Abstract
A split NanoLuc assay system consisting of two fragments, large N-terminal and small C-terminal regions (NanoBiT), was developed to investigate protein-protein interactions within living cells. Interestingly, the replacement of five amino acids among 11 C-terminal amino acids dramatically increased affinity against the large N-terminal fragment, LgBiT, and the complex had NanoLuc luciferase activity. In this study, we first applied this small fragment, HiBiT, to elucidate the expression of ATF4 protein by transient overexpression of HiBiT-tagged ATF4. According to the regulation of intrinsic ATF4 protein, stabilization of HiBiT-tagged ATF4 with a proteasome inhibitor, MG132, was observed by detecting luciferase activity in cell lysate and after SDS-PAGE and transfer onto a PVDF membrane. Next, we knocked-in the HiBiT-epitope tag into the ATF4 gene using the CRISPR/Cas9 system and rapidly selected positive clones by measuring luciferase activity in an aliquot of each cell suspension. Using a selected clone, we observed that the expression of HiBiT-tagged ATF4 in the selected cells varied in response to treatment with protein synthesis inhibitors or proteasome inhibitors and tunicamycin. Altogether, this novel HiBiT tag is a useful tool to evaluate the endogenous expression levels of proteins of interest.
Keywords: ATF4, CRISPR/Cas9, HiBiT, NanoLuc
Highlights
-
•
A novel small epitope-tag, HiBiT, was applied to detect ATF4 protein expression.
-
•
ATF4-HiBiT Neuro2a cells were generated with CRISPR/Cas9 system.
-
•
This HiBiT tag is a useful tool to study other proteins.
1. Introduction
NanoLuc (approximately 19 kDa) produces sustained luminescence and is smaller than green fluorescent protein (GFP), which is commonly used to study protein translocation, and the sensitivity of NanoLuc is also higher than that of the commonly used firefly luciferase [1]. Based on the high sensitivity of NanoLuc, we used NanoLuc to evaluate several ER stress responses, including intracellular transport and secretion of ER stress-related factors and the splicing activity of ER stress-dependent transcription factor, XBP1 [2], [3], [4]. Very recently, we also developed a NanoLuc based-CRISPR/Cas9 system and monitored the endogenous promoter activity of GRP78, an ER stress inducible chaperone, in HEK293 cells [5].
On the other hand, protein engineering for NanoLuc developed another promising approach, a split NanoLuc called NanoBiT, to investigate protein-protein interactions within living cells. This NanoBiT is composed of two fragments, large N-terminal (LgBiT) and small C-terminal (SmBiT) regions, that do not spontaneously interact with each other [6]. Using this NanoBiT system, we found that a single amino acid mutation (G85R and G93A) in human SOD1, one of the causal factors in amyotrophic lateral sclerosis (ALS), abolished its homodimerization in living cells [7]. Interestingly, the 11 amino acids in the C-terminal in which five amino acids were replaced, HiBiT, dramatically increased affinity against LgBiT, and the complex showed NanoLuc luciferase activity [6]. In this study, we used this unique feature of HiBiT to elucidate the expression of ATF4, a well-known ER stress-inducible transcription factor [8], [9], [10]. In combination with the CRISPR/Cas9 system [11], [12], we established knock-in cells containing HiBiT-tagged ATF4 and detected changes in ATF4 following treatment with protein synthesis inhibitors, proteasome inhibitors or tunicamycin.
2. Materials and methods
2.1. Materials
Cycloheximide (CHX), MG132 (MG) and tunicamycin (Tm) were obtained from Sigma-Aldrich, Peptide Institute and Abcam, respectively.
