Abstract
Francisella tularensis is the causative agent of the highly, infectious disease, tularemia. Amongst the genes identified as essential to the virulence of F. tularensis was the proposed serine hydrolase FTT0941c. Herein, we purified FTT0941c to homogeneity and then characterized the folded stability, enzymatic activity, and substrate specificity of FTT0941c. Based on phylogenetic analysis, FTT0941c was classified within a divergent Francisella subbranch of the bacterial hormone sensitive lipase (HSL) superfamily, but with the conserved sequence motifs of a bacterial serine hydrolase. FTT0941c showed broad hydrolase activity against diverse libraries of ester substrates, including significant hydrolytic activity across alkyl ester substrates from 2 to 8 carbons in length. Among a diverse library of fluorogenic substrates, FTT0941c preferred α-cyclohexyl ester substrates, matching with the substrate specificity of structural homologues and the broad open architecture of its modeled binding pocket. By substitutional analysis, FTT0941c was confirmed to have a classic catalytic triad of Ser115, His278, and Asp248 and to remain thermally stable even after substitution. Its overall substrate specificity profile, divergent phylogenetic homology, and preliminary pathway analysis suggested potential biological functions for FTT0941c in diverse metabolic degradation pathways in F. tularensis.
Abbreviations: DSF, differential scanning fluorimetry; FTT0941c, virulence associated serine hydrolase from Francisella tularensis; HSL, hormone sensitive lipase; KEGG, Kyoto Encyclopedia of Genes and Genomes; LB, Luria-Bertani broth; MWCO, molecular weight cut-off; Ni-NTA, nickel-nitrilotriacetic acid; PDB, protein data bank
Keywords: Serine hydrolase, Virulence, Fluorogenic substrates, Substrate specificity, Francisella tularensis
Highlights
-
•
FTT0941c is an unstudied virulence associated hydrolase from F. tularensis (74).
-
•
Classified as a bacterial HSL enzyme within a unique subclade of Francisella (85).
-
•
Broad substrate specificity for alkyl, unsaturated, tertiary, and cyclic esters (79).
-
•
Distinct substrate selectivity for α-cyclohexyl esters (54).
-
•
Proposed metabolic role in cyclohexyl and benzoate ester hydrolysis pathways (76).
1. Introduction
Francisella tularensis, the causative agent of the disease tularemia, is a highly toxic, endemic gram-negative bacterium [1]. F. tularensis is the most virulent member of a genus of gram-negative bacterial species, including other weak human and mammalian pathogens (F. holarctica and F. novicida) [1], [2], [3]. Due to its ability to be spread by aerosol and to its high virulence and mortality, F. tularensis has been marked as a category A potential bioweapon by the Centers for Disease Control and Prevention (CDC) [1], [4]. Currently, F. tularensis infection is treated by an antibiotic regiment of streptomycin and fluoroquinolone, but recent outbreaks have made F. tularensis into a re-emerging infectious disease [1], [5].
Multiple large-scale transposon mutagenesis screens in vivo and in vitro have identified a large number of genes (~20% of the total genome) and gene products involved in the pathogenesis and virulence of F. tularensis [6], [7], [8], [9]. Comparison of the overlap of these transposon data sets indicated multiple conserved pathways involved in virulence [5], [8]. These pathways include the 33 kb pathogenicity island, a set of proteins involved in the Type VI secretion system, that was identified in all screens [7]. Gene products involved in metabolism, protein stability and folding, and the bacterial envelope were also reproducibly identified in the screens [8], [10].
Amongst F. tularensis genes identified in multiple screens by different methodologies as essential to its pathogenesis and virulence was the FTT_0941c gene, a predicted esterase/lipase that has been loosely assigned the name lipP based on limited sequence similarity to the lipP-1 protein from Sulfolobus solfataricus (SwissProt: Q97VW1) [6], [7]. Although disruption of the FTT_0941c gene did not ablate virulence, the level of virulence attenuation with disruption of the FTT_0941c gene suggested that this gene was an important component of the virulence machinery [7]. Further analysis of the initial virulence screens confirmed that the FTT_0941c gene was required for virulence in macrophages and specifically for intracellular replication within macrophages [11]. The FTT_0941c gene has been loosely assigned as involved in metabolism, although the gene was not expressed in the presence of high extracellular lipids [8], [12].
Herein, we examined the phylogeny of the FTT0941c protein to confirm its assignment as a serine hydrolase and to highlight its unique sequence conservation within the Francisella genus. We then heterologously expressed and purified the FTT0941c protein and characterized its biochemical features, including protein folding and enzymatic activity. Using libraries of ester substrates, we then determined the comprehensive substrate specificity of FTT0941c to begin to assign its biological function. Based on its substrate specificity profile and pathway analysis of neighboring genes, we propose potential roles for FTT0941c in Francisella specific metabolism that make it essential to the survival and virulence of F. tularensis within its hosts.
2. Materials and methods
2.1. Purification of FTT0941c from F. tularensis
A bacterial expression plasmid (pDEST17) containing the FTT_0941c gene from Francisella tularensis Schu 4 (Genbank: YP_169934; protein name FTT0941c) was obtained from the Harvard Plasmid Repository (Clone ID: FtCD00063741). This bacterial plasmid (pDEST17-FTT0941c) was transformed into E. coli BL21 (DE3) RIPL cells (Agilent, La Jolla, CA). A saturated overnight culture of E. coli BL21 (DE3) RIPL (pDEST17-FTT0941c) in LB media containing ampicillin (100 μg/mL) and chloramphenicol (30 μg/mL) was used to inoculate LB-media (1 L) containing ampicillin (100 μg/mL) and chloramphenicol (30 μg/mL) and the bacterial culture was grown with constant shaking (225 rpm) at 37 °C. When the OD600 reached 0.6–0.8, the temperature of the culture was decreased to 16 °C and isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to a final concentration of 0.5 mM. Protein induction proceeded for 16–20 h at 16 °C. Bacterial cultures were collected by centrifugation at 6000×g for 10 min at 4 °C. The bacterial cell pellet was resuspended in PBS (40 mL) and stored at −20 °C.
To disrupt the bacterial cell wall, lysozyme (250 mg) and Bug Buster solution (4.0 mL of 10X; EMD Millipore) were added and the cell lysis proceeded on a rotating shaker for 2 h at 4 °C. To remove insoluble cell material, lysed cells were centrifuged at 16,000xg for 10 min at 4 °C. Ni-NTA agarose (1.0 mL; Qiagen, Valencia, CA) was added to the soluble fraction and allowed to incubate at 4 °C for 15 min. The resin was washed three times with PBS containing increasing concentrations of ice-cold imidazole (40 mL each of PBS containing 10 mM imidazole, 25 mM imidazole, or 50 mM imidazole) and recollected by centrifugation at 2000×g for 2 min at 4 °C between wash steps. FTT0941c was eluted in PBS containing imidazole (250 mM; 1000 μL) and dialyzed against PBS overnight at 4 °C with constant stirring (10K MWCO; Pierce, Rockford, IL).
