1. INTRODUCTION
Microsporidia were initially described about 150 years ago with the identification of Nosema bombycis as the organism responsible for the disease pébrine in silkworms (1). Microsporidia are ubiquitous in the environment and infect almost all invertebrates and vertebrates, as well as some protists (2). Spores from microsporidia are commonly found in surface water (3). These organisms are eukaryotes with a nucleus with a nuclear envelope, an intracytoplasmic membrane system, chromosome separation on mitotic spindles, vesicular Golgi and a mitochondrial remnant organelle lacking a genome termed a mitosome (4). For insects, fish, laboratory rodents and rabbits microsporidia are important pathogens; and they have been investigated as biological control agents for destructive species of insects (2). Several species of microsporidia have caused significant agricultural economic losses including Nosema apis and Nosema ceranae in honeybees (5), Loma salmonae in salmonid fish (6) and Thelohania spp. in shrimp (7). An excellent review of the history of research on these pathogens was published by Franzen (8) and a recent textbook by Weiss and Becnel (2) provides a comprehensive examination of what is known about these organisms. The class or order Microsporidia was elevated to the phylum Microspora by Sprague in 1977 (9) and a decade latter Sprague and Becnel (10) suggested that the term Microsporidia should instead be used for the phylum name. These organisms were previously considered “primitive” protozoa (11), however, molecular phylogenetic analysis has resulted in the insight that these organisms are not primitive but instead degenerate, and that Microsporidia are related to the Fungi, either as a basal branch of the Fungi or as a sister group (12–16).
The majority of microsporidia initially infect their hosts via the gastrointestinal tract and they have evolved an infection apparatus, the polar tube, which allows them to infect host cells at a distance, enabling them to traverse the space between the lumen of the gastrointestinal tract and the host cells lining the digestive system (2). Many of the microsporidia disseminate following initial infection and there are descriptions of microsporidian infections in almost every organ system with human cases of encephalitis, ocular infection, sinusitis, myositis, and disseminated infection being well documented in published literature (2, 17). Microsporidia can have significant effects on their hosts and host cells with infection resulting in juvenilization, feminization or other changes to host physiology as well as the formation of xenomas or other multinucleate cellular structures (2). Recent work to investigate microsporidia-host interactions have demonstrated the complexity of this interaction and the effects of microsporidia on the host cell transcriptional response in model systems of this interaction including insects and Caenorhabditis elegans (18–20).
Human infection with microsporidia causing encephalitis was initially described in 1959. The species of microsporidia described to date in human infection suggest that most infections in humans are zoonotic and/or water-borne. Microsporidiosis has been a particular problem in immune suppressed hosts, such as those with advanced human immunodeficiency virus infection (AIDS) (21) and patients on immune suppressive drugs such as patients with organ transplantation or those receiving immune modulatory antibodies such as anti-TNFα (21, 22). Microsporidian infections in immune competent mammals are often chronic and asymptomatic whilst immune compromised hosts often develop lethal infections (23). Diarrhea due to microsporidiosis from Enterocytozoon bieneusi was initially reported in patients with AIDS in 1985 (24), and the number of articles describing human disease increased dramatically after 1990 with improvements in diagnostic techniques (2, 21, 25). The development of effective combination antiretroviral therapy (cART) has resulted in a decline in the cases of microsporidiosis in patients with HIV infection (2, 26). As noted above, however, that infection can also occur in immune competent individuals and in particular diarrhea (often self-limited) and keratitis have been important manifestations in patients with intact immune systems (2).
Microsporidian infection in many mammals leads to a chronic infection with persistently high antibody titers and ongoing inflammation that can reactivate leading to acute infection when immune suppression is administered (27, 28). Studies using Encephalitozoon species in mice have helped defined the protective mammalian immune response to microsporidiosis (29, 30). Immunity to infection is mediated by T cells and dependent on interferon-gamma (IFN-γ) and interleukin-12 (IL-12) (31–34). Evidence for this is provided by the protection against lethal microsporidiosis (E. cuniculi infection) afforded by adoptive transfer of sensitized syngenic T-enriched spleen cells into athymic or SCID mice (35, 36). In intraperitoneal E. cuniculi infection models, mice deficient in CD8+ cells or perforin deficient mice have a lethal infection, but there is no lethality in CD4+ deficient mice (32, 37). Oral E. cuniculi infection murine models have shown that CD4+ cells are also involved in the protective immune response for this natural route of infection (33, 38). The most important subset for protective immunity in oral infection models were CD8+αβ T cells, and dendritic cell IFN-γ protection was critical for priming the gut intraepithelial lymphocyte response (39). The ability to trigger a robust T cell response against microsporidian infection is age-dependent (40). Adoptive transfer of immune B lymphocytes into athymic BALB/c (nu/nu) or SCID mice or passive transfer of hyperimmune serum is not sufficient to protect athymic mice from lethal microsporidiosis (41, 42). Antibodies do, however, provide some protection as evidenced by the observations that maternal antibodies protect newborn rabbits from infection with E. cuniculi during the first 2 weeks of life and that antibodies to the polar tube can prevent infection of host cells (43).
There are approximately 1400 described species of microsporidia which are distributed into about 200 genera (2), of which the following have been demonstrated to cause infections in humans (Table 1): Nosema (N. corneum renamed Vittaforma corneae (44); N. algerae reclassified initially as Brachiola algerae (45) and now as Anncaliia algerae (46)), Pleistophora (47), Encephalitozoon (48, 49), Enterocytozoon (24), Septata (50) (reclassified as Encephalitozoon (51)), Trachipleistophora (52–54), Brachiola (45), Anncaliia (46), Tubulonosema (55, 56), Endoreticulatus (57) and Microsporidium (48). Drugs that are used for the treatment of microsporidiosis in various hosts have been reviewed by Costa et al (58). In humans with AIDS, there is reasonable evidence from case reports that treatment with antiretroviral therapy allows the patient’s immune system to control and eradicate microsporidiosis (59, 60). Albendazole, a benzimidazole that binds to β-tubulin, has activity against many species of microsporidia and is used for therapy of microsporidiosis,; however, albendazole is not effective for infections due to Enterocytozoon bieneusi or Vittafomra corneae. The tubulin genes of both E. bieneusi and V. corneae have amino acid residues that are known to be associated with albendazole resistance (61, 62). Studies suggest that E. bieneusi and many other microsporidian infections are responive to fumagillin, a water-insoluble antibiotic made by Aspergillus fumigatus that inhibits methionine aminiopeptidase type 2, or to synthetic analogs of fumagillin, such as TNP-470 (63, 64). Ocular infections caused by microsporidia have been treated with fumagillin bicylohexylammonium in saline. Itraconazole combined with albendazole has been used as therapy in cases of disseminated infections caused by Trachipleistophora or Anncaliia. Nitazoxinide therapy has been effective in stopping diarrhea caused E. bieneusi in patients with organ transplantation (unpublished data); however this effect may be dependent on the immune status of the patient as this drug had mimimal efficacy in patients with AIDS with low CD4 counts (65).
Table 1.
Species of Microsporidia Infecting Humans
| Species | Synonym(s) | Common sites of infection in humans | Other mammalian hosts | Reported Non-mammalian hosts | Disease Manifestations Reported |
|---|---|---|---|---|---|
| Anncaliia algerae |
Brachiola algerae Nosema algerae |
Eye Muscle |
Mosquitoes | myositis, keratoconjunctivitis, cellulitis | |
| Anncaliia connori |
Brachiola connori Nosema connori |
Systemic | disseminated disease | ||
| Anncaliia vesicularum | Brachiola vesicularum | Muscle | myositis | ||
| Encephalitozoon cuniculi | Nosema cuniculi | Systemic | Wide host range | Birds | hepatitis, encephalitis, peritonitis, urethritis, prostatitis, nephritis, sinusitis, keratoconjunctivitis, cystitis, diarrhea, disseminated infection |
| Encephalitozoon hellem | Eyes | Bats | Birds | superficial keratoconjunctivitis, sinusitis, pneumonitis, nephritis, protatitis, urethritits, cystitits, diarreha, disseminated infection | |
| Encephalitozoon intestinalis | Septata intestinalis | Small intestine | Wide host range | Geese | diarrhea, intestinal perforation,, cholangitis, nephritis, superficial keratoconjunctivitis, disseminated infection |
| Enterocytozoon bieneusi | Small intestine Biliary tract |
Wide host range | Birds | diarrhea, malabsorption with wasting syndrome, cholangitis, rhinitis, bronchitis | |
| Microsporidium africanum | Eyes | stromal keratitis | |||
| Microsporidium ceylonensis | Eyes | stromal keratitis | |||
| Microsporium CU (Endoreticulatus-like) | Muscle | myositis | |||
| Nosema ocularum | Eyes | stromal keratitis | |||
| Pleistophora ronneafiei | Muscle | Fish* | myositis | ||
| Trachipleistophora anthropopthera | Eyes Systemic |
Insects* | encephalitis, keratitis, disseminated infection | ||
| Trachipleistophora hominis | Eyes Muscle |
Mosquitoes* | myositis, keratoconjunctivitis, sinusitis, encephalitis | ||
| Tubulinosema acridophagus | Muscle Systemic |
Fruit fly* | myositis, disseminated infection | ||
| Vittaforma corneae | Nosema corneum | Eyes Urinary tract |
Keratoconjunctivitis, urinary tract infection |
Putative host(s) based on phylogeny or host relationships of other species within the genus
2. MORPHOLOGY AND LIFE CYCLES
The microsporidian life cycle consists of a proliferative phase (merogony), the spore production phase (sporogony) and the mature spore or infective phase (2, 66, 67) (Figure 1).. The general features of the life cycle are: (A) Spores are ingested or inhaled and then germinate, resulting in extension of the polar tube, and injection of the sporoplasm into the host cell cytoplasm; (B) Merogony ensues as injected sporoplasm develops into meronts (the proliferative stage), which multiply, depending on the species, by binary fission or multiple fission, forming multinucleate plasmodial forms; (C) Sporogony follows merogony, and the meront cell membranes thicken forming sporonts; (D) After dividing sporonts give rise to sporoblasts that develop into mature spores without additional multiplication; and (E) The host cell becomes distended with mature spores, ruptures, and releases spores into the environment. The combination of multiplication during both merogony and sporogony results in a very large number of spores being produced from a single infection and is the basis of the enormous reproductive potential of these pathogens.
