Abstract
C-repeat binding factors (CBF) are a subfamily of AP2 transcription factors that play critical roles in the regulation of plant cold tolerance and growth in low temperature. In the present work, we sought to perform a detailed investigation into global transcriptional regulation of plant hormone signaling associated genes in transgenic plants engineered with CBF genes. RNA samples from Arabidopsis thaliana plants overexpressing two CBF genes, CBF2 and CBF3, were subjected to Illumina HiSeq 2000 RNA sequencing (RNA-Seq). Our results showed that more than half of the hormone associated genes that were differentially expressed in CBF2 or CBF3 transgenic plants were related to auxin signal transduction and metabolism. Most of these alterations in gene expression could lead to repression of auxin signaling. Accordingly, the IAA content was significantly decreased in young tissues of plants overexpressing CBF2 and CBF3 compared with wild type. In addition, genes associated with the biosynthesis of Jasmonate (JA) and Salicylic acid (SA), as well as the signal sensing of Brassinolide (BR) and SA, were down-regulated, while genes associated with Gibberellin (GA) deactivation were up-regulated. In general, overexpression of CBF2 and CBF3 negatively affects multiple plant hormone signaling pathways in Arabidopsis. The transcriptome analysis using CBF2 and CBF3 transgenic plants provides novel and integrated insights into the interaction between CBFs and plant hormones, particularly the modulation of auxin signaling, which may contribute to the improvement of crop yields under abiotic stress via molecular engineering using CBF genes.
Keywords: Arabidopsis thaliana, auxin, CBF transcription factor, plant hormone signaling, RNA-Seq, transcriptome analysis
Introduction
Environmental stresses such as low or high temperatures not only restrain the growth and yield of agriculturally important crops but also limit their temporal and spatial distribution (Kasuga et al., 1999). Many plants possess the ability to survive freezing temperatures, which depends to a large extent on their capacity of cold acclimation (Thomashow, 2010). In this process, C-repeat binding factors (CBFs) are a group of key regulatory proteins that can activate downstream regulon to enhance cold resistance via binding to drought responsive element (DRE)/c-repeat transcription (CRT) elements (Zhou et al., 2011). CBFs belong to AP2 transcription factor family and six CBF genes have been identified in Arabidopsis thaliana, including CBF1, CBF2, CBF3, CBF4, DDF1 and DDF2 (also known as DREB1b, DREB1c, DREB1a, DREB1d, DREB1f, and DREB1e) (Sakuma et al., 2002; Haake et al., 2002; Magome et al., 2004). Among which CBF1, CBF2, and CBF3 specifically play central roles in cold acclimation (Shinwari et al., 1998; Hannah et al., 2006; Maruyama et al., 2012; Kang et al., 2013). The expression of CBF1, -2, -3 is induced by low temperature in a fast and transient manner in various plant species (Jaglo-Ottosen et al., 1998; Hsieh et al., 2002; Savitch et al., 2005; Benedict et al., 2006; Akhtar et al., 2012). In addition to increased freezing tolerance, overexpression of CBF genes also cause multiple morphological and biochemical changes, including dehydration and high salt stress tolerances (Kasuga et al., 1999), growth retardation, delayed flowering and reduced yields (Liu et al., 1998; Gilmour et al., 2000; Hsieh et al., 2002; Lee et al., 2004; Kasuga et al., 2004; Pino et al., 2008; Tong et al., 2009; Bhatnagar-Mathur et al., 2014), delayed leaf senescence and extended plant longevity (Sharabi-Schwager et al., 2010a,b), and a slight bluish-green tint of leaves (Gilmour et al., 2000). In summary, CBF overexpression widely affects plant physiological and developmental processes involving a complex transcriptional regulation system (Kendall et al., 2011; Park et al., 2015).
Plant hormones are key signal molecules regulating plant growth, development and stress tolerance (Peleg and Blumwald, 2011; Kurepin et al., 2013; Colebrook et al., 2014; Shi et al., 2015). Understanding the correlation between CBF-mediated biochemical changes and plant hormone signaling pathways will be an effective way to improve the plant adaption to environments through genetic engineering. Gibberellins (GAs) are the first class of plant hormones revealed that it can be regulated by CBFs (Hedden and Phillips, 2000; Shan et al., 2007). It has been reported that the dwarfism of CBF1-ox and CBF3-ox plants can be rescued by exogenous GA treatments (Achard et al., 2008; Zhou et al., 2017). CBF3 transcription factor targets the GA deactivation gene GA2ox7 to reduce endogenous GA levels and caused the accumulation of DELLA proteins, leading to limited plant growth (Zhou et al., 2017). In other plant species such as tobacco, bioactive GA levels were also reduced by CbCBF from Capsella (Zhou et al., 2014). Some recent reports showed that CBF2 overexpression might be involved in leaf response to ethylene via ABA signaling (Sharabi-Schwager et al., 2010a) and enhanced abiotic stress tolerance via cytokinin signaling (Huang et al., 2009). Besides, the expression of CBFs is negatively regulated by ethylene (Shi et al., 2012) and SA (Miura and Ohta, 2010), positively regulated by JA (Hu et al., 2013), and can be induced by exogenous ABA (Knight et al., 2004; Yang et al., 2011; Knight and Knight, 2012). These indicate that CBFs interact with different plant hormones, nevertheless little is known about detailed regulatory modes of hormone signaling genes in CBF overexpression plants. The modulation of more hormones such as auxins by CBFs also needs to be clarified.
Recently, two groups generated cbfs double and triple mutants using CRISPR/Cas9 system to reveal that three CBF genes play redundant and crucial roles in cold acclimation (Jia et al., 2016; Zhao et al., 2016). The genetic analysis clearly demonstrated that CBF2 is more important than CBF1 and CBF3 in cold acclimation-mediated freezing resistance (Zhao et al., 2016). For further application of CBF genes in molecular breeding using transgenic technology, detailed investigation into global changes of gene expression pattern elicited by overexpression of CBF genes is required. Here, we used RNA-seq tools to analyze the transcriptome of CBF2-ox and CBF3-ox Arabidopsis plants. The transcriptome profiling is an effective and widely used method to investigate gene expression dynamics in response to abiotic stresses at a global level in many species (Zhu et al., 2013). We explored differentially expressed genes related to plant hormones with emphasis on auxin signaling. The negative regulation of multiple plant hormone signaling pathways in CBF overexpression plants were discussed.
Materials and Methods
Plant Materials
The wild type (WT) Arabidopsis thaliana (L.) Heynh. ecotype Wassileskija (Ws-2) and transgenic plants constitutively expressing CBF2 (CBF2-ox) or CBF3 (CBF3-ox) in this work were previously described (Gilmour et al., 2004). These plants were grown in Metro-Mix 200 soil under cycles of 16-h-light/8-h-dark in a growth chamber at constant 22°C. Two-week-old seedlings were used for detection of relative expression levels of selected genes.