2.2. Construction of plasmids
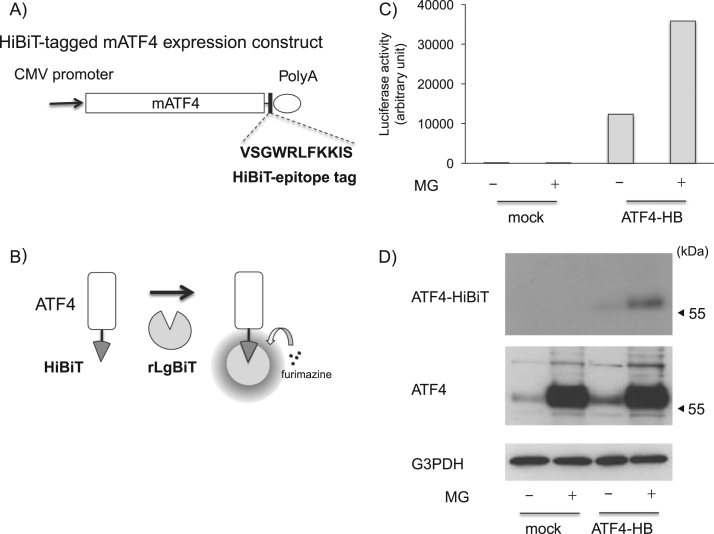
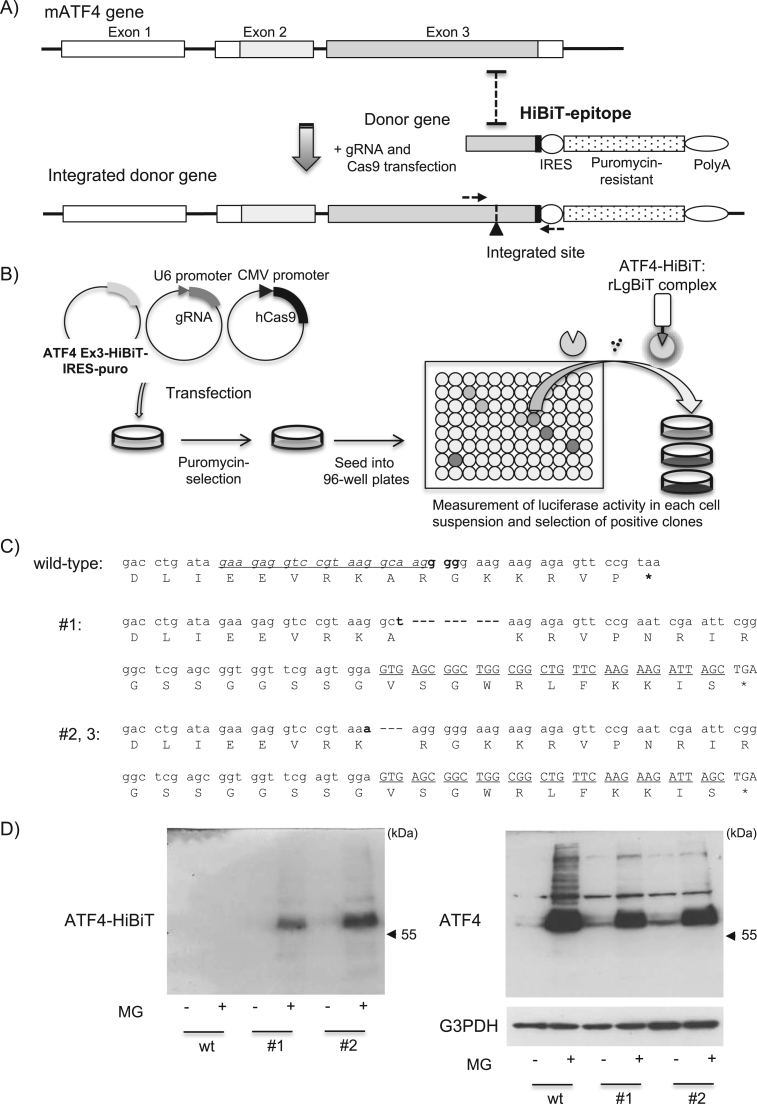
To prepare HiBiT-tagged full-length mouse ATF4, we amplified the full-length ATF4 gene lacking a stop codon using PCR from Neuro2a-derived cDNA and inserted the gene into a pcDNA3.1 vector with a HiBiT epitope, VSGWRLFKKIS (Fig. 1A), at the C-terminus. Twelve amino acids, NRIRGSSGGSSG, were inserted between ATF4 and the HiBiT epitope as a linker sequence. To generate the donor gene for CRISPR/Cas9 gene editing, we amplified ATF4 Ex3-HiBiT, the C-terminal coding region (129 aa) with the HiBiT epitope, from the above full-length ATF4 HiBiT and inserted it into a pGL3-based vector with a puromycin-resistance gene through the IRES sequence (Promega) (Fig. 2A). The gRNA against mouse ATF4 (5′-GAAGAGGTCCGTAAGGCAAG-3′) aligned with tracer RNA was inserted into a pcDNA3.1-derived vector with a U6 promoter. The hCas9 construct (#41815) used in this study was obtained from Addgene [11].
