Abstract
Albumin is thought as an drug carrier for doxorubicin (DOX). The binding of doxorubicin to albumin was studied on the surface of sporopolleninin (SP) to produce a new drug system based natural materials. Human serum albumin (HSA) was immobilized on SPIONs in 20 mM Tris buffer, 7.4 of pH. Data showed that binding amount of HSA has been found to be as 285.53 µg to the 25 mg of Sporopolleninin which also bounded 319.76 µM of DOX. Binding of protein and drug to Sp were clarified by SEM, EDX and FT-IR analysis.
Keywords: Doxorubicin, Magnetic nanoparticles, Protein, Fluorescence spectroscopy
Graphical abstract
Highlights
-
•
Albumin was immobilized on a various amount of sporopollenin (Sp).
-
•
0.79 mg of HSA was bounded on 25 mg of sporopollenin.
-
•
DOX have emission at 480 nm and 560 nm of excitation and emission wavelengths.
-
•
24 μg of the drug was loaded to the HSA immobilized chitosan at 20 mg of sporopollenin.
1. Introduction
Drugs based on proteins, peptides and oligonucleotides tend to attract the greatest attention at present due to their potential to treat chronic diseases [1]. The research is centered on overcoming the main obstacle to the treatment of cancer by chemotherapy, i.e., the destruction of healthy tissue by the therapeutic agents [2]. Biocompatible polymeric based particles (BPs) as effective delivery carriers of drugs has received considerable attention in recent years because they possess the ability to deliver drugs to varied areas of the body for sustained periods of time [3], [4], [5], [6]. Biological polymers with several properties such as good biological compatibility, nontoxicity and high hydrophilicity, comparing with synthetic organic materials when used as drug carriers [5], [6], [7], [8], [9]. Suitable modification and optimization of the form of the biological polymer has to be determined to properly target the selected polymer and its controlled release to the affected organs [10], [11]. Naturally occurring polymers have garnered attention within the scope of obtaining carriers capable of releasing therapeutic molecules to affected sites binding the targeted disease in a living body [12], [13].
Sporopollenin (Sp) represents a most promising biopolymer for such controlled delivery and targeting, due to factors such as its biocompatibility, low toxicity, and feasible biodegradability connected with easy elimination of carrier metabolites from the body [14]. Sporopollenins, derived from the spores of Lycopodium clavatum (Sp), contain carbon, hydrogen and oxygen, and it has the stoichiometry C90H144027. The diameter of a Sp is typically within the range of 5 µm to 250 µm and the material does not swell to any measurable extent in aqueous or organic solvents [15]. Spores which are roughly spherical in shape produced by nonseed bearing plants, for example ferns and mosses, as part of the process of reproduction [16]. The same size and decorations of Sp particles in a single species of plant makes possible to attach small molecules such as ligand and drugs to the outside of complete particles this morphology was regarded as being so applicable to drug delivery [17]. For drug delivery, different particles modified with protein have been widely reported [18] in particular, physical or chemical modification can improve the properties of particles in different ways. Nevertheless, there are some drawbacks related to their toxicity, biocompability and allergic affectivity. In drug delivery applications, naturally occurring Sp has been shown to be non toxic to human endothelial cells and free of allergenic protein epitopes which is crucial [19]. Because Lycopodium clavatum spores have a number of advantages over conventional materials they were chosen as a Np for the drug delivery used for this study.
Albumin is a versatile protein carrier biomaterial for much therapeutics in treating a variety of diseases including cancer, arthritis and diabetes and was considered an attractive drug carrier for hydrophobic drugs [20]. Human serum albumin (HSA) has a molecular weight of 66 kDa and highly soluble globular monomeric protein [21], [22], [23]. HSA is a multiple protein carrier for drug delivery with high stability. It has been shown to be nontoxic, biocompatible and biodegradable, so it can be preferentially taken up by tumors as a nutrition ingredient [24]. Studies have indicated that albumin based on particles have great potential in the smart delivery of therapeutic agents.
