 Flavoenzymes are ubiquitous in biological systems and catalyze a diverse range of chemical transformations.
Flavoenzymes are ubiquitous in biological systems and catalyze a diverse range of chemical transformations.
Abstract
Flavoenzymes are ubiquitous in biological systems and catalyze a diverse range of chemical transformations. The flavoenzyme XiaK from the biosynthetic pathway of the indolosesquiterpene xiamycin A is demonstrated to mediate the in vivo biotransformation of xiamycin A into multiple products, including a chlorinated adduct as well as dimers characterized by C–N and N–N linkages that are hypothesized to form via radical-based mechanisms. Isolation and characterization of XiaK in vitro shows that it acts as a flavin-dependent N-hydroxylase that catalyzes the hydroxylation of xiamycin A at the carbazole nitrogen to form N-hydroxyxiamycin, a product which was overlooked in earlier in vivo experiments because its chemical and chromatographic properties are similar to those of oxiamycin. N-Hydroxyxiamycin is shown to be unstable under aerobic conditions, and characterization by electron paramagnetic resonance spectroscopy demonstrates formation of an N-hydroxycarbazole radical adduct. This radical species is proposed to serve as a key intermediate leading to the formation of the multiple xiamycin A adducts. This study suggests that non-enzyme catalyzed reactions may play a greater role in the biosynthesis of natural products than has been previously recognized.
Introduction
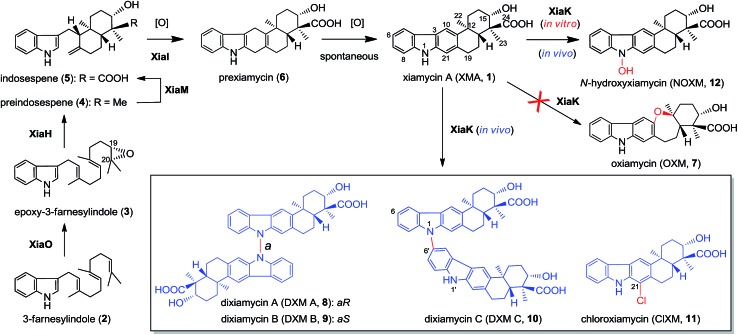
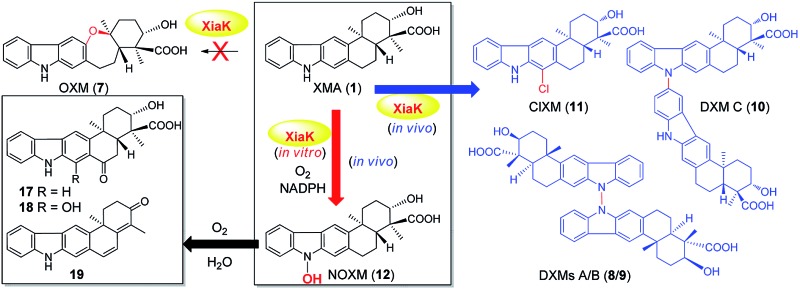
Recent technological advances have accelerated microbial natural product discovery, leading to a renaissance of interest in this field throughout academia as well as in the pharmaceutical industry. The recognition of the isolation of avermectins and artemisin by the Nobel Prize in 2015 has initiated a new wave of natural product drug discovery.1 In recent years, a growing number of indolosesquiterpenes with antibacterial, antitumor and antiviral properties have been isolated from actinomycetes.2–7 The diverse structures and broad activity profiles of indolosesquiterpenes have spurred interest in the chemical synthesis of this class of natural products.8–12 To learn nature's strategy for synthesizing this class of compounds, the biosynthetic gene cluster of the indolosesquiterpene xiamycin A (XMA, 1, Fig. 1) has been independently identified from marine-derived Streptomyces pactum SCSIO 02999 by us,13 and a mangrove-endophyte Streptomyces sp., HKI0576, has been identified by the Hertweck group.14 Further studies have unveiled the enzymatic cyclization cascade leading to the pentacyclic ring structure of 1 (Fig. 1). The pathway is initiated by oxidoreductase XiaO-catalyzed epoxidation (2 → 3),15 followed by membrane protein XiaH-mediated terpene cyclization (3 → 4),15 P450 enzyme XiaM-catalyzed tri-hydroxylations (4 → 5),16 and indole oxygenase XiaI-catalyzed central ring closure (5 → 6 → 1) to yield XMA (1).13 In addition, study of the in vivo biotransformation by the Hertweck group suggested that XMA (1) is the precursor of the cycloether oxiamycin (OXM 7),5,17 N–N and N–C coupled dimeric dixiamycins (DXMs A–C, 8–10),17 and sulfadixiamycins.6 The conversions are proposed to be catalyzed by a flavoenzyme, XiaK13 (equivalent to XiaH, as designated by Hertweck and coworkers14), and are likely radical-induced.
Fig. 1. The proposed biosynthetic pathway of XMA (1) and the XiaK-mediated formation of XMA analogues.
Flavoenzymes are a large family of enzymes that catalyze a diverse range of chemical transformations, including hydroxylations, epoxidations, and halogenations.18 While many radical-based intramolecular19–22 or intermolecular23–30 C–C biaryl formations in natural product biosynthesis are catalyzed by P450 enzymes,31 precedents of flavoenzyme-mediated C–C and N–C couplings are also known. For example, the flavin-dependent halogenases Mpy10 and Mpy11 have been shown to be responsible for the atropo-selective N–C-bipyrrole homo-coupling.32 However, it is unusual for a single flavoenzyme such as XiaK to perform diverse chemical couplings to convert a single precursor 1 to dimeric 8–10 and chloroxiamycin (ClXM, 11), which was also isolated from our in vivo biotransformation of 1 by XiaK. Reported herein is the first in vitro biochemical characterization of XiaK to determine its function in the biosynthesis of XMA (1)-related indolosesquiterpenes. Our results showed that XiaK is a flavin adenosine dinucleotide (FAD)-dependent enzyme, and the product of the XiaK reaction with 1 is merely N-hydroxyxiamycin (NOXM, 12). Interestingly, OXM (7) and other dimeric dixiamycins (8–10) were not found in the in vitro XiaK reaction. Because 12 is unstable and can be transformed into a nitroxyl radical species (16), the latter may serve as the key intermediate in the formation of multiple XMA analogues.
Results and discussion
In vivo expression of the XMA gene cluster and analysis of biosynthetic products
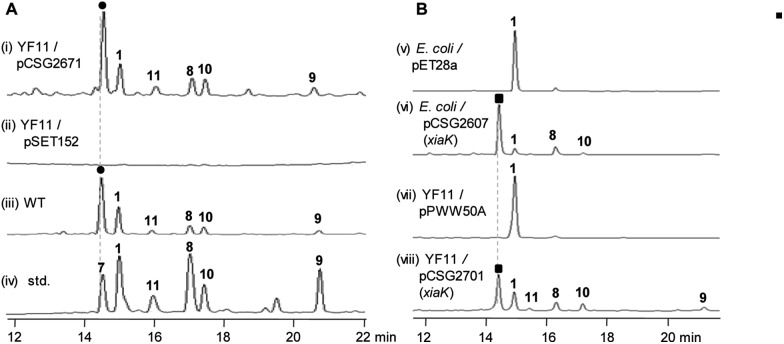
The XMA-gene cluster from S. pactum SCSIO 02999 was introduced into Streptomyces coelicolor YF11 (ref. 33) for heterologous expression studies. XMA (1), OXM (7), DXM A–C (8–10), and ClXM (11) were produced in recombinant S. coelicolor YF11/pCSG2671 (Table S1†) harboring the intact XMA-gene cluster (Fig. 2A, traces i–iv. Please note that the product peak with a symbol of “” contains other species in addition to 7;34 see also Fig. 3 for clarity). None of these products were observed in the control strain S. coelicolor YF11 carrying a void vector (Fig. 2A, trace ii). In addition to 8–10, which were previously observed by the Hertweck group,17 the halogenated metabolite 11 was also detected. This observation is intriguing because no halogenase-encoding genes are found within the XMA-gene cluster.
Fig. 2. HPLC analysis of the production profiles of different strains (A) and the in vivo biotransformation of XMA (1) by XiaK (B). (i) S. coelicolor YF11/pCSG2671 harboring the intact XMA-gene cluster; (ii) control strain YF11/pSET152; (iii) the wild strain S. pactum SCSIO 02999; (iv) standards 1 and 7–11; (v) XMA (1) fed to control strain E. coli BL21(DE3)/pET28a; (vi) XMA (1) fed to E. coli BL21(DE3)/pCSG2607; (vii) XMA (1) fed to control strain S. coelicolor YF11/pPWW50A; (viii) XMA (1) fed to YF11/pCSG2701. Please see note 34 for the symbols “” and “■”.
