Abstract
In order to understand the pathogenic factors that initiate the processes of Alzheimer's disease (AD), a method of inference of multiple differential modules (iMDM) to conduct analysis was performed on the gene expression profile of AD. A total of 11,089 genes and 588,391 interactions were gained based on the gene expression profile and protein-protein interaction network. Subsequently, three differential co-expression networks (DCNs) were constructed with the same nodes but different interactions, and eight multiple differential modules (M-DMs) were identified. Furthermore, by performing Module Connectivity Dynamic Score to quantify the change in the connectivity of component modules, two M-DMs were identified: Module 1 (P=0.0419) and 2 (P=0.0419; adjusted, P≤0.05). Finally, hub genes of MDH1, NDUFAB1, NDUFB5, DDX1 and MRPS35 were gained via topological analysis conducted on the 2 M-DMs. In conclusion, the method of iMDM was suitable for conducting analysis on AD. By applying iMDM, 2 M-DMs were successfully identified and the MDH1, NDUFAB1, NDUFB5, DDX1 and MRPS35 genes were predicted to be important during the occurrence and development of AD.
Keywords: Alzheimer's disease, inference of multiple differential modules, multiple differential co-expression network, hub gene
Introduction
Alzheimer's disease (AD) is a global public health threat that continues to increase as the proportion of the population over the age of 60 rapidly increases (1). As age is the most striking risk factor for AD, the growth of the aging population complicates this public health problem and places a strain on health care systems (2). It is known that AD is associated with perturbations of neural circuitry, which change dynamically as the disease progresses (3). Based on the Mini-Mental State Examination, a useful index to assess cognitive function level, subjects may be categorized into four groups, as follows: Control (score >25), incipient AD (score 20–26), moderate AD (score 14–19), and severe AD (score <14) (4,5). However, the pathogenic factors that initiate the processes of AD remain elusive.
Gene expression data have become an important resource in describing the molecular state associated with various cellular phenotypes and responses to external or internal perturbations (6). However, the probability that a false identification is made may increase when the number of tested genes increases (7). Furthermore, network-based approaches may be used to extract informative genes relying on biomolecular networks, such as the protein-protein interaction (PPI) network, gene regulatory network (8,9) and gene co-expression networks (10) amongst others, rather than individual genes. In co-expression networks, two genes are connected and assumed to functionally interact if there is a correlation in their expression profiles across multiple conditions (11). However, a limitation of previous studies is that networks were only constructed using co-expression information (6,11).
Inference of multiple differential modules (iMDM), a novel computational framework, achieves higher accuracy in inferring gene modules compared with the use of single or multiple co-expression networks (12). More specifically, iMDM enables simultaneous analysis of multiple differential co-expression networks (DCNs), and is able to identify the coherently differentially-expressed genes that are either unique or shared among multiple DCNs as a module. Furthermore, a novel graph-theoretical measure is applied to quantify the dynamic changes in gene modules across conditions, and the gene modules with higher connectivity dynamics may have a greater association with the evolution and progression of the disease. This suggests that iMDM may provide a method to elucidate the dynamics of gene modules and their association with the dynamics of the disease phenotype.
Therefore, in the present study, in order to further identify pathogenic factors that initiate the processes of AD, the iMDM method was applied to conduct an analysis on AD. To achieve this, a gene expression profile of AD and human PPI data were initially obtained. Subsequently, iMDM was performed to identify DCNs and candidate significant multiple differential modules (M-DMs). Finally, a topological analysis of M-DMs was conducted to identify hub genes, which may be important during the occurrence and development of AD.
Materials and methods
Data recruiting
Transcript data
The gene expression profile of AD, with access number GSE1297 (5), was downloaded from the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo/). GSE1297 existed on the A-AFFY-33-Affymetrix GeneChip Human Genome HG-U133A [HG-U133A] platform, which was comprised of 9 normal control samples and 22 AD samples [7 incipient (score 20–26), 8 moderate (score 14–19) and 7 severe (score <14)], and the normal control samples were selected as a baseline for AD in the present study. The microarray data and annotation files were obtained.