The purity of FTT0941c was confirmed by SDS–PAGE on a 4–20% gradient gel and the purity was shown to be greater than 95% (Supplementary Fig. 1). The expected molecular weight of the FTT0941c protein was 35.5 kD. The concentration of FTT0941c was determined by measuring the absorbance at 280 nm and by calculating the extinction coefficient (ε280=46,300 M−1 s−1 with all free cysteines) on ExPASy.
2.2. Site-directed mutagenesis and purification
Variants of FTT0941c were produced by QuikChange II site-directed mutagenesis of the pDEST17-FTT0941c template plasmid DNA using a derivation of the manufacturer's suggested procedure (Agilent, Santa Clara, CA), the only exception being specific annealing temperatures and the mutagenesis primers (Integrated DNA Technologies, Coralville, IA) outlined in Supplementary Table 3. Proper mutations in the FTT0941c DNA sequence were confirmed by DNA sequencing (Genewiz, South Plainfield, NJ) using T7 and/or T7-terminal sequencing primers. Plasmids coding for FTT0941c variants were transformed into E. coli BL21 (DE3) RIPL cells and variants of FTT0941c were overexpressed, purified, quantitated, and characterized using the same procedure as for wild-type FTT0941c.
2.3. Ester hydrolase substrates
p-nitrophenyl substrates were from Sigma-Aldrich. Compounds 1–17 were synthesized as described previously [13], [14], [15], [16].
2.4. Kinetic measurements with p-nitrophenyl substrates
The enzymatic activity of FTT0941c was measured against p-nitrophenyl acetate, p-nitrophenyl butyrate, p-nitrophenyl octanoate, and p-nitrophenyl laurate (Sigma–Aldrich) using a 96-well microplate assay (Fig. 1) [16]. All four substrates, p-nitrophenyl acetate (2 M), p-nitrophenyl butyrate (2 M), p-nitrophenyl octanoate (200 mM), and p-nitrophenyl laurate (200 mM) were prepared as stock solutions in acetonitrile and diluted into PBS containing acetylated BSA (PBS–BSA; 0.1 mg/mL). The starting concentration for p-nitrophenyl acetate and p-nitrophenyl butyrate was 20 mM and for p-nitrophenyl octanoate and p-nitrophenyl laurate was 2 mM. Eight serial dilutions (1:1; 110 μL into 220 μL total volume; 20 mM to 156 μM final concentrations for acetate and butyrate and 2 mM to 15.6 μM for octanoate and laurate) were made using PBS–BSA containing 1% acetonitrile. Substrate dilutions (95 μL) were transferred to a clear 96-well microplate and FTT0941c (5 μL of 125 μg/mL; final concentration FTT0941c=6.25 μg/mL; 176 nM) was added to start the reaction. The absorbance change at 412 nm was measured for 4 min at 25 °C on a Biotek Synergy H1 multimode plate reader (Biotek Instruments; Winooski, VT). The change in absorbance was converted to molar concentrations using the extinction coefficient of p-nitrophenol (Δε412=1.034 mM−1 cm−1) [17]. The initial rates of the reactions were measured in triplicate and plotted versus fluorogenic enzyme substrate concentration. The saturation enzyme kinetic traces were fitted to a standard Michaelis–Menten equation using Origin 6.1 (OriginLab Corp., Northhampton, MA) and values for kcat, KM and kcat/KM calculated.
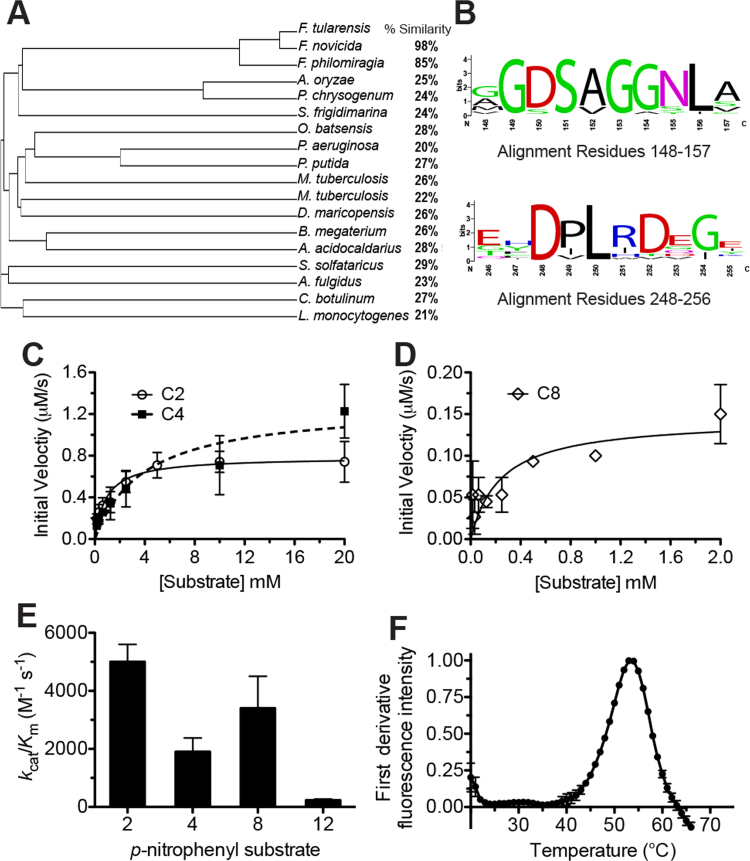
Fig. 1.
Biochemical characterization of FTT0941c. (A) Phylogenetic relationship between FTT0941c and homologous bacterial hydrolases. The amino acid sequence of FTT0941c was aligned with the 17 other bacterial hydrolases and a cladogram of the aligned proteins was constructed with percent similarities from Clustal Omega. Detailed sequence analysis is given in Supplementary Table 1. (B) Sequence conservation of residues adjacent to the proposed catalytic triad. Relative weightings and motif analysis performed using Weblogo [41]. Detailed sequence analysis given in Supplementary Table 1. (C) and (D) Kinetic activity of FTT0941c against p-nitrophenyl acetate (C2), p-nitrophenyl butyrate (C4), and p-nitrophenyl octanoate (C8). The kinetic activity of FTT0941c for C2 and C4 was measured from 20 mM to 156 μM and for C8 from 2 mM to 15.6 μM to account for its lower Km value and solubility. Data points were fitted to the Michaelis-Menten equation and are shown ±SE. Values for kinetic constants are given in Table 1. (E) Catalytic efficiency of FTT0941c against p-nitrophenyl substrates (p-nitrophenyl acetate (2), p-nitrophenyl butyrate (4), p-nitrophenyl octanoate (8), and p-nitrophenyl laurate (12)). Catalytic efficiency values (kcat/Km) are given ±SE. Detailed kinetic values are given in Table 1. (F) Thermal stability of FTT0941c. The folded to unfolded transition for wild-type FTT0941c (0.3 mg/mL in PBS) was observed by DSF. The measurement was completed in triplicate and is shown ±SE.