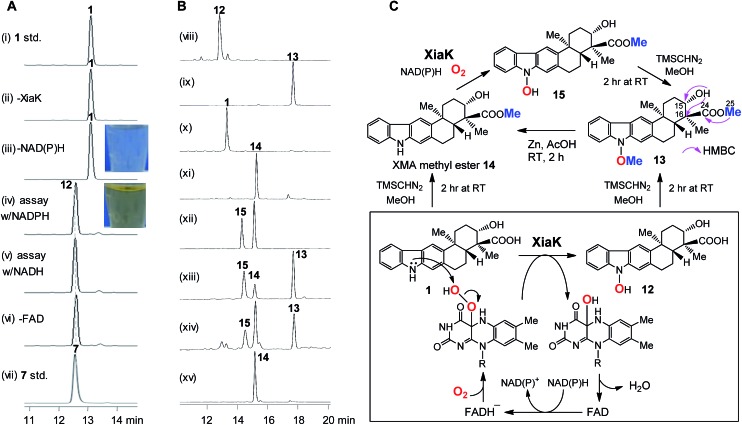
Fig. 3. The in vitro characterization of XiaK. (A) HPLC analysis of the in vitro XiaK enzyme activity assays. A typical XiaK assay was performed in 50 mM Na2HPO4–NaH2PO4 buffer (pH 8.0) for 2 h at 30 °C, comprising 300 μM XMA (1), 1 mM NADPH (or NADH), 1 mM FAD and 5 μM XiaK: (i) XMA (1) standard; (ii) XMA (1) + FAD + NADPH; (iii) XMA (1) + XiaK + FAD; (iv) XMA (1) + XiaK + FAD + NADPH; (v) XMA (1) + XiaK + FAD + NADH; (vi) XMA (1) + XiaK + NADPH; (vii) OXM (7) standard. (B) HPLC analysis of reactions with TMSCHN2. (viii) 12 std; (ix) 12 incubated with TMSCHN2 for 2 h; (x) 1 std; (xi) 1 incubated with TMSCHN2 for 2 h; (xii) an enzyme assay comprising 5 μM XiaK, 0.5 mM 14 and 1 mM NADPH; (xiii) organic extracts from (xii) incubated with TMSCHN2 for 2 h; (xiv) the reaction solution from (xiii) was heated for 2 h, and the conversion of 15 to 14 was observed; (xv) the conversion of 13 to 14 via zinc reduction. (C) Scheme for the chemical transformations of XMA (1) and 12 mediated by XiaK or TMSCHN2.
In vivo expression and characterization of XiaK
Next, we constructed the plasmid pCSG2607 (Table S1†) for the expression of xiaK in E. coli and fed XMA (1) to the resulting E. coli BL21(DE3)/pCSG2607. While XMA (1) remained unchanged in the control strain E. coli BL21(DE3)/pET28a, it appeared that 1 was converted to 7 (please see note 34) as the major product, along with minor amounts of 8 and 10 (Fig. 2B, traces v and vi). We also constructed the plasmid pCSG2701 (Table S1†) for the expression of xiaK in S. coelicolor YF11. Upon feeding 1 to S. coelicolor YF11/pCSG2701, 7 (please see note 34) was found to be the major product, along with minor products 8–11; no change in 1 was observed in the control strain S. coelicolor YF11/pPWW50A15 (Fig. 2B, traces vii and viii). The identity of ClXM (11), isolated either from S. coelicolor YF11/pCSG2671 harboring the intact XMA-gene cluster (Fig. 2A, trace i) or from S. coelicolor YF11/pCSG2701 fed with 1 (Fig. 2B, trace viii), was confirmed by 1H NMR analysis. The failure to convert 1 to 11 in the control strain suggested that the chlorination of 1 (or an activated form of 1) to form 11 in S. coelicolor YF11 is XiaK-dependent.
In vitro characterization of XiaK
To further probe the in vitro function of XiaK, N-His6-tagged XiaK was purified from E. coli BL21(DE3)/pCSG2607 to near homogeneity (Fig. S1†). The purified recombinant XiaK protein was light yellow in color and released the noncovalently-bound flavin adenosine dinucleotide (FAD) cofactor upon heat denaturation (Fig. S1†). Subsequently, an in vitro enzyme activity assay comprising 300 μM XMA (1), 1 mM NADPH (or NADH), 1 mM FAD and 5 μM XiaK was conducted in 50 mM Na2HPO4–NaH2PO4 buffer (pH 8.0) for 2 h at 30 °C. No conversion of XMA (1) was observed in control assays lacking either NAD(P)H or the enzyme XiaK (Fig. 3A, traces i–iii). In the presence of NAD(P)H with or without FAD, XMA (1) was completely converted to a product that was co-eluted with the standard OXM (7) (Fig. 3A, traces iv–vii) and displayed the same molecular mass to OXM (7) (C23H25NO4, m/z [M – H]– 378.1699 for the product, versus [M – H]– 378.1702 for 7, calcd 378.1705; Fig. S2†) and a very similar UV spectrum. As such, we initially hypothesized that OXM (7) was the major product of the in vitro XiaK reaction. Surprisingly, compounds 8–10, observed in XiaK-mediated in vivo biotransformation,17 were not detected in our in vitro XiaK reaction. Moreover, the in vitro assay solution changed from colorless to yellow at the end of the reaction (e.g. Fig. 3A, traces iii & iv; see also Fig. S2†). The yellow color cannot be attributed to OXM (7) because pure 7 is colorless.
The major product of the XiaK reaction is not OXM
To gain more insight into the XiaK-catalyzed reaction, we decided to isolate the major product of the XiaK reaction for 1H NMR analysis to verify its identity. We chose to isolate the product using E. coli BL21(DE3)/pCSG2607-mediated biotransformation of 1 (∼30 mg). Although most of 1 was readily converted to the product (Fig. 2B, trace vi) and the isolation was carefully conducted to collect only the desired product, the major compound retrieved from the isolation process was 1. Only a very minor quantity (around 2 mg) of the desired product was obtained. Although the isolated product was pure and colorless, no 1H NMR signals were discernible in MeOH-d 3, which was used routinely in our NMR characterization of authentic OXM (7).5 Surprisingly, regeneration of 1 from the purified product was noted during NMR characterization, indicating a reverse transformation of the isolated product to 1 in free solution. However, the standard OXM (7) is a stable compound, and the spontaneous conversion of 7 to 1 has never been observed. These cumulative data suggested that while OXM (7) is a metabolite of the producing strain, the major product isolated from the XiaK reaction is not OXM (7) but a closely related compound (designated 12) having a nearly identical HPLC retention time (Fig. S2A†), the same molecular weight, and a similar UV spectrum (Fig. S2B†). The co-elution of 7 and 12 under the HPLC conditions and the spontaneous conversion of 12 to 1 are consistent with an early observation that much less OXM (7, 0.29 mg L–1) than XMA (1, 6.86 mg L–1) was isolated from the wild type S. pactum SCSIO 02999,5 although the production titer of 7 (actually a mixture of 7 and 12, see also note 34) was much higher than that of 1 based on HPLC analysis (Fig. 2A, trace iii).13
Structural determination of the major product of the XiaK reaction
In order to determine the structure of 12, a large scale in vitro XiaK reaction was carried out with 35 mg of XMA (1) for 2 h to ensure 12 as the major product. The 10 mg of 12 isolated from this reaction was used to prepare a more concentrated solution for NMR analysis. However, no 1H NMR signal of 12 was observed in a variety of deuterated solvents (MeOH-d 3, acetone-d 6, CHCl3-d and DMSO-d 6). Recording at low temperature (–20 °C) also did not enable the detection of any signal. An acetyl derivative of 12 was then prepared using acetic anhydride in pyridine. However, the acetylated 12-derivative was unstable and decomposed to multiple products (data not shown).