In order to eliminate the influence of non-specific hybridization, a robust multichip average method (13) was used to perform background correction and a quantiles based algorithm (14) to conduct normalization. Additionally, a Micro Array Suite 5.0 algorithm (Affymetrix, Inc, Santa Clara, CA, USA) (15) was used to revise the perfect match and mismatch value, the value of which was the median value. Finally, the gene expression profile on the probe level was converted into gene symbol level using the annotate package from Bioconductor 3.4 (http://www.bioconductor.org/), duplicated symbols were removed, and a total of 12,493 gene symbols were obtained for further analysis.
PPI data
The global PPI network was integrated from the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING; string-db.org/). In the STRING, there were a total of 787,896 interactions (16,730 genes) of human beings. Based on the preprocessed gene expression profile of AD, a total of 11,089 genes were mapped onto the global PPI network. In addition, the corresponding gene associations were selected from the PPI network, and a total of 588,391 interactions were obtained.
iMDM method
DCN construction
For each of the three stages of AD (incipient, moderate and severe), two steps were performed to construct the DCN. Initially, a binary co-expression network was built. To gain the co-expressed genes, Pearson correlation coefficients (PCC) (16) were implemented to calculate the correlation of each gene pair in the disease. In the present study, for each stage, edges of with absolute values of PCC≥0.9 were separately selected to construct the binary co-expression network.
The second step was that each edge of the binary co-expression network was assigned a weight value based on the differential gene expression between the disease of each stage and the baseline. In the present study, EdgeR V3.16.5 in Bioconductor (17), which is used for differential expression analysis of digital gene expression data, was utilized to detect differential gene expression for the microarray data. Prior to analysis with EdgeR, the P-values of differential gene expression among stages and the baseline condition were obtained using a one-sided Student's t-test. Subsequently, the weight (wx,y) on edge (x,y) in the differential network was calculated as follows:
Where px and py were P-values of differential expression for the × and y genes (P<0.05 was considered to indicate a statistically significant difference), respectively, N was the node set of the co-expression network, and cor (x,y) was the absolute value of PCC between the x, y genes based on their expression profiles.
Under this weighting scheme, genes that were co-expressed and significantly differentially expressed were assigned higher weights, which satisfied the assumption of the present study that those genes likely to participate in a pathway that exhibited differential activities between the two conditions being compared. P<0.05 was considered to indicate a statistically significant difference. Therefore, three DCNs for the three stages of AD were separately built based on the analysis above.
Identification of M-DMs in multiple DCNs
After each edge was given a weight value, candidate M-DMs were mined. The M-module algorithm, which was designed for identifying gene modules with common members but varied connectivity across multiple molecular interaction networks, was applied to identify M-DMs (18). Furthermore, the M-DM search consisted of three steps: Seed prioritization, module search by seed expansion and refinement of candidate modules.
Seed prioritization to seek seed genes
Initially, the genes contained in the DCNs were ranked based on the degree centrality features of the genes in the network. More specifically, for each network Gm=(N, Em) (1≤m≤M) with an adjacency matrix Ai, the importance of the gene × in each DCN was calculated according to the following formula:
Where g(x) denoted the importance of vertex x in the corresponding network, Ni(x) denoted the set of neighbors of x in Gm and A'i denoted the degree normalized weighted adjacency matrix. The product A'-g denoted the information propagation on network via the edges of networks, which meant the importance of a node was associated with the number of its neighbors, strength of connection and importance of its neighbors.
According to the computational formula given above, the importance of the genes in all individual networks was obtained, which were denoted as z-score values. Subsequently, all z-score values were ranked in descending order in all individual networks, and the rank for that gene was obtained across all networks by calculating the mean z-scores across all networks. The top 5% of genes, with the highest z-scores, were selected as seed genes.
M-DMs searching
Initially, each seed gene was selected as a differential module, S. Next, starting from each seed gene, x, the network gene y that was adjacent to x was added to S to form a module S'. The entropy decrease between these two modules was calculated according to the following formula:
∆H(S', S)>0 indicated that the addition of vertex y improved the connectivity of the former M-module, S. Furthermore, genes were iteratively added to module S that were adjacent to x until there was no decrease in the objective function of ∆H. In this case, all genes were connected together to form M-DMs.
