Abstract
Acetohydroxyacid synthase (AHAS) catalyzes the production of acetolactate from pyruvate. The enzyme from the hyperthermophilic bacterium Thermotoga maritima has been purified and characterized (kcat ~100 s−1). It was found that the same enzyme also had the ability to catalyze the production of acetaldehyde and CO2 from pyruvate, an activity of pyruvate decarboxylase (PDC) at a rate approximately 10% of its AHAS activity. Compared to the catalytic subunit, reconstitution of the individually expressed and purified catalytic and regulatory subunits of the AHAS stimulated both activities of PDC and AHAS. Both activities had similar pH and temperature profiles with an optimal pH of 7.0 and temperature of 85 °C. The enzyme kinetic parameters were determined, however, it showed a non-Michaelis-Menten kinetics for pyruvate only. This is the first report on the PDC activity of an AHAS and the second bifunctional enzyme that might be involved in the production of ethanol from pyruvate in hyperthermophilic microorganisms.
Abbreviations: AHAS, acetohydroxyacid synthase; BCAA, branched chain amino acid; CCE, crude cell extract; CFE, cell-free extract; HTCCE, heat-treated crude cell extract; IMAC, immobilized metal affinity chromatography; PDC, pyruvate decarboxylase; TmAHAS, Thermotoga maritima acetohydroxyacid synthase; TPP, thiamine pyrophosphate
Keywords: Thermotoga maritima, Pyruvate decarboxylase, Acetohydroxyacid synthase, Hyperthermophiles
Graphical abstract
Highlights
-
•
The acetohydroxyacid synthase of T. maritima has pyruvate decarboxylase activity
-
•
The AHAS and PDC activities share the same temperature and pH optima
-
•
Reconstitution of the catalytic and regulatory subunits increases both PDC and AHAS activities
1. Introduction
Ethanol is an end-product of fermentation in many extreme thermophiles and some hyperthermophiles [1], [2], [3], [4]. Although some of the enzymes (e.g., alcohol dehydrogenases) involved in the ethanol fermentation pathways have been isolated and characterized from hyper/thermophilic archaea and bacteria, there is limited information available about other enzymes involved in these metabolic pathways. One of the essential questions about the ethanol production by hyper/thermophiles, is the source of the aldehydes which are the substrates for the alcohol dehydrogenases catalyzing the production of alcohols.
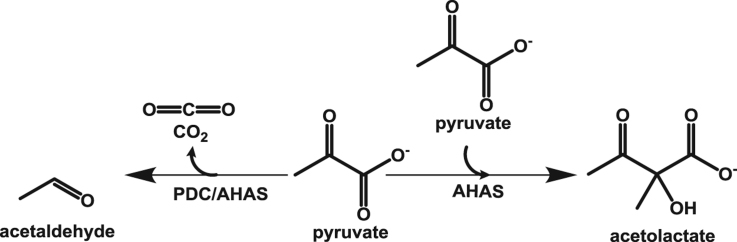
Acetaldehyde is the key intermediate in the ethanol production pathway and it can be produced by either non-oxidative decarboxylation of pyruvate catalyzed by TPP-dependent enzyme pyruvate decarboxylase (PDC, EC 4.1.1.1) or the reduction of acetyl-CoA catalyzed by a CoA-dependent acetaldehyde dehydrogenase (AcDH, EC 1.2.1.10). However, search of the genome sequences of the thermophilic and hyperthermophilic bacteria and archaea against the commonly-known (yeast and bacterial) PDC and AcDH amino acid sequences, failed to detect any orthologues [5]. Consequently, the potential role of alternative enzyme(s) for the catalysis of the acetaldehyde production from pyruvate is proposed. One such enzyme was discovered to be the bifunctional pyruvate decarboxylase/pyruvate ferredoxin oxidoreductase (PDC/POR) and was described the first time in the hyperthermophilic archaeon Pyrococcus furiosus [6]. The bifunctional enzyme is shown to be able to catalyze both non-oxidative and oxidative decarboxylation of pyruvate to produce acetaldehyde and acetyl-CoA, respectively [6], [7], [8] and the bifunctionality seems to be a universal trait of hyperthermophilic (archaeal and bacterial) PORs.
When the genome sequences of various archaea and bacteria were explored for the orthologues of the commonly-known PDCs, the closest hit (when present) is almost exclusively the enzyme acetohydroxyacid synthase (Supplementary Table 1). Like PDC, acetohydroxyacid synthase (AHAS) is a thiamine pyrophosphate (TPP) – and Mg2+-dependent enzyme. Given the high homology levels of the AHAS to the commonly-known PDCs and considering the high frequency of multi-functionality in the superfamily of TPP-dependent enzymes, the question arises: can the enzyme AHAS also catalyze the non-oxidative decarboxylation of pyruvate to acetaldehyde?
It has been shown that Thermotoga maritima does not require the addition of any branched chain amino acids (BCAA) to the growth medium for optimal growth, suggesting the organism is equipped for biosynthesis of these compounds [9]. The properties of an anabolic AHAS from the hyperthermophilic bacterium T. maritima has recently been reported [10]. The purified catalytic subunit of the enzyme is FAD- and TPP-dependent and efficiently catalyzes the production of acetolactate from pyruvate with a kcat of about 100 s−1. Here we report the determination of the ability of this very same enzyme for catalyzing the production of the acetaldehyde via a non-oxidative decarboxylation of pyruvate.