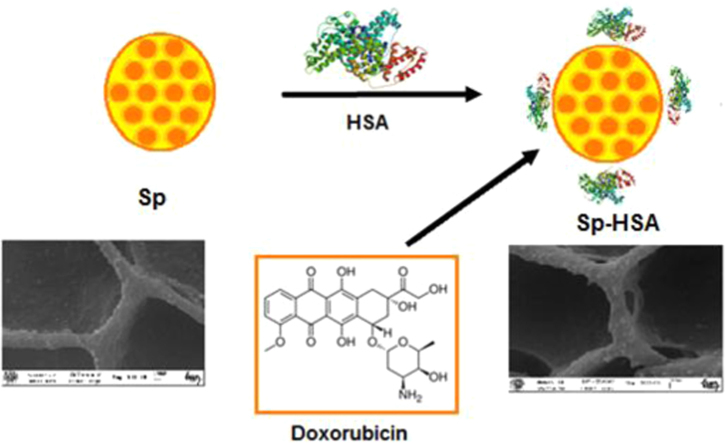
This study focused on the albumin immobilized Sporopollenin (Sp-HSA) for delivery of doxorubicin (DOX) to enhance target ability. DOX as an anti-tumor model drug was loaded to Sp-HSA. As expected, the prepared Sp-HSA system has shown high affinity for DOX.
2. Materials and methods
2.1. Materials and chemicals
Sodium hydroxide (≥97%), hydrochloric acid (37%), tris (hydroxymethyl)-aminomethane (99.8–100.1%), ammonium hydroxide (25% w/w) and sodium chloride were obtained from Merck (Germany). Sporopollenin was obtained from Fluka. Human serum albumin was obtained from Sigma (USA). Doxorubicin was provided from Sigma Aldrich. All aqueous solutions and buffers were prepared with deionized water via Millipore Milli-Q Plus water purification system.
2.2. Immobilization of albumin on sporopolleninin
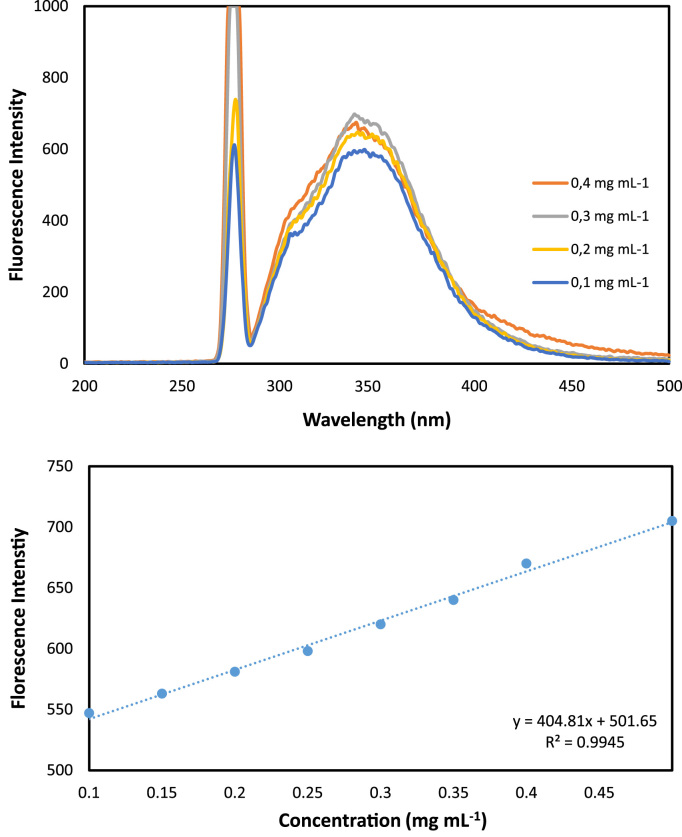
Sp was mixed with a concentration of 1 mg mL−1 of human serum albumin (HSA) in 20 mM Tris, pH 7.4 at a particle concentration range of 5–25 mg mL−1 [14]. Then, the heterogenous solution was mixed for 2 h at 4 °C. Albumin immobilized sporopollenin was washed with Tris buffer for chemical analysis. Supernatant was scanned for determination of binding amount of albumin. The amount of immobilized protein was measured from regression equation of calibration curve of the protein (y=404,81x+501,65, r: 0.99) by using a Perkin-Elmer LS 55 Luminescence Spectrophotometer (Waltham, USA). The emission spectra were scanned at 280 and 342 nm of excitation and emission wavelengths [25].
