Abstract
Background
In the past few years, an increasing number of studies have reported the potential use of microRNAs (miRNA) as circulating biomarkers for diagnosis or prognosis of a wide variety of diseases. There is, however, a lack of reproducibility between studies. Due to the high miRNA content in platelets this may partly be explained by residual platelets in the plasma samples used. When collecting fresh plasma samples, it is possible to produce cell-free/platelet-poor plasma by centrifugation. In this study, we systematically investigated whether biobanked EDTA plasma samples could be processed to be suitable for miRNA analysis.
Materials and methods
Blood samples were collected from ten healthy volunteers and centrifuged to produce platelet-poor-plasma (PPP) and standard biobank plasma. After one week at −80 °C the biobanked EDTA plasma was re-centrifuged by different steps to remove residual platelets. Using RT-qPCR the levels of 14 miRNAs in the different plasma preparations were compared to that of PPP.
Results
We were able to remove residual platelets from biobanked EDTA plasma by re-centrifugation of the thawed samples. Nevertheless, for most of the investigated miRNAs, the miRNA level was significantly higher in the re-centrifuged biobanked plasma compared to PPP, even when the platelet count was reduced to 0–1×109/L.
Conclusion
We found, that pre-storage centrifugation conditions have a significant impact on the measured EDTA plasma level of miRNAs known to be present in platelets. Even for the miRNAs found to be less effected, we showed that a 1.5–3 fold change in plasma levels may possible be caused by or easily overseen due to sample preparation and/or storage.
Abbreviations: PPP, platelet-poor-plasma; RT-qPCR, reverse transcription polymerase chain reaction; °C, degrees Celcius
Keywords: MicroRNA, Preanalytical conditions, Centrifugation, Residual platelets, Biobanking
Highlights
-
•
Platelet count vary significantly in standard biobanked EDTA plasma.
-
•
Residual platelets can be removed by centrifugation of thawed biobanked samples.
-
•
Re-centrifugation of biobanked plasma cannot eliminate contaminating platelet miRNAs.
-
•
Pre-storage centrifugation conditions have large impact on measured miRNA levels.
1. Introduction
MicroRNAs (miRNAs) are short, single-stranded non-coding RNAs acting as posttranscriptional regulators of gene expression [1]. In the circulation cell-free miRNAs are protected against degradation as they are included in microvesicles or exosomes or they are bound to high-density lipoproteins or to the argonaute 2 protein complex [2]. MiRNAs were first identified in Caenorhabditis elegans in 1993 [3], but it took almost a decade before they were discovered in mammals [4], [5], and the first reports of circulating miRNAs came in 2008 [6], [7]. In the past few years, an increasing number of studies have reported the potential use of miRNAs as circulating biomarkers for diagnosis or prognosis of a wide variety of diseases such as cancers [8], [9], cardiovascular diseases [10], diabetes [11] and mental illness [12]. However, in only a minority of studies where miRNAs were reported to be differently expressed in healthy and diseased individuals, the results have been confirmed by others [13]. Pre-analytical conditions are a major source of variation in and between miRNA studies, and have been addressed by several studies in the past few years [2], [14], [15], [16], [17], [18], [19]. An important factor to consider is cellular remnants in the samples, for example in hemolysis where disruption of erythrocytes causes a significant increase in the plasma/serum levels of many miRNAs [15]. Furthermore, due to residual platelets, the levels of some miRNAs are higher in routine plasma samples compared to routine serum samples [14], and certain other miRNAs are found in higher levels in serum compared to plasma, probably due to their release from leucocytes and/or platelets during the coagulation process [20]. When collecting fresh samples, it is possible by additional centrifugation steps to produce cell-free/platelet-poor plasma, but many studies are performed using biobanked routine plasma samples with variable number of residual platelets. In this study we will systematically investigate whether biobanked EDTA plasma samples can be processed to be suitable for miRNA analysis, when investigating the plasma levels of miRNAs known to be present in platelets.
2. Materials and methods
From each of 10 healthy volunteers 3×10 mL of K2-EDTA anticoagulated venous blood were collected using a 21 gauge needle after a minimum of venous stasis, and discard of the first 5 mL of blood drawn (tubes and needles were obtained from Becton-Dickinson, Franklin Lakes, NJ, USA).
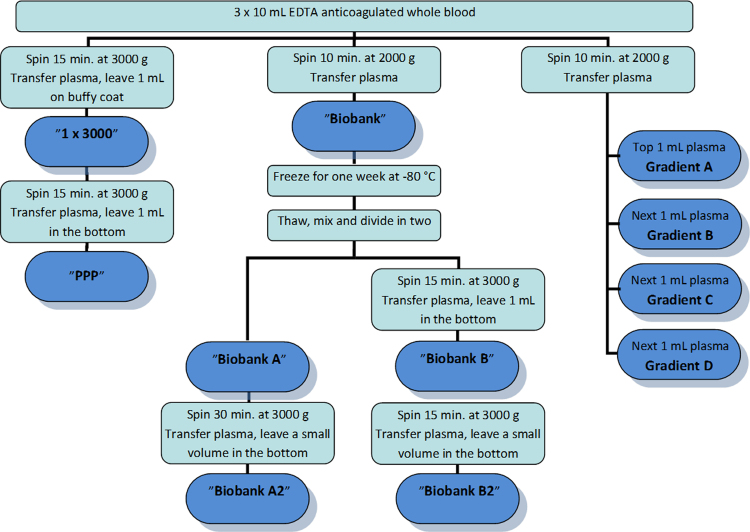
A schematic overview of the sample preparation can be seen in Fig. 1. All the centrifugation steps were carried out at 20 °C.
