Abstract
Hepatitis C virus (HCV) is characterized by considerable genetic variability and, as a consequence, it has 6 genotypes and multitude of subtypes. HCV envelope glycoproteins are involved in the virion formation; the correct folding of these proteins plays the key role in virus infectivity. Glycosylation at certain sites of different genotypes HCV glycoproteins shows substantial differences in functions of the individual glycans (Goffard et al., 2005; Helle et al., 2010) [1], [2]. In this study, differential glycosylation sites of HCV genotype 1b envelope proteins in insect and mammalian cells was demonstrated. We showed that part of glycosylation sites was important for folding of the proteins involved in the formation of viral particles. Point mutations were introduced in the protein N-glycosylation sites of HCV (genotype 1b) and the mutant proteins were analyzed using baculovirus expression system in mammalian and insect cells. Our data showed that, in contrast to HCV 1a and 2a, the folding of HCV 1b envelope proteins E2 (sites N1, N2, N10) and E1 (sites N1, N5) was disrupted, however that did not prevent the formation of virus-like particles (VLP) with misfolded glycoproteins having densities typical for HCV particles containing RNA fragments. Experimental data are supported by mathematical modeling of the structure of E1 mutant variants.
Keywords: Hepatitis C virus, HCV envelope glycoproteins, N-glycans, Baculovirus, Insect Sf9 cells, Mammalian Hek293Tcells, Site-directed mutagenesis
Highlights
-
•
Baculovirus expression system in mammalian and insect cells for.
-
•
Glycosylation at certain sites of HCV genotype 1b.
-
•
Part of glycosylation sites of E1, E2 HCV important for folding of the proteins involved in the formation of viral particles.
1. Introduction
Hepatitis C infection is being called a silent epidemic (HCV currently infected about 3% of the global population). Modern methods of hepatitis C treatment are not perfect, and more detailed knowledge of HCV remains insufficient [3].
The HCV is characterized by considerable genetic variability and, as a consequence, by the presence of 6 genotypes and several dozens of subtypes. One of the main HCV subtypes predominating in many countries including Russia is genotype 1b which is resistant to interferon therapy. Despite the creation of the direct-acting antivirals for the treatment of HCV infection which appeared in the last few years [4], [5], [6], [7] the studies of the molecular mechanisms of viableHCV 1b virion formation that determine the infectivity of the virus, including its morphogenesis and pathogenesis are of great need.
The specific feature of HCV is its high replicative activity leading to high genetic variation and the expressed ability to mutate. The maximal variability is observed in genes encoding the envelope glycoproteins, E1 and E2. As a result, infected patients at various stages of infection may have several dozens of HCV subtypes as a set of closely related genomes within the same genotype. Some viral strains are characterized by a higher degree of pathogenicity, but specific molecular determinants of such phenotype are not clear.
In an infected cell, the formation of infectious viral particles occurs with participation of various HCV components, including envelope glycoproteins. These proteins, E1 and E2, undergo post-translational N-glycosylation, wherein the low-branched oligosaccharide chain consists of nine residues of mannose (Man) and three residual glucose (Glc), attached to specific residues asparagine Asn-X-Ser or Asn-X-Thr sequences (where X – any amino acid except proline) [8]. HCV virion envelope glycoproteins are highly glycosylated; the high degree of conservation of glycosylation sites 4–5 in E1 and 9–11 in E2 indicates their important role in the HCV life cycle [2], [9]. After glycosylation glycoproteins E1 and E2 HCV can either dimerize to form a functional non-covalent complexes with the participation of the cellular chaperone calnexin, or form disulfide bridges with the participation of cellular protein calreticulin resulting in the formation of non-functional aggregates containing incorrectly folded proteins [10], [11]. As a result, the formation of such aggregates may lead to the production of defective viral particles that can possibly inhibit the ability of the virus' input and output of the cell. It is not clear whether the aggregates are formed in the course of HCV infection. Studies related to the glycosylation of the recombinant mutant HCV envelope glycoproteins in HCVpp and HCVcc systems show some glycans that are important in viral assembly, its cell-binding ability and protection from neutralizing antibodies [12], [13]. Furthermore, these oligosaccharides form the N-linked “glycan shield” on the virion surface, which contributes to HCV avoidance of humoral immune response. Thus, glycosylation of HCV envelope protein may play an important role at various stages of the viral life cycle. Investigation of the role of N-glycans in functioning of the HCV envelope proteins of different genotypes is important for understanding of the mechanisms of viral infection and the development of new antiviral drugs. In this work we used the E1 and E2 glycoproteins of hepatitis C virus genotype 1b (strain 274933RU [14]) with point mutations in the N-linked glycosylation sites to determine the role of these glycans in the HCV envelope proteins functioning. Protein mutants were analyzed in the model system of virus-like particles in insect Sf9 and mammalian Hek293T cells using the baculovirus expression system. The data obtained demonstrate the influence of N-glycans localized in specific sites of glycosylation of E1 and E2 on their interactions, E1E2 heterodimer formation and HCV VLP assembly. Mutations at these positions do not prevent the formation of VLPs in insect and mammalian cells. VLPs have a density typical of HCV particles and bind to CD81-hepatoma cell in an independent manner. Computer calculations of 3D models of the mutant E1 protein molecules complement the experimental data.
2. Materials and methods
2.1. Bacterial strains, cells, and plasmids
Escherichia coli, insect Sf9, and mammalian Hek293T and Huh7.0 cells were used. Bacterial cells of the DH5a and DH10Bac strains (Gibco-BRL, USA) were transformed with the plasmid DNA according to the recommendations of the manufacturer (Amersham, USA). Isolation and purification of plasmids, restriction endonucleasedigestion, ligation, agarose gel electrophoresis of DNA, and other genetic engineering experiments were carried out using standard protocols [15].
2.2. Insect cells
The Spodoptera frugiperda Sf9 cells were cultured at 27 °C in a Sf-900 II medium supplemented with 10% fetal bovine serum, following the basic procedures that were previously discussed and described in the protocol [16]. To determine the virus titer, amplification of recombinant virus, infection of Sf9 cells with recombinant baculovirus, and analysis of viral gene expression, the same protocol was used.
2.3. Human embryonic kidney
293T cells (Hek293T) were cultured at 37 °C and 5% CO2 in DMEM media supplemented with 10% fetal bovine serum, 4 mM l-glutamine, 1 mM sodium pyruvate, and streptomycin/penicillin at the concentrations of 100 μg/mL and 100 U/mL, respectively.
2.4. Recombinant constructs
Carrying cDNAs corresponding to the genes of HCV structural proteins, recombinant bacmids, as well as recombinant baculovirus bv-CE1E2, were obtained and analyzed according to standard protocols and previously tried techniques [17].
