Abstract
Background
Expression of the fructose transporter gene SLC2A5 and histone acetylation in the transcribed region are induced by differentiation associated-signals such as glucocorticoids and p44/42 mitogen-activated protein kinase (MAPK) inhibition in small intestinal Caco-2 cells.
Methods
We co-treated with glucocorticoid receptor agonist dexamethasone (Dex) and p44/42 MAPK inhibitor PD98059 (PD) in Caco-2 cells with or without Brd4 small hairpin (sh) RNA expression vector, and the cells were analyzed by qRT-PCR and chromatin immunoprecipitation assays. The small intestine of wild-type mice and Brd4+/− mice during weaning period were analyzed by qRT-PCR.
Results
Co-treatment with Dex and PD increased binding of the bromodomain-containing protein-4 (Brd4)–positive transcriptional elongation factor-b (P-TEFb)–RNA polymerase II complex to acetylated histones in the transcribed region of SLC2A5. Brd4-protein depletion by shRNA revealed that the association of these proteins on the transcribed region of SLC2A5 promoted gene expression in a Brd4-dependent manner. Expression of small-intestine Slc2a5, but not another intestinal gene sucrase-isomaltase, during weaning period, was significantly lower in Brd4+/− mice compared with wild-type mice.
Conclusions
Brd4-P-TEFb plays a crucial role in differentiation-associated transcription of SLC2A5 gene in intestinal Caco-2 cells and in the small intestine of mice during weaning period.
General significance
Histone acetylation and the transcription elongation factor Brd4 are important for SLC2A5 expression in the small intestine.
Keywords: Histone acetylation, Fructose transporter, P44/42 mapk, Glucocorticoid hormone, Small intestine
Highlights
-
•
Brd4 regulates expression of SLC2A5 in human intestinal Caco-2 cells.
-
•
Brd4–P-TEFb and acetylated histones bind transcribed region of SLC2A5.
-
•
Brd4 regulates jejunal Slc2a5 expression during weaning period of mice.
1. Introduction
Stem cells in the crypt of the small intestine rapidly differentiates to absorptive cells in the villus, and start to express many genes related digestion and absorption of nutrients. Indeed, previous studies, including our own, have shown that the expression of the hexose transporter, SLC2A5, generally called as glucose transporter 5 (GLUT5), which is involved in fructose absorption from the lumen, is induced during the transition from the crypt to the villus [1]. It is known that glucocorticoids and inactivation of p44/42 mitogen-activated protein kinase (MAPK), also known as extracellular signal-regulated kinase (ERK)1/2, play important roles in intestinal differentiation and gene expression [2]. Expression of the Slc2a5 gene was increased when serum concentrations of corticosterone were elevated during weaning in rats [3]. Injection of the glucocorticoid receptor agonist dexamethasone (Dex) into suckling rats induced the expression of small-intestine Slc2a5 [4]. p44/42 MAPK was inactivated prior to differentiation of the fetal small intestine in rats and in human intestinal Caco-2 cells [5]. Furthermore, endogenous levels of epidermal growth factor, which activates p44/42 MAPK, were decreased in the rat small intestine during weaning, because it is provided via the mother's milk [3]. Inhibition of p44/42 MAPK was recently shown to enhance glucocorticoid-mediated SLC2A5 expression in human intestinal Caco-2 cells [6]. These evidences suggest that glucocorticoids and p44/42 MAPK inactivation could coordinately enhance SLC2A5 expression in small-intestine cells during the differentiation.
Previous studies showed that hormone-induced gene expression, which frequently occurs in differentiating cells, is mediated by epigenetic memories which are acquired modifications on the chromatin such as modifications of the histone tail, including acetylation, methylation and phosphorylation, and the DNA methylation [7]. Acetylation of histones H3 and H4 is associated with the euchromatin region and transactivation [8]. We previously demonstrated that co-treatment of Caco-2 cells with Dex and PD98059 (PD), which inhibits p44/42 MAPK activation, enhanced the acetylation of histones H3 and H4 around SLC2A5, particularly in the transcribed region of the gene [9]. These results suggest that, under these conditions, induction of SLC2A5 expression is regulated not only by activation of glucocorticoid receptors (GRs), but also by enhancing the histone acetylation on SLC2A5.
Acetylation of histones H3 and H4 in the promoter region of genes facilitates binding of the transcriptional machinery, including transcription factors, co-activators, the SWItch/Sucrose NonFermentable (SWI/SNF) complex, and RNA polymerase II [10]. However, the significance of histone acetylation in transcribed regions remains poorly understood. Bromodomain-containing protein-4 (Brd4), which binds acetylated histones, was recently shown to enhance gene expression by recruiting the mRNA transcription elongation complex to acetylated histones in transcribed regions of genes [11], [12]. This complex, known as positive transcription elongation factor-b (P-TEFb), is a heterodimer of cyclin T1–Cdk9. These results indicate that Brd4 regulates transcription elongation by recruiting P-TEFb to the transcribed region of genes. Brd4 is the only bromodomain-containing protein known to recruit P-TEFb to acetylated histones in transcribed regions. Brd4 is also highly expressed in the small intestine [13], and histone acetylation in the transcribed region of SLC2A5 was enhanced by co-treatment with PD and Dex in Caco-2 cells [9]. However, whether functional genes, including SLC2A5, are regulated by Brd4-P–TEFb in differentiated cells has not yet been elucidated.