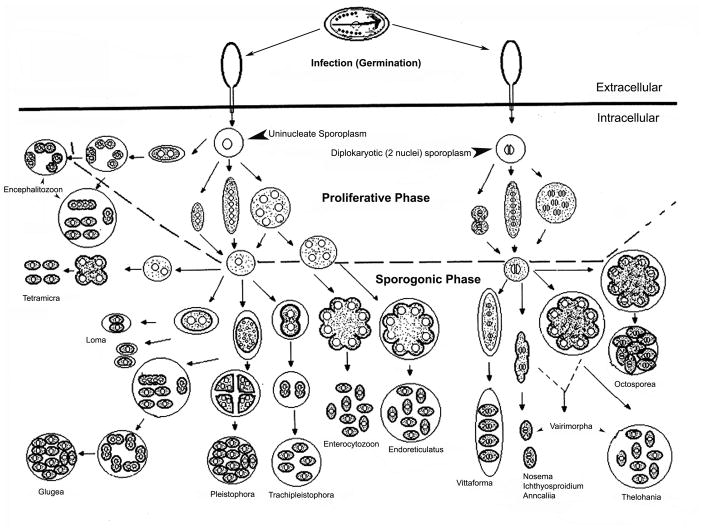
Figure one. Microsporidian Life Cycles.
The initial phase of infection involves spores being exposed to the proper environmental conditions that cause germination of the spores and polar tube extrusion. The polar tube pierces the plasma membrane (solid black line) of the host cell, and the sporoplasm travels through the polar tube into the host cell. The sporoplasm then divides during the proliferative phase and the morphology of this division is used for determination of microsporidian genera. The sporoplasm on the left is uninucleate, and the cells that are produced from it represent the developmental patterns of several microsporidia with isolated nuclei. The sporoplasm on the right is diplokaryotic and it similarly produces the various diplokaryotic developmental patterns. Cells containing either type of nucleation will produce one of three basic developmental forms. Some cycles have cells that divide immediately after karyokinesis by binary fission (e.g. Anncaliia). A second type forms elongated moniliform multinucleate cells that divide by multiple fission (e.g. some Nosema species). The third type forms rounded plasmodial multinucleate cells that divide by plasmotomy (e.g. Endoreticulatus species). Cells may repeat their division cycles one to several times in the proliferative phase. The intracellular stages in this phase are usually in direct contact with the host cell cytoplasm or closely abutted to the host ER; however, the proliferative cells of Encephalitozoon (and probably Tetramicra) are surrounded by a host formed parasitophorous vacuole throughout their development, and the proliferative plasmodium of the genus Pleistophora is surrounded by a thick layer of parasite secretions that becomes the sporophorous vesicle in the sporogonic phase. The sporogonic phase is illustrated below the dashed line. Some of the microsporidian genera maintain direct contact with the host cell cytoplasm during sporogony; i.e. Nosema, Ichthyosporidium, Anncaliia, Enterocyotozoon and probably Tetramicra. The remaining genera form a sporophorous vesicle as illustrated by the circles around developing sporogonial stages. It should be noted that in the Thelohania cycle and the Thelohania-like part of the Vairimorpha cycle, the diplokarya separate and continue their development as cells with isolated nuclei. Adapted with permission from Wittner M and Weiss LM. (Eds.) The Microsporidia and Microsporidiosis. Washington, DC: ASM Press. 1999 (70).
Microsporidia form characteristic unicellular spores (Figure 2) that are environmentally resistant enabling their transmission in food and water (68). Spore structure is characteristic of the phylum, and spores can vary in size and shape depending on the species (9, 69). The spore coat consists of an electron-dense, proteinaceous exospore, an electronlucent endospore and an inner membrane or plasmalemma. A defining characteristic of these pathogenic protists is the presence of an invasion organelle that consists of the polar tube that is attached to the inside of the anterior end of the spore by an anchoring disk, and a series of associated membranes. The polar tube coils around the nucleus and their can be up to 30 coils depending on the species of microsporidia (2, 9, 70). During germination, the polar tube rapidly everts, forming a hollow tube that brings the sporoplasm into intimate contact with the host cell acting as a conduit to deliver the sporoplasm to the host cell, essentially functioning like a hypodermic needle (71–73) (Figure 3). When a spore is phagocytosed by a host cell, germination can occur allowing the polar tube to pierce the phagocytic vacuole permitting the sporoplasm to escape the phagosome and to be delivered into the host cell cytoplasm (74).
Figure two. Diagram of a microsporidian spore.
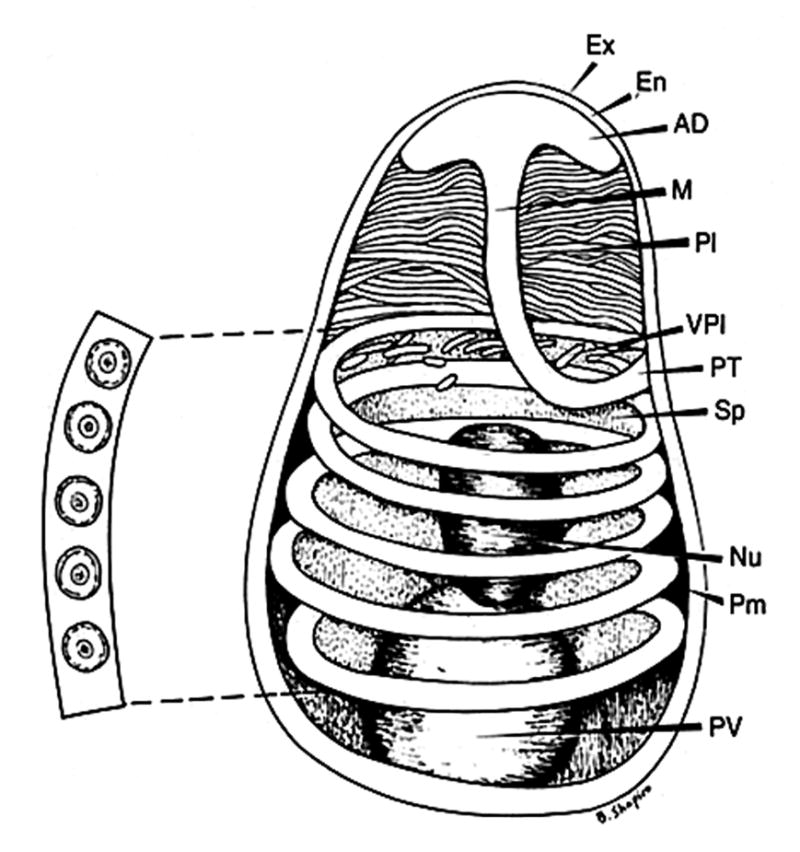
Spores range in size from 1 to 10μm. The spore coat consists of an electron dense exospore (Ex), an electron lucent endospore (En) and plasma membrane (Pm). It is thinner at the anterior end of the spore. The sporoplasm (Sp) contains a single nucleus (Nu), the posterior vacuole (PV) and ribosomes. The polar filament is attached to the anterior end of the spore by an anchoring disc (AD), and is divided into two regions: the manubroid or straight portion (M), and the posterior region forming five coils (PT) around the sporoplasm. The manubroid polar filament is surrounded by the lamellar polaroplast (Pl) and vesicular polaroplast (VPl). The insert depicts a cross section of the polar tube coils (5 coils in this spore), demonstrating the various concentric layers of different electron density and electron dense core present in such cross sections. Reprinted with permission from Wittner M and Weiss LM. (Eds.) The Microsporidia and Microsporidiosis. Washington, DC: ASM Press. 1999 (70).