RNA Extraction, cDNA Library Construction and Illumina Sequencing
Total RNA was extracted from collected 2-week-old seedlings using ZR Plant RNA MiniPrepTM Kit (ZYMO Research Corp., United States) and purified using RNAeasy Plant Mini Kit (Qiagen, Valencia, CA, United States) according to the manufacturer’s protocol. The quality and quantity of total RNA were examined by a 2100 Bioanalyzer (Aligent Technologies, Santa Clara, CA, United States) according to the manufacturer’s instruction. After purification, three biological replicates of RNA samples for each genotype were pooled and digested into 200-nt fragments (Agilent Technologies). The first strand cDNA was generated from RNA fragments by reverse transcriptase using random primers (Invitrogen, San Diego, CA, United States). Second-strand cDNA was synthesized using reaction buffer, dNTPs, RNase H and DNA polymerase I (Amersham Biosciences, Piscataway, NJ, United States). After second strand cDNA synthesis and adaptor ligation, 200-bp cDNA fragments were purified by gel electrophoresis and enriched by 18 cycles of PCR to construct the final sequencing cDNA libraries. Qualities of these libraries were evaluated by an Agilent Bioanalyzer and the ABI Step One Plus Real-Time PCR system (Applied Biosystems, Foster City, CA, United States). The constructed libraries were sequenced using the HiSeqTM 2000 Sequencing System (Illumina, San Diego, CA, United States) with single-end technology in a single run at Shanghai Genergy Biotechnology (Shanghai) Co., Ltd. (Shanghai, China) and subjected to 100 cycles of paired-end (2 × 100 bp) sequencing. RNA-seq data are deposited in the NCBI Sequencing Read Archive database and the accession numbers are SRR5665776, SRR5665777, and SRR5665778.
Identification of Differentially Expressed Genes (DEGs)
Raw reads generated by high-throughput sequencing of Ws-2, CBF2-ox and CBF3-ox samples were cleaned by removing adaptor sequences, empty reads, and low-quality sequences (Ns > 5) (Cock et al., 2010). The sequence reads then were mapped to the Arabidopsis genome downloaded from TAIR101 by TopHat2 (Kim D. et al., 2013)2 using the modified parameters of ‘no mismatch’ and ‘single copy.’ The expression level of each gene was normalized by calculating the Reads Per Kilobase per Million (RPKM) method using Cufflinks (Roberts et al., 2011) with default parameters. The raw read counts for each gene was calculated using HTSEQ v.0.6.0 (Anders et al., 2015). The Differentially expressed genes (DEGs) data were generated with DEGseq2 (Love et al., 2014). DEGs were identified with the criteria of Log2 Fold-Change (Log2FC) ≥ 1 and False Discovery Rate (FDR, Benjamini–Hochberg adjusted P-value) ≤ 0.01.
Functional Annotation and Pathway Analysis
GO functional classifications and enrichment analysis for all DEGs were carried out by agriGO3 (Du et al., 2010). Singular Enrichment Analysis (SEA) tool was used in agriGO to produce pathway signaling. The hypergeometric method of statistical test was used and significance level was 0.05. All parameters were used default settings. The DEGs involve in plant hormone signaling pathways was selected using plant hormone signal transduction map in the database of Kyoto Encyclopedia of Genes and Genomes (KEGG)4.
Relative mRNA Expression Level Analysis
Real-time quantitative reverse transcription-PCR (qRT-PCR) was applied to investigate gene expression patterns. The mRNA levels were measured using SYBR Premix ExTaqTM II (Perfect Real Time) (TaKaRa, Dalian, China) according to manufacturer’s instructions. qRT-PCR was conducted using ABI 7500 (Applied Biosystems, Foster City, CA, United States). The reactions were performed using biological replicates different from RNA-seq samples in three independent experiments, and three technical replicates were used for each run. Values “Ct” obtained for all genes were normalized to that of an internal control EF1α gene from Arabidopsis. The relative expression level of each gene was calculated using the 2-ΔΔCT method (Livak and Schmittgen, 2001). Statistical significances of differences between the sample and control plants were determined by Student’s test. The specific primers were designed using Primer Express software (Applied Biosystems, Foster City, CA, United States) and sequences were listed in Supplementary Table S1.
Measurements of Endogenous Auxin (IAA) Contents of Arabidopsis by Enzyme-Linked Immunosorbent Assay (ELISA)
The endogenous auxin levels of Arabidopsis were estimated on three different types of tissues including young seedlings (S), leaves (L) and roots (R) of 2-week-old plants. Tissues of S, L and R from three plants were combined as one sample and three biological replicates were used for statistical tests. The auxin (IAA) contents were determined by enzyme linked immunosorbent assay (ELISA) as described previously (Chen and Zhao, 2008).
Results
Identification of CBF-ox Regulated Genes
A total number of 2639 genes (995 were up-regulated and 1644 were down-regulated) and 2295 genes (899 were up-regulated and 1396 were down-regulated) were found to be differentially expressed in CBF2-ox and CBF3-ox plants compared to WT (Ws-2) with at least two-fold differences (FDR ≤ 0.01), respectively. Venn diagrams were generated to show an overview of the DEGs (Figure 1A). The numbers of down-regulated genes were larger than up-regulated genes in both CBF2-ox and CBF3-ox plants. In line with previous analysis (Park et al., 2015), a large portion of DEGs (more than 70% for CBF2-ox and more than 80% for CBF3-ox) were coordinately changed in two transgenic lines. According to the expression patterns of DEGs in CBF2-ox and CBF3-ox plants, we generally divided genes regulated by CBF2-ox or CBF3-ox into six groups (Figure 1B). The DEGs specially regulated by CBF2 were around two times as many as DEGs specially regulated by CBF3, indicating that CBF2 may control bigger proportion of downstream CBF regulon than CBF3, which is in line with the conclusion generated from cold tolerance tests using double and triple cbfs mutants (Zhao et al., 2016). For validation of the transcriptome data, we selected three CBF genes and five CBF regulon for qRT-PCR examination in different samples (Figure 1C). The expression changes of these DEGs showed a good agreement between qRT-PCR and RNA-seq data.
FIGURE 1.
Overview of transcriptomes in Ws, CBF2-ox and CBF3-ox plants. (A) Venn diagram representing the total number of differentially expressed genes (DEGs) between CBF2-ox or CBF3-ox and Ws. CBF-ox up-regulated (log2 ≥ 1, FDR ≤ 0.01) or down-regulated (log2 ≤ -1, FDR ≤ 0.01) genes were shown. (B) Hierarchical cluster analysis of the DEGs. The heat-map was drawn with DEGseq of R package. (C) Verification of CBF regulon expression in Ws, CBF2-ox and CBF3-ox plants. Eight genes were selected for comparison of RNA-seq and qRT-PCR results. For RNA-seq data, ∗∗ represents FDR ≤ 0.01. The data of qRT-PCR were the means of three technical replicates ± SD (∗∗P ≤ 0.01; Student’s t-test). Three independent experiments were carried out with similar results.