2.5. Site-directed mutagenesis
Using oligonucleotide-directed mutagenesis with double-stranded plasmid vectors, point mutations were introduced in the N-linked glycosylation sites of the HCV E1 and E2, and the genetic constructs coding the mutant proteins were obtained. To introduce mutations into prescribed positions of the E1 and E2 cDNA sequences, oligonucleotide primers, presented in Table 1, were designed.
Table 1.
Primers used for site-specific mutagenesis.
| Primer | Orientation | Nucleotide sequence 5′→3′ |
|---|---|---|
| 16-Haml | + | ccactacgacaatac g |
| 15-Hamr | – | gag caagtcgacgtg |
| 26-E1N5tl | + | ggactgccaatgctcaatctatcc cg |
| 17-E1N6tl | + | tggtcacttacaacagc |
| 23-E1N5br | – | gag cat tggcagtcctgc act gt |
| 17-E1N6br | – | ctgttgtaagtgaacag |
| 17-E1N7tl | + | gaactggtctaaggttc |
| 17-E1N7br | – | aaccttagaagaccaggt cc |
| 24r-Hpsr | – | cagctgcagttacccgtcaacgcc |
| 29-E1N1m | – | ttatgaagtgcgccaagtgtccgg cat at |
| 30-E1N2m | – | aacgactgctcccaatcaagcattgcg tat |
| 27-E1N3m | – | gttcag gag ggtcaaagctcccgttgc |
| 24-E1N4m | – | cgcggccaggcaagccagcgtccc |
| 19 Haml | – | cactacgacaatacgacg t |
| 18 Hamr | – | agcaagtcgacgtgacgt |
| 28-E2N1m | – | tgaat acc caaggcagctggcac at |
| 30-E2N2m | – | tggcacatccaaagt act gccctaaattgc |
| 30-E2N3m | – | gccctaaattgccaagactccctccaa act |
| 30-E2N4m | – | gcacacaagttccaatcgtccgggtgcccg |
| 25-E2N6m | – | tggggg gag caa gag acagacgtg a |
| 30-E2N7m | – | gtgatgctcctccaaaacacgcgtccgcca |
| 30-E2N8m | – | tgtacatggatgcaaagt act gggttc act |
| 27-E2N9m | – | ggggtcggtcaacgc acc ttgaattgc |
| 30-E2N10m | – | tacccctgc act ctccaatttttc cat cat |
| 27-E2N11m | – | gccgcatgccaatgg act cgagga gag cgc |
| E2 for | + | aggtctagaatgttatgattgttttgctac |
| E2 back | + | ct atagtgtcacctaaatccgaaagcttcggcctcagcttgag |
Each primer consisted of 16–30 nucleotides and contained the sequence encoding the N-glycosylation site: Asn-X-Thr/Ser-X (X-any amino acid, except proline), in which the triplet encoding asparagine (Asn) was substituted with the triplet encoding glutamine (Gln). The method described by Drutsaet al. [18] was used for mutagenesis. PCR was performed on the CycloTemp 107 programmable thermocycler (ResursPribor, Russia). Predetermined base substitutions were verified by sequencing. The DNA fragment containing cDNA sequence of the gene encoding the HCV glycoprotein E1 was cloned into the pBluescriptKS(-) plasmid at the ClaI-PstI sites. The DNA fragment containing cDNA sequence of the gene encoding the HCV glycoprotein E2 was cloned into the pFastBacHTb plasmid at the NcoI–EcoRI sites. This made possible to construct a series of recombinant pBluescriptKS-E1mut and pFastBacHTbE2mut plasmids carrying cDNA of the E1 and E2 proteins with point substitutions at glycosylation sites. As a result, the baculovirus expression vectors carrying the cDNA of E1mut (pFastBacHTbE1mut, pFastBacHTbE1mutE2, and based on the pFastBacMam1GFP, the BacMamCE1mutE2GFP vector DNA) and the cDNA of E2mut (pFastBacHTbE2mut, pFastBacHTbE1E2mut, pBacmamCE1E2mutGFP) were obtained.
2.6. Analysis of total cellular DNA
Total cellular DNA was isolated from insect cells 72 h after infection with the bv-CE1E2, bv-E2mut, bv-E1mut, bv-E1E2mut, bv-E1mutE2, bv-CE1E2mut, bv-CE1mutE2 recombinant baculoviruses (multiplicity of infection being 5 CFU per cell) [19]. The presence of cDNA of HCV structural proteins in total cellular DNA was assessed by PCR with primers for the pFastBacHT baculovirus vector (forward 5′-GTGGTTGGCTACGTATACTCC-3′ and reverse 5′-CCTCTACAAATGTGGTATGGC-3′).
2.7. Analysis of HCV RNA by RT-PCR
The Sf9 cells were infected with the bv-CE1E2mut recombinant baculoviruses (5 CFU per cell) and incubated at 27 °C for 72 h. After 72 h, the medium was eliminated and cell debris was removed by low-speed centrifugation. The supernatant was centrifuged through the 30% sucrose cushion at 23000g for 16 h at 4 °C (Beckman Coulter Optima L-100XP centrifuge, 80Ti rotor). RNA was extracted from the pellet with TRIzol (Invitrogen) according to manufacturer's protocol and treated with DNase I (Promega). Reverse transcription was carried out using the Phusion RT-PCR Kit (Thermo Scientific). The obtained cDNA was amplified using PCR with primers for the genes of structural and nonstructural HCV proteins. Total cellular RNA was extracted from Sf9 cells infected with bv-CE1E2mut or bv-CE1mut E2 (5 CFU per cell) that were incubated at 27 °C for 72 h and washed three times with PBS. RNA isolation, reverse transcription, and amplification were performed using the above-mentioned protocols. The Hek293T cells were transfected with the BacMamCE1E2mut-GFP recombinant plasmids or the BacMamCE1mut E2-GFP (5 CFU per cell) and incubated at 37 °C for 48 h. The medium was removed and RNA isolation, reverse transcription, and amplification were performed using the above-mentioned protocols.
2.8. Anti-HCV antibodies
Mouse monoclonal antibodies to the HCV E1 (Hep C E1 1879: sc-65459) and HCV E2 (Hep C E2 BDI167: sc-57769) (Santa Cruz Biotechnology, USA) proteins, as well as monoclonal antibodies to calnexin (AF18) and calreticulin (FMC75) (Abcam, UK), were used. Rabbit polyclonal anti-protein C antibodies were kindly provided by M.G. Isaguliants (Ivanovsky Institute of Virology, Russian Academy of Sciences, Moscow). Anti-mouse IgG antibodies (AB6706-1EA) conjugated to horseradish peroxidase (Sigma, USA) were used as secondary antibodies.
