Abstract
The manuscript describes the characterization of the interaction between meso-tetrakis(p-sulfonatophenyl)porphyrin (TSPP) and human serum albumin (HSA). TSPP is a candidate for the photosensitization of structural and functional changes in proteins while HSA provides both an excellent protein model and binding and functional characteristics that could be explored in future applications of the approach. A combination of optical spectroscopic techniques (e.g., fluorescence spectroscopy, fluorescence lifetime, circular dichroism, etc.) and computational docking simulations were applied to better characterize the TSPP/HSA interaction. Recent advances have revealed that the complex formed by TSPP and HSA has become potentially relevant to biomedical applications, biomaterials research and protein photosensitized engineering. The study has determined a likely location of the binding site that places TSPP at a site that overlaps partially with the low affinity site of ibuprofen and places one of the groups of the ligand in proximity of the Trp214 residue in HSA. The characterization will enable future studies aimed at photosensitizing non-native functions of HSA for biomedical and biomaterial applications.
Keywords: Albumin, Docking, Porphyrin, Spectroscopy
Graphical abstract

Highlights
-
•
A novel protocol involving extensive dialysis and centrifugation eliminated aggregated protoporphyrins from the solution.
-
•
Reliable FRET between Trp214 and the porphyrin ligands was established.
-
•
FRET and docking simulations converge to a model consistent with experimental X-ray data.
-
•
Photosensitization mediated by the porphyrin ligands prompts localized conformational changes in HSA.
1. Introduction
The importance of porphyrins in living organisms is underscored by their ubiquitous biochemical functions and by their potential biomedical and biotechnological applications [1], [2]. Heme, for instance, occupies a central role in a myriad of biological functions [2]. The combination of the natural occurrence of many porphyrin-like molecules and their photophysical properties (e.g., relatively large yields for intersystem crossing to the triplet state or the potential for photoinduced electron transfer [3], [4]) has created widespread interest that includes basic photophysical and photochemical characterizations [5], self-assembly [6], [7], and biomedical applications such as their use as a photosensitizers in the phototherapy and photodetection of cancer and other abnormal tissues [8], [9], [10]. The nuance of the photosensitization of porphyrins in biomedical applications has recently highlighted the role of the direct interaction between porphyrins and protein targets [11]. Many naturally occurring porphyrins often present the significant challenge of poor aqueous solubility and their consequent tendency to form a very polydispersed family of aggregates [13] which, despite having insignificant photochemical characteristics [13], greatly affect the spectroscopic characterizations. A possible circumvention of the problem is provided by cationic and anionic porphyrins. These are not-naturally occurring, but provide the advantage of easy solubility in aqueous solution. Among them is the anionic tetra-meso-phenylsulfonato-porphyrin (TSPP). At larger concentrations (>10−4 M) and lower pH (<5) this porphyrin is also characterized by interesting self-assembly properties [12], [14], [15], [16]. However at μM concentration and pH >5.5 TSPP is substantially monomeric in aqueous solution [17]. Anionic (and cationic) porphyrins are expected to interact with proteins through different electrostatic mechanisms and are expected to dock at different locations of the protein [15] compared to heme or other hydrophobic tetrapyrroles.
As mentioned above, the focus of our research is to employ photosensitizing ligands to mediate changes in the structure and function of proteins. Our group has demonstrated that low-irradiance optical excitation of TSPP, non-covalently docked to proteins, causes conformational changes of some polypeptide [12], [16]. These initial results prompted us to investigate the effects on proteins that are more relevant to biophysical and biomedical applications. One of these is represented by human serum albumin (HSA) which is the major carrier of exogenous and endogenous products in the bloodstream, including heme and other porphyrin products [18], [19]. Its hypothesized role in the elimination of heme products, led to the discovery and characterization of a binding site for hemin (the Fe3+ chelate of heme), as well as other tetrapyrroles [20], [21] and has produced much interest in the physiological and non-physiological properties of heme-HSA complexes [22], [23]. The heme binding pocket has been experimentally identified in the primary cleft of subdomain IB of HSA and is defined by three basic residues at the pocket entrance and a Tyr residue (Tyr161) which coordinates with the metal at the center of the porphyrins ring [20]. In comparison with heme and other protoporphyrins, the size and charges on TSPP suggest potentially different mechanisms and sites of interaction. We have therefore investigated the non-covalent docking of TSPP to HSA with the goal of estimating the location of its binding site. Since this porphyrin has shown to be more effective at producing conformational changes of proteins [14], [24], it holds the potential to prompt photosensitized changes in a biomedically crucial protein such as HSA. The possibility to trigger conformational changes using the combination of laser irradiation of a porphyrin ligand, could enable us to artificially enhance or modify the binding properties of HSA. This could have repercussions in future developments of the use of porphyrin/albumin complexes as an artificial enzyme [25], or a substitute for hemoglobin or myoglobin [26].
2. Materials and methods
2.1. Chemicals
Globulin-free HSA (A3782), warfarin (A2250), ibuprofen (I4883) and hemin (51,280) were purchased from Sigma-Aldrich (St. Louis, MO). Spectroscopic grade DMSO was also purchased from Sigma-Aldrich (St. Louis, MO) while TSPP was obtained from Frontier Scientific (Logan, UT). All chemicals were used without further purification.
2.2. Sample preparation
Concentrated stock solutions of the mono-dispersed TSSP were prepared at least 24 h prior to the experiments by dissolving the solid porphyrin in deionized water (DI). The stock solutions were subsequently kept in the dark at room temperature and were stable for up to two weeks. Before each experiment the stock solution was diluted in phosphate buffer at physiological pH, to obtain a final porphyrin concentration in the 2–8 μM range. The concentration was determined spectroscopically using the Beer-Lambert relationship [27] () with ϵ413=510,000 M−1 cm−1 for TSPP [28].
Stock solutions of HSA were prepared fresh before each experiment by dissolving the lyophilized protein in phosphate buffer at pH 7.4. Buffers were prepared at least 24 h in advance by dissolving a saline tablet (P4417, Sigma-Aldrich, St. Louis, MO) in 200 mL of DI water (Ω>18 Ω). All buffers were kept at 6 °C, and only the volume necessary for each experiment was withdrawn and allowed to equilibrate at room temperature before the start of the measurements. Buffers were discarded and made fresh every 7 days.