Fig. 1.
Transient overexpression of HiBiT-tagged ATF4 in Neuro2a cells. A) Schematic structure of a HiBiT-tagged ATF4 construct. B) A mechanism of HiBiT-derived luciferase activity. C) Twenty-four hours after transfection with HiBiT-tagged ATF4 or pcDNA3.1 empty vector, cells were treated with MG132 (MG, 10 μM) or vehicle for an additional 12 h. After the cells were harvested and lysed with homogenization buffer, each lysate containing 1 μg protein was mixed with the same amount of reaction mixture containing recombinant LgBiT (rLgBiT) and furimazine in diluted HiBiT lytic buffer. After an incubation at 37 °C for 10 min, each luciferase activity in each sample was measured as described in the Materials and methods section. D) Equal amounts of cell lysate prepared in (C) were separated with SDS-PAGE and transferred onto PVDF membranes. Expression levels of HiBiT-derived signals, ATF4 and G3PDH were detected as described in the Materials and methods section.
Fig. 2.
Establishment of HiBiT knock-in cells to monitor intrinsic ATF4 protein expression in Neuro2a cells. A, B) The schematic structure of the donor gene coding the C-terminal region of ATF4 fused with the HiBiT epitope and the strategy for establishing the HiBiT knock-in cells. An arrowhead indicates the integrated site. Dashed arrows indicate the PCR primer pairs used for the amplification of gene sequences around the HiBiT-integrated mouse ATF4 gene in the knock-in cells. C) Nucleotide sequences corresponding to the C-terminal region of the mouse ATF4 gene (top, NM_009716.3) and those edited by the present CRISPR/Cas9 system. The underlined italic letters and the three bold letters indicate the target sequence of the gRNA and PAM site, respectively. The bold letters in the second and third nucleotide sequence (#1, #2 and #3) indicates substituted nucleotide, respectively. The underlined large letters indicate the nucleotide sequences of the HiBiT epitope. The sequences of #2 and #3 around the ATF4 C-terminal region were identical. D) Wild-type (wt) Neuro2a and 1 and 2 (#1 and #2) clone cells in 12-well plates were treated with MG132 (MG, 10 μM) or vehicle for 12 h. After determination of the protein concentration of each lysate, equal amounts of cell lysate were separated with SDS-PAGE and transferred onto a PVDF membrane. Expression levels of HiBiT-derived signals (left), ATF4 and G3PDH (right) were detected as described in the Materials and methods section.
2.3. Cell culture and treatment
Neuro2a cells were maintained in Dulbecco's Modified Eagle's Minimum Essential Medium containing 5% fetal bovine serum. Transfection of the indicated constructs was performed using PEI-MAX reagent (Polysciences) as previously described [3]. For treatment with the indicated reagents, Neuro2a cells were incubated with cyclohiximide (CHX, 10 μg/ml), MG132 (MG, 10 μM) and tunicamycin (Tm, 1 μg/ml) for the indicated times.