2.9. Western blotting and immunoprecipitation
After 72 h of infection with the bv-E2mut, bv-E1mut, bv-E1E2mut, bv-E1mutE2, bv-CE1E2mut, bv-CE1mutE2 recombinant baculoviruses (multiplicity of infection of 5 CFU per cell), the Sf9 cells were harvested, washed three times with PBS (1.47 mM KH2PO4, 4.29 mMNa2HPO4·7H2O, 137 mM NaCl, 2.68 mM KCl), resuspended in the TNC lysis buffer containing 0.25% digitonin, and disrupted by sonication. Cell debris was removed by centrifugation (15,000g, 15 min, 4 °C). Cell lysate was loaded into a 12% denaturing gel (each sample contained 10 μg of the protein). After electrophoresis, the proteins were transferred onto the Hybond-ECL nitrocellulose membrane (Amersham Biosciences, USA) using semi-dry electrophoretic transfer. The membranes were washed with PBS containing 5% nonfat dry milk, incubated with primary antibodies to the HCV structural proteins E1 or E2 (dilution 1:1500 for E1 and 1:2000 for E2), to calnexin or calreticulin (dilutions 1:1000 and 1:2000, respectively), and then to secondary antibodies (dilution 1: 20,000). Protein complexes on immunoblots were detected using the ECL and ECL Plus chemiluminescent reagents (Western blotting detection reagents and analysis systems, Amersham Biosciences, USA) as recommended by the manufacturer. For immunoprecipitation, the cells infected with the recombinant baculoviruses were harvested after 72 h of infection; the cells were lysed, and the cell debris and nuclei were removed. Structural proteins and their complexes were precipitated by monoclonal antibodies to HCV E1 and HCV E2, calnexin and calreticulin in a dilution of 1:1000 (as recommended by the manufacturer). The precipitated proteins were separated by electrophoresis in 12% PAGE, transferred onto a nitrocellulose membrane, incubated with primary antibodies in the above-mentioned dilutions, and then, treated with secondary antibodies.
2.10. Endoglycosidase H (Endo H) treatment
The proteins of cell lysate were incubated with the corresponding monoclonal antibodies at 4 °C. The obtained complex was precipitated with protein G sepharose (BioVision, USA). The precipitated protein (20 μg) was mixed with 1 μl of the 10× denaturing buffer (5%SDS, 0.4 M DTT), the volume of the mixture was adjusted to 10 μl with water, and the mixture was boiled for 10 min. Next, the total volume was adjusted to 20 μl by adding 2 μl of the 10×G5 reaction buffer (50 mM sodium citrate), 3 μl of water, and 5 μl of the Endo H solution (5 units) (P0702S BioLabs Inc., United Kingdom). The mixture was incubated for 15 min at 37 °C and analyzed by electrophoresis in 12% PAGE.
2.11. Preparation and purification of virus-like particles (VLP)
The cells growing in monolayer at 27 °C infected with the bv-CE1E2 recombinant baculovirus (10 CFU per cell). After 72 h, the cells (2×108) were harvested, washed three times with PBS, resuspended in TNC lysis buffer (10 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1 mM CaCl2, 1 mM PMSF), protease inhibitors cocktail II (Calbiochem, USA) (1:200) containing 0.25% digitonin, and disrupted by sonication. VLPs extracted from homogenized lysates were purified from cell debris with the help of low-speed centrifugation (1200g, 15 min, 4 °C). Then, VLP were concentrated by centrifugation through the 30% sucrose cushion at 23,000g for 16 h at 4 °C. The VLP precipitate was re-suspended in TNC buffer containing 1 mM PMSF, protease inhibitors (1:200), and analyzed using sucrose gradient centrifugation.
2.12. Sucrose gradient centrifugation
The VLP precipitate re-suspended in a 100 μl of TNC buffer containing 1 mM PMSF and protease inhibitors (1:200) was placed layer after layer on the sucrose solutions of different concentration (from 10 to 60% in 50 mM Tris–HCl, 100 mM NaCl, pH 7.4) and centrifuged at 20,000g for 2.5 h at 4 °C (Beckman Coulter Optima L-100XP centrifuge, 80Ti rotor). Ten 1-mL fractions were collected, and each fraction was concentrated by high-speed ultracentrifugation at 230,000g for 16 h at 4 °C. The precipitate was dissolved in 100 μl of TNC buffer containing 1 mM PMSF and protease inhibitors (1:200) and analyzed using Western blotting [15], [19].
2.13. Analysis of the VLP binding to CD-81 receptor
Huh7 cells were incubated in the presence of VLP, produced in Sf9 cells, for 1 h at 4 °C. Huh7 cells pre-incubated with anti-CD81 antibodies (20 μg/mL) for 1 h at 4 °C to block the CD81 receptor were used as a control. The cells were harvested, washed twice with PBS to remove unbound VLPs, and analyzed using Western blotting with anti-E2 antibodies.
2.14. Fluorescent microscopy and flow cytometry
After transfection for 48 h with recombinant BacMamCE1E2GFP plasmids, Hek293T cells were trypsinized, collected by centrifugation, washed twice in phosphate-buffered saline, and analyzed by flow cytofluorometry (Beckman Coulter EPICS, USA) and Western blotting.