2.2.1. Instrumentation and methods
2.2.1.1. Steady state measurements
Absorption spectra were recorded using a dual beam Evolution 300 spectrophotometer (Thermo Fisher Scientific, Waltham, MA) at 1.5 nm resolution and 240 nm/min sampling speed. Steady state fluorescence spectra were recorded with an Aminco Bowman-2 double monochromator fluorimeter (Thermo Fisher Scientific, Waltham, MA) at 1 nm/s with a 4 nm bandwidth in excitation and emission. All spectra were corrected for the spectral response of the instrument as well as absorption and inner filter effects (Eq. (2)a and b).
Circular Dichroism spectra (CD) were recorded using a DSM 16 CD spectroplarimeter with a modified Cary 118 monochromator (Olis Inc., Bogart, GA). The monochromator, lamp-housing and sample-compartment were thoroughly purged with nitrogen gas from a liquid-N2 tank prior to and during data acquisition. All spectra were recorded in the Soret band region and blank subtracted.
2.2.1.2. Time-resolved experiments
Fluorescence decay lifetimes were measured using a time-correlated single photon counting (TCSPC) instrument (Fluorocube, Horiba Scientific, Edison, NJ) with a 294 nm pulsed LED (NanoLED-293, Horiba JobinYvon, Edison NJ) source of ~1 ns pulse width and 1 MHz repetition rate. Decays were recorded at the emission maximum of HSA with a 12 nm bandwidth.
2.2.1.3. Fluorescence quenching
Binding between porphyrins and proteins can be investigated by recording the quenching of the intrinsic fluorescence of the protein as a function of the concentration of the porphyrins (i.e., quencher). From the analysis of the quenching, one can extrapolate whether the mechanism is collisional or static and in the second case establish the binding constant [29]. Fluorescence quenching of HSA by TSPP was measured by recording the emission of the protein upon addition of increasing aliquots of an aqueous stock of the porphyrin to the solution containing HSA (~ 4 μΜ). The concentration of TSPP is determined from the value of its absorption maximum using the molar extinction coefficient reported above. Emission spectra of HSA were recorded between 300 and 450 nm with λex=295 nm (where one can assume exclusive excitation of the lone Trp214 residue [30]). The selected protein concentration ensures an OD295<0.1.
In order to distinguish between the contributions of dynamic and static quenching, we examined the effect of temperature by performing quenching experiments at 22 °C as well as 30 °C and 40 °C using aliquots of the same porphyrin and protein stocks. Temperature was changed using a Peltier temperature control element (SPG 1A, Thermo Fisher Scientific, Waltham, MA) in absorption, and a water circulator (Polystat, Cole Parmer, Vernon Hills, Il) in fluorescence. The setting in both devices were kept as to ensure the same equilibration temperature in the two instruments.
Throughout the sample preparation and measurement, the lights in the laboratory were dimmed in order to ensure minimal ambient light exposure of the sample (measured at ~ 50 μW/cm2 in the areas where samples were handled) as to avoid photochemical effects on the porphyrin and the protein [24].
2.2.1.4. Fluorescence quenching analysis
Fluorescence quenching constants were calculated using the Stern-Volmer (S-V) Eq. (1) [31].
| (1) |
where is the total fluorescence intensity of HSA and indicates the fluorescence before addition of the quencher, is the quenching constant, and [Q] is the concentration of the porphyrin determined spectrophotometrically as described above. is obtained from the slope of vs. [Q]. Additional information of the quenching experiments are provided in the Supplementary Material.
2.2.1.5. Porphyrin fluorescence
In addition to probing the fluorescence quenching of the protein, interactions of porphyrins with proteins can be investigated by recording the changes in the emission spectra of the ligands as a function of the concentration of the polypeptide [28], [32]. Such changes can be formalized and quantified through the Benesi-Hildebrand model that can be summarized by Eq. (2).
| (2) |
In this equation and represent the absorption of free and docked TSPP (see below), respectively and is the absorption signal of the porphyrin extrapolated at infinite protein concentration, is the protein concentration (in molarity) and is the binding constant.
Additional details of the method are provided in the Supplementary Material. In order to determine the binding of TSPP to HSA, increasing aliquots of the albumin stock (40–60 μM, determined spectrophotometrically using ϵ280=35296 M−1 cm−1 [33]) were added to a diluted solution of the porphyrin buffer stock. The absorption and emission spectra of TSPP were recorded within ~2–5 min after each addition (see below). Throughout the experiment the OD of TSPP (free or bound to HSA) remained <0.15 at the excitation wavelength (405 nm). Such OD ensures a substantially uniform light distribution within the 1 cm optical pathlength of the quartz cells and minimizes artifacts from inner filter or other non-linear effects. The molar extinction coefficient of TSPP at the maximum of absorption (278 nm) and excitation wavelength of the protein (294 nm) was estimated to be smaller than the one of the protein (ϵ < 20,000 cm−1 M−1).
Typically Eq. (2) is applied using the fluorescence of the ligand. However, since the emission intensity of TSPP does not increase with the addition of HSA, we used the absorption spectrum of the porphyrin as a function of the protein to extrapolate the amount of free and bound ligand as explained in the Supplementary Content.
2.2.1.6. Competitive binding: protocol
Competitive binding between TSPP and well-characterized HSA ligands (i.e., warfarin, ibuprofen and hemin) were carried out using both the fluorescence of the porphyrins (i.e., using the B-H method) and the quenching of HSA emission (i.e., using the S-V method). Binding competition can be used to establish whether TSPP competes for any of the docking locations of warfarin, ibuprofen and hemin which have been thoroughly characterized [19], [34], [35], [36]. For all competitive binding experiments, HSA stocks were prepared dividing a large volume into smaller volumes. Each smaller-volume was incubated with an excess of competing ligand (3:1 warfarin/ibuprofen/hemin: HSA), while the control volume contained no competing ligand. The concentrations of the competitive ligands were determined spectrophotometrically using ϵ308=12500 M−1 cm−1 for warfarin, ϵ222=9900 M−1 cm−1 for ibuprofen [37], [38] and ϵ405=40573 M−1 cm−1 for hemin (first dissolved in DMSO then added to buffer with final aqueous DMSO concentration under 2% by volume) derived by comparison with identically concentrated solutions in DMSO where ϵ403=1.7×105 M−1 cm−1 [39]. All competition experiments were carried out at room temperature.