In this study, we examined the possible function of Brd4 downstream of glucocorticoids and p44/42 MAPK inhibition in regulating SLC2A5 expression in Caco-2 cells. We also investigated the role of Brd4 in SLC2A5 induction in the small intestine during the suckling–weaning transition using Brd4 heterogeneous gene targeting in mice. Our results in current study suggest that epigenetic regulation via histone acetylation and the Brd4 play vital roles in induction of SLC2A5 expression during the intestinal differentiation.
2. Materials and methods
2.1. Cell culture
Caco-2 cells (American Type Culture Collection, Rockville, MD, USA) were seeded at a density of 0.6×104 cells/cm2 in 10-cm culture collagen plates (Iwaki, Tokyo, Japan) in Dulbecco's Modified Eagle's Medium (DMEM) containing 10% fetal calf serum (FCS), 1% non-essential amino acids (Invitrogen, Carlsbad, CA, USA), 20 mM HEPES (pH 7.4), 1× antibiotic–antimycotic mixed stock solution (Nakaraitesk, Kyoto, Japan), and 2 mM l-glutamate (Invitrogen) at 37 °C in a humidified atmosphere of 5% CO2.
Control short hairpin (sh) RNA- or Brd4-shRNA-expressing Caco-2 cells were constructed by inserting control or Brd4 shRNA into the pSUPERRNAi vector (Oligoengine, Seattle, WA, USA) [14]. The Brd4 shRNA sequence was: 5′-GATCCCCGAAAAGAGGAAGTGGAAGAGATTCAAGAGATCTCTTCCACTTCCTCTTTCTTTTTA-3′, and the control shRNA sequence was: 5′-GATCCCCATGCACGTGCACATATCCCTTCAAGAGAGGGATATGTGCACGTGCATTTTTTGGAAA-3′. These constructs were separately transfected with the plasmid vectors pGag-pol and pAmpho into HEK293T packaging cells, and the supernatants were collected as virus-containing medium 2 days after transfection. Cells were transfected with virus-containing medium mixed with 6 μg polybrene by centrifugation (1000g) for 2 h. Transfected cells were selected with puromycin (Sigma Aldrich, St. Louis, MO, USA) for 7 days and cultured in the same conditions described above. After 2 days, cells had reached approximately 70% confluence and normal Caco-2 and control/Brd4-shRNA-expressing cells were continuously cultured in DMEM containing 10% FCS stripped of glucocorticoids by treatment with charcoal/dextran, with 1 µM Dex (glucocorticoid receptor agonist) and 50 µM PD (p44/42 MAPK kinase inhibitor), or vehicle (DMSO) alone for 48 h.
2.2. Targeted disruption of Brd4 gene in mice
Brd4 heterozygous allele (Brd4+/−) mice were generated by our group in an NIH animal facility. The mice were derived from C57BL/6J mice and bred with wild-type mice purchased from Japan Oriental Yeast Co. Ltd. (Shizuoka, Japan). Brd4+/− mice were divided into four groups analyzed at the following times after birth: 10 days (n=20 mice; control, 10; Brd4+/–, 10), 14 days (n=16 mice; control, 11; Brd4+/–, 5), 21 days (n=14 mice; control, 7; Brd4+/–, 7), and 26 days (n=17 mice; control, 9; Brd4+/–, 8). Animals were genotyped by polymerase chain reaction (PCR) using the following primers: WT forward: 5′-GGACTAGAAACCTCCCAAATGTCTACAA-3′; neo forward: 5′-TGAAGAGCTTGGCGGCGAATGGG-3′; both reverse: 5′-CCTGTGTGCACTTGCTCCCGAGGAGAGA-3′. The PCR conditions for WT were 94 °C for 1 min, and 34 cycles of 94 °C for 20 s, 60 °C for 20 s, and 72 °C for 2 min, and 72 °C for 7 min. The real-time reverse transcription (RT)-PCR conditions for neo were 95 °C for 5 min, and 30 cycles of 95 °C for 10 s, 60 °C for 10 s, and 72 °C for 1 min 30 s, and 95 °C for 10 s, 65 °C for 15 s, and 50 °C for 10 s. Mice were maintained in an air-conditioned room at 23±2 °C and a humidity of 55±5% with a 12-h light–dark cycle (7:00–19:00) at the Japan Oriental Yeast Co. Ltd. Mice were killed by decapitation between 12:00 and 15:00. All experimental procedures were approved by the Institutional Animal Care and Use Committee of the University of Shizuoka.