To circumvent this obstacle, conversion of 12 to its methylated derivative was carried out in the presence of trimethylsilyldiazomethane (TMSCHN2). To our great delight, a stable product 13 was generated and isolated (Fig. 3B, traces viii and ix). The molecular formula of 13 (C25H29NO4, m/z [M + Na]+ 430.1986, calcd 430.1989) was established by HRESIMS analysis (Fig. S3†). Comparison of the NMR data of 13 with XMA (1) revealed the presence of two additional OCH3 groups (δ C/δ H: 51.8/3.62, C-25 and 63.4/4.04, C-26) and the absence of the proton signal for NH (δ H 10.9 in DMSO-d 6). The HMBC correlations from H3-25 to C-24 implied that one of the OCH3 groups is connected to C-24 in 13 (Fig. 3C). The COSY correlation of the OH at C-15 to H-15 and the HMBC correlations from OH at C-15 to C-15 and C-16 indicated that the OH remains free at C-15 in 13. Clearly, one of the OCH3 groups arises from methylation of the C-24 carboxyl group, and the other OCH3 is linked to N-1 in 13. Thus, 13 can be assigned as N-methoxy-xiamycin methyl ester (Fig. S3, Tables S2 and S3†). In separate experiments, we chemically prepared xiamycin methyl ester 14 by reaction of 1 with TMSCHN2 (Fig. 3B, traces x and xi; Fig. 3C).7 Upon incubation with XiaK, compound 14 was converted to an unstable product 15, which could react with TMSCHN2 to afford 13 (Fig. 3B, traces xii–xiv). Furthermore, 13 could be completely converted to 14 by zinc reduction (Fig. 3B, trace xv), which is indicative of the presence of an N–O bond in 13. These data unequivocally established the structure of 12 as N-hydroxyxiamycin (NOXM, Fig. 3C).
Electron paramagnetic resonance of NOXM (12)
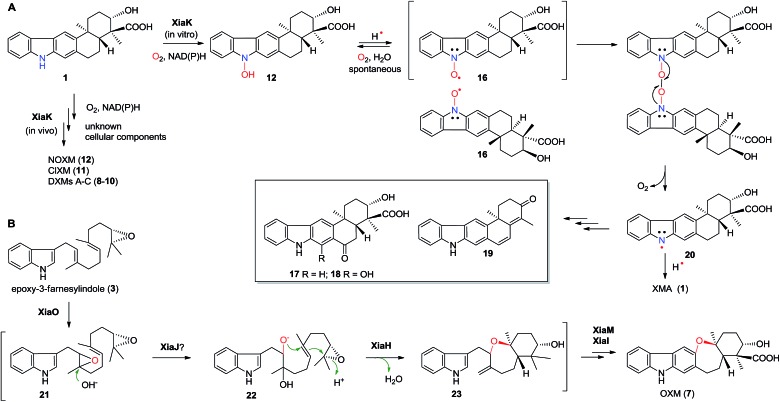
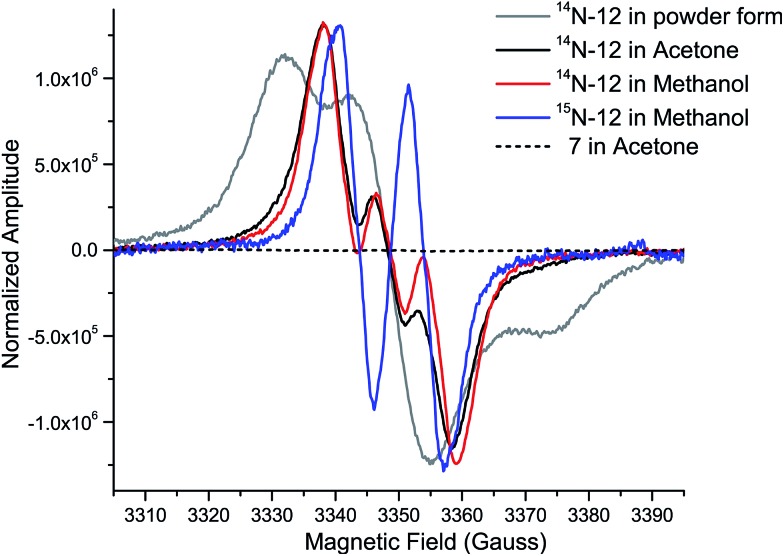
While the product of the XiaK reaction was determined to be NOXM (12), it remained puzzling that no signal was detected when the purified 12 was subjected to NMR analysis. A possible scenario is that 12 may co-exist with a radical species (such as 16, see Fig. 7) whose paramagnetic properties may complicate the NMR characterization. To test this hypothesis, the EPR (electron paramagnetic resonance) spectra of 12 were recorded together with the reference compounds XMA (1) and OXM (7) under three conditions: as a powder, in acetone and in MeOH. The results showed that XMA (1) and OXM (2) are EPR silent (Fig. S4†); however, 12 displayed a triplet EPR signal with a g-value of 2.0051 (Fig. 4), indicating the presence of a nitroxyl radical species (such as 16).
Fig. 7. The proposed mechanism for the formation of diverse chemical bonds and the biosynthesis of OXM (7).
Fig. 4. EPR spectra of 12 (as a powder, in acetone, and in MeOH), OXM (7) (in acetone), and 15N-12 (in MeOH), measured at room temperature.
To confirm this assignment, 15N-labled 12 was prepared from 15N-labeled 3-farnesylindole (2) (Fig. S5†), which was chemically synthesized from 15N-labeled indole employing reported methods.15 15N-Labeled 2 was fed to Streptomyces sp. SCSIO 02999 XM47i, where the essential xiaP gene encoding the 3-farnesylindole (2) synthase was in-frame deleted13,35 (Fig. S6†). 15N-labeled 2 appeared to be converted to 1, 12, 8 and 10 (Fig. S5†); however, our efforts to isolate 15N-labeled 12 were futile. Instead, only pure 15N-labeled 1 could be isolated. An in vitro XiaK reaction with 15N-labeled 1 to prepare 15N-labeled 12 was then attempted. Careful purification at low temperature and under a nitrogen atmosphere eventually led to the isolation of 15N-labeled 12 (Fig. S5†). The EPR spectrum of 15N-labeled 12 is consistent with a nitrogen-centered radical species (16, see Fig. 7) because the triplet splitting was changed to a doublet signal due to the substitution of the 14N nucleus with 15N at the radical center (Fig. 4). A theoretical simulation also supported this conclusion (Fig. S7†). However, the spin concentration in the 12 sample was estimated to be only around 3.6% by calculation using MTSL (1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl methanethiosulfonate) as a standard (Fig. S8†). Despite the low concentration of the nitroxyl radical in the sample, the unpaired electron spin associated with compound 16 was sufficient to broaden the NMR signals of 12 through spin relaxation. An attempt to quench the radical by the addition of ascorbic acid was ineffective, as the NMR spectrum of the resulting sample remained featureless. Compounds exhibiting similar “NMR silent” characteristics have been previously reported for intermediates (e.g. kinobscurinone) in kinamycin biosynthesis.36 Treatment with ion-exchange resin and washing with aqueous EDTA also failed to produce a visible NMR spectrum. Likewise, the structure of kinobscurinone (naturally isolated36 or chemically synthesized37) could only be determined by chemical derivatizations.
Characterization of XiaK as an N-hydroxylase
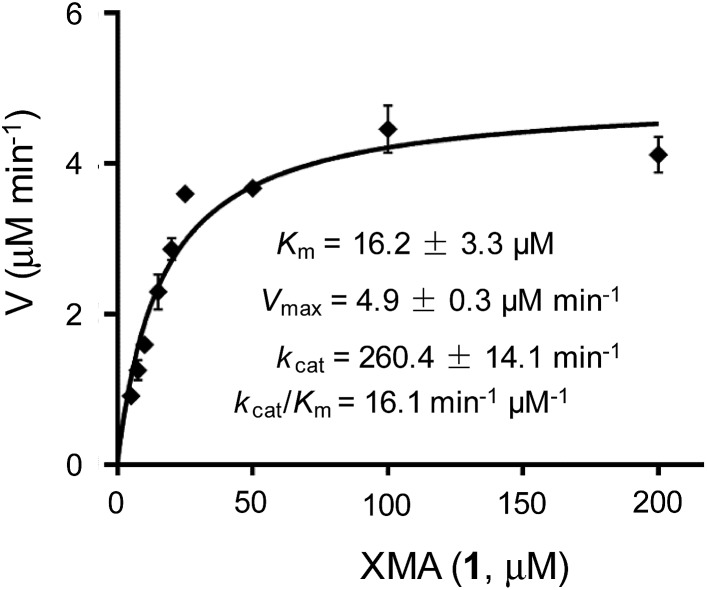
The above results indicated that XiaK is an N-hydroxylase that catalyzes the hydroxylation of 1 at N-1 to generate an unstable product, NOXM (12). Phylogenetic analysis (Fig. S9†) and structural modelling (Fig. S10†) indicate that XiaK is a member of the class A flavoprotein monooxygenase family.38,39 Sequence comparison of XiaK with other known class A flavoprotein monooxygenases reveals that XiaK consists of a conserved motif of glycine residues GXGXXG and a βαβ-Rossmann fold secondary structure, which are characteristic of FAD binding domains (Fig. S11†). To further understand the XiaK-catalyzed N-hydroxylation, the kinetic parameters were determined (Fig. 5). XiaK displayed a K m value of 16.2 μM and a k cat value of 260.4 min–1 toward 1, with a k cat/K m value of 16.1 μM–1 min–1. The efficiency of the XiaK-catalyzed N-hydroxylation is much higher than those catalyzed by other flavoenzymes40–44 and P450 enzymes.45–47 The typical k cat/K m values of a variety of N-hydroxylation reactions range from 0.0053 to 1.21 μM–1 min–1 (Table S4†).