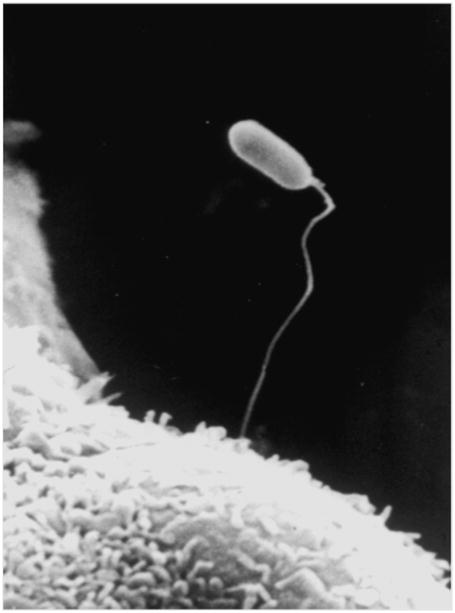
Figure three. Scanning electron micrograph of microsporidia infection of a host cell.
Scanning electron micrograph of extruded polar tube of a spore of Encephalitozoon intestinalis piercing and infecting Vero E6 green monkey kidney cells in tissue culture. Reprinted with permission from Wittner M and Weiss LM. (Eds.) The Microsporidia and Microsporidiosis. Washington, DC: ASM Press. 1999 (70) and with the kind permission of Kock, N.P., C. Schmetz, J. Schottelius, Bernhard Nocht Institute for Tropical Medicine, Hamburg, Germany; published in Kock N.P. 1998. Diagnosis of human pathogen microsporidia (dissertation).
Polar tube discharge from the anterior pole of the spore is a rapid event, and the conditions that promote germination vary widely between different microsporidia and probably reflect the adaptation of a particular microsporidia to a specific host and external environment during transmission of these pathogens (reviewed by Keohane and Weiss (75)). Conditions that promote spore discharge include pH shifts, dehydration followed by rehydration, various cations and anions, mucin or polyanions, hydrogen peroxide, ultraviolet irradiation, and calcium flux (75). In response to their particular activation signal, microsporidia have an increase in intrasporal osmotic pressure resulting in an influx of water into the spore that is accompanied by swelling of the polaroplasts and posterior vacuole prior to spore discharge (76, 77). In A. (N.) algerae, it has been suggested that this is mediated by the breakdown of trehalose by trehalase (76, 78).
3. PHYLOGENETIC ANALYSIS OF THE MICROSPORIDIA
Microsporidia have several apomorphic characteristics including the polar tube, the posterior vacuole, the polaroplast, and the diplokaryon (in some species) that distinguish them as a taxon. These ultrastructural features and phenotypic, developmental and ecological characters have been the basis of the classical classification of these organisms. Tuzet et al (79), Sprague (9);, Larsson (80), Issi (81), Weiser (82), Sprague et al (83), and Weiss and Becnel (2) have all provided useful overviews of the history, ultrastructural and structural characteristics, and life cycle differences for the various microsporidian taxon. Microsporidia can be divided into three main groups: (A.) The “primitive” (Metchnikovellidae) hyperparasites of gregarines in annelids that have a rudimentary polar filament and a spore without a polaroplast.; (B) The Chytridiopsidae, Hesseidsae, and Burkeidae which have a short polar filament and minimal development of the polaroplast and endospore; and (C) The “higher” Microsporidia that have a well-developed polar filament, polaroplast, and posterior vacuole. The various modern classification systems for the microsporidia focus on the characteristics used to divide the third group into subgroups. These systems, however, have problems as many of these characteristics may be convergent. For example, the characteristics of being diplokaryotic during the microsporidian life cycle or the presence of a pansporoblastic membrane are not defining characteristics, as the genus Nosema, defined as being diplokaryotic throughout its life cycle, is actually a polyphyletic assemblage of unrelated taxa. To this end, Nosema locustae has been redefined as both Paranosema (Sokolova et al., 2003, Sokolova et al., 2005) and Antonospora, (Slamovits et al, 2004), Nosema kingi is now Tubulinosema kingi (Franzen et al., 2005), Nosema algerae has been redefined as Anncaliia algerae (Franzen et al., 2006), and Nosema cristatella (Canning et al., 1997) is now Pseudonosema cristatella (Canning, 2002).
Currently over 3000 partial and complete small subunit ribosomal RNA (ssrRNA) genes of various microsporidia species and isolates are accessible on Genbank (http://www.ncbi.nlm.nih.gov/genbank/) and MicrosporidiaDB (http://microsporidiadb.org/micro/). This ssrRNA sequence data has been useful in the development of diagnostic polymerase chain reaction (PCR) primers as well as for investigations into phylogenetic relationships (84, 85) (reviewed by Weiss and Vossbrinck (25)). The ssrRNA sequence of microsporidia diverges greatly from that of other eukaryotes, it is significantly shorter than other eukaryotic ssRNAs and microsporidian ssRNA lacks loops that are present in either eukaryotic and/or prokaryotic ssrRNA (11, 86–88). This divergence of ssRNA sequence structure while once thought to be evidence of microsporidia being an early branch in evolution instead is due to the specialization and extremely reduced genome size reduction of these obligate intracellular pathogens (11). The genome size of the microsporidia varies from 2.3 to 51.3 Mb (89, 90). The genome size of microsporidia in the family Encephalitizoonidae (all of which are human pathogens) are all under 3.0 Mb, making these genomes among the smallest eukaryotic nuclear genomes (91–96). There are almost no introns in these compact genomes, gene density is high, proteins are shorter than the corresponding genes in Saccharomyces cerevisiae, and there is a high degree of gene composition conservation among the various microsporidia (91–96). Chromosomal analysis of E. cuniculi (97, 98) and studies on the heterogeneity of gene loci in both E. cuniculi (99) and Nematocida parisii (100) indicate that microsporidia are probably diploid. Data on Nosema ceranae has further suggested that this microsporidium is tetraploid (101). Several microsporidian genomes have been sequenced and annotated and this data is available on MicrosporidiaDB (http://microsporidiadb.org/micro/) (102).
The development of molecular data, e.g. ssrRNA and now genomic sequencing has resulted in the reassignment of many genera as well as the erection of new genera. It is clear that descriptions of new microsporidian species should now include detailed microscopic and ultrastructural images along with supporting molecular data, e.g. ssrRNA sequence (2, 89, 103). Studies using ssRNA have demonstrated that some microsporidia thought to be separate species are synonymous, e.g. Nosema bombycis and N. tichoplusiae (104), and confirmed that other microsporidia, e.g. Encephalitozoon hellem and E. intestinalis, are indeed separate species (105). A reasonable approach for integration of molecular data and other characters used for microsporidian classification is to create molecular phylogenies of the microsporidia and then to place the non-molecular data upon the tree (84). This type of analysis should facilitate studies that seek to understand the evolution of these organisms, including features such as the loss and gain of alternate hosts, host switching, loss and gain of sexual recombination, use of the pansporoblastic membrane, changes in the numbers of coils in the polar filament versus tissues infected, strategies of generalist versus specialist parasites, and changes in genome size and composition (103). Studies using this technique have demonstrated a significant degree of homoplasy (convergence, parallelism and reversal) among the microsporidia, which probably reflects an effect of the reduced genome size seen in these pathogenic protists. Host-parasite co-speciation may have occurred with microsporidia and has been examined in Nosema species from bees (106, 107), and in the Amblyosporidae from mosquitoes (108, 109). Several other studies have examined the phylogeny of microsporidia isolated from various host types including fish (110, 111) insects (2, 112–114) and crustaceans (115, 116). It should be noted that discrepancies exist between phylogenies based on traditional characters and those based on molecular analysis and that these types of discrepancies have not been fully resolved (103).
Molecular phylogenetic data provide several lines of strong evidence that microsporidia are related to the Fungi as either a basal branch of the Fungi or a sister group (12, 13, 117)}. Analysis of α-tubulin, β-tubulin, glutamyl-tRNA synthetase, seryl-tRNA synthetase, vacuolar ATPase, TATA box binding protein, transcription initiation factor IIB, subunit A of vacuolar ATPase, guanosine triphosphate (GTP)-binding protein, heat shock protein gene (hsp70), the largest sub unit of RNA polymerase II (RPB1) and transcription factor IIB sequences demonstrate that microsporidia are not primitive eukaryotes but instead are related to fungi (117–127). Microsporidian ssrRNA genes lack a paromomycin binding site, similar to fungi (128). Microsporidian genomes display two syntenic ribosomal genes (RPL21 and RPS9) that are also syntenic throughout the fungi, but are linked in other eukarytoic lineages (129, 130). The microsporidian EF-1α gene has an insertion that is found only in fungi and animals, not in protozoa (12, 128, 131, 132). Analysis of the E. cuniculi genome and other microsporidian genomes demonstrates that most microsporidian proteins are most similar to fungal homologues.(13, 14, 91, 95, 96, 133). Biological evidence also supports the relationship of microsporidia to fungi. Microsporidia have chitin in their spore wall and store trehalose, as do fungi (69, 78). Microsporidia display similarities to the fungi during mitosis (e.g., closed mitosis and spindle pole bodies (134)) and meiosis (135). Data suggests that trehalose is probably the major sugar reserve in microsporidia, as is seen in many fungi (2). O-mannosylation, as seen in fungi, occurs in microsporidia as demonstrated by studies on major polar tube protein PTP1 (136).