2.4. Nucleotide sequence analysis of the HiBiT-tagged ATF4 gene
For determination of the integrated HiBiT-epitope sequences, the DNA fragment was extracted from genomic DNA using a mouse ATF4 sense primer, 5′-GTATGAGCCCAGAGTCCT-3′, and an IRES antisense primer, 5′-AAGCGGCTTCGGCCAGTAACGTTA-3, and the resulting DNA sequence was determined using an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems).
2.5. Luciferase assay
After preparation of cell lysate using HiBiT lytic buffer or homogenization buffer (20 mM Tris-HCl (pH 8.0) containing 137 mM NaCl, 2 mM EDTA, 10% glycerol, 1% TritonX-100, 1 mM PMSF, 10 μg/ml leupeptin and 10 μg/ml pepstatin A), the lysate was mixed with an equal amount of reaction mixture containing recombinant LgBiT (rLgBiT) and furimazine in diluted HiBiT lytic buffer and incubated at 37 ℃ for the indicated time. The luciferase activity in each sample was measured using a GloMAX20/20 luminometer (Promega). For the selection of knock-in cells, an aliquot of cell suspensions from each well was mixed with an equal amount of reaction mixture containing rLgBiT and furimazine in diluted HiBiT lytic buffer, and the luciferase activity of each sample was measured as described above.
2.6. Protein expression analysis
We detected the amount of each protein in the cell lysate as previously described with slight modifications [2]. The cells were lysed with homogenization buffer as described above. After the protein concentration was determined, each cell lysate was dissolved with an equal amount of 2× sodium dodecyl sulfate (SDS)-Laemmli sample buffer (62.5 mM Tris-HCl (pH 6.8), 2% SDS and 10% glycerol). Equal amounts of cell lysate were separated on 10% SDS-polyacrylamide gels, immunoblotted onto polyvinylidene difluoride membranes (PVDF) (GE Healthcare) and identified by enhanced chemiluminescence (GE Healthcare) using antibodies against ATF4 (Santa Cruz Biotechnology), actin (Calbiochem) and G3PDH (Acris). To detect HiBiT-tagged protein on PVDF membranes, we incubated the protein-transferred membrane with TTBS for more than 15 min and soaked it with HiBiT reaction mixture containing rLgBiT and furimazine in diluted HiBiT lytic buffer for 5 min. The membrane was exposed to ECL film (GE Healthcare) for an appropriate time, and the images were obtained in the same manner as the immunoblot analysis.
3. Results and discussion
A unique, 11-amino-acid fragment was designed during the development of a novel protein-protein interaction assay, NanoBiT, which was derived from a highly active luciferase, NanoLuc [6]. This fragment, HiBiT had high specific affinity for the N-terminal large fragment of NanoLuc, LgBiT. Interestingly, five amino acids in SmBiT (VTGYRLFEEIL) corresponding to the C-terminal region of NanoLuc were replaced in this HiBiT (VSGWRLFKKIS). The length of this HiBiT epitope was comparable to that of the well-used epitopes, FLAG (DYKDDDDK) and Myc (EQKLISEEDL). On the other hand, it was proposed that restoring NanoLuc activity through the complex formation between HiBiT and LgBiT provides a novel alternative to conventional immunoblot analysis for the detection of protein expression. To elucidate the availability of this novel HiBiT epitope, we used it to detect the expression of ATF4 protein, a proteasomal substrate [13], [14] and a well-known ER stress-inducible transcription factor [8], [9], [10], [15].