Fig. 1.
Schematic overview over the different plasma preparations. A total of 10 mL of whole blood was used for the preparation of platelet poor plasma (PPP). Two tubes of 10 mL whole blood were handled according to our laboratory protocol for preparation of biobank plasma. Plasma from one of the tubes was frozen for one week, thawed and further processed to eliminate platelet contamination (biobank A, A2, B and B2). As illustrated, after freezing and thawing of the biobank plasma we denote it biobank A. Plasma from the other tube was pipetted from the top of the tube mL by mL to investigate the distribution of platelets in the plasma phase (gradient A–D).
In order to obtain platelet-poor-plasma (PPP) 10 mL of EDTA anticoagulated whole blood was centrifuged at 3000 g for 15 min. After centrifugation the plasma phase was carefully transferred to another tube, leaving approximately one mL of plasma on top of the buffy coat. The centrifugation step was repeated on the remaining plasma, and again approximately one mL was left in the bottom of the tube when the platelet-poor-plasma was transferred into cryo-tubes (labeled PPP) for storage at −80 °C.
Ten mL of EDTA anticoagulated whole blood was centrifuged at 2000 g for 10 min, which is standard procedure in our laboratory when collecting plasma for biobanking. After centrifugation the total amount of plasma from the 10 mL collection tube was transferred to another tube (labeled biobank), and stored at −80 °C. After one week the biobank-plasma was thawed and gently mixed. A volume of 1.5 mL was transferred to a cryotube (labeled biobank A). The remaining material was centrifuged at 3000 g for 15 min, and the plasma was transferred to a cryotube (labeled biobank B) leaving approximately one mL in the bottom of the centrifuge tube.
From each of the two plasmas (biobank A/B) 300 μL of plasma was collected for miRNA-isolation, before the remaining plasma was further centrifuged. Biobank A samples were centrifuged at 3000 g for 30 min and biobank B samples at 3000 g for 15 min. The prolonged centrifugation of the biobank A samples was performed, to investigate whether a longer one-step centrifugation could eliminate platelet contamination as efficient as a shorter two-step centrifugation. After centrifugation the plasma was transferred to new tubes (labeled biobank A2 and B2, respectively), leaving approximately 200 μL in the bottom of the centrifuged tubes. From each of the two plasmas (biobank A2/B2) 300 μL of plasma was collected for miRNA-isolation. MiRNA was isolated, as described below, immediately following the plasma preparation procedure without any further freezing/thawing of the samples.
In order to investigate the distribution of platelets in the plasma phase 10 mL of EDTA anticoagulated whole blood was centrifuged at 2000 g for 10 min. After centrifugation the plasma was transferred to new tubes one mL at a time from the top of the collection tubes. In total, 4 mL of plasma (labeled gradient A to D) was collected from each 10 mL whole blood.
After each centrifugation step and from each gradient-sample 200 μL of plasma was collected for platelet count analysis which was performed using the fully automated Sysmex XE 5000 analyzer (Sysmex, Kobe, Japan).
2.1. MiRNA analysis
MiRNA was isolated from 300 μL of each of the plasma preparations PPP, Biobank A, Biobank B, Biobank A2 (only the six samples with a platelet count of 0–1×109/L) and Biobank B2 using Nucleospin®miRNA Plasma (Macherey-nagel, Germany) and according to protocol supplied by the manufacturer. As a mean of normalization all samples were spiked with 5 μL Cel-miR-39 (2.75×10−12 M) (RiboTask, Odense, Denmark). The spike was added after removal of the plasma proteins in the same step as isopropanol. MiRNA was eluted using 30 μL of RNAse free water, and the samples were frozen at −20 °C. MiRNA-analysis was performed within one month from the time of miRNA-isolation.
cDNA synthesis was performed using TaqMan®MicroRNA Reverse Transcription Kit and Custom TaqMan MIR RT Pool (both from Applied Biosystems, Life Sciences). The RT-primer pool, which was designed for a previous study of platelet derived miRNA in our laboratory, contained miRNA-specific stem-loop primers for 14 miRNAs known to be present in platelets and cel-miR-39. The reaction was performed using 3 μL of RNA-sample in a total volume of 10 μL.
As the miRNA level in plasma is relatively low, a preamplification step was performed using 5 μL of the RT-product in a total reaction volume of 25 μL containing TaqMan®PreAmp Master Mix and Custom TaqMan MIR PreAmp Pool (Applied Biosystems).
The final quantification was performed using Custom TaqMan®Array MicroRNA Cards (TLDA) from Applied Biosystems. A total of 1.2 μL of the preamplification products were diluted with TaqMan®Universal Master Mix II, no UNG and loaded onto the array. The miRNAs were analyzed in triplets using the ABI Prism 7900HT, and single outliers were removed before calculating the mean Ct-values for each miRNA.