2.2.1.7. Competitive binding
HSA fluorescence quenching. Quenching experiments were performed by completing the same S-V protocols described above. Aliquots of the concentrated porphyrin stocks were added to the HSA or HSA/competing-ligand solutions (described above). The emission of the protein (ex=295 nm) was recorded between 300 and 450 nm as a function of the total porphyrin concentration.
2.2.1.8. Competitive binding
Benesi-Hildebrand method. Binding experiments were completed with the procedure described earlier. Aliquots of concentrated stocks of HSA, or HSA/competing-ligand were added to aqueous porphyrin stocks. The region of the Soret band of TSPP as a function of total protein concentration was recorded and the spectrum analyzed using the Gaussian fitting described in the Supplementary Content.
2.2.1.9. Analysis of fluorescence decay
The DAS6.2 fluorescence decay analysis software (Horiba Scientific, Edison, NJ) yielded fluorescence lifetime values and their relative amplitudes (τi and αi respectively) using re-convolution and least-square fitting methods under the assumption that the decay kinetics can be represented by the equation
| (3) |
as extensively discussed in the literature (for example Ref. [31] and citations within). The instrument response function needed for the re-convolution algorithm was collected using a 1 mg/mL scattering solution of silica gel (Sigma Aldrich, H0761) in deionized water. Fitting was performed with various numbers of exponential components, and in all cases the best fit with the fewest free parameters was obtained using 3 exponentials, which is consistent with previous results obtained from HSA in solution [40]. From the values retrieved by the fitting procedure, the average decay lifetimes were calculated as [31].
| (4) |
2.2.1.10. Docking simulations-structure preparation
The Protein Data Bank file ID 1O9X, which includes HSA complexed with tetradecanoic acid (myristic acid) and hemin [20], provided the initial protein coordinates. Using ArgusLab 4.0.1 (Planaria Software, Seattle, WA) hemin and all myristic acid coordinates were removed and hydrogen atoms were added based on the hybridization and bonding pattern of the heavy atoms. Kollman charges were then added to the protein using AutoDock Tools 1.5.4 [41]. Initial coordinates for hemin were directly isolated from 1O9X and hydrogen atoms were added using ArgusLab. TSPP was created using Spartan 10 (Wavefunction Inc., Irvine, CA). Gasteiger partial charges were assigned to the porphyrin, and all non-polar hydrogen atoms were merged (i.e., deleted with their charges added to their closest bonded non-hydrogen atom) in AutoDock Tools.
2.2.1.11. Docking simulations-docking procedure
Binding of TSPP to HSA was simulated using AutoDock 4.2 [42] following a procedure described in details elsewhere [43]. Coarse sampling was initially performed to survey the region(s) of the protein that yielded the most energetically favorable binding areas. Coarse sampling was run with larger grid spacing of.375 Å and one order of magnitude fewer maximum number of evaluations (2.5×106 energy evaluations). Refined runs were then carried out at the locations that returned the best energy conformations, by using a smaller grid spacing (see below) and a larger maximum number of evaluations in the genetic algorithm. All rotatable bonds of the ligand were allowed to rotate during the docking simulation while the structure of HSA was held rigid. Grid maps were generated through the program AutoGrid which recalculates atomic affinity, electrostatic, and desolvation potentials for each atom type in the ligand molecule being docked. The energy of interaction of each ligand atom with the protein is assigned to a point in a grid box which was typically set at 126×126×126 Å, except for the binding of PPIX at the cleft-site where the grid box was 110×110×110 Å. In the refined docking simulations the grid points were separated by 0.186 Å. AutoGrid uses the distance-dependent dielectric of Mehler and Solmajer [44]. The standard docking protocol for rigid and flexible ligand docking consisted of 10 independent runs per ligand, using a population of 150 randomly placed individuals, with 2.5×107 energy evaluations, a maximum number of 2.5×106 generations, a mutation rate of 0.02, a crossover rate of 0.80, and an elitism value of 1. The probability of performing a local search on an individual in the population was 0.06, using a maximum of 300 iterations per local search. During the AutoDock calculation, the energy of a particular configuration is evaluated from the values of the grid using the Lamarckian genetic algorithm for minimization with default parameters that include random starting positions, orientations, and torsions that were generated before each run. The most likely docked configuration was retrieved from the binding energy which is derived from the grid score and the internal energy value of the ligand (which accounts for the energy penalty of distorting the molecule).
3. Results and discussion
3.1. Binding
Experiments were carried out to investigate the formation of the non-covalent complex between TSPP and the native form (N) of HSA [45]. In comparison to earlier studies [28] the current investigation places an emphasis on the interaction mechanisms and the location of the docking site which were not characterized previously.
3.1.1. TSPP fluorescence
The addition of HSA causes the broadening and the shift of the absorption peak of TSPP from 413 nm to 420 nm accompanied by a decrease in intensity (Fig. S1). The same addition causes the fluorescence peaks to shift from 643 nm and 699 nm to 648 nm and 713 nm, respectively (Fig. S4, Supplementary Content). These features are consistent with the binding of TSPP to proteins [14], [28]. Unlike the emission of hydrophobic porphyrins [46], the shift in fluorescence is not accompanied by a substantial increase of the intensity. Therefore, one cannot construct B-H plots from the emission spectra of TSPP and an alternative method must be used. One such method employs the Gaussian fitting of the Soret band region in the absorption spectra as described in the “Methods Section” (Fig. S3, Supplementary Content). The data from the fitting yield a linear B-H plot with a Kb=2.8×105 M−1 (Fig. 1 and Table 1).
Fig. 1.
Benesi-Hildebrand plot of TSPP binding affinity to HSA (▲).