2.15. Mathematical modeling of the 3D structure of HCV E1 mutant protein
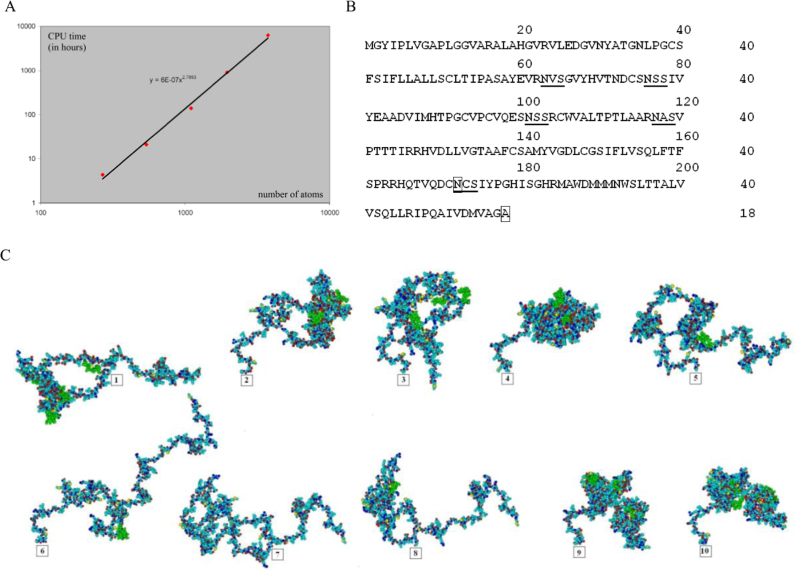
Computer calculations for different fragments of the E1 protein molecules in vacuum and in aqueous environment with the methods of molecular mechanics (energy optimization) and either molecular, or Langevin dynamics were used. The calculations were performed in the HyperChem v.6.01 computational chemistry program (Hypercube, Inc USA) [20], [21], which contains the databases for introduction of amino acids using the 1800 MHz AMD processor. To estimate the shape of the E1 molecule and the effect of N-glycans and water, ten variants of HCV E1 mutant protein (Fig. 1A) with the deleted C-terminal sequence were used. Test calculations were carried out on the E1 fragments of different length and extrapolated to the whole E1 molecule. Five fragments of the protein molecule of different lengths, including 19 amino acids, 268 atoms; 38 amino acids, 538 atoms; 76 amino acids, 1098 atoms; 133 amino acids, 1952 atoms; the whole E1 molecule, devoid of glycans; and 250 amino acids, 3739 atoms were used. Calculations of the protein shape were performed as follows: protein sequence was formed using amino acid residues from the HyperChem database with the angles between the individual atoms within each amino acid residue in the E1 molecules of different length. Then, the angles between atoms of the amino acid residues were set. As a result, the preliminary three-dimensional structure of the E1 protein molecule was obtained. Next, the procedure of the molecule energy minimization was carried out. To optimize the geometry of the E1 protein structure, the positions of all atoms of the amino acid residues were selected to minimize the energy at the time of determining the total energy of the molecule. Free energy, dipole moment, and the geometric dimensions of each structure were calculated, which showed how the molecule under investigation was folded. Then, the calculation of either molecular or Langevin dynamics was run, and then geometric optimization and the process were repeated minimizing the energy of the molecule to the optimum geometry, whereby coiled molecules acquired specific conformation.
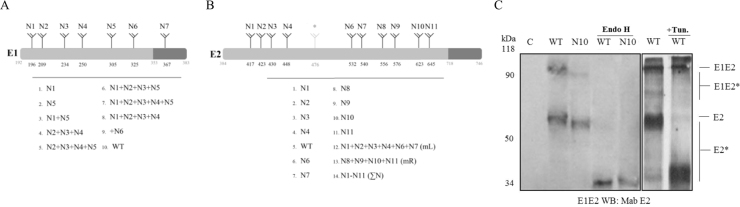
Fig. 1.
N-glycosylation sites of the HCV E1 and E2 structural proteins and their mutant variants. A, schematic representation of the mutant variants of glycoprotein E1 with modified glycosylation sites: 1, N1; 2, N5; 3, N1 and N5; 4, N2, N3, and N4; 5, N2, N3, N4, and N5; 6, N1, N2, N3, and N5; 7, N1—N5; 8, N1, N2, N3 and N4; 9, with additionally introduced N6 glycosylation site; 6, initial wildtype E1 variant. B, schematic representation of the mutant variants of glycoprotein E2 with modified glycosylation sites: 1, N1; 2, N2; 3, N3; 4, N4; 5, initial wildtype E2 variant, containing in 4% of genotype 1b strains glycosylation site N5; 6, N6; 7, N7; 8, N8; 9, N9; 10, N10; 11, N11; 12, N1, N2, N3, N4, N6, N7(mL); 13, N8, N9, N10, N11(mR); 14, N1-N11(ΣN). C, expression analysis of the HCV envelope-encoding genes synthesized in insect cells infected with the bv-E1E2 baculovirus. Western blot analysis of glycoprotein expression in the presence and absence of tunicamycin and glycoproteins treated with EndoH endonuclease, in denaturating 12% PAGE with anti-E2 antibodies. Lysates of the cells infected with recombinant baculoviruses synthesizing E1 and E2: WT, wildtype E1E2; N10, E2 with mutations at the N10 glycosylation site; C, negative control (Hsp90); kDa, protein molecular size markers.
3. Results and discussion
The study of conformational changes and the interactions between HCV envelope proteins and their glycosylation in the model system of virus-like particles formed in the cells of insects and mammals makes it possible to understand the role of glycosylation of the genotype 1b HCV in viral morphogenesis. The glycosylation pattern of viral genotype 1b differs from the extensively studied genotypes 1a and 2a. This difference consists in the presence of the N4 glycosylation site in glycoprotein E1, and the absence of N5 glycosylation site in the HCV E2 protein. There are considerable differences in the functions of individual glycans within different genotypes of hepatitis C virus. In this study, the role of glycans associated with the envelope proteins of HCV genotype 1 b (strain 274933RU) in the protein folding and assembly of viral particles in two different cell types were analyzed in the baculovirus expression system, using site-directed mutagenesis of glycoproteins [14], [17]. Point mutations were introduced into the predetermined positions of the E1 and E2 cDNA nucleotide sequences harboring N- glycosylation sites, using the appropriate primers (Table 1). Based on the baculovirus expression vectors, pFastBacHTb and pFastBacMam1GFP, carrying the E1 and E2 sequences with point substitutions at glycosylation sites (see Section 2), in insect and mammalian cells, mutant HCV envelope proteins were synthesized (Fig. 1A and B).
3.1. The effect of N-glycans on expression of viral envelope protein genes in insect and mammalian cells
It is known that N-glycosylation pathways in the cells of Spodoptera frugiperda, which may lack the appropriate glycosyltransferases, are not always completely the same as in mammalian cells. Fig. 1С shows that the synthesis of HCV envelope proteins, as well as their effective post-translational glycosylation takes place in insect cells. This concerns both wildtype proteins and their mutant variants, sensitive to the presence of tunicamycin and EndoH endoglycosidase.
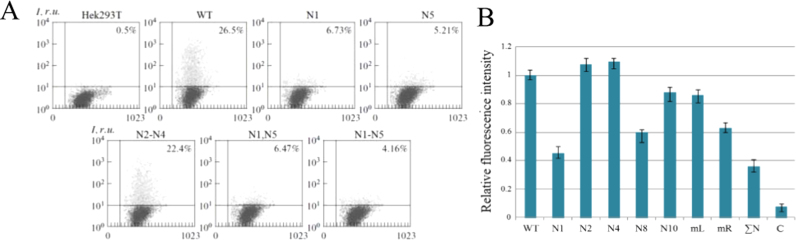
In Sf9 insect cells the disruption of glycosylation sites in various combinations of E1 and E2 glycoproteins (except for the N6 site), had no effect on the synthesis efficiency of these proteins, which follows from the results of electrophoresis in PAGE and immunoblotting with antibodies to these proteins (data not shown). At the same time, flow cytometry data showed that in Hek293T mammalian cells, the absence of carbohydrate chains in the E1 protein N1 and N5 sites and in the E2 protein N1, N8, and N10 sites resulted in considerable reduction of the expression level of these glycoproteins as the parts of CE1E2GFP polypeptides, compared to the initial polypeptides. These data are supported by the decrease in the GFP fluorescence (Fig. 2A and B).