2.3. RNA analysis
Total RNA was subjected to RT using Superscript III reverse transcriptase (Invitrogen). Test genes (Caco-2 cells: SLC2A5; mice: Slc2a5, Si, and Brd4) and housekeeping genes (Caco-2 cells: β-actin [ACTB], mice: 16 S rRNA) were amplified by real-time RT-PCR using a Light Cycler System (Roche Molecular Biochemicals, Bavaria, Germany) and SYBR Green I (Takara Bio, Shiga, Japan). The cycle threshold (CT) values detected by real-time RT-PCR were converted into signal intensities using the delta-delta method [15]. The sequences of the primers for real-time RT-PCR are listed in Supplemental Table 1.
2.4. Immunoblotting
Total cell proteins were extracted in RIPA buffer (1% Nonidet P-40, 0.1% sodium dodecyl sulfate [SDS], 20 mM Tris–HCl pH 8.0, 5 mM ethylenediamine tetraacetic acid, 150 mM NaCl, protease inhibitor tablet (Complete Mini, Roche Molecular Biochemicals)/10 ml, and phosphatase inhibitors [1 mM NaMoO4, 50 mM NaF, and 1 mM Na3VO4]). Lysates were centrifuged at 10,000g for 10 min at 4 °C. The protein concentration of the soluble supernatants was determined by the Lowry method, and samples were stored at −20 °C.
Total proteins (60 μg, Fig. 1A; 70 μg, Fig. 2A) were separated by 10% SDS–polyacrylamide gel electrophoresis and transferred to Immobilon membranes (Millipore; Billerica, MA, USA) at 80 V for 120 min in Tris/glycine/methanol transfer buffer. The membranes were blocked for 30 min in 3% skim milk in phosphate-buffered saline (PBS) with 0.05% Tween 20, pH 7.4 (PBS–Tween) at room temperature. The membranes were then incubated in 3% skim milk in PBS–Tween at 4 °C for >7 h with primary antibodies against Brd4 [16], cyclin T1 (Abcam, Cambridge, MA, USA), Cdk9 (Santa Cruz Biotechnology; Santa Cruz, CA, USA), and TFIIB (Santa Cruz Biotechnology). After washing in PBS–Tween, the membranes were incubated with biotin-conjugated anti-rabbit IgG (GE Healthcare, Little Chalfont, UK) in 3% PBS–Tween. The membranes were then washed in PBS–Tween and incubated with horseradish peroxidase-conjugated anti-biotin third antibody (Cell Signaling Technology, MA, USA). Signals were detected by chemiluminescence (ECL Plus, GE Healthcare), according to the manufacturer's instructions.
Fig. 1.
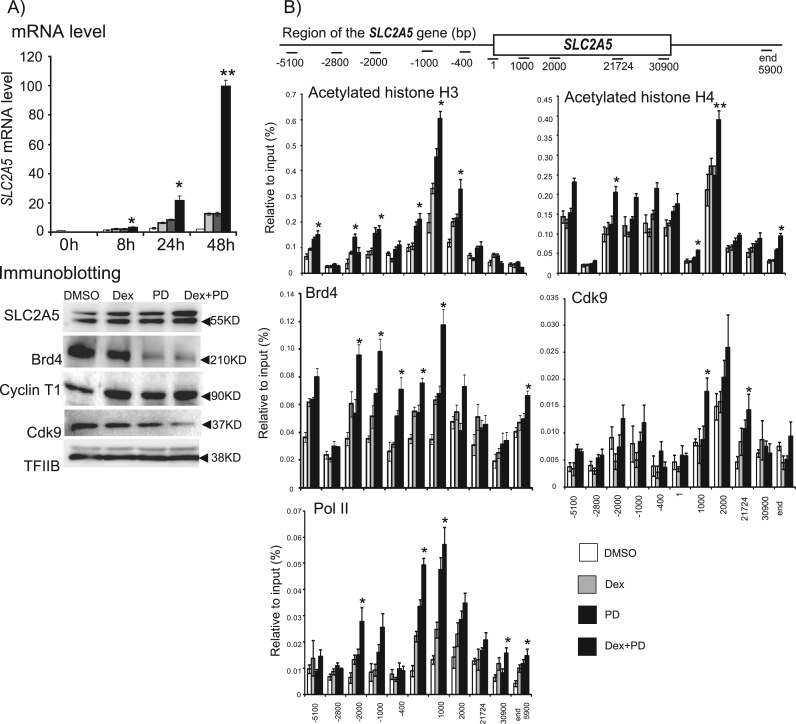
Expression of SLC2A5 in Caco-2 cells treated with Dex and/or PD. (A) SLC2A5 mRNA in cells treated with Dex and/or PD for 8, 24, or 48 h, and protein levels of SLC2A5, Brd4, Cyclin T1, Cdk9, and TFIIB in cells treated with Dex and/or PD for 48 h. (B) ChIP assays for acetylated histone H3 at K9/14, acetylated histone H4 at K5/8/12/16, Brd4, Cdk9, and Pol II around the SLC2A5 gene in cells treated with Dex and/or PD for 48 h. Means±SEM of six (RNA) or five (ChIP assays) experiments are shown. *P <0.05 and **P <0.01 compared with control cells (DMSO).