Functional Enrichment Analysis of CBF-ox Regulated Genes
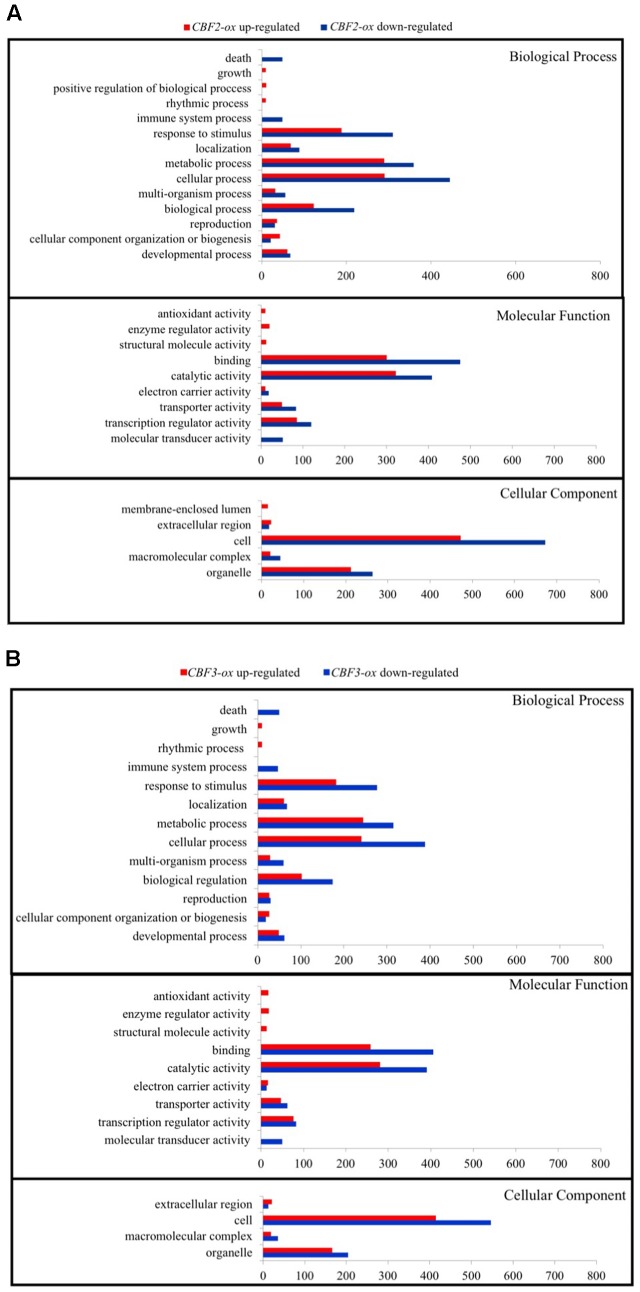
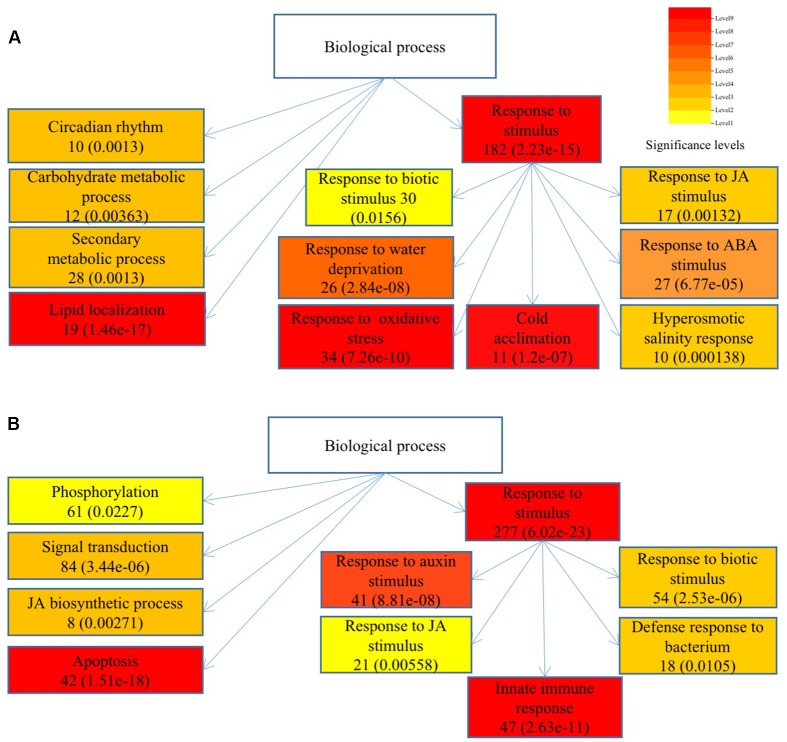
To understand the functions of DEGs and elucidate the metabolic or signal transduction pathways formed by these genes, we used SEA tool in agriGO5. All of the DEGs between Ws-2 and CBF2-ox or CBF3-ox plants were assigned to functional categories by GO analysis. DEGs were categorized into biological process, cellular component, and molecular functions, which included 14, 5, and 9 functional groups in CBF2-ox and 13, 4, and 9 functional groups in CBF3-ox, respectively (Figure 2). In CBF2-ox plants, the up-regulated genes were significantly enriched in GO terms of response to oxidative stress, lipid localization, cold acclimation, water deprivation, salinity response, JA response, and ABA response. The down-regulated genes were significantly enriched in GO terms of apoptosis, auxin response, and innate immune response (Figure 3). Enriched signaling pathways of up and down regulated genes of CBF3-ox showed high similarities with CBF2-ox (Figure 4). Notably, phosphorylation and secondary metabolic process were only detected in CBF3-ox while heat acclimation, amino acid derivative biosynthetic process and response to light stimulus were only detected in CBF2-ox plants.
FIGURE 2.
Signaling pathway enrichment of differentially expressed genes (DEGs) in CBF2-ox and CBF3-ox Arabidopsis plants. (A) GO functional classification of DEGs in CBF2-ox transcriptome; (B) GO functional classification of DEGs in CBF3-ox transcriptome.
FIGURE 3.
Significantly enriched biological process of differentially expressed genes in CBF2-ox Arabidopsis plants. (A) Enriched biological process of up-regulated genes. (B) Enriched biological process of down-regulated genes. Gene numbers are shown in the boxes and e-values are shown in parenthesis.
FIGURE 4.
Significantly enriched biological process of differentially expressed genes in CBF3-ox Arabidopsis plants. (A) Enriched biological process of up-regulated genes. (B) Enriched biological process of down-regulated genes. Gene numbers are shown in the boxes and e-values are shown in parenthesis.
