with Bacillus subtilis LS 1–2 fermentation biomass on growth performance, nutrient digestibility, cecal microbiota and intestinal morphology of weanling pig. Anim Feed Sci & Tech. 2014;188(1-2):102–110. doi: 10.1016/j.anifeedsci.2013.12.001. [DOI] [Google Scholar]
- 19.Choi SC, et al. An antimicrobial peptide-A3: effects on growth performance, nutrient retention, intestinal and faecal microflora and intestinal morphology of broilers. Brit Poultry Sci. 2013;54(6):738–46. doi: 10.1080/00071668.2013.838746. [DOI] [PubMed] [Google Scholar]
- 20.Peng Z, et al. Use of recombinant porcine β-defensin 2 as a medicated feed additive for weaned piglets. Sci Rep. 2016;6:26790. doi: 10.1038/srep26790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jami E, White BA, Mizrahi I. Potential role of the bovine rumen microbiome in modulating milk composition and feed efficiency. Plos One. 2014;9(1):e85423. doi: 10.1371/journal.pone.0085423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kittelmann S, et al. Two Different Bacterial Community Types Are Linked with the Low-Methane Emission Trait in Sheep. Plos One. 2014;9(7):e103171. doi: 10.1371/journal.pone.0103171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kittelmann S, et al. Simultaneous Amplicon Sequencing to Explore Co-Occurrence Patterns of Bacterial, Archaeal and Eukaryotic Microorganisms in Rumen Microbial Communities. Plos One. 2013;8(2):1112–26. doi: 10.1371/journal.pone.0047879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malmuthuge N, Griebel PJ, Guan LL. Taxonomic identification of commensal bacteria associated with the mucosa and digesta throughout the gastrointestinal tracts of preweaned calves. Appl Environ Microbiol. 2014;80(6):2021–2028. doi: 10.1128/AEM.03864-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Omoniyi LA, et al. An analysis of the ruminal bacterial microbiota in West African Dwarf sheep fed grass- and tree-based diets. J Appl Microbiol. 2014;116(5):1094–1105. doi: 10.1111/jam.12450. [DOI] [PubMed] [Google Scholar]
- 26.Rosewarne CP, Pope PB, Cheung JL, Morrison M. Analysis of the bovine rumen microbiome reveals a diversity of Sus-like polysaccharide utilization loci from the bacterial phylum Bacteroidetes. J Ind Microbiol & Biot. 2014;41(3):601–6. doi: 10.1007/s10295-013-1395-y. [DOI] [PubMed] [Google Scholar]
- 27.Williams, A. G., Coleman, G. S. The rumen protozoa. Brock/springer. 73–139 (1992).
- 28.Fuente GDL, Belanche A, Abecia L, Dehority BA, Fondevil M. Rumen protozoal diversity in the Spanish ibex (Capra pyrenaica hispanica) as compared with domestic goats (Capra hircus) Eur J Protistol. 2009;45(45):112–20. doi: 10.1016/j.ejop.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 29.Kittelmann S, Janssen PH. Characterization of rumen ciliate community composition in domestic sheep, deer, and cattle, feeding on varying diets, by means of PCR-DGGE and clone libraries. Fems Microbiol Ecol. 2011;75(3):468–81. doi: 10.1111/j.1574-6941.2010.01022.x. [DOI] [PubMed] [Google Scholar]
- 30.Booyse DG, Dehority BA, Reininghaus B. Rumen ciliates in the African (Cape) buffalo (Syncerus caffer caffer) living in the vicinity of the Orpen Gate entrance into Kruger National Park, South Africa. Zootaxa. 2014;3846(1):138–144. doi: 10.11646/zootaxa.3846.1.8. [DOI] [PubMed] [Google Scholar]
- 31.Martinele I, et al. Abundance and diversity of rumen protozoa in lambs fed Gliricidia sepium silage. R bras zootec. 2014;43(8):436–9. doi: 10.1590/S1516-35982014000800006. [DOI] [Google Scholar]
- 32.Kittelmann S, et al. Phylogeny of intestinal ciliates, including Charonina ventriculi, and comparison of microscopy and 18 S rRNA gene pyrosequencing for rumen ciliate community structure analysi. s. Appl & Environ Microbiol. 2015;81(7):2433–44. doi: 10.1128/AEM.03697-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu S, et al. Effects of the antimicrobial peptide cecropin AD on performance and intestinal health in weaned piglets challenged with Escherichia coli. Peptides. 2012;35(2):225–30. doi: 10.1016/j.peptides.2012.03.030. [DOI] [PubMed] [Google Scholar]