Fig. 2.
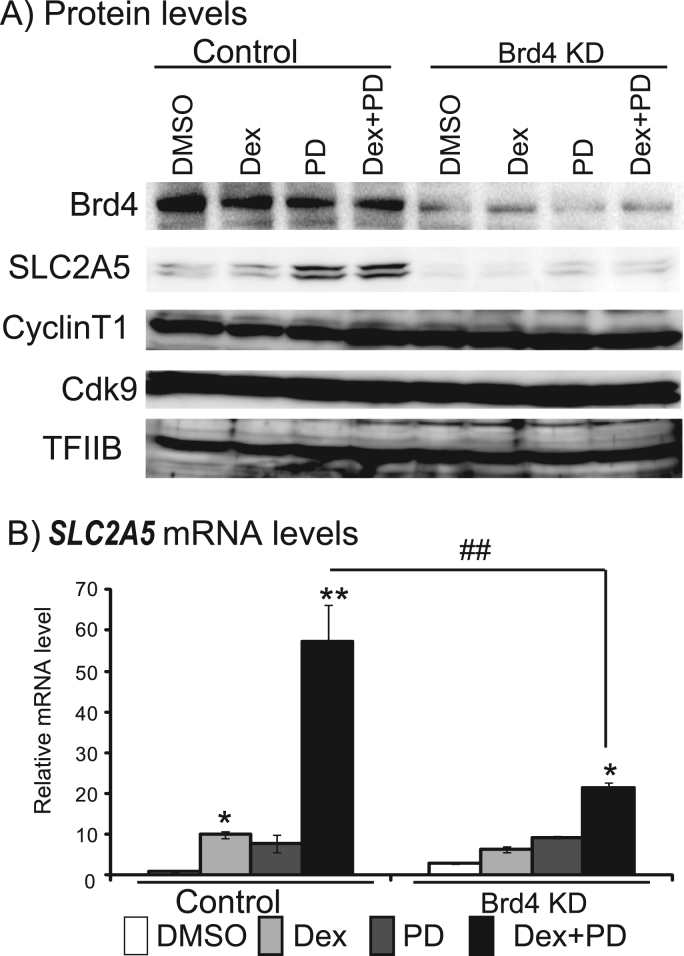
SLC2A5 mRNA expression in Dex and/or PD-treated Brd4-depleted cells. Protein levels of SLC2A5, Brd4, Cyclin T1, Cdk9, and TFIIB (A) and SLC2A5 mRNA (B) in cells treated with Dex and/or PD for 48 h. Means±SEM of six mRNA determinations are shown. *P <0.05 and **P <0.01 compared with control cells (DMSO). ##P <0.01 compared with control cells.
2.5. Chromatin immunoprecipitation (ChIP) assay
Cells were incubated in fixation solution (1% formaldehyde, 4.5 mM HEPES pH 8.0, 9 mM NaCl, 0.09 mM EDTA, 0.04 mM ethylene glycol tetraacetic acid) in 10% FCS/DMEM for 30 min at 37 °C. Reactions were terminated by adding glycine to a final concentration of 150 mM. After washing in fluorescence-activated cell sorting solution (1× PBS, 2% bovine serum, 0.05% NaN3), the samples were sonicated in SDS lysis buffer (50 mM Tris–HCl pH 8.0, 10 mM EDTA pH 8.0, 1% SDS, 0.5 mM phenylmethanesulfonylfluoride) to generate DNA fragments of 200–500 bp. The ChIP assay was performed as described previously [16], using 1 μg antibodies against acetyl histone H3 at K9/14 (Millipore), acetyl histone H4 at K5/8/12/16 (Millipore), Brd4 (custom antibody generated by the Sigma Custom Antibody service using the Brd4C-terminal peptide CFQSDLLSIFEENLF), Cdk9 (Santa Cruz Biotechnology), RNA polymerase II (Pol II) (Covance, Princeton, NJ, USA), or normal rabbit IgG. The precipitated DNA was subjected to real-time PCR using primers corresponding to the indicated sites in the promoter/enhancer and transcribed regions. The CT values of the ChIP signals detected by real-time PCR were converted to percentages of the signal for the input DNA using the delta-delta method [15], with the formula 100×[2(CT input–CT IP sample)]. The primer sequences used in ChIP assays are listed in Supplemental Table 1.
2.6. Statistical analysis
Results were expressed as mean±standard error of the mean (SEM). The significance of differences among groups was determined by Tukey's multiple range test based on one-way analysis of variance (ANOVA) (Fig. 1), two-way ANOVA (Fig. 2, Fig. 3), or Student's t-test (Fig. 4). A P value <0.05 was considered statistically significant.
Fig. 3.