Table 1.
Static quenching constants (KSV) retrieved from Eq. (1). Binding constants (Kb) extrapolated from the Benesi-Hildebrand plots retrieved from the fluorescence of the porphyrin upon addition of HSA.
| T | KSV(M−1) | Kb(M−1) | |
|---|---|---|---|
| 22 °C | 99.3×104 | 2.8×105 | |
| 22 °C | + ibuprofen | 85.2×104 | No apparent binding |
| 30 °C | 87.6×104 | ||
| 40 °C | 78.3×104 |
3.1.2. Competitive binding
The presence of ibuprofen bound to HSA prevents TSPP from docking to HSA as shown by the absence of the red-shift of the absorption maximum of the porphyrin as a function of increasing HSA/ibuprofen complex (Fig. 2A). Binding competition with warfarin and hemin could not yield quantitative results from B-H plots. These competing ligands present photophysical characteristics that interfere with the optical measurements needed to investigate competition with porphyrins. Hemin has a strong quenching effect on TSPP in addition to a substantial overlap in the absorption spectrum. Warfarin also absorbs and emits in a region that interferes with the measurements, thus preventing the calculation of quantitative results. Qualitatively, however, the experiments established (Fig. 2B and C) that TSPP is still able to dock to HSA in the presence of either competing ligands, because the spectrum of the porphyrin shows a shift to longer wavelength consistent with binding to the protein.
Fig. 2.
(A) Absorption of TSPP as a function of increasing concentration of HSA/ibuprofen complex. (B) Absorption of TSPP as a function of increasing concentration of HSA/warfarin complex. (C) Absorption of TSPP as a function of increasing concentration of HSA/hemin complex. In each figure the arrow indicates the direction of the spectral changes with increasing protein concentration. The initial and final concentrations are also indicated next to the arrow.
3.1.3. HSA fluorescence quenching
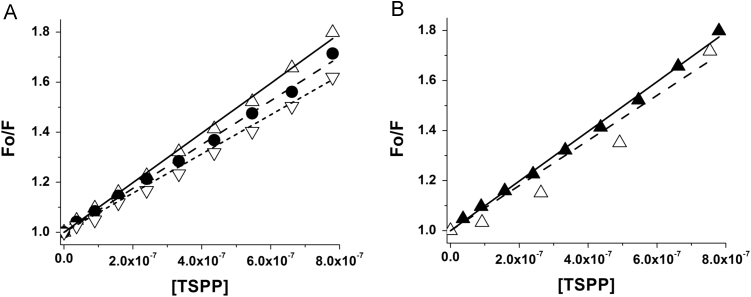
Adding TSPP to HSA (4 μM solution) causes the fluorescence intensity of the protein to decrease without peak shift (Fig. S2, Supplementary Content). This is consistent with the fact that HSA has a single Trp residue, thus, its quenching cannot lead to a shift in emission because there are no other non-quenched residues that can contribute to the fluorescence. S-V plots (Fig. 3A) provide KQ=9.9×105 M−1. S-V plots can be ambiguous since a linear trend is common to both collisional and static quenching. However, the two mechanisms describe different physical situations and yield a different meaning of the quenching constant [31]. One approach that can distinguish static and collisional mechanisms employs the temperature dependence of the quenching constant. Raising the temperature tends to dissociate a non-covalent ligand-protein complex, therefore decreases the static quenching constant. Conversely, higher temperature, increases the dynamic quenching constant, due to a larger frequency of thermally-driven collisions [29]. The data show that KQ decreases with increasing temperature (Fig. 3A, Table 1) which is consistent with static quenching due to the formation of the TSPP/HSA complex. Static quenching can be confirmed also theoretically. In fact from [31] one retrieves a value of the bimolecular quenching rate assuming as the average fluorescence lifetime of HSA without TSPP (Table 2). The value of is related to the diffusion coefficients and of the quencher and the chromophore (HSA in this case) through [31].
| (5) |
where
| (6) |
Fig. 3.
(A) Temperature dependent Stern-Volmer plots of HSA fluorescence quenched by TSPP at 22 °C (△, —), 30 °C (•, ― ―), and 40 °C (▽, ‑ ‑).(B) Fluorescence quenching by porphyrin in the absence and presence of competing ibuprofen. Quenching of HSA by TSPP (▲,—); quenching of HSA/ibuprofen complex by TSPP (△,― ―).The solid lines represent the linear regression of the individual plots.
Table 2.
Fluorescence lifetimes before and after irradiation in air and deoxygenated samples.
| Sample (conditions) | Laser dose |
Fluorescence decay parameters |
Average lifetime | |||||
|---|---|---|---|---|---|---|---|---|
| HSA | J/cm2 | τ1(ns) | A1(%) | τ2(ns) | A1(%) | τ3(ns) | A1(%) | <τ> |
| Air | 0 | 6.20 | 47.73 | 2.59 | 41.89 | 0.41 | 10.68 | 4.09 |
| TSPP/HSA | J/cm2 | τ1(ns) | A1(%) | τ2(ns) | A1(%) | τ3(ns) | A1(%) | <τ> |
| Air | 0 | 6.25 | 43.70 | 2.16 | 39.25 | 0.28 | 17.04 | 3.63 |
| 44 | 6.32 | 38.44 | 2.06 | 38.74 | 0.22 | 22.82 | 3.02 | |
| Deoxygenated | 0 | 6.03 | 38.35 | 1.85 | 36.07 | 0.16 | 25.58 | 3.02 |
| 44 | 6.48 | 32.21 | 2.03 | 34.25 | 0.17 | 33.54 | 2.84 | |
In this last relation ~ [31], while and are the radii of the protein and the quencher respectively. From the size of HSA and TSPP [47] ~ ~ m [48] and from the diffusion coefficients at room temperature for the two molecules ~ m2s−1 [49], [50] one estimates a value ~ which is 10 orders of magnitude smaller than the one obtained from our experiments. The estimated value of would actually be even smaller since the one we calculated considers a quenching efficiency, =1. Therefore collisional quenching between TSPP and HSA can be confidently ruled out and Eq. (2) yields ==9.9×105 M−1 which is in agreement with the binding of porphyrins to proteins [28].
