Abstract
The 14-3-3 protein family interacts with more than 2000 different proteins in mammals, as a result of its specific phospho-serine/phospho-threonine binding activity. Seven paralogs are strictly conserved in mammalian species. Here, we show that during adipogenic differentiation of 3T3-L1 preadipocytes, the level of each 14-3-3 protein paralog is regulated independently. For instance 14-3-3β, γ, and η protein levels are increased compared to untreated cells. In contrast, 14-3-3ε protein levels decreased after differentiation while others remained constant. In silico analysis of the promoter region of each gene showed differences that explain the results obtained at mRNA and protein levels.
Abbreviations: IBMX, 3-isobutyl-1-methylxanthine; PPARγ, Peroxisome Proliferator-Activated Receptor γ; TAZ, transcriptional regulator with PDZ binding motif; C/EBPβ, CCAAT/Enhancer-Binding Protein-β
Keywords: 3T3-L1, Differentiation, Adipogenesis, 14-3-3
Graphical abstract
Highlights
-
•
14-3-3 paralogs are regulated independently during 3T3-L1 adipogenesis.
-
•
14-3-3β, γ, and η are specifically accumulated during adipogenesis.
-
•
Promoter sequence differences may explain this specific regulation.
1. Introduction
Mouse 3T3-L1 preadipocytes are a well characterized model for adipogenic differentiation. Undifferentiated cells possess a typical fibroblast morphology, and undergo differentiation to adipocytes in response to a cocktail of adipogenic factors containing insulin, dexamethasone (a glucocorticoid receptor agonist), and 3-isobutyl-1-methylxanthine (IBMX, a phosphodiesterase inhibitor resulting in elevated cyclic AMP levels [1]). Zebisch et al., [2] described a detailed protocol for the efficient differentiation of 3T3-L1 cells to adipocytes. In this protocol they added the Peroxisome Proliferator-Activated Receptor γ (PPARγ) agonist rosiglitazone to the classical cocktail based on a previous work by Wabitsch et al. [3], who found that rosiglitazone stimulated adipogenic differentiation in a dose-dependent manner with a maximum effect at concentration of 2 μM. This drug works as an insulin sensitizer, by binding to the PPARγ receptors and making the cells more responsive to insulin. Approximately 4 days after adding the adipogenic cocktail to 3T3-L1 preadipocytes, the cells start to accumulate lipids in the form of cytosolic lipid droplets that grow in number and size over cultivation time [2], and adipogenic differentiation is achieved within 5–7 days [4]. At the molecular level, this process is finely tuned through a transcriptional cascade in which many factors are involved [5]. Among other proteins that affect adipogenesis, Lipin1 and TAZ (Transcriptional regulator with PDZ binding motif) share the mode of regulation of their subcelular localization through cytoplasmic retention induced by phosphorylation and binding to the regulatory 14-3-3 proteins [6], [7]. Lipin-1 is a bifunctional protein involved in both adipogenesis, in the biosynthesis of triacylglycerols through its phosphatidate phosphatase activity, and lipid metabolism, acting as a transcriptional co-activator of genes involved in oxidative metabolism [7]. TAZ was reported to interact with a variety of transcription factors that control cell differentiation and organ development [8], [9]. In mesenchymal stem cells, it co-represses PPARγ dependent gene expression [8], thus inhibits adipogenesis. The 14-3-3s are phospho-serine/threonine binding proteins that have a pivotal role in the regulation of metabolism and signal transduction networks. This family, ubiquitous among eukaryotic organisms, is composed of ~30 kDa acidic proteins. Seven highly conserved 14-3-3 paralogs are present in mammals (β, γ, ε, η, θ, ζ, and σ), and their expression pattern varies in different cell types and tissues [10], [11]. These paralogs are homo- and hetero-dimers, regulating a diverse array of cellular proteins [12]. Although initially thought as redundant, the number of studies showing specialization of these paralogs is constantly growing. The 14-3-3 proteins bind to specific phosphorylated motifs on their target proteins and regulate their subcelular localization, stability and/or activation [13]. Although serine/threonine phosphorylation plays a crucial role in metabolic regulation, the effects of 14-3-3s and particularly the involvement of the different paralogs on the regulation of metabolic processes are poorly understood. Together with kinases and phosphatases, the regulatory 14-3-3 proteins are essential components of phosphorylation-mediated signaling [15].
Péterfy and coworkers [7] demonstrated that insulin stimulation of 3T3-L1 adipocytes results in increased lipin-1 phosphorylation, enhanced interaction with 14-3-3, and predominantly cytoplasmic localization. Similarly, 14-3-3 proteins regulate the subcelular localization of the Hippo effectors TAZ and YAP by retention in the cytoplasm as a consequence of their phosphorylation in specific residues by the kinase cascade [6]. These and other interactions of 14-3-3 with adipogenesis related proteins (see also Seipin in the discussion, [14]) show the importance of this family in the regulation of this differentiation process. Despite this, to our knowledge there is no available data from a systematic study analyzing the entire 14-3-3 family at both mRNA and protein levels during differentiation. Although Pal et al. [4] performed a proteomic analysis of rosiglitazone treated 3T3-L1 preadipocytes, this study only mentions one of the seven 14-3-3 mammalian isoforms. The strict conservation of 7 paralogs throughout all mammalian species, and evidences of their tissue specificity and protein-protein interaction networks differences [15] makes it interesting to investigate in detail the relative levels of the 14-3-3 isoforms before and after 3T3-L1 adipogenic differentiation. In this study, we have shown differential expression and accumulation of specific 14-3-3 isoforms, both at the mRNA as protein levels. We also performed a bioinformatics analysis of their promoter regions and discussed the results in the context of the regulation of this family in response to adipogenic signals and its possible role in the process.
