SUMMARY
Reverse phase protein arrays (RPPAs), also called reverse phase lysate arrays (RPLAs), involve immobilizing cell or tissue lysates, in small spots, onto solid supports which are then probed with primary antibodies specific for proteins or post-translational modifications of interest. RPPA assays are well suited for large-scale, high-throughput measurement of protein and PTM levels in cells and tissues. RPPAs are affordable and highly multiplexable, as a large number of arrays can readily be produced in parallel and then probed separately with distinct primary antibodies. This article describes a procedure for treating cells and preparing cell lysates, as well as a procedure for generating RPPAs using these lysates. A method for probing, imaging, and analyzing RPPAs is also described. These procedures are readily adaptable to a wide range of studies of cell signaling in response to drugs and other perturbations.
Key Terms: reverse phase protein arrays, antibodies, fluorescence, melanoma, cell signaling, BRAF
INTRODUCTION
Quantitative protein measurements are fundamental to all areas of biology. The analysis of proteins in cells and tissues involves determination of both total levels of proteins and the post-translational modification (PTM) states of proteins, such as phosphorylation on specific amino acid residues. An important application of quantitative protein measurements is the characterization of the response of cellular signaling networks to external perturbations such as nutrients and growth factors. System-wide studies of these cellular response networks are becoming increasingly important in understanding human diseases such as cancer and inflammatory disorders. Since these studies can involve the collection of thousands of data points resulting from combinations of cell lines, perturbations, and time points, it is frequently necessary to measure protein levels in an automated high-throughput format (Albeck et al., 2006).
Large scale protein profiling experiments are also important in understanding the mechanism of action and basis for clinical response of the drugs used to treat disease. For example, the response of tumors to kinase inhibitors involves variations or adaptations in multiple signaling pathways that frequently cause primary or acquired resistance to drug treatment (Zhang, Yang, & Gray, 2009). Methods for profiling these adaptations by quantifying protein and PTMs are important in understanding the basis for resistance and identifying possible targets for co-drugging. Furthermore many drugs, especially kinase inhibitors, exhibit significant polypharmacology and affect proteins other than their principal target (Knight, Lin, & Shokat, 2010). Protein profiling methods allow the identification of pathways affected by these off-target activities, which can guide the development of co-drugging strategies and the repurposing of drugs for new therapeutic indications. In addition to existing drugs, multiplex protein profiling methods are useful in characterizing the cellular activities and specificity of newly discovered lead and tool compounds.
A number of technologies have been developed for collecting quantitative protein measurements in a high-throughput format. These include high-throughout Western blotting, ELISAs, high-throughput mass spectrometry, Luminex xMAP assays, various label-free technologies, and protein microarrays (Wolf-Yadlin, Sevecka, & MacBeath, 2009). The three major classes of protein microarray are functional arrays, forward phase or antibody arrays, and lysate or reverse phase protein arrays (RPPAs). Functional arrays involve immobilization of peptides or whole proteins and are used for applications such as discovery of protein-protein interactions (Stiffler, Grantcharova, Sevecka, & MacBeath, 2006). In forward phase or antibody arrays, specific antibodies are immobilized on a surface and capture specific antigens from a cell lysate or biological fluid (Nielsen, Cardone, Sinskey, MacBeath, & Sorger, 2003). The bound antigens are then quantified by either direct labeling of analytes or by a labeled detection antibody that recognizes a separate epitope from the capture antibody. Reverse phase protein arrays (RPPAs), or reverse phase lysate arrays (RPLAs), involve immobilizing cell or tissue lysates onto solid supports, which are then probed with primary antibodies specific for proteins or PTMs of interest (Luckert et al., 2012). These primary antibodies can be either directly labeled or probed with a labeled secondary antibody, with chemiluminescent, colorimetric, or fluorescent readouts used to quantify the proteins of interest.
RPPA assays are well suited for large-scale, high-throughput format measurement of protein and PTM levels in cells and tissues. The technology can readily accommodate large number of samples, as up to several thousand individual lysate samples can be printed in the area of a standard 75 by 25 mm slide. The process of generating and analyzing RPPAs can be automated to a significant extent, using robotic instrumentation for preparing and spotting samples and specialized software for imaging and extracting data from the arrays. RPPA is also a highly multiplexable technology, as a large number of arrays can readily be produced in parallel and probed with distinct primary antibodies. Production of each array requires only a minute amount of lysate, with as little as 5 µl of 1 mg/ml solution of lysate being sufficient to print hundreds of arrays (Luckert et al., 2012). Thus, the RPPA technology is compatible with small initial sample sizes, and lysates for arraying can readily be produced by growing and treating cells in multiwell plates. The denaturing conditions used in RPPA lysate buffers allow detection of antigens that may be masked in other types of assay. Furthermore, lysates prepared for an RPPA experiment can be stored long term at −80°C for future use in validating the earlier findings or testing new antibodies. A final advantage of RPPA methods is their affordability (particularly for large scale experiments) as cost per data point is significantly lower than other technologies such as bead-based sandwich assays.