3.1.4. Fluorescence lifetime
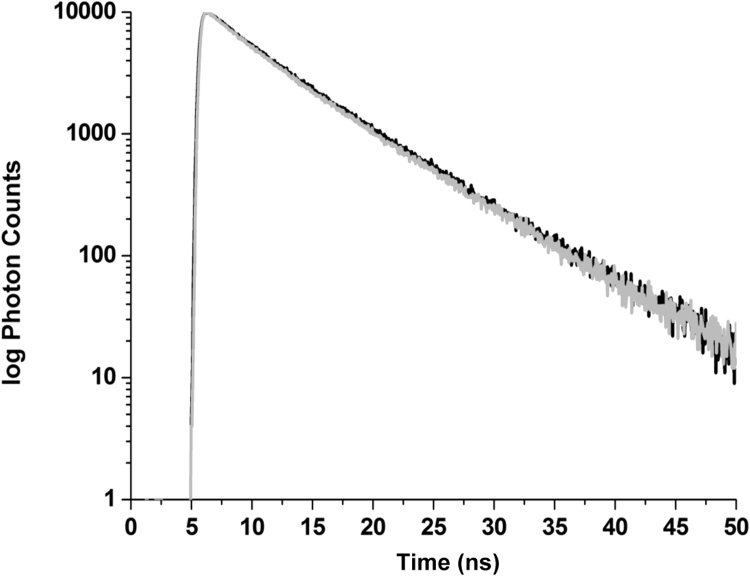
Finally, the fluorescence decay of HSA as a function of added TSPP confirms the static mechanism. As shown in Fig. 4, addition of the porphyrin does not change the emission lifetime of the protein in the same samples where there is instead a large quenching of the emission intensity. This evidence confirms that TSPP, under the conditions of our experiments, does not cause collisional quenching. In addition the nearly constant fluorescence decay of HSA in the presence of TSPP also rules out the occurrence of fluorescence resonance energy transfer (FRET) between Trp214 and bound TSPP despite the overlap between the emission of the aromatic residue and the Soret band of the porphyrin. The absence of FRET indicates that the quenching of Trp214 might be caused by the proximity of one of the groups to the indole of the Trp residue, rather than the stacking of the porphyrin macrocycle (where the Soret band transition is localized [51]).
Fig. 4.
Fluorescence decay of HSA in the absence of TSPP (black line) and in the presence of 3 μM TSPP (gray line).
Thus, demonstrated that the origin of the quenching mechanism is static, the value of KSV reduces to the one of Kb [31] and the discrepancy between the magnitude of the binding constant obtained through the B-H method and the one retrieved via the S-V plots must be explained. The value of Kb retrieved through quenching is consistent (actually identical within the experimental error) to the one reported by another group [28]. Thus we are inclined to believe that the value obtained from the B-H plot is underestimated. The reason for underestimating Kb may be due to the methodology used. Since we retrieved the amount of free and bound TSPP from Gaussian fitting of overlapping spectra the cross-correlation of the fitting parameters is likely to be large as to affect the ratio of the areas of the free and bound TSPP to the overall area of the Soret band.
Competition studies show that the quenching constant is reduced by the presence of ibuprofen, while quenching competition with warfarin and hemin did not provide reliable results for the reasons explained earlier.
3.1.5. Circular dichroism
Interesting is the dichroic activity of TSPP bound to HSA. The dichroic signal shows a week negative Cotton effect [52], with a trough at 411 nm, for the free form of TSPP (Fig. 5). However, binding to HSA prompts a peak shift (to 418 nm) and an increase in the intensity of the negative Cotton effect (Fig. 5). Fig. 5 therefore suggests that binding increases the distortion of TSPP [53] through interactions with amino acid side chains [52].
Fig. 5.
CD spectra of free (solid lines) and HSA-docked (dotted lines) PPIX (gray) and TSPP (black) in the region of the Soret band.
3.2. Docking simulations
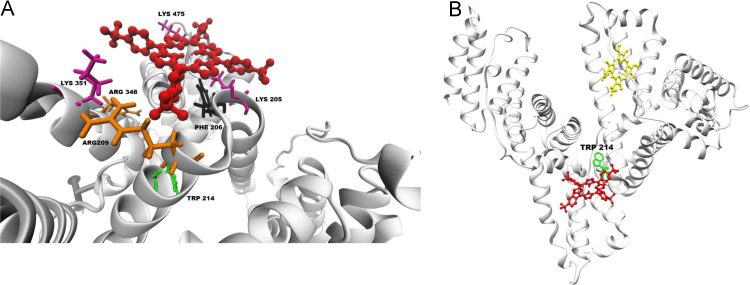
A coarse sampling across the entire HSA molecule, resulted in many possible binding locations with binding energies that are in the order of −5.0 kcal/mol. Based on the binding competition between TSPP and ibuprofen (Fig. 3B), refined searches were completed in and around the two main binding sites of the drug which, according to x-ray diffraction results, are located in subdomain IIIA (higher affinity) and between subdomain IIA and IIB (lower affinity) [19]. The search yielded a viable docked conformation of TSPP near the location of the lower affinity binding site for ibuprofen (Fig. 6A). Though the lowest free energy of binding is only −4.15 kcal/mol at this location, each negatively charged sulfonic group of TSPP appears to interact with one or more positively charged Lys or Arg residues located between 5 Å (Lys 475, Lys 205, Arg 209) and 11 Å (Lys 351 and Arg 348). Closer inspection shows that at this location the “penalty” for the torsional energy of the porphyrin is large and positive (+3.58 kcal/mol) and causes an overall less favorable energy minimum than would be expected from the electrostatic interactions alone. Such “penalty” would be consistent with the Cotton effect observed in CD. The energy “penalty” in the docking simulations is created by the fact that the protein is considered rigid and the ligand has limited flexibility. Therefore the energy penalty is an indication of strain on the structure of the ligand (i.e., a positive contribution to the free energy) produced by the steric clash between atoms of the ligand and atoms of the protein, that cannot be compensated by their structural modifications (e.g., rotation of bonds). However the actual docked TSPP would not have the constrained posed by the computational model and in the presence of steric clashes it could modify its structure. If the steric hindrance is at the macrocycle it may prompt it to “deform” out of plane, thus causing a change of symmetry of the porphyrin ring [54], [55] that may originate the Cotton effect. Additionally, this binding location places TSPP in close proximity to several aromatic residues which is also in agreement with the spectroscopic findings [53]. As discussed earlier, TSPP causes highly efficient quenching of the fluorescence of the lone Trp214 residue of HSA (Table 1). The binding location of Fig. 6B places one of the S atoms of TSPP 12 Å from Trp 214. This distance is approximately half the distance to the site of the heme (Fig. 6B) [20]. Sulfur atoms are efficient fluorescence quenchers [56], [57] and the presence of the in proximity to Trp214 is in agreement with the static quenching of the aromatic residue.