2. Materials and methods
2.1. Chemicals
High-glucose DMEM and antibiotics penicillin and streptomycin were from Gibco Laboratories (Grand Island, NY, USA) and FBS was from GBO (Germany). IBMX and dexamethasone were purchased from Sigma Chemical Co (St. Louis, MO). Bovine insulin and rosiglitazone were from Beta Laboratories (Buenos Aires, Argentina). TRIzol Reagent was from Life technologies (California, USA) and Green Mastermix from Roche (Basel, Switzerland).
2.2. Cell culture and differentiation
3T3-L1 preadipocytes (ATCC, USA) were cultured in 10 cm diam. dishes in complete DMEM (10% FBS, 100 U/ml penicillin and 100 µg/ml streptomycin) at 37 °C and 5% CO2. Different insulin and rosiglitazone concentrations were evaluated by Oil red O staining at days 5, 9, 15 and 20. In further experiments 80% confluence cultures were induced to differentiate in complete DMEM supplemented with 250 nM dexamethasone, 0.5 mM IBMX, 2 µM rosiglitazone and 10 µg/ml insulin during 3 days, followed by 2 days in complete DMEM with 10 µg/ml insulin and 2 final days in complete DMEM. Untreated cells were grown for 3 days in serum free DMEM and then the media was supplemented with 2.5% FBS until the end of the experiment.
2.3. Oil red O staining
Lipid droplets in differentiated and control 3T3-L1 cells were stained using Oil red O as follows. The cells grown on cover slips in 24-wells plates were washed three times with PBS containing 1 mM CaCl2 and 1 mM MgCl2, and then fixed for 20 min with 4% paraformaldehyde. Oil red O (0.35% w/v) in isopropanol was diluted in water (6/4 ratio), filtered by 0.2 µm nitrocellulose membrane, added to the fixed cells for 2 h at room temperature, exhaustively rinsed with MQ water and then mounted in Mowiol (as recommended by the manufacturer) for microscopy analysis. In parallel samples, the extent of fat accumulation (which is proportional to the Oil red O staining), was quantified as in [16]. Briefly, water excess was evaporated by placing the stained cells at 32 °C, 150 µl of isopropyl alcohol was added to each well with stained cells in a 24-well-plate to extract the dye, which was removed after 10 min of incubation and its absorbance monitored spectrophotometrically at 510 nm.
2.4. RNA isolation, cDNA synthesis and quantitative real-time PCR
Total RNA was purified as previously described [17] using 1 ml of TRIzol reagent per 10 cm diam. culture dish containing confluent cells. After verifying the RNA integrity, retrotranscription was achieved using the M-MLV Reverse Transcriptase (Life Technologies). The PCR reactions were performed using three biological samples and three technical replicates in 20 µl volume containing SYBER Green Mastermix with the following steps: denaturation 95 °C 15 s, annealing and elongation 60 °C 30 s. The primers used were designed to be highly specific for each 14-3-3 isoform and are described in Table S1 (supplementary material). Primers sequences for the adipocyte Protein 2 (aP2) gene were taken from [8]. Peptidylprolyl Isomerase A (PPIA) was chosen as a normalizing gene [18].
2.5. Whole cells lysates and Western Blot analysis
Whole cells lysates were prepared in RIPA buffer (50 mM Tris, pH 7.5; 150 mM sodium chloride; 1% NP-40; 0.5% sodium deoxicholate and 0.1% SDS). The proteins were resolved by SDS-PAGE and then electrotransfered to a nitrocellulose membrane in Towbin buffer for WB assay. Specific validated antibodies recognizing the amino-terminal regions of each 14-3-3 mammalian isoform [19] were kindly donated by Dr. A. Aitken (University of Edinburgh, Scotland, UK). Mouse monoclonal anti-α-Tubulin antibody (Sigma) was used as loading control. The secondary antibodies used were anti-rabbit IgG-HRP and anti-mouse IgG-HRP (Promega).
2.6. In silico analysis of 14-3-3 genes promoter regions
The in silico promoter region analysis of the seven genes that codify for the 14-3-3 isoforms was performed following the hypothesis that functional DNA sequences are evolutionary constrained and thus are highly conserved through different species. We used several tools implemented in dcode.org (http://ecrbrowser.dcode.org [20] and http://mulan.dcode.org [21]). First, we searched each gene of the 14-3-3 proteins in the Mus musculus genome. Our selection included from the 3′UTR to the 5′UTR of each gene. To include regulatory sequences we added 1000 bp upstream to the first nucleotide of the 3′UTR intron. Local multiple genome sequence alignments of these regions from four mammalian genomes (mouse, cow, human and macaque) were performed. Finally, we searched for transcription factor binding sites for transcription factors described to have participation in adipogenic differentiation [5], and listed those that are evolutionary conserved across the four species.