- 34.Tang ZR. Effects of dietary supplementation with an expressed fusion peptide bovine lactoferricin-lactoferrampin on performance, immune function and intestinal mucosal morphology in piglets weaned at age 21 d. Brit J Nutr. 2009;101(7):998–1005. doi: 10.1017/S0007114508055633. [DOI] [PubMed] [Google Scholar]
- 35.Jin Z, et al. Potato (Solanum tuberosum L. cv. Gogu valley) protein as a novel antimicrobial agent in weanling pigs. J Anim Sci. 2008;86(7):1562–1572. doi: 10.2527/jas.2007-0414. [DOI] [PubMed] [Google Scholar]
- 36.Yoon JH, et al. Effects of dietary supplementation of synthetic antimicrobial peptide-A3 and P5 on growth performance, apparent total tract digestibility of nutrients, fecal and intestinal microflora and intestinal morphology in weanling pigs. J Sci Food Agr. 2014;93:587–592. doi: 10.1002/jsfa.5840. [DOI] [PubMed] [Google Scholar]
- 37.Jin Z, et al. Effects of potato (Solanum tuberosum L. cv. Golden valley) protein having antimicrobial activity on the growth performance, and intestinal microflora and morphology in weanling pigs. Anim Feed Sci Tech. 2008;140(1):139–54. doi: 10.1016/j.anifeedsci.2007.12.006. [DOI] [Google Scholar]
- 38.Wang YZ, Shan TZ, Xu ZR, Feng J, Wang ZQ. Effects of the lactoferrin (LF) on the growth performance, intestinal microflora and morphology of weanling pigs. Anim Feed Sci Tech. 2007;135(3):263–272. doi: 10.1016/j.anifeedsci.2006.07.013. [DOI] [Google Scholar]
- 39.Ohh SH, et al. Effects of potato (Solanum tuberosum I. cv. golden valley) protein on performance, nutrient metabolizability, and cecal microflora in broilers. Arch Fur Geflugelkd. 2010;74(1):30–35. [Google Scholar]
- 40.Tang Z, et al. Effects of dietary supplementation with an expressed fusion peptide bovine lactoferricin-lactoferrampin on performance, immune function and intestinal mucosal morphology in piglets weaned at age 21 d. Brit J Nutr. 2009;101(7):998–1005. doi: 10.1017/S0007114508055633. [DOI] [PubMed] [Google Scholar]
- 41.Stewart CS, Flint HJ. Bacteroides (Fibrobacter) succinogenes, a cellulolytic anaerobic bacterium from the gastrointestinal tract. Appl Microbiol Biot. 1989;30(5):433–439. doi: 10.1007/BF00263846. [DOI] [Google Scholar]
- 42.Stackebrandt ER, Murray RGE, Truper HG. Proteobacteria classis nov. a name for the phylogenetic taxon that includes the “purple bacteria and their relatives. Int J Syst Bacteriol. 1988;38(3):321–325. doi: 10.1099/00207713-38-3-321. [DOI] [Google Scholar]
- 43.Zhu H, Paradis FW, Krell PJ, Phillips JP, Forsberg CW. Enzymatic specificities and modes of action of the two catalytic domains of the XynC xylanase from Fibrobacter succinogenes S85. J Bacteriol. 1994;176(13):3885–94. doi: 10.1128/jb.176.13.3885-3894.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Béramaillet C, Ribot Y, Forano E. Fiber-degrading systems of different strains of the genus Fibrobacter. App & Environ Microb. 2004;70(4):2172–9. doi: 10.1128/AEM.70.4.2172-2179.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stanton TB, Canaleparola E. Treponema bryantii sp. nov., a rumen spirochete that interacts with cellulolytic bacteria. Arch Microbiol. 1980;127(2):145–56. doi: 10.1007/BF00428018. [DOI] [PubMed] [Google Scholar]
- 46.Mah RA, Hungate RE. Physiological Studies on the Rumen Ciliate, Ophryoscolex purkynei Stein. J Eukaryot Microbiol. 1965;12(1):131. doi: 10.1111/j.1550-7408.1965.tb01825.x. [DOI] [PubMed] [Google Scholar]
- 47.Orpin CG. The role of ciliate protozoa and fungi in the rumen digestion of plant cell walls. Anim Feed Sci Tech. 1984;10(2):121–143. doi: 10.1016/0377-8401(84)90003-8. [DOI] [Google Scholar]
- 48.Liu JH, Bian GR, Zhu WY, Mao SY. High-grain feeding causes strong shifts in ruminal epithelial bacterial community and expression of Toll-like receptor genes in goats. Front Microbiol. 2015;6:167. doi: 10.3389/fmicb.2015.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fan, W. B., Liu, Y. B., Chang-Qing, L. I. Development of a Real-time Quantitative PCR for Anaerovibrio lipolytica in Rumen. Acta Agric Boreali Sin. (2015).