CBF-ox Influences Plant Hormone Related Genes
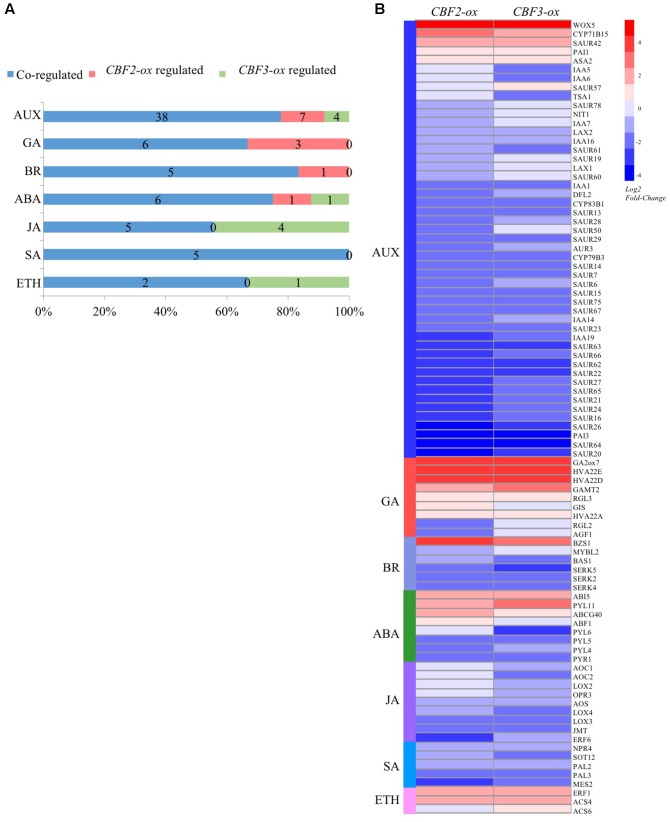
We identified 89 DEGs that participate in plant hormone modulation (Figure 5A). All these 89 genes were reported to play direct or indirect roles in hormone biosynthesis, molecular modification, transportation or degradation. Numerous elements containing “CCGAC,” the core sequence of CRT/DRE motif, were identified in the 3000 bp area upstream from coding regions of these genes (Supplementary Table S2). Figure 5B shows the expression patterns of the 89 genes in different plants. Among them, 17 DEGs were co-up-regulated by CBF2 and CBF3, 5 of which were related to auxin (WOX5, CYP71B15, ASA2, PAI1, SAUR42) and 6 (RGL3, GA2ox7, GAMT2, HVA22, HVA22A and HVA22D) were related to GA. There are 2 DEGs were up-regulated by CBF2-ox only and 2 were CBF3-ox only, respectively. For down-regulation, 10 and 8 DEGs were CBF2-ox only and CBF3-ox only, respectively. Most of which were auxin or JA related genes. Besides, 49 DEGs were co-down-regulated and 32 of which were auxin related genes, indicating that CBFs widely affect plant hormone signaling especially toward auxins.
FIGURE 5.
CBF2-ox and CBF3-ox regulate genes that are associated with auxin (AUX), gibberellin (GA), Brassinolide (BR), abscissic acid (ABA), Jasmonate (JA), Salicylic acid (SA) and ethylene (ETH) signaling. (A) The number of hormone associated genes regulated or co-regulated by CBF2-ox and CBF3-ox. (B) Heat-map showing expression fold change of 89 genes associated with plant hormones.
CBF-ox Shows a Negative Regulation in Auxin Metabolism and Signaling
Auxins are the first class of defined plant hormones, which extensively control cell elongation, division and differentiation (Mano and Nemoto, 2012). In CBF2-ox and CBF3-ox plants, 49 DEGs were associated with auxin, which was more than any other plant hormones. Among these 49 auxin related genes, 5 genes were co-up-regulated and 32 genes were co-down-regulated by CBF2-ox and CBF3-ox. In addition, 7 genes were down-regulated by CBF2-ox only, 4 genes were down-regulated by CBF3-ox only and 1 gene was up-regulated by CBF3 only (Figure 5A). These indicated that CBFs generally showed a negative regulation in auxin associated genes.
Auxin biosynthesis can be grouped as tryptophan dependent and independent pathways (Hull et al., 2000; Tromas and Perrot-Rechenmann, 2010). Two genes down-regulated by CBF2-ox and CBF3-ox, CYP79B3 and CYP83B1, are critical enzymes in tryptophan dependent pathway (Mano and Nemoto, 2012). NIT1 catalyzing the last step of IAA biosynthesis (Bartling et al., 1994; Lehmann et al., 2017) was also down-regulated by CBF2-ox. These suggested that CYP79B3, CYP83B1 and NIT1 are three potential CBF-regulated nodes in auxin biosynthesis. Meanwhile, auxin carrier LAX1 and LAX2 were also down-regulated. Multiple early auxin signaling transduction related genes such as small auxin up RNA genes (SAURs), AUX/IAAs and GH3 family genes were diversely changed (Figure 6). There were 2 up-regulated and 27 down-regulated SAURs out of 59 SAUR genes in Arabidopsis (Li et al., 2015). The 7 down-regulated IAAs are auxin inducible and function as key regulators in auxin responses (Santner et al., 2009). The GH3-like gene DFL2 is not induced by auxin but may play a role in hypocotyl elongation through light-controlled auxin modification (Takase et al., 2003). The repression of the other GH3-family gene AHR3 could be due to a feedback of down-regulated auxin biosynthesis, for AHR3 protein conjugates free IAA to amino acids (Staswick et al., 2005). Twelve genes functioning in auxin metabolism and signaling transduction were subjected to qRT-PCR and their expression changes in CBF2-ox and CBF3-ox plants were in line with the RNA-seq data (Figure 7).
FIGURE 6.

The influence of CBF-ox on auxin signaling pathway. For presumptive IAA biosynthesis pathways in plants, blue arrows indicate the tryptophan synthetic pathway in the chloroplast; a thin dashed black arrow denotes the tryptophan-independent IAA biosynthetic pathway; purple arrows indicate IPA biosynthetic pathway; red arrows indicate the indole alkaloid and serotonin biosynthetic pathway; green arrows indicate the Brassicaceae species-specific pathway. IAA transportation and signal transduction routes are also shown. Genes significantly changed in CBF2-ox or CBF3-ox plants are labeled by red stars.
FIGURE 7.
Expression of auxin pathway related genes in WS, CBF2-ox and CBF3-ox plants. Twenty genes were selected from the 49 auxin pathway genes for comparison of RNA-seq and qRT-PCR. For RNA-seq data, ∗∗ represents FDR ≤ 0.01. The data of qRT-PCR were the means of three technical replicates ± SD (∗∗P ≤ 0.01; Student’s t-test). Three independent experiments were carried out with similar results.
CBF-ox Decreases Endogenous IAA Contents
Since the auxin associated genes were largely repressed, we measured the contents of IAA, a major form of naturally occurring auxins (Mano and Nemoto, 2012), in CBF2-ox and CBF3-ox plants to verify the negative regulation of auxins level. The IAA contents in tissues of whole seedlings, leaves and roots of 2-week-old Arabidopsis were determined. Indeed, all three types of tissues of both CBF2-ox and CBF3-ox plants showed significantly decreased IAA levels (Figure 8). The difference of IAA levels between CBF-ox and WT in roots was bigger than whole seedlings and leaves, indicating that roots are most influenced by CBF-ox in auxin contents.
FIGURE 8.
Auxin (IAA) contents in WS, CBF2-ox and CBF3-ox plants. S, the seedling of 2-week-old plants; L, leaves from 2-week-old plants; R, Roots of 2-week-old plants. The data are the means of three technical replicates ± SD (∗P ≤ 0.05, ∗∗P ≤ 0.01; Student’s t-test). Three independent experiments were carried out with similar results.