MiRNA levels were normalized using either the exogenous cel-miR-39 or the endogenous miR-16, and calculated as 2−∆Ct (∆Ct=Cttarget miRNA−Ctcel-miR-39 or ∆Ct=Cttarget miRNA−CtmiR-16).
The measured miRNA levels in the different plasma preparations were evaluated using PPP as reference, and fold changes were calculated as 2−ΔΔCt.
2.2. Statistical analysis
All statistical calculations were carried out using R version 3.2.3. [21]. As data were not normally distributed we used the Wilcoxon signed rank test for comparison of the miRNA levels in the different plasma preparations and the levels in PPP. P-values ≤0.05 were considered significant.
3. Results
3.1. Residual platelet count
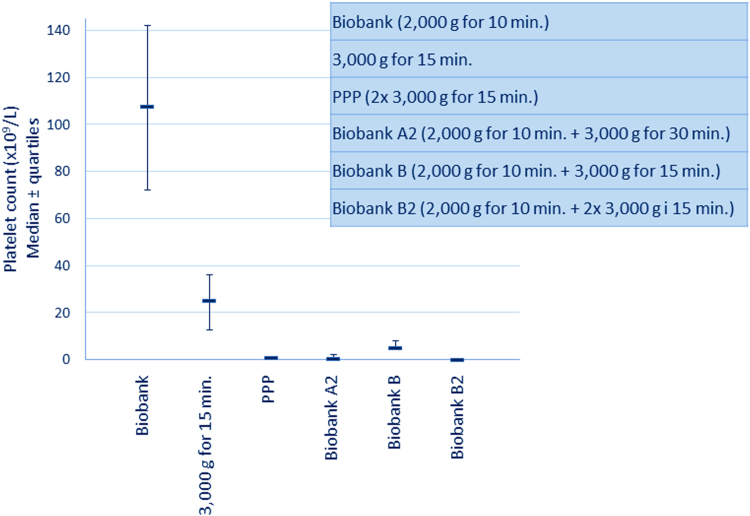
Platelet count was found to be low in all PPP (0–1×109/L) but to vary significantly in the 1×3000 (range 9–49×109/L), biobank (range 53–195×109/L) and biobank B (range 2–16×109/L) preparations (Fig. 2). After an additional centrifugation of the biobank A/B samples the platelet count was 0–1×109/L in all biobank B2 and in six of the biobank A2 samples. The remaining four biobank A2 samples still contained a small number of platelets (range 2–4×109/L).
Fig. 2.
Residual platelet count in the different plasma preparations. Platelet count in the different plasma preparations from the 10 volunteers. It can be seen that the platelet count is relatively high and vary significantly in the biobank (equals biobank A, but has not been frozen) and 3000 g for 15 min preparations, and that the biobank B preparations still contain some platelets. After further processing of the samples the platelet count was 0–1×109/L in all PPP, all biobank B2 and six of the biobank A2 samples.
3.2. Platelet distribution in the plasma phase
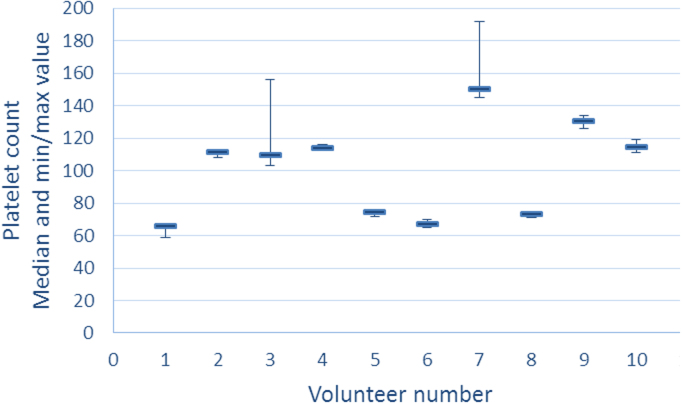
To evaluate a potential platelet gradient through the plasma phase, we investigated the distribution of platelets in the plasma of a 10 mL collection tube after a 10 min centrifugation at 2000 g. We found platelets to be equally distributed in the plasma phase of samples from the 10 volunteers (Fig. 3) except for the last gradient plasma (gradient D) in two samples which contained a higher number of platelets.
Fig. 3.
Platelet count in the four gradient samples from each of ten volunteers. For each of the ten volunteers are plotted the median platelet count of the four gradient samples A–D, and the lines illustrates the minimum and maximum value. The highest platelet count in samples from volunteer 3 and 7 were seen in gradient D, which is the plasma phase just above the buffy coat.
3.3. MiRNA levels
To evaluate the effect on miRNA levels of different residual platelet count in biobanked plasma samples, we compared the miRNA levels in the different plasma preparations with the levels in PPP (Table 1 and Supplementary Figs. 1 and 2). When preparing the biobank A2/B2 samples, an additional centrifugation step was used as an effort to eliminate any remaining platelets from the biobank A/B samples. Despite this additional centrifugation step, four biobank A2 samples still contained a small amount of platelets, and to ensure a sample population with minimal platelet contamination, we chose not to use these four samples for miRNA-analysis. Therefore, miRNA-analysis was performed on PPP, biobank A, biobank B and biobank B2 samples from ten volunteers, and on the six biobank A2 samples with a platelet count of 0–1×109/L.
