Abstract
Near-IR (NIR) emitting semiconducting polymer dots (Pdots) with ultrabright fluorescence have been prepared for specific cellular targeting. A series of π-conjugated polymers were synthesized to form water dispersible multicomponent Pdots by an ultrasonication-assisted co-precipitation method. By optimizing cascade energy transfer in Pdots, high-intensity NIR fluorescence (φ = 0.32) with tunable excitations, large absorption–emission separation (up to 330 nm), and narrow emission bands (FWHM = 44 nm) have been achieved. Single-particle fluorescence imaging show that the as-prepared NIR Pdots were more than three times brighter than the commercially available Qdot705 with comparable sizes under identical conditions of excitation and detection. Because of the covalent introduction of carboxylic acid groups into polymer side chains, the bioconjugation between NIR-emitting Pdots and streptavidins can be readily completed via these functional groups on the surface of Pdots. Furthermore, through flow cytometry and confocal fluorescence microscopy the NIR-emitting Pdot–streptavidin conjugates proved that they could effectively label EpCAM receptors on the surface of MCF-7 cells, via specific binding between streptavidin and biotin.
Introduction
Optical imaging with fluorescence microscopy is a vital tool in the study of living systems owing to its attractive features, such as high sensitivity and spatiotemporal resolution.1 Among different fluorescent labels, water-soluble fluorescent dyes tend to suffer from rapid photobleaching, which limits their broad applicability in long-term monitoring of live cells with high sensitivity. Fluorescent inorganic nanoparticles, particularly, quantum dots (Qdots) have gained much attention in the past decades owing to their unique optical features such as higher photostability and brightness, tunable emission wavelengths, narrow emission bands, as well as lower susceptibility to cellular efflux mechanisms when compared with small-molecule labels.2–5 Recently, semiconducting polymer nanoparticles (Pdots) have emerged as a new class of promising fluorescent nanoprobes. Their superior characteristics such as non-toxic features, ultrabright photoluminescence, high photostability, non-blinking property, and fast emission rates make them well-suited to bioimaging and biosensing applications.6–21
When biological applications are targeted, fluorescence in the deep red and near-infrared (NIR) is highly desirable, because this wavelength region offers maximal penetration into biological tissues, good spectral separation from autofluorescence, and less scattering in turbid media.22,23 However, because most of the NIR emitters are flat molecules with extended π-conjugations or strong donor–acceptor charge transfer chromophores, self-quenching is often a serious issue when they are condensed into a nanoparticle or solid form.24–28 As self-quenching is difficult to overcome, the development of bright NIR-emitting Pdots is a significant challenge. Recently some strategies, such as Pdots doped with NIR dyes14,21 and hybrid Pdots–Qdots,29 have been demonstrated to realize NIR-emitting Pdots, which is usually applied to improve the sensitivity and specificity of bio-imaging and tracking, since the strong autofluorescence of living tissues, as well as the scattering and the absorption of short-wavelength light in tissues can greatly decrease sensitivity. However, the leakage of the dyes from Pdots13,14,19 and the toxicity of the Qdots29 are important limitations for using dye-doped NIR-emitting Pdots or hybrids as fluorescent probes. In particular, the leakage of NIR dyes from Pdots results in poor optical properties, such as reduced emission color purity and quantum yields, because the donor–acceptor ratio in Pdots significantly affects the energy transfer efficiency. Another drawback of current Pdots is a broad fluorescence emission band, which can lead to constraints in multiplex applications because of spectral interference with other fluorophores, especially for Pdots with high brightness, where leakage of emitted photons into other detection channels can easily overwhelm the fluorescence from other non-Pdot probes. To address these problems, we report here the preparation and application of bright NIR-emitting Pdots with narrow emission band and large absorption–emission separation (Scheme 1) via cascade Förster resonance energy transfer (FRET).
Scheme 1.
Schematic illustration of NIR fluorescence in Pdots obtained by cascade FRET.
Results and discussion
Polyfluorene (P1 in Scheme 1) and poly[(9,9-dioctylfluorenyl-2,7-diyl)-co-(1,4-benzo{2,1′,3}thiadiazole)] (P2) were used as primary donors in cascade FRET systems because of their high brightness and absorption profiles. In the synthesis, monomer 2,7-dibromo-9,9-bis(3-(tert-butyl propanoate))fluorene was co-polymerized (molar ratios of 5 and 9%, respectively, for P1 and P2), which enabled further covalent functionalization and bioconjugation after removal of protecting tert-butyl groups by trifluoroacetic acid. Boron dipyrrin (BODIPY)-containing polymer P4 was designed as terminal acceptor because BODIPY derivatives exhibit favorable optical properties, such as high extinction coefficients and fluorescence quantum yields, negligible photobleaching, and narrow emission peaks.30 Our purpose is to combine the beneficial signature properties of both donor and acceptor in light-harvesting system, such as the strong absorption of primary donor and narrow emission band and high fluorescence quantum yield of terminal acceptor.
Here, P4 can be doped into P1 or P2 Pdots by co-precipitation. However, the absorption of P4 (peak at 690 nm) does not overlap well with the emission spectrum of P1 (peak at 440 nm) or P2 (peak at 540 nm), which leads to poor energy transfer from primary donors to P4. This issue was also observed in our recent work, where we achieved NIR fluorescence by encapsulation of NIR emitting dye silicon 2,3-naphthalocyanine bis(trihexylsilyl oxide) into Pdots.14 Therefore, in this work we introduced polymer P3 to facilitate efficient energy transfer from the primary donor to the terminal acceptor P4. P1 exhibits an absorption peak at 380 nm and fluorescence peak at 440 nm. The absorption spectrum of P2 (peak at 460 nm) overlaps well with the emission spectrum of P1. The absorption spectrum of P3 (peak at 540 nm) also perfectly overlaps with the emission spectrum of P2 (peak at 540 nm). Likewise, the absorption of P4 (peak at 690 nm) overlaps well with the emissions of P3 (peak at 680 nm). These optimized spectral overlaps (Scheme 1) ensure an efficient Förster-type cascade energy transfer, which is further established by the time-resolved fluorescence results (Fig. 1f). Owing to these perfect spectral overlaps, a minimum amount of P4 is needed to quench the donor emissions, and thus greatly reduced self-quenching of the final NIR fluorescence from P4.