The major disadvantage of RPPAs is their dependence on antibodies with very high selectivity. Unlike Western blots, in RPPA assays there is no separation of the protein of interest from off-target proteins by molecular weight prior to detection. While sandwich-style immunoassays such as the Luminex xMAP technology achieve high specificity through the use of two antibodies that recognize distinct nonoverlapping epitopes on the target antigen, RPPA assays use a single antibody per analyte and are more susceptible to high background caused by nonspecific antibody binding. The availability of high quality primary antibodies is thus the major limiting factor in data quality and the number of targets that can be measured in RPPA experiments. A common strategy is to use RPPA in a large scale screen combining multiple cell lines, perturbations (such as drug or growth factors), and time points. For each data point in this matrix, RPPAs enable collection of a large number of measurements of levels of proteins and PTM states. After analysis of this dataset, lower-throughput and/or more expensive methods such as quantitative Western blotting and high-content microscopy can be used to validate the findings of the RPPA assays.
STRATEGIC PLANNING
Scale of the Experiment
The first consideration in designing an RPPA study is the scale of the experiment. RPPAs can be produced on scales ranging from fewer than a hundred to thousands of spots per array. In order to obtain data of the highest possible quality, it is recommended to design experiments so that each data point has multiple biological replicates (corresponding to unique lysate samples) and technical replicates (corresponding to spots printed on the same or different arrays from the same lysate sample). Depending on the size of the experiment, slides or other solid supports can be used with multiple arraying surfaces (each which can be probed with a distinct primary antibody) or a single arraying surface. In designing a study it will be important to make sure that the desired number of spots (incorporating sufficient biological and technical replicates) can fit in the arraying surface that will be used.
Antibodies
A second component of RPPA study design is the selection of antibodies to be used. A relatively small subset of all antibodies has sufficient selectivity to work in this assay format, due to the problem of high background caused by off-target antigen binding. Several efforts have screened large collections of antibodies to identify those that can perform well in RPPA experiments (Hennessy et al., 2010; Sevecka, Wolf-Yadlin, & MacBeath, 2011). An approach used in these studies is to generate RPPAs containing samples from multiple “biological contexts”, representing specific combinations of cell types and treatment conditions. For those antibodies that give a significant signal in one or more contexts using RPPA, quantitative Western blotting is used to confirm antibody performance. An antibody is considered to be validated if the data from the RPPA measurements is in agreement with the Western blotting data (defined as having a Pearson correlation coefficient of greater than 0.75; Sevecka et al., 2011). For this analysis, the signal for two or more replicate array spots is averaged and compared to the signal in a western blot band corresponding to the protein of interest. The antibodies that have been validated in these studies include a large number of proteins and PTM states that include growth factor receptors, transcription factors, cell cycle regulatory proteins, and components of the major signaling networks such as the MAPK and PI3K/AKT/mTOR pathways.
Choice of Equipment
Additional considerations in the planning of RPPA experiments are the instruments used in producing and imaging arrays. When choosing an arraying method one option is a contact printer, in which pins are dipped into samples and transfer a defined volume by directly touching the arraying surface. Examples of contact printers that have been successfully used for RPPA production include the Aushon 2470, theSpotBot and NanoPrint (ArrayIT Corporation), the OmniGrid and MicroGrid (Genomics Solutions), and the Q-array (Genetix). The other main type of arraying instrument is a non-contact printer, which may use syringes or piezoelectric technology: examples of these that have been used for generation of RPPAs include the NanoPlotter (GeSiM) and sciFLEXARRAYER (Scienion Ag). In general, contact printers are faster, less expensive, and simpler in design than non-contact arrayers. In choosing the instrument to be used in imaging arrays, the most important consideration is the detection method to be used whether it is luminescence, fluorescence, or colorimetric detection. For fluorescence detection, it is important that the array scanner be capable of measuring signal at the desired excitation and emission wavelengths with adequate sensitivity.
Detection Method
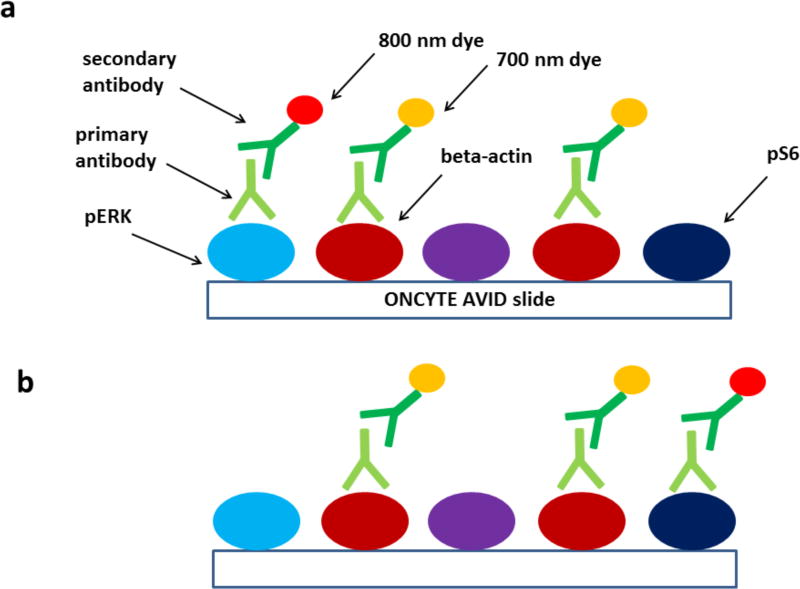
A detection method with several key advantages is the use of dyes that fluoresce in the near-IR region of the spectrum (at 700 nm or 800 nm). These labels can provide higher signal-to-noise ratios than shorter-wavelength visible fluorescent dyes as they exhibit less light scattering, lower signal loss due to absorbance, and reduced autofluorescence from proteins, lysate buffer components, and array materials. Instruments that can measure fluorescence at the near-IR wavelengths and have sufficient resolution to image RPPAs include the Odyssey (LI-COR) and the InnoScan 710-IR (Innopsys). A further advantage of using far-red fluorophores is that the measurement of fluorescence at 700 and 800 nm allows the simultaneous detection of the protein or PTM state of interest and a second signal (typically a constitutively expressed housekeeping protein) that can be used to normalize the signal of interest to total protein content in the lysate. Figure 1 shows a diagram of this detection scheme in RPPA experiments.