Table 1.
MiRNA levels in the different plasma preparations relative to PPP.
| MicroRNA | Plasma type | Normalized to cel-mir-39 |
Normalized to miR-16 |
||
|---|---|---|---|---|---|
| Average fold change from PPP | P-value | Average fold change from PPP | P-value | ||
| miR-142–3p | Biobank A | 37.3 | <0.01 | 11.0 | <0.01 |
| BiobankA2 | 5.2 | <0.05 | 2.8 | <0.05 | |
| Biobank B | 4.7 | <0.01 | 5.8 | <0.01 | |
| BiobankB2 | 4.1 | <0.01 | 2.4 | <0.01 | |
| miR-145 | Biobank A | 68.4 | <0.01 | 22.5 | <0.01 |
| BiobankA2 | 4.9 | <0.05 | 2.3 | <0.05 | |
| Biobank B | 6.6 | <0.01 | 6.2 | <0.01 | |
| BiobankB2 | 3.0 | <0.01 | 1.8 | <0.01 | |
| miR-16 | Biobank A | 2.6 | <0.01 | ||
| BiobankA2 | 1.9 | <0.05 | |||
| Biobank B | 1.2 | 0.43 | |||
| BiobankB2 | 1.6 | <0.01 | |||
| miR-26a | Biobank A | 92.3 | <0.01 | 24.1 | <0.01 |
| BiobankA2 | 2.4 | <0.05 | 0.9 | 0.35 | |
| Biobank B | 9.2 | <0.01 | 7.2 | <0.01 | |
| BiobankB2 | 1.4 | 0.56 | 0.7 | <0.05 | |
| miR-28 | Biobank A | 102.5 | <0.01 | 34.2 | <0.01 |
| BiobankA2 | 13.2 | <0.05 | 6.8 | <0.05 | |
| Biobank B | 9.3 | <0.01 | 7.9 | <0.01 | |
| BiobankB2 | 6.3 | <0.01 | 3.0 | <0.01 | |
| miR-301 | Biobank A | 84.0 | <0.01 | 24.4 | <0.01 |
| BiobankA2 | 14.2 | <0.05 | 6.2 | <0.05 | |
| Biobank B | 11.8 | <0.01 | 8.4 | <0.01 | |
| BiobankB2 | 13.0 | <0.01 | 5.9 | <0.01 | |
| miR-30a-5p | Biobank A | 9.8 | <0.01 | 4.7 | <0.01 |
| BiobankA2 | 3.7 | <0.05 | 2.2 | 0.17 | |
| Biobank B | 2.5 | <0.05 | 2.8 | <0.01 | |
| BiobankB2 | 2.4 | <0.01 | 1.7 | <0.01 | |
| miR-30d | Biobank A | 14.9 | <0.01 | 5.7 | <0.01 |
| BiobankA2 | 2.8 | <0.05 | 1.4 | 0.25 | |
| Biobank B | 3.8 | <0.01 | 4.0 | <0.01 | |
| BiobankB2 | 2.4 | <0.01 | 1.4 | <0.01 | |
| miR-328 | Biobank A | 94.1 | <0.01 | 30.7 | <0.01 |
| BiobankA2 | 368 | <0.05 | 233 | <0.05 | |
| Biobank B | 23.0 | <0.01 | 16.9 | <0.01 | |
| BiobankB2 | 97.4 | <0.01 | 53.6 | <0.01 | |
| miR-331 | Biobank A | 63.5 | <0.01 | 20.9 | <0.01 |
| BiobankA2 | 113.1 | <0.05 | 48.9 | <0.05 | |
| Biobank B | 8.4 | <0.01 | 6.6 | <0.01 | |
| BiobankB2 | 55.8 | <0.01 | 31.3 | <0.01 | |
| miR-335 | Biobank A | 95.1 | <0.01 | 28.4 | <0.01 |
| BiobankA2 | 26.9 | <0.05 | 11.4 | <0.05 | |
| Biobank B | 17.1 | <0.01 | 14.4 | <0.01 | |
| BiobankB2 | 19.1 | <0.01 | 9.3 | <0.01 | |
| miR-340 | Biobank A | 53.7 | <0.01 | 18.4 | <0.01 |
| BiobankA2 | 8.3 | <0.05 | 4.1 | <0.05 | |
| Biobank B | 11.4 | <0.01 | 10.5 | <0.01 | |
| BiobankB2 | 5.2 | <0.01 | 3.8 | <0.01 | |
| miR-92a | Biobank A | 8.4 | <0.01 | 2.7 | <0.01 |
| BiobankA2 | 13.8 | <0.05 | 6.5 | <0.05 | |
| Biobank B | 2.5 | <0.05 | 2.1 | <0.01 | |
| BiobankB2 | 8.6 | <0.01 | 3.9 | <0.01 | |
| miR-93 | Biobank A | 13.1 | <0.01 | 4.3 | <0.01 |
| BiobankA2 | 4.9 | <0.05 | 2.6 | <0.05 | |
| Biobank B | 3.5 | <0.01 | 2.8 | <0.01 | |
| BiobankB2 | 3.8 | <0.01 | 1.9 | <0.01 | |
Average fold change in miRNA level relative to PPP are given for biobank A, A2, B and B2 samples. MiRNA levels are normalized to cel-miR-39 and miR-16, respectively. Biobank A preparations represent the standard biobank plasma (centrifuged at 2000 g for 10 min) after one freezing/thawing cycle. P-values were calculated using the Wilcoxon signed rank test, and were considered significant if <0.05.