Keeling (137), in an analysis of β-tubulin data that included microsporidia and representatives of many fungal phyla, found evidence that microsporidia were a sister group to the Zygomycota. Comparative analysis utilizing the complete genomes of several microsporidia also supports a similar relationship of microsporidia with the Fungi (13, 92, 130) Analysis of a six gene dataset (16) that included a large diversity of Fungi suggested that microsporidia branched deeply forming a sister clade to the Fungi with Rozella, a hyper-parasitic chytrid that infects other chytrids. Analyses of individual gene trees, supertrees, and trees based on concatenation of hundreds of genes also supported a deep-branching position as a sister group to the Fungi (13, 138). However, these data, while suggestive, do not definitively distinguish if the microsporidia (and Rozella) are within the Fungi or sisters to the Fungi. Environmental microbiome studies have defined a previously unrecognized group of organisms termed the Cryptomycota (139–143), which are a large and diverse lineage, including aphelids and Rozella, of Fungi or fungal relatives that are poorly described at the biological level, but that are closely related to the microsporidia. The most recent genome-scale phylogenies place microsporidia together with Cryptomycota as the basal branch of the fungal kingdom (or alternatively as a sister phylum). These, and other, recently described microsporidia-like organisms, e.g. Mitosporidium daphniae (144), Nucleophaga terricolae and Paramicrosporidium (15, 145, 146), illustrate that more environmental sampling and genome sequencing will be needed to fully resolve the relationship of microsporidia and the Fungi, as well as to provide a better understanding of the Cryptomycota and reveal what this group has to teach us about origin and diversification of the microsporidia.
4. STRUCTURE AND COMPOSITON OF THE MICROSPORIDIAN SPORE
The spore is the infectious stage of the microsporidian life cycle stage and the only stage that can survive outside the host cell. The shape of the microsporidian spore is usually pyriform or oval, and the spores vary in size from approximately 1 to 12μm, although some needle-like spores can be as long as 20μm (9). The spore has three general features: the spore coat, the sporoplasm, and the invasion apparatus (147). The spore coat contains three layers: an electron-dense outer exospore layer, an electron-lucent inner endospore layer, and the plasma membrane (147). The endospore layer is composed predominantly of chitin and glycoproteins and it is thinnest in the region of anterior anchoring disk complex where rupture occurs during extrusion of the polar tube. Chitin, a major component of the endospore, forms bridges across the endospore and is part of the fibrillar system of the exospore (69). The presence of chitin in the endospore enables microsporidian spores to be stained by fluorescent brighteners such as calcofluor white or Uvitex 2B (148). Both chitin deactylase, in several microsporidia (149), and a Nosema bombycis GH19 chitinase were found to localize to the spore wall (150).
The spore wall protects the organism from harsh environmental conditions permitting transmission of the organism via water or food. It is probable that proteins in the spore wall are involved in processes such as spore adherence, signaling or other interactions with host cells (151). Increased hydrostatic pressure in the spore has been demonstrated to cause discharge of polar tube (152). Several spore wall proteins have been identified using proteomic techniques and the localization of these proteins has been validated by immunofluorescence (IFA) and ultrastructural (immunoEM) studies. The first SWP to be identified was E. cuniculi spore wall protein 1 (EcSWP1) (153). Ultrastructural analysis has demonstrated that this protein is found in the exospore. During spore development EcSWP1 was absent in meronts, first seen in early sporonts, and the amount of this protein increased in the exospore during spore maturation (153). Homologs of SWP1 have been identified in E. intestinalis and E. hellem (154, 155). In E. hellem two SWP1 proteins (EhSWP1a, EhSWP1b) were identified. Using monoclonal antibodies to various SWP1 proteins, EiSWP1, EhSWP1a, EhSWP1b were all found to localize to the exospore in mature spores (154, 155). A second SWP protein (SWP2) was identified in E. intestinalis along with EiSWP1. This 150kDa protein shared a high similarity with the N-terminal region of EiSWP1, immuno-EM demonstrated that EiSWP2 localized to the spore surface, and that EiSWP2 became much more abundant as spores continued development into sporoblasts and mature spores. Homologs of EiSWP2 have not been found in E. hellem or E. cuniculi, suggesting that this protein may have a unique role in E. intestinalis. A third spore wall protein (SWP3/EnP2) was identified from E. cuniculi using a proteomic strategy (156, 157). EcSWP3, also called endospore protein 2 (EnP2), has been demonstrated to localize to the endospore adjacent to the plasma membrane (156). Another endospore protein (EnP1) was identified at the same time as SWP3/EnP2 in E. cuniculi (156). EnP1, which is also called SWP4, is a cysteine rich protein, and contains disulfide linkages that may facilitate its binding in the endospore. It has been found that host cell surface sulfated glycosaminoglycans (GAGs) play an important role in the adherence of E. intestinalis and E. cuniculi to host cell surfaces (158). EnP1 has been found to be involved in host cell adherence by its interacting with host cell membrane GAGs through the heparin binding motifs (HBMs) found in EnP1 (151). Homologs of these proteins have been found in other microsporidia. These SWP adhesion domains may facilitate the binding of microsporidian spores to either the cell surface or mucus of the gastrointestinal tract prior to germination. Many of the SWPs that have been identified in various microsporidia have posttranslational glycosylation, often involving mannosylation, which is likely important for spore wall adherence to host cells during invasion. Using various proteomic techniques additional SWPs such as SWP5 (159), SWP6 and SWP7 (L.M. Weiss manuscript in preparation) have also been identified in the Encephalitozoonidae and other putative SWPs have been identified in Nosema bombycis (160–164). Some N. bombycis SWPs have been suggested to function as scaffolding proteins that support other proteins to form the integrated spore wall of microsporidia (162). Many of these newly identified N. bombycis SWPs have not been found in the Encephalitozoonidae suggesting (not surprisingly) that some SWPs may be species (or genus) specific and related to the environment and the specific interactions that are needed for infection between different hosts (e.g. insects versus mammalian hosts). SWPs appear to be specific to microsporidia as homologs of these various SWPs have not been found in other organisms.
5. STRUCTURE AND COMPOSITION OF THE POLAR TUBE
It has been almost 125 years since the initial description of polar tubes and their discharge from spores (165, 166). The polar tube is an invasion organelle that is unique to the microsporidia. While the polar tube superficially resembles the structures seen in Myxosporea (which are related to cnidarians), the microsporidian polar tube has a distinct function acting as a tube rather than as nematocyst. During infection this highly specialized structure is discharged from the spore, forming a hollow tube that delivers the infectious sporoplasm into its host cell’s cytoplasm. The polar tube in different species of microsporidia is variable in diameter, length and arrangement in the spore. The number of coils in a spore can range from 3 to 50 depending on the species of microsporidia (167). Polar tubes can be from 50 to 500μm in length, and its length can be two to three times longer after the polar tube has completely discharged from the spore (73). The diameter of the tube is 0.1 to 0.2μm and during sporoplasm passage the diameter can increase significantly illustrating the flexibility of this structure (73, 168, 169).
The mechanism of spore germination still remains to be definitively determined. It is generally believed that spore germination occurs in several phases that may overlap: (1) activation; (2) an increase in intrasporal osmotic pressure; (3) eversion of the polar tube; and (4) passage of the sporoplasm through the everted polar tube (2). Before germination, the polar tube is coiled inside the mature spores and filled with material when visualized by TEM. After activation, the conditions of which are often specific to a particular species of microsporidia, the polar tube will burst through the anterior pole of spore at the location of the anchoring disk (73, 170) and form a hollow tube (170, 171). This discharge process occurs rapidly with full eversion occurring within 2 seconds (71). Different species of microsporidia require different conditions for germination and this probably depends on the host species target as well as environmental conditions related to transmission of a particular organism. While a universal stimulant for spore activation is not known, many species can be induced to germinate in distilled water containing 1 to 5% hydrogen peroxide (75). Various reports using Anncaliia (Nosema) algerae have demonstrated that successful germination is associated with an increase in intrasporal osmotic pressure and that this osmotic pressure is the force that drives polar tube extrusion, rather than this requiring ATP or another energy source (152). These studies on A. algerae found that the increase in osmotic pressure was related to a decrease in trehalose levels in germinating spores. This suggests that upon activation of spores, the breakdown in compartmentalization brings trehalose in contact with the enzyme trehalase resulting in its digestion into simple sugars increasing spore osmotic pressure (172).