First, we constructed an ATF4 expression construct in which the HiBiT epitope was fused at the C-terminus with a twelve-amino-acid linker, NRIRGSSGGSSG (Fig. 1). After preparation of lysate from mock or ATF4-HiBiT-overexpressing cells, each lysate was mixed and incubated with rLgBiT and furimazine. As shown in Fig. 1C, NanoLuc luciferase activity was detected only in the ATF4-HiBiT-overexpressing cells, and its activity was remarkably higher after treatment with a proteasome inhibitor, MG132. To verify the molecular size of ATF4-HiBiT, we separated each lysate using SDS-PAGE and transferred the separated proteins onto a PVDF membrane. After incubation with rLgBiT and furimazine, specific signals were detected at approximately 55 kDa, and the signal from MG132-treated cells was more intense that from the control cell lysates (Fig. 1D). Consistently, the bands were well overlapped with those detected using conventional immunoblot analysis with an antibody against ATF4 protein.
Next, we attempted to knock-in HiBiT into the mouse ATF4 gene in Neuro2a cells and detect NanoLuc luciferase activity. Based on our previous report in which we knocked-in the NanoLuc gene into the human GRP78 gene in HEK293 cells using the CRIPSR/Cas9 system [5], the HiBiT epitope was integrated into the C-terminal end of ATF4 (Fig. 2). A donor gene coding the C-terminal ATF4 with the HiBiT epitope was aligned with the puromycin-resistance gene through the IRES sequence. Several days after transfection with each construct coding the donor gene, gRNA and hCas9, an appropriate amount of puromycin was added to the culture dish to select the knock-in cells. After puromycin-resistant cells proliferated, one cell was seeded and cultured into each well of a 96-well plate. In this approach, the HiBiT epitope worked as a useful tool to rapidly select positive clones from 96-well plates. As shown in Fig. 2, aliquots of cell suspensions from each well were mixed with rLgBiT and substrate in the diluted HiBiT lytic buffer and incubated for approximately 5 min at 37 ℃. After measurement of the NanoLuc luciferase activity, we quickly obtained more than 10 clones showing remarkable HiBiT-derived NanoLuc luciferase activity. In this study, we sequenced the nucleotide sequences around the C-terminus of ATF4 gene from three clones and found that the HiBiT epitope was actually integrated into the ATF4 gene in Neuro2a cells (Fig. 2C). Since the nucleotide sequences around the HiBiT epitope in two clones (#2, 3) were identical, we tested the expression of HiBiT-tagged ATF4 protein in two different clone types, #1 and #2. As shown in Fig. 2D, HiBiT-derived signals were specifically detected at approximately 55 kDa in MG132-treated cloned cells, but the signals were negligible under the resting condition. In addition to the above experiment involving the transient overexpression of ATF4-HiBiT in Neuro2a cells, the HiBiT-derived signals were well overlapped with that detected in the immunoblot analysis.
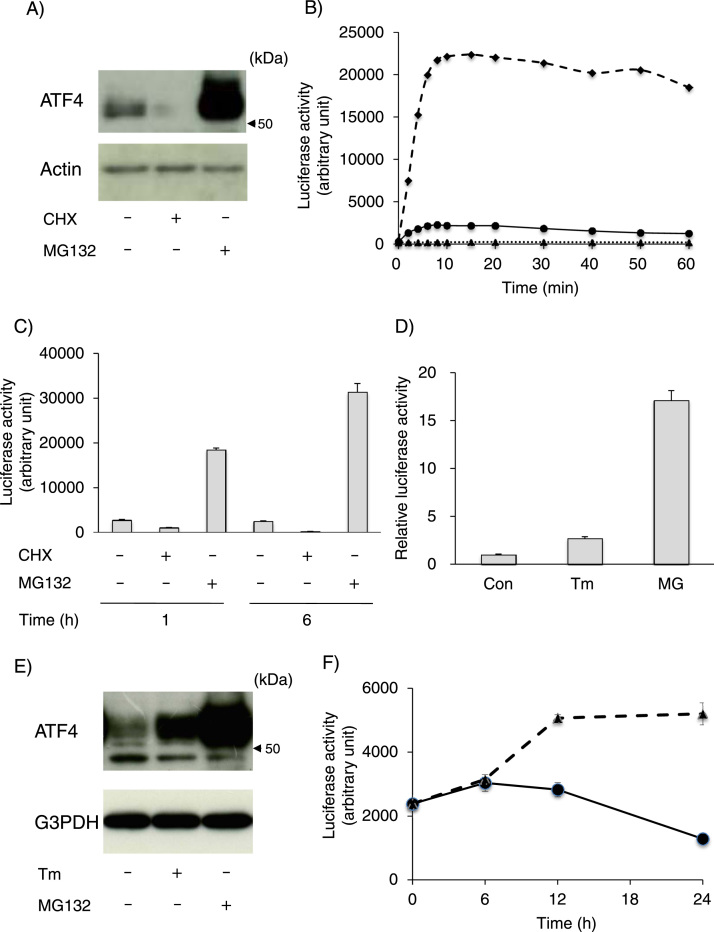
Next, we examined the expression of ATF4 protein in wild-type Neuro2a cells in the presence of MG132 and a protein synthesis inhibitor CHX, and observed that expression of ATF4 in this cell-line was unstable and continuously degraded by the proteasome pathway (Fig. 3 A), which is consistent with other reports [13], [14]. We then examined cell lysate from #2 clone cells in 96-well plates 6 h after treatment with MG132 or CHX to detect HiBiT-derived NanoLuc luciferase activities. In this assay, the cells in each well were lysed with equal amounts of diluted HiBiT