2.5. Kinetic measurements with fluorogenic ester substrates
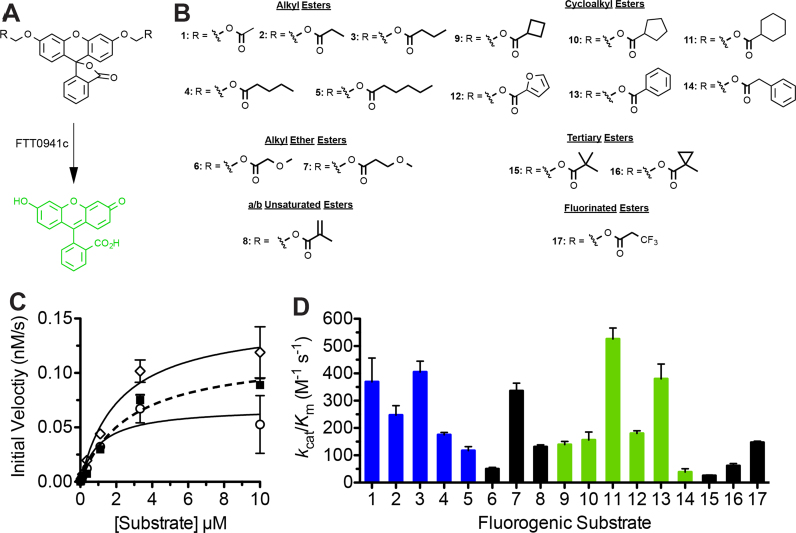
The enzymatic activity of FTT0941c was measured against the fluorogenic ester substrates (Fig. 2) using a 96-well microplate assay [16], [18]. Fluorogenic substrates were prepared as stock solutions in DMSO (10 mM) and were diluted into PBS containing acetylated BSA (PBS–BSA; 0.1 mg/mL) to starting concentrations between 10–1000 μM, depending on the Km value of FTT0941c for the substrate. The majority of the substrates (substrates 1–5; 9–17) had the same starting concentration (10 μM) with substrates 6–8 (100 μM) requiring higher starting concentrations. Eight serial dilutions (1:2; 60 μL into 180 μL total volume) of each substrate were made using PBS–BSA. Fluorogenic substrate dilutions (95 μL) were then transferred to a black 96-well microplate (Corning, Lowell, MA).
Fig. 2.
Substrate specificity of FTT0941c against fluorogenic hydrolase substrates. (A) Activation of fluorogenic substrates by FTT0941c. Hydrolysis of the ester bond on the diacyloxymethyl ether fluorescein substrates by FTT0941c converts the fluorescein core from the nonfluorescent lactone form to the highly fluorescent quinoid form. The rate of fluorophore activation is measured at a range of substrate concentrations to determine the kinetic constants for fluorophore activation. (B) Fluorogenic substrate library. Each of the substrates is composed of diacyloxymethyl ether fluorescein (1A) with varying R-groups. The differing R-groups have been organized into classes based on chemical functionality. All of the substrates were synthesized as described previously [13], [15], [16]. (C) Kinetic activity of FTT0941c against substrates 1 (open circles), 2 (closed squares), and 3 (open diamonds). All measurements completed in triplicate and shown ±SE. (D) Global comparison of the catalytic specificity (kcat/KM) of FTT0941c against each of the 17 substrates (structures and numbering given in Fig. 2B). The substrate specificity against alkyl ester substrates (blue) and against cycloalkyl ester substrates (green) illustrates the substrate selectivity of FTT0941c based on alkyl chain length, the size of the cyclic ring, and the distance from the ester carbonyl. Detailed kinetic results for each substrate are provided in Table 1.
FTT0941c protein (5 μL of 125 μg/mL; final concentration FT941c=6.25 μg/mL; 176 nM) was added to the diluted fluorogenic substrates in the 96-well microplate (100 μL final volume) and the fluorescence change (λex=485 nm, λem=528 nm) was measured for 4 min at 25 °C on a Biotek Synergy H1 Multimode plate reader (Biotek Instruments; Winooski, VT). For FTT0941c variants, the concentration of enzyme was increased by 3-fold to detect even low enzymatic activity for the variants (5 μL of 375 μg/mL; final concentration FT941c variants=18.75 μg/mL; 528 nM). The fluorescence change was converted to molar concentrations using a fluorescein standard curve (30–0.23 nM for 10 μM starting substrate concentrations and 300–2.3 nM for higher starting concentrations), whose fluorescence was measured simultaneously. The initial rates of the reactions were measured in triplicate and plotted versus fluorogenic enzyme substrate concentration. The saturation enzyme kinetic traces were fitted to a standard Michaelis–Menten equation using Origin 6.1 (OriginLab Corp., Northhampton, MA) and values for kcat, KM and kcat/KM calculated.
2.6. Thermal stability measurement
Similar to previously published methods, the thermal stability of FTT0941c and variants of FTT0941c was determined using differential scanning fluorimetry (DSF) [16], [19]. Wild-type FTT0941c (0.3 mg/mL) and variants of FTT0941c (0.30–0.50 mg/mL) were diluted in at least triplicate in PBS containing a 1:250 dilution of Sypro Orange (Invitrogen, Carlsbad, CA). The samples was heated from 15 °C to 85 °C at 1.0 °C/min in a thermocycler (Bio-rad C1000 Thermocycler with CFX96 Real-time System, Hercules, CA) and the change in Sypro Orange fluorescence followed over time (λex=450–490 nm, λem=610–650 nm). The melting temperature (Tm) was determined by plotting the first derivative of fluorescence versus temperature and finding the temperature at the midpoint of the transition. As in previous analyses [20], all graphs were normalized so that minimum fluorescence was set to 0 and maximum fluorescence set to 1.