Fig. 6.
(A) Docking simulation results for PPIX in the central cleft of HSA. The figure shows the remaining proximity with the Arg114 residue already participating in the binding to the heme pocket. (B) Docking simulation results for TSPP near the ibuprofen binding site. The figure shows the proximity of the 4 negatively charged sulfonic groups of the porphyrin with the positively charged Lys and Arg residues. (C) Overall view of HSA with docked PPIX (blue) and TSPP (red) and the crystal structure heme (yellow) at the sites shown in part A and B. The figure shows the position of each porphyrin relative to the lone Trp214 residue.
4. Conclusions
The binding of TSPP to HSA was first demonstrated over a decade ago [28] however the mechanisms of the binding or the possible location of the docking site was not established. The work presented here confirms the formation of a ground-state TSPP/HSA complex and validates the binding constant observed previously (~106 M−1) [28]. Our results however shed additional light on the location of the binding and the nature of the interaction. The combination of spectroscopic data and docking simulation converge into a model that establishes that TSPP docks HSA at a location that overlaps with the region of the low affinity site for ibuprofen and is consistent not only with the quenching of the Trp214 residue but also with the induced negative Cotton effect detected with CD spectroscopy. The lack of FRET indicates that the conformation of TSPP places one of the groups of the porphyrin in proximity of Trp214 instead of the porphyrin macrocycle. However, the presence of the group of aromatic amino acids (e.g., Phe206) near the macrocycle could prompt the negative Cotton effect. These amino acids would likely force the porphyrin ring to deform, thus relaxing the energetic penalties recorded in the docking simulations. The results we obtained are important to undergoing studies to determine the photosensitization of conformational changes in HSA as they allow us to more precisely investigating the possible conformational changes and correlate them with functional changes of the protein.
Acknowledgments
The research was in part funded by an AFOSR grant (FA8650-07-D-6800) to R. J. T. and an NIH grant (G12RR013646-1) to L. B. Support for S. C. R is provided by the Consortium Research Fellows Program (FA8650-13-2-6366). The authors wish to thank Mr. David Stolarski of TASC, Inc. for assistance in editing the manuscript.
Footnotes
Transparency document associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.bbrep.2016.07.014.
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.bbrep.2016.07.014.
Transparency document. Supplementary material
Supplementary material
.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Guallar V., Wallrapp F.H. QM/MM methods: Looking inside heme proteins biochemistry. Biophys. Chem. 2010;149:1–11. doi: 10.1016/j.bpc.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 2.Srajer V., Royer W.E. Time-resolved x-ray crystallography of heme proteins. Methods Enzymol. 2008;437:379–395. doi: 10.1016/S0076-6879(07)37019-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kennedy J.C., Marcus S.L., Pottier R.H. Photodynamic therapy (PDT) and photodiagnosis (PD) using endogenous photosensitization induced by 5-aminolevulinic acid (ALA): mechanisms and clinical results. J. Clin. Las. Med. Surg. 1996;14:289–304. doi: 10.1089/clm.1996.14.289. [DOI] [PubMed] [Google Scholar]
- 4.Komatsu T., Wang R.-M., Zunszain P.A., Curry S., Tsuchida E. Photosensitized reduction of water to hydrogen using human serum albumin complexed with zinc-protoporphyin IX. J. AM Chem. Soc. 2006;128:16297–16301. doi: 10.1021/ja0656806. [DOI] [PubMed] [Google Scholar]
- 5.Croney J.C., Helms M.K., Jameson D.M., Larsen R.W. Conformational dynamics and temperature dependence of photoinduced electron transfer within self-assembled coproporphyrin: cytochrome c complexes. Biophys. J. 2003;84:4135–4143. doi: 10.1016/S0006-3495(03)75138-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balaban T.S. Tailoring porphyrins and chlorins for self-assembly in biomimetic artificial antenna systems. Acc. Chem Res. 2005;38:612–623. doi: 10.1021/ar040211z. [DOI] [PubMed] [Google Scholar]
- 7.Elemans J.A.A.W., van Hameren R., Nolte R.J.M., Rowan A.E. Molecular materials by self-assembly of porphyrins, phthalocyanines, and perylenes. Adv. Mater. 2006;18:1251–1266. [Google Scholar]
- 8.Fukuda H., Casas A., Batlle A. Aminolevulinic acid: from its unique biological function to its star role in photodynamic therapy. Int. J. Biochem. Cell Biol. 2005;37:272–276. doi: 10.1016/j.biocel.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 9.Murahari M.S., Yergeri M.C. Identification and usage of fluorescent probes as nanoparticle contrast agents in detecting cancer. Curr. Pharm. Des. 2013;19:4622–4640. doi: 10.2174/1381612811319250009. [DOI] [PubMed] [Google Scholar]