Fig. 5. Determination of XiaK kinetic parameters for the substrate 1 ranging from 5 to 200 μM.
The XiaK-catalyzed reaction likely involves nucleophilic attack at the peroxy oxygen of a flavin-4a-hydroperoxy intermediate by the carbazole nitrogen of 1 to cleave the O–O bond and expel 4a-hydroxyflavin. Subsequent elimination of water from flavin to regenerate the oxidized coenzyme thereby completes the catalytic cycle (Fig. 3C). Although the flavin-4a-hydroperoxy species has been proposed as an intermediate in reactions catalysed by other flavoprotein monooxygenases,38 it is only transiently formed during turnover. Because XiaK is an efficient catalyst of N-hydroxylation (Table S4†), it is difficult to directly detect the flavin-4a-hydroperoxy intermediate shown in Fig. 3C. Further characterization of this intermediate will require stop-flow spectroscopic and/or rapid kinetic experiments, which will be pursued in the near future.
Time course of the XiaK reaction
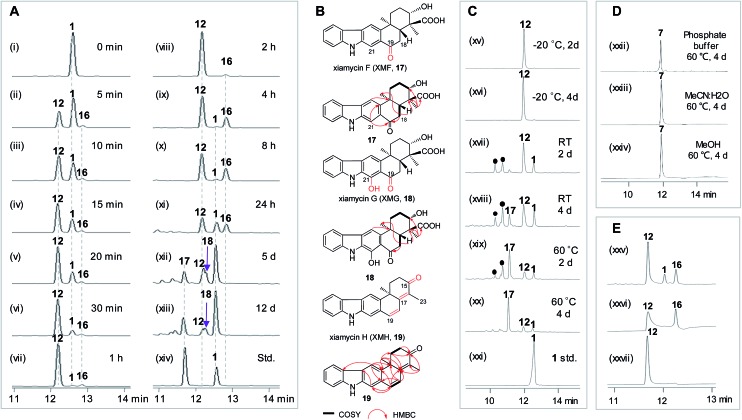
To learn more about the XiaK catalysis, a time course analysis of the in vitro XiaK reaction was also performed. As shown in Fig. 6A, XMA (1) was converted to 12 in a time-dependent manner, achieving near completion in 2 h (Fig. 6A, traces i–viii). During the period from 5 min to 2 h, another product 16 was also detected, albeit in minute quantity (Fig. 6A, traces ii–viii). The molecular weight of 16 was determined by LC-HRESIMS analysis as m/z 378 (379.1770 [M + H]+, 377.1607 [M – H]–, Fig. S12†), which is one mass unit less than that of 12. Given the fact that 16 absorbs in the visible region (Fig. S12†), the yellow color of the XiaK reaction may be attributed to the nitroxyl radical 16. Interestingly, the ratio of 16 to 12 steadily increased from 4 h to 8 h, during which time a small amount of 1 was detected (Fig. 6A, traces ix and x). After 24 h, more 12 was converted to 1 and 16 (Fig. 6A, trace xi); however, prolonged incubation (5 d or 12 d) (Fig. 6A, traces xii and xiii) resulted in depletion of 16, with 1 as the predominant product. The appearance of additional products, such as 17 and 18, was also noted; both exhibited similar UV spectra to 1 (Fig. S13†). Thus, the nascent product of the XiaK reaction is 12, which is the precursor of 16–19. However, formation of 16–19 may not be XiaK-catalyzed.
Fig. 6. Time course of the XiaK reaction and stabilities of XiaK products. (A) HPLC analysis of the time course assay of an in vitro XiaK reaction: (i) 0 min (before adding XiaK); (ii) 5 min; (iii) 10 min; (iv) 15 min; (v) 20 min; (vi) 30 min; (vii) 1 h; (viii) 2 h; (ix) 4 h; (x) 8 h; (xi) 24 h; (xii) 5 days; (xiii) 12 days; (xiv) 1 and 17 standards. (B) Chemical structures of XMA analogues. (C) Stability of 12 dissolved in 50 mM Na2HPO4–NaH2PO4 buffer (pH 8.0): (xv) –20 °C for 2 days; (xvi) –20 °C for 4 days; (xvii) room temperature (RT) for 2 days; (xviii) RT for 4 days; (xix) 60 °C for 2 days; (xx) 60 °C for 4 days; (xxi) XMA (1) standard. The filled black circles () denote multiple XMA-related products which were not isolated for structure elucidation. (D) Stability of OXM (7) treated under various conditions: (xxii) OXM (7) dissolved in 50 mM Na2HPO4–NaH2PO4 buffer (pH 8.0) and incubated at 60 °C for 4 days; (xxiii) 7 dissolved in H2O/MeCN (1 : 1, v/v) and incubated at 60 °C for 4 days; (xxiv) 7 dissolved in MeOH and incubated at 60 °C for 4 days. (E) Stability of 16: (xxv) a complete XiaK assay at 28 °C for 6 h; (xxvi) 16 isolated from an analytical XiaK assay (in trace xxv) immediately used for HPLC analysis; (xxvii) NOXM (12) isolated from an analytical XiaK assay (in trace xxv) immediately used for HPLC analysis.
To characterize the products generated during prolonged incubation with XiaK, a large scale XiaK reaction with 80 mg of XMA (1) for 7 d was performed. From this reaction, 3.3 mg of 17 (designated xiamycin F, XMF), 8.0 mg of 18 (designated xiamycin G, XMG) and 1.0 mg of 19 (designated xiamycin H, XMH) were isolated (Fig. 6B). HRESIMS analyses established molecular formulas of C23H23NO4 for 17 (Fig. S14†), C23H23NO5 for 18 (Fig. S15†), and C22H19NO for 19 (Fig. S16†). Comparison of the NMR data of 1 and 17 (Fig. S14, Tables S2 and S3†) revealed that 17 contains a C-19 carbonyl group (δ C 198.3) instead of a C-19 methylene group (δ C 31.8, δ H 3.09) as in 1. This assignment is supported by HMBC correlations from H-21 and H-18 to C-19 in 17 (Fig. S14†). Inspection of the NMR data of 18 indicated that its structure closely resembles that of 17 (Fig. S15, Tables S2 and S3†). The signal for H-21 (δ H 8.08) in 17 is absent in 18, and the chemical shift of C-21 is deshielded from δ C 110.6 in 17 to δ C 152.2 in 18, indicating a hydroxy substitution at C-21 in 18 (Fig. 6B). Detailed NMR analysis also established the structure of 19 (Fig. 6B, S16, Tables S2 and S3†) as a highly oxidized congener of 1.
Stabilities of XiaK reaction products
The stabilities of the products generated in the XiaK reaction were examined under different conditions. Compound 12 was found to be stable when stored in organic or mixed solvents such as MeOH or H2O/MeCN (1 : 1). Only minor conversion of 12 to 1 was observed under harsh conditions, such as heating at 60 °C for 4 d (Fig. S17†). When stored in 50 mM Na2HPO4–NaH2PO4 buffer (pH 8.0), compound 12 was stable for at least 4 d at low temperature (–20 °C, Fig. 6C, traces xv and xvi); however, it decomposed to multiple products at room temperature or upon heating at 60 °C (Fig. 6C, traces xvii–xx). In contrast, the reference OXM (7) remained unchanged after the same treatments, even when heated at 60 °C for 4 d (Fig. 6D, traces xxii–xxiv). These data further validated that 12 and OXM (7) are structurally distinct compounds. In addition, although OXM (7) could be isolated from the recombinant strain S. coelicolor YF11/pCSG2671 carrying the XMA gene cluster (Fig. 2A, trace i), it was absent in the S. coelicolor YF11/pCSG2701-mediated biotransformation of 1 (Fig. 2B, trace vi).34 This observation indicated that the production of OXM (7) cannot be catalyzed by XiaK alone but requires additional proteins encoded by the XMA gene cluster. It is also possible that OXM (7) is not directly derived from XMA (1) but from another intermediate in the pathway. Interestingly, compound 16 could be isolated from a XiaK assay using HPLC, and indeed was yellow in color. However, it is highly labile and quickly converted to 12, as shown by immediate re-submission to HPLC analysis (Fig. 6E, traces xxv and xxvii).