Discussion
Ectopic expression of CBFs are useful tools for genetic engineering of stress tolerant plants (Zhou et al., 2011). The key point is how to eliminate the plant growth retardation caused by high expression of CBF genes. Previous studies surveyed around one-third of genes in Arabidopsis transcriptome and indicated that CBF1-, CBF2- and CBF3-ox shared similar affected gene sets (Fowler and Thomashow, 2002; Gilmour et al., 2004; Maruyama et al., 2004). In this study, we presented a genome-wide transcriptome analysis in CBF2-ox and CBF3-ox plants and identified 89 genes involved in plant hormone signaling and metabolism. Consistent with previous reports, DEGs regulated by CBF2-ox and CBF3-ox are largely overlapped and pathways enriched in GO analysis are quite similar, verifying the conclusion that CBF2 and CBF3 have redundant functions revealed by cbfs triple mutants (Jia et al., 2016). Meanwhile, CBF2-ox and CBF3-ox diversely regulate a series of genes. DEGs specifically regulated by CBF2-ox were mainly related to lipid localization, starch metabolic process, light stimulus response and regulation of transcription, while CBF3-ox specific DEGs were mainly related to oxidative stress response. These suggest that different strategies with respect to physiological and metabolic controls can be applied when using CBF2 and CBF3 to engineer crops.
It was shown that cold stress disturbed multiple auxin related genes (Lee et al., 2005). Those genes, such as auxin-inducible SAUR family genes, were also largely impacted in CBF2-ox and CBF3-ox plants (Figure 5 and Supplementary Table S1). The fact that IAA levels were significantly reduced in CBF2-ox and CBF3-ox young tissues, especially in roots, is another agreement with previous study showing that DR5 gene promoter activity, a reporter of endogenous auxin level, was downregulated in roots under cold treatments (Lee et al., 2005). The genes suppressed by CBFs are mainly related to the first and last step of IAA biosynthesis originated from tryptophan, as well as auxin transportation and signal transduction. Moreover, the gene with biggest changes in the list of 89 hormone related ones is WUSCHEL HOMEOBOX 5 (WOX5), a transcription factor playing a central role in stem cell maintenance in different meristem types (Pi et al., 2015). The up-regulation of WOX5 suggests that CBF can inhibit the stem cell differentiation, which is consistent with the contribution of CBFs to delayed leaf senescence and extended longevity (Sharabi-Schwager et al., 2010a). Although it is not clear whether these genes are direct targets of CBF proteins yet, our transcriptome data provide a relatively comprehensive investigation of effects on auxin signaling from CBFs. The repression of auxin signaling can be another reason of plant growth retardation widely observed in CBF-ox plants in addition to influence on GA signaling (Achard et al., 2008; Zhou et al., 2017), and can also explain the reason why dwarf phenotype of CBF-ox Arabidopsis plants could only be partially rescued by GA application (Zhou et al., 2017).
Compared with old microarray data (Fowler and Thomashow, 2002), we detected more GA associated genes changed in CBF2-ox and CBF3-ox plants. Besides the reported GA2-oxidase gene GA2ox7 and DELLA gene RGL3 (Zhou et al., 2017), a GA methyl transferase gene GAMT2 was significantly up-regulated in CBF3-ox plants. GAMT enzyme transfers active GA into inactive form (Varbanova et al., 2007; Doherty et al., 2009; Kim Y. et al., 2013) and can be a novel regulatory node of GA controlled by CBF3. Further, HVA22, HVA22A and HVA22D were up-regulated by both CBF2-ox and CBF3-ox. These three genes serve as repressors of programmed cell death mediated by GA (Guo and Ho, 2008). Surprisingly, another GA signaling repressor DELLA gene RGL2 was down-regulated by CBF2, suggesting that CBF2 may possess new functions that are different from CBF1 and CBF3 in GA regulation. For other hormones, although the regulation of CBFs and cold responses from ABA, JA and ethylene have been widely investigated, few information of CBF-caused genome wide influence in these hormones are reported. Here we show that CBF2-ox and CBF3-ox also impacted multiple aspects of plant hormones other than auxin and GA (Figure 7). The genes associated with biosynthesis (PAL2 and PAL3) (Huang et al., 2010) and modification (MES2 and SOT12) (Baek et al., 2010) of SA as well as the SA receptor (NPR4) (Fu et al., 2012) were coordinately repressed in CBF2-ox and CBF3-ox plants. Genes that are similarly down-regulated by CBF2 and CBF3 also include receptor kinase genes SERK2, -4, -5 involved in BR signaling (Gou et al., 2012), key enzyme genes in JA biosynthesis such as AOC1, -2, LOX2, -3, -4 and OPR3 (Schaller et al., 2000), positive regulator gene ERF6 in JA signaling and MeJA formation associated gene JMT (Seo et al., 2001). For ABA, receptor genes (Park et al., 2011) PYL4, -5, -6 were down-regulated while PYL11 was up-regulated. Regulator genes ABF1 and ABI5 as well as transportation gene ABCG40 (Choi et al., 2000; Finkelstein et al., 2005) were all up-regulated. The ethylene biosynthesis gene ACS4, ACS6 (Tsuchisaka et al., 2009) and positive regulator gene ERF1 (Moffat et al., 2012) were up-regulated, which is in agreement with the study of positive correlation between ethylene biosynthesis and cold tolerance (Catalá et al., 2014). The recent transcriptome analyses performed in cbfs triple mutants also identified a series of hormone related genes (Jia et al., 2016; Zhao et al., 2016). Jia et al. reported 26 CBF-regulated genes associated with plant hormones and Zhao et al. showed 11 CBF-activated hormone related genes. Similar groups of genes were presented in our data, such as GAMT2, RGL3 and HVA genes related to GA signaling, BZS1 related to BR signaling, and ABCG40 related to ABA signaling. Notably, a big proportion of hormone related genes detected in our work, including GA2ox7, WOX5 and some of SAUR genes, are not detected in these two data sets of cbfs triple mutants, which can be due to different plant growth conditions and experimental procedures. More important, gene overexpression produced by transgenic technologies can trigger more extensive molecular changes and will help to identify pathway components that might remain undetected using loss-of-function analysis especially for functionally redundant genes (Prelich, 2012). Thus, our investigation would be a fine supplement to the research on CBF-dependent regulatory network and could guide the utilization of plant hormones in CBF transgenic plants.
In summary, CBFs negatively regulate auxin, GA, SA, JA and BR signaling, and disturb auxin, GA, SA and JA metabolism including biosynthesis, modification and transportation (Figure 9). All these hormones are associated with plant growth and development, implying a key role of CBFs in the balance of enhanced stress tolerance and restrained growth for plants to survive the environmental changes. Our data revealed more potential targets for crop improvement using genetic engineering approaches. The comprehensive modulation of CBFs and downstream regulons will hopefully realize the production of hardy crops without reduced growth.
FIGURE 9.
Negative regulation of multiple plant hormone pathways elicited by CBF overexpression. CBF2 and CBF3 repress biosynthesis of auxin, SA and JA; negatively regulate receptors of BR and SA; promote deactivation of GA.
Author Contributions
JL, MZ, and AL were responsible for the overall design and conduct of experiments. MZ prepared plant tissues and performed RNA extraction and sequencing. AL and CY conducted the seq data analysis. DW and HC carried out the qRT-PCR and auxin content quantification. JL, AL, and MZ took the lead on manuscript development. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (31170287) for JL, the Major Program for the Fundamental Research of Shanghai, China (09JC1401700) and the National High Technology Research and Development Program of China (2008AA10Z105).