3. Results
3.1. 3T3-L1 differentiation
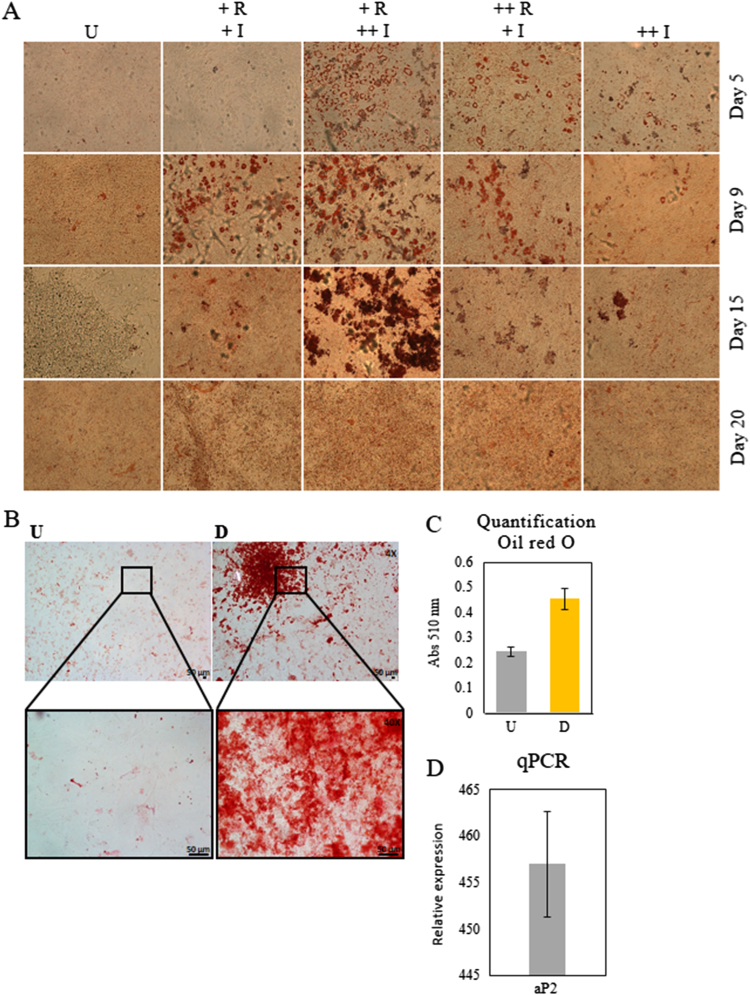
Several adipogenic cocktails with variable compositions are reported in the literature to induce differentiation of 3T3-L1 cells. However, because of differences in their efficiency, we first tested four different formulations including two different insulin (1 and 10 µg/ml) and three different rosiglitazone (0, 2 and 4 µM) concentrations. The efficiency of these cocktails was evaluated by Oil red O staining at 5, 9, 15 and 20 days after induction. We found that the most efficient induction cocktail contained 0.5 mM IBMX, 250 nm dexamethasone, 2 µM rosiglitazone and 10 µg/ml insulin (Fig. 1(A)). Lipid droplets accumulation inside the differentiating cells increased during 7 days following induction. Later, many lipid droplets released their content, and the cell layer started to detach at the well margins progressing through the center until complete detachment. For this reason we decided to finish the experiments at day 7 after induction. Microscopy images of differentiated and control 3T3-L1 cells stained with Oil red O evidenced groups of differentiated cells (Fig. 1(B)) spread among undifferentiated cells. Oil red O quantification was significantly higher (p=0.03) in differentiated cells (Fig. 1(C)). Consistently, the adipogenic marker aP2 gene relative expression measured by qPCR was more than 450 folds higher at 7 days of differentiation (Fig. 1(D)) compared to untreated cells.
Fig. 1.
Adipogenic differentiation of 3T3-L1 cells. A) Bright-field microscopy images (10×) of 3T3-L1 preadipocytes (U, untreated) and subjected to adipogenic differentiation using four different formulations including 0.5 mM IBMX, 250 nM dexamethasone and variable concentrations of insulin and rosiglitazone. + I, 1 µg/ml insulin;++ I, 10 µg/ml insulin;+ R, 2 µM rosiglitazone;++ R, 4 µM rosiglitazone. Cells were fixed and lipid droplets stained with Oil red O at 5, 9, 15 and 20 days after induction. B) Same as in A, with 10 µg/ml insulin and 2 µM rosiglitazone, fixed and Oil red O stained at day 7 after induction. Magnification 4×, inset 40×. C) Fat quantification through absorbance of Oil red O at 510 nm recuperated from discoloration of 3T3-L1 stained cells untreated (U) and after 7 days of adipogenic induction (D). A, B and C) Data were similar in two other experiments. D) aP2 gene relative expression measured by qPCR at 7 days of differentiation of 3T3-L1 cells compared to untreated cells. Bars represent averaged mean data from three biological and technical replicates of qPCR.