Proteomic studies on the polar tube have begun to define the full complement of proteins that make up this structure which should lead to a better understanding of how the polar tube is formed and functions during invasion (136, 173–188) Polar tube proteins (PTPs) are highly immunogenic in both experimental and natural infections. In a large serosurvey, antibodies reacting to E. cuniculi were present in 5% of pregnant French women and 8% of Dutch blood donors (189). Polar tubes have been found to be insoluble in 1% SDS and 9 M urea and soluble in 2% DTT allowing the purification of potential polar tube proteins (PTPs). Weidner used these solubility properties to isolate potential PTPs from A. michaelis and demonstrated that this protein mixture could self polymerize (169). Using a similar extraction procedure and reverse-phase high performance liquid chromatography (HPLC), a 43kDa PTP designated PTP1, based on it being the major protein by mass in the Glugea americanus DTT solubilized material by (174). Subsequently, additional PTP1s were purified from several microsporidia of the genus Encephalitozoonidae using the same purification protocol (175, 177, 190). PTP1 is a proline-rich protein that contains many cysteine residues in its N-terminal and C-terminal domains. Based on the solubilization of the polar tube by DTT, disulfide bridges probably play an essential role in maintaining this structure. The abundance of proline in PTP1 is consistent with properties seen in the intact polar tube as this amino acid has been reported to increase the tensile strength and elasticity of proteins. Analysis of the glycosylation of PTP1 demonstrated that there are a significant number of O-linked mannosylation sites in this protein (136, 183, 186). Mannose pretreatment of host cells was found to decrease E. hellem infection which was consistent with an interaction between mannosylated PTP1 and some unknown host cell mannose-binding receptor protein (136, 184, 185, 191, 192). Examination of published microsporidian genomes (www.MicrosporidiaDB.org) has demonstrated that PTP1 homologues are not easily identified in all genomes which may reflect the high evolution rates in microsporidia that often make the identification of orthologous proteins difficult. For example, the putative PTP1 of Antonospora locustae, Paranosema grylli and Nosema ceranae were identified based upon a combination of features including the presence of a predicted signal peptide, their similar length, conserved acidic pI and predicted amino acid compositions along with their close physical juxtaposition to another PTP named PTP2 (180); but would not have been identified by a simple BLAST search of the genomes (2, 193–195).
PTP2 was also identified using proteomic and antibody based approaches (180, 184, 196).. Interestingly PTP2 is found at the same genomic locus as PTP1. Preservation of gene order may be due to constraints of evolution on these small eukaryotic genomes (197). PTP2 is more conserved in size than PTP1 and the identified PTP2s from various microsporidia share common characteristics, despite a high degree of sequence divergence, e.g. they have a basic isoelectric point (pI), high lysine content and a conservation of cysteine residues that are most likely involved in intra- and/or inter-protein disulfide bridges (180, 184, 196). PTP2 has been identified in the genome of many microsporidia including Antonospora (Paranosema) locustae, Trachipleistophora hominis, E. cuniculi, E. hellem, E. intestinalis, P. grylli N. cerana, N. bombycis, Enterocytozoon bieneusi, Anncaliia algerae, Tubulinosema ratisbonensis, Nematocida parisii, Vittaforma cornea, and Edhazardia aedis (2, 184, 194, 196).
Immunoscreening of a cDNA library of E. cuniculi led to the identification of a third polar tube protein, PTP3 (182). Unlike both PTP1 and PTP2, it was found that PTP3 is solubilized in the presence of SDS alone without the need for a reducing agent such as DTT (180), explaining why it was not seen in standard polar tube preparations. PTP3 along with PTP1 and PT2 was, however, found in studies examining cross linked polar tube complexes and these three PTPs were also demonstrated to interact in yeast two hybrid assays (182, 185). It has been suggested that PTP3 may act as a scaffolding protein for the assembly of other PTPs in the formation of the polar tube during development. Genome analysis has confirmed the presence of PTP3 orthologs in the genomes of E. intestinalis, E. hellem, E. romalae, N. ceranae, N. bombycis, A. locustae, P. grylli, T. hominis, E. bieneusi, A. algerae, N. parisii, Octosporea bayeri, T. ratisbonensis and Vavria culicis floridensis (2). The sequence of these PTP3 orthologs is more conserved than the sequences of the PTP1 and PTP2 orthologs.
Two new PTPs (PTP4 and PTP5) have been identified in the polar tube using proteomic and antibody based approaches (198). Orthologous PTP4 and PTP5 are present in many microsporidian genomes including E. cuniculi, E. hellem, E. intestinalis, P. grylli, N. ceranae, N. bombycis, A. algerae, O. bayeri, E. aedis, V. cornea, and T. ratisbonensis (2). Similar to the PTP1/PTP2 the genes for PTP4 and PTP5 usually are in a cluster in the genome (2). Monoclonal antibody to PTP4 stains the extruded tip of the polar tube, whereas, the other PTPs (e.g. PTP1, PTP2, PTP3, PTP5) are located uniformly along the entire polar tube (Han and Weiss, paper in submission) (2, 196). This specialized location of PTP4 suggests that this protein is probably involved in the interaction of the polar tube and host cell at the invasion synapse. Recently, employing PTP4 for an immunoprecipitation assay followed by proteomic analysis we have been able to identify a potential host cell receptor for PTP4 and mutant host cells without this receptor have a markedly reduced infection rate (Han and Weiss, paper in submission). The mechanism by which the polar tube interacts with the host cell membrane resulting in penetration is currently unknown; however, there is some evidence that host cell actin may be involved in microsporidian penetration of the host cell within the invasion synapse (199). Furthermore, proteomic studies of the polar tube that are ongoing suggest that there are additional PTPs that remain to be characterized in this structure (Weiss LM unpublished)
6. SUMMARY
Microsporidia are unicellular eukaryotes that develop as obligate intracellular parasites. They have a very unique invasion organelle, the polar tube, which upon appropriate environmental stimulation rapidly discharges out of the spore, pierces a cell membrane, and serves as a conduit for sporoplasm passage into a host cell. Phylogenetic analysis suggests that microsporidia are related to the Fungi, being either a basal branch or sister group. Despite the description of microsporidia over 150 years ago, we still lack an understanding of the mechanism of invasion including the role of various PTPs and SWPs as well as host cell proteins in the formation and function of the invasion synapse. Over the past two decades, proteomic approaches have helped define PTPs and SWPs as well as the importance of post translational modifications such as glycosylation in the functioning of these proteins, but the absence of genetic techniques for the manipulation of microsporidia has hampered research on the function of these various proteins. Recent advances in ultrastructural techniques are, however, helping to better define the role of various PTPs and SWPs in the formation and functioning of the invasion synapse. The study of the mechanism of invasion should provide fundamental insights into the biology of these ubiquitous intracellular pathogens that can be integrated into studies aimed at treating or controlling microsporidiosis.
Acknowledgments
This work was supported by an NIH-NIAID grant AI124753. Bing Han was supported by a Chinese Scholarship Council (File Number 201406990033).
References
- 1.Pasteur L. Études sur la maladie des vers à soie [M] Paris, Gauthier-Villars, successeur de Mallet-Bachelier. 1870;1870:148–168. [Google Scholar]
- 2.Weiss LM, Becnel JJ. Microsporidia: Pathogens of Opportunity. Wiley-Blackwell; Oxford: 2014. [Google Scholar]
- 3.Izquierdo F, Castro Hermida JA, Fenoy S, Mezo M, Gonzalez-Warleta M, del Aguila C. Detection of microsporidia in drinking water, wastewater and recreational rivers. Water Res. 2011;45:4837–43. doi: 10.1016/j.watres.2011.06.033. [DOI] [PubMed] [Google Scholar]