Figure 1. Principle of RPPA experiments.
a. Simplified diagram of the detection scheme in a typical RPPA assay. A lysate deposited slide coated with a nitrocellulose membrane (ONCYTE AVID) contains a complex mixture of denatured proteins including phosphorylated ERK (light blue), phosphorylated S6 (dark blue), and beta-actin (dark red). The array is probed with primary antibodies (light green) specific for phosphorylated ERK and beta-actin but that do not bind to phosphorylated S6 or off-target proteins (purple). Following primary antibody incubation the array is probed with secondary antibodies conjugated to near-IR fluorescent dyes that emit near 700 nm (orange) or 800 nm (red).
b. A view of an identical array as in panel a, but probed with primary antibodies specific for phosphorylated S6 and beta-actin.
The protocols in this unit describe a RPPA study designed to characterize alterations in cell signaling networks in melanoma during treatment with BRAF kinase inhibitors. At least 50% of human melanomas contain an activating mutation in BRAF (most frequently V600E) that drives constitutively activated proliferation and survival signaling. The development of small molecule inhibitors that specifically inhibit the mutated form of BRAF such as PLX-4032 (vemurafenib) has shown great potential in treatment of melanoma, with most patients exhibiting rapid initial tumor regression (Bollag et al., 2012). However, the development of lethal drug resistance in the majority of patients has posed a significant challenge for this type of therapy (Flaherty, 2010). A comprehensive profile of the effect of BRAF inhibitors on the state of cell signaling pathways is necessary to understand the molecular basis for resistance and develop co-drugging strategies that can overcome resistance.
In this study, a panel of BRAF mutant melanoma cell lines was treated with a panel of BRAF inhibitors: cells were treated with 8 different doses of each inhibitor and analyzed at 5 time points post-treatment (1, 5, 10, 24, and 48 hours). For each cell line, arrays were printed using 4 biological replicates and 2 technical replicates for each drug/concentration/time point combination, and each probed with a different primary antibody for a specific protein or PTM state of a protein. The arrays in this study were generated using a contact printing method on an Aushon 2470 arrayer, and imaged using near-IR wavelengths on an InnoScan 710-IR slide scanner. To illustrate the expected results of RPPA experiments, representative data is shown in this unit for the response of the BRAF V600E melanoma line RVH-421 to increasing doses of PLX-4032. This data includes measurements of phosphorylated ERK (which is downstream of BRAF and MEK in the MAPK signaling cascade), and phosphorylated ribosomal protein S6 (which is activated by both the MAPK pathway and parallel signaling pathways). Basic Protocol 1 describes a procedure for treating cells and preparing cell lysates, and Basic Protocol 2 describes a procedure for generating RPPAs using these lysates. In Basic Protocol 3 a method for probing, imaging, and analyzing RPPAs is described. We expect that the procedures described in this unit will be readily adaptable to a wide range of studies of cell signaling in response to drugs and other perturbations.
BASIC PROTOCOL 1: GENERATION OF LYSATES FOR REVERSE PHASE PROTEIN ARRAYS
This protocol describes a method for preparation of lysates from cells treated with drugs or tool compounds that is suitable for preparation of RPPAs and was used in a study of BRAF kinase inhibitors in human melanoma cell lines. The protocol was used for dose-response measurements in BRAF V600E mutant cell lines. Cells were treated with compounds in 96-well tissue culture plates, with each well used to produce a unique lysate sample for arraying. For each compound, quadruplet samples of 8 concentrations were collected at 5 time points post-treatment (1, 5, 10, 24, and 48 hours), along with 16 DMSO-treated control wells at each time point and 96 pre-treatment control wells for a total of 1056 unique biological samples.
Materials
Dimethyl sulfoxide (DMSO)
Compounds being studied: e.g., PLX-4032
Melanoma cells grown in dishes: e.g., RVH-421
Trypsin/EDTA
RPMI
Fetal bovine serum (FBS)
Penicillin/streptomycin (Invitrogen)
Plasmocin (Invivogen)
Tissue culture hood
Polypropylene 384-well compound storage plates (Thermo Scientific, cat. no. AB-1056)
Pin transfer apparatus (V&P Scientific, http://www.vp-scientific.com) (Rudnicki and Johnston, 2009)
Multichannel pipette
Matrix WellMate (or other automated liquid dispenser for multiwall plates) (Rudnicki and Johnson, 2009) (optional)
WellMate manifold (sterile, optional)
20 °C incubator
Standard tissue culture incubator capable of maintaining an environment of 37°C and 5 CO2
-
1
Before the experiment, prepare source plates of small molecules dissolved in DMSO for the determination of dose-response curves for each compound of interest.
Compound stocks are prepared in polypropylene 384-well storage plates.
The stock plate should contain compounds at 720× the desired final concentration in each well (assuming a pin transfer volume of 250 nl and an assay volume of 180 nl). A number of wells should contain only DMSO as a control. Small molecule stock plates can be stored long-term at −20 C.