2. Materials and methods
2.1. Recombinant protein expression and purification
The recombinant plasmids for the expression of genes encoding the catalytic and regulatory subunits were constructed and propagated in Escherichia coli strain DH5α. The recombinant proteins overexpressed under the control of T7 polymerase of the plasmid pET30a (+) (Novagen, WI, USA) in E. coli BL21 (DE3) Rosetta 2 [F− ompT hsdSB (rB− mB−) gal dcm pRARE27 (CamR)] (Novagen, WI, USA) as described elsewhere [10], [11]. The recombinant catalytic subunit was purified by heat-induced precipitation of the host proteins (1 h at 80 °C) followed by DEAE and HAP column chromatography as described previously [10], [11]. The AHAS activity was followed during the purification and the purity of the final active fractions were tested by SDS-PAGE. The inactive small subunit was also overexpressed in the same host and purified by denaturing IMAC and on column refolding of the denatured protein using a previously described procedure [10]. The purity of the desired protein in the fractions was checked on SDS-PAGE. When necessary, anaerobic procedures were followed as described elsewhere [7].
2.2. Enzyme assays
A discontinuous colorimetric assay was used to measure the AHAS activity via the production of acetolactate from pyruvate as described elsewhere [10].
PDC activity was assayed by measuring the rate of the production of acetaldehyde from pyruvate [7], [8]. The final concentration of acetaldehyde and isobutyraldehyde were determined using a calibration curve prepared by linear regression plotting of known concentrations of each product processed under the same assay conditions. Unless specified, both PDC and AHAS activities were measured using the purified large (catalytic) subunit of TmAHAS.
All enzyme assays were carried out in duplicates, in 8 ml vials under anaerobic conditions at 80 °C, unless otherwise specified. When specified, sodium pyruvate was replaced with 50 mM of 2-ketoisovalerate.
2.3. Biochemical and biophysical characterization
The pH dependency of both activities of PDC and AHAS were determined in a range between 4.0 and 11.0. The pH values expressed throughout this manuscript were adjusted and measured at room temperature. In each case, assays were carried out at 80 °C, under anaerobic conditions using 100 mM buffers degassed as described previously. The following list of buffers was used for the various pH values: sodium acetate buffer (pKa=4.76, ΔpKa/°C=0.0002) was used for the pH range between 4 and 5.6, sodium phosphate buffer (pKa 7.20, ΔpKa/°C=−0.0028) for pH values 6.0, 7.0, and 7.5, HEPES buffer (pKa 7.39, ΔPKa/°C=−0.014) for pH values 7.5, 8.0, 8.5 and 9.0 and glycine buffer (pKa 9.55, ΔpKa/°C=−0.0025) the pH values 9.0, 9.5, 10.0, and 10.5. Finally, CAPS buffer (pKa 10.40, ΔpKa/°C=−0.009) was used 10.0, 10.5, and 11.0.
The steady state kinetics was determined under optimal assay conditions and at 80 °C. The kinetic parameters were determined for pyruvate (5–125 mM), TPP (0.05–4 mM), and FAD (0.05–40 µM) by applying various concentrations of each component and keeping the concentration of other assay components constant. All of the assays were repeated in triplicate. The activity then was assayed for various concentrations, and the kinetic parameters were calculated from the best fit of the results to the Michaelis-Menten equation by non-linear regression using SigmaPlot® software (SYSTAT Software Inc., CA, USA). The oxygen sensitivity of each activity was tested by exposing an enzyme aliquot to ambient atmosphere at 4 °C by gentle stirring and comparing the PDC and AHAS activities at different time courses with the control preparation kept under anaerobic conditions. Both enzyme samples were protected from light through the experiments.
The temperature dependence of enzymes was determined by measuring the enzyme activity at different temperatures ranging from 30 °C to 95 °C under anaerobic conditions and in 100 mM EPPS buffer, pH 8.4 containing 10 mM MgCl2. Thermal stability of the enzyme was determined by incubation of an anaerobic enzyme preparation and determining the residual activity at different time points compared to unheated control.
3. Results and discussion
AHAS is a member of the decarboxylase family that catalyzes the reactions whose initial step is decarboxylation of 2-keto acids. The decarboxylase family is one of the five members of TPP-dependent enzymes superfamily [12]. AHAS shares a common ancestry with pyruvate decarboxylase [13], [14], [15] as is indeed apparent from its sequence homology levels to the PDCs from various organisms (Supplementary Table 1) and pyruvate oxidase (POX) [16]. Considering the catalytic mechanism of the enzyme, it is also believed that the enzyme is very close to pyruvate decarboxylase.
The recombinant AHAS from T. maritima is shown to catalyze the production of the acetolactate from pyruvate in a TPP- and FAD-dependent manner [10]. The cell free extracts (CFEs) prepared from the clones expressing the large (catalytic) subunits showed PDC activity and no detectable PDC activity was observed in the CFEs prepared from the clones expressing the small (regulatory) subunit. The purified TmAHAS exhibited a specific activity of 17.7 U mg−1 for PDC at 80 °C.
The ability of a catabolic AHAS (ALS) in catalyzing non-oxidative decarboxylation of 2-ketoisobutyrate (3-methyl-2-oxobutyrate) is reported [17]. To investigate the ability of the recombinant anabolic TmAHAS for non-oxidative decarboxylation of other 2-keto acids, the decarboxylase activity assays were conducted with 2-ketoisovalerate instead of pyruvate. A standard curve was prepared with the product of decarboxylation reaction, isobutyraldehyde. Although with lower affinity, the recombinant TmAHAS was able to non-oxidatively decarboxylate the 2-ketoisovalerate to isobutyraldehyde with a rate about 10% of that when pyruvate was used.
