Abstract
Somatic cells can be reprogrammed to a pluripotent state by nuclear transfer into oocytes, yet developmental arrest often occurs. While incomplete transcriptional reprogramming is known to cause developmental failure, reprogramming also involves concurrent changes in cell cycle progression and nuclear structure. Here we study cellular reprogramming events in human and mouse nuclear transfer embryos prior to embryonic genome activation. We show that genetic instability marked by frequent chromosome segregation errors and DNA damage arise prior to, and independent of, transcriptional activity. These errors occur following transition through DNA replication and are repaired by BRCA1. In the absence of mitotic nuclear remodelling, DNA replication is delayed and errors are exacerbated in subsequent mitosis. These results demonstrate that independent of gene expression, cell-type-specific features of cell cycle progression constitute a barrier sufficient to prevent the transition from one cell type to another during reprogramming.
Reprogramming of somatic cells to a pluripotent state by either nuclear transfer (NT) or ectopic expression of transcription factors entails genome-wide changes in gene expression1,2. Unlike induced pluripotent stem cell reprogramming, the first cell cycle following NT does not involve transcriptional activity. Yet, NT embryos often arrest at, or before the onset of transcription3, suggesting that barriers other than changes to gene expression can prevent cell-type transitions. However, these barriers have not been identified. NT provides an experimental system to dissect reprogramming events in the absence of transcriptional changes.
Here, we used NT to elucidate the roles of mitotic remodelling and DNA replication during reprogramming. We observed frequent genetic instability in both human and mouse NT embryos. The formation of DNA damage and chromosome segregation errors were replication dependent, and independent of transcription. Segregation errors carried the hallmarks of replication-dependent damage, including chromosome bridges and acentric chromosome fragments. Chromosome segregation error rates depended on the origin of the donor cell used for NT. Thus, cell-type-specific features of the chromatin affect progression through S phase and subsequently, genetic stability. Nuclear remodelling following mitotic chromosome condensation was required for normal cell cycle progression, as a lack thereof delayed DNA replication and induced severely abnormal mitosis. Furthermore, DNA damage, marked by γH2AX, was found primarily in late replicating regions, and was elevated in the absence of BRCA1, indicating replication stress. Therefore, cell-type-specific aspects of DNA replication present a barrier to cellular transitions by affecting genetic stability.
RESULTS
Genomic instability after human somatic cell NT
We have previously derived diploid stem cell lines by somatic cell NT4. While several studies reported the derivation of NT embryonic stem cell (ESC) lines from human somatic cells5,6, developmental arrest remains common. As we had previously reported a cell line containing a chromosomal translocation3, we considered genetic instability as a possible cause for the failure to reprogram.
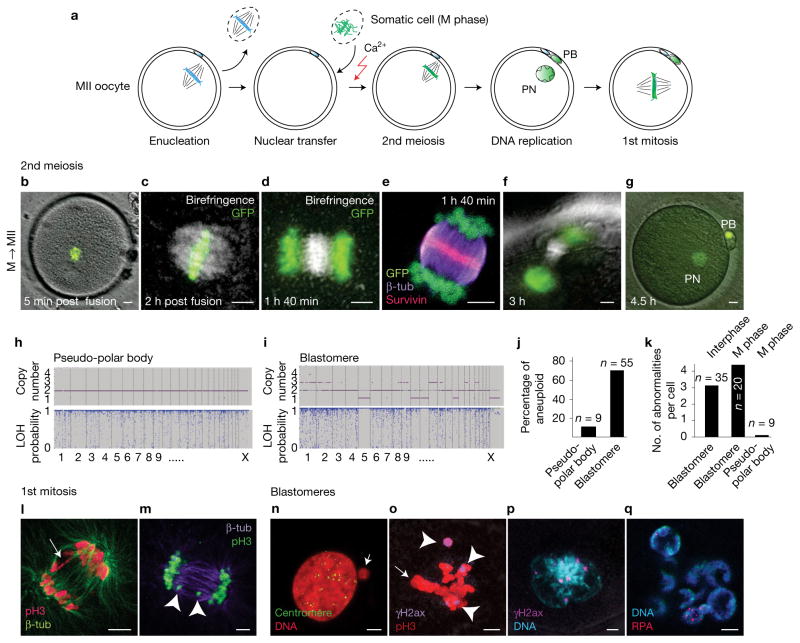
To determine whether and when karyotypic abnormalities occur in human NT, we examined spindle assembly and chromosome segregation immediately after NT, during the second meiosis. We transferred mitotic somatic fibroblast genomes marked with H2B-GFP into enucleated human oocytes and observed the second meiotic division after artificial activation. Somatic cells were arrested in mitosis using nocodazole, to prevent spindle formation prior to transfer (Fig. 1a). Within 1–2 h post-transfer (Fig. 1b), chromosomes were aligned on the metaphase plate of birefringent spindles in 6 of 7 oocytes observed (Fig. 1c). After artificial activation using calcium ionophore and puromycin, 16 of 19 oocytes observed segregated somatic cell chromatin into a polar body and formed a single pronucleus within the activated oocyte (Fig. 1d–g and Supplementary Fig. 1a). Polar bodies (n = 9) were biopsied and analysed using polymorphism arrays, and 8/9 showed a balanced heterozygous genome, indicating that a diploid genome of 46 sister chromatids was segregated into the polar body (Fig. 1h). 1/9 polar bodies contained a single karyotypic abnormality, an error rate comparable to that seen following transfer of an oocyte spindle–chromosome complex7. Spindle assembly also occurred when somatic nuclei at G1 were transferred (Supplementary Fig. 1b). Therefore, after NT, the human oocyte retains the ability to nucleate a spindle and segregate somatic chromatin, allowing a normal second meiosis.
Figure 1.
Genomic instability after human somatic NT. (a) Schematic representation of human NT. Mitotic H2B-GFP-expressing fibroblast genomes were transferred into enucleated MII human oocytes. PB, polar body; PN, pronucleus. (b,c) H2B-GFP fibroblast genome at 5 min (b) and 2 h (c) after NT into MII human oocytes (post fusion). (d–g) Chromosome segregation after oocyte activation using calcium ionophore and puromycin. (h,i) Representative samples of single-nucleotide polymorphism array analysis of copy number and heterozygosity in polar body (h) and NT blastomeres (i). LOH, loss of heterozygosity. (j) Percentage of karyotypically abnormal pseudo-polar bodies (n = 9) and blastomeres (n=55). (k) Average number of abnormalities per blastomere in NT embryos (interphase blastomere, n = 35; M-phase blastomere, n = 20; pseudo-polar body, n = 9). Top label indicates the cell cycle of the transferred genome. (l–q) Immunostaining for DNA damage and genomic instability. (l) Bridge formation at the first anaphase (arrow). (m) Chromosome fragments (arrowheads) at the first anaphase. (n) Centromere-negative micronucleus (arrow) in a blastomere. (o) Immunostaining for γH2AX (arrowheads) indicating DNA damage at mitosis in blastomeres of NT embryos and failure of integration of all chromosomes on metaphase plate (arrow). (p) γH2AX foci at interphase. (q) Multinucleation and replication protein A (RPA) foci in interphase blastomere. Scale bars, 5 μm.
To investigate genetic stability in diploid human NT embryos, we performed NT using either M-phase genomes, or G1 somatic genomes8. Following artificial activation and culture for 3–5 days, we analysed the karyotypes of blastomeres from cleavage stage embryos using a single-nucleotide polymorphism array, and found a large number of abnormalities (Fig. 1i,j and Supplementary Fig. 2). Of 55 total blastomeres (from 11 embryos of 4 different donors), 39 (71%) were abnormal (Fig. 1j). Blastomeres contained multiple abnormalities, and were found in NT embryos reconstituted using M-phase genomes or G1 genomes (Fig. 1k). No completely normal embryos were found. In addition to numerical chromosome abnormalities, 87 of 138 blastomeres (63%) contained micronucleation, more often than in IVF (in vitro fertilization) embryos (Supplementary Fig. 1c).
To identify the origin of these abnormalities, we observed the first mitosis using immunocytochemistry. NT embryos were fixed and stained as they cleaved to the 2-cell stage. On entry into anaphase, we observed bridges, chromosome fragments, incompletely condensed chromatin (Fig. 1l,m and Supplementary Fig. 1d–g), and the segregation of chromosomes into micronuclei (Fig. 1n and Supplementary Fig. 1e,h). Of 10 dividing cells analysed at the first mitosis, all contained at least one of these abnormalities. Abnormal chromosome segregation was also observed at a later stage in blastomeres, with mitotic chromosomes failing to integrate into the metaphase plate (Fig. 1o). These mitotic figures contained γH2AX foci, evidence of DNA damage. In 14/19 embryos of 5 independent experiments, DNA damage was seen in blastomeres at interphase, marked by the presence of γH2AX and replication protein A 32 (RPA) foci (Fig. 1p,q and Supplementary Fig. 1i–r). RPA is a single-strand DNA-binding protein9, suggesting resection of DNA breaks.
Chromosome segregation errors depend on the origin of the donor nucleus
To investigate the mechanisms of chromosome segregation errors and DNA damage, we performed NT in mouse oocytes (Fig. 2a). We first transferred ESC or fibroblast genomes, arrested in mitosis by nocodazole, into MII oocytes and observed spindle assembly and chromosome segregation at anaphase of the second meiosis. The frequency of chromosomal segregation errors at the second meiosis was low in both ESCs and fibroblasts-reconstituted embryos (Fig. 2b,i). The error rate in un-manipulated activated oocytes (parthenotes) was 1.8% (4/227), and 0% (0/18) after transfer of an oocyte genome using Sendai virus. We conclude that replicated somatic cell genomes can be segregated normally in mouse meiotic oocytes after NT.