Plating of cells
-
2Prepare melanoma cell suspension for plating as follows:
- Trypsinize the contents of one or more dishes of cells by first aspirating the medium from the plates and briefly washing with sterile PBS. After the wash add 2 mL .25% trypsin/EDTA and incubate 5 min at 37 °C. Add 10 ml complete medium (to stop the trypsinization), centrifuge 5 min at 1000 × g, aspirate the supernatant, and resuspend the cell pellet in fresh medium.
- Resuspend the cells at a final density of 111.111 cells/ml in RPMI, containing 10% FBS, 1% penicillin/streptomycin, and 5 µg/mL plasmocin.
-
3
Using a multichannel pipette, add 180 uL of cell suspension to each well of the clear tissue 96-well microplates. Prepare duplicate plates for each time point, and an additional plate for the pre-treatment controls (for 11 plates total). Transfer the plates into the tissue culture incubator and incubate at 37 °C for 24 hr to allow the cells to adhere to the plates.
If other assays will be run in parallel with the RPPAs, it is convenient to seed additional plates for these at the same time. It may be advantageous to use an automated liquid dispenser to prepare plates if the total number is large.
Compound treatment
-
4
If necessary, thaw the small molecule stock plates 1 hr prior to transferring compounds to plates.
-
5
After 24 hrs, remove the plates from the incubator, and pin transfer the compounds to the plate using the pin transfer robot and a 250-nl pin array.
At this point if additional assays will be run in parallel it is convenient to treat these plates with compounds at the same time.
-
6
For the drug treatment time point plates, return the plates to the tissue culture incubator and incubate for 1, 5, 10, 24, and 48 hrs.
-
7
For the untreated plate, aspirate tissue culture media using the manifold. Transfer 100 µl of ice cold PBS to each well using a multichannel pipette, rinse briefly, and aspirate again using the manifold.
-
8
Add 30 µL of lysis buffer equilibrated to RT to each well of the tissue culture plate, and incubate on an orbital shaker for 5 min.
-
9
Optionally, seal plates with opaque foil and store at −80 °C until ready to array samples.
-
10
Proceed to Basic Protocol 2.
BASIC PROTOCOL 2: PRODUCTION OF REVERSE PHASE PROTEIN ARRAYS
This protocol describes a procedure for producing RPPAs from cell lysates generated in Basic Protocol 1. This procedure was used to print a set of replicate nitrocellulose coated slides, each containing an array of 2112 lysate spots generated by printing duplicates of the 1056 unique lysates produced in the previous procedure. The combination of 2 technical replicates and 4 biological replicates yielded 8 array spots for each compound/concentration/time point combination (1600 total), 64 array spots for DMSO controls at each time point (320 total), and 192 array spots corresponding to untreated cells at the start of the experiment. Prior to printing, it is necessary to filter the lysates generated in Basic Protocol 1 to remove nucleic acids and cell debris
Materials
Multichannel pipettor
Centrifuge
96-well filter plates (Pall, cat. no. 5042)
96-well lysate collection plates (Corning, cat. no. 3870)
Polypropylene 384-well lysate storage plates (Thermo Scientific, cat. no. AB-1056)
ONCYTE AVID nitrocellulose coated single-pad slides (Grace Biolabs, cat. no. 305177)
Arraying robot (e.g., Aushon 2470)
Filtration and formatting of cell lysates
-
1
If necessary, thaw the lysate plates generated in Basic Protocol 1.
-
2
For each 96-well lysate plate, prepare a 96-well filtration plate by placing it on top of a 96-well collection plate. Use a small piece of small plastic tape on each side to hold the two plates together
-
3
Using a multichannel pipettor, carefully transfer the 30 uL of lysates from each 96-well source plate into a 96-well filter plate stacked on top of a collection plate.
The lysates will be highly viscous prior to filtration. Thus, if a pipetting robot is used instead of a multichannel pipettor, it will be important to confirm that is capable of handling viscous solutions.
-
4
Spin the stacked filtration/collection plates for 10 min at 2000 g in a centrifuge to filter the lysates.
-
5
Remove the clarified lysates in collection plates from the centrifuge, detaching and discarding the filtration plates.
-
6
Using a multichannel pipettor, transfer the clarified lysates into 384-well Abgene plates. The volume will be reduced after centrifugation and filtration. At this stage, the clarified lysate plates can be stored long-term at −80 °C until arraying.
Printing of arrays
-
7
Prepare the arrayer to be used for printing slides (e.g., Aushon 2470) by adjusting the pin configuration in the print head, emptying the waste container, and filling the wash container with ddH20.
-
8
Set up the desired printing configuration within the arrayer software.
-
7
Transfer the 384-well lysate source plates and ONCYTE AVID slides into the appropriate racks inside the arrayers
-
8
Print the desired number of slides on the arraying robot. For the configuration used in this study on an Aushon 2470, each slide will took approximately 30 minutes to print. If necessary, the printing of slides can be run overnight.
-
9
Transfer the printed arrays to a sealed container and store at room temperature, until ready to proceed with Basic Protocol 3.