Implications for indolosesquiterpene diversification
Flavoenzymes play important roles in N-oxidations of natural products to introduce N-hydroxyl,40–44,46,48,49 oxime,50–54 nitro,45,47,55–60 nitrone,61,62 and nitrile functional groups.63,64 In this study, XiaK was biochemically characterized as a FAD-dependent N-hydroxylase that catalyzes the conversion of XMA (1) into NOXM (12). The assembly of the dimeric DXMs A–C (8–10) was previously proposed to proceed through an enzyme-mediated N-radical mechanism on the basis that DXMs A–C (8–10) could be generated from 1 by treatment with the radical-forming agent benzoyl peroxide.17 However, our current data demonstrated that the nascent product of the XiaK reaction is NOXM (12), indicating that the by-products 8–11 detected in the cell culture are unlikely to be generated directly by XiaK. However, if by-product formation is indeed radically induced as hypothesized,17 NOXM (12) may serve as a precursor for the multiple XMA analogues. Given that 12 is stable under nitrogen atmosphere and in organic solvent, the conversion of 12 into radical species must occur in an oxygen- and buffer-dependent manner. Although the detailed mechanism of radical formation awaits further investigation, we propose that the radical derived from 12 is most likely a nitroxyl radical 16 (Fig. 7A). This is based on the facts that 16 is yellow in color (consistent with its UV-vis spectrum) and its molecular mass is one unit less than that of 12. In addition, the EPR analysis of the purified 12 and its 15N-labeled isotopologue provided strong evidence supporting the transient occurrence of a nitroxyl radical species, such as 16, in the sample of 12 (Fig. 4). In buffer solution, 16 may be subjected to single electron reduction to regenerate 12 (Fig. 6E, trace xxvi and Fig. 7A). Alternatively, 16 may undergo dimerization followed by deoxygenation to yield nitrogen radical species 20 (Fig. 7A). The observed spontaneous conversion of 12 to 1 may also be explained by a proton-coupled single electron reduction of 20 (Fig. 7A). Regardless, the source of the reducing equivalents and the mechanism of the reduction are not immediately apparent.
Once formed, the radical species 20 could react with another 20 or its resonance congeners to afford N–N-coupled atropo-diastereomers DXMs A (8) and B (9) and the N–C-coupled DXM C (10) (Fig. S18A†). Alternatively, DXMs A–C (8–10) could be formed by a non-radical mechanism by coupling 12 and 1 directly (Fig. S18B†). Because production of 8–11 from 1 was observed in vivo but not in the in vitro XiaK reaction, it is conceivable that the formation of these compounds may require some necessary but unknown cellular components in vivo. The conversion of 12 to 17–19 under prolonged incubation in aqueous solution also occurs in the absence of XiaK. However, the reaction mechanisms are not clear (Fig. 7A). Likewise, it is shown here that formation of OXM (7) from 1 is not a XiaK-catalyzed event, as previously proposed.17 The fact that OXM (7) could be isolated from a recombinant strain expressing the intact XMA-gene cluster but not from in vitro incubation with XiaK alone prompted us to propose an alternative route for the biosynthesis of OXM (7). As depicted in Fig. 7B, the oxygenase XiaO may be responsible for the double epoxidation of 3 to yield 21. This is similar to the PaxM-catalyzed reaction in paxilline biosynthesis.65 Hydrolysis of the internal oxirane of 21 by the limonene-1,2-epoxide hydrolase XiaJ13 could lead to 22. The final steps include formation of the ether ring catalyzed by XiaH,15 a stepwise dehydration to generate an exocyclic methylene moiety to form 23, the oxidation of C-24 in 23 to a carboxyl group by XiaM,16 and formation of the central ring catalyzed by XiaI to furnish OXM (7).
Conclusions
This work is significant for three reasons. First, XiaK has been unequivocally established to be a flavoenzyme functioning as an N-hydroxylase in the biosynthesis of XMA (1)-related indolosesquiterpenes. Second, although in vivo biotransformation results suggested that XiaK is a versatile catalyst facilitating the formation of diverse chemical bonds in the assembly of a variety of indolosesquiterpene derivatives, our in vitro biochemical characterizations of XiaK demonstrated that this enzyme only catalyzes the N-hydroxylation of 1 to afford NOXM (12), which decomposes to a variety of products, likely in an enzyme-free and radical-mediated manner. Third, this work clearly reveals that many compounds isolated from natural sources are not true secondary metabolites but are derivatives generated during post-biosynthetic, non-enzyme catalyzed reactions.
Experimental
General HPLC analysis
General HPLC analysis was carried out using a reversed phase column (Luna C18, 5 μm, 150 × 4.6 mm, Phenomenex) with UV detection at 254 nm on a Varian Prostar Station (or Agilent series 1200) using the following program: solvent system (solvent A, water supplemented with 0.05% formic acid; solvent B, MeCN with 0.05% formic acid); 5% B to 100% B (0 to 14 min), 100% B (14 to 19 min), 100% B to 5% B (19 to 20 min), 5% B (20 to 25 min); flow rate 1 mL min–1. HPLC-MS analysis was conducted on a Bruker Daltonics amaZon SL instrument.
Heterologous expression of the XMA-gene cluster
The cosmid pCSG2407 (ref. 13) was digested by EcoRI, and the ∼30 kb DNA fragment containing the intact XMA-gene cluster was ligated to the integrative vector pSET152 (EcoRI), yielding the plasmid pCSG2671 (Table S1†). Then, pCSG2671 was transconjugated into S. coelicolor YF11 through a conventional method66 to yield the recombinant strain S. coelicolor YF11/pCSG2671 for heterologous expression. Three independent clones were randomly chosen for small scale fermentation and subjected to HPLC analyses.
Biotransformation of XMA
The xiaK gene was PCR amplified from genomic DNA of S. pactum SCSIO 02999 using primers XiaK-3F and XiaK-3R (Table S1†) with high fidelity DNA polymerase Pyrobest (Takara). The PCR products were digested with NdeI/BglII and inserted into the vector pET28a, affording the plasmid pCSG2607. A single transformant of the E. coli strain BL21(DE3)/pCSG2607 was inoculated into 3 mL of LB media containing 50 μg mL–1 kanamycin and was grown to an A 600 of around 0.7 at 37 °C. Then, 0.1 mM IPTG (isopropyl β-d-thiogalactopyranoside) and 20 μM XMA (1) were added to the culture, with another 20 h of growth at 16 °C. The cells were collected by centrifugation, resuspended in 200 μL 50 mM Tris–HCl (pH 8.0), and extracted with butanone. The organic extracts were then subjected to HPLC analysis. E. coli BL21(DE3)/pET28a was treated in the same manner to serve as a negative control. In an attempt to isolate 12, E. coli BL21(DE3)/pCSG2607 was incubated in 1 L LB media containing 50 μg mL–1 kanamycin at 37 °C for 2 h (A 600 around 0.7). Subsequently, 0.1 mM IPTG and 30 mg of XMA 1 (dissolved in 5 mL DMSO) were added to the culture, with another 20 h of growth at 16 °C. The products were extracted 4 times with an equal volume of butanone. After evaporation under vacuum to remove the organic solvents, the residue was resuspended in MeOH (2 mL) and separated by semi-preparative HPLC using a reversed phase column (Gemini C18, 5 μm, 250 × 10 mm, Phenomenex) on an Agilent series 1200 station (solvent A, 0.1% formic acid in water; solvent B, 90% MeCN in water; eluted with constant 85% solvent B; flow rate 2.5 mL min–1) to yield 12 (2 mg). The PCR product of the xiaK gene was digested by NdeI/BglII and inserted into pPWW50A to afford pCSG2701. Then, pCSG2701 was introduced into S. coelicolor YF11 by conjugation via E. coli ET12567/pUZ8002. The strain S. coelicolor YF11/pCSG2701 was inoculated into 3 mL of AM6-4 media containing 35 μg mL–1 apramycin and 20 μM XMA 1 and was grown at 28 °C for 3 d. The products were extracted with an equal volume of butanone and were subjected to general HPLC analysis. S. coelicolor YF11/pPWW50A was treated in the same manner to serve as a negative control.