2.7. Phylogenetic analysis of FTT0941c
The amino acid sequence of FTT0941c was aligned using Clustal Omega (EMBL EBI). The catalytic triad amino acids were then extracted from the alignment based on sequence conservation and the presence of the catalytic motif (G-x-S-x-G). A cladogram of the aligned proteins was then constructed using Drawgram from the Mobyle Pasteur (Fig. 1 and Supplementary Table 2). The sequences used in the alignment were from Francisella tularensis subsp. tularensis SCHU S4 (56708038), Francisella tularensis subsp. novicida U112 (118497410), Francisella philomiragia subsp. philomiragia ATCC 25017 (167598025), Aspergillus oryzae RB40 (169776543), Penicillium rubens Wisconsin 54-1255 (255949072), Shewanella frigidimarina NCIMB 400 (114564803), Oceanicola batsensis (497492433), Pseudomonas aeruginosa (489180078), Pseudomonas putida KT2440 (26990910), Mycobacterium tuberculosis H37Rv (15610107), Mycobacterium tuberculosis H37Rv (15608537), Deinococcus maricopensis DSM 21211 (320333828), Bacillus megaterium QM B1551 (294499059), Alicyclobacillus acidocaldarius (47168664), Sulfolobus solfataricus P2 (15899234), Archaeoglobus fulgidus DSM 4304 (11499305), Clostridium botulinum A str. ATCC 3502 (148380141) and Listeria monocytogenes EGD-e (16804128). Sequences for alignment were chosen based on protein BLAST analysis of FTT0941c and extracting unique protein sequences with significant percent similarity (>20%).
2.8. Homology modeling
Structural models for FTT0941c were constructed using three validated web server based protein modeling programs (Swiss-Model, RaptorX, and Phyre2) [21], [22], [23], as FTT0941c has been recalcitrant to ongoing structural determination efforts. In each model, the complete protein sequence for FTT0941c was submitted to the web based server and homology structures were evaluated using the provided evaluation statistics. For Swiss-Model, the homology model was constructed based on the structure of an enantioselective esterase from Pyrobaculum calidifontis (PDB: 2YH2) with 34% sequence similarity to FTT0941c. For RaptorX, the model was constructed based on a template of a unique carboxylesterase from Ferroplasma (PDB: 3WJ1) with 50% sequence similarity to FTT0941c, a p-value of 2.23×10−11, and all 305 residues modeled without disorder. For Phyre2, the heuristic model was constructed using six different hydrolase templates (1LZL, 3GA7, 4OB7, 2ZSH, 4KRX, and 4C87) with between 16–26% sequence identity to FTT041c. For this model, 100% of the 305 residues in FTT041c were modeled with greater than 90% confidence.
3. Results and discussion
3.1. Francisella specific enzyme family
Phylogenetic analysis of FTT0941c based on amino acid similarity and the presence of conserved motifs placed FTT0941c into group H of four hydrolase subclasses in the ESTHER database and Group IV of seven subfamilies of bacterial lipolytic enzymes [24], [25]. In each of these subclassifications, FTT0941c was assigned to the subgroup of bacterial hormone sensitive lipases (HSL; Pfam: PF07859), an evolutionarily conserved enzyme superfamily with high sequence similarity to the catalytic subdomain of human HSL [24], [25]. Structurally, bacterial HSL enzymes are grouped into the α/β hydrolase fold III subclass and commonly contain an N-terminal CAP domain helix, determining the substrate specificity, and a C-terminal α/β hydrolase fold, containing the catalytic triad [24]. Detailed sequence analysis of the FTT0941c protein to a wide range of bacterial hydrolases, however, showcased the divergence of FTT0941c within the bacterial HSL family (Fig. 1A). Across gram-negative and gram-positive bacterial species, FTT0941c had less than 30% overall sequence similarity to each bacterial hydrolase (Fig. 1A). FTT0941c instead shared high sequence similarity with other Francisella genus proteins (98% to F. novicida and 85% to F. philomiragia) and together these three homologs formed a separate subbranch within the phylogenetic tree (Fig. 1A).
Although the overall sequence identity of FTT0941c to similar bacterial proteins is low, specific subregions of the amino acid sequence are highly conserved, including the regions adjacent to the proposed catalytic amino acids (Fig. 1B and Supplementary Table 2). The G-x-S-x-G motif centered on the nucleophilic serine is nearly universally conserved with GDSAG being the dominant motif (Fig. 1B). Bacterial HSL members containing this GDSAG motif are the largest subclass of bacterial HSL enzymes, which has been subdivided based on the sequence motif surrounding the catalytic serine [24], [26]. Conservation of this catalytic motif around the nucleophilic serine places FTT0941c firmly into the HSL subfamily [26]. Yet, wide-variation in catalytic reactivity exists within the HSL subfamily from true lipases to carboxylesterases to arylamide hydrolases, making predictions of cellular activity difficult [24], [25]. Overall, FTT0941c is a unique Francisella specific member of the diverse bacterial HSL family with the conserved sequence motifs of a bacterial serine hydrolase.
3.2. Biochemical characterization of FTT0941c
To confirm the assignment of FTT0941c as a bacterial hydrolase, we developed an expression and purification system in Escherichia coli for FTT0941c that produced >95% pure enzyme in sufficient yields (~1 mg/L) (Supplementary Fig. 1). The final FTT0941c protein was well-folded with a clear folded to unfolded transition with increasing temperature and a Tm value of 53.3±1.0 °C (Fig. 1F), which is well above the normal growth temperature of F. tularensis, but less than many bacterial HSL proteins that are highly thermally stable [26], [27]. The serine hydrolase activity of FTT0941c was initially characterized using a range of straight-chain p-nitrophenyl ester substrates from acetate to laurate esters to subclassify FTT0941c into the general esterase versus lipase subclasses. (Fig. 1C–E). FTT0941c showed measurable activity across the full range of substrates with higher turnover rates (kcat values) against shorter substrates (C2 kcat=4.6±0.3 s−1; C4 kcat=7.6±1.2 s−1), but lower Km values for longer carbon substrates (C8 Km value=0.11±0.07 μM) (Table 1). Weighted together as the overall catalytic efficiency (kcat/Km), FTT0941c showed broad substrate specificity with nearly identical activity toward p-nitrophenyl acetate and octanoate and only a less than 3-fold decrease with p-nitrophenyl butyrate (Fig. 1E). Only the longest carbon substrate (p-nitrophenyl laurate) had significantly decreased catalytic efficiency. The increased Km value (1.8±0.6 μM) for the longest substrate and decreased overall catalytic efficiency indicated that FTT0941c was not a lipase with interfacial activation with large hydrophobic substrates [25], [28], [29], [30]. Instead, this broad substrate specificity across alkyl ester substrates subdivided FTT0941c into the carboxylesterase classification of serine hydrolases, encompassing a range of metabolic hydrolysis reactions.
Table 1.