Fig. 2.
Gene expression analysis of mutant E1 and E2 proteins within the HCV CE1E2 polypeptide in mammalian cells.A, Assessment of the GFP fluorescence in HEK293T cells transfected with the pFastBacMamCE1E2GFP plasmids with different mutations in the E1 gene by flow cytofluorometry. Linear sizes of the examined particles (cells) in relative units are marked on the X-axis of the histogram, and relative values of the fluorescence intensity are marked along the Y-axis. Hek293T, initial cells; cells transfected with the pFastBacMamCE1E2GFP with the E1 gene: WT, wildtype E1; E1 with mutation at the N1 glycosylation site; N5; N2, N3, and N4; N1 and N5; N1—N5. B, Assessment of the GFP fluorescence in HEK293T cells transfected with pFastBacMamCE1E2GFP carrying mutations in the E2 gene by flow cytofluorometry. Relative values of the fluorescence intensity are plotted on the Y-axis; On the X-axis, cells expressing: WT, wildtype E2; E2 with mutations at N1, N2, N4, N8, N10, mL(N1—N7), mR(N8—N11), and ΣN(N1—N11) glycosylation sites are marked; C, negative control (Hsp90).
3.2. The effect of N-glycans of HCV envelope glycoproteins on the formation of productive E1E2 complex in insect cells
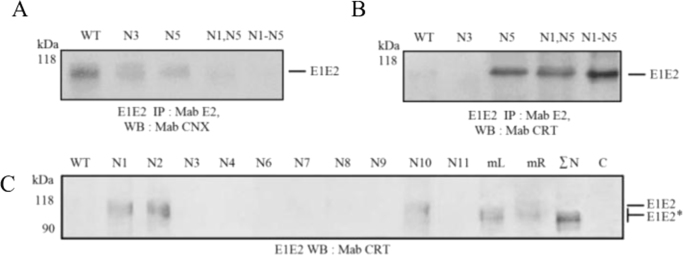
It is suggested, that glycans associated with the viral envelope, affect the correct folding of glycoproteins with the participation of ER chaperones [10]. Glycoproteins are then combined into functional complexes defining productive assembly of viral particles in the cells. The effect of mutations in glycosylation sites of HCV E1 and E2 proteins on their folding and the formation of E1E2 complexes in insect cells was assessed by their interaction with cellular chaperones calnexin and calreticulin. Apparently, in insect cells, the mutant glycoproteins E1 and E2 specifically interact with calnexin, indicating the formation of correctly folded functional proteins and the E1E2 heterodimers. At the same time, aggregates of misfolded E1E2 dimers interact with calreticulin [1]. It was found that assembly of noncovalently bound HCV E1E2 complex in insect cells depended on the disruption of N1 and N5 glycosylation sites of E1 and was broken as a result of disruption of one of the N1, N2, or N10 sites of the HCV glycoprotein E2 (Fig. 3A and B).
Fig. 3.
Gene expression analysis of mutant E1 and E2 proteins within the HCV E1E2 in Sf9 cells. A, Western blotting in 10% PAGE with anti-calnexin antibodies and B, with anti-calreticulin antibodies, after preliminary immunoprecipitation with anti-E2 antibodies. Lysates of the cells infected with recombinant baculoviruses expressing E1: WT, wildtype E1; E1 with mutations at the glycosylation sites N3; N5; N1 and N5; N1 and N5; N1—N5. kDa, protein molecular size markers. C, Gene expression analysis of mutant E2 proteins of HCV in Sf9 cells.Western blotting in 10% PAGE with anti-calreticulin antibodies. Lysates of the cells infected with recombinant baculoviruses expressing E2: WT, wildtype E2; E2 with mutations at N1, N2, N3, N4; N6; N7, N8; N9; N10; N11; N1—N7(mL); N8—N11(mR); N1—N11(ΣN), at all E2 glycosylation sites; C, negative control (Hsp90); kDa, protein molecular size markers. Mutant proteins are designated as E2*.
It seems likely that mutations at these E1 and E2 glycosylation sites prevent the proteins constituting functional E1E2 complex from assuming their proper conformation, while mutations at other sites do not disturb this process. As the number of damaged sites increases (N1–N7 (mL) and N8–N11 (mR)), assembly of the productive E1E2 complex reduces with the formation of the aggregates of the misfolded E1E2 dimers, interacting with calreticulin (Fig. 3C).
Thus, it is obvious that the glycans associated with the HCV E1 glycosylation sites N1 and N5, and E2 glycosylation sites N1, N2, and N10 are important to stabilize the structure of these proteins, and play an important role in their correct conformation when forming a functional complex and, accordingly, in the productive assembly of virus particles in Sf9 cells.
3.3. The effect of N-glycans of HCV envelope glycoproteins on the formation of virus-like particles in insect and mammalian cells
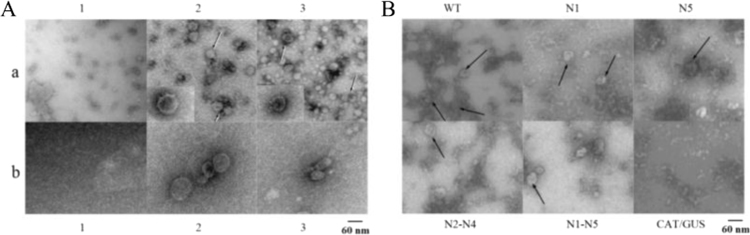
It is known that efficient expression of the HCV structural proteins in insect cells is accompanied by the formation of virus-like particles. We showed that in these cells infected with the bv-CE1E2 recombinant baculovirus the synthesized VLP were localized on the ER membranes and were not secreted into the culture medium. Electron microscopy of microsomal and purified VLP preparations is demonstrated in Fig. 4A, and electron microscopy of VLP in the microsomal fractions of Sf9 cells infected with the bv-CE1E2 baculoviruses synthesizing mutant E1 is shown in Fig. 4B. (see Section 2).
Fig. 4.
A, electron microscopy of microsomal preparations (a) and purified VLP preparations (b). The preparations were obtained from the Sf9 cells infected with recombinant baculoviruses: 1, bv-CAT/GUS (control); 2, bv-CE1E2; 3, bv-C+E1+E2 (co-transfection); B, electron microscopy of VLPs in microsomal fractions of the Sf9 cells infected with bv-CE1E2 baculoviruses, synthesizing: WT, wildtype E1; E1 with mutation at the N1; N5; N2, N3, and N4; N1—N5 glycosylation site; CAT/GUS, negative control. The position of VLP is indicated by the arrows.