- 4.Williams BA, Hirt RP, Lucocq JM, Embley TM. A mitochondrial remnant in the microsporidian Trachipleistophora hominis. Nature. 2002;418:865–9. doi: 10.1038/nature00949. [DOI] [PubMed] [Google Scholar]
- 5.Fries I. Nosema apis—a parasite in the honey bee colony. Bee World. 1993;74:5–19. [Google Scholar]
- 6.Kent ML, Elliott DG, Groff JM, Hedrick RP. Loma salmonae (Protozoa: Microspora) infections in seawater reared coho salmon Oncorhynchus kisutch. Aquaculture. 1989;80:211–222. [Google Scholar]
- 7.Overstreet RM. Parasites of some penaeid shrimps with emphasis on reared hosts. Aquaculture. 1973;2:105–140. [Google Scholar]
- 8.Franzen C. Microsporidia: a review of 150 years of research. The Open Parasitology Journal. 2008;2:1–34. [Google Scholar]
- 9.Sprague V. In: Systematics of the Microsporidia, vol. 2 Biology of the Microsporidia, Series: Comparative Biology. Bulla LA, Cheng TC, editors. Plenum Press; New York: 1977. [Google Scholar]
- 10.Sprague VV, Becnel JJ. Note on the Name-Author-Date Combination for the Taxon MICROSPORIDIES Balbiani, 1882, When Ranked as a Phylum. J Invertebr Pathol. 1998;71:91–4. doi: 10.1006/jipa.1997.4702. [DOI] [PubMed] [Google Scholar]
- 11.Vossbrinck CF, Maddox JV, Friedman S, Debrunner-Voxxbrinck BA, Woese CR. Ribosomal RNA sequence suggests microsporidia are extremely ancient eukaryotes. Nature. 1987;326(6111):411–414. doi: 10.1038/326411a0. [DOI] [PubMed] [Google Scholar]
- 12.Weiss LM, Edlind TD, Vossbrinck CR, Hashimoto T. Microsporidian molecular phylogeny: the fungal connection. J Eukaryot Microbiol. 1999;46:17S–18S. [PubMed] [Google Scholar]
- 13.Capella-Gutierrez S, Marcet-Houben M, Gabaldon T. Phylogenomics supports microsporidia as the earliest diverging clade of sequenced fungi. BMC Biol. 2012;10:47. doi: 10.1186/1741-7007-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee SC, Corradi N, Byrnes EJ, 3rd, Torres-Martinez S, Dietrich FS, Keeling PJ, Heitman J. Microsporidia evolved from ancestral sexual fungi. Curr Biol. 2008;18:1675–9. doi: 10.1016/j.cub.2008.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corsaro D, Michel R, Walochnik J, Venditti D, Muller KD, Hauroder B, Wylezich C. Molecular identification of Nucleophaga terricolae sp. nov. (Rozellomycota), and new insights on the origin of the Microsporidia. Parasitol Res. 2016;115:3003–3011. doi: 10.1007/s00436-016-5055-9. [DOI] [PubMed] [Google Scholar]
- 16.James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, Celio G, Gueidan C, Fraker E, Miadlikowska J, Lumbsch HT, Rauhut A, Reeb V, Arnold AE, Amtoft A, Stajich JE, Hosaka K, Sung GH, Johnson D, O’Rourke B, Crockett M, Binder M, Curtis JM, Slot JC, Wang Z, Wilson AW, Schussler A, Longcore JE, O’Donnell K, Mozley-Standridge S, Porter D, Letcher PM, Powell MJ, Taylor JW, White MM, Griffith GW, Davies DR, Humber RA, Morton JB, Sugiyama J, Rossman AY, Rogers JD, Pfister DH, Hewitt D, Hansen K, Hambleton S, Shoemaker RA, Kohlmeyer J, Volkmann-Kohlmeyer B, Spotts RA, Serdani M, Crous PW, Hughes KW, Matsuura K, Langer E, Langer G, Untereiner WA, Lucking R, Budel B, Geiser DM, Aptroot A, Diederich P, Schmitt I, Schultz M, Yahr R, Hibbett DS, Lutzoni F, McLaughlin DJ, Spatafora JW, Vilgalys R. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature. 2006;443:818–22. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- 17.Weber R, Bryan RT, Schwartz DA, Owen RL. Human microsporidial infections. Clinical Microbiology Reviews. 1994;7:426–461. doi: 10.1128/cmr.7.4.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Troemel ER, Félix MA, Whiteman NK, Barrière A, Ausubel FM. Microsporidia are natural intracellular parasites of the nematode Caenorhabditis elegans. PLoS Biol. 2008;6:e309. doi: 10.1371/journal.pbio.0060309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma Z, Li C, Pan G, Li Z, Han B, Xu J, Lan X, Chen J, Yang D, Chen Q, Sang Q, Ji X, Li T, Long M, Zhou Z. Genome-wide transcriptional response of silkworm (Bombyx mori) to Infection by the microsporidian Nosema bombycis. PLoS One. 2013;8:e84137. doi: 10.1371/journal.pone.0084137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Antúnez K, Martín-Hernández R, Prieto L, Meana A, Zunino P, Higes M. Immune suppression in the honey bee (Apis mellifera) following infection by Nosema ceranae (Microsporidia) Environmental microbiology. 2009;11:2284–2290. doi: 10.1111/j.1462-2920.2009.01953.x. [DOI] [PubMed] [Google Scholar]
- 21.Weber R, Deplazes P, Schwartz D. Diagnosis and clinical aspects of human microsporidiosis. Contrib Microbiol. 2000;6:166–92. doi: 10.1159/000060360. [DOI] [PubMed] [Google Scholar]
- 22.Coyle CM, Weiss LM, Rhodes LV, 3rd, Cali A, Takvorian PM, Brown DF, Visvesvara GS, Xiao L, Naktin J, Young E, Gareca M, Colasante G, Wittner M. Fatal myositis due to the microsporidian Brachiola algerae, a mosquito pathogen. N Engl J Med. 2004;351:42–7. doi: 10.1056/NEJMoa032655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Texier C, Vidau C, Viguès B, El Alaoui H, Delbac F. Microsporidia: a model for minimal parasite–host interactions. Current opinion in microbiology. 2010;13:443–449. doi: 10.1016/j.mib.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 24.Desportes I, Charpentier YL, Galian A, Bernard F, Cochand-Priollet B, Lavergne A, Ravisse P, Modigliani R. Occurrence of a new microsporidan: Enterocytozoon bieneusi ng, n. sp., in the enterocytes of a human patient with AIDS. The Journal of Protozoology. 1985;32:250–254. doi: 10.1111/j.1550-7408.1985.tb03046.x. [DOI] [PubMed] [Google Scholar]
- 25.Weiss LM, Vossbrinck CR. Microsporidiosis: molecular and diagnostic aspects. Adv Parasitol. 1998;40:351–95. doi: 10.1016/s0065-308x(08)60127-x. [DOI] [PubMed] [Google Scholar]
- 26.Didier ES, Weiss LM. Microsporidiosis: not just in AIDS patients. Current opinion in infectious diseases. 2011;24:490. doi: 10.1097/QCO.0b013e32834aa152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Didier ES, Varner PW, Didier PJ, Aldras AM, Millichamp NJ, Murphey-Corb M, Bohm R, Shadduck JA. Experimental microsporidiosis in immunocompetent and immunodeficient mice and monkeys. Folia Parasitol (Praha) 1994;41:1–11. [PubMed] [Google Scholar]
- 28.Ghosh K, Weiss LM. T cell response and persistence of the microsporidia. FEMS Microbiol Rev. 2012;36:748–60. doi: 10.1111/j.1574-6976.2011.00318.x. [DOI] [PubMed] [Google Scholar]
- 29.Khan IA, Moretto M, Weiss LM. Immune response to Encephalitozoon cuniculi infection. Microbes and infection. 2001;3:401–405. doi: 10.1016/s1286-4579(01)01397-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mathews A, Hotard A, Hale-Donze H. Innate immune responses to Encephalitozoon species infections. Microbes and Infection. 2009;11:905–911. doi: 10.1016/j.micinf.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 31.Achbarou A, Ombrouck C, Gneragbe T, Charlotte F, Renia L, Desportes-Livage I, Mazier D. Experimental model for human intestinal microsporidiosis in interferon gamma receptor knockout mice infected by Encephalitozoon intestinalis. Parasite Immunol. 1996;18:387–92. doi: 10.1046/j.1365-3024.1996.d01-128.x. [DOI] [PubMed] [Google Scholar]
- 32.Khan IA, Schwartzman JD, Kasper LH, Moretto M. CD8+ CTLs are essential for protective immunity against Encephalitozoon cuniculi infection. The Journal of Immunology. 1999;162:6086–6091. [PubMed] [Google Scholar]
- 33.Moretto MM, I, Khan A, Weiss LM. Gastrointestinal cell mediated immunity and the microsporidia. PLoS Pathog. 2012;8:e1002775. doi: 10.1371/journal.ppat.1002775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moretto MM, Lawlor EM, Khan IA. Lack of interleukin-12 in p40-deficient mice leads to poor CD8+ T-cell immunity against Encephalitozoon cuniculi infection. Infect Immun. 2010;78:2505–11. doi: 10.1128/IAI.00753-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hermanek J, Koudela B, Kucerova Z, Ditrich O, Travnicek J. Prophylactic and therapeutic immune reconstitution of SCID mice infected with Encephalitozoon cuniculi. Folia Parasitol (Praha) 1993;40:287–91. [PubMed] [Google Scholar]
- 36.Schmidt EC, Shadduck JA. Mechanisms of resistance to the intracellular protozoan Encephalitozoon cuniculi in mice. J Immunol. 1984;133:2712–9. [PubMed] [Google Scholar]
- 37.Wong P, Pamer EG. CD8 T cell responses to infectious pathogens. Annu Rev Immunol. 2003;21:29–70. doi: 10.1146/annurev.immunol.21.120601.141114. [DOI] [PubMed] [Google Scholar]