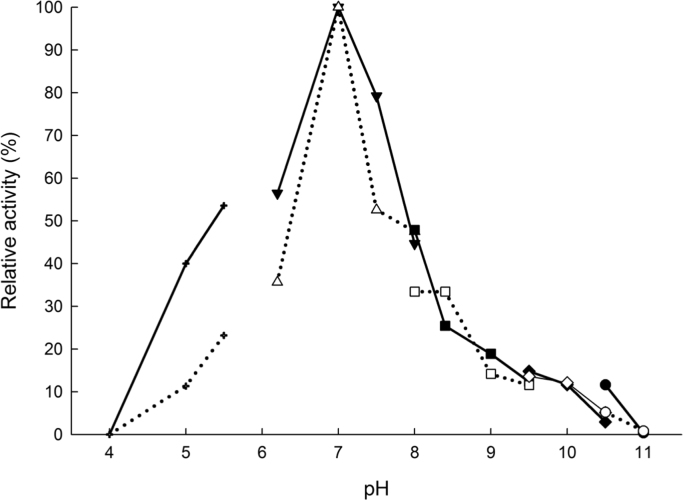
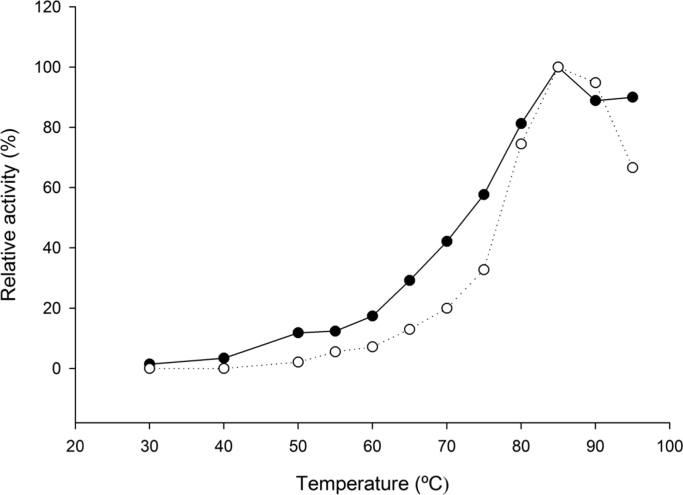
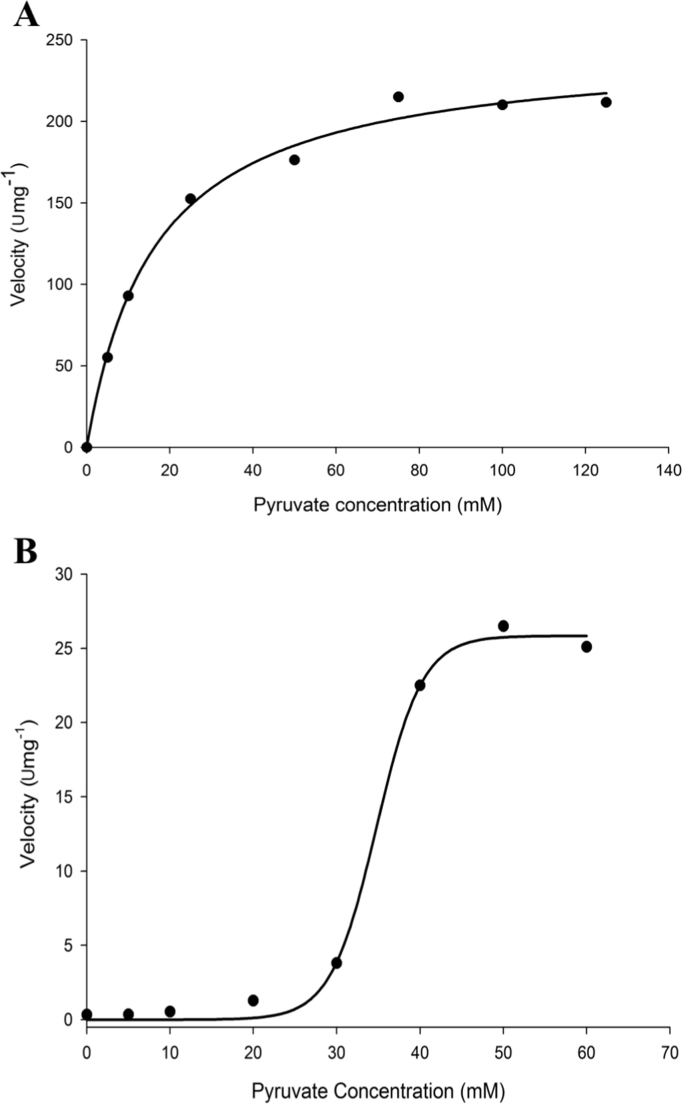
The optimal pH and temperature for both PDC and AHAS activities were 7.0 (Fig. 1) and 85 °C (Fig. 2), respectively. Both PDC and AHAS activities were quite stable and only about 40% of each activity were lost after five days incubation at 4 °C (without stirring). The kinetic parameters were determined for each substrate (pyruvate, TPP, and FAD) at pH 7.0 and 80 °C for both PDC and AHAS activities (Table 1). Interestingly the effect of the pyruvate concentration on the velocity of the PDC reaction catalyzed by TmAHAS did not follow a usual Michaelis-Menten kinetics, but displayed a sigmoid cooperative effect (Fig. 3). Several repeats of the same experiment resulted in the similar curve shapes. The non-Michaelis-Menten kinetics for AHAS substrate pyruvate is a common phenomenon and the positive cooperativity is reported from the AHAS activities of Lactococcus lactis [18], Serratia marcescens [19], Haemophilus influenza [20], [21], Mycobacterium tuberculosis [22], [23], and the mesophilic archaeon Methanococcus aeolicus [24]. However, AHAS activity of the TmAHAS followed the Michaelis-Menten kinetics, indicating a possible change of enzyme conformation induced by the substrate pyruvate affects its PDC activity only. It is assumed that the active site for catalyzing both PDC and AHAS activities should be the same. But how such conformational change could only affect the PDC not AHAS activity remains unknown. A structural study of the enzyme by using X-ray crystallography maybe helpful to provide answers to the question in the future. The difference in catalysis by PDC and AHAS may also be shown by slightly higher Km values measured for AHAS activity, because the apparent Km values for FAD of both activities were 0.12 µM and 0.15 µM for PDC and AHAS, respectively, and the apparent Km value for TPP were 35 µM and 57 µM for PDC and AHAS, respectively. No apparent Km value for pyruvate due to the non-Michaelis-Menten kinetics of the PDC activity (Table 1).