MiRNA levels were normalized using either the exogenous cel-miR-39 or the endogenous miR-16 as reference gene. When normalized to cel-miR-39, levels of miR-16 were found to be significantly (p<0.05) higher in biobank A, A2 and B samples compared to PPP samples (1.6–2.6 fold in average). Therefore, the fold changes in miRNA levels between the different plasma preparations and PPP found for the other investigated miRNAs were lower when miRNA levels were normalized using miR-16 as compared to normalization using cel-miR-39 (Table 1). Still, regardless of the choice of normalization, we found significantly higher levels of all the investigated miRNAs in biobank A/B samples compared to PPP samples (p<0.05), which was expected as these plasmas had high/medium residual platelets content.
Furthermore, when using cel-miR-39 for normalization we found significantly higher levels (P<0.05) of all investigated miRNAs in the six biobank A2 samples (1.9–368 fold on average) and of all investigated miRNAs except miR-26a in the biobank B2 samples (1.6–97.4 fold on average) as compared to PPP samples. Alongside that, miRNA levels normalized to miR-16 were found to be significantly higher (p<0.05) for 10 of the investigated miRNAs in biobank A2 samples (2.3–233 fold on average) and for all of the investigated miRNAs except miR-26a in biobank B2 samples (1.4–53.6 fold on average). The level of miR-30a-5p and miR-30d were found to be higher (1.4–2.2 fold on average), but not significantly different between biobank A2 and PPP samples (p>0.05). Levels of miR-26a were found to be slightly lower in biobank A2/B2-samples compared to PPP, and in A2 samples the difference was not significant.
4. Discussion
Circulating miRNA may have great potential as biomarkers for many diseases, and many studies have addressed associations between disease status and miRNA levels in biobanked plasma or serum samples [22], [23], [24]. As platelets contain large amounts of miRNA [25], residual platelets may contribute significantly to the miRNA levels in plasma, and therefore platelets should be removed prior to miRNA analysis. During freezing and/or thawing of the plasma, miRNAs may be released from residual platelets and contribute to the miRNA levels even despite of an additional centrifugation step after thawing of the plasma. To evaluate biobanked plasma as a suitable material for miRNA-analysis, or whether it can be transformed into such, we performed a range of centrifugation steps on biobanked plasma samples, after which we analyzed the plasma preparations for their residual platelet contents as well as miRNA levels.
When using our standard procedure for biobanking (centrifugation of whole blood for 10 min at 2000 g) plasma contained a relatively high number of platelets (Fig. 2). A further centrifugation of the biobanked plasma for 15 min at 3000 g reduced, but did not fully eliminate residual platelets, whereas a two-step centrifugation at these conditions or a 30 min centrifugation at 3000 g, for most samples resulted in plasma with minimal residual platelet content (0–1×109/L). A total of four samples still contained a small number of platelets after the 30 min centrifugation, probably due to disturbance of the pellet when plasma was transferred.
In order to evaluate whether these centrifugation steps could also eliminate contamination with miRNAs derived from residual platelets during sample storage, we used RT-qPCR analysis to compare the level of 14 selected miRNAs in the different plasma preparations to that of PPP. We found the level of most investigated miRNAs to be significantly higher in all plasma preparations compared to PPP (Table 1 and Supplementary Figs. 1 and 2), except for miR-26a levels in biobank B2 samples (with either normalization strategy) and the levels of miR-26, miR-30a-5p and miR-30d in the biobank A2 samples (when normalized to miR-16). For miR-328, miR-331 and to a minor extent miR-92a the level was significantly higher in the biobank A2/B2 samples compared to the biobank A/B samples, though the biobank A2/B2 samples have very low platelet content. The plasma level of miR-328 is very low, and analytical limitations could explain our findings for this miRNA, but this is not the case for miR-331 or miR-92a. Zheng et.al. also found a few miRNAs that were present in plasma after high speed centrifugation, but not detectable in plasma centrifuged at only 820 g [16]. A possible explanation for these findings could be the content of different miRNAs in microvesicles of different sizes, but further studies are needed to ascertain the reason and importance of this difference between miRNAs.