- 50.Miki I. 110 Effect of rations on the composition of milk and rumen VFAs. High pressure res. 2010;30(4):643–652. doi: 10.1080/08957959.2010.533664. [DOI] [Google Scholar]
- 51.Kochapakdee S, Pralomkarn W, Saithanoo S, Lawpetchara A, Norton BW. Grazing management studies with Thai goats. I. Productivity of female goats grazing newly established pasture with varying levels of supplementary feeding. Asian Austral J Anim Sci. 1994;7(2):289–93. doi: 10.5713/ajas.1994.289. [DOI] [Google Scholar]
- 52.Mahajan, J. M., Chauhan, D. S., Tomar, V. P. S. Effect of supplementary feeding to grazing on growth and wool production in sheep. Indlan J Anim Res. (1976).
- 53.Salim, H. M., Shahjalal, M., Tareque, A. M. M., Kabir, F. Effects of Concentrate Supplementation on Growth and Reproductive Performance of Female Sheep and Goats under Grazing Condition. Pakistan Journal of Nutrition. (2002).
- 54.Franzolin R, Wright AG. Microorganisms in the rumen and reticulum of buffalo (Bubalus bubalis) fed two different feeding systems. BMC Res Notes. 2016;9(1):1–5. doi: 10.1186/s13104-016-2046-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wetzels SU, et al. Pyrosequencing reveals shifts in the bacterial epimural community relative to dietary concentrate amount in goats. J Dairy Sci. 2015;98(8):5572–87. doi: 10.3168/jds.2014-9166. [DOI] [PubMed] [Google Scholar]
- 56.Khafipour E, Li S, Plaizier JC, Krause DO. Rumen Microbiome Composition Determined Using Two Nutritional Models of Subacute Ruminal Acidosis. Appl & Environ Microb. 2009;75(22):7115–24. doi: 10.1128/AEM.00739-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Metzler-Zebeli BU, et al. Grain-rich diets differently alter ruminal and colonic abundance of microbial populations and lipopolysaccharide in goats. Anaerobe. 2013;20(2):65–73. doi: 10.1016/j.anaerobe.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 58.Lillis L, et al. The effect of dietary concentrate and soya oil inclusion on microbial diversity in the rumen of cattle. J Appl Microbiol. 2011;111(6):1426–35. doi: 10.1111/j.1365-2672.2011.05154.x. [DOI] [PubMed] [Google Scholar]
- 59.Luo, C. et al. editors. Study on Accurate Determination of Volatile Fatty Acids in Rumen Fluid by Capillary Gas Chromatography. International Conference on Information Engineering for Mechanics and Materials; (2015).
- 60.Broderick GA, Kang JH. Automated simultaneous determination of ammonia and total amino acids in ruminal fluid and in vitro media. J Dairy Sci. 1980;63(1):64. doi: 10.3168/jds.S0022-0302(80)82888-8. [DOI] [PubMed] [Google Scholar]
- 61.Horwitz W. Official methods of analysis of the Association of Official Analytical Chemists. Am J Public Health. 1984;46(7):916. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