Fig. 1.
pH dependency of PDC and AHAS activities of recombinant TmAHAS. The relative activities of 100% equals to highest measured specific activity for both enzymes at 80 °C and in sodium phosphate buffer, pH 7.0 (17.7 U mg−1 for PDC and 114 U mg−1 for AHAS activity). The open symbols represent the PDC activity and the filled symbols represent the AHAS activity. The plus symbols represent the sodium acetate buffer (pH values 4.0, 5.0, and 5.5); the triangles and inverted triangles represent the sodium phosphate buffer (pH 6.2, 7.0, 7.5, and 8.0); the square symbols represent the EPPS buffer (pH 8.0, 8.4, 9.0, and 9.5); the diamonds represent glycine buffer (pH 9.5 and 10); and circles represent the CAPS buffer (pH 10.5 and 11.0). Each point is an average of experimental values in duplicates.
Fig. 2.
Determination of optimal temperatures of PDC and AHAS activities of recombinant TmAHAS. The reaction mixtures were equilibrated at each temperature for 4 min before adding the enzyme. The relative activities of 100% equals to highest measured specific activity for both enzymes at 85 °C (18.6 U mg−1 for PDC and 235 U mg−1 for AHAS activity). The open symbols indicate the PDC and the filled symbols indicate the AHAS activity. Each point is an average of experimental values in duplicates.
Table 1.
Apparent kinetic parameters for PDC and AHAS activities of recombinant TmAHAS.
| Substrate | AHAS activitya |
PDC activity |
||||
|---|---|---|---|---|---|---|
| Apparent Km (µM) | Apparent Vmax (U mg−1) | kcat (S−1) | Apparent Km (µM) | Apparent Vmax (U mg−1) | kcat (S−1) | |
| Pyruvate | 16,400±1900 | 246±7 | 99 | ND | ND | ND |
| TPP | 57±6.0 | 242±4 | 97 | 35±0.6 | 25.9±0.6 | 8 |
| FAD | 0.15±0.07 | 134±11 | 54 | 0.12±0.05 | 18.1±1.7 | 7 |
ND, not determined as the saturation curve was not following the Michaelis-Menten kinetics.
Kinetic parameters are from [10].
Fig. 3.
Dependence of PDC (B) and AHAS (A) activities on pyruvate concentration. Each point is an average of experimental values in duplicates (three independent repeats). The curves are the theoretical fit of data to the hyperbolic and sigmoid plot equations for PDC and AHAS activities, respectively.
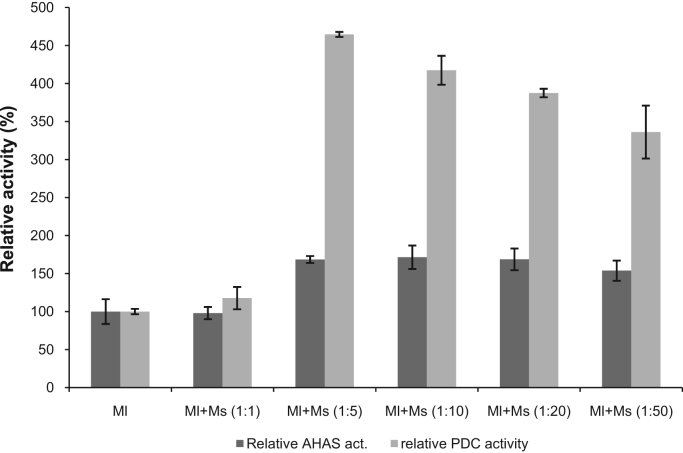
Activation of the enzyme by a rapid and cooperative reconstitution of the catalytic and regulatory subunit is shown for various mesophilic [22], [25], [26], [27] and hyperthermophilic AHAS [10]. The reconstitution of the enzyme TmAHAS resulted in considerable and reproducible increase of both PDC and AHAS activities. Both activities followed the same increment trend for most part, however, it appeared that when 1:5 ratio of catalytic and regulatory subunits were mixed, and the stimulatory effect of the regulatory subunit became significant as there were a 3-fold increase in PDC activity and only a slight increase in AHAS activity; But, no further increase of both activities were observed by mixing increasing ratios of catalytic to regulatory subunit up to 1:50 (Fig. 4). The small (regulatory) subunit MS in crude extract, cell-free extract and as a purified protein showed no activity of PDC and AHAS. Therefore, the increase in activities shown in Fig. 4 can be only resulted from the interaction of MS with the large (catalytic) subunit MI. However, only MI was used for measuring the activities shown in Fig. 3, indicating a conformational change induced by the increase of the substrate concentration (pyruvate), which affected the PDC activity only (not AHAC). Further studies including X-ray crystallography are required for understanding the mechanism involved.
Fig. 4.