It was found that the absence of glycans in the E1 glycosylation sites had no effect on the VLP formation in insect cells (Fig. 4B). These results were also confirmed by PAGE electrophoresis and Western blotting with the antibody to E2 (data not shown).
The effect of the glycosylation sites disruption in E2 glycoprotein as the component of CE1E2 on the assembly of functional glycoprotein complexes, and respectively, the CE1E2 packaging in VLP in Sf9 and Hek293T cells, was assessed by specific interaction of mutant glycoproteins with calnexin and calreticulin. Gene expression analysis of mutant E2 proteins as the components of HCV CE1E2 in mammalian Hek293T cells is demonstrated in Fig. 5A and B.
Fig. 5.
Gene expression analysis of mutant E2 proteins as parts of the HCV CE1E2 in mammalian Hek293T cells. A, Western blotting in denaturating 10% PAGE with anti-calnexin antibodies and B, with anti-calreticulin antibodies. Lysates of the cells transfected with the recombinant pFastBacMamCE1E2mutGFP plasmids synthesizing E2 with mutations at glycosylation sites: N1; N2; N4; N8; N10; N1—N7(mL); N8—N11(ΣN), at all glycosylation sites; WT, wildtype E2; C- negative control (Hsp90). C, with anti - β– actin antibodies; kDa, protein molecular size markers; Mutant proteins are designated as E2*.
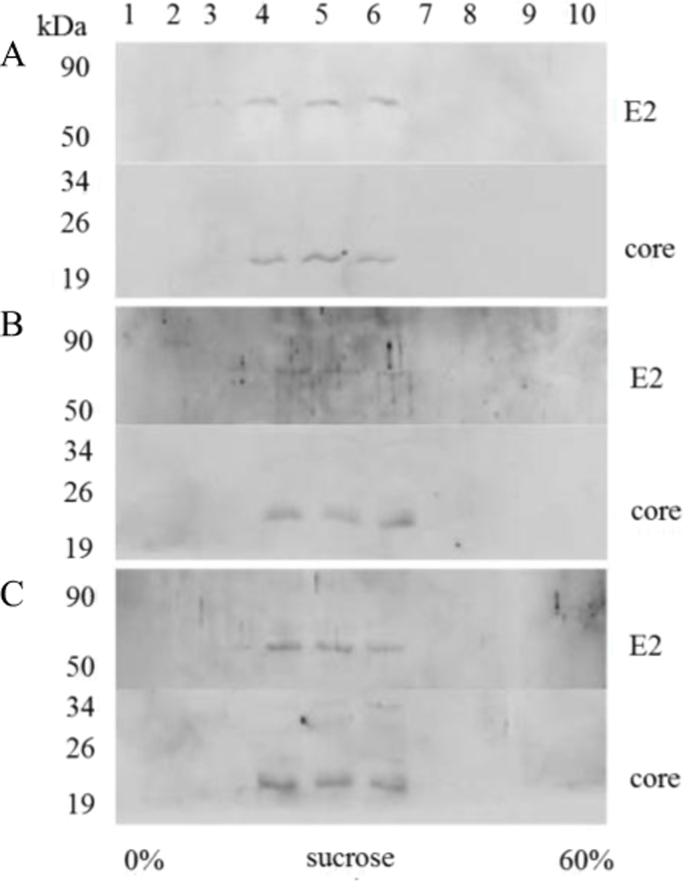
Analysis of gene expression of mutant E2 proteins as parts of CE1E2 in insect (data not shown) and mammalian cells showed that disruption of glycosylation sites in different combinations (except for the N6 site) had no effect on formation of the functional E1E2 heterodimers, and correspondingly, on the CE1E2 packaging within HCV. At the same time, disruption of the E2 N1, N2, and N10 sites led to formation of the aggregates of misfolded glycoproteins in the E1E2 complex that also did not prevent the formation of VLPs. Obviously, the produced VLP contain nonfunctional E1E2 complexes with misfolded mutant glycoproteins. The HCV-like particles formed in Hek293T cells, transfected with recombinant pFastBacMamCE1E2mutGFP, were also evaluated for their physical characteristics. For these purposes, VLPs were purified and concentrated by centrifugation through the cushion of 30% sucrose at 23,000g, and the VLP precipitate was then analyzed using centrifugation in sucrose gradient. The collected fractions were compared by sedimentation at a density typical for the hepatitis C virus particles (from 1.14 to 1.16 g/cm3) and analyzed by Western blotting using antibodies to structural proteins (Fig. 6A, B, and C).
Fig. 6.
Analysis of the HCV-like particles isolated from Hek293T cells by centrifugation in sucrose gradient. Western blotting of ten VLP fractions, top-to-bottom, in denaturating 10% PAGE, with antibodies to HCV C (Core) and E2. Hek293T cells were transfected with recombinant pFastBacMamCE1E2mutGFP: A, intact DNA; B, DNA with mutation at the E2 N1 glycosylation site; C, with mutations at all E2 glycosylation sites; Fractions 1—10; kDa, protein molecular size markers.
Immunoblotting using antibodies to the C (Core) and E2 proteins revealed that VLPs were present in all fractions with the density of 1.14–1.16 g/cm3. This finding can be an indication that RNA fragments are present in VLPs [22].
As follows from the data obtained, the introduction of mutations at glycosylation sites of virus envelope glycoproteins does not prevent the formation of virus-like particles of the hepatitis C virus in insect and mammalian cells. However, it seems likely that HCV VLPs formed with the participation of E1N1, E1N5, E2N1, E2N2, and E2N10 mutant glycoproteins may contain the protein E1E2 complex with the disrupted conformation that prevent the formation of the proper virus particles in the cells. To confirm our results and the conclusion on defective conformation of certain mutant HCV envelope glycoproteins synthesized in insect cells, computer models of the spatial structure of mutant E1 proteins, with and without N-glycans were constructed.