2.3. DOX loading and releasing
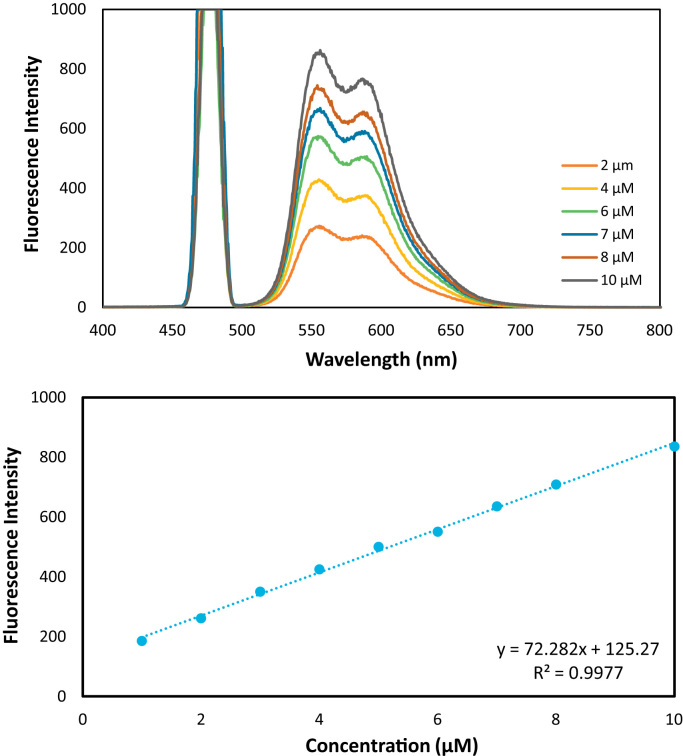
1 mM of doxorubicin was dissolved in mL of 20 mM Tris–HCl buffer (pH 7.4) [26]. The DOX solution was added to the 5–25 mg of protein immobilized sporopollenin in 20 mM Tris–HCl buffer (pH 7.4) in order to measure DOX loading. DOX release from Sp-HSA was studied via different buffer with a various pH at 60 rpm and 37 °C. The release behaviors of DOX-loaded albumin particles under different pH (154 mM NaCl, pH:2.2; and 6; 20 mM Tris buffer, pH: 7.4) were evaluated. The excitation and emission wavelengths of the drug were also scanned at 480 nm and 560 nm by fluorescence spectroscopy, respectively. The excitation and emission slit width were both set to 10 nm. Loading amount of DOX to albumin-Sp and release of the DOX from albumin-Sp were calculated from calibration curve of DOX (y=72, 282x +125, 27, r: 0.99).
2.4. Instrumentation
FT-IR spectra were scanned on a Perkin Elmer spectrum 100 FTIR spectrometer (ATR). SEM images were obtained using a Zeiss LS-10 field emission SEM instrument equipped with an Inca Energy 350 X-Max (Oxford Instruments) spectrometer. Samples were sputter-coated with Au (60%) and Pd (40%) alloy using a Q150R (Quorum Technologies) instrument.
3. Results and discussions
3.1. Immobilization of albumin
A broad scale of materials such as chitosan, sporopollenin, magnetic nanoparticles and polymers by different functional groups have been widely used in drug delivery systems [27], [28], [29], [30], [31], [32], [33], [34]. Our group has focus on development of drug carrier systems having proteins [26], [27], [28], [29], [30], [31], [32], [33], [34]. One of our previous studies has been on immobilization of hemoglobin onto sporopollenin which was showed to be high affinity to the protein, so it is possible to develop a new drug delivery system based on sporopollenin-protein [14]. Because, the use of protein in drug carrier systems was very effective on bioavailability and reduction of side effects of the drugs in vivo [32], [33], [34].