Refinement of candidate modules
In the refinement step, only those M-DMs with sizes ≥5 were kept. In addition, the ratio of intersection over union for two sets was measured by the Jaccard index, and the two sets were merged into a module with a Jaccard index ≥0.5.
Calculation of the statistical significance of candidate M-DMs
To further analyze the statistical significance of candidate M-DMs, randomized networks were constructed based on the null score distribution of M-DMs. A module search was performed on the randomized networks according to the methods mentioned above. Each network was completely randomized 100 times. Furthermore, the empirical P-value of an M-DM was calculated as the probability of the module having the observed score or smaller by chance. Furthermore, the Benjamini and Hochberg (19) method was performed to conduct multiple testing on the P-values. The modules with P≤0.05 were considered as M-DMs.
Quantification of connectivity dynamics of shared M-DMs
A graph-theoretical measure, the Module Connectivity Dynamic Score (MCDS) was utilized for the M-DMs that had multiple component modules from different DCNs in order to quantify the change in the connectivity of the component modules. Briefly, given a dynamic module (C) whose weighted adjacent matrices of the corresponding induced subgraphs were denoted by (1≤x≤M), the MCDS between two adjacent component modules was defined as the L2 norm of the matrix subtraction normalized by the number of genes in the M-DM, for example,
where || ||2 was the matrix L2 norm. Furthermore, the overall MCDS of an M-DM was defined as the mean MCDS of all pairwise comparisons:
The statistical significance of MCDS for an M-DM was computed in a similar way as that for M-DMs, and Benjamini-Hochberg was used to conduct multiple testing corrections. The modules were adjusted and values with P≤0.05 were considered as differential modules with significant connectivity dynamics.
Topological analysis of M-DMs
Each M-DM with higher quantification of connectivity dynamics was considered as a sub-network, and topological analysis of differential modules was conducted in order to further investigate the biological functions and significance of nodes in the differential modules, indices of topological analysis [degree (20), closeness (21), betweenness (22) and shortest path length (23)], and were performed to characterize the importance of the nodes. Traditionally, nodes with a high degree (highly connected) are called ‘hubs’, which interact with several other genes, suggesting a central role in the interaction network. In the present study, the top 10% of the genes according to the degree centralities were considered as hub genes.
Results
DCN constructions
In the present study, an analysis was conducted on the gene expression profile of AD to further elucidate its molecular mechanism. After having selected the gene expression profile that contained the interactions of the PPI network, the absolute value of the PCC of the expression profiles of each gene interaction in each stage was calculated. In total, three DCNs, one for each stage of AD, were built separately. It was revealed that the nodes in these DCNs were the same, whereas the interactions were different from each other in the three stages of AD. Immediately after, the EdgeR package was utilized to assign weight values based on differential gene expression between the different stages of AD and the baseline, respectively. In this case, three DCNs that had the same nodes but different interactions were identified.
Identification of M-DMs in multiple DCNs
Initially, the genes contained in the DCNs were ranked according to the degree centrality values in each DCN. Degree centrality values were subsequently converted into z-score values, and ranked for that gene across all networks by calculating the mean z-score across all networks. Furthermore, the top 5% genes with the highest z-scores were selected as seed genes, and 28 seed genes were obtained (Table I).
Table I.
Z-score values of the 28 seed genes.