lytic buffer and OPTI-MEM culture medium. After the lysate was harvested, each lysate without protein normalization was mixed with rLgBiT and substrate, and the HiBiT-derived NanoLuc luciferase activity of each sample was periodically monitored for 60 min. As shown in Fig. 3B, the luciferase activities from cells treated with MG132 or vehicle peaked within almost 5 min and maintained these activities for 20 min. After 20 min, luciferase activity slightly declined during incubation. On the other hand, luciferase activities from CHX-treated cells hardly changed and were quite low. Therefore, the luciferase activities of CHX-treated cells were background values. Using this approach, we measured luciferase activity 1 or 6 h after treatment with MG132, CHX or vehicle (Fig. 3 C). Interestingly, only 1-h CHX treatment decreased luciferase activity by more than 50%, and 6-h CHX treatment resulted in luciferase activity in treated cells less than one-tenth of that in control cells. On the other hand, MG132 treatment for 1 h increased luciferase activity by nearly 5 times, and treatment for 6 h further elevated luciferase activity.
Fig. 3.
Characterization of HiBiT-tagged ATF4 expression in Neuro2a cells. A) Wild-type Neuro2a cells in 12-well plates were treated with cycloheximide (CHX, 10 μg/ml), MG132 (MG, 10 μM) or vehicle for 6 h, and the expression levels of the indicated proteins were measured by immunoblot analysis as described in the Materials and methods section. B) After seeding #2 clone cells into a 96-well plate, the culture medium was replaced with OPTI-MEM, and the cells were treated with CHX (10 μg/ml), MG132 (10 μM) or vehicle for 6 h. After treatment, the cells were lysed with equal amounts of diluted HiBiT lytic buffer. Each lysate was mixed with equal amounts of reaction mixture containing rLgBiT and furimazine in diluted HiBiT lytic buffer and incubated at 37 °C. The HiBiT-derived luciferase activity of each sample was measured at the indicated time. The representative luciferase activities from control and MG132- or CHX-treated cells are indicated as circles, squares and triangles, respectively. C) #2 clone cells in a 96-well plate were treated with CHX (10 μg/ml), MG132 (10 μM) or vehicle for 1 or 6 h, and the cells were lysed as described above. After incubation of each lysate with reaction mixture at 37 °C for 10 min, luciferase activity was measured. Each value represents the mean ± SEM from 3 independent cultures. D, E) #2 clone cells in a 12-well plate were treated with tunicamycin (Tm, 1 μg/ml), MG132 (10 μM) or vehicle for 12 h. D) After the cells were harvested and lysed with homogenization buffer, each lysate containing 1 μg protein was mixed with the same amount of reaction mixture containing rLgBiT and furimazine in diluted HiBiT lytic buffer. After the lysates were incubated at 37 °C for 10 min, luciferase activity in each lysate was measured. Each value represents the mean ± SEM from 6 independent cultures. E) Equal amounts of cell lysate prepared in (D) were separated with SDS-PAGE and transferred onto PVDF membranes. The expression levels of ATF4 and G3PDH were detected by immunoblot analysis. F) After seeding #2 clone cells into a 96-well plate, the culture medium was replaced with OPTI-MEM. The cells were treated with Tm (1 μg/ml, triangles) or vehicle (circles) for the indicated times, and they were lysed as described above. After incubation of each lysate with reaction mixture at 37 °C for 10 min, the luciferase activity of each lysate was measured. Each value represents the mean ± SEM from 3 independent cultures.