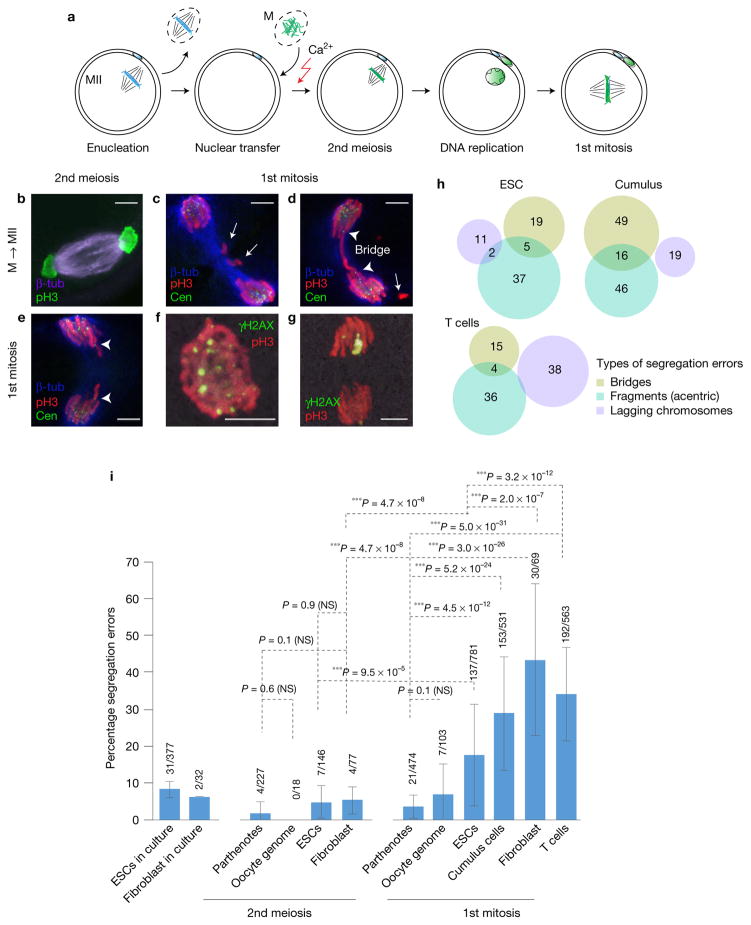
Figure 2.
Segregation errors are pre-mitotic in origin. (a) Schematic of mitotic cell genome transfer. The oocyte genome is removed and replaced with a somatic genome in mitosis. (b) Immunostaining for chromosome segregation at the second meiosis. (c–g) Immunostaining for chromosome segregation at the first mitosis. (c) Absence of centromeres in chromosomal fragments within the spindle (arrows). (d) Arrowheads point to centromeres at both ends of the mitotic bridge and the arrow to a centromere-less chromosomal fragment. (e) Lagging chromosomes containing centromeres (arrowheads). (f,g) Immunostaining for the DNA damage marker γH2AX at the first mitosis, observed in pro-metaphase (f) and anaphase (g). (h) Venn diagram representing types of chromosomal segregation errors in each affected embryo. G1/G0 ESCs, cumulus and naive T cells were used as donors of the nuclear genome. Overlapping regions indicate embryos with both types of defects. (i) Frequency of chromosomal segregation errors in cultured cells, and in parthenotes or NT embryos made with the indicated genome, including the genome of oocytes, ESCs, cumulus cells, naive T cells and fibroblasts. Analysis was performed during the second meiosis or first mitosis after activation. n=377 (ESCs), 32 (fibroblasts), 227 (parthenotes, 2nd meiosis), 18 (oocyte genome NT, 2nd meiosis), 146 (ESC NT, 2nd meiosis), 77 (fibroblast NT, 2nd meiosis), 474 (parthenotes, 1st mitosis), 103 (oocyte genome NT, 1st mitosis), 781 (ESC NT, 1st mitosis), 531 (cumulus cell NT, 1st mitosis), 69 (fibroblast NT, 1st mitosis) and 563 (T-cell NT, 1st mitosis) embryos. The bars represent the mean ± s.d., data are pooled from 3 or more independent experiments. ***P < 0.005; NS, not significant (χ2 test). Scale bars, 5 μm.
To determine whether mitotic abnormalities are dependent on the somatic cell of origin, we transferred different murine donor genomes, obtained from oocytes, ESCs, cumulus cells, fibroblasts or T cells, into mouse oocytes. Somatic NT embryos showed a significant increase in abnormally segregating chromosomes at the first mitosis relative to parthenotes and oocyte genome NT embryos (Fig. 2i). We observed chromosomal defects similar to those of human NT embryos (Fig. 2c–e and Supplementary Fig. 3a). Unlike a previous report10, we did not find differences in mitotic segregation errors between two different drugs used to inhibit actin polymerization during artificial activation, cytochalasin B and latrunculin A (Supplementary Fig. 3b). Next, we compared the frequency of segregation errors using different somatic cells and ESCs. NT embryos reconstituted using donor ESC nuclei showed lower frequencies of mitotic abnormalities compared with embryos reconstituted with somatic nuclei (cumulus cells, T cells and fibroblasts) at the first mitosis (Fig. 2i). Therefore, segregation abnormalities are reprogramming dependent; low with oocyte genomes, intermediate with ESC genomes, and highest with terminally differentiated somatic cell genomes.
Most segregation errors are pre-mitotic in origin
To investigate the mechanisms underlying the mitotic segregation errors, we stained segregating chromosomes for centromeres, tubulin and phospho-H3 (pH3). We observed that all chromosomes (146/146) failing to segregate lacked centromeres (Fig. 2c and Supplementary Fig. 3a). We also observed chromosome bridges that contained a centromere mark pointing towards the spindle pole (Fig. 2d and Supplementary Fig. 3a). Lagging chromosomes were found in pairs, protruding from each side of a group of anaphase chromosomes (Fig. 2e and Supplementary Fig. 3a). The DNA damage marker, γH2AX, was observed on partially condensed pro-metaphase chromosomes (Fig. 2f) and anaphase chromosomes (Fig. 2g), indicating the presence of DNA damage at early mitosis.
Among the different errors, acentric chromosome fragments that did not incorporate into the spindle were most frequent, followed by bridges and lagging chromosomes (Fig. 2h). The presence of acentric chromosomal fragments, accounting for 146/297 (49.2%) of the total defects, as well as bridges (108/297, 36.4%), suggests that the chromosomal aberrations stem primarily from pre-mitotic defects, not from abnormal spindle–chromosome contacts. This led us to investigate whether DNA replication could lead to abnormal chromosome segregation at mitosis.
Chromosome segregation errors are replication, not transcription, dependent
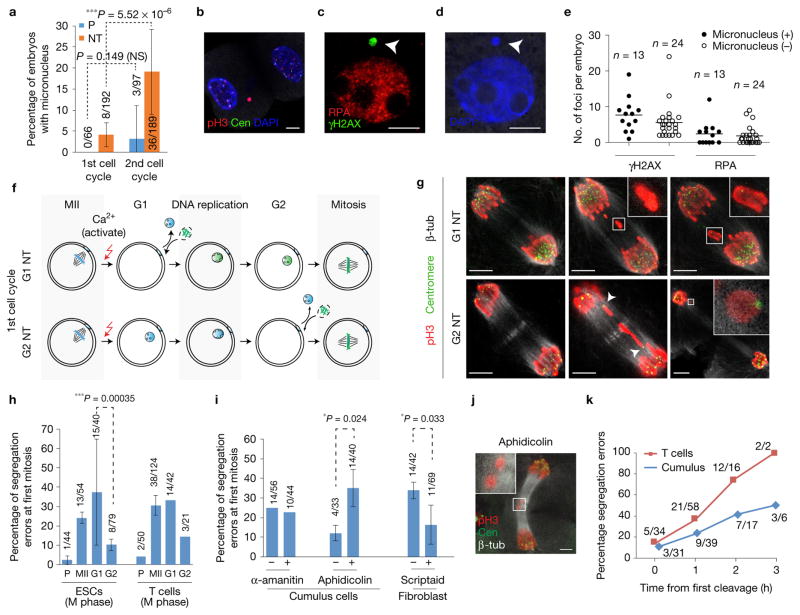
We first explored whether the formation of micronuclei after exit from meiosis could be responsible for chromosome segregation errors at the first mitosis. Micronuclei are known to cause replication-dependent chromosomal segregation errors at mitosis11. Micronuclei in the first cell cycle were rare with only 4.2% (8/192; Fig. 3a), and cannot fully account for the frequent errors (>20–30%) observed at the first mitosis (Fig. 2i). At the second cell cycle, the number of NT embryos with micronucleation was greatly increased (Fig. 3a). Micronuclei in the second cell cycle exhibited persistent pH3 staining, indicating delayed exit from mitosis (Fig. 3b). The lack of centromeres in the micronuclei in the second cell cycle (G2, 28 h postactivation) suggested that the micronuclei arose from chromosomal fragments rather than entire chromosomes (Fig. 3b). Micronuclei were positive for γH2AX and RPA, indicators of DNA damage (Fig. 3c). Notably, embryos with and without micronuclei showed DNA damage in the second cell cycle (Fig. 3e). Therefore, micronucleation is a consequence and not the primary cause for the genetic instability after NT.