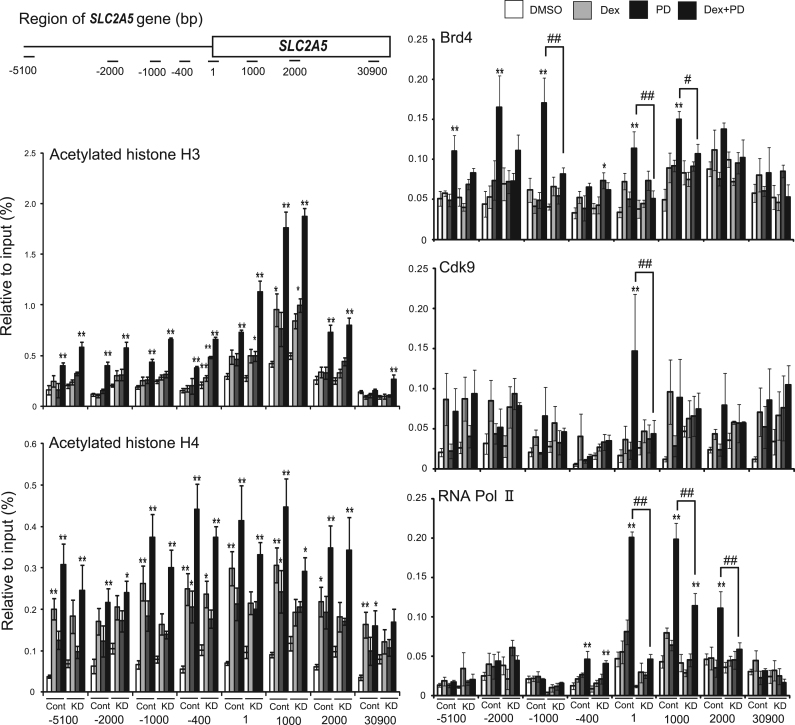
Binding of acetylated histone- and chromatin-remodeling factors around SLC2A5 in Brd4-depleted cells. ChIP assays for acetylated histone H3 at K9/14, acetylated histone H4 at K5/8/12/16, Brd4, Cdk9, and Pol II around the SLC2A5 gene in control-shRNA- and Brd4-shRNA-expressing cells treated with Dex and/or PD for 48 h. Means±SEM of five experiments are shown. *P <0.05 and **P <0.01 compared with control cells (DMSO). #P <0.05 and ##P <0.01 compared with control cells.
Fig. 4.
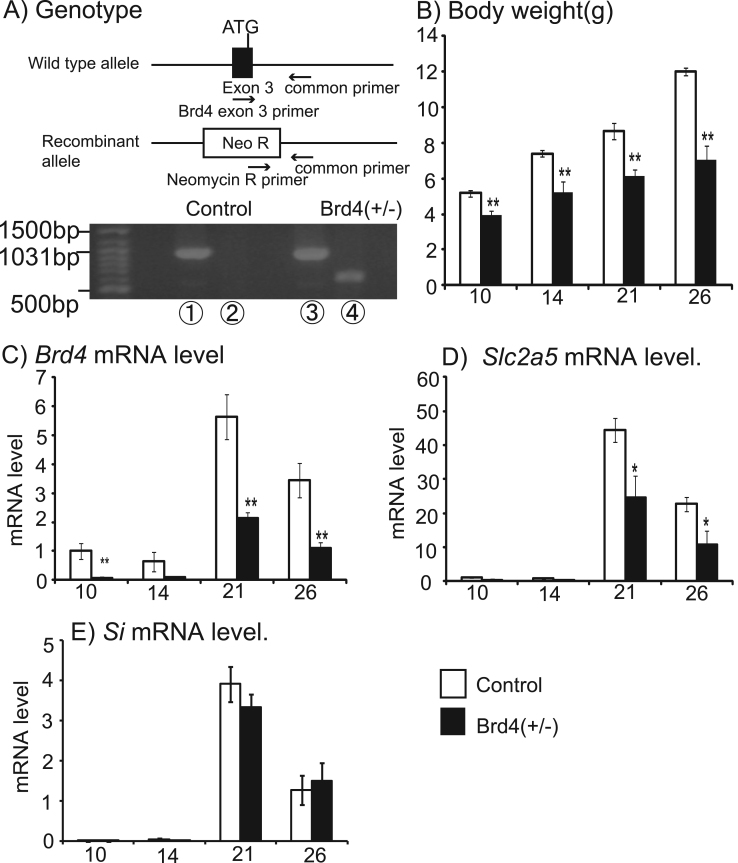
Effects of Brd4 deficiency on mouse small-intestine SLC2A5 expression during the suckling–weaning period. (A) Mice were genotyped by PCR using DNA isolated from the tail. Body weights (B), mRNA levels for Brd4 (C), Slc2a5 (D), and Si (E) in control and Brd4+/− mice at 10, 14, 21, and 26 days after birth. Numbers of X-axis in (B)–(E) indicate days after birth of mice. Means±SEM for between five and 11 samples are shown. *P <0.05 and **P <0.01 compared with control mice.