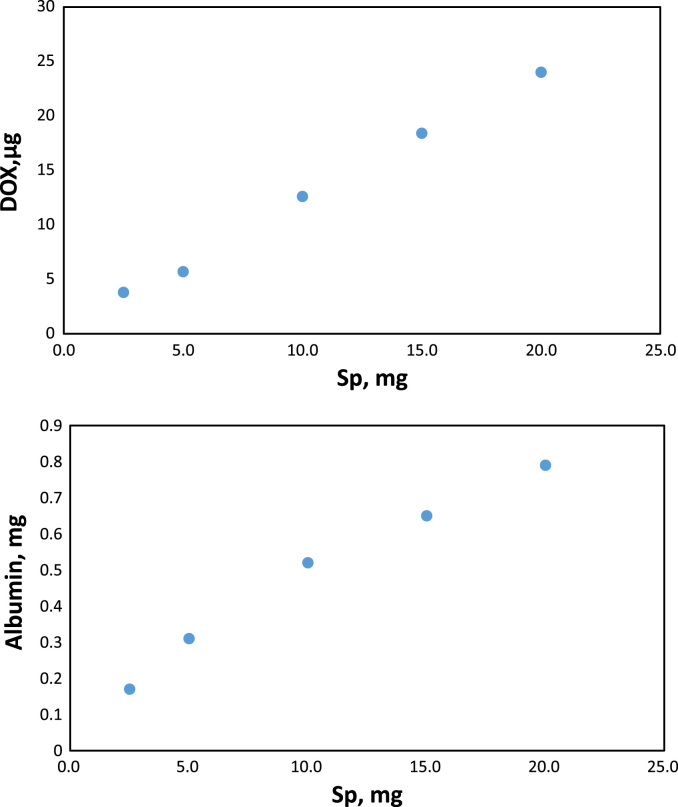
The standard albumin solution was mixed with different amount of sporopollenin particles. After binding of albumin to particles, supernatant from the heterogeneous system was removed and scanned with spectrofluorometer at 280 nm and 342 nm of excitation and emission wavelengths where exhibited intrinsic fluorescence of protein. The intrinsic fluorescence of protein comes from tryptophan and tyrosine residues in the albumin's structure [35], [36], [37]. The emission spectrum for albumin were shown in Fig. 1. The binding amount of the albumin was calculated via the regression equation at a particles amount range of 0.1 mg and 0.5 mg (Fig. 1). The binding amount of albumin was increased with increase in the particles amount. Data showed that 285.53 µg of albumin was bounded to the 25 mg of Sp in Tris buffer (Fig. 2).
Fig. 1.
Emission spectra and calibration curve of albumin from human serum at a concentration range of 0,1–0,5 mg mL−1.
Fig. 2.
Binding amount of HSA and DOX versus amount of Sp.
3.2. Releasing and loading of doxorubicin
DOX is a anticancer drug which exhibits maximum fluorescence intensity at 480 nm and 560 nm of excitation and emission wavelengths, respectively (Fig. 3). The binding amount of DOX was measured via regression equation of the drug as viewed from inset of Fig. 3. at a concentration range of 1 µg and10 µg. Data showed that 20 mg and 5 mg of sporopollenin bounded 24 µg and 319.76 µM of DOX (Fig. 2).
Fig. 3.
Emission spectra and calibration curve of doxorubicin at a concentration range of 1–10 µM.
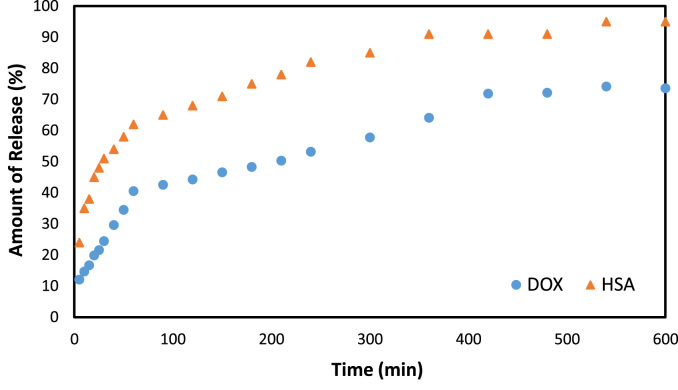
Release of HSA from Sp particles was studied at different pH as 7,4, 6 and 2,2. According to Table 1, maximum release of the protein from sporopollenin particles has obviously been viewed at 7.4 of pH as % 95.06 at 25 mg of particles. According to Table 2, release of DOX from Sp-HSA particles was studied at different pH as 7,4, 6 and 2,2. Data showed that amount of released DOX has been found as % 51.24 at 25 mg of particles. Fig. 4 showed DOX and BSA release at optimum pH of 2.2 related to the time during 10 h being stable.