Figure 3.
Mitotic segregation defects due to DNA damage arising from progression through DNA replication. (a) Percentage of embryos with micronuclei at the first and second cell cycle. n = 66 (parthenotes (P), 1st cell cycle), 192 (NT, 1st cell cycle), 97 (parthenotes, 2nd cell cycle), 189 (NT, 2nd cell cycle) embryos. The bars represent the mean ± s.d., data are pooled from 3 independent experiments. ***P < 0.005; NS, not significant (χ2 test). (b–d) Micronucleus (arrowheads) with DNA damage in blastomeres at the interphase of the second cell cycle, 28 h post oocyte activation. (e) Quantification of DNA damage in embryos with and without micronuclei at 28 h postactivation. Lines represent the median number of foci per embryo. n=13 (micronucleus (+)) and n=24 (micronucleus (−)) embryos. (f) Schematic of NT in G1 (1 h postactivation) or G2 (13 h postactivation) using mitotic donor cells. G2 NT embryos bypass S phase, undergoing mitosis without DNA replication in the oocyte, in contrast to G1 NT embryos. Oocytes were activated with ionomycin, puromycin, cytochalasin B and 6-DMAP. (g) Representative mitotic segregation defects in G1 and G2 NT embryos. Presence of centromeres in lagging chromosomes in G2 NT embryos (arrowheads and inset), in contrast to centromere-negative fragments in G1 NT embryos (insets). (h) Frequency of mitotic segregation errors in parthenotes (P) and NT embryos where mitotic donor nuclei from ESCs or T cells are transferred into MII, G1 or G2 oocytes. Activation was performed with DMAP. The bars represent the mean ± s.d. from n = 54 (MII oocytes, ESCs), 40 (G1 oocytes, ESCs), 79 (G2 oocytes, ESCs), 124 (MII oocytes, T cells), 42 (G1 oocytes, T cells) and 21 (G2 oocytes, T cells) embryos. ***P < 0.005 (χ2 test). (i) Frequency of segregation errors at the first mitosis in NT embryos after treatment with α-amanitin (RNA polymerase II and III inhibitor), aphidicolin (DNA polymerase inhibitor) and scriptaid (HDAC inhibitor). The bars represent the mean ± s.d. from n=56(−α-amanitin), 44 (+α-amanitin), 33 (−aphidicolin), 40 (+aphidicolin), 42 (−scriptaid) and 69 (+scriptaid) embryos. *P < 0.05; NS, not significant (χ2 test). (j) Representative image of chromosome segregation defects (acentric fragments, inset) at the first mitosis in NT embryos treated with 0.025 μM aphidicolin. (k) Frequency of mitotic segregation defects as a function of timing of the first cleavage (the first cleavage within the cohort of NT embryos is set as time = 0 h). T-cell NT: n = 34 (0 h), 58 (1 h), 16 (2 h) and 2 (3 h); cumulus cell NT: n=31 (0 h), 39 (1 h), 17 (2 h) and 6 (3 h) embryos. Scale bars, 5 μm.
To directly assess whether DNA replication is responsible for mitotic segregation errors, we transferred mitotic donor cells (ESCs and activated T cells) into G1 and G2 oocytes and analysed chromosomal segregation at the first mitosis. Unlike transfer into G1 and MII oocytes, transfer of mitotic nuclei into G2 oocytes bypasses S phase (Fig. 3f). In contrast to the transfer into G1 or MII oocytes, transfer of somatic nuclei into G2 oocytes results in significantly reduced levels of chromosomal segregation errors at the first mitosis (Fig. 3g,h). Some G2 NT embryos exhibited segregation defects, but the nature of these defects was different from G1 NT embryos. While G1 NT embryos showed centromere-less chromosome fragments and bridges, G2 NT embryos showed lagging chromosomes with centromeres and no bridges (Fig. 3g), indicating that the errors occurred only during mitosis. These results directly demonstrate that the formation of chromosome fragments and bridges is dependent on the replication of somatic cell chromatin within the oocyte.
To exclude a role of transcriptional reprogramming in chromosome segregation, we incubated NT embryos in the transcriptional inhibitor α-amanitin. Although the zygotic genome is largely inactive during the first cell cycle12 and transcription is not required for progression to the 2-cell stage13, we considered the possibility that the somatic nucleus remains transcriptionally active after transfer. No changes in segregation error frequency were observed in the presence of α-amanitin (Fig. 3i). Therefore, chromosome segregation errors at the first mitosis are independent of gene expression and transcriptional reprogramming. In contrast, we observed increased segregation errors when aphidicolin, a DNA polymerase inhibitor, was added (Fig. 3i). The predominant type of errors were chromosome fragments (75.0%, Fig. 3j), consistent with observations in somatic cells undergoing DNA replication stress14,15.
Earlier studies had shown that scriptaid and other histone deacetylase inhibitors in mouse and human cells increased reprogramming efficiency and developmental potential4,16–18. When scriptaid was added to the embryo culture medium during the first cell cycle prior to transcription activation, the frequency of segregation errors decreased significantly (Fig. 3i), suggesting a positive effect of scriptaid on DNA replication. This is consistent with previous data showing that HDAC inhibitors can stimulate DNA replication and advance the timing of DNA replication after NT18. To determine whether the timing of cell cycle progression affected the frequency of segregation errors, we quantified segregation errors dependent on the timing of cleavage. While embryos with early cleavage rarely showed segregation errors, embryos with delayed cleavage almost always did (Fig. 3k).
Nuclear remodelling by chromosome condensation is required for normal DNA replication and chromosome segregation
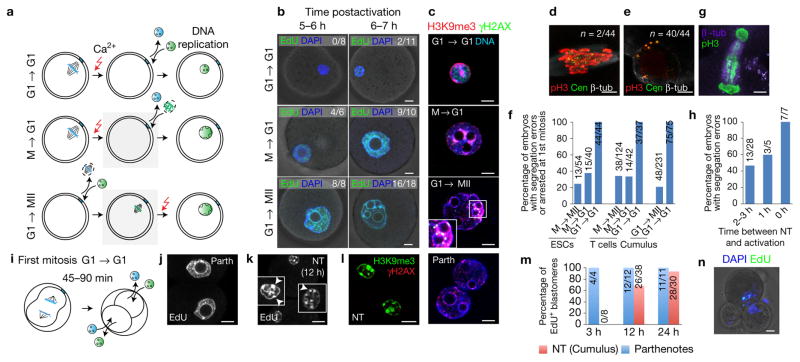
We previously showed that progression through mitosis facilitates transcriptional reprogramming by NT19. To investigate the role of mitotic chromosome condensation in S phase progression after NT, we transferred interphase somatic cell nuclei (naive T cells in G1, Supplementary Fig. 4) immediately prior to enucleation of the oocyte genome at meiotic telophase/early G1 (G1 → G1, Fig. 4a) following oocyte activation. In this group, chromosome condensation did not occur. As controls, NT was also performed either with a mitotic donor genome (activated T cells in M phase) into telophase/G1 oocytes (M → G1) or by the transfer of interphase nuclei into MII oocytes (G1 → MII). In both of these groups, the somatic chromatin underwent chromosome condensation. In the G1 → G1 group, we did not detect morphological changes in the nucleus associated with somatic to embryonic transition (Fig. 4b). In contrast, in both the M → G1 and G1 → MII groups, the somatic genome re-established an embryonic nuclear morphology (Fig. 4b).
Figure 4.
Nuclear remodelling is necessary for normal mitotic segregation. (a) Schematic of interphase transfer. To prevent chromosomal condensation, G0/G1 donor nuclei were transferred into interphase oocytes (G1 → G1, top panel). As controls, mitotic donor nuclei were transferred into G1 oocytes (M → G1, middle panel) and G1 donor nuclei into MII oocytes (G1 → MII, bottom panel), respectively. Grey boxes indicate somatic chromosome condensation. (b) DNA replication in NT embryos incubated with EdU (green) for 1 h at the indicated times after activation. (c) Immunostaining for H3K9me3 distribution and γH2AX in NT embryos at G2 (13 h postactivation). (d) Mitotic chromosome segregation following G1 → G1 NT. (e) Arrested G1 → G1 NT embryo. (f) Quantification of chromosomal segregation errors after transfer of interphase nuclei from ESCs, T cells or cumulus cells at the indicated cell cycle stages. The bars represent the mean from n=54 (M → MII, ESCs), 40 (M → G1, ESCs), 44 (G1 → G1, ESCs), 124 (M → MII, T cells), 42 (M → G1, T cells), 37 (G1 → G1, T cells), 48 (G1 → MII, cumulus) and 75 (G1 → G1, cumulus) number of embryos. (g) Immunostaining at the first mitosis in a cumulus cell NT embryo using oocytes activated immediately after transfer. (h) Percentage of embryos with abnormal chromosomal segregation at the first mitosis when activated at the indicated time points after transfer of naive T cells. n = 28 (2–3 h), 5 (1 h) and 7 (0 h) embryos. (i) Schematic of NT at G1 of the 2-cell stage. (j,k) Replicated DNA (EdU labelling) in blastomeres of control parthenotes (j) and 2-cell NT embryos (k) at 12 h post-transfer. Insets show nuclei at greater magnification and arrowheads point to somatic heterochromatin. (l) Immunostaining for H3K9me3 in 2-cell G1 NT embryos. (m) Percentage of EdU-positive blastomeres in parthenotes and G1 2-cell NT embryos at the indicated time points after the first cleavage. n=4 (parthenotes, 3 h), 8 (NT, 3 h), 12 (parthenotes, 24 h), 38 (NT, 24 h), 11 (parthenotes, 48 h) and 30 (NT, 48 h) blastomeres. (n) Abnormal cleavage of 2-cell NT embryo. (Parth, parthenote). Scale bars, 10 μm.