3. Results
3.1. Co-treatment with Dex and PD enhances SLC2A5 mRNA and binding of acetylated histones and Brd4–P-TEFb–Pol II around the SLC2A5 gene in Caco-2 cells
We performed real-time RT-PCR and ChIP assays in Caco-2 cells treated with 1 µM Dex, an agonist for glucocorticoid receptor, with or without 50 µM PD, a p44/42 MAPK inhibitor. SLC2A5 expression was significantly increased by co-treatment with Dex and PD for 8, 24, and 48 h, and SLC2A5 protein levels were significantly elevated by co-treatment with Dex and PD for 48 h. Brd4, Cyclin T1, Cdk9, and TFIIB were expressed in Caco-2 cells treated with DMSO, Dex, PD, or Dex and PD. However, Brd4 protein levels were decreased by PD or co-treatment with Dex plus PD (Fig. 1A).
All ChIP signals for IgG were <0.02% relative to input signals. ChIP signals from upstream/transcribed regions associated with acetylated H3 (K9/14) were markedly enhanced by co-treatment with Dex and PD for 48 h, particularly the transcribed regions of SLC2A5 at −5100 bp and from −1000 bp to 2000 bp. Signals throughout the upstream/transcribed region of SLC2A5 associated with acetylated histone H4 (K5/8/12/16) were slightly enhanced by co-treatment with Dex and PD for 48 h, and signals from 2000 bp of SLC2A5 were significantly higher. Co-treatment also notably increased the ChIP signals associated with Brd4 on the promoter/enhancer region and the transcribed region close to the initiation site of SLC2A5 from −2000 bp to 1000 bp. Co-treatment with Dex and PD for 48 h strongly increased the ChIP signals associated with Cdk9 on the transcribed region close to the initiation site of SLC2A5 at 1000 bp and 22,000 bp. ChIP signals were also significantly increased in association with Pol II on the promoter/enhancer region and the transcribed region close to the initiation site of SLC2A5 at −2000 bp, 1 bp, and 1000 bp (Fig. 1B).
3.2. Effects of Brd4 depletion on SLC2A5 expression and binding of acetylated histones and Brd4–P-TEFb–Pol II around the SLC2A5 gene in Caco-2 cells co-treated with Dex and PD
We generated Caco-2 cell lines stably expressing Brd4 shRNA to determine if Brd4 was necessary for the induction of SLC2A5 in Caco-2 cells co-treated with Dex and PD. Brd4 protein levels were reduced in these cells compared with controls. Levels of Cyclin T1, Cdk9, and TFIIB proteins were similar in cells subjected to different treatments in both control- and Brd4-shRNA-expressing cells. In contrast, co-treatment with Dex and PD for 48 h increased SLC2A5 levels in control cells, but not in Brd4-shRNA-expressing cells (Fig. 2A). We obtained similar results for SLC2A5 mRNA levels, with a significant difference between co-treated control- and Brd4-shRNA-expressing cells (Fig. 2B).
Acetylation of histone H3 K9/14 on the promoter/enhancer and transcribed regions of SLC2A5 (control: from −5100 bp to 2000 bp; Brd4 shRNA: from −5100 bp to 30,900 bp) increased after co-treatment with Dex and PD for 48 h in both control- and Brd4-shRNA-expressing cells (Fig. 4B). Acetylation of histone H4 K5/8/12/16 on the promoter/enhancer and transcribed regions of SLC2A5 (control: from −5100 bp to 30,900 bp; Brd4 shRNA: from −5100 bp to 2000 bp) also increased after co-treatment with Dex and PD in both cell lines. We obtained similar results in terms of the associations of Brd4, Cdk9, and Pol II with SLC2A5 in control cells and in WT cells in Fig. 1. However, this association was significantly reduced in Brd4-depleted cells (Fig. 3).
3.3. Effects of Brd4 depletion on small-intestine Slc2a5 gene expression during postnatal development in mice
We determined if Brd4 regulated the expression of Slc2a5 in the mouse small intestine by real-time RT-PCR using total RNA extracted from WT or Brd4+/− mice at 10, 14, 21, and 26 days after birth. The genetic background of Brd4 heterozygous (Brd4+/−) mice is shown in Fig. 4. This covered the suckling–weaning transition period when serum glucocorticoid levels increase and p44/42 MAPK is inactivated in rodents. Body weight was lower in Brd4+/− mice compared with WT mice at each day after birth. We confirmed that Brd4 expression was significantly lower in Brd4+/− mice compared with WT mice. Slc2a5, but not Si expression was also significantly lower in Brd4+/− mice compared with WT mice at 21 and 26 days after birth (Fig. 4.).
4. Discussion
In this study, we demonstrated that co-treatment with Dex and PD induced SLC2A5 expression as well as acetylation of histones around the SLC2A5 gene in Caco-2 cells. Furthermore, co-treatment with Dex and PD also promoted the association of Brd4 around SLC2A5. Notably, although Brd4 protein levels were reduced by PD and co-treatment with PD and Dex, Brd4 binding around SLC2A5 was strongly enhanced by co-treatment with PD and Dex. Furthermore, we demonstrated that co-treatment with Dex and PD promoted the association of P-TEFb and Pol II on the transcribed region close to the transcription initiation site of SLC2A5 in Caco-2 cells. These results were confirmed by RNA interference, which showed that these associations and the expression of SLC2A5 were Brd4-dependent. Recent studies have also suggested that Brd4 recruits the P-TEFb mRNA transcription elongation factor (Cyclin T1–Cdk9 complex), which regulates mRNA transcription elongation by regulating Pol II activity [11]. Our results thus suggest that binding of Brd4 to the transcribed region of SLC2A5 may enhance gene expression by recruiting P-TEFb and Pol II to the acetylated histones in the transcribed region, thus enhancing mRNA transcription elongation.