Table 1.
Amount of loaded and released DOX.
| Sp, mg | 5 | 10 | 15 | 20 | 25 | |
| Loaded HSA, µg | 119.62 | 142.27 | 177.49 | 216.08 | 285.53 | |
| Loaded HSA, % | 12.59 | 14.94 | 18.63 | 22.75 | 30.06 | |
| Released HSA, µg | pH: 7.4 | 78.28 | 101.13 | 142.58 | 198.19 | 270.17 |
| pH: 6 | 68.74 | 86.59 | 112.71 | 158.70 | 236.26 | |
| pH: 2.2 | 48.65 | 66.08 | 92.63 | 125.58 | 178.05 | |
| Released HSA, % | pH: 7.4 | 65.54 | 71.12 | 80.22 | 91.66 | 95.06 |
| pH: 6 | 57.14 | 60.56 | 63.27 | 73.15 | 82.88 | |
| pH: 2.2 | 40.33 | 46.48 | 51.98 | 57.87 | 62.57 |
Table 2.
Amount of loaded and released DOX.
| Sp, mg | 5 | 10 | 15 | 20 | 25 | |
| Loaded DOX, µM | 116.58 | 222.29 | 267.71 | 304.89 | 319.76 | |
| Loaded DOX, % | 30.58 | 58.65 | 70.64 | 80.45 | 84.37 | |
| Released DOX, µM | pH: 7.4 | 34.83 | 42.43 | 44.66 | 47.78 | 56.27 |
| pH: 6 | 40.64 | 51.80 | 54.49 | 56.72 | 58.06 | |
| pH: 2.2 | 66.10 | 92.00 | 130.41 | 141.58 | 163.46 | |
| Released DOX, % | pH: 7.4 | 30.03 | 19.11 | 16.72 | 15.71 | 17.64 |
| pH: 6 | 35,03 | 23.33 | 20.40 | 18.66 | 18.20 | |
| pH: 2.2 | 56.98 | 41.44 | 48.84 | 46.57 | 51.24 |
Fig. 4.
Released HSA and doxorubicin expressed as % against time (min) at pH 2.2.
3.3. FT-IR analysis
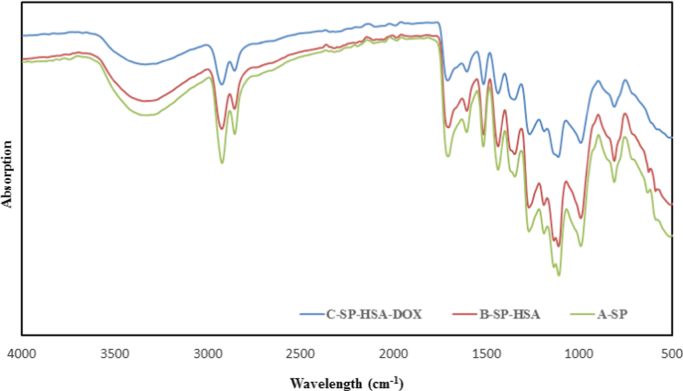
Binding procedure of albumin to sporopollenin was given at Scheme 1. Fig. 5 illustrates FT-IR spectra of Sp and Sp-albumin before and after binding to doxorubicin. The FT-IR spectrum of Sp shows typical bands around at 3300 cm−1 which comes from hydroxyl groups (–OH) [38]. The peaks at around 2921 cm−1 and 2858 cm−1 are belong to C–H stretching vibration. The peak which has been observed in Sp spectrum belongs to C–O stretching vibrations (1706 cm−1) [39], [40], [41]. As seen in Fig. 5, these spectrum are not much different from each other due to many FT-IR peaks with similar wavelengths in Sp molecules [14]. But, several bands at 1191, 2925, 1710 and 1610 cm−1 in FT-IR of Sp slightly shifted to 1218, 2950, 1729 and 1589 cm−1 in spectra of Sp-HSA and 1303, 2921, 1741 and 1606 cm−1 in spectra of Sp-HSA-DOX.