In the G1 → G1 embryos, we observed a significant delay in the initiation of DNA replication, and the punctate pattern of replication did not resemble that of an embryonic nucleus (Fig. 4b, top panel). When the transferred donor nucleus is in mitosis instead of interphase (M → G1, Fig. 4b, middle panel), DNA replication occurred in an embryonic pattern throughout the nucleus at 6–7 h postactivation. In the absence of chromosome condensation, the somatic heterochromatin pattern, marked by H3K9me3, is retained in all embryos and seen at the periphery of the nucleus (15/15) (Fig. 4c, top panel). M → G1 or G1 → MII NT embryos showed foci of γH2AX particularly in regions within H3K9me3-enriched heterochromatic regions (Fig. 4c, middle panels). At the first mitosis, all dividing embryos in the G1 → G1 NT group showed abnormal chromosome segregation at mitosis, with lagging chromatin between the segregating spindle poles (Fig. 4d). Most of the embryos (40/44) remained arrested at the 1-cell stage (Fig. 4e). In contrast, in the M → MII and M → G1 controls, significantly fewer embryos displayed abnormalities at the first mitosis across donor nuclei of different origins (Fig. 4f). Mitotic segregation defects were also frequent (7/7, 100%) when MII oocytes were activated immediately after transferring the G1 somatic nucleus, thereby reducing the time for nuclear remodelling to occur (Fig. 4g,h).
We next determined whether the requirement for chromosome condensation is limited to the 1-cell stage. A previous report demonstrated that the transfer of interphase somatic nuclei into 2-cell interphase zygotes could result in reprogramming and development to the blastocyst stage20. We transferred the genome from interphase cumulus cells into G1 interphase 2-cell embryos as they exited the first mitosis, at 45 min to 1.5 h after initiation of cleavage (Fig. 4i). We found this to be the earliest possible time point that does not lead to reversion to a 1-cell embryo when enucleated in the presence of cytochalasin B. After NT, embryos were incubated in EdU to assess for replicated DNA at 3 h, 12 h and 24 h post-transfer. At 3 h, all 4 nuclei of the parthenotes had undergone DNA replication (Fig. 4j,m), whereas all 8 nuclei of the 2-cell NT embryos were negative for EdU staining (Fig. 4k,m). DNA replication was initiated with significant delay in the 2-cell NT embryos and was observed only at 12 h (Fig. 4k,m). In contrast to the parthenotes, analysis of nuclear morphology revealed abundance of pericentric heterochromatin foci (Fig. 4k), marked by H3K9me3 (Fig. 4l), which is typical of somatic nuclei (compare with parthenotes in Fig. 4c). Both the size of the nucleus and heterochromatic foci indicate lack of transition from a somatic to an embryonic nuclear structure. While parthenote controls progressed in development, NT embryos failed to develop past the 2-cell stage, with the exception of one 3-cell embryo (Fig. 4n) containing micronucleation, consistent with abnormal mitosis. A possible difference between the two studies is that parthenotes instead of IVF embryos were used.
Therefore, incomplete nuclear remodelling results in delayed DNA replication and abnormal chromosomal segregation at the following mitosis. This principle applies to transfer at the 1-cell or at the 2-cell stage. These results highlight an interdependence between cell-type-specific features of the nuclear structure, timing of DNA replication and genetic stability.
DNA damage following DNA replication in NT embryos
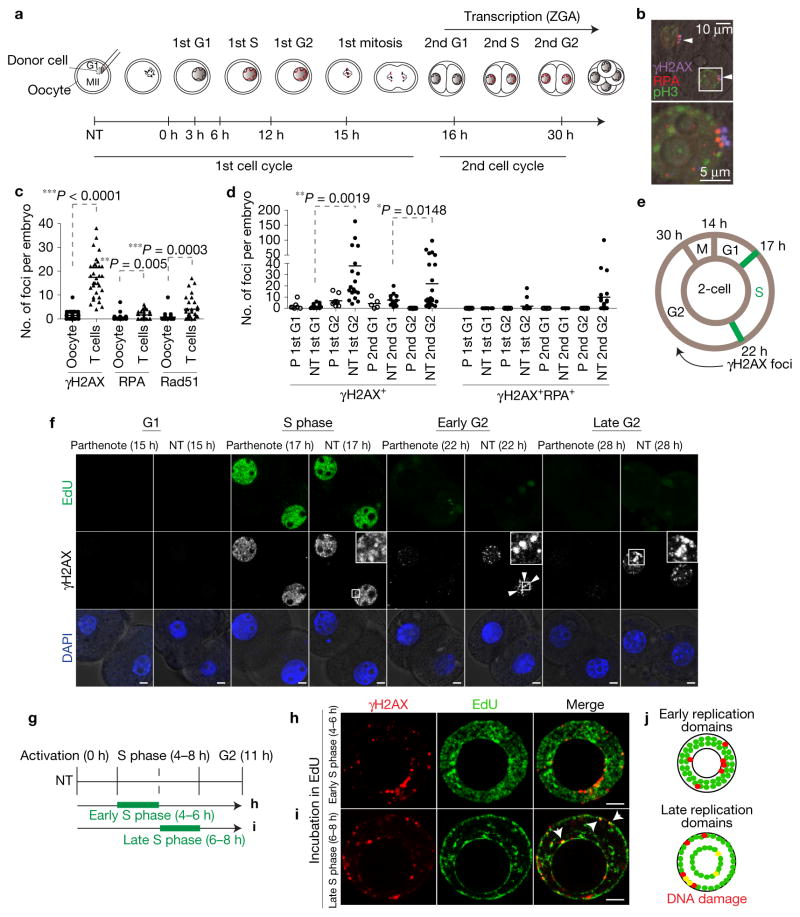
To determine the timing and frequency of DNA damage after NT, we quantified γH2AX and RPA foci at different stages of the cell cycle (Fig. 5a). To distinguish DNA damage due to the reprogramming process from damage caused by the manipulation, we first compared embryos reconstituted using mitotic T cells with embryos reconstituted from G1 oocyte genomes at the second interphase (G2) of the second cell cycle. DNA damage is significantly higher in somatic cell NT embryos (Fig. 5b) than in the embryos reconstituted from oocyte genomes (Fig. 5c), indicating that DNA damage was due to reprogramming.
Figure 5.
DNA damage following progression through DNA replication in the second cell cycle. (a) Schematic representation of the first and second cell cycle after NT. ZGA, zygotic genome activation. (b) DNA damage in interphase nuclei of a NT embryo (G2 of the second cell cycle). The arrowheads point to γH2AX + RPA + DNA damage foci. The two channels are shifted for visibility. Scale bars, 10 μm (top panel), 5 μm (bottom panel). (c) Quantification of DNA damage foci at G2 of the second cell cycle (28 h postactivation) in embryos reconstituted with oocyte nuclei versus somatic nuclei. The lines represent the mean number of foci in n=31 (oocytes as nuclear donor) and n=33 (T cells) embryos. (d) Quantification of γH2AX+ and γH2AX+ RPA+ foci in parthenotes and NT embryos at the indicated time points. The lines represent the mean number of foci in n=7 (P, 1st G1), 25 (NT, 1st G1), 9 (P, 1st G2), 30 (NT, 1st G2), 5 (P, 2nd G1), 13 (NT, 2nd G1), 23 (P, 2nd G2), 24 (NT, 2nd G2) embryos. *P < 0.05; **P < 0.01; ***P < 0.005 (Student’s t-test). (e) Schematic of cell cycle progression and appearance of DNA damage in NT embryos in the second cell cycle. Hours after activation are indicated. (f) Representative images of DNA damage (γH2AX foci, arrowheads) at the indicated time points. Experiments were repeated 3 times for γH2AX and DAPI and once for EdU. Multiple representative images of different embryos were captured per experiment, except for the parthenotes 15 h time point, for which just one image was captured. NT embryos were reconstituted using T cells. (g) Schematic representation of EdU labelling at early or late S phase in the first cell cycle after NT, followed by fixing and immunostaining in G2. (h,i) Staining for EdU and γH2AX, a marker of DNA damage, after incubation with EdU at early (h), or late (i) S phase. Nuclei from cumulus cells were transferred into enucleated MII oocyte. Arrowheads point to DNA damage in late replicating regions. (j) Schematic representation of early and late replicating regions. Co-localization of DNA damage with the replication domain is marked by yellow dots. Scale bars, 5 μm.
We next performed a detailed analysis of the timing of DNA damage. DNA damage was not detectable at the first G1 interphase. In S phase, diffuse distribution of γH2AX was observed in both parthenotes and NT embryos (Fig. 5f). The frequency of both γH2AX and RPA foci was elevated at the end of each S phase in both the first and the second cell cycles (Fig. 5d–f). In contrast, the number of foci was reduced at G1 of the second cell cycle compared with G2 of the first cycle (Fig. 5d), indicating that DNA damage had been resolved. As in the first cell cycle, the number of γH2AX and RPA foci was again increased at G2 of the second cell cycle. Notably, the number of foci decreased immediately before mitosis at 28–30 h postactivation (Supplementary Fig. 5a,b), and following prolonged arrest (42 h postactivation, Supplementary Fig. 5c). These observations suggest that the progression through S phase results in DNA damage that may be partially resolved prior to entry into mitosis.