Kinetic characterization of FTT0941c.
| Substrate | kcat (s−1) | Km (mM) | kcat/Km (M−1 s−1) |
|---|---|---|---|
| C2a | 4.6±0.3 | 0.92±0.22 | 5000±700 |
| C4 | 7.6±1.2 | 4.0±1.9 | 1900±600 |
| C8 | 0.82±0.16 | 0.24±0.15 | 3400±1100 |
| C12 | 0.42±0.08 | 1.8±0.6 | 230±50 |
| kcat (10−3 s−1) | Km (µM) | kcat/Km (M−1 s−1) | |
| 1b | 0.39±0.05 | 1.1±0.5 | 370±100 |
| 2 | 0.67±0.07 | 2.7±0.7 | 250±40 |
| 3 | 0.85±0.06 | 2.2±0.4 | 400±50 |
| 4 | 0.21±0.01 | 1.2±0.1 | 180±10 |
| 5 | 0.049±0.003 | 0.42±0.10 | 120±20 |
| 6 | 4.2±0.4 | 83±16 | 50±6 |
| 7 | 3.6±0.2 | 8.3±1.4 | 340±30 |
| 8 | 0.064±0.002 | 0.48±0.05 | 130±10 |
| 9 | 0.053±0.002 | 0.38±0.06 | 140±10 |
| 10 | 0.76±0.03 | 4.2±0.4 | 180±10 |
| 11 | 8.7±0.7 | 17±2 | 530±50 |
| 12 | 0.93±0.15 | 5.9±2.0 | 160±60 |
| 13 | 0.43±0.04 | 1.1±0.3 | 380±60 |
| 14 | 0.33±0.20 | 8.6±0.9 | 39±13 |
| 15 | 0.012±0.002 | 0.48±0.04 | 26±3 |
| 16 | 0.026±0.006 | 0.41±0.04 | 62±9 |
| 17 | 110±10 | 780±40 | 150±10 |
Kinetic constants for p-nitrophenyl substrates were determined by measuring the change in A412 due to ester hydrolysis. Substrates represent different carbon chain lengths: p-nitrophenyl acetate (C2), p-nitrophenyl butyrate (C4), p-nitrophenyl octanoate (C8), and p-nitrophenyl laurate (C12).
Kinetic constants for substrates 1–17 were determined by measuring the increase in fluorogenic enzyme substrate fluorescence over time. Data were fitted to a standard Michaelis-Menten equation to determine the values for kcat, KM, and kcat/KM. Kinetic measurements for each substrate were repeated three times and the values are given ±SE.
3.3. Comprehensive substrate specificity of FTT0941c
To clarify the substrate specificity of FTT0941c, we utilized a diverse library of fluorogenic ester substrates (Fig. 2A and B). These ester substrates based on acyloxymethyl ether fluorescein are nonfluorescent when ester protected at their phenolic oxygens, but rapidly produce highly fluorescent fluorescein upon incubation with a hydrolase such as FTT0941c (Fig. 2A and C) [14], [16], [31]. The low background fluorescence and high sensitivity of these substrates allow kinetic measurements for even low activity hydrolases against non-optimal substrates [13], [15]. The library also focuses on simple alcohol ester scaffolds of common biological ester classes and allows general assignment of hydrolases into subclasses of hydrolase activity [13], [16], [32].
Similar to the p-nitrophenyl substrates, FTT0941c showed classic Michaelis-Menten kinetics across a wide-range of fluorogenic substrates (Fig. 2C). Comparison of catalytic efficiency values across the full range of fluorogenic substrates again reinforces the broad substrate specificity of FTT0941c with significant measurable activity against all 17 substrates (Fig. 2D). FTT0941c showed highest enzymatic activity against substrates 11 and 13, which both have six membered cyclic rings appended to their ester bond (Fig. 2B), but retained comparable activity against other substrates (3 and 7) with similar overall lengths. Other bacterial hydrolases tested with the same fluorogenic library have shown only minimal activity toward these cycloalkyl substrates, suggesting a high degree of selectivity for these substrates by FTT0941c (Fig. 2D) [13], [31], [32].
Within the alkyl ester subclass of fluorogenic substrates (Substrates 1–5; blue), FTT0941c showed general enzymatic activity against all five alkyl esters with no clear size selectivity (Fig. 2D), matching with the lack of substrate selectivity for short alkyl esters (<10 carbons) observed for p-nitrophenyl ester substrates (Fig. 1E). This lack of strict length selectivity within alkyl esters also differentiated FTT0941c from all previous hydrolases characterized against these substrates (Fig. 2D) [13], [16], [31], [32]. The current substrate library does not, however, differentiate based on degrees of saturation, which may influence the selectivity for these short alkyl ester substrates. In comparison to alkyl esters, higher chemical selectivity was observed with cycloalkyl substrates, where six membered cyclic ring substrates (11 and 13) had higher activity than four or five-membered cyclic rings (9, 10, and 12) (Fig. 2D; green). The relative attachment distance from the ester bond to the cycloalkyl ring also influenced the relative catalytic activity of FTT0941c, as insertion of a methylene carbon between the carbonyl carbon and the phenyl ring decreased the relative catalytic activity of FTT0941c by almost 10-fold (13 versus 14; Fig. 2D). The catalytic activity of FTT0941c toward six membered cyclic substrates and its substrate selectivity for small structural differences in cycloalkyl substrates provided preliminary evidence that six membered cycloalkyl substrates may represent the natural substrate of FTT0941c.
Homology models of the binding pocket of FTT0941c structurally reinforced the ability of FTT0941c to accept a range of ester substrates, including bulky cycloalkyl substrates (Fig. 3). Three different homology models were constructed, as each validated program identified a separate structural template for FTT0941c, but each structure converged on a similar architecture for the binding pocket and overall fold for FTT0941c (Fig. 3 and Supplementary Fig. 2). The surface accessible binding pocket of the three different modeled FTT0941c structures showed a broad, open binding pocket that can accommodate a range of esters, including bulky fluorescein derivatives (Fig. 2A). The modeled binding pockets begin to narrow around the catalytic serine, but still maintained sufficient width to accommodate the full range of ester substrates (Fig. 3). Similar to FTT0941c (Figs. 1E and 2D), the homologous bacterial HSLs used to build the structural homology models of FTT0941c in Fig. 3A and B also showed broad substrate specificity toward a range of ester substrates [27], [33]. Interestingly, one close homologue, the enantioselective esterase from Pyrobaculum calidifontis, displayed highest catalytic activity against six-membered aromatic esters with selectivity for cyclic ring attachment (Fig. 3A) [33]. The substrate specificity and enantioselectivity of this esterase from P. calidifontis was explained based on broad substrate access tunnels, similar to the broad access channels modeled for FTT0941c [33].
Fig. 3.