- 10.Tetard M.C., Vermandel M., Mordon S., Lejeune J.P., Reyns N. Experimental Use of Photodynamic Therapy in High Grade Gliomas: a Review focused on 5-aminolevulinic Acid. Photo. Photo. Ther. 2014;11:319–330. doi: 10.1016/j.pdpdt.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Acedo P., Zawacka-Pankau J. p53 family members - important messangers in cell death signalin in photodynamic therapy of cancer? Photchem. Photobiol. Sci. 2015;14:1390–1396. doi: 10.1039/c5pp00251f. [DOI] [PubMed] [Google Scholar]
- 12.Andrade S.M., Teixiera R., Costa S.M.B., Sobral A.J.F.N. Self-aggregation of free base porphyrins in aqueous solution and in DMPC vescicles. Biophys. Chem. 2008;133:1–10. doi: 10.1016/j.bpc.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 13.Monsu’ Scolaro L., Castriciano M., Romeo A., Patane’ S., Cefali E., Allegrini M. Aggregation behavior of protoporphyrin IX in aqueous solutions: Clear evidence of vescivle formation. J. Phys. Chem. B. 2002;106:2453–2459. [Google Scholar]
- 14.McMicken B., Thomas R.J., Brancaleon L. Photoinduced partial unfolding of tubulin bound to meso-tetrakis(sulfonatophenyl) porphyrin leads to inhibition of microtubule formation in vitro. J. Biophot. 2014;7:874–888. doi: 10.1002/jbio.201300066. [DOI] [PubMed] [Google Scholar]
- 15.Silva I., Sansone S., Brancaleon L. An Anionic Porphyrin Binds b-Lactoglobulin A at a Superficial Site Rich in Lysine Residues. Protein J. 2009;28:1–13. doi: 10.1007/s10930-008-9158-8. [DOI] [PubMed] [Google Scholar]
- 16.Tian F., Johnson E.M., Zamarripa M., Sansone S., Brancaleon L. Binding of porphyrins to tubulin heterodimers. Biomacromolecules. 2007;8:3767–3778. doi: 10.1021/bm700687x. [DOI] [PubMed] [Google Scholar]
- 17.Kelbauskas L., Bagdonas S., Dietel W., Rotomskis R. Excitation relaxation and structure of TPPS4 J-aggregates. J. Lumin. 2003;101:253–262. [Google Scholar]
- 18.Ascenzi P., Fasano M. Serum heme-albumin: an allosteric protein. IUBMB Life. 2009;61:1118–1122. doi: 10.1002/iub.263. [DOI] [PubMed] [Google Scholar]
- 19.Curry S. Lessons from the crystallographic analysis of small molecule binding to human serum albumin. Drug. Metab. Pharmacokinet. 2009;24:342–357. doi: 10.2133/dmpk.24.342. [DOI] [PubMed] [Google Scholar]
- 20.Zunszain P.A., Ghuman J., Komatsu T., Tsuchida E., Curry S. Crystal structural analysis of human serum albumin complexed with hemin and fatty acid. BMC Struct. Biol. 2003;3:6–15. doi: 10.1186/1472-6807-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zunszain P.A., Ghuman J., McDonagh A.F., Curry S. Crystallographic analysis of human serum albumin complexed with 4Z,15E-Bilirubin-IXa. J. Mol. Biol. 2008;381:394–406. doi: 10.1016/j.jmb.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wardell M., Wang Z., Ho J.X., Robert J., Ruker F., Ruble, Carter D.C. , The atomic structure of methemalbumin at 1.9 A. Biochem Biophys. Res Commun. 2002;291:813–819. doi: 10.1006/bbrc.2002.6540. [DOI] [PubMed] [Google Scholar]
- 23.Fernandez N.F., Sansone S., Mazzini A., Brancaleon L. Irradiation of the porphyrin causes unfolding of the protein in the Protoporphyrin IX/ b-lactoglobulin non covalent complex. J. Phys. Chem. B. 2008;112:7592–7600. doi: 10.1021/jp710249d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belcher J., Sansone S., Fernandez N.F., Haskins W.E., Brancaleon L. Photoinduced Unfolding of Beta-Lactoglobulin Mediated by a Water-Soluble Porphyrin. J. Phys. Chem B. 2009;113:6020–6030. doi: 10.1021/jp900957d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bocedi A., De Sanctis G., Ciaccio C., Tundo G.R., di Masi A., Fanali G., Nicoletti F.P., Fasano M., Smulevich G., Ascenzi P., Coletta M. Reciprocal allosteric modulation of carbon monoxide and warfarin binding to ferrous human serum heme-albumin. PLoS One. 2013;8:e58842. doi: 10.1371/journal.pone.0058842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas C.M., Ward T.R. Artificial metalloenzymes: proteins as hosts for enantioselective catalysis. Chem Soc. Rev. 2005;34:337–346. doi: 10.1039/b314695m. [DOI] [PubMed] [Google Scholar]
- 27.Cantor C.R., Schimmel P.R. W.H. Freeman and Company; New York: 1980. Biophysical Chemistry (part II): Techniques for the Study Of Biological Structure and Function. [Google Scholar]
- 28.Andrade S.M., Costa S.M.B. Spectroscopic studies on the interaction of a water soluble porphyrin and two drug carrier proteins. Biophys. J. 2002;82:1607–1619. doi: 10.1016/S0006-3495(02)75512-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van de Weert M., Stella L. Fluorescence quenching and ligand binding: a critical discussion of a popular methodology. J. Mol. Struct. 2011;998:144–150. [Google Scholar]
- 30.He X.M., Carter D.C. Atomic structure and chemistry of human serum albumin. Nature. 1992;358:209–215. doi: 10.1038/358209a0. [DOI] [PubMed] [Google Scholar]
- 31.Lakowicz J.R. Third ed., Springer,; New York: 2006. Principles of Fluorescence Spectroscopy. [Google Scholar]
- 32.Tian F., Johnson K., Lesar A.E., Moseley H., Ferguson J., Samuel I.D.W., Mazzini A., Brancaleon L. The pH-Dependent Conformational Transition of b-Lactoglobulin Modulates the Binding of Protoporphyrin IX. Biochim. Biophys. Acta. 1760;2005:38–46. doi: 10.1016/j.bbagen.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 33.Gill S.C., von Hippel P.H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 34.Baroni S., Mattu M., Vannini A., Cipollone R., Aime S., Ascenzi P., Fasano M. Effect of ibuprofen and warfarin on the allosteric properties of haem-human serum albumin. Eur. J. Biochem. 2001;268:6214–6220. doi: 10.1046/j.0014-2956.2001.02569.x. [DOI] [PubMed] [Google Scholar]