As embryonic transcription is highly active at the second cell cycle12, we considered a role of transcription in the generation of DNA damage, such as the collision between replication and transcription complexes21, or transcription-coupled DNA damaged repair22. We inhibited transcription with α-amanitin in NT embryos (donor T cells) during S phase (15–22 h), without affecting DNA replication (Supplementary Fig. 5d). Quantification of DNA damage foci revealed a slight increase in the median number of γH2AX foci (Supplementary Fig. 5e,f), but no increase in RPA or Rad51 foci (Supplementary Fig. 5e,g,h) after transcription inhibition. Therefore, transcription is not primarily responsible for the generation of, or the resolution of DNA damage during reprogramming.
We next examined whether DNA damage is related to replication timing and whether it arises in early or late replicating regions. To this end, we labelled NT embryos at early and late S phase using EdU and analysed both groups at G2 (Fig. 5g). We observed that γH2AX foci co-localized primarily, but not exclusively with late replicating regions (Fig. 5h–j). The repair of damaged DNA in G2, and the delay in mitotic entry in cells with genetic instability (Fig. 3k), suggest a role of replication checkpoint proteins in the maintenance of genetic integrity during reprogramming.
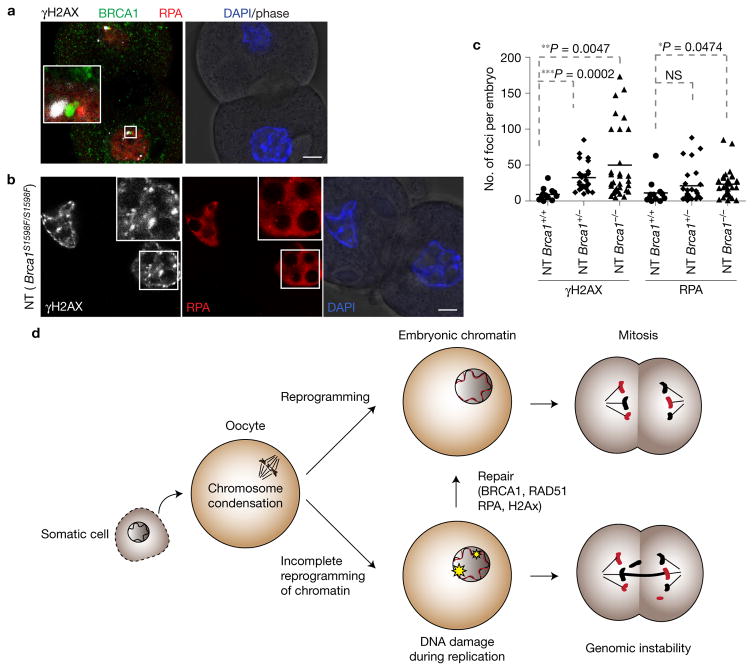
DNA repair after NT is mediated through BRCA1
To assess the involvement of factors implicated in the repair of replication-associated DNA damage, we first examined the co-localization of RAD51, RPA and BRCA1. RPA binds to single-stranded DNA, while RAD51 and BRCA1 are involved in homology-directed repair during DNA replication. We found that RAD51 co-localized with RPA (Supplementary Fig. 5e), and γH2AX foci co-localized with both RPA and BRCA1 (Fig. 6a). To determine whether DNA damage repair after NT is dependent on BRCA1, we transferred Brca1S1598/S1598 cumulus cells into enucleated MII oocytes from Brca1S1598/S1598 mice. This S1598F mutation of BRCA1 disrupts the DNA damage response23. Notably, we observed a significant increase in DNA damage in the BRCA1-deficient NT embryos (Fig. 6b,c). Interestingly, heterozygous Brca1S1598/+ mutant embryos also showed significantly increased numbers of γH2AX foci relative to wild-type embryos (Fig. 6c), suggestive of haploinsufficiency. Therefore, we conclude that DNA damage after NT can be resolved through BRCA1-dependent repair pathways (Fig. 6d).
Figure 6.
DNA damage is repaired by BRCA1. (a) Co-localization of BRCA1, γH2AX and RPA in NT embryos at G2 of the second cell cycle (28 h postactivation). The three channels were shifted relative to each other for visibility. (b) DNA damage (insets) in NT embryos reconstituted from Brca1S1598F/S1598F oocytes. (c) Quantification of DNA damage in Brca1S1598F/S1598F oocyte NT embryos. The lines represent the mean number of foci in n = 13 (Brca1+/+), 26 (Brca1+/−) and 35 (Brca1−/−) number of embryos. *P < 0.05; **P < 0.01; ***P < 0.005; NS, not significant (Student’s t-test). (d) Schematic representation of DNA replication reprogramming and its significance in maintaining genomic stability during reprogramming and potentially other cell fate transitions. Red chromatin represents replicated somatic chromatin after NT. Black chromatin represents parental somatic chromatin. Scale bars, 10 μm.
DISCUSSION
Derivation of pluripotent stem cell lines by NT into human oocytes allows the generation of patient-matched stem cells4,6. Here we show that developmental arrest after human NT is associated with genetic instability, including DNA damage and chromosome segregation errors at mitosis. These errors are developmentally consequential, as had been reported in both frog and mouse24,25. Using mice as a model system, we find that the frequency of DNA damage and chromosome segregation errors is dependent on the developmental origin of the transferred nucleus, with fibroblast and T-cell genomes showing the highest error rate, and oocyte genomes the lowest, while ESC genomes were intermediate. These errors occur during cell-type transition, dependent on DNA replication and independent of transcription. A major difference in the replication of embryonic cells and somatic cells is their temporal pattern of DNA replication26–28. Indeed, DNA damage was found primarily, but not exclusively, in late replicating chromatin and regions marked by H3K9me3. Although the experimental system is not currently amenable to map replication timing, we provide evidence that the reprogramming process results in replication-dependent DNA damage.
The DNA replication profiles of different cell types are closely associated with nuclear organization29. Interestingly, cell-type-specific organization is largely lost as the chromatin condenses during mitosis, and is then re-established in a cell-type-specific manner at interphase30. Our results show that when a somatic cell nucleus is not remodelled through chromosome condensation prior to entry into S phase, an embryonic DNA replication pattern cannot be established, resulting in mitotic chromosome segregation errors. Therefore, chromosomal condensation is required to establish an embryonic pattern of DNA replication and allow cell cycle progression. In frog oocytes, chromosome condensation was shown to mediate the transition between different DNA replication programs31, although the functional and developmental consequences were not examined.
The Waddington epigenetic landscape32 and the barriers to reprogramming are widely interpreted in the context of changes in gene expression1,33–35. However, parallel observations that DNA damage36, p53 activation37,38, and senescence39, as well as the findings that mutations in replication proteins BRCA1 and the Fanconi group proteins, dramatically reduce reprogramming efficiency40,41, collectively point to problems in DNA replication during the transition to induced pluripotent stem cells. Our findings that reprogramming-associated DNA damage is both replication dependent and is increased in BRCA1-deficient cells are consistent with the finding that BRCA1 is involved in the resolution of DNA replication stress42.
Our results demonstrate that an induced transition between different cell types can compromise the integrity of the genome. Therefore, genomic stability during DNA replication is intimately tied to cellular identity. On the basis of our studies, we propose that cell-type-specific aspects of DNA replication and nuclear structure provide a mechanism to restrict cell proliferation to defined cellular states43,44. We further posit that tumour suppressors such as BRCA1 or p53 not only safeguard genomic stability, but are also ‘gatekeepers of cell identity’ by sensing replication-dependent DNA damage and in response to the damage, suppress the proliferation of abnormal cells. In contrast, oncogenes with a prominent role in DNA replication, such as c-myc45, enable cell transformation by reducing the constraints of a cell-type-specific DNA replication program. Exploring this hypothesis will require further investigation.
METHODS
Human oocytes manipulations
Human oocytes were donated by women enrolled in the oocyte donation programme at Columbia University after providing informed consent. All human subjects research was reviewed and approved by the Columbia University Medical Center Institutional Review Board and stem cell committees. Human somatic cell NT was performed as described previously4. In brief, human oocytes were enucleated in G-MOPS (Vitrolife) plus containing 5 μg ml−1 cytochalasin B (Sigma Aldrich). A somatic cell was incubated in diluted Sendai virus (GenomeOne, Cosmo bio) for 5–10 s and then inserted below the zona pellucida of the enucleated oocyte. After 1–3 h post manipulation, artificial activation was performed using either a calcium ionophore or an electrical pulse, followed by incubation in puromycin (ThermoFisher Scientific) and cytochalasin B for 4h. Oocytes were then cultured in Global total (IVFonline) in an incubator at 37°C, 5% CO2 and cultured for 20–30 h for analysis of the first mitosis, or for 3–5 days until analysis of blastomeres.
Karyotyping and cell-line analysis
Karyotyping of blastomeres and cell lines was performed by Reproductive Medicine Associates of New Jersey as previously described20. Dissociated blastomeres were placed into tubes containing 1 μl KOH solution and shipped on dry ice for analysis. Whole-genome amplification and 262K NspI Affymetrix single-nucleotide polymorphism microarray analysis was performed as previously described46.