Cheng et.al. reported that an additional centrifugation step after thawing of biobanked samples could minimize the significance of the plasma preparation prior to freezing [17]. They found that centrifugation of thawed plasma samples at 1940 g for 10 min reduced the average platelet count from 80×109/L to 10×109/L and the plasma level of a number of miRNAs by several fold, as example miR-142–3p was reduced by 5 fold, miR-16 by 1.8 fold and miR-92a by 2.0 fold. These results are a little lower, but comparable with our findings when comparing biobank A and biobank B samples, normalizing miRNA levels to cel-miR-39 and taking into account the differences in centrifugation time and speed (our centrifugation was 3000 g for 15 min). In the study by Cheng et.al. the investigators did not include samples from which all platelets was removed prior to freezing, and our present study illustrates that even when residual platelets are almost completely removed after thawing of biobanked samples, they still contain a significantly higher level of platelet derived miRNA compared to PPP. As an example, when using cel-miR-39 for normalization we found the plasma level of miR-142-3p to be 5.2 fold higher in the biobank A2 samples and 4.1 fold higher in the biobank B2 samples compared to PPP. Using miR-16 for normalization the fold changes were 2.8 and 2.4, respectively. In recent publications miR-142–3p has been reported to be significantly up- or down- regulated in different diseases/conditions, in some studies by 1.5–3 fold [26], [27], [28], [29], [30], [31] and in other studies by 4.8–19 fold [32], [33], [34]. Our results indicate that a 1.5–3 fold change in plasma levels of this miRNA may possibly be caused by or could easily be overseen due to differences in sample preparation and/or storage. We found miR-26a to be only slightly affected by sample preparation prior to freezing, nevertheless, the observed differences between the biobank A2/B2 samples and PPP of up to 2.4 fold, still reaches the level of changes in miRNA levels reported by a handful of recent studies [35], [36], [37], [38], [39]. We did not examine the impact of the platelet count before centrifugation, but since the platelet count ranges across approximately 3-fold in healthy patients and up to 100-fold in diseased patients, preanalytical conditions might be of even greater importance and different sample preparation may be needed in case of very high platelet count.
When preparing PPP we first centrifuged the whole blood samples, and then performed a re-centrifugation of the plasma, both centrifugations at 3000 g for 15 min. A study by Chandler found that a two-step centrifugation of platelet-rich plasma at 1500 g for 20 min removed 99% of the platelets, but also 58–93% of platelet-derived microparticles [40]. Therefore, as miRNA released from activated cells are often included in microparticles [41], [42], an effort to eliminate platelet contamination, may affect the measured level of many circulating miRNAs due to a reduction in the number of microparticles. This dilemma illustrates the need of a standardized protocol for sample preparation, in order to compare results obtained in different laboratories.
Since a routine centrifugation at 2000 g does not eliminate all platelets from the plasma, a platelet gradient may exist that could influence measured miRNA levels. To evaluate this, we investigated the platelet distribution in the plasma phase of 10 mL blood collection tubes from 10 volunteers. We found the platelets to be equally distributed throughout the plasma phase in most tubes, only the plasma closest to the buffy coat (gradient D) from two volunteers contained a higher number of platelets. When collecting 4 mL of plasma from 10 mL of whole blood, the volume of plasma left on the buffy coat vary depending on the hematocrit, and the higher platelet count observed in the two gradient D plasmas may possible be due to a higher hematocrit in these volunteers. Our results indicate that it is not important from which plasma phase the samples for miRNA analysis origin, but care should be taken to avoid the buffy coat.
The present study has some limitations, including the relatively small sample size, the fact that we did not investigate other matrices than EDTA plasma and that we did not include miRNAs not present in platelets.
In conclusion, our results show that the pre-storage centrifugation conditions have a significant impact on the measured EDTA plasma level of miRNAs known to be present in platelets. Even for the miRNAs found to be less effected, we found that a 1.5–3 fold change in plasma levels may possible be caused by or easily overseen due to sample preparation and/or storage. Thus, in order to compare miRNA levels of plasma samples within or between studies, it is essential to use standardized sampling and processing protocols and to report these in details in future publications.
Funding
This research did not receive any specific Grant from funding agencies in the public, commercial, or not-for-profit sectors.
Transparency document. Supplementary material Transparency data associated with this article can be found in the online version at 10.1016/j.bbrep.2016.06.005.
Footnotes
Supplementary data associated with this article can be found in the online version at 10.1016/j.bbrep.2016.06.005.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary Figure 1: Average fold change in miRNA level relative to PPP are given for biobank A, A2, B and B2 samples. MiRNA levels are normalized to cel-miR-39. Biobank A preparations represent the standard biobank plasma (centrifuged at 2000 g for 10 min) after one freezing/thawing cycle. The miRNA-328 level in biobank A2 was 368 fold higher than in PPP, so it is out of the scale of the figure. P-values were calculated using the Wilcoxon signed rank test, and were considered significant if <0.05.
.
Supplementary Figure 2: Average fold change in miRNA level relative to PPP are given for biobank A, A2, B and B2 samples. MiRNA levels are normalized to miR-16. Biobank A preparations represent the standard biobank plasma (centrifuged at 2000 g for 10 min) after one freezing/thawing cycle. The miRNA-328 level in biobank A2 was 233 fold higher than in PPP, so it is out of the scale of the figure. P-values were calculated using the Wilcoxon signed rank test, and were considered significant if <0.05.
.