Construction of in-frame deletion mutant XM47i (ΔxiaPi)
The cosmid pCSG2517,13 in which the xiaP gene was partially replaced by a gene cassette comprising aac(3)IV and oriT flanked by FRT (flippase recognition target) sites, was used to transform E. coli DH5α/BT340. The temperature-sensitive FLP recombination plasmid BT340 was induced at 42 °C to express FLP-recombinase; the inserted aac(3)V cassette was removed, leaving an 81 bp “scar” in the xiaP gene, to afford the cosmid pCSG2547 (Table S1†). The generation and screening of the xiaP in-frame deletion mutant XM47i (Fig. S6†) was carried out according to previously described methods.15
Overexpression and purification of XiaK
N-(His)6-tagged XiaK proteins were purified from E. coli BL21(DE3)/pCSG2607 via nickel affinity chromatography. Cells were disrupted by sonification on ice after washing and were resuspended in the lysing buffer (50 mM Tris–HCl, pH 8.0). The cellular lysates were centrifuged at 13 500g for 0.5 h. The proteins were further purified by nickel–nitrilotriacetic acid (Ni2+–NTA) agarose (Invitrogen) according to the manufacturer's protocols. The purified protein was desalted through a PD-10 column (GE Healthcare) and was concentrated using a Vivaspin concentrator (10 kD, Sartorius). The protein concentration was determined by the Bradford method. Purified N-(His)6-tagged XiaK was aliquoted and stored in 50 mM phosphate buffer (pH 8.0) containing 1 mM dithiothreitol (DTT) and 20% glycerol at –80 °C until use.
XiaK assays
A standard XiaK assay comprising 300 μM XMA (1), 1 mM NADPH, 1 mM FAD and 5 μM XiaK in 50 mM Na2HPO4–NaH2PO4 buffer (pH 8.0) was incubated at 30 °C for 2 h. The XiaK reaction was stopped by adding an equal volume of 100% cold MeOH. An assay with boiled XiaK was performed as a control. For the XiaK time course assay, the reaction solution was monitored by HPLC at the time points of 0 min, 5 min, 10 min, 15 min, 20 min, 30 min, 1 h, 2 h, 4 h, 8 h, 24 h, 5 d and 12 d.
Isolation of XiaK reaction products
To isolate NOXM (12), a 200 mL of enzyme reaction was made with 35 mg XMA (1), 0.2 mM NADPH and 15 μM XiaK. After incubation at 30 °C for 2 h, the products were extracted 4 times with an equal volume of butanone. After evaporating the organic solvents under vacuum, the residue was resuspended in MeOH (2 mL) and separated by semi-preparative HPLC to yield 12 (10 mg) as previously described. To isolate the XiaK reaction products, 80 mg of XMA (1) was reacted with XiaK at room temperature (RT) for 7 d, and the products were extracted with an equal volume of butanone 4 times. After evaporation of the organic solvents under vacuum, the crude extract was subjected to a normal phase silica gel column (100–200 mesh, 12 g) and eluted with CHCl3/CH3OH (100/0, 99/1, 98/2, 96/4, 92/8, 84/16, 68/32 and 36/64 v/v, each 72 mL) to yield eight fractions (Fr. 1 to Fr. 8). Fr. 1 was further purified by semi-preparative HPLC (solvent A, 0.1% formic acid in water; solvent B, 90% MeCN in water; eluted with constant 85% solvent B; flow rate 2.5 mL min–1) to yield 19 (1.0 mg). Fr. 6 and Fr. 7 were combined and further separated by semi-preparative HPLC as described in the section “Biotransformation of XMA” to afford 17 (3.3 mg), 18 (8.0 mg) and NOXM (12) (5.0 mg).
Acetylation of NOXM (12)
A mixture of 12 (0.02 mM) and acetic acid anhydride (100 μL) in pyridine (1 mL) was stirred overnight at RT; then, the mixture was extracted with EtOAc and dried by evaporation under vacuum. The residue was redissolved in MeOH and checked by HPLC.
Methylation with TMSCHN2
TMSCHN2 (2.0 M solution in hexanes, 0.16 mmol, 80 μL) was slowly added to a solution of 12 (0.012 mmol, 4.6 mg) in MeOH (0.5 mL). After stirring the mixture for 2 h at room temperature, AcOH (0.17 mmol, 10 μL) was added, and the mixture was stirred for another 15 min. Afterwards, the solvent was removed under vacuum and the residue was subjected to semi-preparative HPLC using a reversed phase column (Luna C18, 5 μm, 250 × 20 mm, Phenomenex) with an elution gradient (solvent A: H2O + 0.1% formic acid, solvent B: MeCN 90%) to afford 13 (1.8 mg). Similarly, reaction of XMA 1 (0.055 mM, 20 mg) with TMSCHN2 afforded 14 (13.7 mg), which was subjected to a large scale (100 mL) in vitro transformation by XiaK for 4 h at 30 °C. The solution was extracted 4 times with an equal volume of EtOAc, and the extracts were evaporated under vacuum to remove the organic solvents. The residue was resuspended in MeOH (2 mL) and was subjected to semi-preparative HPLC purification using a reversed phase column (Gemini C18, 5 μm, 250 × 10 mm, Phenomenex) on an Agilent series 1200 station (solvent A, 0.1% formic acid in water; solvent B, 90% MeCN in water; eluted with isocratic 85% solvent B; flow rate 2.5 mL min–1) under nitrogen gas protection and on an ice bath to obtain 15 (5.3 mg). Finally, 15 (0.014 mM, 5.3 mg) was subjected to reaction with TMSCHN2 under the same conditions described above to afford 13 (3.4 mg) after purification.
Reduction of 12 or 13 using zinc powder in AcOH
A mixture of 12 (0.02 mM) or 13 (0.02 mM), activated Zn powder (0.4 mM) and AcOH (0.12 mL) in MeOH (0.5 mL) was stirred at RT for 2 h. The reaction was stopped by adding 1 mL of saturated aqueous NaHCO3 solution. The mixture was then extracted with 1.5 mL EtOAc and dried under vacuum. The residue was redissolved in MeOH and checked by HPLC using a reversed phase column (Luna C18, 5 μm, 150 × 4.6 mm, Phenomenex) with UV detection at 254 nm using the following program: solvent system (solvent A, 10% MeCN in water supplemented with 0.1% formic acid; solvent B, 90% MeCN in water); 5% B to 100% B (linear gradient, 0 to 20 min), 100% B (20 to 25 min), 100% B to 5% B (25 to 26 min), 5% B (26 to 30 min); flow rate 1 mL min–1.
Preparation of 15N-12
15N-3-Farnesylindole (15N-2, 680 mg) was prepared from 15N-indole (1 g) according to previously reported methods.15 Then, 15N-2 (147 mg) was fed to S. pactum XM47i (where xiaP was inactivated to block the synthesis of 2) cultured in AM6-416 media (2 L). The production of 1 and 12 in S. pactum XM47i cultured at 28 °C was monitored by HPLC. The cells were harvested at 10 d. The metabolites were subsequently isolated following a reported protocol16 to obtain 15N-labeled XMA 1 (5.1 mg). Incubation of XiaK with 15N-1 was carried out in a 30 mL volume, and the 15N-12 (2.4 mg) product was isolated according to a previously described semi-preparative HPLC method under the protection of N2.
Determination of kinetic parameters of XiaK
To determine the kinetic parameters of XiaK-catalyzed N-hydroxylation, XMA (1) was set as a variable substrate in concentrations of 5, 7.5, 10, 15, 20, 25, 50, 100, and 200 μM. Enzyme assays (a total volume of 100 μL) were performed in Na2HPO4–NaH2PO4 buffer (50 mM, pH 8.0) containing 1 mM NADPH, 18.8 nM XiaK and variable concentrations of 1 at 30 °C for 5 min. The reaction was quenched by adding ice-cold MeOH (100 μL) and was centrifuged at 8600g for 10 min. The sample was then subjected to HPLC analysis to determine the conversion rate. Five parallel assays were carried out for each concentrate of XMA (1). K m and V max were calculated by nonlinear regression (Michaelis–Menten) analysis using Prism 6.0 software.