Basic Protocol 3: IMMUNOSTAINING, IMAGING, AND ANALYSIS OF REVERSE PHASE PROTEIN ARRAYS
Materials
Phosphate-buffered saline (PBS; see recipe)
PBS-T wash buffer (see recipe)
RPPA array wash buffer (see recipe)
Arrays to be immunostained and imaged (see Basic Protocol 2)
Odyssey Blocking Buffer (LI-COR, cat. no. 927-40010)
Rabbit anti-Erk1/2 phospho-T204/T205 antibody (Cell Signaling Technologies, cat. no. 4370)
Rabbit anti-rpS6 phospho-S235/S236 antibody (Cell Signaling Technologies, cat. no. 4848)
Mouse Anti-beta actin antibody (Sigma, cat. no. A1978)
Goat anti-mouse IgG antibody, DyLight 680 conjugated (Pierce, cat. no. 35518)
Goat anti-rabbit IgG antibody, DyLight 800 conjugated (Pierce, cat. no. 35571)
Perfect Western 6 Chamber Incubation Tray (GenHunter, cat. no. B131)
Orbital shaker
Centrifuge
50 mL Falcon tubes
Forceps
Near-infrared-capable scanner (e.g., InnoScan 710-IR, Innopsys)
Microarray analysis software (e.g., Mapix, Innopsys)
Spreadsheet software (e.g., Microsoft Excel)
Multidimensional data visualization software (e.g., Matlab and DataPflex)
Washing of slides
-
1
Transfer the array slides to the 6 chamber incubation trays, and wash 3 × 5 minutes with 10 mL PBS-T in each chamber using an orbital shaker at room temperature. Remove the buffer between washes by careful decantation from the incubation tray.
-
2
Add 10 mL per slide of RPPA wash buffer to the trays, and incubate on an orbital shaker at room temperature. Wash the slides for a total of 48 hours, with at least 4 changes of RPPA wash buffer during this time.
-
3
After the 48 hours wash the slides 3 × 5 minutes with 10 mL PBS-T at room temperature.
-
4
Carefully transfer the slides to 50 mL Falcon tubes using a forceps. Spin the slides dry in the falcon tubes by centrifugation at 1000 rpm for 5 minutes.
-
5
Remove the slides from the Falcon tubes and briefly air dry on a flat, clean surface. Optionally, at this time scan the slides for residual autofluorescence from the lysate buffer by scanning at 700 nm on an available instrument.
Blocking the arrays
-
6
Transfer the washed slides to 6 chamber incubation trays, and add at least 5 mL of LI-COR Odyssey blocking buffer to each slides.
-
7
Incubate the slides in blocking buffer on an orbital shaker for 1 hour at room temperature.
-
8
Just before the end of the blocking period, prepare appropriate dilutions of primary antibodies in LI-COR Odyssey blocking buffer using 15 mL Falcon tubes, pooling each rabbit primary antibody against a protein or PTM of interest with a 1:1000 dilution of mouse anti-beta actin antibody. Each dilution should be prepared so there is at least 4 mL total volume per slide.
-
9
Carefully remove the blocking buffer by decantation without washing.
Probing arrays with antibodies
-
10
Add primary antibody dilutions to the slide chambers, and incubate overnight on an orbital shaker at 4 °C
-
11
The next day, wash the slides 4 × 5 minutes with 10 mL PBS-T at room temperature
-
12
During the final wash, prepare dilutions of secondary antibodies. Dilute both the DyLight 680 conjugated goat anti-mouse IgG antibody and the DyLight 800 conjugated goat anti-rabbit IgG antibody 1:7500 in LI-COR Odyssey buffer using 15 mL Falcon tubes, preparing at least 4 mL total volume per slide.
-
13
After decanting the final wash add secondary antibody dilutions to slides and incubate for 2 hours on an orbital shaker at room temperature.
-
14
Wash the slides 4 × 5 minutes with 10 mL PBS-T at room temperature
-
15
Briefly rinse the slides 2–3 times with 10 mL each ddH2O to remove residual salt from the PBS-T wash buffer.
-
16
Carefully transfer the slides to 50 mL Falcon tubes using a forceps. Spin the slides dry in the falcon tubes by centrifugation at 1000 rpm for 5 minutes.
-
17
Remove the slides from the Falcon tubes and briefly air dry on a flat, clean surface. Keep the probed slides in a sealed container protected from light until ready to image.
Imaging slides and data analysis
-
18
Scan the slides using a fluorescence-scanning instrument capable of measuring fluorescence at 700 and 800 nm (e.g., an Innopsys InnoScan 710-IR). Figure 2 shows representative images of arrays scanned at these 2 wavelengths.
To obtain optimal signal, it is important to optimize the focus offset position of the scanner, the resolution of the scanning, and the intensity settings for both the 700- and 800-nm channels. The details will depend on the instrument used.
-
19
Using the instrument software, export the raw images of the scanned arrays to a format that is compatible with the image analysis software to be used.
On some scanners (such as the InnoScan 710-R), image analysis is integrated with the instrument software, and can be performed automatically as soon as slides are imaged.
-
20
Using the image analysis software (e.g., Mapix from Innopsys), extract the fluorescence intensity values for the arrays from the raw images.
-
21
Check the results of the analysis to make sure no parts of the arrays are misassigned, and re-do the analysis if necessary.
-
22
Export the data from the image analysis into a format usable by Microsoft Excel or other suitable spreadsheet software.
-
23
For each spot in the array, calculate the normalized signal intensity for the protein or PTM state of interest by dividing the mean spot intensity measured in the 800 nm channel by the mean spot intensity measured in the 700 nm channel (to control for total protein content).
-
24
Plot the data using the spreadsheet software or a software package for multidimensional data visualization (e.g., DataPFlex).