3.2. Relative levels of 14-3-3 proteins in untreated and differentiated 3T3-L1 cells
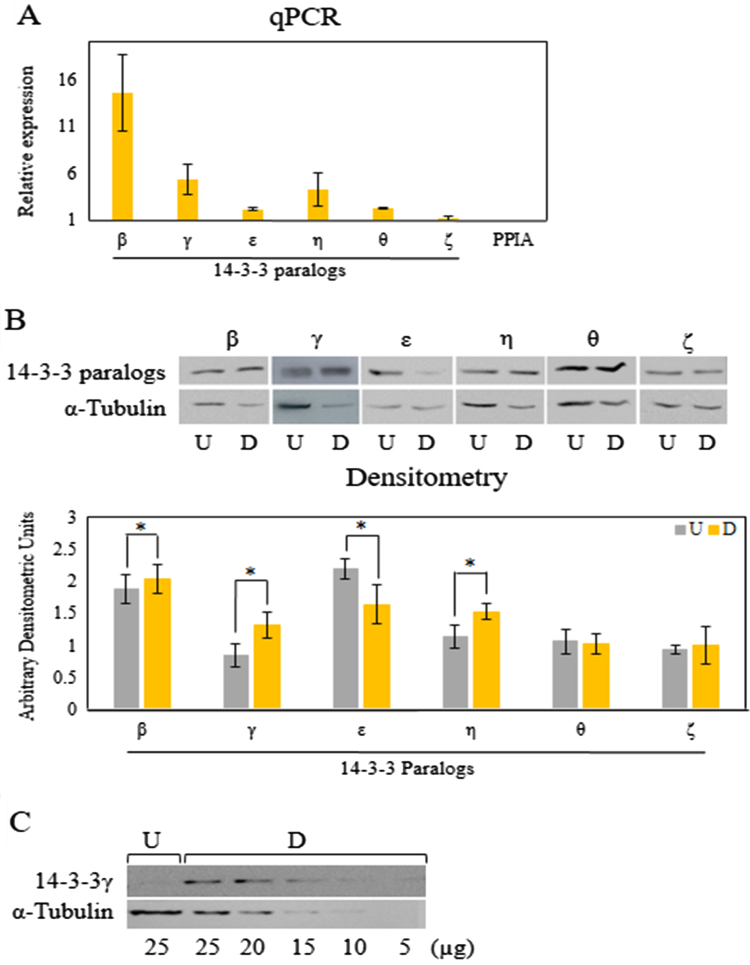
Because 14-3-3 proteins regulate several other proteins known to affect cell differentiation, we evaluated the mRNA and protein levels of the different 14-3-3 paralogs in untreated and differentiated 3T3-L1 cells. We found significant differences in the mRNA levels of three from six isoforms evaluated by qPCR (Fig. 2(A)). 14-3-3β presented the highest difference, with an average of more than 14 folds increase in 7 days differentiated cells compared to untreated 3T3-L1 preadipocytes. 14-3-3γ and η paralogs also showed higher mRNA levels in differentiated cells, although the difference was less pronounced (5.3 and 4.2 folds respectively) that the one observed for 14-3-3β (Fig. 2(A)). 14-3-3β, γ and η protein levels were also higher in differentiated compared to untreated cells, in this case being γ the isoform with the highest difference. In contrast, 14-3-3ε protein levels decreased after differentiation, whereas θ and ζ levels remained unaltered (Fig. 2(B)). 14-3-3ζ mRNA was also invariable upon differentiation (Fig. 1(A)). Because 14-3-3γ showed the highest difference at the protein level, in order to quantify its relative increase in differentiated cells, we analyzed by WB decreasing amounts of total lysates from differentiated cells, from 25 to 5 µg. We verified a 2.5 folds increase in differentiated compared to control cells (Fig. 2(C)). 14-3-3σ protein levels were undetectable by WB in 3T3-L1 cells.
Fig. 2.
Relative levels of 14-3-3 proteins paralogs in untreated and differentiated 3T3-L1 cells 7 days after adipogenic induction. A) 14-3-3 paralogs expression measured by qPCR of differentiated 3T3-L1 cells relative to untreated cells. PPIA was used as normalizing gene. Bars represent averaged mean data from three biological and technical replicates of qPCR. B) 40 µg of whole cell lysate from untreated and differentiated 3T3-L1 cells were loaded per well and separated by SDS/PAGE. Blots were divided for reaction with specific antibodies against each of the different 14-3-3 paralogs and α-Tubulin as loading control. Bars in the graphs represent mean±SD, of values from 3 experiments quantified by densitometry and expressed relative to that of α-Tubulin values from the same western blot line. Asterisks indicate significant differences between samples (using the non-parametric Wilcoxon rank-sum test, p<0.05). C) Same as in B using antibodies against 14-3-3γ and α-Tubulin. Decreasing amounts of whole cell lysate of differentiated cells were loaded per well. B and C) Data were similar in two other experiments.