Fig. 5.
IR spectrum of Sp; Sp-HSA and Sp-HSA-DOX.
3.4. SEM and EDX
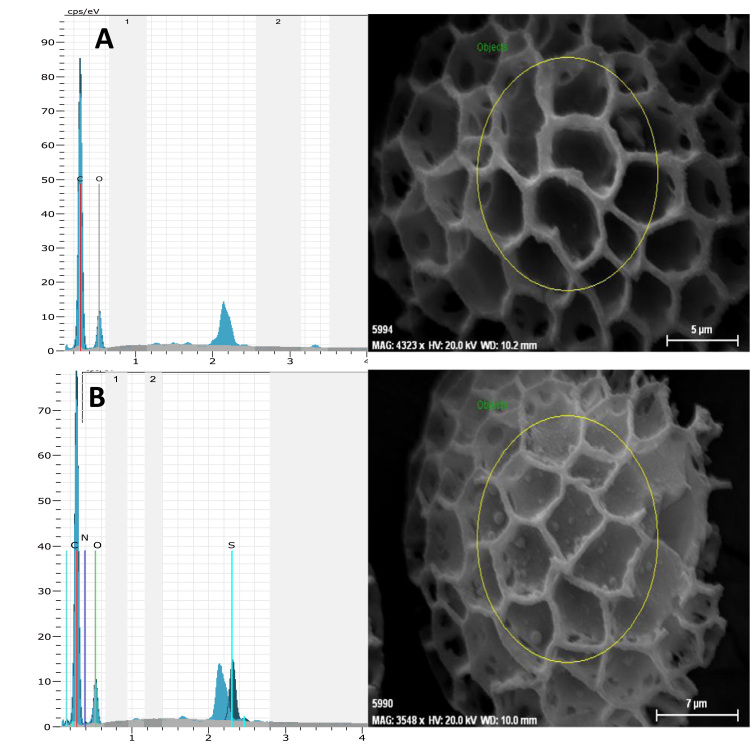
Scanning electron microscopy allowed the verification of morphological differences between sporopollenin and albumin. SEM images of Sp at 2 and 10 µm were shown in Fig. 6a-b. As wieved from shape of bare sporopollenin, the gaps on the Sp-HSA were detected. After albumin immobilized on the particles, the globular structure on Sp surface was observed at 5 µm as shown in Fig. 7a-b because of protein binding.
Fig. 6.
SEM images of sporopollen at 2 and 10 µm of scale.
Fig. 7.
EDX graphs and SEM images of Sp and Sp-HSA at 5 and 7 µm of scale.
EDX graphs of Sp related to SEM images that were given in 5 µm of scales (Fig. 7a-b). According to the results, C and O ratios of Sp were found to be as 57.53% and 42.47%, respectively. After binding of albumin to sporopollenin, nitrogen (8.98%) and sulfur (3.14%) were introduced to EDX graphs of Sp due to protein structure.
4. Conclusion
Albumin from human serum is responsible for transport of the drug. Therefore, albumin may be used in drug delivery systems based on nanoparticles and polymers. In this study, Sp-HSA particles were prepared successfully with a chemical method. It is the first time that we have developed albumin-sporopollenin system for doxorubicin loading in order to use drug delivery. As a result of this study, biological macromolecules are bioavailable and biocompatible materials as drug carrier due to release of the drug at different physiological pH. Therefore, newly developed Sp-HSA is efficient platform for new drugs.
Acknowledgement
We would like to thank The Research Foundation of Selcuk University (BAP) for financial support of this work.
Footnotes
Transparency document associated with this article can be found in the online version at doi:http://dx.doi.org/10.1016/j.bbrep.2016.06.012.
Appendix A. Transparency document
Supplementary material
.