Effect of enzyme reconstitution on PDC and AHAS activities. Ml, the purified catalytic subunit of TmAHAS; Ms, the purified regulatory subunit of TmAHAS. The specific activity of 100% was considered for PDC and AHAS activities of catalytic subunit alone and was corresponding to 9.5 U mg−1 for PDC and 145 U mg−1 for AHAS activity.
As a well-known side reaction, AHASs are also capable of catalyzing the carboligation reactions and producing α-hydroxy ketones. During the reaction, decarboxylation of pyruvate generates a hydroxyethyl-TPP (HETDP), also known as “active acetaldehyde”, intermediate [28] and the stereospecific condensation of this intermediate with aldehydes produces α-hydroxy ketones [29]. The reaction has been extensively studied in pyruvate decarboxylases [30], and there have been some studies demonstrating higher efficiency of AHASs than the pyruvate decarboxylases, presumably because the condensation reaction is an intrinsic property of the AHASs [31], [32]. Similar to PDC [29], [33] AHAS I from E. coli is shown to be able to use a wide range of aldehydes as the second substrate, including benzaldehydes, heterocyclic, hetero-aromatic and non-aromatic aldehydes [34]. As another side reaction, it is reported that some AHASs can catalyze oxidative decarboxylation of pyruvate or 2-oxobutyrate, yielding peracetic acid or peroxypropionic acid, respectively. During this reaction, the enzyme bond hydroxyethyl-TPP (E-HETPP) reacts with the molecular oxygen, leading to the formation of hydroperoxide-HETPP, which is then decomposed to peracetic acid and TPP. The rate of this oxygen-consuming reaction is approximately 1% of the AHAS activity in case of AHAS II from E. coli [35], [36]. Within the members of the TPP-dependent enzymes, AHAS and pyruvate decarboxylases are the only enzymes that can catalyze this oxygenase side reaction [37]. The PDC activity that is being reported here can be considered to be another “side reaction” of AHASs, though other possible side reactions of the enzyme were not tested.
The principal catalytic mechanism of AHAS, follows the activation step of the enzyme is similar to those of the other pyruvate utilizing TPP-dependent enzymes especially the classic example of this group, PDC [38]. Protonation of the N1 atom of pyrimidine ring by the highly conserved glutamine residue results in induction of a 1,4-iminotautomer. Due to the common “V-conformation” the 4-imino group is located in the proximity of the C2 catalytic center of the AHAS, which results in de-protonation of the C2 atom. The proton abstraction step gives rise to a highly reactive ylide [39], [40]. The nucleophilic attack of the ylide to the first molecule of pyruvate, results in production of 2-(2-lactyl)-TPP (also known as L-TPP or L-TDP). Later, decarboxylation of L-TPP gives the resonating carbanion/enamine forms of 2-(1-hydroxyethyl)-TPP (HETPP, also known as hydroxyethylidene-TPP), which acts as a central and highly reactive intermediate [6], [41], [42], [43].
During the normal course of the catalysis, HETPP intermediate reacts in an enantio-specific manner with the second molecule of 2-keto acid (usually pyruvate or 2-ketobutyrate), resulting in the release of the product. The ability of heterologously expressed TmAHAS to generate acetaldehyde is most likely the result of protonation of the HETDP/enamine intermediate in C2 position. This step, results in the releases of acetaldehyde from active site of the enzyme. Unfortunately, with the available data on recombinant TmAHAS, it is not possible to deduce further mechanistic details of PDC activity of the enzyme. Complete and accurate understanding of the mechanisms involved in acetaldehyde formation demands more in-depth study of the reaction intermediates and more importantly crystal structure of enzyme. Other than a preliminary X-ray crystal structure for catalytic subunit of E. coli AHAS II [44], so far, no crystal structure has been provided for any catalytic (large) subunit of AHAS [45].
Catabolic AHAS (ALS) has been isolated and characterized mainly from enteric bacteria, including Enterobacter, certain Klebsiella and Serratia species, some lactic acid bacteria, and Bacillus subtilis. A typical ALS is an FAD-independent enzyme, composed of only one subunit of ~60 kDa (no regulatory subunit) and the gene is generally located in the butanediol operon (when present). Despite catalyzing same reactions, anabolic and catabolic AHASs are very divergent, showing less than 30% amino acid sequence identity with each other [37], [45], [46]. Different factors can trigger activation of butanediol operon in bacteria, including low pH, presence of excess acetate and/or pyruvate, as well as growth during the stationary phase [46], [47], [48], [49], [50], [51]. As an important development, it is shown that besides its synthase activity, the catabolic ALS of B. subtilis can also catalyze the non-oxidative decarboxylation of the 2-ketoisovalerate both in vitro and in vivo [17]. Considering the major mechanistic similarities between the anabolic and catabolic AHAS, the involvement of the anabolic AHAS in the similar non-oxidative decarboxylation reaction is not very surprising. However, understanding the mechanisms of differential non-Michaelis-Menten kinetics of TmAHAS may provide further insight into enzyme multifunctionality.
Besides the bifunctional PDC/POR enzymes characterized in different hyperthermophilic archaea [6], [7] and bacteria [8], PDC/AHAS of T. maritima is the second type of acetaldehyde producing enzymes characterized in hyperthermophiles. Similar to the archaeal PDC/POR bifunctionality (and unlike the bacterial PDC/POR activity), the PDC to AHAS activity ratio was high. Thermotogales have been shown to be able to use both carbohydrates (simple and complex) and peptides as a source of carbon and energy, and many of them are capable of de novo synthesis of amino acids required for growth [10]. Presumably, the “extra” amino acids imported and/or synthesized inside the cells would be channeled to the energy conservation process, during which they are decarboxylated after a transamination step. The reduced ferredoxin produced during the POR reaction can be used via different hydrogenase enzymes for energy conservation and acetyl-CoA is used for both biosynthesis and energy conservation.