3.3. In silico promoter analysis
In order to search for functional highly conserved DNA sequences containing binding sites for transcription factors described to have participation in adipogenic differentiation [5], we performed local multiple genome sequence alignments of the seven 14-3-3 genes including their 3′ and 5′ UTR, and 1000 bp upstream to the first nucleotide of the 3′UTR intron, from four mammalian genomes (mouse, cow, human and macaque). We listed those transcription factor binding sites that are evolutionary conserved across the four species and summarized their frequencies in Table 1. Their sequences were specified in a note below Table 1, and their locations on 14-3-3 genes were given in Table S2 (Supplementary material). 14-3-3γ contained the highest number (17) of total adipogenesis related transcription binding sites, compared to the other 14-3-3 paralogs. 14-3-3γ was also the isoform that included DNA binding sequences for a larger array of transcription factors related to adipogenesis (Table 1). Although 14-3-3β number of adipogenesis related transcription factors binding sites was relatively low compared to other paralogs, it includes 2 binding sites for CCAAT/ Enhancer-Binding Protein-β (C/EBPβ), which is critical during differentiation induction (Section 4), and also includes one for the master regulator of adipogenesis PPARγ. The opposite was observed in 14-3-3ζ promoter regions, which even though it has a higher number of adipogenesis factors binding sites than 14-3-3β, c/EBPβ and PPARγ are absent. It is to be noted that 14-3-3σ promoter region does not contain any of the transcription factors binding sites related to adipogenesis differentiation.
Table 1.
Frequency of adipogenesis related transcription factor binding sitesa in 14-3-3 genes promoter regions.
| 14-3-3β | 14-3-3γ | 14-3-3ε | 14-3-3η | 14-3-3θ | 14-3-3ζ | 14-3-3σ | |
| C/EBP α | 0 | 4 | 0 | 0 | 0 | 1 | 0 |
| C/EBPβ | 2 | 1 | 0 | 1 | 1 | 0 | 0 |
| C/EBPδ | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| CREB | 0 | 4 | 0 | 1 | 1 | 1 | 0 |
| GATA2 | 1 | 1 | 1 | 1 | 0 | 0 | 0 |
| GATA3 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| GR | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| KROX | 0 | 2 | 6 | 0 | 0 | 1 | 0 |
| PPARγ | 1 | 2 | 1 | 0 | 0 | 0 | 0 |
| SMAD | 1 | 1 | 0 | 1 | 0 | 1 | 0 |
| STAT5A | 0 | 1 | 1 | 0 | 0 | 1 | 0 |
| Total | 5 | 17 | 10 | 5 | 3 | 7 | 0 |
The sequences (strictly conserved nucleotides in capital letters, less conserved in lower case letters) of the transcriptions factors are as follows: C/EBPα, ccaTTgcttaatct; C/EBPβ, TTTAGCAA; C/EBPδ ATTGCTTAA; CREB, TGACACCA/TGAAGTC; GATA2, GATCAGA; GATA3, GATCAAA; GR, TGTTC; KROX, CTCCCCC; PPARGγ, TGGCCTTTGCCC; SMAD, AGACagctc; STAT5A, TTCCTTGAACACCACTGCTTTG. The locations of these sequences on 14-3-3 genes are given in Table S2 (Supplementary material).
4. Discussion
The 14-3-3 proteins are important regulators of many phosphorylation-mediated cellular functions. Here we focused on the analysis of 14-3-3 paralogs at both mRNA and protein levels in 3T3-L1 differentiating cells. First, we optimized adipogenesis induction cocktails based on previous works [2], [8]. The most effective cocktail was similar to the one used by Zebisch et al. [2] with a higher insulin concentration (10 µg/ml). We observed groups of differentiated 3T3-L1 cells scattered among undifferentiated cells, which was previously observed [22], possibly due to intercellular contact and signaling.