- 38.Moretto M, Weiss LM, Khan IA. Induction of a rapid and strong antigen-specific intraepithelial lymphocyte response during oral Encephalitozoon cuniculi infection. The Journal of Immunology. 2004;172:4402–4409. doi: 10.4049/jimmunol.172.7.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moretto MM, Weiss LM, Combe CL, Khan IA. IFN-γ-producing dendritic cells are important for priming of gut intraepithelial lymphocyte response against intracellular parasitic infection. The Journal of Immunology. 2007;179:2485–2492. doi: 10.4049/jimmunol.179.4.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moretto MM, Lawlor EM, Khan IA. Aging mice exhibit a functional defect in mucosal dendritic cell response against an intracellular pathogen. The Journal of Immunology. 2008;181:7977–7984. doi: 10.4049/jimmunol.181.11.7977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmidt EC, Shadduck JA. Murine encephalitozoonosis model for studying the host-parasite relationship of a chronic infection. Infect Immun. 1983;40:936–42. doi: 10.1128/iai.40.3.936-942.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sak B, Salat J, Horka H, Sakova K, Ditrich O. Antibodies enhance the protective effect of CD4+ T lymphocytes in SCID mice perorally infected with Encephalitozoon cuniculi. Parasite Immunol. 2006;28:95–9. doi: 10.1111/j.1365-3024.2005.00813.x. [DOI] [PubMed] [Google Scholar]
- 43.Bywater JE, Kellett BS. Humoral immune response to natural infection with Encephalitozoon cuniculi in rabbits. Lab Anim. 1979;13:293–7. doi: 10.1258/002367779780943305. [DOI] [PubMed] [Google Scholar]
- 44.Silveira H, Canning EU. Vittaforma corneae n. comb. for the human microsporidium Nosema corneum Shadduck, Meccoli, Davis & Font, 1990, based on its ultrastructure in the liver of experimentally infected athymic mice. J Eukaryot Microbiol. 1995;42:158–65. doi: 10.1111/j.1550-7408.1995.tb01557.x. [DOI] [PubMed] [Google Scholar]
- 45.Cali A, Takvorian PM, Lewin S, Rendel M, Sian CS, Wittner M, Tanowitz HB, Keohane E, Weiss LM. Brachiola vesicularum, n. g., n. sp., a new microsporidium associated with AIDS and myositis. J Eukaryot Microbiol. 1998;45:240–51. doi: 10.1111/j.1550-7408.1998.tb04532.x. [DOI] [PubMed] [Google Scholar]
- 46.Franzen C, Nassonova ES, Scholmerich J, Issi IV. Transfer of the members of the genus Brachiola (microsporidia) to the genus Anncaliia based on ultrastructural and molecular data. J Eukaryot Microbiol. 2006;53:26–35. doi: 10.1111/j.1550-7408.2005.00066.x. [DOI] [PubMed] [Google Scholar]
- 47.Cali A, Weiss LM, Takvorian PM. A review of the development of two types of human skeletal muscle infections from microsporidia associated with pathology in invertebrates and cold-blooded vertebrates. Folia Parasitol (Praha) 2005;52:51–61. doi: 10.14411/fp.2005.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weber R, Bryan RT. Microsporidial infections in immunodeficient and immunocompetent patients. Clin Infect Dis. 1994;19:517–21. doi: 10.1093/clinids/19.3.517. [DOI] [PubMed] [Google Scholar]
- 49.Didier ES, Didier PJ, Friedberg DN, Stenson SM, Orenstein JM, Yee RW, Tio FO, Davis RM, Vossbrinck C, Millichamp N, et al. Isolation and characterization of a new human microsporidian, Encephalitozoon hellem (n. sp.), from three AIDS patients with keratoconjunctivitis. J Infect Dis. 1991;163:617–21. doi: 10.1093/infdis/163.3.617. [DOI] [PubMed] [Google Scholar]
- 50.Cali A, Kotler DP, Orenstein JM. Septata intestinalis n. g., n. sp., an intestinal microsporidian associated with chronic diarrhea and dissemination in AIDS patients. J Eukaryot Microbiol. 1993;40:101–12. doi: 10.1111/j.1550-7408.1993.tb04889.x. [DOI] [PubMed] [Google Scholar]
- 51.Hartskeerl RA, Van Gool T, Schuitema AR, Didier ES, Terpstra WJ. Genetic and immunological characterization of the microsporidian Septata intestinalis Cali, Kotler and Orenstein, 1993: reclassification to Encephalitozoon intestinalis. Parasitology. 1995;110(Pt 3):277–85. doi: 10.1017/s0031182000080860. [DOI] [PubMed] [Google Scholar]
- 52.Vavra J, Yachnis AT, Shadduck JA, Orenstein JM. Microsporidia of the genus Trachipleistophora--causative agents of human microsporidiosis: description of Trachipleistophora anthropophthera n. sp. (Protozoa: Microsporidia) J Eukaryot Microbiol. 1998;45:273–83. doi: 10.1111/j.1550-7408.1998.tb04536.x. [DOI] [PubMed] [Google Scholar]
- 53.Yachnis AT, Berg J, Martinez-Salazar A, Bender BS, Diaz L, Rojiani AM, Eskin TA, Orenstein JM. Disseminated microsporidiosis especially infecting the brain, heart, and kidneys. Report of a newly recognized pansporoblastic species in two symptomatic AIDS patients. Am J Clin Pathol. 1996;106:535–43. doi: 10.1093/ajcp/106.4.535. [DOI] [PubMed] [Google Scholar]
- 54.Field AS, Marriott DJ, Milliken ST, Brew BJ, Canning EU, Kench JG, Darveniza P, Harkness JL. Myositis associated with a newly described microsporidian, Trachipleistophora hominis, in a patient with AIDS. J Clin Microbiol. 1996;34:2803–11. doi: 10.1128/jcm.34.11.2803-2811.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meissner EG, Bennett JE, Qvarnstrom Y, da Silva A, Chu EY, Tsokos M, Gea-Banacloche J. Disseminated microsporidiosis in an immunosuppressed patient. Emerg Infect Dis. 2012;18:1155–8. doi: 10.3201/eid1807.120047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choudhary MM, Metcalfe MG, Arrambide K, Bern C, Visvesvara GS, Pieniazek NJ, Bandea RD, Deleon-Carnes M, Adem P, Zaki SR, Saeed MU. Tubulinosema sp. microsporidian myositis in immunosuppressed patient. Emerg Infect Dis. 2011;17:1727–30. doi: 10.3201/eid1709.101926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suankratay C, Thiansukhon E, Nilaratanakul V, Putaporntip C, Jongwutiwes S. Disseminated infection caused by novel species of Microsporidium, Thailand. Emerg Infect Dis. 2012;18:302–4. doi: 10.3201/eid1802.111319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Costa SF, Weiss LM. Drug treatment of microsporidiosis. Drug Resist Updat. 2000;3:384–399. doi: 10.1054/drup.2000.0174. [DOI] [PubMed] [Google Scholar]
- 59.Maggi P, Larocca AM, Quarto M, Serio G, Brandonisio O, Angarano G, Pastore G. Effect of antiretroviral therapy on cryptosporidiosis and microsporidiosis in patients infected with human immunodeficiency virus type 1. Eur J Clin Microbiol Infect Dis. 2000;19:213–7. doi: 10.1007/s100960050461. [DOI] [PubMed] [Google Scholar]
- 60.Goguel J, Katlama C, Sarfati C, Maslo C, Leport C, Molina JM. Remission of AIDS-associated intestinal microsporidiosis with highly active antiretroviral therapy. Aids. 1997;11:1658–9. [PubMed] [Google Scholar]
- 61.Akiyoshi DE, Weiss LM, Feng X, Williams BA, Keeling PJ, Zhang Q, Tzipori S. Analysis of the beta-tubulin genes from Enterocytozoon bieneusi isolates from a human and rhesus macaque. J Eukaryot Microbiol. 2007;54:38–41. doi: 10.1111/j.1550-7408.2006.00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Franzen C, Salzberger B. Analysis of the beta-tubulin gene from Vittaforma corneae suggests benzimidazole resistance. Antimicrob Agents Chemother. 2008;52:790–3. doi: 10.1128/AAC.00928-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Didier PJ, Phillips JN, Kuebler DJ, Nasr M, Brindley PJ, Stovall ME, Bowers LC, Didier ES. Antimicrosporidial activities of fumagillin, TNP-470, ovalicin, and ovalicin derivatives in vitro and in vivo. Antimicrob Agents Chemother. 2006;50:2146–55. doi: 10.1128/AAC.00020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Molina JM, Tourneur M, Sarfati C, Chevret S, de Gouvello A, Gobert JG, Balkan S, Derouin F. Fumagillin treatment of intestinal microsporidiosis. N Engl J Med. 2002;346:1963–9. doi: 10.1056/NEJMoa012924. [DOI] [PubMed] [Google Scholar]
- 65.Bicart-See A, Massip P, Linas MD, Datry A. Successful treatment with nitazoxanide of Enterocytozoon bieneusi microsporidiosis in a patient with AIDS. Antimicrob Agents Chemother. 2000;44:167–8. doi: 10.1128/aac.44.1.167-168.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Canning EU. Transmission of microsporida. Proc Int Colloq Insect Pathol. 1970;4:415–424. [Google Scholar]
- 67.Canning EU. Phylum Microspora. Jones and Bartlett; Boston: 1991. [Google Scholar]
- 68.Didier ES. Microsporidiosis: an emerging and opportunistic infection in humans and animals. Acta Trop. 2005;94:61–76. doi: 10.1016/j.actatropica.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 69.Vavra J, Sprague V. In: Biology of the Microsporidia, Series: Comparative Biology. Bulla LA, Cheng TC, editors. Vol. 1. Plenum Press; New York: 1976. [Google Scholar]