It seems that keto acids produced by catabolism of different amino acids and/or carbohydrates can be decarboxylated both oxidatively and non-oxidatively via either bifunctional PDC/POR or, when present, the bifunctional PDC/AHAS. The aldehydes resulted from the non-oxidative decarboxylation of keto acids can be further reduced to produce corresponding alcohols. This reduction is accompanied by the regeneration of oxidized NADP+.
Acknowledgments
The authors are grateful to Mr. Alton Wong, Benozir Sarafuddin, and Frank Gong for their helps with growth of the organisms and some purification aspects. This work was supported by research grant from the Natural Sciences and Engineering Research Council of Canada (NSERC), (grant number 227233) to KM.
Footnotes
Transparency document associated with this article can be found in the online version at 10.1016/j.bbrep.2016.07.008.
Appendix A. Transparency document
Supplementary material
.
Supplementary material
.
Supplementary material
.
References
- 1.Ma K., Loessner H., Heider J., Johnson M., Adams M. Effects of elemental sulfur on the metabolism of the deep-sea hyperthermophilic archaeon Thermococcus strain ES-1: characterization of a sulfur-regulated, non-heme iron alcohol dehydrogenase. J. Bacteriol. 1995;177:4748–4756. doi: 10.1128/jb.177.16.4748-4756.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balk M., Weijma J., Stams A.J.M. Thermotoga lettingae sp. nov., a novel thermophilic, methanol-degrading bacterium isolated from a thermophilic anaerobic reactor. Int. J. Syst. Evol. Microbiol. 2002;52:1361–1368. doi: 10.1099/00207713-52-4-1361. [DOI] [PubMed] [Google Scholar]
- 3.de Vrije T., Bakker R., Budde M., Lai M., Mars A., Claassen P. Efficient hydrogen production from the lignocellulosic energy crop Miscanthus by the extreme thermophilic bacteria Caldicellulosiruptor saccharolyticus and Thermotoga neapolitana. Biotechnol. Biofuels. 2009;2:12. doi: 10.1186/1754-6834-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ying X., Ma K. Characterization of a zinc-containing alcohol dehydrogenase with stereoselectivity from the hyperthermophilic archaeon Thermococcus guaymasensis. J. Bacteriol. 2011;193:3009–3019. doi: 10.1128/JB.01433-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eram M.S., Ma K. Decarboxylation of pyruvate to acetaldehyde for ethanol production by hyperthermophiles. Biomolecules. 2013;3:578–596. doi: 10.3390/biom3030578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma K., Hutchins A., Sung S.-J.S., Adams M.W.W. Pyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon, Pyrococcus furiosus, functions as a CoA-dependent pyruvate decarboxylase. Proc. Natl. Acad. Sci. USA. 1997;94:9608–9613. doi: 10.1073/pnas.94.18.9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eram M.S., Oduaran E., Ma K. The bifunctional pyruvate decarboxylase/pyruvate ferredoxin oxidoreductase from Thermococcus guaymasensis. Archaea. 2014 doi: 10.1155/2014/349379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eram M.S., Wong A., Oduaran E., Ma K. Molecular and biochemical characterization of bifunctional pyruvate decarboxylases and pyruvate ferredoxin oxidoreductases from Thermotoga maritima and Thermotoga hypogea. J. Biochem. 2015 doi: 10.1093/jb/mvv058. [DOI] [PubMed] [Google Scholar]
- 9.Rinker K.D., Kelly R.M. Effect of carbon and nitrogen sources on growth dynamics and exopolysaccharide production for the hyperthermophilic archaeonThermococcus litoralis and bacterium Thermotoga maritima. Biotechnol. Bioeng. 2000;69:537–547. doi: 10.1002/1097-0290(20000905)69:5<537::aid-bit8>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 10.Eram M.S., Sarafuddin B., Gong F., Ma K. Characterization of acetohydroxyacid synthase from the hyperthermophilic bacterium Thermotoga maritima. Biochem. Biophys. Rep. 2015;4:89–97. doi: 10.1016/j.bbrep.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eram M.S., Sarafuddin B., Gong F., Ma K. Optimization of expression and properties of the recombinant acetohydroxyacid synthase of Thermotoga maritima. Data in Brief. 2015;5:489–497. doi: 10.1016/j.dib.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duggleby R.G. Domain relationships in thiamine diphosphate-dependent enzymes. Acc. Chem. Res. 2006;39:550–557. doi: 10.1021/ar068022z. [DOI] [PubMed] [Google Scholar]
- 13.Neale A.D., Scopes R.K., Wettenhall R.E.H., Hoogenraad N.J. Nucleotide sequence of the pyruvate decarboxylase gene from Zymomonas mobilis. Nucleic Acids Res. 1987;15:1753–1761. doi: 10.1093/nar/15.4.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green J.B.A. Pyruvate decarboxylase is like acetolactate synthase (ILV2) and not like the pyruvate dehydrogenase E1 subunit. FEBS Lett. 1989;246:1–5. doi: 10.1016/0014-5793(89)80241-8. [DOI] [PubMed] [Google Scholar]