| Gene symbol | Z-score 1 (incipient AD) | Z-score 2 (moderate AD) | Z-score 3 (severe AD) | Average |
|---|---|---|---|---|
| NDUFB5 | 1,003.556 | 1,437.713 | 2,098.125 | 1,513.132 |
| DDX1 | 905.415 | 1,359.723 | 1,809.015 | 1,358.051 |
| NDUFAB1 | 854.492 | 1,262.754 | 1,610.130 | 1,242.459 |
| ACTR6 | 802.895 | 1,034.827 | 1,646.069 | 1,161.264 |
| C14orf166 | 794.725 | 1,102.397 | 1,426.624 | 1,107.916 |
| ACTR10 | 694.259 | 1,063.502 | 1,541.512 | 1,099.757 |
| MRPS35 | 694.404 | 1,000.464 | 1,508.590 | 1,067.819 |
| HSPE1 | 650.006 | 1,122.219 | 1,431.178 | 1,067.801 |
| MDH1 | 602.146 | 1,125.608 | 1,417.861 | 1,048.538 |
| NDUFB3 | 713.060 | 1,035.019 | 1,391.509 | 1,046.529 |
| SLIRP | 688.469 | 1,007.552 | 1,440.883 | 1,045.634 |
| DLD | 617.417 | 913.604 | 1,460.811 | 997.277 |
| COPS5 | 661.617 | 978.652 | 1,332.187 | 990.819 |
| PSMD10 | 635.760 | 927.430 | 1,383.522 | 982.237 |
| LAMTOR3 | 753.325 | 913.979 | 1,254.756 | 974.020 |
| ATP5F1 | 618.705 | 942.688 | 1,325.607 | 962.333 |
| SNRNP27 | 628.393 | 882.229 | 1,349.686 | 953.436 |
| EIF3J | 730.916 | 800.174 | 1,284.984 | 938.691 |
| COX7C | 596.897 | 883.640 | 1,234.575 | 905.037 |
| PTGES3 | 742.810 | 752.692 | 1,160.379 | 885.293 |
| DYNLL1 | 531.537 | 934.580 | 1,125.114 | 863.744 |
| PCMT1 | 570.177 | 785.947 | 1,145.326 | 833.816 |
| PSMB1 | 587.862 | 799.568 | 1,106.171 | 831.200 |
| SRP9 | 581.777 | 759.436 | 1,136.389 | 825.867 |
| DPM1 | 572.269 | 725.154 | 1,092.859 | 796.761 |
| METTL5 | 516.985 | 830.050 | 999.440 | 782.158 |
| POMP | 526.684 | 757.540 | 996.599 | 760.275 |
| GHITM | 474.148 | 787.383 | 988.797 | 750.110 |
Z-score 1 corresponds to incipient AD; Z-score 2 corresponds to moderate AD; Z-score 3 corresponds to severe AD.
Beginning with each seed gene, modules were searched for each stage. By removing those M-DMs with sizes <5, as well as merging the two sets with Jaccard index ≥0.5, 17 modules were identified. These modules were regarded as candidate M-DMs. To further analyze the statistical significance of candidate M-DMs, a randomized test method was applied to conduct the analysis, and the Benjamini-Hochberg method was performed to conduct multiple testing on the P-values. By setting the threshold value of adjusted P≤0.05, eight M-DMs were gained.
Quantification of connectivity dynamics of shared M-DMs
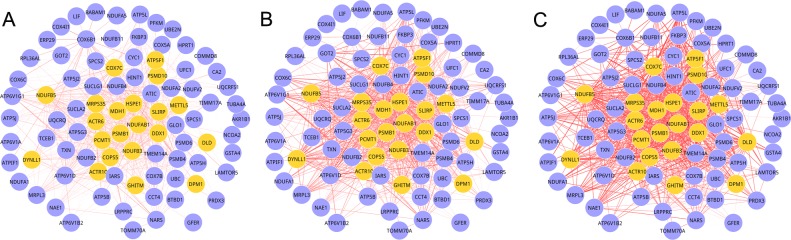
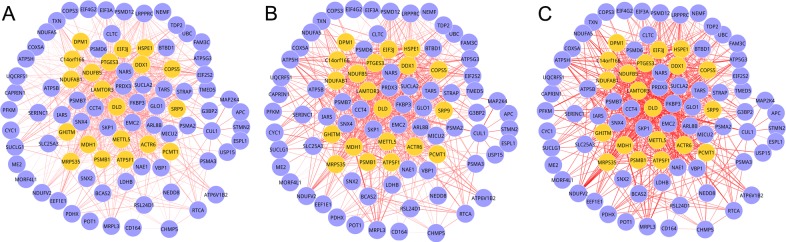
For the eight M-DMs that had multiple component modules from different DCNs, MCDS was performed to quantify the change in the connectivity of the component modules. Under adjusted P≤0.05, two M-DMs: Module 1 (P=0.0419) and 2 (P=0.0419) exhibited significant connectivity dynamics across multiple conditions (P<0.05). These two M-DMs were regarded to be closely associated with AD. As shown in Figs. 1 and 2, there were 88 nodes (884 edges) in module 1 and 89 nodes (731 edges) in module 2. As observed from Figs. 1 and 2, the gene composition of each module in the three stages was the same, but the edges varied. As the edge weight in DCNs was a measure of differential gene expression between the disease and normal control conditions, a larger mean edge weight of an M-DM indicated a greater difference in the expression of module genes. Therefore, after investigating the root tracing, the dynamics of the M-DMs resulted from the gene expression value of the genes in different stages of the AD.