Cell culture
Mouse embryonic stem cells (ESCs) were maintained and passaged in Dulbecco’s modified Eagle’s medium (DMEM) with 4,500mg l−1 glucose supplemented with GlutaMAX (ThermoFisher Scientific), 15% fetal bovine serum (FBS), non-essential amino acid (NEAA, ThermoFisher Scientific), recombinant mouse LIF (EBioscience), β-mercaptoethanol (Sigma Aldrich) and cultured on inactivated mouse embryonic fibroblasts (MEFs, GlobalStem). Cultured cells were tested for mycoplasma with MycoAlert (Lonza) and were found mycoplasma free. Human fibroblasts were cultured in DMEM supplemented with GlutaMAX, 15% FBS, NEAA and β-mercaptoethanol. Cell lines used were previously reported and authenticated3,4. No cell lines used in this study were found in the database of commonly misidentified cell lines that is maintained by ICLAC and NCBI Biosample. To induce G0/G1 arrest, fibroblasts were cultured in MEF medium lacking FBS for 24 h. G1 ESCs were isolated by flow cytometry using the FUCCI reporters47. Mitotic ESCs and fibroblasts were obtained as previously described20. Briefly, cells were cultured in medium containing 0.1 μg ml−1 nocodazole (Sigma Aldrich) for 3 h. Mitotic cells were then collected by washing with PBS, brief centrifugation, and manually selected under the microscope for NT.
Isolation of naive T cells
Six- to eight-week-old BDF1 (C57BL/6xDBA F1) male mice were purchased from Jackson Laboratories. Spleen and lymph nodes were harvested and mechanically disrupted to obtain a single-cell suspension. After filtering through a 70 μm cell strainer (ThermoFisher Scientific), red blood cells were lysed for 3 min at room temperature using multispecies RBC lysis buffer (eBiosciences). Cells were stained with CD8 (53-6.7)-FITC, CD4 (RM4-5)-APC-Alexa788, CD3e (145-2C11)-PerCPCy5.5, CD62L (MEL-14)-APC, CD44 (IM7)-eFluor450, MHC class II (I-A/I-E)-eFluor450 and CD69 (H1.2F3)-PE (all from eBiosciences) in 2% BSA (ThermoFisher) 2 mM EDTA (ThermoFisher) PBS (Life Technologies). Anti-mouse CD16/CD32 (93) in 2% BSA 2 mM EDTA PBS was used prior to stain for blocking. Naive CD8+ T cells were gated as MHCII− CD3e+ CD8a+ CD4− CD62L+ CD44− and CD69−; naive CD4 T cells were gated as MHCII- CD3e+ CD4+ CD8a− CD62L+ CD44− and CD69−. They were flow cytometry-sorted with a purity > 99% by using an Aria 4LS (BD) and collected in 10%-FBS-supplemented DMEM (all from Life Technologies). Naive T cells were cultured in RPMI 1640 media supplemented with 10% FBS, GlutaMAX and 30 U ml−1 of recombinant mouse IL-2 (RnD). Naive T cells were activated by incubation CD3/CD28 Dynabeads (Thermofisher Scientific) according to the manufacturer’s instructions.
Mouse NT
Six- to eight-week-old BDF1 female mice were obtained from Charles River Laboratories and were superovulated as previously described48. Five female mice were used per experiment and a total of 1,440 mice were used in this study. Mouse oocytes were retrieved from the dissected oviducts at 13 h post hCG injection and incubated in EmbryoMax KSOM (Emdmillipore) at 37°C, 5% CO2 until further use. Cumulus cells were removed by incubating the cumulus–oocyte complex in hyaluronidase (0.1% w/v, Sigma) at 37°C for approximately 5 min. Dissociated cumulus cells were collected, spun briefly and incubated in G-MOPS plus medium and kept on ice for NT. Mouse oocytes were enucleated in G-MOPS plus medium containing 5 μg ml−1 cytochalasin B, on a stage kept at 32°C. The inner diameter of transfer needles for cumulus cells was 5–6 μm, for fibroblasts 10 μm, for ESCs 8–10 μm, and for T cells 5 μm. The somatic cell was given 1–2 piezo pulses (intensity 1 or 2) while at the tip of the transfer needle to break the plasma membrane. The somatic cell was then injected without delay into an enucleated oocyte, in G-MOPS plus medium containing 5 μg ml−1 cytochalasin B. The plasma membrane of the oocyte was broken with a single piezo pulse (Prime Tech, Japan), the cell injected and the membrane sealed by aspiration of a small amount of cytoplasm. Oocytes were activated using a strontium chloride in calcium-free MCZB, in the presence of 5 μg ml−1 cytochalasin B for interphase nuclei, and no cytochalasin B for M-phase nuclei, to allow the extrusion of a pseudo-polar body. Alternatively, oocytes were activated with a 5 min pulse of 3 μM ionomycin (Sigma Aldrich) followed by 3 h incubation at 37°C in the presence of 2 mM 6-DMAP (Sigma Aldrich), 10 μM puromycin, and 5 μg ml−1 cytochalasin B. Different activation protocols were tested on the different types of embryos/eggs (see statistics source data). Oocytes were cultured during and after activation in EmbryoMax KSOM or Global Total (Life Global). NT of somatic cells or ESCs into activated G1 oocytes was done at 1–3 h after activating oocytes with ionomycin, 6-DMAP, puromycin and cytochalasin B. For G1 transfer, enucleation was performed in the presence of 5 μg ml−1 cytochalasin B, 0.1 μg ml−1 nocodazole and 10 μM puromycin. For G2 transfer, enucleation and NT was performed between 11–13 h after activation, in the presence of nocodazole and cytochalasin B, without puromycin. Approximately 13–16 h postactivation, embryos were monitored regularly and fixed in 4% paraformaldehyde (Santa Cruz Biotechnology) when they assumed an elongated shape (mitosis), rather than a round shape. To inhibit transcription or DNA replication, NT embryos were treated with 5 μg ml−1 α-amantin (Sigma Aldrich) or 0.025 μM aphidicolin (Sigma Aldrich), respectively. To inhibit histone deacetylase, scriptaid (Sigma Aldrich) was added to the embryo culture at a concentration of 250 nM. For 2-cell interphase transfer, oocytes were activated with ionomycin, 6-DMAP, puromycin and cytochalasin B. Enucleation was performed at 45 min to 1.5 h after the first mitotic cleavage, in G-MOPS plus medium containing 0.1 μg ml−1 nocodazole and 5 μg ml−1 cytochalasin B. Oocytes were incubated for half an hour in EmbryoMax KSOM at 37°C to allow recovery, followed by transfer of cumulus cells (majority in G1) into oocytes, in the presence of 5 μg ml−1 cytochalasin B, at 32°C. Animal experiments were approved by the Institutional Animal Care and Use Committee of Columbia University.
Immunocytochemistry
Oocytes and NT embryos were fixed with 4% paraformaldehyde, washed in phosphate-buffered saline (PBS), permeabilized in 0.25% Triton X-100 (Sigma Aldrich), blocked in PBS containing 2% donkey serum (Sigma Aldrich) and 0.01% Tween 20 (Sigma Aldrich), and stained and analysed using the following antibodies recognizing survivin (Cell Signaling Technologies 2808), beta tubulin (Millipore 05-661, clone AA2, 1:1,000), Cenp-A (Cell Signaling Technologies 2186 and 2048), phosho-γH2AX (Millipore 05-636, clone JBW301, 1:1,000), anti-centromere (15-235-0001 Antibodies Inc., 1:50), phospho-histone H3 Ser10 (Millipore 06-570, 1:1,000), phospho-histone H3 Ser28 (Millipore MABE76, 1:1,000), RPA32 (Cell Signaling Technologies 2208, 1:200), Rad51 (EMD Millipore PC130, 1:1,000), H3K9me3 (Abcam ab8898, 1:1,000) and BRCA1 (ref. 23) (1:5,000). Primary antibodies were diluted in blocking buffer and oocytes/embryos were incubated at 4°C overnight. They were washed thrice in blocking buffer the following day, and incubated in the appropriate secondary antibodies (donkey Alexa488/555/647-conjugated antibodies, Thermofisher Scientific), diluted at 1 in 1,000 in blocking buffer, for at least 1 h at room temperature. The embryos were then washed thrice in blocking buffer and imaged using a Zeiss LSM710 confocal microscope. Nascent DNA synthesis is labelled with Click-iT EdU Alexa Fluor 488 Imaging Kit (Life Technologies C10337) and RNA is labelled with Click-iT Nascent RNA Capture Kit (Life Technologies C10365) according to the manufacturer’s instructions. Labelled embryos were then washed in PBS, blocked and incubated in the appropriate primary antibodies. Embryos were subsequently processed as described above.
Statistical analysis and reproducibility
No statistical method was used to predetermine sample size and the experiments were not randomized. Investigators were not blinded to allocation during experiments and outcome assessment. Statistics for data was calculated using the Chi square (χ2) function in Microsoft Excel. GraphPad Prism 5.0 software was used to calculate statistical significance for Student’s t-test for relevant figures, as specified in the figure legends. All data with error bars consist of at least 2 independent NT experiments, and commonly 3 or more. Figures with representative images were repeated at least twice, independently. Multiple embryos were stained and for all embryos all stains were performed simultaneously. All embryos were examined under the microscope and used for quantification. Representative embryos were imaged for all stains and images of the same embryo were used for display in the figures. Source data for figures with representative experiments of multiple repeats can be found in the statistics source data (Supplementary Table 1).