SLC2A5 is induced in the small intestine of rodents during the suckling–weaning transition period when serum glucocorticoid levels increase and p44/42 MAPK is inactivated. Interestingly, we found that small-intestine expression of Slc2a5 was significantly lower in Brd4+/− mice compared with WT mice at 21 and 26 days after birth, suggesting that Brd4 regulates the induction of small-intestine Slc2a5 expression during the suckling–weaning transition in mice. It should be noted that body weight was lower in Brd4+/− mice compared with WT mice, and the lower Slc2a5 mRNA levels in Brd4+/− mice may thus be associated with delayed growth. However, mRNA levels of another intestinal gene, Si, were similar in Brd4+/− and WT mice, implying that the decreased Slc2a5 mRNA levels in Brd4+/− mice were likely related to reduction of Brd4 as a result of gene targeting, rather than delayed growth. Further studies are needed to determine if acetylated histone-Brd4–P-TEFb is recruited to the transcribed region of rat jejunal Slc2a5 during the suckling–weaning transition.
We previously demonstrated that both GR translocation from the cytosol to the nucleus, and GR binding to the promoter/enhancer region of SLC2A5 were enhanced by co-treatment with Dex and PD in Caco-2 cells [6]. These and the current results suggest that Brd4–P-TEFb may enhance the rate of mRNA synthesis associated with triggering of the transcription initiation reaction by GR binding. Both transcription initiation and transcription elongation may thus be important for mRNA production of functional genes. However, expression of the Si gene during the suckling–weaning transition in mice may not be affected by Brd4 reduction via gene targeting. It is possible that the rates of expression of several genes are controlled by Brd4-associated transcription elongation and/or transcription initiation, while other genes may be controlled by Brd4. This issue should be examined in future studies.
It is still unclear which cascade promotes Brd4–P-TEFb recruitment to the transcribed region of SLC2A5. Because the enhancement of histone acetylation around SLC2A5 by co-treatment with Dex and PD was closely associated with Brd4–P-TEFb binding, co-treatment may enhance histone acetylation, which may in turn promote Brd4–P-TEFb recruitment. However, the specific histone acetyltransferase (HAT) that regulates histone acetylation around SLC2A5 has not yet been identified. GCN5 is a HAT that controls amino acid synthesis and is recruited around the transcription initiation site of intestinal genes such as Si and Slc5a1 in response to a high-carbohydrate diet in rats [17]. Given that GCN5 enhances the mRNA transcriptional elongation step by acetylating histones around the transcribed region [18], it may also contribute to the induction of SLC2A5. A high-carbohydrate diet has also been shown to promote the association of another HAT, CREB-binding protein (CBP), with the Mgam gene, leading to increased Mgam expression in mice [19]. HATs, including GCN5 and CBP, may thus be involved in the observed regulation of SLC2A5 expression. However, although many HATs have been identified [20], the specific HAT that is activated by Dex and PD remains unknown and should be investigated in future studies.
In conclusion, we demonstrated that Brd4 enhances the induction of SLC2A5 in human intestinal Caco-2 cells after co-treatment with a glucocorticoid hormone agonist and a MAPK inhibitor. This induction is mediated by recruiting P-TEFb to the transcribed region and enhancing mRNA polymerase II transcription elongation. We further demonstrated that Brd4 regulated the induction of Slc2a5 in the small intestine during the suckling–weaning transition in mice. These results suggest that Brd4–P-TEFb plays a crucial role in differentiation-associated transcription of SLC2A5 gene in intestinal Caco-2 cells and in the small intestine of mice during weaning period.
Author contributions
Conceived and designed the experiments: YI TG KM.
Performed the experiments and analyzed the data: YI KM.
Generated the Brd4 heterogeneous mice: AD AN KO.
Wrote the paper: YI TG TK KO KM.
Funding
This work was supported by a Grant-in-Aid for JSPS Researcher Fellows for Young Scientists (23-9920), Grants-in-Aid for Young Scientists (22680054), a Grant-in-Aid for Scientific Research (26282023) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), and a Grant-in-Aid, and for Development of Core Technologies for Innovative Drug Development based upon IT from the Japan Agency for Medical Research and Development (AMED), and the Takeda Science Foundation.
Conflict of interest
The authors declare that they have no conflicts of interest.