Fig. 1.
(a and b) TEM images and size distribution measured by DLS of 3-NIR Pdots; (c and d) TEM images and size distribution measured by DLS of 4-NIR Pdots; (e) absorbance (solid line) and fluorescence spectra (dashed line) of 4-NIR (blue, λexc = 380 nm) and 3-NIR Pdots (red, λexc = 450 nm); (f) time-resolved fluorescence decay of P1 Pdots, and 4-NIR Pdots; (g) time-resolved fluorescence decay of P2 Pdots, and 3-NIR Pdots.
We have also found an interesting result. As polymer P3 is an intermediary in the cascade FRET system described above, the molar ratio between monomer fluorene and 2,3-bisphenylthieno[3,4-b]pyrazine in the P3 polymer chain significantly influenced the final optical properties. As we know, the thieno[3,4-b]pyrazine (TP) unit has a high electron affinity, and the degree of intramolecular charge transfer from fluorene monomer to TP varies with TP molar ratio in the polymer chain.31 Therefore, by optimizing the optical property and energy transfer, a ratio of 13% of monomer TP in P3 polymer was found as optimal for P3 as an effective intermediary for cascade FRET. We also discovered that the molar ratio of BODIPY monomer in P4 is also crucial to the final NIR-emission brightness of the NIR Pdots. Experimental results showed that in the prepared Pdots, even though the doping ratio of polymer P4 (doped into P1 or P2) is very low (weight percentage < 1%), the high molar ratio of BODIPY units in the P4 polymer chain induced significant self-quenching of final NIR emission due to the high local concentration of BODIPY monomers contained in a single polymer chain. In this work, 8% molar ratio of BODIPY monomer contained in P4 was determined to ensure the best emission performance in the cascade FRET system of Pdots.
Pdots composed of different polymers described above could be readily prepared by a co-precipitation method (details shown in Experimental section). The prepared Pdots were water dispersible and were stable for several months. Generally, the prepared Pdots had an average size of about 16 nm, which are comparable to commercial Qdots705 (ESI, Fig. S1†), as reported by TEM analysis and dynamic light scattering (DLS) (Fig. 1a–d). Here, multicomponent Pdots formed from P1, P2, P3 and P4 are denoted as 4-NIR Pdots and those formed from P2, P3 and P4 polymers are denoted as 3-NIR Pdots.
By careful optimization of the weight ratios of premixed polymers P1, P2, P3 and P4 (50 : 50 : 70 : 8) in THF solution, the emissions of P1, P2 and P3 in Pdots were quenched and the final 4-NIR Pdots exhibited a high-intensity fluorescence peak at 710 nm with very large absorption-fluorescence separation (330 nm) and a high quantum yield of 32% (Fig. 1e). The emission peak is very narrow with a full width at half maximum (FWHM) of 44 nm, which is even narrower than that of commercially available Qdots705. Similarly efficient FRET was also achieved in 3-NIR Pdots with optimized weight ratio of P2 : P3 : P4 = 100 : 50 : 6.2 and the final Pdots gave an intense fluorescence peaking at 720 nm (FWHM = 44 nm) with a quantum yield of 30%. Because NIR emission was derived from semiconducting polymers that comprised the Pdots, there was no leakage issue, which often plagues dye-doped Pdots.
Time-resolved fluorescence decay was measured by time-correlated single-photon counting (TCSPC) to study the FRET process. As shown in Fig. 1f, the lifetime of P1 Pdots at 440 nm is 0.65 ns, but the lifetime of P1 in 4-NIR Pdots was much too fast to be measured because the decay curve was comparable with the instrument response function (IRF) (black curve in Fig. 1f). The lifetime of P4 in 4-NIR Pdots was 3.48 ns. Similarly efficient energy transfer and very short lifetime of P2 was also observed in 3-NIR Pdots (Fig. 1g). These results agree well with the total quenching of P1 or P2 emission by energy transfer (in 4-NIR and 3-NIR Pdots, respectively), which were also proved by the emission spectra of these Pdots in Fig. 1e showing efficient energy transfer in both 4-NIR and 3-NIR Pdots.
To further develop and test ultrabright Pdots more suitable for bioimaging, we decided to focus on 3-NIR Pdots because their absorbance (peak at 460 nm) conveniently matches the commonly available laser wavelengths at 473 and 488 nm for fluorescence microscopy and detection. A useful estimate of fluorescence brightness is given by the product of the absorption cross-section and the fluorescence quantum yield. We carried out single-particle imaging to experimentally evaluate and compare the brightness of 3-NIR Pdots and commercially available Qdot705. Fig. 2a–c showed typical single-particle images of Qdot705 and 3-NIR Pdots, respectively, which were obtained under identical excitation and acquisition conditions. With a low excitation power from the 488 nm laser, very bright, near-diffraction-limited spots were clearly observed for single 3-NIR Pdots (Fig. 2b). In addition, 3-NIR Pdots were much brighter than Qdot705 under fluorescence microscopy. From the distribution curves of single-particle brightness of several thousand Qdot705 and 3-NIR Pdots, we determined 3-NIR Pdots were >3 times brighter than Qdot705 (Fig. 2c). Such a prominent contrast is primarily due to the high per-particle absorption cross-section of Pdots, which would be particularly useful for long-term fluorescence detection or tracking requiring low excitation powers.