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01647/full#supplementary-material
References
- Achard P., Gong F., Cheminant S., Alioua M., Hedden P., Genschik P. (2008). The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20 2117–2129. 10.1105/tpc.108.058941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar M., Jaiswal A., Taj G., Jaiswal J. P., Qureshi M. I., Singh N. K. (2012). DREB1/CBF transcription factors: their structure function and role in abiotic stress tolerance in plants. J. Genet. 91 385–395. 10.1007/s12041-012-0201-3 [DOI] [PubMed] [Google Scholar]
- Anders S., Pyl P. T., Huber W. (2015). HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D., Pathange P., Chung J. S., Jiang J., Gao L., Oikawa A., et al. (2010). A stress-inducible sulphotransferase sulphonates salicylic acid and confers pathogen resistance in Arabidopsis. Plant Cell Environ. 33 1383–1392. 10.1111/j.1365-3040.2010.02156.x [DOI] [PubMed] [Google Scholar]
- Bartling D., Seedorf M., Schmidt R. C., Weiler E. W. (1994). Molecular characterization of two cloned nitrilases from Arabidopsis thaliana: key enzymes in biosynthesis of the plant hormone indole-3-acetic acid. Proc. Natl. Acad. Sci. U.S.A. 91 6021–6025. 10.1073/pnas.91.13.6021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict C., Skinner J. S., Meng R., Chang Y., Bhalerao R., Huner N. P., et al. (2006). The CBF1-dependentlow temperature signaling pathway, regulon and increase in freeze tolerance are conserved in Populus spp. Plant Cell Environ. 29 1259–1272. 10.1111/j.1365-3040.2006.01505.x [DOI] [PubMed] [Google Scholar]
- Bhatnagar-Mathur P., Rao J. S., Vadez V., Dumbala S. R., Rathore A., Yamaguchi-Shinozaki K., et al. (2014). Transgenic peanut overexpressing the DREB1A transcription factor has higher yields under drought stress. Mol. Breed. 33 327–340. 10.1007/s11032-013-9952-7 [DOI] [Google Scholar]
- Catalá R., López-Cobollo R., Castellano M. M., Angosto T., Alonso J. M., Ecker J. R., et al. (2014). The Arabidopsis 14-3-3 protein RARE COLD INDUCIBLE 1A links low-temperature response and ethylene biosynthesis to regulate freezing tolerance and cold acclimation. Plant Cell 26 3326–3342. 10.1105/tpc.114.127605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Zhao J. (2008). Free IAA in stigmas and styles during pollen germination and pollen tube growth of Nicotiana tabacum. Physiol. Plant. 134 202–215. 10.1111/j.1399-3054.2008.01125.x [DOI] [PubMed] [Google Scholar]
- Choi H., Hong J., Ha J., Kang J., Kim S. Y. (2000). ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 275 1723–1730. 10.1074/jbc.275.3.1723 [DOI] [PubMed] [Google Scholar]
- Cock P. J., Fields C. J., Goto N., Heuer M. L., Rice P. M. (2010). The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 38 1767–1771. 10.1093/nar/gkp1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colebrook E. H., Thomas S. G., Phillips A. L., Hedden P. (2014). The role of gibberellin signaling in plant responses to abiotic stress. J. Exp. Biol. 217(Pt 1), 67–75. 10.1242/jeb.089938 [DOI] [PubMed] [Google Scholar]
- Doherty C. J., Van Buskirk H. A., Myers S. J., Thomashow M. F. (2009). Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21 972–984. 10.1105/tpc.108.063958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Zhou X., Ling Y., Zhang Z., Su Z. (2010). agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38 W64–W70. 10.1093/nar/gkq310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R., Gampala S. S., Lynch T. J., Thomas T. L., Rock C. D. (2005). Redundant and distinct functions of the ABA response loci ABA-INSENSITIVE (ABI) 5 and ABRE-BINDING FACTOR (ABF) 3. Plant Mol. Biol. 59 253–267. 10.1007/s11103-005-8767-2 [DOI] [PubMed] [Google Scholar]
- Fowler S., Thomashow M. F. (2002). Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14 1675–1690. 10.1105/tpc.003483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z. Q., Yan S., Saleh A., Wang W., Ruble J., Oka N., et al. (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486 228–232. 10.1038/nature11162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour S. J., Fowler S. G., Thomashow M. F. (2004). Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol. Biol. 54 767–781. 10.1023/B:PLAN.0000040902.06881.d4 [DOI] [PubMed] [Google Scholar]
- Gilmour S. J., Sebolt A. M., Salazar M. P., Everard J. D., Thomashow M. F. (2000). Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 124 1854–1865. 10.1104/pp.124.4.1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou X., Yin H., He K., Du J., Yi J., Xu S., et al. (2012). Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLOS Genet. 8:e1002452 10.1371/journal.pgen.1002452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W. J., Ho T. H. (2008). An abscisic acid-induced protein, HVA22, inhibits gibberellin-mediated programmed cell death in cereal aleurone cells. Plant Physiol. 147 1710–1722. 10.1104/pp.108.120238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake V., Cook D., Riechmann J. L., Pineda O., Thomashow M. F., Zhang J. Z. (2002). Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol. 130 639–648. 10.1104/pp.006478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah M. A., Wiese D., Freund S., Fiehn O., Heyer A. G., Hincha D. K. (2006). Natural genetic variation of freezing tolerance in Arabidopsis. Plant Physiol. 142 98–112. 10.1104/pp.106.081141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P., Phillips A. L. (2000). Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci. 5 523–530. 10.1016/S1360-1385(00)01790-8 [DOI] [PubMed] [Google Scholar]
- Hsieh T. H., Lee J. T., Yang P. T., Chiu L. H., Charng Y. Y., Wang Y. C., et al. (2002). Heterology expression of the Arabidopsis C-repeat/dehydration response element binding factor 1 gene confers elevated tolerance to chilling and oxidative stresses in transgenic tomato. Plant Physiol. 129 1086–1094. 10.1104/pp.003442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Jiang L., Wang F., Yu D. (2013). Jasmonate regulates the INDUCER OF CBF EXPRESSION-C-REPECT BINDING FACTOR/DRE BINGDING FACTOR1 cascade and freezing tolerance in Arabidopsis. Plant Cell 25 2907–2924. 10.1105/tpc.113.112631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Gu M., Lai Z., Fan B., Shi K., Zhou Y. H., et al. (2010). Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 153 526–538. 10.1104/pp.110.157370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J. G., Yang M., Liu P., Yang G. D., Wu C. A., Zheng C. C. (2009). GhDREB1 enhances abiotic stress tolerance, delays GA-mediated development and represses cytokinin signaling in transgenic Arabidopsis. Plant Cell Environ. 32 1132–1145. 10.1111/j.1365-3040.2009.01995.x [DOI] [PubMed] [Google Scholar]