Figure 1.
Multiple differential modules of module 1 for the three stages of AD. (A) Incipient, (B) moderate and (C) severe AD. Yellow nodes represent the seed genes, and the thickness of the edges represented the strength of the interactions between the nodes. AD, Alzheimer's disease.
Figure 2.
Multiple differential modules of module 2 for the three stages of AD. (A) Incipient, (B) moderate and (C) severe AD. Yellow nodes represent the seed genes, and the thickness of the edges represented the strength of the interactions between the nodes. AD, Alzheimer's disease.
Topological analysis of M-DMs
Since it was indicated that the dynamics of the M-DMs resulted from the genes, M-DMs with higher quantification of connectivity dynamics were separately considered as a network in order to further disclose the molecular mechanism of the AD. Topological analysis was conducted on them to further to investigate the biological functions and significance of the nodes in the differential modules. In these DMs, the nodes and the number of edges were the same in all stages of AD, and only the edge values were different. Therefore, the degree centralities were the same in different stages of AD. As the genes were ranked in descending order according to the degree centralities, the top 10% of the genes were considered as hub genes. For module 1, 8 hub genes were obtained as shown in Table II: MDH1 (degree=78), NDUFAB1 (degree=65), NDUFB5 (degree=64), SLIRP (degree=52), DDX1 (degree=51), HSPE1 (degree=50), NDUFB3 (degree=47) and MRPS35 (degree=44). A total of 8 hub genes were also obtained for module 2 as shown in Table III: DLD (degree=80), NDUFB5 (degree=55), ACTR6 (degree=50), NDUFAB1 (degree=46), MRPS35 (degree=45), DDX1 (degree=43), PTGES3 (degree=39) and MDH1 (degree=37). Additionally, it was identified that all hub genes were also seed genes. Furthermore, MDH1, NDUFAB1, NDUFB5, DDX1 and MRPS35 were hub genes in both modules 1 and 2, and it was predicted that they may have close association during the occurrence and development of AD.
Table II.
Topological properties of the hub genes in module 1.
| Gene | Degree | Closeness | Betweenness | Shortest Path Length |
|---|---|---|---|---|
| MDH1 | 78 | 0.906250 | 0.25422146 | 1.10344828 |
| NDUFAB1 | 65 | 0.776786 | 0.06621343 | 1.28735632 |
| NDUFB5 | 64 | 0.769912 | 0.05491728 | 1.29885057 |
| SLIRP | 52 | 0.696000 | 0.02372897 | 1.43678161 |
| DDX1 | 51 | 0.690476 | 0.02436002 | 1.44827586 |
| HSPE1 | 50 | 0.685039 | 0.02535913 | 1.45977011 |
| NDUFB3 | 47 | 0.669231 | 0.01829438 | 1.49425287 |
| MRPS35 | 44 | 0.654135 | 0.01638579 | 1.52873563 |
Table III.