3.4. The effect of N-glycans of hepatitis C virus E1 envelope glycoprotein on its 3D structure
One of the possible ways to confirm the results obtained in a chemical experiment is the mathematical modeling of the 3D protein structure with or without glycans. For these purposes, in this study, using the method of molecular dynamics as implemented in the HyperChem software program, optimal geometry of the E1 molecules were calculated and a computer model of the spatial structure of the mutant E1 protein was constructed. Since in the HyperChem program the geometry optimization procedure results in the loss of time in comparison with the computation of the same molecule, for example, in the NAMD and GROMACS programs, we compensated this drawback by increasing the processing time spent for the calculation. We examined the molecular conformations of ten HCV E1 protein variants with the deleted C-terminal sequence (Fig. 7) and with different number of glycans and their arrangement. Test calculations were carried out using fragments of the molecules of different lengths and extrapolated to the whole E1 molecule. To select the criteria necessary for the full calculation of the optimum geometry of E1 molecules, both wildtype and mutant, test calculation of the E1 protein fragment of 38 aa (538 atoms), devoid of all glycans was performed. The computation showed that the total energy and geometrical characteristics simultaneously came to the value characterizing optimal configuration of the E1 fragment (graphs not shown). Determination of the effect of the molecular dynamics computation time on the geometry of molecule indicated that the time step had no effect on the final geometry of the molecule. The achievement of the optimum geometry of the molecule depends on the total time duration. The most informative criterion for the optimum geometry of the protein molecule is that the molecular energy has reached its plateau value. To find the time-dependence between the Langevin dynamics counts and the number of atoms in a molecule, required to reach a plateau of minimum energy, we conducted a test calculations for five E1 protein fragments of varying length (see Section 2). The total energy and geometric characteristics were determined in each of five E1 fragments. For all five E1 molecules, the time-to-reach a plateau was about 75, 150, 300, 500, and 1200 ps. Using these data, we determined the time required for the full calculation of the optimum geometry of the E1 molecules on the PC used. The dependency between the computer processing time and the number of atoms in the E1 molecule required for the full calculation of the optimum geometry of the molecules is shown in Fig. 7A.
Fig. 7.
A, Dependency between the computer processing time and the number of atoms in the E1 molecule required for the full calculation of the optimum geometry of the molecule. On abscissa, the number of atoms; on ordinate, CPU time in hours. Three-dimensional structure of mutant HCV E1 protein molecules: B, Amino acid sequence of wildtype HCV E1 protein with 218 aa. The glycan attachment sites are underlined.  , the first amino-acid residue protruding from the membrane.
, the first amino-acid residue protruding from the membrane.  , the closest to the membrane asparagine to which glycan is attached; C, Computer models of mutant E1 glycoprotein variants with the disrupted glycosylation sites (N1—N6): 1, N1; 2, N5; 3, N1, N5; 4, N2, N3, and N4; 5, N2, N3, N4, and N5; 6, N1, N2, N3, and N5; 7, N1—N5; 8, N1, N2, N3, and N4; 9, with the additional glycosylation site N6; 10, initial variant of wildtype E1;
, the closest to the membrane asparagine to which glycan is attached; C, Computer models of mutant E1 glycoprotein variants with the disrupted glycosylation sites (N1—N6): 1, N1; 2, N5; 3, N1, N5; 4, N2, N3, and N4; 5, N2, N3, N4, and N5; 6, N1, N2, N3, and N5; 7, N1—N5; 8, N1, N2, N3, and N4; 9, with the additional glycosylation site N6; 10, initial variant of wildtype E1;  –
–  , the E1 membrane-anchoring site. The E1 protein is oriented so that the
, the E1 membrane-anchoring site. The E1 protein is oriented so that the  locates at the origin of the axis Oy, and
locates at the origin of the axis Oy, and  lies in the picture plane or in the plane Oxy. The scale of each protein variant was adjusted so that the sizes of all carbon atoms (light blue), or nitrogen (blue), or oxygen (red) or sulfur (yellow) were the same for all ten variants. Hydrogen is not shown. Glycans are in green color.
lies in the picture plane or in the plane Oxy. The scale of each protein variant was adjusted so that the sizes of all carbon atoms (light blue), or nitrogen (blue), or oxygen (red) or sulfur (yellow) were the same for all ten variants. Hydrogen is not shown. Glycans are in green color.
Interestingly, extrapolation of power dependence for 3D protein structure on the graph shows the exponent=2.7853, close to three. A similar relationship is shown in [23]. The amino acid sequence of the wildtype HCV E1 genotype 1b protein (strain 274933RU) with 218 aa and the calculated forms of ten HCV E1 structural protein variants with mutations are shown in Fig. 7B and C.
The data obtained demonstrate various mutant protein structures that differ from the structure of the original wildtype HCV E1 protein (Fig. 7C, No. 10). The absence of carbohydrate chains at the N1 or N5E1 glycosylation sites leads to misfolding of these proteins and formation of the similar spatial structures (Fig. 7C, No.1, N1; No.2, N5; No.3, N1, N5), that considerably differ from the structure of the wildtype protein. These simulations favor the suggestion that glycans, associated with the N1 and N5 sites, play the most important role in proper E1 folding, required for the formation of functional E1E2 complex. Three-dimensional structures of the other mutant protein variants showed the differences in the conformation of the molecules lacking one or more glycosylation sites. However, these differences, as it was demonstrated in our in vitro biochemical experiments, did not lead to the formation of unproductive E1E2 complexes. Thus, 3D structure models of the E1 protein with mutations show that the N-linked glycans directly affect the E1 folding, the shape of the molecule, and, apparently, can play an important role in functioning of these proteins. Molecular modeling data obtained in this study are generally consistent with our experimental data.
4. Conclusion
In this study, we showed that the HCV (genotype 1b, strain 274933RU) structural proteins are synthesized in the infected Sf9 insect and Hek293T mammalian cells using the baculovirus expression system. These proteins are subjected to post-translational modifications and form virus-like particles that are localized in ER membranes of the infected cells. The characterization of these particles by sucrose gradient centrifugation has revealed physical properties similar to virus-like particles with the density of 1.14–1.16 g/cm3 and containing RNA fragments. These results indicate that glycosylation of viral envelope proteins expressed in Sf9 and Hek293T cells affect their expression, processing and formation of virus particles. Introduction of mutations at the glycoprotein glycosylation sites does not prevent the formation of virus-like particles in insect and mammals cells. However, the disruption of the HCV 1b E2 protein N1, N2, and N10 glycosylation sites, as well as E1 N1 and N5 sites, results in the formation of mostly unproductive E1E2 complexes. Earlier Goffard et al. and Helle et al. showed that N1- and N5-linked glycans of the HCV genotype 1a E1 protein also affected folding of viral envelope proteins and the correct folding of viral envelope glycoproteins of HCV genotypes 1a and 2a was affected by the disruption of N-linked glycans of the E2 protein, different from those examined in our study [1], [2]. It is known that some E2 glycans, depending on the strain of the virus, are critical for entry into the cell and its infection. The E2 glycosylation sites are highly conservative, and that indicates their important role in the HCV life cycle. Binding of the virus particle to the hepatocyte cell-surface receptors and its uptake into the cell is determined by viral envelope glycoproteins. It was demonstrated that the effect of the CD81 receptor on binding of the HCV-like particles of different genotypes to various cells (HepG2, Huh7, NKNT-3, Molt-4) with the participation of envelope proteins could be different [24], [25]. We showed that HCV genotype 1b virus-like particles, formed in the model baculovirus expression system, bound to Huh-7 cells irrespective of the presence of the CD81 receptor on their surface and that mutations in the HCV E2 glycosylation sites did not affect binding of viral particles to the CD81 receptor (publication in press). The obtained mathematical models of 3D structure of the mutant HCV E1 protein with different number of glycans and their arrangement agree with the experimental data of our study. They confirm that the N-linked glycans have a direct effect on the protein folding, demonstrating considerable differences in the configuration of such molecules.