Data availability
Microarray data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under the accession code GSE93123. Source data for Figs 2i, 3a,h, 4f and 5c,f–h have been provided in Supplementary Table 1.
All other data supporting the findings of the study are available from the corresponding author on request.
Supplementary Material
Supplementary Figure 1 Genetic instability in human nuclear transfer embryos. a) Normal chromosome segregation at the second meiotic division upon transfer of a somatic cell genome. b–d) Anaphase at the first mitosis. b) The arrow points to a chromosome fragment. c) The arrows point the direction of chromosome movement. Note the splitup of chromosomes into different groups, and a chromosome fragment (arrowhead). n=142 (IVF), 138 (NT), 111 (NT, caffeine pulse) embryos. P=0.0000 (Student’s t-test). d) lagging chromosomes occurring in pairs (arrows). e) A single confocal section of the cell shown in (d). Note the indentation (arrowhead) in the anaphase chromosomes, likely as a result of strain from a resolving chromosome bridge (arrow). f) Formation of nuclei at telophase of the first mitosis. Note the formation of multiple nuclei. g) 6-cell NT embryo with four blastomeres (one not visible) in mitosis on day 4 after transfer. Arrows point to the metaphase plate, arrowheads to chromosomes that fail to integrate. h) Blastomere arrested in mitosis. Abnormal chromosome condensation prevents the formation of a metaphase plate. DNA damage is evident from phosphorylated γH2AX staining. The three channels were shifted relative to each other for visibility. i, j) Examples of multinucleation in blastomeres. pH3 indicates phosphorylation on serine 10 of histone H3. k) Another example of an embryo arrested with DNA damage, at entry into mitosis. l, m) RPA foci in an interphase nucleus, and centromere staining. n–p) Examples of RPA and phosphorylated γH2AX in NT blastomeres. Scale bar: 10μm.
Supplementary Figure 2 SNP array analysis of copy number and heterozygosity in NT blastomeres. Each of the four clusters is derived from a different embryo. Only cells with successful biopsy and array analysis are shown.
Supplementary Figure 3 Different types of mitotic chromosomal defects in nuclear transfer embryos. a) Immunostaining of NT embryos at the first mitosis reconstituted using different donor cell types (ES cells, T-cells and cumulus cells). b) Percentage of embryos (parthenotes and NT embryos), activated with either latrunculin A or cytochalasin B, with chromosomal segregation defects at the first mitosis. n= 64 (Parthenotes, Latrunculin A), 202 (Parthenotes, Cytochalasin B), 170 (NT, Latrunculin A) and 129 (NT, Cytochalasin B) embryos. Scale bar: 10μm.
Supplementary Figure 4 Cell cycle of somatic donor cells. Cell cycle profile by flow cytometry of naïve T-cells used for transfer. Note that most cells are in G1 of the cell cycle.
Supplementary Figure 5 DNA damage is reduced in G2, following exit from S-phase, and is independent of transcription. a) NT embryos and parthenotes were incubated in EdU for 1h at the following time points (17h, 22h and 28h postactivation). Bars represent the mean percentage of NT embryos that are undergoing DNA replication (EdU positive) at the respective time points. n=12 (P, 17h), 13 (NT, 17h), 12 (P, 22h), 12 (NT, 22h), 5 (P, 28h) and 12 (NT, 28h) embryos.] b) DNA damage (no. of γH2AX) in NT embryos and parthenotes at the respective time points in (a). Line represents mean number of foci in n=12 (P, 17h), 13 (NT, 17h), 12 (P, 22h), 12 (NT, 22h), 5 (P, 28h) and 12 (NT, 28h) number of embryos. **p<0.01; ns=not significant (Student’s t-test). c) Quantification of γH2AX, RPA and Rad51 foci at 30h versus 42h after nuclear transfer and activation. Lines represent the median no. of foci in n=23 (untreated, γH2AX) and 25 (α-amanitin, γH2AX), n=56 (untreated, RPA), 69 (α-amanitin, RPA), 34 (untreated, Rad51) and 42 ((α-amanitin, Rad51) number of embryos. Arrowhead indicates outlier with significant DNA damage. d) DNA replication (EdU incorporation, top panel) and nascent RNA transcription (5-ethynyluridine, EU incorporation, bottom panel) in untreated and α-amanitin treated NT embryos. Arrowheads point to EU labelled foci indicating high transcriptional activity. e) RPA, Rad51 and γH2AX staining in untreated and α-amanitin treated NT embryos. f–h) Quantification of γH2AX, RPA and Rad51 foci in untreated and α-amanitin treated embryos. Line represents median number of foci. n= 23 (γH2AX, untreated), 25 (γH2AX, α-amanitin treated), 56 (RPA32, untreated), 69 (RPA32, α-amanitin treated), 34 (Rad51, untreated) and 42 (Rad5, α-amanitin treated) embryos. Scale bar: 5μm and 10μm (inserts).
Supplementary Table 1 Source data for figures with representative experiments with multiple repeats, Source data for Fig. 2i, 3a, 3h, 4f, 5c, and 5f–h, have been provided in Supplementary Table 1.
Acknowledgments
This research was supported by the New York Stem Cell Foundation (NYSCF) and a New York State Stem Cell Science (NYSTEM) IIRP Award no. C026184 and the John M. Driscoll, Jr, M. D. Children’s Fund. D.E. is a NYSCF-Robertson Investigator. We thank W. Kearns (Shady Grove Center for Preimplantation Genetic Diagnosis) for initial help with karyotyping. G.C. is supported by the Agency for Science, Technology and Research (A*STAR) International Fellowship. We thank A. Ciccia and W.-W. Tee for critical comments on the manuscript.
Footnotes
AUTHOR CONTRIBUTIONS
G.C. and D.E. designed and performed the experiments, analysed the data and wrote the manuscript with input from all authors. D.E. conceived and supervised the studies and performed NT. J.A. and B.D.B. provided T cells. N.T. performed karyotype analysis of blastomeres. D.B. and R.B. provided Brca1 mutant mice and helped with data interpretation and manuscript writing. M.V.S. was involved in all aspects of oocyte donation, including consent and oocyte retrieval.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Ladewig J, Koch P, Brustle O. Leveling Waddington: the emergence of direct programming and the loss of cell fate hierarchies. Nat Rev Mol Cell Biol. 2013;14:225–236. doi: 10.1038/nrm3543. [DOI] [PubMed] [Google Scholar]
- 2.Mikkelsen TS, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noggle S, et al. Human oocytes reprogram somatic cells to a pluripotent state. Nature. 2011;478:70–75. doi: 10.1038/nature10397. [DOI] [PubMed] [Google Scholar]
- 4.Yamada M, et al. Human oocytes reprogram adult somatic nuclei of a type 1 diabetic to diploid pluripotent stem cells. Nature. 2014;510:533–536. doi: 10.1038/nature13287. [DOI] [PubMed] [Google Scholar]
- 5.Chung YG, et al. Human somatic cell nuclear transfer using adult cells. Cell Stem Cell. 2014;14:777–780. doi: 10.1016/j.stem.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 6.Ma H, et al. Metabolic rescue in pluripotent cells from patients with mtDNA disease. Nature. 2015;524:234–238. doi: 10.1038/nature14546. [DOI] [PubMed] [Google Scholar]
- 7.Paull D, et al. Nuclear genome transfer in human oocytes eliminates mitochondrial DNA variants. Nature. 2013;493:632–637. doi: 10.1038/nature11800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- 9.Fairman MP, Stillman B. Cellular factors required for multiple stages of SV40 DNA replication in vitro. EMBO J. 1988;7:1211–1218. doi: 10.1002/j.1460-2075.1988.tb02933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Terashita Y, et al. Latrunculin a treatment prevents abnormal chromosome segregation for successful development of cloned embryos. PLoS ONE. 2013;8:e78380. doi: 10.1371/journal.pone.0078380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crasta K, et al. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flach G, Johnson MH, Braude PR, Taylor RA, Bolton VN. The transition from maternal to embryonic control in the 2-cell mouse embryo. EMBO J. 1982;1:681–686. doi: 10.1002/j.1460-2075.1982.tb01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rambhatla L, Latham KE. Strain-specific progression of α-amanitin-treated mouse embryos beyond the two-cell stage. Mol Reprod Dev. 1995;41:16–19. doi: 10.1002/mrd.1080410104. [DOI] [PubMed] [Google Scholar]
- 14.Burrell RA, et al. Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013;494:492–496. doi: 10.1038/nature11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol. 2009;11:753–760. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- 16.Kishigami S, et al. Significant improvement of mouse cloning technique by treatment with trichostatin A after somatic nuclear transfer. Biochem Biophys Res Commun. 2006;340:183–189. doi: 10.1016/j.bbrc.2005.11.164. [DOI] [PubMed] [Google Scholar]
- 17.Rybouchkin A, Kato Y, Tsunoda Y. Role of histone acetylation in reprogramming of somatic nuclei following nuclear transfer. Biol Reprod. 2006;74:1083–1089. doi: 10.1095/biolreprod.105.047456. [DOI] [PubMed] [Google Scholar]
- 18.Bui HT, et al. Effect of trichostatin A on chromatin remodeling, histone modifications, DNA replication, and transcriptional activity in cloned mouse embryos. Biol Reprod. 2010;83:454–463. doi: 10.1095/biolreprod.109.083337. [DOI] [PubMed] [Google Scholar]
- 19.Egli D, Rosains J, Birkhoff G, Eggan K. Developmental reprogramming after chromosome transfer into mitotic mouse zygotes. Nature. 2007;447:679–685. doi: 10.1038/nature05879. [DOI] [PubMed] [Google Scholar]