Fig. 2.
(a, b) Single-particle fluorescence images of Qdots and 3-NIR Pdots obtained under identical excitation and detection conditions. (c) Histograms of intensity distribution of single-particle fluorescence for Qdots and 3-NIR Pdots (log-normal fitting on the histograms resulted in the black and blue fitting curves, as well as mean values of 1200 and 3900 CCD counts for Qdots and 3-NIR Pdots, respectively). (d) Flow-cytometry measurements of the intensity distributions of MCF-7 cells labeled with Qdot705–streptavidin (negative labeling, cyan curve; positive labeling, orange curve) and 3-NIR Pdot–streptavidin (negative labeling, green curve; positive labeling, red curve). All the positive and negative labeling was completed and measured under identical experimental conditions. In the negative labeling, primary biotinylated antibodies were absent.
To evaluate the NIR-emitting Pdots for bioimaging, we performed bioconjugation between the Pdots and streptavidin (SA) for specific targeting towards cell surface receptors. Streptavidin was used ubiquitously because of its remarkable binding affinity to biotin. The final streptavidin-conjugated Pdots were used to label the specific cell-surface receptor, EpCAM, which is widely used for the detection of circulating tumor cells. MCF-7 cells were sequentially incubated with biotinylated primary anti-EpCAM antibody and 3-NIR Pdot–SA. Flow-cytometry measurements of negative and positive labeling (Fig. 2d and S2, ESI†) revealed that both 3-NIR Pdot–SA and 4-NIR Pdot–SA could specifically bind to the biotinylated antibody on the surface of the MCF-7 cells like commercial Qdot705–SA. However, the fluorescence intensity of cells labeled with 3-NIR Pdots and 4-NIR Pdots are >3 times higher than that labeled with Qdot705. This ensemble result of 3-NIR Pdots agrees well with that based on single-particle brightness measurements.
To further evaluate the NIR emitting Pdots for potential bioimaging, MCF-7 cells labeled with 3-NIR Pdots–SA conjugate were studied using confocal fluorescence microscopy. As shown in Fig. 3a, the 3-NIR Pdot–SA probes effectively labeled EpCAM receptors on the MCF-7 cell surface. In the control experiment, where the biotinylated antibody on the surface of the MCF-7 cells was absent, no fluorescence on the cell surface was detected (Fig. 3b), which further confirmed that the covalent bioconjugation of streptavidin to Pdots was successful and the cellular targeting was specific through biotin–streptavidin interaction.
Fig. 3.
Confocal fluorescence images of MCF-7 cells labeled with 3-NIR Pdot–SA probes. (a) Positive labeling using 3-NIR Pdot–SA probe. (b) Negative labeling carried out under the same condition as (a) but in the absence of biotinylated antibody on the surface of the MCF-7 cells. Images from left to right: blue fluorescence from the nuclear stain Hoechst 34580; deep red fluorescence images from 3-NIR Pdot–SA probes; Nomarski (DIC) images; combined fluorescence images. Scale bars: 20 µm.
Conclusions
To sum up, we demonstrated a simple approach for the design and preparation of ultrabright NIR-emitting Pdots with narrow fluorescence band and large absorption–emission separation with the aid of cascade FRET and careful design of polymers. Single particle fluorescence brightness measurements and flowcytometric analysis of Pdots-labeled MCF-7 cells showed that these NIR emitting dots are >3 times brighter than Qdot705. This work has successfully addressed the self-quenching issue of NIR-emitting Pdots by employing the cascade FRET strategy, which combined the beneficial signature properties of both donor and acceptor in a light-harvesting system. The preparation method presented here is general and can be extended to the preparation of other NIR-emitting nanoparticles for different biological applications.
Experimental section
Materials
All of the chemicals and solvents were purchased from Sigma-Aldrich unless indicated elsewhere. Monomer C13 and monomer E,32,33 and 2-hydroxy-4′-iodoacetophenone34 were synthesized by the methods reported before.
Synthesis
Polymers were synthesized by Suzuki coupling using a series of monomers shown in Chart 1.
Chart 1.
Molecular structure of the monomers used in polymer synthesis.