Thus, for the HCV genotype 1b (strain 274933RU) it was demonstrated that in Sf9 and Hek293T cells glycosylation of viral envelope glycoproteins combined into functional glycoprotein complexes affects the virion formation, which depends on correct glycoprotein folding. According to our results, a combination of experimental and computational approaches can be used in further studies, associated with the effect of glycoprotein mutations on viral replication, secretion of virus particles, as well as in those, assessing the role of individual glycans of the HCV glycoproteins in the interaction with cell receptors and viral entry into the cell.
Acknowledgements
This research was supported by grants from the Russian Foundation for Basic Research (projects nos. 11-04-00231; 13-04-01837; 14-04-00792), the Ministry for Science and Education of the Russian Federation (state contract no. 16.512.11.2266) and program “Molecular and Cellular Biology” RAS.
Footnotes
Supplementary data associated with this article can be found in the online version at 10.1016/j.bbrep.2016.05.019.
Appendix A. Transparency document
Transparency document
.
References
- 1.Goffard A., Callens N., Bartosch B., Wychowski C., Cosset F.L., Montpellier C., Dubuisson J. J. Virol. 2005;79:8400–8409. doi: 10.1128/JVI.79.13.8400-8409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Helle F., Vieyres G., Elkrief L., Popescu C.I., Wychowski C., Descamps V., Castelain S., Roingeard P., Duverlie G., Dubuisson J. J. Virol. 2010;84:11905–11915. doi: 10.1128/JVI.01548-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chou R., Clark E., Helfand M. Ann. Intern Med. 2004;140:465–479. doi: 10.7326/0003-4819-140-6-200403160-00014. [DOI] [PubMed] [Google Scholar]
- 4.Karayiannis P., Main J., Thomas H.C. Br. Med. Bull. 2004;70:29–49. doi: 10.1093/bmb/ldh024. [DOI] [PubMed] [Google Scholar]
- 5.Griffin S., Stgelais C., Owsianka A.M., Patel A.H., Rowlands D., Harris M. Hepatology. 2008;48:1779–1790. doi: 10.1002/hep.22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steinmann E., Penin F., Kallis S., Patel A.,H., Bartenschlager R., PietschmannT PLoS Pathog. 2007;3:e103. doi: 10.1371/journal.ppat.0030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiao X., Wang R.Y., Feng Z., Alter H.J., Shih J.W. Hepatology. 2003;37:452–460. doi: 10.1053/jhep.2003.50051. [DOI] [PubMed] [Google Scholar]
- 8.Burda P., Aebi M. Biochim. Biophys. Acta. 1999;1426:239–257. doi: 10.1016/s0304-4165(98)00127-5. [DOI] [PubMed] [Google Scholar]
- 9.Bas T., Gao G.Y., Lvov A., Chandrasekhar K.D., Gilmore R., Kobertz W.R. J. Biol. Chem. 2011;286:28150–28159. doi: 10.1074/jbc.M111.235168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Op de Beeck A., Cocquerel L., Dubuisson J. J. Gen. Virol. 2001;82:2589–2595. doi: 10.1099/0022-1317-82-11-2589. [DOI] [PubMed] [Google Scholar]
- 11.Montreuil J., Vliegenthart J.F., Schachter H. Elsevier; Amsterdam: 1995. Glycoproteins; pp. 1–12. [Google Scholar]
- 12.Suzuki R., Saito K., Kato T., Shirakura M., Akazawa D., Ishii K., Aizaki H., Kanegae Y., Matsuura Y., Saito I., Wakita T., Suzuki T. Virology. 2012;432:29–38. doi: 10.1016/j.virol.2012.05.033. [DOI] [PubMed] [Google Scholar]
- 13.Steinmann E., Brohm C., Kallis S., Bartenschlager R., Pietschmann T. J. Virol. 2008;82(14):7034–7046. doi: 10.1128/JVI.00118-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.V.V. Mokhonov, D.V. Novikov, E.I. Samokhvalov, A.G. Shatalov, N.A. Selivanov, A.G Prilipov, D.K. L'vov, VoposiVirusology, 47, 2002, pp. 9–12 [PubMed]
- 15.Sambrook J., Fritsch E.F., Maniatis T. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 16.Bac-to-BacBaculovirus Expression System, Instruction Manual. Monsanto Corporate Research, Life Technologies Inc., St. Louis, 1993.
- 17.Belzhelarskaya S.N., Koroleva N.N., Popenko V.I., Drutza V.L., Orlova O.V., Rubtzov P.M., Kochetkov S.N. Mol. Biol. 2010;44(1):97–108. [PubMed] [Google Scholar]
- 18.I.L. Drutsa, V.R. Kaberdin, O.N. Koroleva, I.O. Shilov, BioorganicheskayaKhimiya, 17, 1991, pp. 1487–1493 [PubMed]
- 19.Methods in Molecular Biology, Baculovirus Expression Protocols, Humana press Inc, Totowa, New Jersey, 1995.
- 20.Froimowitz M. Biotechniques. 1993;4(6):1010–1013. [PubMed] [Google Scholar]
- 21.Rostamian M., Mousavy S.J., Ebrahimi F., Ghadami S.A., Sheibani N., Minaei A.E., Torabi M.A. Iran. Biomed. J. 2012;16(4):185–192. doi: 10.6091/ibj.1076.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McEwen C.R. Anal. Biochem. 1967;20(1):114–149. doi: 10.1016/0003-2697(67)90271-0. [DOI] [PubMed] [Google Scholar]
- 23.A.A. Granovsky, Presentation on the large-scale QC and QM/MM modeling Using PC GAMESS, 2006, Available at URL: 〈http://classic.chem.msu.su/gran/gamess/largescale.pdf〉.
- 24.Steinmann D., Barth H., Gissler B., Schurmann P., Adah M.I., Gerlach J.T., Pape G.R., Depla E., Jacobs D., Maertens G., Patel A.H., Inchauspe G., Liang T.J., Blum H.E., Baumert T.F. J. Virol. 2004;78(17):9030–9040. doi: 10.1128/JVI.78.17.9030-9040.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Triyatni M., Saunier B., Maruvada P., Davis A.R., Ulianich L., Heller T., Patel A., Kohn L.D., Liang T.J. J. Virol. 2002;76(18):9335–9344. doi: 10.1128/JVI.76.18.9335-9344.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transparency document