Electron paramagnetic resonance measurements
EPR measurements were performed on 12 (including 15N-12), XMA (1) and OXM (7) using a Bruker X-band (9.4 GHz) EMX plus 10/12 spectrometer at room temperature. A cylindrical resonator (ER4119hs TE011) was used for data collection. Different forms of NOXM (12) (powder, dissolved in MeOH, dissolved in acetone) were prepared and measured for comparison. The EPR data acquisition parameters for free radical analysis were as follows: frequency, 9.390 GHz; microwave power, 1 mW; modulation frequency, 100 kHz; modulation amplitude, 0.5 Gauss; sweep time, 40 s per scan. Different numbers of scans were accumulated for each sample until a reasonable S/N ratio was achieved. The double-integration method was used to estimate the concentration of radicals in 12. A set of dilutions with different concentrations (20, 50, 100, 200, and 500 μM) of MTSL (1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl methanethiosulfonate, Toronto Research Chemicals, Ontario, Canada) was used as a standard for estimating the spin concentration. EPR spectra of the MTSL standards and 12 were collected using the same acquisition parameters. Then, the double integration of the first derivative EPR spectrum (to correspond to the area under a simple absorption spectrum) of 12 was compared against those of the MTSL standards to determine the spin concentration of 12. The EPR spectra of 12 (actually 16) (including 15N-12) in different forms were simulated using biomolecular EPR spectroscopy software (Isotropic Radicals program).67
Acknowledgments
This work is supported in part by the Chinese Academy of Sciences (XDA11030403, QYZDJ-SSW-DQC004), National Natural Science Foundation of China (41406183, 31300084, 31630004, 31290233, 31125001), the Administration of Ocean and Fisheries of Guangdong Province (GD2012-D01-002), and the PPP program between the China Scholarship Council (CSC) and Deutscher Akademischer Austausch Dienst (DAAD). Q. Z. is a “Pearl New Star” scholar and is supported by the Science and Technology Program of Guangzhou. H. L. is a recipient of the K. C. Wong Education Foundation, Hong Kong. H.-W. L. is funded by the National Institutes of Health (GM040541) and the Welch Foundation (F-1511). We are grateful for the use of the analytical facilities in SCSIO.
Footnotes
References
- Shen B. Cell. 2015;163:1297–1300. doi: 10.1016/j.cell.2015.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada K., Kajiwara H., Imamura N. J. Nat. Prod. 2010;73:698–701. doi: 10.1021/np1000522. [DOI] [PubMed] [Google Scholar]
- Ding L., Munch J., Goerls H., Maier A., Fiebig H. H., Lin W. H., Hertweck C. Bioorg. Med. Chem. Lett. 2010;20:6685–6687. doi: 10.1016/j.bmcl.2010.09.010. [DOI] [PubMed] [Google Scholar]
- Ding L., Maier A., Fiebig H. H., Lin W. H., Hertweck C. Org. Biomol. Chem. 2011;9:4029–4031. doi: 10.1039/c1ob05283g. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Mándi A., Li S., Chen Y., Zhang W., Tian X., Zhang H., Li H., Zhang W., Zhang S., Ju J., Kurtán T., Zhang C. Eur. J. Org. Chem. 2012;2012:5256–5262. [Google Scholar]
- Baunach M., Ding L., Willing K., Hertweck C. Angew. Chem., Int. Ed. 2015;54:13279–13283. doi: 10.1002/anie.201506541. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Ha T. K., Oh W. K., Shin J., Oh D. C. J. Nat. Prod. 2016;79:51–58. doi: 10.1021/acs.jnatprod.5b00634. [DOI] [PubMed] [Google Scholar]
- Sun Y., Chen P., Zhang D., Baunach M., Hertweck C., Li A. Angew. Chem., Int. Ed. 2014;53:9012–9016. doi: 10.1002/anie.201404191. [DOI] [PubMed] [Google Scholar]
- Rosen B. R., Werner E. W., O'Brien A. G., Baran P. S. J. Am. Chem. Soc. 2014;136:5571–5574. doi: 10.1021/ja5013323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Z., Yu H., Li L., Tao W., Chen H., Wan M., Yang P., Edmonds D. J., Zhong J., Li A. Nat. Commun. 2015;6:6096. doi: 10.1038/ncomms7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotta A. H. Org. Lett. 2015;17:3358–3361. doi: 10.1021/acs.orglett.5b01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Meng Z. C., Chen P. X., Zhang D. L., Baunach M., Hertweck C., Li A. Org. Chem. Front. 2016;3:368–374. [Google Scholar]
- Li H., Zhang Q., Li S., Zhu Y., Zhang G., Zhang H., Tian X., Zhang S., Ju J., Zhang C. J. Am. Chem. Soc. 2012;134:8996–9005. doi: 10.1021/ja303004g. [DOI] [PubMed] [Google Scholar]
- Xu Z., Baunach M., Ding L., Hertweck C. Angew. Chem., Int. Ed. 2012;51:10293–10297. doi: 10.1002/anie.201204087. [DOI] [PubMed] [Google Scholar]
- Li H., Sun Y., Zhang Q., Zhu Y., Li S. M., Li A., Zhang C. Org. Lett. 2015;17:306–309. doi: 10.1021/ol503399b. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Li H., Li S., Zhu Y., Zhang G., Zhang H., Zhang W., Shi R., Zhang C. Org. Lett. 2012;14:6142–6145. doi: 10.1021/ol302782u. [DOI] [PubMed] [Google Scholar]
- Baunach M., Ding L., Bruhn T., Bringmann G., Hertweck C. Angew. Chem., Int. Ed. 2013;52:9040–9043. doi: 10.1002/anie.201303733. [DOI] [PubMed] [Google Scholar]
- Walsh C. T., Wencewicz T. A. Nat. Prod. Rep. 2013;30:175–200. doi: 10.1039/c2np20069d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikezawa N., Iwasa K., Sato F. J. Biol. Chem. 2008;283:8810–8821. doi: 10.1074/jbc.M705082200. [DOI] [PubMed] [Google Scholar]
- Gesell A., Rolf M., Ziegler J., Chavez M. L. D., Huang F. C., Kutchan T. M. J. Biol. Chem. 2009;284:24432–24442. doi: 10.1074/jbc.M109.033373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino M., Sugimoto H., Shiro Y., Asamizu S., Onaka H., Nagano S. Proc. Natl. Acad. Sci. U. S. A. 2007;104:11591–11596. doi: 10.1073/pnas.0702946104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff D., Pelzer S., Holtzel A., Nicholson G. J., Stockert S., Wohlleben W., Jung G., Sussmuth R. D. Angew. Chem., Int. Ed. 2001;40:1693–1696. doi: 10.1002/1521-3773(20010504)40:9<1693::aid-anie16930>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Ma J., Wang Z., Huang H., Luo M., Zuo D., Wang B., Sun A., Cheng Y. Q., Zhang C., Ju J. Angew. Chem., Int. Ed. 2011;50:7797–7802. doi: 10.1002/anie.201102305. [DOI] [PubMed] [Google Scholar]
- Funa N., Funabashi M., Ohnishi Y., Horinouchi S. J. Bacteriol. 2005;187:8149–8155. doi: 10.1128/JB.187.23.8149-8155.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Guengerich F. P., Bellamine A., Lamb D. C., Izumikawa M., Lei L., Podust L. M., Sundaramoorthy M., Kalaitzis J. A., Reddy L. M., Kelly S. L., Moore B. S., Stec D., Voehler M., Falck J. R., Shimada T., Waterman M. R. J. Biol. Chem. 2005;280:11599–11607. doi: 10.1074/jbc.M410933200. [DOI] [PubMed] [Google Scholar]
- Prag A., Gruning B. A., Hackh M., Ludeke S., Wilde M., Luzhetskyy A., Richter M., Luzhetska M., Gunther S., Muller M. J. Am. Chem. Soc. 2014;136:6195–6198. doi: 10.1021/ja501630w. [DOI] [PubMed] [Google Scholar]
- Saruwatari T., Yagishita F., Mino T., Noguchi H., Hotta K., Watanabe K. ChemBioChem. 2014;15:656–659. doi: 10.1002/cbic.201300751. [DOI] [PubMed] [Google Scholar]