Synthesis of polymer P1
To a 100 mL flask was added monomer A (0.9 mmol, 493 mg), monomer B (1.02 mmol, 569 mg), monomer C (0.1 mmol, 58 mg), Bu4NBr (15 mg), toluene (20 mL) and Na2CO3 (2 M, 10 mL). The mixture was stirred at room temperature and the flask was degassed and recharged with N2, which was repeated four times before and after addition of Pd(PPh3)4 (0.02 mmol, 23 mg). The reactants were stirred at 90 °C for 48 h and then phenylboronic acid (100 mg) dissolved in THF (1 mL) was added. After 2 h, bromobenzene (1 mL) was added and further stirred for 3 h. The mixture was poured into methanol (200 mL). The precipitate was filtered off, washed with methanol, water and acetone to remove monomers, small oligomers and inorganic salts. The crude product was dissolved in DCM (15 mL), filtered through a 0.2 µm membrane and re-precipitated in methanol (150 mL). The powder was then stirred in acetone (200 mL) overnight. then collected by filtration, and dried in vacuum. Yield: 580 mg (74%). 1H NMR (500 MHz, CDCl3): δ 7.84 (d, J = 7.8 Hz, 2H), 7.63–7.73 (m, 4H), 2.12 (br, 4H), 1.32 (s, 1H), 1.10–1.25 (m, 20H), 0.68–0.92 (m, 10H). GPC: Mn 5400, Mw 11 400, PDI 2.11. The tert-butyl protecting group was removed by TFA in DCM at room temperature.13
Synthesis of polymer P2
To a 100 mL flask was added monomer B (1.02 mmol, 569 mg), monomer C (0.18 mmol, 104 mg), monomer D (0.82 mmol, 241 mg), Bu4NBr (15 mg), toluene (20 mL) and Na2CO3 (2 M, 10 mL). The mixture was stirred at room temperature and the flask was degassed and recharged with N2, which was repeated four times before and after addition of Pd(PPh3)4 (0.02 mmol, 23 mg). The reactants were stirred at 90 °C for 48 h and then phenylboronic acid (100 mg) dissolved in THF (1 mL) was added. After 2 h, bromobenzene (1 mL) was added and further stirred for 3 h. The mixture was poured into methanol (200 mL). The precipitate was filtered off, washed with methanol, water and acetone to remove monomers, small oligomers and inorganic salts. The crude product was dissolved in DCM (15 mL), filtered through a 0.2 µm membrane and re-precipitated in methanol (150 mL). The powder was then stirred in acetone (200 mL) overnight and collected by filtration, and dried in vacuum. Yield: 436 mg (75%). 1H NMR (500 MHz, CDCl3): δ 7.94–8.12 (m, 7.08 H), 7.73 (d, J = 6 Hz, 1.64H), 2.40–2.60 (m, 0.72H), 2.04–2.22 (br, 4H), 1.678 (br, 0.72H), 1.33 (s, 3.24H), 1.45–1.21 (m, 20H), 0.96 (br, 4H), 0.81 (t, J = 4.2 Hz, 6H). GPC: Mn 6000, Mw 13 200, PDI 2.2. The tert-butyl protecting group was removed by TFA in DCM at room temperature.13
Synthesis of polymer P3
To a 100 mL flask was added monomer A (0.7 mmol, 384 mg), monomer B (1.02 mmol, 569 mg), monomer E (0.3 mmol, 134 mg), Bu4NBr (15 mg), toluene (20 mL) and Na2CO3 (2 M, 10 mL). The mixture was stirred at room temperature and the flask was degassed and recharged with N2, which was repeated four times before and after addition of Pd(PPh3)4 (0.02 mmol, 23 mg), respectively. The reactants were stirred at 90 °C for 48 h and then phenylboronic acid (100 mg) dissolved in THF (1 mL) was added. After 2 h, bromobenzene (1 mL) was added and further stirred for 3 h. The mixture was poured into methanol (200 mL). The precipitate was filtered off, washed with methanol, water and acetone to remove monomers, small oligomers and inorganic salts. The crude product was dissolved in DCM (15 mL), filtered through a 0.2 µm membrane and re-precipitated in methanol (150 mL). The powder was then stirred in acetone (200 mL) overnight and collected by filtration, and dried in vacuum. Yield: 532 mg (71%). 1H NMR (300 MHz, CDCl3): δ 7.84 (d, J = 8.1 Hz, 2H), 7.53–7.62 (br, 4H), 7.48 (t, J = 7.2 Hz, 0.28H), 7.36–7.43 (br, 1.07H), 2.12 (br, 4H), 1.10–1.22 (m, 20H), 0.76–0.89 (m, 10H). The molar fractions of fluorene and thieno[3,4-b]pyrazine (TP) units were estimated by the elemental analysis data for [fluorene]–[TP] = 0.87 : 0.13. Calc. (%) C, 88.3, H, 9.78, N, 0.88; found, C, 88.5, H, 9.9, N, 0.89. GPC: Mn 6200, Mw 14 200, PDI 2.29.
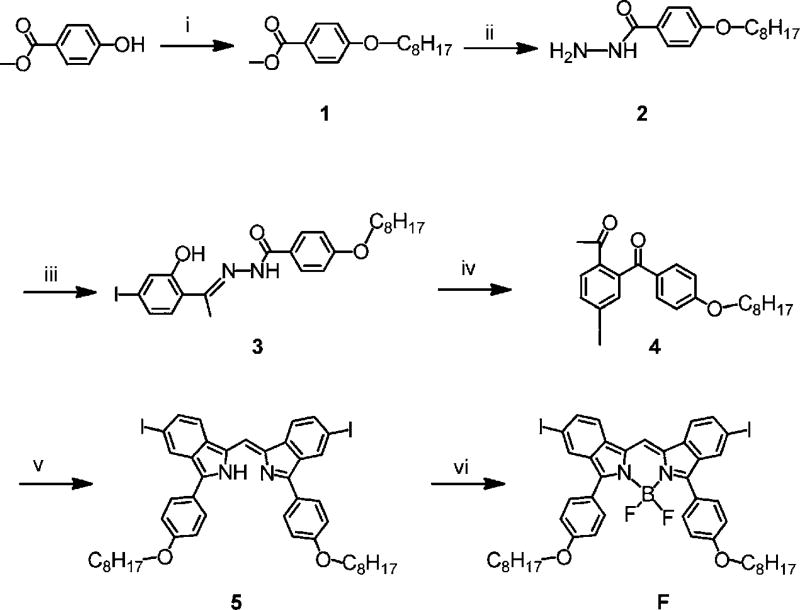
The synthesis route of monomer F is shown in Scheme 2.
Scheme 2.
Synthesis route of monomer F. Reagents and conditions: (i) 1-Bromooctane, Na2CO3/DMF, 80 °C, 48 h; (ii) hydrazine hydrate, reflux in ethanol overnight; (iii) 2-hydroxy-4′-iodoacetophenone, reflux in ethanol for 4 h; (iv) lead tetraacetate, THF, r.t. for 2 h; (v) NH3 (aq)–acetic acid, PEG300, 100 °C, 2 h; (vi) Et3N/BF3·OEt2, CHCl3, 50 °C, 12 h.