Finally, we measured this HiBiT-derived luciferase activity in #2 clone cells in response to tunicamycin (Tm) treatment because the expression of ATF4 protein under ER stress conditions is reportedly regulated in a translational manner [16], [17]. As shown in Fig. 3DE, increased expression of ATF4 protein in #2 clone cells after 12-h Tm treatment was detected by immunoblot analysis. Similarly, the measurement of HiBiT-derived luciferase activity in each lysate used for immunoblotting analysis showed that Tm treatment increased luciferase activity. We then measured the Tm-induced elevation of the luciferase activity of #2 clone cells in 96-well plates as performed above for the CHX and MG132 treatments. Under this condition, we observed an increase in HiBiT-derived luciferase activity following treatment with Tm for 12 h and 24 h (Fig. 3 F).
In the past few years, we have employed NanoLuc and its derivative to more sensitively and conveniently monitor several cellular events associated with ER-stress responses [2], [3], [4], [5], [7]. Based on this knowledge, we first applied a novel NanoLuc-derived 11-amino-acid epitope, HiBiT, to detect the expression of ATF4 protein and showed the ability of this HiBiT system to detect protein expression after SDS-PAGE and transfer onto a PVDF membrane. Detecting protein expression without considering antibody specificity is advantageous, but the sensitivity of this HiBiT approach seems to be lower than that of conventional immunoblot analysis in our experimental conditions. Furthermore, we knocked-in this HiBiT epitope into the ATF4 gene together with the puromycin-resistance gene through the IRES sequence to simplify the screening of positive clones and to evaluate endogenous ATF4 expression even in cells in 96-well plates. However, this HiBiT system is very novel and has rarely been used to elucidate protein expression in various cellular issues. We therefore conclude that further utilization of this novel system for many other proteins will result in more unique approaches and demonstrate its ability to measure protein expression during various developmental and pathophysiological conditions.
Acknowledgements
We thank Dr. George Church and Promega Corporation for providing the hCas9 gene and the HiBiT system. This work is, in part, supported by Grant-in-Aid for Challenging Exploratory Research (no. 17K19901 to K.O.) and the OGAWA Science and Technology Foundation (to K.O.).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2017.08.002.