Binding pocket models for FTT0941c. Binding pocket models for FTT0941c were constructed using three validated protein modeling programs. In each model, the surface accessible binding pocket is colored with the nucleophilic serine shown in sticks and labeled. (A) Homology model of FTT0941c from Swiss-Model. The homology model was constructed based on the structure of an enantioselective esterase from Pyrobaculum calidifontis (PDB:2YH2) with 34% sequence similarity to FTT0941c. For Swiss-Model, the homology model with the lowest Qmean4 score was chosen for analysis. (B) Homology model of FTT0941c from RaptorX. The model was constructed based on a template of a unique carboxylesterase from Ferroplasma (PDB:3WJ1) with 50% sequence similarity to FTT0941c, a p-value of 2.23×10−11, and all 305 residues of FTT0941c modeled without disorder. (C) Phyre2 heuristic model of FTT0941c. Model was constructed using six different hydrolase templates (1LZL, 3GA7, 4OB7, 2ZSH, 4KRX, and 4C87) with between 16–26% sequence identity to FTT041c. For this model, all 305 residues of FTT041c, including the active site and binding pocket, were modeled with greater than 90% confidence.
Among bacterial pathways utilizing six membered ester substrates similar to substrates 11 and 13 and matching with the binding pocket structure of FTT0941c, multiple metabolic hydrolysis steps involving cyclic ester substrates have been hypothetically assigned to FTT0941c in the KEGG pathway database. These proposed pathways were hypothetically assigned to FTT0941c based on analogy to metabolic pathways in similar bacteria and neighboring gene operons to FTT0941c and include bisphenol (KEGG entry ftu00363) and aminobenzoate (KEGG entry ftu00627) degradation pathways [34], [35], [36]. These substrates mimic the ester substrates of 11 and 13 based on ring size and attachment distance and the selectivity for the hydroxylation on the phenyl ring may explain the increased activity of FTT0941c toward substrates 7 and 8 (Fig. 2D). Such degradation pathways are not main metabolic pathways for these bacteria, but have been connected to metabolic pathways required for growth under opportunistic metabolic conditions [10], [36], [37], [38]. The variable identification of FTT0941 as a virulence gene in F. tularensis based on the screening conditions may also be reflective of a similar niche metabolic role for FTT0941c [6], [7], [8], [11].
3.4. Assignment of key catalytic amino acids
To begin to define the substrate selectivity profile of FTT0941c, potential catalytic amino acids were analyzed using substitutional analysis. Based on sequence alignments and sequence motif analysis, the nucleophilic serine and general base in the catalytic triad of FTT0941c were identified as Ser151 and His278, respectively (Fig. 1B and Supplementary Table 1). The third member of the catalytic triad, an acidic amino acid involved in stabilizing the general base, was more difficult to pinpoint, as multiple aspartates were highly conserved across bacterial homologs. Three different aspartates (Asp90, Asp248, Asp252) were chosen as potential catalytic triad members based on sequence conservation and placement in different structural models of FTT0941c.
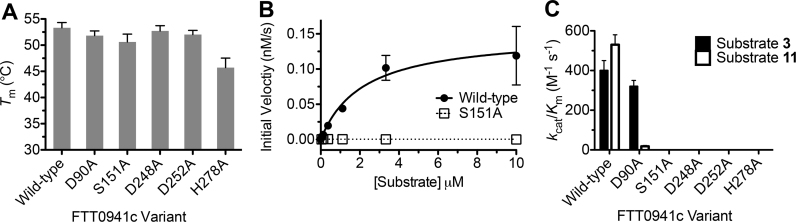
Each of these five amino acids was individually substituted with alanine and the resulting FTT0941c variants were purified to homogeneity and characterized for thermal stability and enzymatic activity. All five variants were well-folded with thermal stabilities within 7 °C of wild-type FTT0941c (Fig. 4) with only the substitution of His278 for the smaller alanine residue causing any deviation in stability. As expected, substitution of the nucleophilic serine (Ser151) or general base (His278) completely ablated the catalytic activity of FTT0941c toward each fluorogenic substrate (Fig. 4B and C). Multiple aspartate variants showed complete loss of catalytic activity, as substitution of each of the two closely spaced aspartates (Asp248 and Asp252) completely abolished the catalytic activity of FTT0941c, but substitution of a control aspartate residue (D90A) did not change the activity toward substrate 3. Based on comparison of FTT0941c to the recent structure and alignment of a bacterial HSL enzyme from Lactobacillus plantarum, Asp248 is likely the catalytic aspartate and is conserved to human HSL [39]. The second Asp252 may, however, play a role in structural stabilization, as observed in a recent structure of a metagenomic bacterial HSL enzyme [26]. Affirming the importance of Asp248 and Asp252 to the catalytic activity of FTT0941c, the D90A variant of FTT0941c maintained near wild-type catalytic activity toward substrate 3. The D90A variant also showed unexpected substrate selectivity with reduced activity toward the cycloalkyl ester substrate (11), as compared to the straight-chain alkyl substrate (3). Homology models for FTT0941c all placed Asp90 near the edge of the substrate-binding pocket of FTT0941c with Asp90 potentially playing an interesting role in determining the substrate selectivity of FTT0941c (Supplementary Fig. 2). Thus, FTT0941c catalyzed the hydrolysis of a range of ester substrates, using the action of a classic serine (Ser151), histidine (His278), and aspartate (Asp252) catalytic triad with catalytic assistance from other conserved aspartates.
Fig. 4.
Identification of the catalytic amino acids for FTT0941c. (A) Thermal stability of FTT0941c variants. The thermal stability of each variant was determined by DSF. The measurement was completed in triplicate and is shown ±SE. (B) Kinetic activity of FTT0941c variants. The kinetic activity of wild-type FTT0941c (closed circles) and the S115A variant (open squares) were determined against substrate 3. All measurements were completed in triplicate and are shown ±SE. Data were fitted to the Michaelis-Menten equation using Graphpad Prism 5.0. (C) Relative catalytic activity of FTT0941c variants. The catalytic activity of each of the FTT0941c variants was determined against substrates 3 and 11. Detailed kinetic and thermal stability analysis for FTT0941c variants are given in Supplementary Table 1.
4. Conclusions
FTT0941c represents a potentially key enzyme involved in the virulence of the highly deadly bacteria, F. tularensis. Sequence analysis of FTT0941c grouped this enzyme with other bacterial HSL enzymes and highlighted the unique amino acid structure of FTT0941c amongst diverse bacterial hydrolases (Fig. 1A). Purification and characterization of FTT0941c confirmed its hydrolase activity with broad specificity toward alkyl ester substrates from 2 to 8 carbons in length and significant hydrolase activity toward a wide-range of simple, ester scaffolds, irrespective of the carboxylic acid moiety off the ester bond (Fig. 1C–E and Fig. 2). FTT0941c had a classic catalytic triad of Ser115, His278, and Asp248 and maintained high folded stability even after substitution (Fig. 1B and C and Fig. 4). Detailed substrate specificity analysis identified cyclohexyl esters, as the highest activity substrate class for FTT0941c, matching with the identified substrates of modeled homologs and the modeled, broad binding pocket of FTT0941c. With the proposed involvement of FTT0941c in metabolic pathways and with the potential utility for prodrug targeting in F. tularensis treatment [40], our biochemical characterization and substrate specificity profile of FTT0941c – one of only five hydrolases identified in F. tularensis – might also be used to refine the chemical structure of ester based prodrugs for F. tularensis infection.