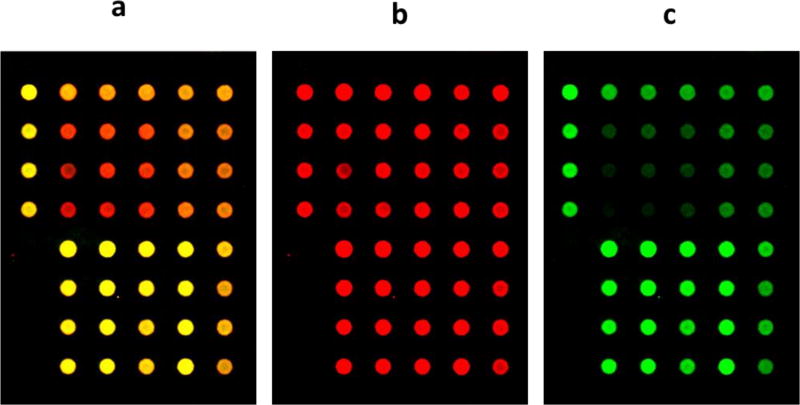
Figure 2. Fluorescent Images of RPPA slides.
a. Fluorescent image of part of an RPPA slide scanned on the InnoScan 710-R (Innopsys) at 700 nm (red) and 800 nm (green), showing the two channels merged. The slide was probed for phosphorylated S6 and for beta-actin as a normalization control.
b. Same image as part a, but only showing the 700 nm channel.
c. Same image as part a, but only showing the 800 nm channel.
REAGENTS AND SOLUTIONS
Unless a different solvent is specified, use Milli-Q purified water or equivalent in all recipes and protocols steps.
Phosphate-buffered saline (PBS)
37 mM NaCl
2 mM KCl
10 mM sodium phosphate dibasic
2 mM potassium phosphate monobasic
Adjust to pH 7.4 using NaOH and/or HCl
Store indefinitely at room temperature if sterile-filtered or autoclaved
PBS-T washing solution
To PBS (see recipe), add Tween-20 to a final concentration of .2%. Store up to 1 month at room temperature
RPPA cell lysis buffer
Make the following buffer stock:
50 mM TrisHCl
2% SDS
5% Glycerol
5 mM EDTA
NaF
Adjust pH to 6.8 using NaOH and/or HCl
Can store this stock at 4 °C
To the above stock, add:
1× protease inhibitor tablets (Complete Mini EDTA-Free Protease Inhibitor Cocktail, Roche cat. no. 11836170001)
1× phosphatase inhibitors (Phosphatase Inhibitor Cocktail 2, Sigma cat. no. P5726)
10 mM β-glycerophosphate
1 mM PMSF
1 mM Na3VO4
1 mM DTT
Divide the buffer into aliquots, and freeze at −80 °C
RPPA wash buffer
100 mM Tris
Adjust to pH 9.0 using NaOH and/or HCl
COMMENTARY
Background Information
RPPAs have been successfully used in numerous studies of human disease and cellular pharmacology. In experiments using cultured mammalian cell lines, RPPAs have facilitated cell signaling applications such as classification of receptor tyrosine kinases (Wagner et al., 2013), characterization of the rewiring of apoptotic pathways in response to EGFR inhibition (Lee et al., 2012), and a study of canonical and non-canonical Wnt signaling (Luckert et al., 2012). The technology is also suitable for lysates prepared from tissue samples: for example, RPPAs have been used to profile signaling networks that are altered in breast cancer using samples from tumor biopsies and matched normal tissue (Gujral et al., 2013). Thus, biomarkers of disease state or drug response identified in RPPA experiments with cultured cell lines can readily be monitored in clinical studies using the same technology. RPPAs can also be used as a method for screening compound libraries by identifying molecules that induce a specific state of cell signaling, as determined by measuring the phosphorylation state of multiple proteins (Sevecka & MacBeath, 2006).
Critical Parameters
Antibody selection
When possible, antibodies to be used in should be selected from those previously validated for RPPA. In addition to several published studies (Hennessy et al., 2010; Sevecka et al., 2011), the RPPA Core Facility at the MD Anderson Cancer Center maintains a list of validated antibodies (see https://www.mdanderson.org/research/research-resources/core-facilities/functional-proteomics-rppa-core/antibody-information-and-protocols.html). It is important to note that an antibody validated for RPPA in one or more biological contexts may not perform as well in other cell lines, or under different growth and treatment conditions. This is due to the fact that the expression levels of proteins responsible for off-target interactions with a given antibody can vary significantly between cell lines (or in some cases, under different growth conditions in the same cell line). In large studies including many cell lines and perturbations, it is likely not feasible to re-validate antibodies in every new biological context. Thus, the data from these studies must be interpreted with care, and should be validated by follow-up experiments using complementary methods such as high-content microscopy and Western blotting.
Choice of normalization method
A key parameter in RPPA experiments is the specific method used to normalize the signal of interest to the total amount of protein in each spot in the array. Normalization is commonly accomplished by incubation with a primary antibody specific for a generally expressed housekeeping protein, followed by an appropriate fluorescently-conjugated secondary antibody. The detection of a reference protein in RPPA experiments can be considered analogous to the use of a loading control in a Western blot. Beta-actin is a popular and commonly used normalization signal for RPPA experiments, and a number of highly specific primary antibodies from multiple species including mouse and rabbit are available for this protein. Other housekeeping proteins that can be used for normalization include α and β-tubulin, COX IV, and GAPDH. The choice of normalization protein will be influenced by the specific biology being studied: for example, GAPDH would be preferable to β-actin in biological contexts where actin stability is expected to be altered. In most cases, the protein used for normalization will be more abundant in lysates than the protein or PTM of interest. Since the 700 nm channel is less sensitive than the 800 nm channel in fluorescence scanners, this channel is generally used to detect the normalization signal. A potential alternative to antibody staining for normalization is the use of a total protein stain that fluoresces in the 700 nm channel, such as IRDye Blue Protein Stain (LI-COR).