Our results showed that both at the mRNA and protein level, 14-3-3β and γ were the isoforms that most changed after adipogenic differentiation. Their higher mRNA levels may be due to adipogenesis related transcription factor binding sites present in their promoter regions. Even though many transcription factors have a role in adipogenesis, it is the expression of PPARγ, C/EBPα, C/EBPβ and C/EBPδ, as well as the epigenomic coordination between these factors, that are the primary drivers of adipocyte gene induction during terminal differentiation [5]. PPARγ and these C/EBPs also regulate each other in a positive feedback circuit, which is central to terminal differentiation. This may explain the higher mRNA levels of 14-3-3β and γ, which are the only 14-3-3 paralogs that contain a combination of binding sites in their promoter regions including both types of transcription factors. Furthermore, genome-wide studies have shown that C/EBPβ is present at low levels in committed preadipocytes before the addition of adipogenic stimuli, and is bound to quiescent adipogenic hotspots. These are regions of the genome that display marks of active enhancers and recruit other adipogenic transcription factors after the addition of the adipogenic cocktail [5]. 14-3-3β promoter region contains 2 binding sites for C/EBPβ, which may explain its upregulation upon adipogenic stimuli. Aghazadeh et al. [10] summarized the tissue specific expression of human 14-3-3 isoforms based on data collected from two online human microarray databases. Notably, human adipose tissue is -after the brain- the second in relative expression of 14-3-3γ, whereas for other isoforms this tissue ranks 5th or more. The other 14-3-3 isoform that we found to raise both mRNA and protein levels in differentiated 3T3-L1 cells compared to preadipocytes is 14-3-3β. This was previously observed by Yang et al. [14] who reported in 3T3-L1 cells a maximum of 14-3-3β mRNA at day 8 (fully differentiated cells) and at the protein level a plateau reached at day 4 after adipogenic induction. Indeed, 14-3-3β has been recently identified as an interacting partner of the ER-membrane Seipin protein, an important regulator in adipocyte development [14]. Seipin regulates lipid homeostasis by preventing lipid droplet (LD) formation in non-adipocytes but promoting it in developing adipocytes. Yang et al. [14] proposed a model in which 14-3-3β is recruited by Seipin after detection of high lipid levels in the cell. 14-3-3β transduces the Seipin-guided lipid signal from the ER to the cytoplasm, directing cofilin-1-mediated cytoskeleton reorganization, which is necessary for preadipocytes to differentiate [14].
Most recently, Park et al. [23] reported that 14-3-3β and γ differentially regulate the transcriptional activity of the nuclear receptor PPARγ2 in mouse primary hepatocytes and HepG2 cells. Although 14-3-3β and γ competitively interact with the same serine residue on PPARγ2, 14-3-3β increased while 14-3-3γ decreased the transcriptional activity of PPARγ2. In adipogenic differentiated 3T3-L1 cells, both 14-3-3γ and β increased their levels, suggesting that 14-3-3γ may have, as Seipin, opposites roles on lipid metabolism in non-adipose vs. adipose cells [14], or it may be up-regulated as part of a negative feedback loop regulation mechanism.
To our knowledge, there are no reports about 14-3-3η and ε roles in lipid metabolism. Our results showed accumulation at the mRNA and protein levels of 14-3-3η, and a specific depletion of 14-3-3ε protein levels. This suggests that both may have probably opposed regulatory roles in this differentiation process.
After adipogenesis of 3T3-L1 cells, we observed unchanged 14-3-3ζ mRNA and protein levels. Indeed, the gene coding for 14-3-3ζ (YWHAZ) was identified among the best performers to be used as a housekeeping gene for normalization for qPCR of mesenchymal stem cells in adipogenic, osteogenic and chondrogenic differentiation experiments [24]. 14-3-3ζ promoter region contrasts with that of its paralogs 14-3-3β and γ in the absence of both types of transcription factors that are critical for the adipogenesis transcription program (C/EBPs and PPARγ), which may be the cause of its unchanged mRNA levels in differentiated cells. 14-3-3σ was the only paralog that we could not detect in 3T3-L1 cells, both in preadipocytes and after differentiation, thus it was not surprising to find that it was the only one not containing any of the adipogenesis factors binding sites analyzed (Table 1). This was expected as it is known to be specifically expressed in epithelial cells [25].
The fact that seven 14-3-3 isoforms have been evolutionary retained throughout the entire mammalian class has been an intriguing issue driving research for years in the 14-3-3 field. Using a computational system approach we have previously found that the protein-protein interaction networks of the 14-3-3 paralogs are statistically different. Among other differences, certain domains are over-represented in partners of one specific isoform but absent in others. That is the case for the Low density Lipoprotein Receptor LDLR (A and B) domains, which are exclusive to the 14-3-3γ network [15]. Skogsberg et al. [26] showed that the binding of apoB100-LDL to adipocytes via the LDL receptor inhibits intracellular noradrenaline-induced lipolysis in adipocytes. Although the relationship between 14-3-3γ and the LDLR remains unknown, the specific up-regulation of 14-3-3γ upon adipogenic differentiation may suggest that this 14-3-3 isoform, whose interaction network is specifically enriched in LDLR domains, may possibly have a role in adipogenesis through interaction with the LDLR.
5. Conclusions
Our results clearly show an independent regulation of 14-3-3 family members during 3T3-L1 adipogenesis. Specifically, 14-3-3β, γ, and η are accumulated while 14-3-3ε is down regulated. The frequency and distribution of adipogenesis related transcription factor binding sites on the different 14-3-3 paralogs promoter sequences may explain these differences. Although some previous articles described the relative expression and/or levels of isolated 14-3-3 isoforms and their response to a differentiation process, to our knowledge, this is the first study to systematically analyze the complete 14-3-3 family at both protein and mRNA levels before and after a differentiation process. Our analysis supports the currently growing concept of specific molecular functions for the different 14-3-3 paralogs.
A global understanding of the regulation of each member of this important regulatory protein family will contribute to predict what could occur when regulatory circuits become dysfunctional or are modified in response to external stimuli.