- 35.Petersen M.J., Hansen C., Craig S. Ultraviolet A irradiation stimulates collagenase production in cultured human fibroblasts. J. Invest. Dermatol. 1992;99:440–444. doi: 10.1111/1523-1747.ep12616142. [DOI] [PubMed] [Google Scholar]
- 36.Petitpas I., Bhattacharya A.A., Twine S., East M., Curry S. Crystal structure analysis of warfarin binding to human serum albumin: Anatomy of drug site I. J. Biol. Chem. 2001;276:22804–22809. doi: 10.1074/jbc.M100575200. [DOI] [PubMed] [Google Scholar]
- 37.Perry J.L., Goldsmith M.R., Williams T.R., Radack K.P., Christensen T., Gorham J., Pasquinelli M.A., Toone E.J., Beratan D.N., Simon J.D. Binding of warfarin influences the acid-base equilibrium of H242 in sudlow site I of human serum albumin. Photochem Photobiol. 2006;82:1365–1369. doi: 10.1562/2006-02-23-RA-811. [DOI] [PubMed] [Google Scholar]
- 38.Tang J.H., Luan F., Chen X.G. Binding analysis of glycyrrhetinic acid to human serum albumin: fluorescence spectroscopy, FTIR, and molecular modeling. Bioorg. Med. Chem. 2006;14:3210–3217. doi: 10.1016/j.bmc.2005.12.034. [DOI] [PubMed] [Google Scholar]
- 39.Muryoi N., Tiedemann M.T., Pluym M., Cheung J., Heinrichs D.E., Stillman M.J. Demonstration of the Iron-regulated Surface Determinant (Isd) Heme Transfer Pathway in Staphylococcus aureus. J. Biol. Chem. 2008;283:28125–28136. doi: 10.1074/jbc.M802171200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amiri M., Jankeje K., Albani J.R. orgin of fluorescnce lifetimes in human serum albumin. Studies on native and denatured protein. J. Fluor. 2010;20:651–656. doi: 10.1007/s10895-010-0597-1. [DOI] [PubMed] [Google Scholar]
- 41.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morris G.M., Goodsell D.S., Halliday R.S., Huey R., Hart W.E., Belew R.K., Olson A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comp. Chem. 1998;19:1639–1662. [Google Scholar]
- 43.Farooqi M.J., Penick M.A., Negrete G.R., Brancaleon L. Interaction of Human Serum Albumin with Novel 3,9-Disubstituted Perylenes. Protein J. 2013;32:493–504. doi: 10.1007/s10930-013-9508-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mehler E.L., Solmajer T. Electrostatic effects in proteins: Comparison of dielectric and charge models. Protein Eng. 1991;4:903–910. doi: 10.1093/protein/4.8.903. [DOI] [PubMed] [Google Scholar]
- 45.Peters T. Genetics, and Medical Applications, Academic; San Diego: 1996. All About Albumin: Biochemistry. [Google Scholar]
- 46.Brancaleon L., Moseley H. Effects of Photoproduct on the Binding Properties of Protoporphyrin IX. Biophys. Chem. 2002;96:77–87. doi: 10.1016/s0301-4622(02)00035-2. [DOI] [PubMed] [Google Scholar]
- 47.Nogales E., Wolf S.G., Downing K.H. Structure of the ab tubulin dimer by electron crystallography. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- 48.Armstrong J.K., Wenby R.B., Meiselman H.J., Fisher T.C. The Hydrodynamic Radii of Macromolecules and Their Effect on Red Blood Cell Aggregation. Biophys. J. 2004;87:4259–4270. doi: 10.1529/biophysj.104.047746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.D. Brune, Kim, S., Predicting protein diffusion coefficients, Proc. Natl. Acad. Sci., 90, 1993, pp. 3835–3839. [DOI] [PMC free article] [PubMed]
- 50.da Costa V.C.P., Ribeiro A.C.F., Sobral A.J.F.N., Lobo V.M.M., Annunziata O., Santos C.I.A.V., Willis S.A., Price W.S., Esteso M.A. Mutual and self-diffusion of charged porphyrines in aqueous solutions. J. Chem. Thermodyn. 2012;47:312–319. [Google Scholar]
- 51.Milgrom L.R. Academic Press; New York: 1978. The Colours of Life: An Introduction to the Chemistry of Porphyrins and Related Compounds. [Google Scholar]
- 52.Hsu M.-C., Woody R.W. The origin of the heme Cotton effects in myoglobin and hemoglobin. J. Am. Chem. Soc. 1971;93:3515–3525. doi: 10.1021/ja00743a036. [DOI] [PubMed] [Google Scholar]
- 53.Konishi K., Miyazaki K., Aida T., Inoue S. Photoinduced Conformational Ruffling of Distorted Porphyrin. Optical Resolution and Photochemical Behavior of Chiral “Single-Armed” Porphyrin Complexes. J. AM Chem. Soc. 1990;112:5639–5640. [Google Scholar]
- 54.Hu S., Smith K.M., Spiro T.G. Assignment of protoheme resonance raman spectrum by heme labeling in myoglobin. J. Am. Chem. Soc. 1996;118:12638–12646. [Google Scholar]
- 55.Kozlowski P.M., Rush III T.S., Jarzecki A.A., Zgierski M.Z., Chase B., Piffat C., Ye B.-H., Li X.-Y., Pulay P., Spiro T.G. DFT-SQM force field for nickel porpine: intrinsic ruffling. J. Phys. Chem. A. 1999;103:1357–1366. [Google Scholar]
- 56.Cho Y., Batt C.A., Sawyer L. Probing the retinol-binding site of bovine b-lactoglobulin. J. Biol. Chem. 1994;269:11102–11107. [PubMed] [Google Scholar]
- 57.Merola F., Rigler R., Holmgren A., Brochon J.C. Picosecond tryptophan fluorescence of thioredoxin: evidence for discrete species in slow exchange. Biochemistry. 1989;28:3383–3398. doi: 10.1021/bi00434a038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material