- 15.Kellermann E., Seeboth P.G., Hollenberg C.P. Analysis of the primary structure and promoter function of a pyruvate decarboxylase gene (PDCI) from Saccharomyces cerevisiae. Nucleic Acids Res. 1986;14:8963–8977. doi: 10.1093/nar/14.22.8963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang Y.Y., Cronan J.E., Jr. Common ancestry of Escherichia coli pyruvate oxidase and the acetohydroxy acid synthases of the branched-chain amino acid biosynthetic pathway. J. Bacteriol. 1988;170:3937–3945. doi: 10.1128/jb.170.9.3937-3945.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Atsumi S., Li Z., Liao J.C. Acetolactate synthase from Bacillus subtilis serves as a 2-Ketoisovalerate decarboxylase for isobutanol biosynthesis in Escherichia coli. Appl. Environ. Microbiol. 2009;75:6306–6311. doi: 10.1128/AEM.01160-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snoep J.L., Teixeira de Mattos M.J., Starrenburg M.J., Hugenholtz J. Isolation, characterization, and physiological role of the pyruvate dehydrogenase complex and alpha-acetolactate synthase of Lactococcus lactis subsp. lactis bv. diacetylactis. J. Bacteriol. 1992;174:4838–4841. doi: 10.1128/jb.174.14.4838-4841.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang J.H., Kim S.S. Purification and characterization of the valine sensitive acetolactate synthase from Serratia marcescens ATCC 25419. Biochim. Biophys. Acta (BBA) 1993;1157:178–184. doi: 10.1016/0304-4165(93)90062-d. [DOI] [PubMed] [Google Scholar]
- 20.Gedi V., Moon J.-Y., Lim W.-M., Lee M.-Y., Lee S.-C., Koo B.-S., Govindwar S., Yoon M.-Y. Identification and characterization of inhibitors of Haemophilus influenzae acetohydroxyacid synthase. Enzym. Microb. Technol. 2011;49:1–5. doi: 10.1016/j.enzmictec.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 21.Choi K.-J., Noh K.M., Kim D.-E., Ha B.H., Kim E.E., Yoon M.-Y. Identification of the catalytic subunit of acetohydroxyacid synthase in Haemophilus influenzae and its potent inhibitors. Arch. Biochem. Biophys. 2007;466:24–30. doi: 10.1016/j.abb.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 22.Choi K.-J., Yu Y.G., Hahn H.G., Choi J.-D., Yoon M.-Y. Characterization of acetohydroxyacid synthase from Mycobacterium tuberculosis and the identification of its new inhibitor from the screening of a chemical library. FEBS Lett. 2005;579:4903–4910. doi: 10.1016/j.febslet.2005.07.055. [DOI] [PubMed] [Google Scholar]
- 23.Singh V., Chandra D., Srivastava B.S., Srivastava R. Biochemical and transcription analysis of acetohydroxyacid synthase isoforms in Mycobacterium tuberculosis identifies these enzymes as potential targets for drug development. Microbiology. 2011;157:29–37. doi: 10.1099/mic.0.041343-0. [DOI] [PubMed] [Google Scholar]
- 24.Xing R., Whitman W.B. Purification and characterization of the oxygen-sensitive acetohydroxy acid synthase from the archaebacterium Methanococcus aeolicus. J. Bacteriol. 1994;176:1207–1213. doi: 10.1128/jb.176.5.1207-1213.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Porat I., Vinogradov M., Vyazmensky M., Lu C.-D., Chipman D.M., Abdelal A.T., Barak Ze. Cloning and characterization of acetohydroxyacid synthase from Bacillus stearothermophilus. J. Bacteriol. 2004;186:570–574. doi: 10.1128/JB.186.2.570-574.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vinogradov V., Vyazmensky M., Engel S., Belenky I., Kaplun A., Kryukov O., Barak Z.E., Chipman D.M. Acetohydroxyacid synthase isozyme I from Escherichia coli has unique catalytic and regulatory properties. Biochim. Biophys. Acta (BBA) 2006;1760:356–363. doi: 10.1016/j.bbagen.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 27.Pang S.S., Duggleby R.G. Expression, purification, characterization, and reconstitution of the large and small subunits of yeast acetohydroxyacid synthase. Biochemistry. 1999;38:5222–5231. doi: 10.1021/bi983013m. [DOI] [PubMed] [Google Scholar]
- 28.Hübner G., Tittmann K., Killenberg-Jabs M., Schäffner J., Spinka M., Neef H., Kern D., Kern G., Schneider G., Wikner C., Ghisla S. Activation of thiamin diphosphate in enzymes. Biochim. Biophys. Acta (BBA) – Protein Struct. Mol. Enzymol. 1998;1385:221–228. doi: 10.1016/s0167-4838(98)00070-3. [DOI] [PubMed] [Google Scholar]
- 29.Iding H., Siegert P., Mesch K., Pohl M. Application of alpha-keto acid decarboxylases in biotransformations. Biochim. Biophys. Acta. 1998;1385:307–322. doi: 10.1016/s0167-4838(98)00076-4. [DOI] [PubMed] [Google Scholar]
- 30.Ward O.P., Singh A. Enzymatic asymmetric synthesis by decarboxylases. Curr. Opin. Biotechnol. 2000;11:520–526. doi: 10.1016/s0958-1669(00)00139-7. [DOI] [PubMed] [Google Scholar]