- Mazzaferro L. S., Huttel W., Fries A., Muller M. J. Am. Chem. Soc. 2015;137:12289–12295. doi: 10.1021/jacs.5b06776. [DOI] [PubMed] [Google Scholar]
- Gil Girol C., Fisch K. M., Heinekamp T., Gunther S., Huttel W., Piel J., Brakhage A. A., Muller M. Angew. Chem., Int. Ed. 2012;51:9788–9791. doi: 10.1002/anie.201203603. [DOI] [PubMed] [Google Scholar]
- Griffiths S., Mesarich C. H., Saccomanno B., Vaisberg A., De Wit P. J., Cox R., Collemare J. Proc. Natl. Acad. Sci. U. S. A. 2016;113:6851–6856. doi: 10.1073/pnas.1603528113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldemir H., Richarz R., Gulder T. A. Angew. Chem., Int. Ed. 2014;53:8286–8293. doi: 10.1002/anie.201401075. [DOI] [PubMed] [Google Scholar]
- Yamanaka K., Ryan K. S., Gulder T. A., Hughes C. C., Moore B. S. J. Am. Chem. Soc. 2012;134:12434–12437. doi: 10.1021/ja305670f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Wang Y., Yu Y., Bai T., Chen L., Liu P., Guo H., Zhu C., Tao M., Deng Z. Curr. Microbiol. 2012;64:185–190. doi: 10.1007/s00284-011-0048-5. [DOI] [PubMed] [Google Scholar]
- In Fig. 2, traces i and iii, the product peak () contains both 12 (major) and 7 (minor); while in traces vi and viii, the product peak (■) contains merely 12
- Saha S., Zhang W., Zhang G., Zhu Y., Chen Y., Liu W., Yuan C., Zhang Q., Zhang H., Zhang L., Zhang W., Zhang C. Chem. Sci. 2017;8:1607–1612. doi: 10.1039/c6sc03875a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S. J., Melville C. R. Tetrahedron Lett. 1997;38:1473–1476. [Google Scholar]
- Gould S. J., Melville C. R. Bioorg. Med. Chem. Lett. 1995;5:51–54. [Google Scholar]
- van Berkel W. J., Kamerbeek N. M., Fraaije M. W. J. Biotechnol. 2006;124:670–689. doi: 10.1016/j.jbiotec.2006.03.044. [DOI] [PubMed] [Google Scholar]
- Huijbers M. M., Montersino S., Westphal A. H., Tischler D., van Berkel W. J. Arch. Biochem. Biophys. 2014;544:2–17. doi: 10.1016/j.abb.2013.12.005. [DOI] [PubMed] [Google Scholar]
- Meneely K. M., Lamb A. L. Biochemistry. 2007;46:11930–11937. doi: 10.1021/bi700932q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield J. A., Frederick R. E., Streit B. R., Wencewicz T. A., Ballou D. P., DuBois J. L. J. Biol. Chem. 2010;285:30375–30388. doi: 10.1074/jbc.M110.157578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binda C., Robinson R. M., Martin Del Campo J. S., Keul N. D., Rodriguez P. J., Robinson H. H., Mattevi A., Sobrado P. J. Biol. Chem. 2015;290:12676–12688. doi: 10.1074/jbc.M114.629485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugai Y., Katsuyama Y., Ohnishi Y. Nat. Chem. Biol. 2016;12:73–75. doi: 10.1038/nchembio.1991. [DOI] [PubMed] [Google Scholar]
- Huang Z. D., Wang K. K. A., van der Donk W. A. Chem. Sci. 2016;7:5219–5223. doi: 10.1039/c6sc01389a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Zhao H. M. Angew. Chem., Int. Ed. 2006;45:622–625. doi: 10.1002/anie.200502903. [DOI] [PubMed] [Google Scholar]
- Johnson H. D., Thorson J. S. J. Am. Chem. Soc. 2008;130:17662–17663. doi: 10.1021/ja807557a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y. S., Zhang H., Brunzelle J. S., Nair S. K., Zhao H. Proc. Natl. Acad. Sci. U. S. A. 2008;105:6858–6863. doi: 10.1073/pnas.0712073105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry R. J., Li W., Cooper H. N. J. Bacteriol. 1997;179:409–416. doi: 10.1128/jb.179.2.409-416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. K., Abdelwahab H., Martin Del Campo J. S., Mehra-Chaudhary R., Sobrado P., Tanner J. J. J. Biol. Chem. 2016;291:21553–21562. doi: 10.1074/jbc.M116.745315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibbesen O., Koch B., Halkier B. A., Moller B. L. J. Biol. Chem. 1995;270:3506–3511. doi: 10.1074/jbc.270.8.3506. [DOI] [PubMed] [Google Scholar]
- Kelly W. L., Townsend C. A. J. Am. Chem. Soc. 2002;124:8186–8187. doi: 10.1021/ja025926g. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Zhang Q., Li S., Lin Q., Fu P., Zhang G., Zhang H., Shi R., Zhu W., Zhang C. J. Am. Chem. Soc. 2013;135:18750–18753. doi: 10.1021/ja410513g. [DOI] [PubMed] [Google Scholar]
- Liu N., Song L. J., Liu M. H., Shang F., Anderson Z., Fox D. J., Challis G. L., Huang Y. Chem. Sci. 2016;7:482–488. doi: 10.1039/c5sc03021h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Minami A., Igarashi Y., Izumikawa M., Umemura M., Nagano N., Machida M., Kawahara T., Shin-Ya K., Gomi K., Oikawa H. Angew. Chem., Int. Ed. 2016;55:8072–8075. doi: 10.1002/anie.201602611. [DOI] [PubMed] [Google Scholar]
- He J., Hertweck C. J. Am. Chem. Soc. 2004;126:3694–3695. doi: 10.1021/ja039328t. [DOI] [PubMed] [Google Scholar]
- Zhang H., White-Phillip J. A., Melancon III C. E., Kwon H. J., Yu W. L., Liu H. W. J. Am. Chem. Soc. 2007;129:14670–14683. doi: 10.1021/ja0744854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Al-Mestarihi A., Grimes C. L., Kahne D., Bachmann B. O. J. Am. Chem. Soc. 2008;130:15756–15757. doi: 10.1021/ja8051415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke J., Ishida K., Ishida-Ito M., Hertweck C. Angew. Chem., Int. Ed. 2013;52:8271–8275. doi: 10.1002/anie.201303196. [DOI] [PubMed] [Google Scholar]
- Li S., Xiao J., Zhu Y., Zhang G., Yang C., Zhang H., Ma L., Zhang C. Org. Lett. 2013;15:1374–1377. doi: 10.1021/ol400342e. [DOI] [PubMed] [Google Scholar]
- Setser J. W., Heemstra Jr J. R., Walsh C. T., Drennan C. L. Biochemistry. 2014;53:6063–6077. doi: 10.1021/bi500655q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Qian G., Ye Y., Wright S., Chen H., Shen Y., Liu F., Du L. Org. Lett. 2016;18:2495–2498. doi: 10.1021/acs.orglett.6b01089. [DOI] [PubMed] [Google Scholar]
- Newmister S. A., Gober C. M., Romminger S., Yu F., Tripathi A., Parra L. L., Williams R. M., Berlinck R. G., Joullie M. M., Sherman D. H. J. Am. Chem. Soc. 2016;138:11176–11184. doi: 10.1021/jacs.6b04915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olano C., Moss S. J., Brana A. F., Sheridan R. M., Math V., Weston A. J., Mendez C., Leadlay P. F., Wilkinson B., Salas J. A. Mol. Microbiol. 2004;52:1745–1756. doi: 10.1111/j.1365-2958.2004.04090.x. [DOI] [PubMed] [Google Scholar]
- Lane A. L., Nam S. J., Fukuda T., Yamanaka K., Kauffman C. A., Jensen P. R., Fenical W., Moore B. S. J. Am. Chem. Soc. 2013;135:4171–4174. doi: 10.1021/ja311065v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami K., Liu C., Minami A., Noike M., Isaka T., Fueki S., Shichijo Y., Toshima H., Gomi K., Dairi T., Oikawa H. J. Am. Chem. Soc. 2013;135:1260–1263. doi: 10.1021/ja3116636. [DOI] [PubMed] [Google Scholar]
- Kieser T., Bibb M. J., Buttner M. J., Chater K. F. and Hopwood D. A., Practical Streptomyces Genetics, Norwich, 2000. [Google Scholar]
- Hagen W. R., Biomolecular EPR Spectroscopy, CRC Press, Taylor and Francis Group, 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.