Synthesis of 1
Methyl 4-hydroxybenzoate (0.1 mol, 15.2 g), 1-bromooctane (0.13 mol, 25 g) and Na2CO3 (0.28 mol, 30 g) were stirred in DMF (100 mL) at 80 °C for 48 h under N2. After cooling to room temperature, water 100 mL and DCM (100 mL) were added and stirred for 5 min. The DCM phase was washed with water (100 mL) and Na2CO3 solution (1 M, 100 mL), and dried over MgSO4. After removal of the solvent, the residue was purified by column chromatography (CHCl3–hexane = 1 : 1) to gave colorless crystals. Yield: 76%. 1H NMR (500 MHz, CDCl3): δ 7.98 (d, J = 8.5 Hz, 2H), 6.90 (d, J = 8.5 Hz, 2H), 4.00 (t, 2H), 3.88 (s, 3H), 1.79 (m, 2H), 1.24–1.40 (m, 10H), 0.89 (t, 3H). 13C NMR (500 MHz, CDCl3): δ 167.12, 163.15, 131.75, 122.50, 114.25, 68.41, 52.03, 32.01, 29.54, 29.43, 29.37, 29.33, 26.20, 22.87, and 14.31. ESI-MS: m/z 264.40. Calc. for C16H24O3 (%): C, 72.69; H, 9.15. Found: C, 72.82; H, 8.99.
Synthesis of 2
Methyl 4-(octyloxy)benzoate (1) (0.1 mol, 26.4 g) and hydrazine hydrate (30 mL, excess) were refluxed in ethanol (80 mL) overnight. After cooling to room temperature, the crystals were filtered off, washed with ethanol, and dried in vacuum. The filtrate was concentrated to 50 mL and water (60 mL) was added under vigorous stirring. The white crystals were filtered off and washed with ethanol–H2O (v/v = 1 : 1) and dried in a vacuum desiccator. Overall yield: 91%. 1H NMR (500 MHz, DMSO-d6): δ 9.61 (s, 1H), 7.79 (d, J = 8.5 Hz, 2H), 6.96 (d, J = 8.5 Hz, 2H), 4.42 (s, 2H), 4.01 (t, 2H), 1.72 (m, 2H), 1.22–1.46 (m, 10H), 0.87 (t, 3H). 13C NMR (500 MHz, DMSO-d6): δ 165.76, 161.04, 128.86, 125.48, 114.09, 67.76, 31.42, 28.90, 28.84, 28.76, 25.66, 22.27, 14.14. ESI-MS: m/z 265.01. Calc. for C15H24N2O2 (%): C, 68.15; H, 9.15; 10.60. Found: C, 68.22; H, 9.04; N, 10.48.
Synthesis of 3
2-Hydroxy-4′-iodoacetophenone (10 mmol, 2.62 g) and 2 (10 mmol, 2.64 g) were dissolved in ethanol and heated to reflux for 4 h. After cooling to r.t. the powder was filtered off and washed with ethanol and dried in vacuum. Yield: 4.72 g, 93%. 1H NMR (500 MHz, DMSO-d6): δ 13.63 (s, 1H), 11.20 (s, 1H), 7.92 (d, J = 8.5 Hz, 2H), 7.39 (d, J = 8.5 Hz, 1H), 7.29 (s, 1H), 7.25 (d, J = 8.5 Hz, 1H), 7.06 (d, J = 8.5 Hz, 2H), 4.06 (t, 2H), 2.45 (s, 3H), 1.74 (m, 2H), 1.42 (m, 2H), 1.26–1.33 (m, 8H), 0.87 (t, 3H). 13C NMR (500 MHz, DMSO-d6): δ 163.58, 161.60, 159.01, 156.36, 129.98, 129.76, 127.15, 125.59, 124.33, 119.06, 113.92, 97.12, 67.61, 31.09, 28.58, 28.51, 28.40, 21.93, 13.81, 13.77. ESI-MS: m/z 508.84. Calc. for C23H29IN2O3 (%): C, 54.34; H, 5.75; N, 5.51. Found: C, 54.08; H, 5.63; N, 5.72.
Synthesis of 4
Intermediate 4 was synthesized by a previously reported method.35 Lead tetraacetate (5.38 g, 12.13 mmol) was added to a suspension of 3 (5.08 g, 10 mmol) in dry THF (150 mL) in small portions over a period of 5 min. After stirring at room temperature for 2 h, the resulting solid was removed by filtration. The filtrate was concentrated by rotary evaporator and purified by silica gel chromatography (hexane–ethyl acetate = 4 : 1). 1H NMR (500 MHz, CDCl3): δ 7.94 (d, J = 8.5 Hz, 1H), 7.74 (s, 1H), 7.71 (d, J = 9.0 Hz, 2H), 7.59 (d, J = 8.5 Hz, 1H), 6.92 (d, J = 8.5 Hz, 2H), 4.03 (t, 2H), 2.50 (s, 3H), 1.81 (m, 2H), 1.47 (m, 2H), 1.26–1.39 (m, 8H), 0.91 (t, 3H). 13C NMR (500 MHz, CDCl3): δ 197.76, 194.48, 163.42, 142.66, 138.53, 136.93, 136.71, 131.70, 130.50, 129.43, 114.34, 99.48, 68.34, 31.82, 29.32, 29.23, 29.10, 27.53, 26.00, 22.67, 14.15. ESI-MS: m/z 478.24. Calc. for C23H27IO3 (%): C, 57.75; H, 5.69. Found: C, 57.88; H, 5.54.