Arraying conditions
Proper selection of arraying substrates and printing conditions are necessary for obtaining high quality data using the RPPA method. For the array surface, two key parameters are the protein binding capacity and the background autofluorescence of the material. One of the most commonly used arraying substrates is nitrocellulose due to its high binding capacity and history of successful use in other methods such as Western blotting. The total surface area available for protein binding can be enhanced by using nitrocellulose surfaces with a porous three-dimensional structure, as in ONCYTE AVID substrates (Grace Biolabs). ONCYTE AVID slides also have relatively low background fluorescence compared to other materials. When designing arrays, it is necessary to optimize the spacing and configuration of spots to allow analysis of the maximum number of samples without overlap. In addition the size of array spots, which is affected by the number of depositions and the protein concentration of the lysates, must be optimized to ensure adequate signal. A general rule of thumb for imaging arrays is that the pixel size should be no more than 1/10th of the spot diameter. Therefore, the resolution of the instrument to be used in scanning arrays should be kept in mind when adjusting the spot size.
TROUBLESHOOTING
Missing or misaligned spots in arrays
Problems with the printing process can result in missing or misaligned array spots. The quality of arrays can be assessed immediately after printing by scanning in a fluorescent imager using the 700 nm channel, as autofluorescence of buffer components will permit visualization of the spots. If spots are missing, the source plates should be checked to make sure no wells are empty or have very low volume of lysate. Assuming there are no issues with the source plates, a common cause of missing or misaligned spots for contact arrayers is damage to the pins. The integrity of pins can be checked using a microscope or stereoscope, and any that are bent or broken should be replaced in the print head and sent to the manufacturer for repair if possible. Missing array spots can also result from the deposition of salts or other materials onto pins during the printing process. This problem can be resolved by thoroughly cleaning the pins and optimizing the washing protocol during print runs, following the procedures recommended by the instrument manufacturer.
Overlapping spots in arrays
If the diameter of printed spots is too large relative to their spacing, overlapping will interfere with analysis of the array images. This problem can be resolved by using a smaller number of pin touches/depositions, diluting samples with additional lysate before printing, or reprinting arrays with a larger spacing between spots.
Improper alignment of print head
Improper alignment of the print head for a contact arrayer can cause less lysate than expected to be spotted on arrays (if the head is too high), or damage the arraying surface causing misformed spots (if the height is too low). If either issue is suspected the alignment of the print head should be recalibrated as specified by the manufacturer of the instrument.
Weak antibody staining on arrays
When low signal is observed when scanning one or more of the arrays in an experiment, the first step to take should be increasing the intensity settings on the scanner. If this does not improve the signal, it may be necessary to optimize the primary and secondary antibody concentrations to improve the staining. In the case the low signal is observed for multiple arrays using different antibodies, it may be necessary to increase the amount of protein in each array spot. This can be achieved by using a higher number of depositions when printing, or preparation of lysates using a larger starting number of cells in each well.
High array background staining
In the case of high background staining on arrays, it may be necessary to further optimize the blocking and washing procedures, or adjust the concentrations of antibodies used.
Saturated fluorescent signal
If any array spots show saturated pixels in either the 700 or 800 nm fluorescence channels (saturated pixels are colored white on many instruments), the arrays should be to rescanned using lower intensity settings until there is no longer saturation. In some cases it may be necessary to use a lower concentration of labeled secondary antibody and/or primary antibody.
Errors in image analysis
Occasionally the algorithms used by array analysis software will incorrectly assign individual spots or entire regions of the array, leading to bad data points or systematic errors in the data. Errors in the image analysis process are more likely in cases of high background, overlapping spots, or irregularities in the arraying pattern. If these are observed, it will be necessary to optimize parameters in the algorithm used to detect spots and the array pattern. In the case that only a few spots are incorrectly assigned in an area, it may be possible to manually correct these using the analysis software.
Anticipated Results
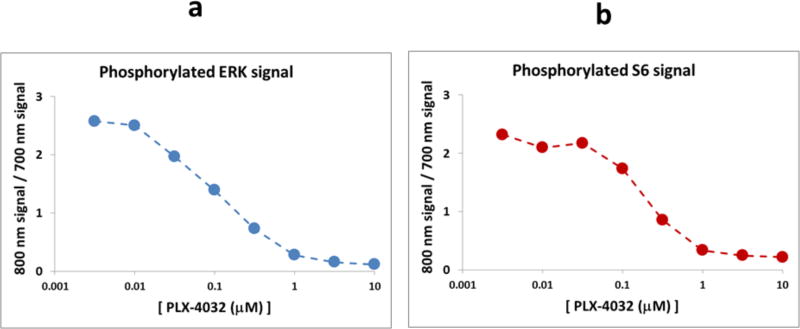
The protocols described in this unit will produce a set of protein and post-translational state measurements, as illustrated in Figure 3 for phosphorylated ERK and S6 ribosomal protein in RVH-421 cells treated with PLX-4032.