Author contributions
DMB and MU conceived and supervised the study; ADG, DMB and MU designed experiments; ADG and DMB performed experiments; ADG, DMB and MU analyzed data, wrote the manuscript and made manuscript revisions.
Acknowledgements
ADG is a fellow from CONICET; DMB and MU are members of the Investigator Career of the National Research Council (CONICET, Argentina). Funding: This work was supported by CONICET [grant numbers PIP 0304 and PIP 0439]; and ANPCYT, Argentina [grant numbers PICT’12–2498 and PICT’11–1450]. The funding institutions had no role in study design, data collection, analysis and interpretation, decision to publish, or preparation of the manuscript.
Footnotes
Transparency Document associated with this article can be found in the online version at doi:10.1016/j.bbrep.2016.05.020.
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2016.05.020.
Contributor Information
Aldana D. Gojanovich, Email: agojanovich@fcm.uncu.edu.ar.
Diego M. Bustos, Email: dbustos@mendoza-conicet.gob.ar.
Marina Uhart, Email: muhart@mendoza-conicet.gob.ar.
Transparency Document
Transparency Document
.
Transparency Document
.
Transparency Document
.
Transparency Document
.
Transparency Document
.
Appendix A. Supplementary material
Supplementary material: Table 1. Information of the dataset files. Table 2. The peptide quantification file combines the four search result files as listed in Supplementary Table 1. Network 1. Network built based on the filtered proteins of interest. Under the PSIMI25 visual style, the red (positive value) to blue (negative value) shade is applied to the nodes according to the ratio of protein abundance on the inner membrane of the mutant. The thickness of the edges is drawn according to the mentha scores. Network 2. Network of MinD and MinE. Under the PSIMI25 visual style, the red (positive value) to blue (negative value) shade is applied to the nodes according to the ratio of protein abundance on the inner membrane of the mutant. The thickness of the edges is drawn according to the mentha scores.
References
- 1.Martini C.N., Plaza M.V., Vila M.C. PKA-dependent and independent cAMP signaling in 3T3-L1 fibroblasts differentiation. Mol. Cell Endocrinol. 2009;298:42–47. doi: 10.1016/j.mce.2008.10.023. [DOI] [PubMed] [Google Scholar]
- 2.Zebisch K., Voigt V., Wabitsch M., Brandsch M. Protocol for effective differentiation of 3T3-L1 cells to adipocytes. Anal. Biochem. 2012;425:88–90. doi: 10.1016/j.ab.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 3.Wabitsch M., Brenner R., Melzner I., Braun M., Möller P., Heinze E. Characterization of a human preadipocyte cell strain with high capacity for adipose differentiation. Int. J. Obes. Relat. Metab. Disord. J. Int. Assoc. Study Obes. 2001;25:8–15. doi: 10.1038/sj.ijo.0801520. [DOI] [PubMed] [Google Scholar]
- 4.Pal P., Kanaujiya J.K., Lochab S., Tripathi S.B., Sanyal S., Behre G. Proteomic analysis of rosiglitazone and guggulsterone treated 3T3-L1 preadipocytes. Mol. Cell. Biochem. 2013;376:81–93. doi: 10.1007/s11010-012-1551-0. [DOI] [PubMed] [Google Scholar]
- 5.Cristancho A.G., Lazar M.A. Forming functional fat: a growing understanding of adipocyte differentiation. Nat. Rev. Mol. Cell Biol. 2011;12:722–734. doi: 10.1038/nrm3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanai F., Marignani P.A., Sarbassova D., Yagi R., Hall R.A., Donowitz M. TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J. 2000;19:6778–6791. doi: 10.1093/emboj/19.24.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Péterfy M., Harris T.E., Fujita N., Reue K. Insulin-stimulated interaction with 14-3-3 promotes cytoplasmic localization of lipin-1 in adipocytes. J. Biol. Chem. 2010;285:3857–3864. doi: 10.1074/jbc.M109.072488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hong J.-H., Hwang E.S., McManus M.T., Amsterdam A., Tian Y., Kalmukova R. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309:1074–1078. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- 9.Park K.-S., Whitsett J.A., Di Palma T., Hong J.-H., Yaffe M.B., Zannini M. TAZ interacts with TTF-1 and regulates expression of surfactant protein-C. J. Biol. Chem. 2004;279:17384–17390. doi: 10.1074/jbc.M312569200. [DOI] [PubMed] [Google Scholar]
- 10.Aghazadeh Y., Papadopoulos V. The role of the 14-3-3 protein family in health, disease, and drug development. Drug Discov. Today. 2015:1681–1690. doi: 10.1016/j.drudis.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 11.Kilani R.T., Medina A., Aitken A., Jalili R.B., Carr M., Ghahary A. Identification of different isoforms of 14-3-3 protein family in human dermal and epidermal layers. Mol. Cell. Biochem. 2008;314:161–169. doi: 10.1007/s11010-008-9777-6. [DOI] [PubMed] [Google Scholar]