Synthesis of 5
Synthesis of derivatives of 5 have been reported before.35 In this work, we used a modified method with much shorter reaction time using low molecular-weight poly-(ethylene glycol) (PEG) as solvent. Concentrated aqueous ammonia (NH3 content 28–30%, 9 mL) was added to a solution of 4 (2 mmol, 0.96 g) in acetic acid (15 mL) and PEG300 (25 mL). The mixture was stirred at 100 °C for 2 h. The resulting solid was filtered off and washed thoroughly with ethanol. The crude product was purified by silica gel chromatography (DCM). Yield: 0.43 g, 47.6%. 1H NMR (500 MHz, CDCl3): δ 8.30 (s, 2H), 7.87 (d, J = 8.5 Hz, 4H), 7.55–7.62 (m, 4H), 7.37 (s, 1H), 7.07 (d, J = 8.5 Hz, 4H), 4.07 (t, 4H), 1.86 (m, 4H), 1.25–1.56 (m, 20H), 0.91 (t, 6H). 13C NMR (500 MHz, CDCl3): δ 159.99, 145.28, 134.63, 134.18, 132.64, 131.03, 130.68, 128.75, 125.62, 120.65, 115.19, 110.80, 100.14, 89.31, 68.42, 32.05, 29.64, 29.54, 29.48, 26.31, 22.87, 14.30. ESI-MS: m/z 905.36. Calc. for C45H50I2N2O2 (%): C, 59.74; H, 5.57; N, 3.10. Found: C, 59.57; H, 5.64; N, 3.02.
Synthesis of monomer F
Dry triethylamine (1 mL, 7.2 mmol) was added to a solution of 5 (0.90 g, 1 mmol) in chloroform (250 mL), followed by addition of BF3·OEt2 (2.51 mL, 20.3 mmol). After the reaction mixture was stirred at 50 °C for 12 h, it is washed with water. The organic layer is separated, dried over anhydrous magnesium sulfate, and concentrated by a rotary evaporator to give a blue solid. The crude product was purified by silica gel chromatography (hexane–chloroform = 1 : 1) to give F as a deep blue solid. Yield: 0.7 g, 73.5%. 1H NMR (500 MHz, CDCl3): δ 7.94 (s, 2H), 7.74 (d, J = 8.5 Hz, 4H), 7.64 (s, 1H), 7.61 (d, J = 8.5 Hz, 2H), 7.55 (d, J = 8.5 Hz, 2H), 7.02 (d, J = 8.5 Hz, 4H), 4.03 (t, 4H), 1.83 (m, 4H), 1.49 (m, 4H), 1.29–1.41 (m, 16H), 0.91 (t, 6H). 13C NMR (500 MHz, CDCl3): δ 160.47, 150.78, 137.00, 132.51, 132.37, 132.18, 131.59, 127.30, 122.33, 120.32, 114.36, 113.91, 89.51, 67.92, 31.70, 29.23, 29.12, 25.95, 22.54, 13.99. ESI-MS: m/z 952.43. Calc. for C45H49BF2I2N2O2 (%): C, 56.74; H, 5.19; N, 2.94. Found: C, 57.01; H, 4.96; N, 3.05.
Synthesis of polymer P4
To a 100 mL flask was added monomer B (0.1 mmol, 55.8 mg), monomer F (0.02 mmol, 19 mg), monomer G (0.08 mmol, 51.4 mg), toluene (4 mL), Na2CO3 (2 M, 3 mL) and ethanol (0.7 mL). The mixture was stirred at room temperature and the flask was degassed and recharged with N2, which was repeated four times before and after addition of Pd(PPh3)4 (0.004 mmol, 4.6 mg), respectively. The reactants were stirred at 83 °C for 30 h and then phenylboronic acid (20 mg) dissolved in THF (0.5 mL) was added. After two hours, bromobenzene (0.5 mL) was added and further stirred for 2 h. The mixture was poured into methanol (100 mL). The precipitate was filtered off, washed with methanol, water and acetone to remove monomers, small oligomers and inorganic salts. The crude product was dissolved in DCM (5 mL), filtered through a 0.2 µm membrane and re-precipitated in methanol (60 mL). The powder was collected by filtration, and dried in vacuum. Yield: 64.8 mg (77%). 1H NMR (300 MHz, CDCl3): δ 7.55–7.95 (m, 6.72H), 4.06 (t, J = 6.9 Hz, 0.32H), 2.09 (br, 3.66 H), 1.06–1.40 (m, 12H), 0.61–0.95 (m, 18H). GPC: Mn 9200, Mw 26 300, PDI 2.86.
Preparation of Pdots
The semiconducting polymer dots were prepared by co-precipitation method. All polymers were dissolved into anhydrous THF, respectively, to form a 1 mg mL−1 THF solution, then as the volume ratio of 50 : 50 : 70 : 8 to mix polymers P1, P2, P3, P4 (for 4-NIR Pdots), and ratio of 100 : 50 : 6.2 to mix P2, P3, P4 (for 3-NIR Pdots) in THF. Then 0.2 mL mixed solution was added into 1.8 mL anhydrous THF, which was further injected directly into 10 mL DI water under ultrasonication. THF was removed by N2 flow at room temperature. The sizes of these two kinds of particles (4-NIR Pdots and 3-NIR Pdots) were characterized as 15.7 nm by DLS analysis.