Footnotes
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.bbrep.2016.05.016.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
References
- 1.Suzuki T., Mochizuki K., Goda T. Localized expression of genes related to carbohydrate and lipid absorption along the crypt-villus axis of rat jejunum. Biochim Biophys. Acta. 2009;1790:1624–1635. doi: 10.1016/j.bbagen.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 2.McDonald M.C., Henning S.J. Synergistic effects of thyroxine and dexamethasone on enzyme ontogeny in rat small intestine. Pediatr. Res. 1992;32:306–311. doi: 10.1203/00006450-199209000-00012. [DOI] [PubMed] [Google Scholar]
- 3.Koldovsky O. Response of the gastrointestinal tract to premature weaning in experimental animals. Pediatrics. 1985;75:199–206. [PubMed] [Google Scholar]
- 4.Douard V., Cui X.L., Soteropoulos P., Ferraris R.P. Dexamethasone sensitizes the neonatal intestine to fructose-induction of GLUT5 transport function. Endocrinology. 2008;149:409–423. doi: 10.1210/en.2007-0906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gauthier R., Harnois C., Drolet J.F., Reed J.C., Vezina A., Vachon P.H. Human intestinal epithelial cell survival: differentiation state-specific control mechanisms. Am. J. Physiol. Cell Physiol. 2001;280:C1540–C1554. doi: 10.1152/ajpcell.2001.280.6.C1540. [DOI] [PubMed] [Google Scholar]
- 6.Takabe S., Mochizuki K., Goda T. De-phosphorylation of GR at Ser203 in nuclei associates with GR nuclear translocation and GLUT5 gene expression in Caco-2 cells. Arch. Biochem. Biophys. 2008;475:1–6. doi: 10.1016/j.abb.2008.03.036. [DOI] [PubMed] [Google Scholar]
- 7.Rice J.C., Briggs S.D., Ueberheide B., Barber C.M., Shabanowitz J., Hunt D.F., Shinkai Y., Allis C.D. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell. 2003;12:1591–1598. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- 8.Yan C., Boyd D.D. Histone H3 acetylation and H3 K4 methylation define distinct chromatin regions permissive for transgene expression. Mol. Cell Biol. 2006;26:6357–6371. doi: 10.1128/MCB.00311-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inamochi Y., Mochizuki K., Osaki A., Ishii T., Nakayama T., Goda T. Histone H3 methylation at lysine 4 on the SLC2A5 gene in intestinal Caco-2 cells is involved in SLC2A5 expression. Biochem. Biophys. Res. Commun. 2010;392:16–21. doi: 10.1016/j.bbrc.2009.12.136. [DOI] [PubMed] [Google Scholar]
- 10.Yang X.J. Lysine acetylation and the bromodomain: a new partnership for signaling. Bioessays. 2004;26:1076–1087. doi: 10.1002/bies.20104. [DOI] [PubMed] [Google Scholar]
- 11.Jang M.K., Mochizuki K., Zhou M., Jeong H.S., Brady J.N., Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 12.Mochizuki K., Nishiyama A., Jang M.K., Dey A., Ghosh A., Tamura T., Natsume H., Yao H., Ozato K. The bromodomain protein Brd4 stimulates G1 gene transcription and promotes progression to S phase. J. Biol. Chem. 2008;283:9040–9048. doi: 10.1074/jbc.M707603200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dey A., Ellenberg J., Farina A., Coleman A.E., Maruyama T., Sciortino S., Lippincott-Schwartz J., Ozato K. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G(2)-to-M transition. Mol. Cell Biol. 2000;20:6537–6549. doi: 10.1128/mcb.20.17.6537-6549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo J.J., Jang R., Louder A., Cluxton R.J. Acute pancreatitis associated with different combination therapies in patients infected with human immunodeficiency virus. Pharmacotherapy. 2005;25:1044–1054. doi: 10.1592/phco.2005.25.8.1044. [DOI] [PubMed] [Google Scholar]
- 15.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Dey A., Chitsaz F., Abbasi A., Misteli T., Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc. Natl. Acad. Sci. USA. 2003;100:8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inoue S., Mochizuki K., Goda T. Jejunal induction of SI and SGLT1 genes in rats by high-starch/low-fat diet is associated with histone acetylation and binding of GCN5 on the genes. J. Nutr. Sci. Vitaminol. 2011;57:162–169. doi: 10.3177/jnsv.57.162. [DOI] [PubMed] [Google Scholar]
- 18.Johnsson A.E., Wright A.P. The role of specific HAT-HDAC interactions in transcriptional elongation. Cell Cycle. 2010;9:467–471. doi: 10.4161/cc.9.3.10543. [DOI] [PubMed] [Google Scholar]
- 19.Mochizuki K., Honma K., Shimada M., Goda T. The regulation of jejunal induction of the maltase-glucoamylase gene by a high-starch/low-fat diet in mice. Mol. Nutr. Food Res. 2010;54:1445–1451. doi: 10.1002/mnfr.200900467. [DOI] [PubMed] [Google Scholar]
- 20.Marmorstein R., Roth S.Y. Histone acetyltransferases: function, structure, and catalysis. Curr. Opin. Genet. Dev. 2001;11:155–161. doi: 10.1016/s0959-437x(00)00173-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material