Acknowledgments
We thank Perry Rabin for assistance with protein analysis and Luke Lavis for guidance with the synthesis of fluorogenic substrates. A.M.F. was supported by a Holcomb Grant Scholarship. R.J.J. was supported by start-up funds and a Holcomb grant from Butler University and a Senior Research Grant from the Indiana Academy of Sciences.
Footnotes
Supplementary data associated with this article can be found in the online version at 10.1016/j.bbrep.2016.07.006.
Appendix A. Supplementary data
Transparency document
.
Transparency document
.
References
- 1.McLendon M.K., Apicella M.A., Allen L.A. Francisella tularensis: taxonomy, genetics, and Immunopathogenesis of a potential agent of biowarfare. Annu. Rev. Microbiol. 2006;60:167–185. doi: 10.1146/annurev.micro.60.080805.142126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rohmer L., Fong C., Abmayr S., Wasnick M., Larson Freeman T.J., Radey M., Guina T., Svensson K., Hayden H.S., Jacobs M., Gallagher L.A., Manoil C., Ernst R.K., Drees B., Buckley D., Haugen E., Bovee D., Zhou Y., Chang J., Levy R., Lim R., Gillett W., Guenthener D., Kang A., Shaffer S.A., Taylor G., Chen J., Gallis B., D’Argenio D.A., Forsman M., Olson M.V., Goodlett D.R., Kaul R., Miller S.I., Brittnacher M.J. Comparison of Francisella tularensis genomes reveals evolutionary events associated with the emergence of human pathogenic strains. Genome Biol. 2007;8:R102. doi: 10.1186/gb-2007-8-6-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beckstrom-Sternberg S.M., Auerbach R.K., Godbole S., Pearson J.V., Beckstrom-Sternberg J.S., Deng Z., Munk C., Kubota K., Zhou Y., Bruce D., Noronha J., Scheuermann R.H., Wang A., Wei X., Wang J., Hao J., Wagner D.M., Brettin T.S., Brown N., Gilna P., Keim P.S. Complete genomic characterization of a pathogenic A.II strain of Francisella tularensis subspecies tularensis. PLoS One. 2007;2:e947. doi: 10.1371/journal.pone.0000947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pohanka M., Skladal P. Bacillus anthracis, Francisella tularensis and Yersinia pestis. The most important bacterial warfare agents – review. Folia Microbiol. 2009;54:263–272. doi: 10.1007/s12223-009-0046-1. [DOI] [PubMed] [Google Scholar]
- 5.Pechous R.D., McCarthy T.R., Zahrt T.C. Working toward the future: insights into Francisella tularensis pathogenesis and vaccine development. Microbiol. Mol. Biol. Rev. 2009;73:684–711. doi: 10.1128/MMBR.00028-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su J., Yang J., Zhao D., Kawula T.H., Banas J.A., Zhang J.R. Genome-wide identification of Francisella tularensis virulence determinants. Infect. Immun. 2007;75:3089–3101. doi: 10.1128/IAI.01865-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiss D.S., Brotcke A., Henry T., Margolis J.J., Chan K., Monack D.M. In vivo negative selection screen identifies genes required for Francisella virulence. Proc. Natl. Acad. Sci. USA. 2007;104:6037–6042. doi: 10.1073/pnas.0609675104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meibom K.L., Charbit A. The unraveling panoply of Francisella tularensis virulence attributes. Curr. Opin. Microbiol. 2010;13:11–17. doi: 10.1016/j.mib.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 9.Lindemann S.R., Peng K., Long M.E., Hunt J.R., Apicella M.A., Monack D.M., Allen L.A., Jones B.D. Francisella tularensis Schu S4 O-antigen and capsule biosynthesis gene mutants induce early cell death in human macrophages. Infect. Immun. 2011;79:581–594. doi: 10.1128/IAI.00863-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meibom K.L., Charbit A. Francisella tularensis metabolism and its relation to virulence. Front. Microbiol. 2010;1:140. doi: 10.3389/fmicb.2010.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Llewellyn A.C., Jones C.L., Napier B.A., Bina J.E., Weiss D.S. Macrophage replication screen identifies a novel Francisella hydroperoxide resistance protein involved in virulence. PLoS One. 2011;6:e24201. doi: 10.1371/journal.pone.0024201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kingry L.C., Cummings J.E., Brookman K.W., Bommineni G.R., Tonge P.J., Slayden R.A. The Francisella tularensis FabI enoyl-acyl carrier protein reductase gene is essential to bacterial viability and is expressed during infection. J. Bacteriol. 2013;195:351–358. doi: 10.1128/JB.01957-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellis E.E., Adkins C.T., Galovska N.M., Lavis L.D., Johnson R.J. Decoupled roles for the atypical, bifurcated binding pocket of the ybfF hydrolase. ChemBioChem. 2013;14:1134–1144. doi: 10.1002/cbic.201300085. [DOI] [PubMed] [Google Scholar]
- 14.Lavis L.D., Chao T.-Y., Raines R.T. Synthesis and utility of fluorogenic acetoxymethyl ethers. Chem. Sci. 2011;2:521–530. doi: 10.1039/C0SC00466A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tian L., Yang Y., Wysocki L.M., Arnold A.C., Hu A., Ravichandran B., Sternson S.M., Looger L.L., Lavis L.D. Selective esterase-ester pair for targeting small molecules with cellular specificity. Proc. Natl. Acad. Sci. USA. 2012;109:4756–4761. doi: 10.1073/pnas.1111943109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hedge M.K., Gehring A.M., Adkins C.T., Weston L.A., Lavis L.D., Johnson R.J. The structural basis for the narrow substrate specificity of an acetyl esterase from Thermotoga maritima. Biochim. Biophys. Acta. 2012;1824:1024–1030. doi: 10.1016/j.bbapap.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 17.Kim S., Lee S.B. Thermostable esterase from a thermoacidophilic archaeon: purification and characterization for enzymatic resolution of a chiral compound. Biosci. Biotechnol. Biochem. 2004;68:2289–2298. doi: 10.1271/bbb.68.2289. [DOI] [PubMed] [Google Scholar]
- 18.Johnson R.J., Hoops G.C., Savas C.J., Kartje Z., Lavis L.D. A sensitive and robust enzyme kinetic experiment using microplates and fluorogenic ester substrates. J. Chem. Educ. 2014;92:385–388. [Google Scholar]