Appendix A. Transparency document
Supplementary material
References
- 1.Hall M.P., Unch J., Binkowski B.F., Valley M.P., Butler B.L., Wood M.G., Otto P., Zimmerman K., Vidugiris G., Machleidt T., Robers M.B., Benink H.A., Eggers C.T., Slater M.R., Meisenheimer P.L., Klaubert D.H., Fan F., Encell L.P., Wood K.V. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazione substrate. ACS Chem. Biol. 2012;7:1848–1857. doi: 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Norisada J., Hirata Y., Amaya F., Kiuchi K., Oh-hashi K. A sensitive assay for the biosynthesis and secretion of MANF using NanoLuc activity. Biochem. Biophys. Res. Commun. 2014;449:483–489. doi: 10.1016/j.bbrc.2014.05.031. [DOI] [PubMed] [Google Scholar]
- 3.Hikiji T., Norisada J., Hirata Y., Okuda K., Nagasawa H., Ishigaki S., Sobue G., Kiuchi K., Oh-hashi K. A highly sensitive assay of IRE1 activity using the small luciferase NanoLuc: evaluation of ALS-related genetic and pathological factors. Biochem. Biophys. Res. Commun. 2015;463:881–887. doi: 10.1016/j.bbrc.2015.05.132. [DOI] [PubMed] [Google Scholar]
- 4.Oh-hashi K., Irie N., Sakai T., Okuda K., Nagasawa H., Hirata Y., Kiuchi K. Elucidation of a novel phenformin derivative on glucose-deprived stress responses in HT-29 cells. Mol. Cell. Biochem. 2016;419:29–40. doi: 10.1007/s11010-016-2747-5. [DOI] [PubMed] [Google Scholar]
- 5.Oh-hashi K., Furuta E., Norisada J., Amaya F., Hirata Y., Kiuchi K. Application of NanoLuc to monitor the intrinsic promoter activity of GRP78 using the CRISPR/Cas9 system. Genes Cells. 2016;21:1137–1143. doi: 10.1111/gtc.12401. [DOI] [PubMed] [Google Scholar]
- 6.Dixon A.S., Schwinn M.K., Hall M.P., Zimmerman K., Otto P., Lubben T.H., Butler B.L., Binkowski B.F., Machleidt T., Kirkland T.A., Wood M.G., Eggers C.T., Encell L.P., Wood K.V. NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem. Biol. 2016;11:400–408. doi: 10.1021/acschembio.5b00753. [DOI] [PubMed] [Google Scholar]
- 7.Oh-hashi K., Hirata Y., Kiuchi K. SOD1 dimerization monitoring using a novel split NanoLuc. NanoBit. Cell Biochem. Func. 2016;34:497–504. doi: 10.1002/cbf.3222. [DOI] [PubMed] [Google Scholar]
- 8.Harding H.P., Zhang Y., Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 9.Harding H.P., Zhang Y., Zeng H., Novoa I., Lu P.D., Calfon M., Sadri N., Yun C., Popko B., Paules R., Stojdl D.F., Bell J.C., Hettmann T., Leiden J.M., Ron D. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 10.Blais J.D., Filipenko V., Bi M., Harding H.P., Ron D., Koumenis C., Wouters B.G., Bell J.C. Activating transcription factor 4 is translationally regulated by hypoxic stress. Mol. Cell. Biol. 2004;24:7469–7482. doi: 10.1128/MCB.24.17.7469-7482.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esvelt K.M., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O., Cradick T.J., Marraffini L.A., Bao G., Zhang F. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lassot I., Ségéral E., Berlioz-Torrent C., Durand H., Groussin L., Hai T., Benarous R., Margottin-Goguet F. ATF4 degradation relies on a phosphorylation-dependent interaction with the SCF(βTrCP) ubiquitin ligase. Mol. Cell. Biol. 2001;21:2192–2202. doi: 10.1128/MCB.21.6.2192-2202.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milani M., Rzymski T., Mellor H.R., Pike L., Bottini A., Generali D., Harris A.L. The role of ATF4 stabilization and autophagy in resistance of breast cancer cells treated with Bortezomib. Cancer Res. 2009;69:4415–4423. doi: 10.1158/0008-5472.CAN-08-2839. [DOI] [PubMed] [Google Scholar]
- 15.Lange P.S., Chavez J.C., Pinto J.T., Coppola G., Sun C.W., Townes T.M., Geschwind D.H., Ratan R.R. ATF4 is an oxidative stress-inducible, prodeath transcription factor in neurons in vitro and in vivo. J. Exp. Med. 2008;205:1227–1242. doi: 10.1084/jem.20071460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harding H.P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M., Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 17.Vattem K.M., Wek R.C. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. USA. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material