Topological properties of the hub genes in module 2.
| Gene | Degree | Closeness | Betweenness | Shortest Path Length |
|---|---|---|---|---|
| DLD | 80 | 0.916667 | 0.326576 | 1.090909 |
| NDUFB5 | 55 | 0.709677 | 0.060582 | 1.409091 |
| ACTR6 | 50 | 0.682171 | 0.078818 | 1.465909 |
| NDUFAB1 | 46 | 0.661654 | 0.028408 | 1.511364 |
| MRPS35 | 45 | 0.656716 | 0.029722 | 1.522727 |
| DDX1 | 43 | 0.661654 | 0.034592 | 1.511364 |
| PTGES3 | 39 | 0.624113 | 0.02588 | 1.602273 |
| MDH1 | 37 | 0.619718 | 0.02162 | 1.613636 |
Discussion
Network biology has been proven to be a powerful tool for analyzing and representing complex molecular networks. From a systems biology point of view, diseases are caused by perturbations to the gene network, which change dynamically during the progression of the disease (24). As genes present in the same pathway tend to have correlated expression, analyzing a co-expression network is an effective strategy for pathway inference (25). Furthermore, complex diseases such as AD are associated with a continuum of molecular events that begin with the early initiation events through progression and catastrophic end-stage events. Although analyzing and understanding disease-stage-specific molecular events are critical for understanding disease etiology and development of therapeutic interventions, the current knowledge on the dynamics of gene networks during disease progression is rather limited. Many previous studies on connectivity dynamics have been focused on the dynamics of hub genes, such as using integrative network analysis to both identify candidate driver genes and candidate pathways (26). Vandin et al (27) have used a network diffusion algorithm to identify sub-networks enriched for mutations within a large gene interaction network. Additionally, studies have also investigated the network properties of complex disease alterations, and have noted that complex disease alterations tend to cluster within closely knit network modules or communities, and that altered modules are associated with specific biological pathways (28,29). However, there were two problems for understanding network dynamics; identifying the sub-networks that exhibit structural changes in response to different growth or developmental events, or different spatial location and quantifying the dynamic changes in the sub-network structure (18). Using iMDM, only sets of genes that are differentially expressed as well as those that exhibit correlated expression patterns under diseased states will be identified as a module, so that the entire pathway consists of perturbed genes under disease condition.
In the present study, iMDM was applied as a novel method to conduct analysis on the gene expression profile of AD, and two M-DMs were identified: Module 1 (P=0.0419) and 2 (P=0.0419), which had higher quantification of connectivity dynamics. In order to understand the association between the genes and AD and because the dynamics of the M-DMs resulted from the gene expression value of the genes at different stages of the AD, a topological analysis was conducted on these two M-DMs. It was revealed that MDH1, NDUFAB1, NDUFB5, DDX1 and MRPS35 were hub genes in both modules 1 and 2. Furthermore, it was predicted that these genes may have a close association during the occurrence and development of AD. In order to further expose the correlation between these genes and AD, the MDH1 gene was selected as an example.
MDH1, which encodes soluble cytosolic malate dehydrogenase (MDH1), is important in transporting nicotinamide adenine dinucleotide equivalents across the mitochondrial membrane, controlling the tricarboxylic acid cycle pool size and providing contractile function (30). The enzyme that functionally connects glutamate dehydrogenase (GDH) and MDH1 is aspartate aminotransferase (AAT). GDH is co-localized in cells with AAT, which participates in the malate-aspartate shuttle with MDHs (31). Previous studies have indicated that, under AD conditions, the activity of MDH is increased in the brain (32), and the activity also increases with age in a rat brain (33). Furthermore, Korolainen et al (34) have indicated that the total amount of soluble MDH1 is unchanged in AD when compared with controls, indicating differences in the amounts of distinctive isoforms. The most likely explanations are aberrant post-translational modification of MDH1 in AD leading to a differential quantitative distribution of isoforms when compared with controls. However, the most notable result, from the study of Korolainen et al (34), was that GDH and MDH1, which were closely associated with neurotransmitter release and energy metabolism, were less oxidized in AD, compared with controls. In the present study, MDH1 was identified as a hub gene in both M-DMs, which suggests that it serves an important role in AD.