References
- 1.Martino A.D., Kucharczyk P., Zednik J., Sedlarik V. Int. J. Pharm. 2015;496:912–921. doi: 10.1016/j.ijpharm.2015.10.017. [DOI] [PubMed] [Google Scholar]
- 2.Montha W., Maneeprakorn W., Buatong N., Tang I.-M., Pon-On W. Mater. Sci. Eng. C. 2016;59:235–240. doi: 10.1016/j.msec.2015.09.098. [DOI] [PubMed] [Google Scholar]
- 3.Tua H., Lua Y., Wua Y., Tian J., Zhan Y., Zeng Z., Deng H., Jianga L. Int. J. Pharm. 2015;493:426–433. doi: 10.1016/j.ijpharm.2015.07.063. [DOI] [PubMed] [Google Scholar]
- 4.Liu S., Zhang J., Cui X., Guo Y., Zhang X., Hongyan W. Coll. Surf. A: Physicochem. Eng. Asp. 2016;490:91–97. [Google Scholar]
- 5.Montha W., Maneeprakorn W., Buatong N., Tang I.-M., Pop-On W. Mater. Sci. Eng. C. 2016;59:235–240. doi: 10.1016/j.msec.2015.09.098. [DOI] [PubMed] [Google Scholar]
- 6.Shagholania H., Ghoreishib S.M., Mousazadeh M. Int. J. Biol. Macromol. 2015;78:130–136. doi: 10.1016/j.ijbiomac.2015.02.042. [DOI] [PubMed] [Google Scholar]
- 7.Yan E., Cao M., Wang Y., Hao X., Pei S., Gao J., Wang Y., Zhang Z., Zhang D. Mater. Sci. Eng. C. 2016;58:1090–1097. doi: 10.1016/j.msec.2015.09.080. [DOI] [PubMed] [Google Scholar]
- 8.Delmara K., Bianco-Peled H. Carbohyd. Polym. 2016;136:570–580. doi: 10.1016/j.carbpol.2015.09.072. [DOI] [PubMed] [Google Scholar]
- 9.Anirudhan T.S., Divya P.L., Nima J. Chem. Eng. J. 2016;284:1259–1269. [Google Scholar]
- 10.Ding X., Wang Y., Wang Y., Pan Q., Chen J., Huang Y., Xu K. Analy. Chim. Acta. 2015;861:36–46. doi: 10.1016/j.aca.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 11.Lee E.-J., Kim H.-E. Mater. Sci. Eng. C. 2016;59:339–345. doi: 10.1016/j.msec.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 12.Liu H., He J. Mater. Lett. 2015;161:415–418. [Google Scholar]
- 13.Balan V., Dodi G., Tudorachi N., Ponta O., Simon V., Butnaru M., Verestiuc L. Chem. Eng. J. 2015;279:188–197. [Google Scholar]
- 14.Gubbuk I.H., Maltas E., Ozmen M. Int. J. Biol. Macromol. 2012;50:1346–1352. doi: 10.1016/j.ijbiomac.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 15.Shaw G., Sykes M., Humble R.W., Mackenzie G., Marsden D., Pehlivan E. React. Pol. 2016;1988:211–217. [Google Scholar]
- 16.Souza S.P., Bassut J., Marquez H.V., Junior I.I., Miranda L.S.M., Huang Y., Mackenzie G., Boa A.N., Souza R.O.M.A. Catal. Sci. Technol. 2015;5:3130–3136. [Google Scholar]
- 17.Taboada A.D., Beckett S.T., Atkin S.L., Mackenzie G. Pharmaceutics. 2014;6:80–96. doi: 10.3390/pharmaceutics6010080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paunov V.N., Mackenzie G., Stoyanov S.D. J. Mater. Chem. 2007;17:609–612. [Google Scholar]
- 19.Taboada A.D., Maillet L., Banoub J.H., Lorch M., Rigby A.S., Boa A.N., Atkin S.L., Mackenzie G. J. Mater. Chem. B. 2013;1:707. doi: 10.1039/c2tb00228k. [DOI] [PubMed] [Google Scholar]
- 20.Thao L.Q., Byeon H.J., Lee C., Lee S., Lee E.S., Choi H.-G., Park E.-S., Youn Y.S. Int. J. Pharm. 2016;497:268–276. doi: 10.1016/j.ijpharm.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 21.Yu X., Jin C. J. Mater. Sci. Mater. Med. 2016;27:4. doi: 10.1007/s10856-015-5618-9. [DOI] [PubMed] [Google Scholar]