- 20.Kang E, et al. Nuclear reprogramming by interphase cytoplasm of two-cell mouse embryos. Nature. 2014;509:101–104. doi: 10.1038/nature13134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helmrich A, Ballarino M, Tora L. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol Cell. 2011;44:966–977. doi: 10.1016/j.molcel.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 22.Mellon I, Hanawalt PC. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989;342:95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- 23.Shakya R, et al. BRCA1 tumor suppression depends on BRCT phosphoprotein binding, but not its E3 ligase activity. Science. 2011;334:525–528. doi: 10.1126/science.1209909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Berardino MA, King TJ. Development and cellular differentiation of neural nuclear-transplants of known karyotype. Dev Biol. 1967;15:102–128. doi: 10.1016/0012-1606(67)90009-7. [DOI] [PubMed] [Google Scholar]
- 25.Mizutani E, et al. Abnormal chromosome segregation at early cleavage is a major cause of the full-term developmental failure of mouse clones. Dev Biol. 2012;364:56–65. doi: 10.1016/j.ydbio.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 26.Ryba T, et al. Replication timing: a fingerprint for cell identity and pluripotency. PLoS Comput Biol. 2011;7:e1002225. doi: 10.1371/journal.pcbi.1002225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shufaro Y, et al. Reprogramming of DNA replication timing. Stem Cells. 2010;28:443–449. doi: 10.1002/stem.303. [DOI] [PubMed] [Google Scholar]
- 28.Hiratani I, et al. Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res. 2010;20:155–169. doi: 10.1101/gr.099796.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pope BD, et al. Topologically associating domains are stable units of replication-timing regulation. Nature. 2014;515:402–405. doi: 10.1038/nature13986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naumova N, et al. Organization of the mitotic chromosome. Science. 2013;342:948–953. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemaitre JM, Danis E, Pasero P, Vassetzky Y, Mechali M. Mitotic remodeling of the replicon and chromosome structure. Cell. 2005;123:787–801. doi: 10.1016/j.cell.2005.08.045. [DOI] [PubMed] [Google Scholar]
- 32.Waddington CH. The epigenotype. Int J Epidemiol. 2012;41:10–13. doi: 10.1093/ije/dyr184. [DOI] [PubMed] [Google Scholar]
- 33.Bortvin A, et al. Incomplete reactivation of Oct4-related genes in mouse embryos cloned from somatic nuclei. Development. 2003;130:1673–1680. doi: 10.1242/dev.00366. [DOI] [PubMed] [Google Scholar]
- 34.Buganim Y, et al. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Polo JM, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marion RM, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hong H, et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawamura T, et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Utikal J, et al. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gonzalez F, et al. Homologous recombination DNA repair genes play a critical role in reprogramming to a pluripotent state. Cell Rep. 2013;3:651–660. doi: 10.1016/j.celrep.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muller LU, et al. Overcoming reprogramming resistance of Fanconi anemia cells. Blood. 2012;119:5449–5457. doi: 10.1182/blood-2012-02-408674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pathania S, et al. BRCA1 haploinsufficiency for replication stress suppression in primary cells. Nat Commun. 2014;5:5496. doi: 10.1038/ncomms6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Egli D, Le Bin GC. Tying replication to cell identity. Nat Rev Mol Cell Biol. 2013;14:326. doi: 10.1038/nrm3593. [DOI] [PubMed] [Google Scholar]
- 44.Chia G, Egli D. Connecting the cell cycle with cellular identity. Cell Reprogram. 2013;15:356–366. doi: 10.1089/cell.2013.0041. [DOI] [PubMed] [Google Scholar]
- 45.Dominguez-Sola D, et al. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448:445–451. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- 46.Treff NR, Su J, Tao X, Levy B, Scott RT. Jr Accurate single cell 24 chromosome aneuploidy screening using whole genome amplification and single nucleotide polymorphism microarrays. Fertil Steril. 2010;94:2017–2021. doi: 10.1016/j.fertnstert.2010.01.052. [DOI] [PubMed] [Google Scholar]
- 47.Sakaue-Sawano A, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 48.Hogan B, Costantini F, Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. Vol. 500. Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 Genetic instability in human nuclear transfer embryos. a) Normal chromosome segregation at the second meiotic division upon transfer of a somatic cell genome. b–d) Anaphase at the first mitosis. b) The arrow points to a chromosome fragment. c) The arrows point the direction of chromosome movement. Note the splitup of chromosomes into different groups, and a chromosome fragment (arrowhead). n=142 (IVF), 138 (NT), 111 (NT, caffeine pulse) embryos. P=0.0000 (Student’s t-test). d) lagging chromosomes occurring in pairs (arrows). e) A single confocal section of the cell shown in (d). Note the indentation (arrowhead) in the anaphase chromosomes, likely as a result of strain from a resolving chromosome bridge (arrow). f) Formation of nuclei at telophase of the first mitosis. Note the formation of multiple nuclei. g) 6-cell NT embryo with four blastomeres (one not visible) in mitosis on day 4 after transfer. Arrows point to the metaphase plate, arrowheads to chromosomes that fail to integrate. h) Blastomere arrested in mitosis. Abnormal chromosome condensation prevents the formation of a metaphase plate. DNA damage is evident from phosphorylated γH2AX staining. The three channels were shifted relative to each other for visibility. i, j) Examples of multinucleation in blastomeres. pH3 indicates phosphorylation on serine 10 of histone H3. k) Another example of an embryo arrested with DNA damage, at entry into mitosis. l, m) RPA foci in an interphase nucleus, and centromere staining. n–p) Examples of RPA and phosphorylated γH2AX in NT blastomeres. Scale bar: 10μm.
Supplementary Figure 2 SNP array analysis of copy number and heterozygosity in NT blastomeres. Each of the four clusters is derived from a different embryo. Only cells with successful biopsy and array analysis are shown.
Supplementary Figure 3 Different types of mitotic chromosomal defects in nuclear transfer embryos. a) Immunostaining of NT embryos at the first mitosis reconstituted using different donor cell types (ES cells, T-cells and cumulus cells). b) Percentage of embryos (parthenotes and NT embryos), activated with either latrunculin A or cytochalasin B, with chromosomal segregation defects at the first mitosis. n= 64 (Parthenotes, Latrunculin A), 202 (Parthenotes, Cytochalasin B), 170 (NT, Latrunculin A) and 129 (NT, Cytochalasin B) embryos. Scale bar: 10μm.
Supplementary Figure 4 Cell cycle of somatic donor cells. Cell cycle profile by flow cytometry of naïve T-cells used for transfer. Note that most cells are in G1 of the cell cycle.
Supplementary Figure 5 DNA damage is reduced in G2, following exit from S-phase, and is independent of transcription. a) NT embryos and parthenotes were incubated in EdU for 1h at the following time points (17h, 22h and 28h postactivation). Bars represent the mean percentage of NT embryos that are undergoing DNA replication (EdU positive) at the respective time points. n=12 (P, 17h), 13 (NT, 17h), 12 (P, 22h), 12 (NT, 22h), 5 (P, 28h) and 12 (NT, 28h) embryos.] b) DNA damage (no. of γH2AX) in NT embryos and parthenotes at the respective time points in (a). Line represents mean number of foci in n=12 (P, 17h), 13 (NT, 17h), 12 (P, 22h), 12 (NT, 22h), 5 (P, 28h) and 12 (NT, 28h) number of embryos. **p<0.01; ns=not significant (Student’s t-test). c) Quantification of γH2AX, RPA and Rad51 foci at 30h versus 42h after nuclear transfer and activation. Lines represent the median no. of foci in n=23 (untreated, γH2AX) and 25 (α-amanitin, γH2AX), n=56 (untreated, RPA), 69 (α-amanitin, RPA), 34 (untreated, Rad51) and 42 ((α-amanitin, Rad51) number of embryos. Arrowhead indicates outlier with significant DNA damage. d) DNA replication (EdU incorporation, top panel) and nascent RNA transcription (5-ethynyluridine, EU incorporation, bottom panel) in untreated and α-amanitin treated NT embryos. Arrowheads point to EU labelled foci indicating high transcriptional activity. e) RPA, Rad51 and γH2AX staining in untreated and α-amanitin treated NT embryos. f–h) Quantification of γH2AX, RPA and Rad51 foci in untreated and α-amanitin treated embryos. Line represents median number of foci. n= 23 (γH2AX, untreated), 25 (γH2AX, α-amanitin treated), 56 (RPA32, untreated), 69 (RPA32, α-amanitin treated), 34 (Rad51, untreated) and 42 (Rad5, α-amanitin treated) embryos. Scale bar: 5μm and 10μm (inserts).
Supplementary Table 1 Source data for figures with representative experiments with multiple repeats, Source data for Fig. 2i, 3a, 3h, 4f, 5c, and 5f–h, have been provided in Supplementary Table 1.
Data Availability Statement
Microarray data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under the accession code GSE93123. Source data for Figs 2i, 3a,h, 4f and 5c,f–h have been provided in Supplementary Table 1.
All other data supporting the findings of the study are available from the corresponding author on request.