Figure 3. Representative RPPA data for RVH-421 Melanoma Cells Treated With PLX-4032.
a. RPPA dose response curve of phosphorylated ERK at 10 hrs post-treatment with increasing doses of the BRAF kinase inhibitor PLX-4032, as determined by the fluorescence intensity ratio of the 800 nm channel (used for the anti-pERK primary antibody) and the 700 nm channel (used for the anti-beta-actin primary antibody).
b. RPPA dose response curve of phosphorylated S6 at 24 hrs post-treatment with increasing doses of the BRAF kinase inhibitor PLX-4032, as determined by the fluorescence intensity ratio of the 800 nm channel (used for the anti-pS6 primary antibody) and the 700 nm channel (used for the anti-beta-actin primary antibody).
Time Considerations
Once all of the necessary reagents are in hand, the procedures described in this unit can be completed in approximately two standard work weeks: the first week to seed cells, treat with compounds, prepare lysates, and print arrays; and the second week to wash, probe, and scan arrays followed by analysis of the images. In practice, the length of time required to complete an RPPA experiment will depend on the scope of the data set, the time points for which data is collected, the number of protein levels and PTM states measured, and the type of data analysis required.
Acknowledgments
We would like to thank Innopsys for valuable technical discussions as well as early use of a demo model of their InnoScan 710-IR scanner to generate the data in this study, and Grace Bio-Labs for valuable discussions regarding the use of ONCYTE AVID slides for arraying. We also thank Cyril Benes and Anahita Dastur of the Center for Molecular Therapeutics at MGH for providing the melanoma cell lines used in this study. Finally, we thank Caroline Shamu and the members of the ICCB-Longwood Screening Facility for laboratory automation support. This work was supported by NIH LINCS grant U54 HG006097.
LITERATURE CITED
- Albeck JG, MacBeath G, White FM, Sorger PK, Lauffenburger DA, Gaudet S. Collecting and organizing systematic sets of protein data. Nat Rev Mol Cell Biol. 2006;7(11):803–812. doi: 10.1038/nrm2042. [DOI] [PubMed] [Google Scholar]
- Bollag G, Tsai J, Zhang J, Zhang C, Ibrahim P, Nolop K, Hirth P. Vemurafenib: the first drug approved for BRAF-mutant cancer. Nat Rev Drug Discov. 2012;11(11):873–886. doi: 10.1038/nrd3847. [DOI] [PubMed] [Google Scholar]
- Flaherty KT. Narrative review: BRAF opens the door for therapeutic advances in melanoma. Ann Intern Med. 2010;153(9):587–591. doi: 10.7326/0003-4819-153-9-201011020-00008. [DOI] [PubMed] [Google Scholar]
- Gujral TS, Karp RL, Finski A, Chan M, Schwartz PE, MacBeath G, Sorger P. Profiling phospho-signaling networks in breast cancer using reverse-phase protein arrays. Oncogene. 2013;32(29):3470–3476. doi: 10.1038/onc.2012.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy BT, Lu Y, Gonzalez-Angulo AM, Carey MS, Myhre S, Ju Z, Mills GB. A Technical Assessment of the Utility of Reverse Phase Protein Arrays for the Study of the Functional Proteome in Non-microdissected Human Breast Cancers. Clin Proteomics. 2010;6(4):129–151. doi: 10.1007/s12014-010-9055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight ZA, Lin H, Shokat KM. Targeting the cancer kinome through polypharmacology. Nat Rev Cancer. 2010;10(2):130–137. doi: 10.1038/nrc2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MJ, Ye AS, Gardino AK, Heijink AM, Sorger PK, MacBeath G, Yaffe MB. Sequential application of anticancer drugs enhances cell death by rewiring apoptotic signaling networks. Cell. 2012;149(4):780–794. doi: 10.1016/j.cell.2012.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckert K, Gujral TS, Chan M, Sevecka M, Joos TO, Sorger PK, Potz O. A dual array-based approach to assess the abundance and posttranslational modification state of signaling proteins. Sci Signal. 2012;5(206):pl1. doi: 10.1126/scisignal.2002372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen UB, Cardone MH, Sinskey AJ, MacBeath G, Sorger PK. Profiling receptor tyrosine kinase activation by using Ab microarrays. Proc Natl Acad Sci U S A. 2003;100(16):9330–9335. doi: 10.1073/pnas.1633513100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevecka M, MacBeath G. State-based discovery: a multidimensional screen for small-molecule modulators of EGF signaling. Nat Methods. 2006;3(10):825–831. doi: 10.1038/nmeth931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevecka M, Wolf-Yadlin A, MacBeath G. Lysate microarrays enable high-throughput, quantitative investigations of cellular signaling. Mol Cell Proteomics. 2011;10(4):M110 005363. doi: 10.1074/mcp.M110.005363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiffler MA, Grantcharova VP, Sevecka M, MacBeath G. Uncovering quantitative protein interaction networks for mouse PDZ domains using protein microarrays. J Am Chem Soc. 2006;128(17):5913–5922. doi: 10.1021/ja060943h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner JP, Wolf-Yadlin A, Sevecka M, Grenier JK, Root DE, Lauffenburger DA, MacBeath G. Receptor tyrosine kinases fall into distinct classes based on their inferred signaling networks. Sci Signal. 2013;6(284):ra58. doi: 10.1126/scisignal.2003994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf-Yadlin A, Sevecka M, MacBeath G. Dissecting protein function and signaling using protein microarrays. Curr Opin Chem Biol. 2009;13(4):398–405. doi: 10.1016/j.cbpa.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer. 2009;9(1):28–39. doi: 10.1038/nrc2559. [DOI] [PubMed] [Google Scholar]