- Hull A. K., Vij R., Celenza J. L. (2000). Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 97 2379–2384. 10.1073/pnas.040569997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglo-Ottosen K. R., Gilmour S. J., Zarka D. G., Schabenberger O., Thomashow M. F. (1998). Arabidopsis CBF1 over expression induces COR genes and enhances freezing tolerance. Science 280 104–106. 10.1126/science.280.5360.104 [DOI] [PubMed] [Google Scholar]
- Jia Y., Ding Y., Shi Y., Zhang X., Gong Z., Yang S. (2016). The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol. 212 345–353. 10.1111/nph.14088 [DOI] [PubMed] [Google Scholar]
- Kang J., Zhang H., Sun T., Shi Y., Wang J., Zhang B., et al. (2013). Natural variation of C-repeat-binding factor (CBFs) genes is a major cause of divergence in freezing tolerance among a group of Arabidopsis thaliana populations along the Yangtze River in China. New Phytol. 199 1069–1080. 10.1111/nph.12335 [DOI] [PubMed] [Google Scholar]
- Kasuga M., Liu Q., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. (1999). Improving plant drought, salt and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17 287–291. 10.1038/7036 [DOI] [PubMed] [Google Scholar]
- Kasuga M., Miura S., Shinozaki K., Yamaguchi-Shinozaki K. (2004). A combination of the Arabidopsis DREB1a gene and stress-inducible rd29a promoter improved drought- and low-temperature stress tolerance in tobacco by gene transfer. Plant Cell Physiol. 45 346–350. 10.1093/pcp/pch037 [DOI] [PubMed] [Google Scholar]
- Kendall S. L., Hellwege A., Marriot P., Whalley C., Graham I. A., Penfield S. (2011). Induction of dormancy in Arabidopsis summer annuals requires parallel regulation of DOG1 and hormone metabolism by low temperature and CBF transcription factors. Plant Cell 23 2568–2580. 10.1105/tpc.111.087643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Park S., Gilmour S. J., Thomashow M. F. (2013). Roles of CAMTA transcription factors and salicylic acid in configuring the low-temperature transcriptome and freezing tolerance of Arabidopsis. Plant J. 75 364–376. 10.1111/tpj.12205 [DOI] [PubMed] [Google Scholar]
- Knight H., Zarka D. G., Okamoto H., Thomashow M. F., Knight M. R. (2004). Abscisic acid induces CBF gene transcription and subsequent induction of cold-regulated genes via the CRT promoter element. Plant Physiol. 135 1710–1717. 10.1104/pp.104.043562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight M. R., Knight H. (2012). Low-temperature perception leading to gene expression and cold tolerance in higher plants. New Phytol. 195 737–751. 10.1111/j.1469-8137.2012.04239.x [DOI] [PubMed] [Google Scholar]
- Kurepin L. V., Dahal K. P., Savitch L. V., Singh J., Bode R., Ivanov A. G., et al. (2013). Role of CBFs as integrators of chloroplast redox, phytochrome and plant hormone signaling during cold acclimation. Int. J. Mol. Sci. 14 12729–12763. 10.3390/ijms140612729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B. H., Hendenson D. A., Zhu J. K. (2005). The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17 3155–3175. 10.1105/tpc.105.035568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. C., Huh K. W., An K., An G., Kim S. R. (2004). Ectopic expression of a cold-inducible transcription factor, CBF1/DREB1b, in transgenic rice (Oryza sativa L). Mol. Cells 18 107–114. [PubMed] [Google Scholar]
- Lehmann T., Janowitz T., Sánchez-Parra B., Alonso M. P., Trompetter I., Piotrowski M., et al. (2017). Arabidopsis NITRILASE 1 contributes to the regulation of root growth and development through modulation of auxin biosynthesis in seedlings. Front. Plant Sci. 8:36 10.3389/fpls.2017.00036.eCollection [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. G., Chen H. W., Li Q. T., Tao J. J., Bian X. H., Ma B., et al. (2015). Three SAUR proteins SAUR76, SAUR77 and SAUR78 promote plant growth in Arabidopsis. Sci. Rep. 5:12477 10.1038/srep12477 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S., Yamaguchi-Shinozaki K., et al. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature -responsive gene expression, respectively, in Arabidopsis. Plant Cell 10 1391–1406. 10.1105/tpc.10.8.1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magome H., Yamaguchi S., Hanada A., Kamiya Y., Oda K. (2004). Dwarf and delayed-flowering 1, a novel Arabidopsis mutant deficient in gibberellins biosynthesis because of overexpression of a putative AP2 transcription factor. Plant J. 37 720–729. 10.1111/j.1365-313X [DOI] [PubMed] [Google Scholar]
- Mano Y., Nemoto K. (2012). The pathway of auxin biosynthesis in plants. J. Exp. Bot. 63 2853–2872. 10.1093/jxb/ers091 [DOI] [PubMed] [Google Scholar]
- Maruyama K., Sakuma Y., Kasuga M., Ito Y., Seki M., Goda H., et al. (2004). Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J. 38 982–993. 10.1111/j.1365-313X.2004.02100.x [DOI] [PubMed] [Google Scholar]
- Maruyama K., Todaka D., Mizoi J., Yoshida T., Kidokoro S., Matsukura S., et al. (2012). Identification of cis-acting promoter elements in cold- and dehydration-induced transcriptional pathways in Arabidopsis, rice, and soybean. DNA Res. 19 37–49. 10.1093/dnares/dsr040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Ohta M. (2010). SIZ1, a small ubiquitin-related modifier ligase, controls cold signaling through regulation of salicylic acid accumulation. J. Plant Physiol. 167 555–560. 10.1016/j.jplph.2009.11.003 [DOI] [PubMed] [Google Scholar]
- Moffat C. S., Ingle R. A., Wathugala D. L., Saunders N. J., Knight H., Knight M. R. (2012). ERF5 and ERF6 play redundant roles as positive regulators of JA/Et-mediated defense against Botrytis cinerea in Arabidopsis. PLOS ONE 7:e35995 10.1371/journal.pone.0035995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Lee C. M., Doherty C. J., Gilmour S. J., Kim Y., Thomashow M. F. (2015). Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. Plant J. 82 193–207. 10.1111/tpj.12796 [DOI] [PubMed] [Google Scholar]
- Park S. Y., Fung P., Nishimura N., Jensen D. R., Fujii H., Zhao Y., et al. (2011). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324 1068–1071. 10.1126/science.1173041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg Z., Blumwald E. (2011). Hormone balance and abiotic stress tolerance in crop plants. Curr. Opin. Plant Biol. 14 290–295. 10.1016/j.pbi.2011.02.001 [DOI] [PubMed] [Google Scholar]
- Pi L., Aichinger E., van der Graaff E., Llavata-Peris C. I., Weijers D., Hennig L., et al. (2015). Organizer-derived WOX5 signal maintains root columella stem cells through chromatin-mediated repression of CDF4 expression. Dev. Cell 33 576–588. 10.1016/j.devcel.2015.04.024 [DOI] [PubMed] [Google Scholar]