References
- 1.Zampetaki A., Mayr M. Analytical challenges and technical limitations in assessing circulating miRNAs. Thromb. Haemost. 2012;108:592–598. doi: 10.1160/TH12-02-0097. [DOI] [PubMed] [Google Scholar]
- 2.Sourvinou I.S., Markou A., Lianidou E.S. Quantification of Circulating miRNAs in Plasma: effect of preanalytical and analytical parameters on their isolation and stability. J. Mol. Diagn. 2013;15:827–834. doi: 10.1016/j.jmoldx.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 3.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 4.Min P.-K., Chan S.Y. The biology of circulating microRNAs in cardiovascular disease. Eur. J. Clin. Investig. 2015;45:860–874. doi: 10.1111/eci.12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pasquinelli A.E., Reinhart B.J., Slack F. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 6.Chim S.S.C., Shing T.K.F., Hung E.C.W. Detection and characterization of placental MicroRNAs in maternal plasma. Clin. Chem. 2008;54:482–490. doi: 10.1373/clinchem.2007.097972. [DOI] [PubMed] [Google Scholar]
- 7.Lawrie C.H., Gal S., Dunlop H.M. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 8.J. Yu, L. Jin, W. Li, et al., Serum miR-372 is a diagnostic and prognostic biomarker in patients with early colorectal cancer, Anticancer Agents Med. Chem., 2015. [DOI] [PubMed]
- 9.Jia W., Wu Y., Zhang Q. Expression profile of circulating microRNAs as a promising fingerprint for cervical cancer diagnosis and monitoring. Mol. Clin. Oncol. 2015;3:851–858. doi: 10.3892/mco.2015.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sayed A.S.M., Xia K., Li F. The diagnostic value of circulating microRNAs for middle-aged (40–60-year-old) coronary artery disease patients. Clin. São Paulo Braz. 2015;70:257–263. doi: 10.6061/clinics/2015(04)07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higuchi C., Nakatsuka A., Eguchi J. Identification of circulating miR-101, miR-375 and miR-802 as biomarkers for type 2 diabetes. Metabolism. 2015;64:489–497. doi: 10.1016/j.metabol.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Wang X., Sundquist K., Hedelius A. Circulating microRNA-144–5p is associated with depressive disorders. Clin. Epigenetics. 2015;7:69. doi: 10.1186/s13148-015-0099-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Witwer K.W. Circulating MicroRNA biomarker studies: pitfalls and potential solutions. Clin. Chem. 2015;61:56–63. doi: 10.1373/clinchem.2014.221341. [DOI] [PubMed] [Google Scholar]
- 14.McDonald J.S., Milosevic D., Reddi H.V. Analysis of circulating microRNA: preanalytical and analytical challenges. Clin. Chem. 2011;57:833–840. doi: 10.1373/clinchem.2010.157198. [DOI] [PubMed] [Google Scholar]
- 15.MacLellan S.A., MacAulay C., Lam S., Garnis C. Pre-profiling factors influencing serum microRNA levels. BMC Clin. Pathol. 2014;14:27. doi: 10.1186/1472-6890-14-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng X.-H., Cui C., Zhou X.-X. Centrifugation: an important pre-analytic procedure that influences plasma microRNA quantification during blood processing. Chin. J. Cancer. 2013;32:667–672. doi: 10.5732/cjc.012.10271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng H.H., Yi H.S., Kim Y. Plasma processing conditions substantially influence circulating microRNA biomarker levels. PLoS One. 2013;8 doi: 10.1371/journal.pone.0064795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirschner M.B., Edelman J.J.B., Kao S.C.-H. The impact of hemolysis on cell-free microRNA biomarkers. Front. Genet. 2013;4 doi: 10.3389/fgene.2013.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blondal T., Jensby Nielsen S., Baker A. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods San Diego Calif. 2013;59:S1–S6. doi: 10.1016/j.ymeth.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 20.Wang K., Yuan Y., Cho J.-H. Comparing the MicroRNA spectrum between serum and plasma. PLoS One. 2012;7 doi: 10.1371/journal.pone.0041561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.R Core Team, R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria, 2015 (n.d.). 〈https://www.R-project.org/〉.
- 22.Permuth-Wey J., Chen D.-T., Fulp W.J. Plasma MicroRNAs as novel biomarkers for patients with intraductal papillary mucinous neoplasms of the pancreas. Cancer Prev. Res. 2015;8:826–834. doi: 10.1158/1940-6207.CAPR-15-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho G.Y.F., Jung H.J., Schoen R.E. Differential expression of circulating microRNAs according to severity of colorectal neoplasia. Transl. Res. 2015;166:225–232. doi: 10.1016/j.trsl.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J., Mao Q., Liu Y. Analysis of miR-205 and miR-155 expression in the blood of breast cancer patients. Chin. J. Cancer Res. 2013;25:46–54. doi: 10.3978/j.issn.1000-9604.2012.11.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osman A., Fälker K. Characterization of human platelet microRNA by quantitative PCR coupled with an annotation network for predicted target genes. Platelets. 2011;22:433–441. doi: 10.3109/09537104.2011.560305. [DOI] [PubMed] [Google Scholar]