Bioconjugation
We performed bioconjugation by utilizing the EDC-catalyzed reaction between carboxyl groups on Pdots surface and amine groups on biomolecules. In a typical bioconjugation reaction, 80 µL of polyethylene glycol (5% w/v PEG, Mw 3350) and 80 µL of concentrated HEPES buffer (1 M) were added to 4 mL of functionalized Pdot solution (50 µg mL−1 in MilliQ water), resulting in a Pdot solution in 20 mM HEPES buffer with a pH of 7.4. Then, 240 µL of mg mL−1 streptavidin (purchased from Invitrogen (Eugene, OR, USA)) was added to the solution and mixed well on a vortex. 80 µL of freshly prepared EDC solution (5 mg mL−1 in MilliQ water) was added to the solution, and the above mixture was kept under stirring. After 3 h at room temperature, 80 µL 10 wt% BSA solution was added, the mixed solution was stirred for 20 min, then 80 µL 2.5 wt% Triton-X 100 (0.25% (w/v), 20 µL) were added. Finally, the resulting Pdot bioconjugates were transferred to a centrifugal ultrafiltration tube (Amicon® Ultra-4, MWCO: 100 kDa), and concentrated to 0.5 mL by using the centrifuge, and then were separated from free biomolecules by gel filtration using Sephacryl HR-300 gel media.
Cell culture
The breast cancer cell line MCF-7 was ordered from American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were cultured at 37 °C, 5% CO2 in Eagles minimum essential medium supplemented with 10% Fetal Bovine Serum (FBS), 50 U mL−1 penicillin, and 50 µg mL−1 streptomycin. The cells were pre-cultured prior to experiments until confluence was reached. The cells were harvested from the culture flask by briefly rinsing with culture media followed by incubation with 5 mL of trypsin–EDTA solution (0.25 w/v% trypsin, 0.53 mM EDTA) at 37 °C for 5–15 min. After complete detachment, the cells were rinsed, centrifuged and resuspended in 1× PBS buffer. The cell concentration was determined by microscopy using a hemocytometer.
Flow cytometry
For specific cell labeling with the narrow-band emissive Pdot–streptavidin (Pdot–SA) and Qdot705–SA, a million cells were blocked with BlockAid blocking buffer (Invitrogen, Eugene, OR, USA) and then were incubated sequentially with biotinylated primary anti-EpCAM antibody (used to label the cell-surface EpCAM receptors on MCF-7 cells) and 2 nM Pdot–SA or Qdot705–SA for 30 min each, followed by two washing steps using labeling buffer. Finally, the specifically labeled cells were fixed in 0.5 mL 1% (v/v) paraformaldehyde solution. For the control labeling, no biotinylated primary anti-EpCAM antibody was added. Flow cytometry measurements were performed on fresh samples with 106 cells per 0.5 mL, prepared following the procedure described previously. Flow cytometers BD FACScan and FACS Canto II (BD Bioscience, San Jose, CA USA) were used for cells labeled with 3-NIR Pdots–SA (or Qdot705–SA) and 4-NIR Pdots–SA, respectively. Excitation sources of BD FACScan and FACS Canto II were 488-nm laser and 405-/488-nm lasers, respectively. Corresponding detection channels for fluorescence emission were filtered by a 655 nm long pass and by a 670 nm long pass filter for both BD FACScan and FACS Canto II. Scattered light and fluorescence emission were detected by PMT arrays. Representative populations of cells were chosen by selection of appropriate gates. Detection of cell scattered and fluorescence light was continued until at least 104 events had been collected in the active gate. Data were analyzed using FlowJo Software (Tree Star, Inc., Ashland, OR, USA).
Cell labeling
For labeling cell-surface proteins with the narrow-band emissive Pdot–SA conjugates, live MCF-7 cells in a glass-bottomed culture dish were blocked with BlockAid blocking buffer (Invitrogen, Eugene, OR, USA). Then the MCF-7 cells were incubated sequentially with biotinylated primary anti-EpCAM antibody (used to label the cell-surface EpCAM receptors on MCF-7 cells) and 5 nM Pdot–SA for 30 min each, followed by two washing steps after each incubation. For the control, no biotinylated primary anti-EpCAM antibody was added. The Pdot-tagged cells were then counterstained with Hoechst 34580 and imaged immediately on a fluorescence confocal microscope (Zeiss LSM 510), excited by a 405 nm diode laser for the Hoechst fluorescence channel (BP filer: 420–480 nm) and by a 488 nm Argon laser for 3-NIR Pdots fluorescence channel (650 nm Long Pass filter). An EC Plan-Neofluar 40/1.30 Oil DIC objective lens was utilized for cellular surface imaging.
Supplementary Material
Acknowledgments
We gratefully acknowledge support from the Swedish Foundation for Strategic Research (SSF) within the Nano-X program (Grant no. SSF [A3 05:204] and Swedish government strategic faculty grant in material science (SFO, MATLIU) in Advanced Functional Materials (AFM); and the National Institutes of Health (CA147831 and GM085485) of USA and the University of Washington.