- Pino M. T., Skinner J. S., Jeknić Z., Hayes P. M., Soeldner A. H., Thomashow M. F., et al. (2008). Ectopic AtCBF1 over-expression enhances freezing tolerance and induces cold acclimation-associated physiological modifications in potato. Plant Cell Environ. 31 393–406. 10.1111/j.1365-3040.2008.01776.x [DOI] [PubMed] [Google Scholar]
- Prelich G. (2012). Gene overexpression: uses, mechanisms, and interpretation. Genetics 190 841–854. 10.1534/genetics.111.136911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Pimentel H., Trapnell C., Pachter L. (2011). Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 27 2325–2329. 10.1093/bioinformatics/btr355 [DOI] [PubMed] [Google Scholar]
- Sakuma Y., Liu Q., Dubouzet J. G., Abe H., Shinozaki K., Yamaguchi-Shinozaki K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 3 998–1009. 10.1006/bbrc.2001.6299 [DOI] [PubMed] [Google Scholar]
- Santner A., Calderon-Villalobos L. I. A., Estelle M. (2009). Plant hormones are versatile chemical regulators of plant growth. Nat. Chem. Biol. 5 301–307. 10.1038/nchembio.165 [DOI] [PubMed] [Google Scholar]
- Savitch L. V., Allard G., Seki M., Robert L. S., Tinker N. A., Hüner N. P. A., et al. (2005). The effect of overexpression of two Brassica CBF/DREB1-like transcription factors on photosynthetic capacity and freezing tolerance in Brassica napus. Plant Cell Physiol. 46 1525–1539. 10.1093/pcp/pci165 [DOI] [PubMed] [Google Scholar]
- Schaller F., Biesgen C., Müssig C., Altmann T., Weiler E. W. (2000). 12-Oxophytodienoate reductase 3 (OPR3) is the isoenzyme involved in jasmonate biosynthesis. Planta 210 979–984. 10.1007/s004250050706 [DOI] [PubMed] [Google Scholar]
- Seo H. S., Song J. T., Cheong J. J., Lee Y. H., Lee Y. W., Hwang I., et al. (2001). Jasmonic acid carboxyl methyltransferase: a key enzyme for jasmonate-regulated plant responses. Proc. Natl. Acad. Sci. U.S.A. 98 4788–4793. 10.1073/pnas.081557298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan D. P., Huang J. G., Yang Y. T., Guo Y. H., Wu C. A., Yang G. D., et al. (2007). Cotton GhDREB1 increases plant tolerance to low temperature and is negatively regulated by gibberellic acid. New Phytol. 176 70–81. 10.1111/j.1469-8137.2007.02160.x [DOI] [PubMed] [Google Scholar]
- Sharabi-Schwager M., Lers A., Samach A., Guy C. L., Porat R. (2010a). Overexpression of the CBF2 transcriptional activator in Arabidopsis delays leaf senescence and extends plant longevity. J. Exp. Bot. 61 261–273. 10.1093/jxb/erp300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharabi-Schwager M., Samach A., Porat R. (2010b). Overexpression of the CBF2 transcriptional activator in Arabidopsis suppresses the responsiveness of leaf tissue to the stress hormone ethylene. Plant Biol. 12 630–638. 10.1111/j.1438-8677.2009.00255.x [DOI] [PubMed] [Google Scholar]
- Shi Y., Ding Y., Yang S. (2015). Cold signal transduction and its interplay with phytohormones during cold acclimation. Plant Cell Physiol. 56 7–15. 10.1093/pcp/pcu115 [DOI] [PubMed] [Google Scholar]
- Shi Y., Tian S., Hou L., Huang X., Zhang X., Guo H., et al. (2012). Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 24 2578–2595. 10.1105/tpc.112.098640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinwari Z. K., Nakashima K., Miura S., Kasuga M., Seki M., Yamaguchi-Shinozaki K., et al. (1998). An Arabidopsis gene family encoding DRE/CRT binding proteins involved in low- temperature-responsive gene expression. Biochem. Biophys. Res. Commun. 250 161–170. 10.1006/bbrc.1998.9267 [DOI] [PubMed] [Google Scholar]
- Staswick P. E., Serban B., Rowe M., Tiryaki I., Maldonado M. T., Maldonado M. C., et al. (2005). Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17 616–627. 10.1105/tpc.104.026690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takase T., Nakazawa M., Ishikawa A., Manabe K., Matsui M. (2003). DFL2, a new member of the Arabidopsis GH3 gene family, is involved in red light-specific hypocotyl elongation. Plant Cell Physiol. 44 1071–1080. 10.1093/pcp/pcg130 [DOI] [PubMed] [Google Scholar]
- Thomashow M. F. (2010). ). Molecular basis of plant cold acclimation: insights gained from studying the CBF cold response pathway. Plant Physiol. 154 571–577. 10.1104/pp.110.161794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Z., Hong B., Yang Y., Li Q., Ma N., Ma C., et al. (2009). Overexpression of two chrysanthemum DgDREB1 group genes causing delayed flowering or dwarfism in Arabidopsis. Plant Mol. Biol. 71 115–129. 10.1007/s11103-009-9513-y [DOI] [PubMed] [Google Scholar]
- Tromas A., Perrot-Rechenmann C. (2010). Recent progress in auxin biology. C. R. Biol. 333 297–306. 10.1016/j.crvi.2010.01.005 [DOI] [PubMed] [Google Scholar]
- Tsuchisaka A., Yu G., Jin H., Alonso J. M., Ecker J. R., Zhang X., et al. (2009). A combinatorial interplay among the 1-aminocyclopropane-1-carboxylate isoforms regulates ethylene biosynthesis in Arabidopsis thaliana. Genetics 183 979–1003. 10.1534/genetics.109.107102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varbanova M., Yamaguchi S., Yang Y., McKelvey K., Hanada A., Borochov R., et al. (2007). Methylation of gibberellins by Arabidopsis GAMT1 and GAMT2. Plant Cell 19 32–45. 10.1105/tpc.106.044602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Liu X. D., Chi X. J., Wu C. A., Li Y. Z., Song L. L., et al. (2011). Dwarf apple MbDREB1 enhances plant tolerance to low temperature, drought, and salt stress via both ABA-dependent and ABA-independent pathways. Planta 233 219–229. 10.1007/s00425-010-1279-6 [DOI] [PubMed] [Google Scholar]
- Zhao C., Zhang Z., Xie S., Si T., Li Y., Zhu J. K. (2016). Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis. Plant Physiol. 171 2744–2759. 10.1104/pp.16.00533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Chen H., Wei D., Ma H., Lin J. (2017). Arabidopsis CBF3 and DELLAs positively regulate each other in response to low temperature. Sci. Rep. 7:39819 10.1038/srep39819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Xu M., Wu L., Shen C., Ma H., Lin J. (2014). CbCBF from Capsella bursa-pastoris enhances cold tolerance and restrains growth in Nicotiana tabacum by antagonizing with gibberellin and affecting cell cycle signaling. Plant Mol. Biol. 85 259–275. 10.1007/s11103-014-0181-1 [DOI] [PubMed] [Google Scholar]
- Zhou M. Q., Shen C., Wu L. H., Tang K. X., Lin J. (2011). CBF-dependent signaling pathway: a key low temperature responder in plants. Crit. Rev. Biotechnol. 31 186–192. 10.3109/07388551.2010.505910 [DOI] [PubMed] [Google Scholar]
- Zhu Y. N., Shi D. Q., Ruan M. B., Zhang L. L., Meng Z. H., Liu J., et al. (2013). Transcriptome analysis reveals crosstalk of responsive genes to multiple abiotic stresses in cotton (Gossypium hirsutum L.). PLOS ONE 8:e80218 10.1371/journal.pone.0080218 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.