- 19.Johnson R.J., Savas C.J., Kartje Z., Hoops G.C. Rapid and adaptable measurement of protein thermal stability by differential scanning fluorimetry: updating a common biochemical laboratory experiment. J. Chem. Educ. 2014;91:1077–1080. [Google Scholar]
- 20.Froese D.S., Healy S., McDonald M., Kochan G., Oppermann U., Niesen F.H., Gravel R.A. Thermolability of mutant MMACHC protein in the vitamin B12-responsive cblC disorder. Mol. Genet. Metab. 2010;100:29–36. doi: 10.1016/j.ymgme.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Cassarino T.G., Bertoni M., Bordoli L., Schwede T. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kallberg M., Wang H.P., Wang S., Peng J., Wang Z.Y., Lu H., Xu J.B. Template-based protein structure modeling using the RaptorX web server. Nat. Protoc. 2012;7:1511–1522. doi: 10.1038/nprot.2012.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lenfant N., Hotelier T., Velluet E., Bourne Y., Marchot P., Chatonnet A. ESTHER, the database of the alpha/beta-hydrolase fold superfamily of proteins: tools to explore diversity of functions. Nucleic Acids Res. 2013;41:D423–D429. doi: 10.1093/nar/gks1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arpigny J.L., Jaeger K.E. Bacterial lipolytic enzymes: classification and properties. Biochem. J. 1999;343(Pt 1):177–183. [PMC free article] [PubMed] [Google Scholar]
- 26.Li P.Y., Ji P., Li C.Y., Zhang Y., Wang G.L., Zhang X.Y., Xie B.B., Qin Q.L., Chen X.L., Zhou B.C., Zhang Y.Z. Structural basis for dimerization and catalysis of a novel esterase from the GTSAG motif subfamily of the bacterial hormone-sensitive lipase family. J. Biol. Chem. 2014;289:19031–19041. doi: 10.1074/jbc.M114.574913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ejima K., Liu J., Oshima Y., Hirooka K., Shimanuki S., Yokota Y., Hemmi H., Nakayama T., Nishino T. Molecular cloning and characterization of a thermostable carboxylesterase from an archaeon, Sulfolobus shibatae DSM5389: non-linear kinetic behavior of a hormone-sensitive lipase family enzyme. J. Biosci. Bioeng. 2004;98:445–451. doi: 10.1016/S1389-1723(05)00310-5. [DOI] [PubMed] [Google Scholar]
- 28.Burdette R.A., Quinn D.M. Interfacial reaction dynamics and acyl-enzyme mechanism for lipoprotein lipase-catalyzed hydrolysis of lipid p-nitrophenyl esters. J. Biol. Chem. 1986;261:12016–12021. [PubMed] [Google Scholar]
- 29.van Tilbeurgh H., Egloff M.P., Martinez C., Rugani N., Verger R., Cambillau C. Interfacial activation of the lipase-procolipase complex by mixed micelles revealed by X-ray crystallography. Nature. 1993;362:814–820. doi: 10.1038/362814a0. [DOI] [PubMed] [Google Scholar]
- 30.Schrag J.D., Li Y., Cygler M., Lang D., Burgdorf T., Hecht H.J., Schmid R., Schomburg D., Rydel T.J., Oliver J.D., Strickland L.C., Dunaway C.M., Larson S.B., Day J., McPherson A. The open conformation of a Pseudomonas lipase. Structure. 1997;5:187–202. doi: 10.1016/s0969-2126(97)00178-0. [DOI] [PubMed] [Google Scholar]
- 31.Filippova E.V., Weston L.A., Kuhn M.L., Geissler B., Gehring A.M., Armoush N., Adkins C.T., Minasov G., Dubrovska I., Shuvalova L., Winsor J.R., Lavis L.D., Satchell K.J., Becker D.P., Anderson W.F., Johnson R.J. Large scale structural rearrangement of a serine hydrolase from Francisella tularensis facilitates catalysis. J. Biol. Chem. 2013;288:10522–10535. doi: 10.1074/jbc.M112.446625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lukowski J.K., Savas C.P., Gehring A.M., McKary M.G., Adkins C.T., Lavis L.D., Hoops G.C., Johnson R.J. Distinct substrate selectivity of a metabolic hydrolase from Mycobacterium tuberculosis. Biochemistry. 2014;53:7386–7395. doi: 10.1021/bi501108u. [DOI] [PubMed] [Google Scholar]
- 33.Palm G.J., Fernandez-Alvaro E., Bogdanovic X., Bartsch S., Sczodrok J., Singh R.K., Bottcher D., Atomi H., Bornscheuer U.T., Hinrichs W. The crystal structure of an esterase from the hyperthermophilic microorganism Pyrobaculum calidifontis VA1 explains its enantioselectivity. Appl. Microbiol. Biotechnol. 2011;91:1061–1072. doi: 10.1007/s00253-011-3337-9. [DOI] [PubMed] [Google Scholar]
- 34.Inoue D., Hara S., Kashihara M., Murai Y., Danzl E., Sei K., Tsunoi S., Fujita M., Ike M. Degradation of bis(4-hydroxyphenyl)methane (bisphenol F) by Sphingobium yanoikuyae strain FM-2 isolated from river water. Appl. Environ. Microbiol. 2008;74:352–358. doi: 10.1128/AEM.01708-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diaz E., Jimenez J.I., Nogales J. Aerobic degradation of aromatic compounds. Curr. Opin. Biotechnol. 2013;24:431–442. doi: 10.1016/j.copbio.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 36.Steele S., Brunton J., Ziehr B., Taft-Benz S., Moorman N., Kawula T. Francisella tularensis harvests nutrients derived via ATG5-independent autophagy to support intracellular growth. PLoS Pathog. 2013;9:e1003562. doi: 10.1371/journal.ppat.1003562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santic M., Abu Kwaik Y. Nutritional virulence of Francisella tularensis. Front. Cell. Infect. Microbiol. 2013;3:112. doi: 10.3389/fcimb.2013.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abu Kwaik Y. Targeting nutrient retrieval by Francisella tularensis. Front. Cell. Infect. Microbiol. 2013;3:64. doi: 10.3389/fcimb.2013.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alvarez Y., Esteban-Torres M., Cortes-Cabrera A., Gago F., Acebron I., Benavente R., Mardo K., de Las Rivas B., Munoz R., Mancheno J.M. Esterase LpEst1 from Lactobacillus plantarum: a novel and atypical member of the alpha beta hydrolase superfamily of enzymes. PLoS One. 2014;9:e92257. doi: 10.1371/journal.pone.0092257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McKenney E.S., Sargent M., Khan H., Uh E., Jackson E.R., San Jose G., Couch R.D., Dowd C.S., van Hoek M.L. Lipophilic prodrugs of FR900098 are antimicrobial against Francisella novicida in vivo and in vitro and show GlpT independent efficacy. PLoS One. 2012;7:e38167. doi: 10.1371/journal.pone.0038167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transparency document
Transparency document