Footnotes
Electronic supplementary information (ESI) available: DLS, flow cytometry result of 4-NIR Pdots and Qdot705, other characterization and measurement methods. See DOI: 10.1039/c3sc50222h
Notes and references
- 1.Suhling K, French PMW, Phillips D. Photochem. Photobiol. Sci. 2005;4:13–22. doi: 10.1039/b412924p. [DOI] [PubMed] [Google Scholar]
- 2.Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Nat. Mater. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 3.Somers RC, Bawendi MG, Nocera DG. Chem. Soc. Rev. 2007;36:579–591. doi: 10.1039/b517613c. [DOI] [PubMed] [Google Scholar]
- 4.Choi HS, Liu W, Misra P, Tanaka E, Zimmer JP, Ipe BI, Bawendi MG, Frangioni JV. Nat. Biotechnol. 2007;25:1165–1170. doi: 10.1038/nbt1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu C, Chiu DT. Angew. Chem., Int. Ed. 2013 doi: 10.1002/anie.201205133. [DOI] [Google Scholar]
- 7.Howes P, Green M, Levitt J, Suhling K, Hughes M. J. Am. Chem. Soc. 2010;132:3989–3996. doi: 10.1021/ja1002179. [DOI] [PubMed] [Google Scholar]
- 8.Kaeser A, Schenning APHJ. Adv. Mater. 2010;22:2985–2997. doi: 10.1002/adma.201000427. [DOI] [PubMed] [Google Scholar]
- 9.Pecher J, Mecking S. Chem. Rev. 2010;110:6260–6279. doi: 10.1021/cr100132y. [DOI] [PubMed] [Google Scholar]
- 10.Tuncel D, Demir HV. Nanoscale. 2010;2:484–494. doi: 10.1039/b9nr00374f. [DOI] [PubMed] [Google Scholar]
- 11.Wu C, Hansen SJ, Hou Q, Yu J, Zeigler M, McNeill J, Olson JM, Chiu DT. Angew. Chem., Int. Ed. 2011;50:3430–3434. doi: 10.1002/anie.201007461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu C, Schneider T, Zeigler M, Yu J, Schiro PG, Burnham DR, McNeill JD, Chiu DT. J. Am. Chem. Soc. 2010;132:15410–15417. doi: 10.1021/ja107196s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, Yu J, Wu C, Jin Y, Yu R, Ye F, Chiu DT. ACS Nano. 2012;6:5429–5439. doi: 10.1021/nn301308w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin Y, Ye F, Zeigler M, Wu C, Chiu DT. ACS Nano. 2011;5:1468–1475. doi: 10.1021/nn103304m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petkau K, Kaeser A, Fischer I, Brunsveld L, Schenning APHJ. J. Am. Chem. Soc. 2011;133:17063–17071. doi: 10.1021/ja2075345. [DOI] [PubMed] [Google Scholar]
- 16.Wu C, Hansen SJ, Hou Q, Yu J, Zeigler M, Jin Y, Burnham DR, McNeill JD, Olson JM, Chiu DT. Angew. Chem., Int. Ed. 2011;50:3430–3434. doi: 10.1002/anie.201007461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye F, Wu C, Jin Y, Chan Y-H, Zhang X, Chiu DT. J. Am. Chem. Soc. 2011;133:8146–8149. doi: 10.1021/ja202945g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye F, Wu C, Jin Y, Wang M, Chan Y-H, Yu J, Sun W, Hayden S, Chiu DT. Chem. Commun. 2012;48:1778–1780. doi: 10.1039/c2cc16486h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu J, Wu C, Zhang X, Ye F, Gallina ME, Rong Y, Wu I-C, Sun W, Chan Y-H, Chiu DT. Adv. Mater. 2012;24:3498–3504. doi: 10.1002/adma.201201245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu J, Wu C, Sahu SP, Fernando LP, Szymanski C, McNeill J. J. Am. Chem. Soc. 2009;131:18410–18414. doi: 10.1021/ja907228q. [DOI] [PubMed] [Google Scholar]
- 21.Xiong L, Shuhendler AJ, Rao J. Nat. Commun. 2012;3:1193. doi: 10.1038/ncomms2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Massin J, Dayoub W, Mulatier J-C, Aronica C, Bretonnière Y, Andraud C. Chem. Mater. 2011;23:862–873. [Google Scholar]
- 23.Raghavachari R. Near-Infrared Applications in Biotechnology. Marcel Dekker, Inc.; WI: 2001. [Google Scholar]
- 24.Massin J, Dayoub W, Mulatier J-C, Aronica C, Bretonnière Y, Andraud C. Chem. Mater. 2011;23:862–873. [Google Scholar]
- 25.Park SY, Kubota Y, Funabiki K, Shiro M, Matsui M. Tetrahedron Lett. 2009;50:1131–1135. [Google Scholar]
- 26.Fenwick O, Sprafke JK, Binas J, Kondratuk DV, Di Stasio F, Anderson HL, Cacialli F. Nano Lett. 2011;11:2451–2456. doi: 10.1021/nl2008778. [DOI] [PubMed] [Google Scholar]
- 27.Qian G, Zhong Z, Luo M, Yu D, Zhang Z, Wang ZY, Ma D. Adv. Mater. 2009;21:111–116. [Google Scholar]
- 28.Lim C-K, Kim S, Kwon IC, Ahn C-H, Park SY. Chem. Mater. 2009;21:5819–5825. [Google Scholar]
- 29.Chan Y-H, Ye F, Gallina ME, Zhang X, Jin Y, Wu I-C, Chiu DT. J. Am. Chem. Soc. 2012;134:7309–7312. doi: 10.1021/ja3022973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ulrich G, Ziessel R, Harriman A. Angew. Chem., Int. Ed. 2008;47:1184–1201. doi: 10.1002/anie.200702070. [DOI] [PubMed] [Google Scholar]
- 31.Rasmussen SC, Schwiderski RL, Mulholland ME. Chem. Commun. 2011;47:11394–11410. doi: 10.1039/c1cc12754c. [DOI] [PubMed] [Google Scholar]
- 32.Wen L, Rasmussen SC. J. Chem. Crystallogr. 2007;37:387–398. [Google Scholar]
- 33.Wen L, Nietfeld JP, Amb CA, Rasmussen SC. J. Org. Chem. 2008;73:8529–8536. doi: 10.1021/jo801632z. [DOI] [PubMed] [Google Scholar]
- 34.Bennet CJ, Caldwell ST, McPhail DB, Morrice PC, Duthie GG, Hartley RC. Bioorg. Med. Chem. 2004;12:2079–2098. doi: 10.1016/j.bmc.2004.02.031. [DOI] [PubMed] [Google Scholar]
- 35.Nagai A, Chujo Y. Macromolecules. 2010;43:193–200. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.