- 22.Hu Y.-J., Liu Y., Shen X.-S., Fang X.-Y., Qu S.-S. J. Mol. Struct. 2015;738:143–147. [Google Scholar]
- 23.Jiang W., Sun Z., Li F., Chen K., Liu T., Liu J., Zhou T., Guo R. J. Magn. Magn. Mater. 2011;323:435–439. [Google Scholar]
- 24.Wan X., Zheng X., Pang X., Zhang Z., Zhang Q. Coll. Surf. B: Biointerfaces. 2015;136:817–827. doi: 10.1016/j.colsurfb.2015.10.018. [DOI] [PubMed] [Google Scholar]
- 25.Maltas E., Ertekin B. Int. J. Biol. Macromol. 2015;72:984–989. doi: 10.1016/j.ijbiomac.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 26.Maltas E., Ozmen M. Mater. Sci. Eng. C. 2015;54:43–49. doi: 10.1016/j.msec.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 27.Manocha B., Margaritis A. J. Nanomater. 2010;2010:1–9. [Google Scholar]
- 28.Xiang Y., Liang L., Wang X., Wang J., Zhang X., Zhang Q. J. Control. Release. 2011;152:402–410. doi: 10.1016/j.jconrel.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 29.Zhang A.X., Shastry S., Bradforth S.E., Nadeau J.L. Nanoscale. 2015;7:240–251. doi: 10.1039/c4nr04707a. [DOI] [PubMed] [Google Scholar]
- 30.Anirudhan T.S., Divya P.L., Nima J. Chem. Eng. J. 2016;284:1259–1269. [Google Scholar]
- 31.Hua M.Y., Yang H.W., Liu H.L., Tsai R.-Y., Pang S.T., Chuang K.L., Chang Y.S., Hwang T.L., Chang Y.H., Chuang H.C., Chuang C.K. Biomaterials. 2011;32:8999–9010. doi: 10.1016/j.biomaterials.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 32.Jahanshahi M., Babaei Z. Afr. J. Biotech. 2008;7:4926–4934. [Google Scholar]
- 33.Chen P.C., Mwakwari S.C., Oyelere A.K. J. Nanotech. Sci. Appl. 2008;1:45–66. doi: 10.2147/nsa.s3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weber C., Coester C., Kreuter J., Langer K. Int. J. Pharm. 2000;194:91–102. doi: 10.1016/s0378-5173(99)00370-1. [DOI] [PubMed] [Google Scholar]
- 35.Tamyurek E., Maltas E., Bas S.Z., Ozmen M., Yildiz S. Int. J. Biol. Macromol. 2015;73:76–83. doi: 10.1016/j.ijbiomac.2014.10.061. [DOI] [PubMed] [Google Scholar]
- 36.Zhang L.N., Wu F.Y., Liu A.H. Spectrochim. Acta Part A: Mol. Biomol. Spectrosc. 2011;79:97. doi: 10.1016/j.saa.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 37.Guo M., Lu W.J., Li M.H., Wang W. Eur. J. Med. Chem. 2008;43:2140–2148. doi: 10.1016/j.ejmech.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 38.Yilmaz E., Sezgin M., Yilmaz M. J. Mol. Catal. B: Enzym. 2010;62:162–168. [Google Scholar]
- 39.Tutar H., Yilmaz E., Pehlivan E., Yilmaz M. Int. J. Biol. Macromol. 2009;45:315–320. doi: 10.1016/j.ijbiomac.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 40.Barth A. Biochim. Biophys. Acta. 2007;1767:1073–1101. doi: 10.1016/j.bbabio.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 41.Kong J., Yu Acta S. Biochim. Biophys. Sin. 2007;39:549–559. doi: 10.1111/j.1745-7270.2007.00320.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material