In conclusion, in the present study a method of iMDM was performed in order to conduct analysis on AD. By applying iMDM, two M-DMs were successfully identified, and the MDH1, NDUFAB1, NDUFB5, DDX1 and MRPS35 genes were predicted to have key roles during the occurrence and development of AD. Further experimental verification will be conducted to verify the results of the present study.
Acknowledgements
The present study was supported by the Office of Medical Social Work and Department of Neurology, Yidu Central Hospital of Weifang (Weifang, China). The authors would like to thank all members of the research group. Furthermore, the authors are grateful to Beijing Springer Medical Research Institute for providing professional translation and manuscript editing services.
References
- 1.Glenner GG, Wong CW. Alzheimer's disease: Initial report of the purification and characterization of a novel cerebrovascular amyloid protein. 1984. Biochem Biophys Res Commun. 2012;425:534–539. doi: 10.1016/j.bbrc.2012.08.020. [DOI] [PubMed] [Google Scholar]
- 2.Alzheimer's Association: 2014 Alzheimer's disease facts and figures. Alzheimers Dement. 2014;10:e47–e92. doi: 10.1016/j.jalz.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Leal SL, Yassa MA. Perturbations of neural circuitry in aging, mild cognitive impairment, and Alzheimer's disease. Ageing Res Rev. 2013;12:823–831. doi: 10.1016/j.arr.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark CM, Sheppard L, Fillenbaum GG, Galasko D, Morris JC, Koss E, Mohs R, Heyman A. Variability in annual mini-mental state examination score in patients with probable Alzheimer disease: A clinical perspective of data from the consortium to establish a registry for Alzheimer's disease. Arch Neurol. 1999;56:857–862. doi: 10.1001/archneur.56.7.857. [DOI] [PubMed] [Google Scholar]
- 5.Blalock EM, Geddes JW, Chen KC, Porter NM, Markesbery WR, Landfield PW. Incipient Alzheimer's disease: Microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci USA. 2004;101:2173–2178. doi: 10.1073/pnas.0308512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter SL, Brechbühler CM, Griffin M, Bond AT. Gene co-expression network topology provides a framework for molecular characterization of cellular state. Bioinformatics. 2004;20:2242–2250. doi: 10.1093/bioinformatics/bth234. [DOI] [PubMed] [Google Scholar]
- 7.Dudoit S, Yang YH, Callow MJ, Speed TP. Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Stat Sin. 2002;12:111–139. [Google Scholar]
- 8.Jeong H, Mason SP, Barabási AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 9.Teichmann SA, Babu MM. Gene regulatory network growth by duplication. Nat Genet. 2004;36:492–496. doi: 10.1038/ng1340. [DOI] [PubMed] [Google Scholar]
- 10.Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 11.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- 12.Ma X, Gao L, Karamanlidis G, Gao P, Lee CF, Garcia-Menendez L, Tian R, Tan K. Revealing pathway dynamics in heart diseases by analyzing multiple differential networks. PLoS Comput Biol. 2015;11:e1004332. doi: 10.1371/journal.pcbi.1004332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma L, Robinson LN, Towle HC. ChREBP*Mlx is the principal mediator of glucose-induced gene expression in the liver. J Biol Chem. 2006;281:28721–28730. doi: 10.1074/jbc.M601576200. [DOI] [PubMed] [Google Scholar]
- 14.Rifai N, Ridker PM. Proposed cardiovascular risk assessment algorithm using high-sensitivity C-reactive protein and lipid screening. Clin Chem. 2001;47:28–30. [PubMed] [Google Scholar]
- 15.Pepper SD, Saunders EK, Edwards LE, Wilson CL, Miller CJ. The utility of MAS5 expression summary and detection call algorithms. BMC Bioinformatics. 2007;8:273. doi: 10.1186/1471-2105-8-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benesty J, Chen J, Huang Y, Cohen I. Pearson correlation coefficient. Noise reduction in speech processing. 2009:1–4. doi: 10.1007/978-3-642-00296-0_7. [DOI] [Google Scholar]