- 26.Steen S.O., Iversen L.V., Carlsen A.L. The circulating cell-free microRNA profile in systemic sclerosis is distinct from both healthy controls and systemic lupus erythematosus. J. Rheumatol. 2015;42:214–221. doi: 10.3899/jrheum.140502. [DOI] [PubMed] [Google Scholar]
- 27.Prats-Puig A., Ortega F.J., Mercader J.M. Changes in circulating microRNAs are associated with childhood obesity. J. Clin. Endocrinol. Metab. 2013;98:E1655–E1660. doi: 10.1210/jc.2013-1496. [DOI] [PubMed] [Google Scholar]
- 28.Kumar P., Dezso Z., MacKenzie C. Circulating miRNA biomarkers for alzheimer's disease. PLoS One. 2013;8 doi: 10.1371/journal.pone.0069807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ellis K.L., Cameron V.A., Troughton R.W. Circulating microRNAs as candidate markers to distinguish heart failure in breathless patients. Eur. J. Heart Fail. 2013;15:1138–1147. doi: 10.1093/eurjhf/hft078. [DOI] [PubMed] [Google Scholar]
- 30.Pivarcsi A., Meisgen F., Xu N. Changes in the level of serum microRNAs in patients with psoriasis after antitumour necrosis factor-α therapy. Br. J. Dermatol. 2013;169:563–570. doi: 10.1111/bjd.12381. [DOI] [PubMed] [Google Scholar]
- 31.Ortega F.J., Mercader J.M., Catalán V. Targeting the circulating microRNA signature of obesity. Clin. Chem. 2013;59:781–792. doi: 10.1373/clinchem.2012.195776. [DOI] [PubMed] [Google Scholar]
- 32.Summerer I., Unger K., Braselmann H. Circulating microRNAs as prognostic therapy biomarkers in head and neck cancer patients. Br. J. Cancer. 2015;113:76–82. doi: 10.1038/bjc.2015.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ortega F.J., Mercader J.M., Moreno-Navarrete J.M. Profiling of circulating MicroRNAs reveals common microRNAs linked to Type 2 diabetes that change with insulin sensitization. Diabetes Care. 2014;37:1375–1383. doi: 10.2337/dc13-1847. [DOI] [PubMed] [Google Scholar]
- 34.Kanaan Z., Roberts H., Eichenberger M.R. A plasma MicroRNA panel for detection of colorectal adenomas: a step toward more precise screening for colorectal cancer. Ann. Surg. 2013;258:400–408. doi: 10.1097/SLA.0b013e3182a15bcc. [DOI] [PubMed] [Google Scholar]
- 35.Ormseth M.J., Solus J.F., Vickers K.C. Utility of select plasma microRNA for disease and cardiovascular risk assessment in patients with rheumatoid arthritis. J. Rheumatol. 2015 doi: 10.3899/jrheum.150232. jrheum.150232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li C., Chen X., Huang J. Clinical impact of circulating miR-26a, miR-191, and miR-208b in plasma of patients with acute myocardial infarction. Eur. J. Med. Res. 2015;20 doi: 10.1186/s40001-015-0148-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holm A., Bang-Berthelsen C.H., Knudsen S. miRNA profiles in plasma from patients with sleep disorders reveal dysregulation of mirnas in narcolepsy and other central hypersomnias. Sleep. 2014;37:1525–1533. doi: 10.5665/sleep.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murata K., Furu M., Yoshitomi H. Comprehensive microRNA analysis identifies miR-24 and miR-125a-5p as plasma biomarkers for rheumatoid arthritis. PLoS One. 2013;8 doi: 10.1371/journal.pone.0069118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu L., Zhou H., Lin H. Circulating microRNAs are elevated in plasma from severe preeclamptic pregnancies. Reproduction. 2012;143:389–397. doi: 10.1530/REP-11-0304. [DOI] [PubMed] [Google Scholar]
- 40.Chandler W.L. Microparticle counts in platelet-rich and platelet-free plasma, effect of centrifugation and sample-processing protocols. Blood Coagul. Fibrinolysis Int. J. Haemost. Thromb. 2013;24:125–132. doi: 10.1097/MBC.0b013e32835a0824. [DOI] [PubMed] [Google Scholar]
- 41.Laffont B., Corduan A., Plé H. Activated platelets can deliver mRNA regulatory Ago2•microRNA complexes to endothelial cells via microparticles. Blood. 2013;122:253–261. doi: 10.1182/blood-2013-03-492801. [DOI] [PubMed] [Google Scholar]
- 42.Jansen F., Yang X., Hoelscher M. Endothelial microparticle-mediated transfer of MicroRNA-126 promotes vascular endothelial cell repair via SPRED1 and is abrogated in glucose-damaged endothelial microparticles. Circulation. 2013;128:2026–2038. doi: 10.1161/CIRCULATIONAHA.113.001720. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary Figure 1: Average fold change in miRNA level relative to PPP are given for biobank A, A2, B and B2 samples. MiRNA levels are normalized to cel-miR-39. Biobank A preparations represent the standard biobank plasma (centrifuged at 2000 g for 10 min) after one freezing/thawing cycle. The miRNA-328 level in biobank A2 was 368 fold higher than in PPP, so it is out of the scale of the figure. P-values were calculated using the Wilcoxon signed rank test, and were considered significant if <0.05.
Supplementary Figure 2: Average fold change in miRNA level relative to PPP are given for biobank A, A2, B and B2 samples. MiRNA levels are normalized to miR-16. Biobank A preparations represent the standard biobank plasma (centrifuged at 2000 g for 10 min) after one freezing/thawing cycle. The miRNA-328 level in biobank A2 was 233 fold higher than in PPP, so it is out of the scale of the figure. P-values were calculated using the Wilcoxon signed rank test, and were considered significant if <0.05.