- 31.Engel S., Vyazmensky M., Berkovich D., Barak Ze, Merchuk J., Chipman D.M. Column flow reactor using acetohydroxyacid synthase I from Escherichia coli as catalyst in continuous synthesis of R-phenylacetylcarbinol. Biotechnol. Bioeng. 2005;89:733–740. doi: 10.1002/bit.20392. [DOI] [PubMed] [Google Scholar]
- 32.Engel S., Vyazmensky M., Geresh S., Barak Z.E., Chipman D.M. Acetohydroxyacid synthase: a new enzyme for chiral synthesis of R-phenylacetylcarbinol. Biotechnol. Bioeng. 2003;83:833–840. doi: 10.1002/bit.10728. [DOI] [PubMed] [Google Scholar]
- 33.Crout D.H.G., Davies S., Heath R.J., Miles C.O., Rathbone D.R., Swoboda B.E.P., Gravestock M.B. Applications of hydrolytic and decarboxylating enzymes in biotransformations. Biocatal. Biotransform. 1994;9:1–30. [Google Scholar]
- 34.Engel S., Vyazmensky M., Berkovich D., Barak Ze, Chipman D.M. Substrate range of acetohydroxy acid synthase I from Escherichia coli in the stereoselective synthesis of α-hydroxy ketones. Biotechnol. Bioeng. 2004;88:825–831. doi: 10.1002/bit.20275. [DOI] [PubMed] [Google Scholar]
- 35.Abell L.M., Schloss J.V. Oxygenase side reactions of acetolactate synthase and other carbanion-forming enzymes. Biochemistry. 2002;30:7883–7887. doi: 10.1021/bi00246a002. [DOI] [PubMed] [Google Scholar]
- 36.Tse J.M.T., Schloss J.V. The oxygenase reaction of acetolactate synthase. Biochemistry. 1993;32:10398–10403. doi: 10.1021/bi00090a015. [DOI] [PubMed] [Google Scholar]
- 37.Chipman D., Barak Ze, Schloss J.V. Biosynthesis of 2-aceto-2-hydroxy Acids: Acetolactate synthases and Acetohydroxyacid synthases. Biochim. Biophys. Acta (BBA) – Protein Struct. Mol. Enzymol. 1998;1385:401–419. doi: 10.1016/s0167-4838(98)00083-1. [DOI] [PubMed] [Google Scholar]
- 38.Bar-Ilan A., Balan V., Tittmann K., Golbik R., Vyazmensky M., Hubner G., Barak Ze, Chipman D.M. Binding and activation of thiamin diphosphate in acetohydroxyacid synthase. Biochemistry. 2001;40:11946–11954. doi: 10.1021/bi0104524. [DOI] [PubMed] [Google Scholar]
- 39.Lie M.A., Celik L., Jorgensen K.A., Schiott B. Cofactor activation and substrate binding in pyruvate decarboxylase. Biochemistry. 2005;44:14792–14806. doi: 10.1021/bi051134y. [DOI] [PubMed] [Google Scholar]
- 40.McCourt J.A., Duggleby R.G. Acetohydroxyacid synthase and its role in the biosynthetic pathway for branched-chain amino acids. Amino Acids. 2006;31:173–210. doi: 10.1007/s00726-005-0297-3. [DOI] [PubMed] [Google Scholar]
- 41.Kluger R., Tittmann K. Thiamin diphosphate catalysis: enzymic and nonenzymic covalent intermediates. Chem. Rev. 2008;108:1797–1833. doi: 10.1021/cr068444m. [DOI] [PubMed] [Google Scholar]
- 42.Candy J.M., Duggleby R.G. Structure and properties of pyruvate decarboxylase and site-directed mutagenesis of the Zymomonas mobilis enzyme. Biochim. Biophys. Acta. 1998;1385:323–338. doi: 10.1016/s0167-4838(98)00077-6. [DOI] [PubMed] [Google Scholar]
- 43.Tittmann K., Golbik R., Uhlemann K., Khailova L., Schneider G., Patel M., Jordan F., Chipman D.M., Duggleby R.G., Hubner G. NMR analysis of covalent intermediates in thiamin diphosphate enzymes. Biochemistry. 2003;42:7885–7891. doi: 10.1021/bi034465o. [DOI] [PubMed] [Google Scholar]
- 44.Niu X., Liu X., Zhou Y., Niu C., Xi Z., Su X.-D. Preliminary X-ray crystallographic studies of the catalytic subunit of Escherichia coli AHAS II with its cofactors. Acta Crystallogr. Sect. F. 2011;67:659–661. doi: 10.1107/S1744309111008839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gedi V., Yoon M.-Y. Bacterial acetohydroxyacid synthase and its inhibitors – a summary of their structure, biological activity and current status. FEBS J. 2012;279:946–963. doi: 10.1111/j.1742-4658.2012.08505.x. [DOI] [PubMed] [Google Scholar]
- 46.Duggleby R.G., Pang S.S. Acetohydroxyacid synthase. J. Biochem. Mol. Biol. 2000;33:1–36. [Google Scholar]
- 47.Störmer F.C. The pH 6 acetolactate-forming enzyme from Aerobacter aerogenes. I. Kinetic studies. J. Biol. Chem. 1968;243:3735–3739. [PubMed] [Google Scholar]
- 48.Störmer F.C. The pH 6 acetolactate-forming enzyme from Aerobacter aerogenes.II. Evidence that it is not a flavoprotein. J. Biol. Chem. 1968;243:3740–3741. [PubMed] [Google Scholar]
- 49.Störmer F.C. Evidence for induction of the 2,3-butanediol-forming enzymes in Aerobacter aerogenes. FEBS Lett. 1968;2:36–38. doi: 10.1016/0014-5793(68)80094-8. [DOI] [PubMed] [Google Scholar]
- 50.Tsau J.-L., Guffanti A.A., Montville T.J. Conversion of pyruvate to acetoin helps to maintain pH homeostasis in Lactobacillus plantarum. Appl. Environ. Microbiol. 1992;58:891–894. doi: 10.1128/aem.58.3.891-894.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Renna M.C., Najimudin N., Winik L.R., Zahler S.A. Regulation of the Bacillus subtilis alsS, alsD, and alsR genes involved in post-exponential-phase production of acetoin. J. Bacteriol. 1993;175:3863–3875. doi: 10.1128/jb.175.12.3863-3875.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material