- 17.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma X, Gao L, Tan K. Modeling disease progression using dynamics of pathway connectivity. Bioinformatics. 2014;30:2343–2350. doi: 10.1093/bioinformatics/btu298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc. 1995;57:289–300. [Google Scholar]
- 20.Haythornthwaite C. Social network analysis: An approach and technique for the study of information exchange. Libr Inf Sci Res. 1996;18:323–342. doi: 10.1016/S0740-8188(96)90003-1. [DOI] [Google Scholar]
- 21.Wasserman S, Faust K. Social network analysis: Methods and applications. Struct Anal Soc Sci. 1994;8 [Google Scholar]
- 22.Barthelemy M. Betweenness centrality in large complex networks. Eur Phys J B. 2004;38:163–168. doi: 10.1140/epjb/e2004-00111-4. [DOI] [Google Scholar]
- 23.Chuang TN, Kung JY. The fuzzy shortest path length and the corresponding shortest path in a network. Comput Oper Res. 2005;32:1409–1428. doi: 10.1016/j.cor.2003.11.011. [DOI] [Google Scholar]
- 24.Ellis JD, Barrios-Rodiles M, Çolak R, Irimia M, Kim T, Calarco JA, Wang X, Pan Q, O'Hanlon D, Kim PM, et al. Tissue-specific alternative splicing remodels protein-protein interaction networks. Mol Cell. 2012;46:884–892. doi: 10.1016/j.molcel.2012.05.037. [DOI] [PubMed] [Google Scholar]
- 25.D'haeseleer P, Liang S, Somogyi R. Genetic network inference: From co-expression clustering to reverse engineering. Bioinformatics. 2000;16:707–726. doi: 10.1093/bioinformatics/16.8.707. [DOI] [PubMed] [Google Scholar]
- 26.Torkamani A, Schork NJ. Identification of rare cancer driver mutations by network reconstruction. Genome Res. 2009;19:1570–1578. doi: 10.1101/gr.092833.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vandin F, Upfal E, Raphael BJ. Algorithms for detecting significantly mutated pathways in cancer. J Comput Biol. 2010;18:506–521. doi: 10.1089/cmb.2010.0265. [DOI] [PubMed] [Google Scholar]
- 28.Cerami E, Demir E, Schultz N, Taylor BS, Sander C. Automated network analysis identifies core pathways in glioblastoma. PLoS One. 2010;5:e8918. doi: 10.1371/journal.pone.0008918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu G, Feng X, Stein L. Research a human functional protein interaction network and its application to cancer data analysis. Genome Biol. 2010;11:R53. doi: 10.1186/gb-2010-11-5-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lo AS, Liew CT, Ngai SM, Tsui SK, Fung KP, Lee CY, Waye MM. Developmental regulation and cellular distribution of human cytosolic malate dehydrogenase (MDH1) J Cell Biochem. 2005;94:763–773. doi: 10.1002/jcb.20343. [DOI] [PubMed] [Google Scholar]
- 31.Kimmich GA, Roussie JA, Randles J. Aspartate aminotransferase isotope exchange reactions: Implications for glutamate/glutamine shuttle hypothesis. Am J Physiol Cell Physiol. 2002;282:C1404–C1413. doi: 10.1152/ajpcell.00487.2001. [DOI] [PubMed] [Google Scholar]
- 32.den Velde Op W, Stam FC. Some cerebral proteins and enzyme systems in Alzheimer's presenile and senile dementia. J Am Geriatr Soc. 1976;24:12–16. doi: 10.1111/j.1532-5415.1976.tb03247.x. [DOI] [PubMed] [Google Scholar]
- 33.Malik P, McKenna MC, Tildon JT. Regulation of malate dehydrogenases from neonatal, adolescent and mature rat brain. Neurochem Res. 1993;18:247–257. doi: 10.1007/BF00969080. [DOI] [PubMed] [Google Scholar]
- 34.Korolainen MA, Goldsteins G, Nyman TA, Alafuzoff I, Koistinaho J, Pirttilä T. Oxidative modification of proteins in the frontal cortex of Alzheimer's disease brain. Neurobiol Aging. 2006;27:42–53. doi: 10.1016/j.neurobiolaging.2004.11.010. [DOI] [PubMed] [Google Scholar]