- 12.Morrison D.K. The 14-3-3 proteins: integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol. 2009;19:16–23. doi: 10.1016/j.tcb.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakiyama H., Wynn R.M., Lee W.-R., Fukasawa M., Mizuguchi H., Gardner K.H. Regulation of nuclear import/export of carbohydrate response element-binding protein (ChREBP): interaction of an α-helix of ChREBP with the 14-3-3 proteins and regulation by phosphorylation. J. Biol. Chem. 2008;283:24899–24908. doi: 10.1074/jbc.M804308200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang W., Thein S., Wang X., Bi X., Ericksen R.E., Xu F. BSCL2/seipin regulates adipogenesis through actin cytoskeleton remodelling. Hum. Mol. Genet. 2014:502–513. doi: 10.1093/hmg/ddt444. [DOI] [PubMed] [Google Scholar]
- 15.Uhart M., Bustos D.M. Human 14-3-3 paralogs differences uncovered by cross-talk of phosphorylation and lysine acetylation. PLoS One. 2013;8:e55703. doi: 10.1371/journal.pone.0055703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramirez-Zacarias J., Castro-Munozledo F., Kuri-Harcuch W. Quantitation of adipose conversion and triglycerides by staining intracytoplasmic lipids with Oil red O. Histochemistry. 1992;97:493–497. doi: 10.1007/BF00316069. [DOI] [PubMed] [Google Scholar]
- 17.Padilla P.I., Uhart M., Pacheco-Rodriguez G., Peculis B.A., Moss J., Vaughan M. Association of Guanine Nucleotide-Exchange Protein BIG1 in HepG2 Cell Nuclei with nucleolin, U3 snoRNA, and fibrillarin. Proc. Natl. Acad. Sci. 2008;105:3357–3361. doi: 10.1073/pnas.0712387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fink T., Lund P., Pilgaard L., Rasmussen J.G., Duroux M., Zachar V. Instability of standard PCR reference genes in adipose-derived stem cells during propagation, differentiation and hypoxic exposure. BMC Mol. Biol. 2008;9:2199–2208. doi: 10.1186/1471-2199-9-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin H., Patel Y., Jones D., Howell S., Robinson K., Aitken A. Antibodies against the major brain isofbrms of 14-3-3 protein: an antibody specific for the N-acetylated ammo-terminus of a protein. FEBS Lett. 1993;331:296–303. doi: 10.1016/0014-5793(93)80356-y. [DOI] [PubMed] [Google Scholar]
- 20.Ovcharenko I., Nobrega M.A., Loots G.G., Stubbs L. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 2004;32:W280–W286. doi: 10.1093/nar/gkh355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ovcharenko I., Loots G.G., Giardine B.M., Hou M., Ma J., Hardison R.C. Mulan: multiple-sequence local alignment and visualization for studying function and evolution. Genome Res. 2005;15:184–194. doi: 10.1101/gr.3007205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nan X., Cheng J.-X., Xie X.S. Vibrational imaging of lipid droplets in live fibroblast cells with coherent anti-Stokes Raman scattering microscopy. J. Lipid Res. 2003;44:2202–2208. doi: 10.1194/jlr.D300022-JLR200. [DOI] [PubMed] [Google Scholar]
- 23.Park S., Yoo S., Kim J., An H.-T., Kang M., Ko J. 14-3-3β and γ differentially regulate peroxisome proliferator activated receptor γ 2 transactivation and hepatic lipid metabolism. Biochim. Biophys. Acta BBA-Gene Regul. Mech. 2015;1849:1237–1247. doi: 10.1016/j.bbagrm.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 24.Ragni E., Vigano M., Rebulla P., Giordano R., Lazzari L. What is beyond a qRT-PCR study on mesenchymal stem cell differentiation properties: how to choose the most reliable housekeeping genes. J. Cell Mol. Med. 2013;17:168–180. doi: 10.1111/j.1582-4934.2012.01660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moreira J.M., Shen T., Ohlsson G., Gromov P., Gromova I., Celis J.E. A combined proteome and ultrastructural localization analysis of 14-3-3 proteins in transformed human amnion (AMA) cells. Mol. Cell. Proteom. 2008;7:1225–1240. doi: 10.1074/mcp.M700439-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Skogsberg J., Dicker A., Rydén M., Astrom G., Nilsson R., Bhuiyan H. ApoB100-LDL acts as a metabolic signal from liver to peripheral fat causing inhibition of lipolysis in adipocytes. PLoS One. 2008;3:e3771. doi: 10.1371/journal.pone.0003771. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transparency Document
Transparency Document
Transparency Document
Transparency Document
Transparency Document
Supplementary material: Table 1. Information of the dataset files. Table 2. The peptide quantification file combines the four search result files as listed in Supplementary Table 1. Network 1. Network built based on the filtered proteins of interest. Under the PSIMI25 visual style, the red (positive value) to blue (negative value) shade is applied to the nodes according to the ratio of protein abundance on the inner membrane of the mutant. The thickness of the edges is drawn according to the mentha scores. Network 2. Network of MinD and MinE. Under the PSIMI25 visual style, the red (positive value) to blue (negative value) shade is applied to the nodes according to the ratio of protein abundance on the inner membrane of the mutant. The thickness of the edges is drawn according to the mentha scores.