- 70.Wittner M, WL . The Microsporidia and Microsporidiosis. ASM Press; Washington, D.C: 1999. [Google Scholar]
- 71.Lom J. On the structure of the extruded microsporidian polar filament. Z Parasitenkd. 1972;38:200–213. [Google Scholar]
- 72.Weidner E. Cell invasion by microsporidian spores: An ultrastructural study. J Protozool. 1971;18(Suppl):13–14. [Google Scholar]
- 73.Weidner E. Ultrastructural study of microsporidian invasion into cells. Zeitschrift für Parasitenkunde. 1972;40:227–242. doi: 10.1007/BF00329623. [DOI] [PubMed] [Google Scholar]
- 74.Franzen C. How do microsporidia invade cells? Folia Parasitol (Praha) 2005;52:36–40. doi: 10.14411/fp.2005.005. [DOI] [PubMed] [Google Scholar]
- 75.Keohane E, Weiss LM. The Structure, Function, and Composition of the Microsporidian Polar Tube. In: Wittner M, Weiss LM, editors. The Microsporidia and Microsporidiosis. ASM Press; Washington, D.C: 1999. pp. 196–224. [Google Scholar]
- 76.Undeen AH, ElGazzar LM, Vandermeer RK, Narang S. Trehalose levels and trehalase activity in germinated and ungerminated spores of Nosema algerae (Microspora: Nosematidae) J Invertebr Pathol. 1987;50:230–237. [Google Scholar]
- 77.Frixione E, Ruiz L, Cerbon J, Undeen AH. Germination of Nosema algerae (Microspora) spores: conditional inhibition by D-2O, ethanol and Hg-2+ suggests dependence of water influx upon membrane hydration and specific transmembrane pathways. Journal Of Eukaryotic Microbiology. 1997;44:109–116. doi: 10.1111/j.1550-7408.1997.tb05946.x. [DOI] [PubMed] [Google Scholar]
- 78.Undeen AH, Vander Meer RK. Microsporidian intrasporal sugars and their role in germination. Journal of invertebrate pathology. 1999;73:294–302. doi: 10.1006/jipa.1998.4834. [DOI] [PubMed] [Google Scholar]
- 79.Tuzet O, Maurand J, Fize JA, Michel R, Fenwick B. Proposition d’un nouveau cadre systematique pour les genres de Microsporidies. C R Acad Sci (Paris) 1971;272:1268–1271. [Google Scholar]
- 80.Larsson JIR. Identification of micrsporidian genera - a guide with comments on the taxonomy. Arch Protistenkd. 1988;136:1–37. [Google Scholar]
- 81.Issi IV. Microsporidia as a phylum of parasitic protozoa. Protozoology. 1986;10:6–135. (in Russian) [Google Scholar]
- 82.Weiser J. A proposal of the basis for microsporidian taxonomy. Proc Int Congr Protozool. 1977;5:267. [Google Scholar]
- 83.Sprague V, Becnel JJ, Hazard EI. Taxonomy of phylum Microspora. Critical reviews in microbiology. 1992;18:285–395. doi: 10.3109/10408419209113519. [DOI] [PubMed] [Google Scholar]
- 84.Baker MD, Vossbrinck CR, Maddox JV, Undeen AH. Phylogenetic relationships among Vairimorpha and Nosema species (Microspora) based on ribosomal RNA sequence data. J Invertebr Pathol. 1994;64:100–6. doi: 10.1006/jipa.1994.1077. [DOI] [PubMed] [Google Scholar]
- 85.Franzen C, Muller A. Molecular techniques for detection, species differentiation, and phylogenetic analysis of microsporidia. Clin Microbiol Rev. 1999;12:243–85. doi: 10.1128/cmr.12.2.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Curgy JJ, Vavra J, Vivares C. Presence of ribosomal RNAs with prokaryotic properties in Microsporidia, eukaryotic organisms. Biol Cell. 1980;38:49–52. [Google Scholar]
- 87.Vossbrinck CR, Woese CR. Eukaryotic ribosomes that lack a 5.8S RNA. Nature. 1986;320:287–288. doi: 10.1038/320287a0. [DOI] [PubMed] [Google Scholar]
- 88.Azevedo C, Corral L, Vivares CP. Ultrastructure of the microsporidian Inodosporus octospora (Thelohaniidae), a parasite of the shrimp Palaemon serratus (Crustacea, Decapoda) Dis Aquat Organ. 2000;41:151–8. doi: 10.3354/dao041151. [DOI] [PubMed] [Google Scholar]
- 89.Weiss LM. Molecular phylogeny and diagnostic approaches to microsporidia. Contrib Microbiol. 2000;6:209–35. doi: 10.1159/000060362. [DOI] [PubMed] [Google Scholar]
- 90.Ndikumana S, Pelin A, Williot A, Sanders JL, Kent M, Corradi N. Genome analysis of Pseudoloma neurophilia: a microsporidian parasite of zebrafish (Danio rerio) J Eukaryot Microbiol. 2016 doi: 10.1111/jeu.12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Katinka MD, Duprat S, Cornillot E, Metenier G, Thomarat F, Prensier G, Barbe V, Peyretaillade E, Brottier P, Wincker P, Delbac F, El Alaoui H, Peyret P, Saurin W, Gouy M, Weissenbach J, Vivares CP. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414:450–3. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- 92.Corradi N. Microsporidia: Eukaryotic Intracellular Parasites Shaped by Gene Loss and Horizontal Gene Transfers. Annu Rev Microbiol. 2015;69:167–83. doi: 10.1146/annurev-micro-091014-104136. [DOI] [PubMed] [Google Scholar]
- 93.Corradi N, Akiyoshi DE, Morrison HG, Feng X, Weiss LM, Tzipori S, Keeling PJ. Patterns of genome evolution among the microsporidian parasites Encephalitozoon cuniculi, Antonospora locustae and Enterocytozoon bieneusi. PLoS One. 2007;2:e1277. doi: 10.1371/journal.pone.0001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Corradi N, Pombert JF, Farinelli L, Didier ES, Keeling PJ. The complete sequence of the smallest known nuclear genome from the microsporidian Encephalitozoon intestinalis. Nat Commun. 2010;1:77. doi: 10.1038/ncomms1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Corradi N, Selman M. Latest progress in microsporidian genome research. J Eukaryot Microbiol. 2013;60:309–12. doi: 10.1111/jeu.12030. [DOI] [PubMed] [Google Scholar]
- 96.Corradi N, Slamovits CH. The intriguing nature of microsporidian genomes. Brief Funct Genomics. 2011;10:115–24. doi: 10.1093/bfgp/elq032. [DOI] [PubMed] [Google Scholar]
- 97.Brugere JF, Cornillot E, Metenier G, Bensimon A, Vivares CP. Encephalitozoon cuniculi (Microspora) genome: physical map and evidence for telomere-associated rDNA units on all chromosomes. Nucleic Acids Res. 2000;28:2026–33. doi: 10.1093/nar/28.10.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Brugere JF, Cornillot E, Metenier G, Vivares CP. Occurence of subtelomeric rearrangements in the genome of the microsporidian parasite Encephalitozoon cuniculi, as revealed by a new fingerprinting procedure based on two-dimensional pulsed field gel electrophoresis. Electrophoresis. 2000;21:2576–81. doi: 10.1002/1522-2683(20000701)21:12<2576::AID-ELPS2576>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 99.Selman M, Sak B, Kvac M, Farinelli L, Weiss LM, Corradi N. Extremely reduced levels of heterozygosity in the vertebrate pathogen Encephalitozoon cuniculi. Eukaryot Cell. 2013;12:496–502. doi: 10.1128/EC.00307-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cuomo CA, Desjardins CA, Bakowski MA, Goldberg J, Ma AT, Becnel JJ, Didier ES, Fan L, Heiman DI, Levin JZ, Young S, Zeng Q, Troemel ER. Microsporidian genome analysis reveals evolutionary strategies for obligate intracellular growth. Genome Res. 2012;22:2478–88. doi: 10.1101/gr.142802.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pelin A, Selman M, Aris-Brosou S, Farinelli L, Corradi N. Genome analyses suggest the presence of polyploidy and recent human-driven expansions in eight global populations of the honeybee pathogen Nosema ceranae. Environ Microbiol. 2015;17:4443–58. doi: 10.1111/1462-2920.12883. [DOI] [PubMed] [Google Scholar]
- 102.Aurrecoechea C, Barreto A, Brestelli J, Brunk BP, Caler EV, Fischer S, Gajria B, Gao X, Gingle A, Grant G, Harb OS, Heiges M, Iodice J, Kissinger JC, Kraemer ET, Li W, Nayak V, Pennington C, Pinney DF, Pitts B, Roos DS, Srinivasamoorthy G, Stoeckert CJ, Jr, Treatman C, Wang H. AmoebaDB and MicrosporidiaDB: functional genomic resources for Amoebozoa and Microsporidia species. Nucleic Acids Res. 2011;39:D612–9. doi: 10.1093/nar/gkq1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vossbrinck CR, Debrunner-Vossbrinck BA. Molecular phylogeny of the Microsporidia: ecological, ultrastructural and taxonomic considerations. Folia Parasitol (Praha) 2005;52:131–42. doi: 10.14411/fp.2005.017. discussion 130. [DOI] [PubMed] [Google Scholar]
- 104.Pieniazek NJ, da Silva AJ, Slemenda SB, Visvesvara GS, Kurtti TJ, Yasunaga C. Nosema trichoplusiae is a synonym of Nosema bombycis based on the sequence of the small subunit ribosomal RNA coding region. J Invertebr Pathol. 1996;67:316–7. doi: 10.1006/jipa.1996.0049. [DOI] [PubMed] [Google Scholar]
- 105.Zhu X, Wittner M, Tanowitz HB, Cali A, Weiss LM. Ribosomal RNA sequences of Enterocytozoon bieneusi, Septata intestinalis and Ameson michaelis: phylogenetic construction and structural correspondence. J Eukaryot Microbiol. 1994;41:204–9. doi: 10.1111/j.1550-7408.1994.tb01498.x. [DOI] [PubMed] [Google Scholar]
- 106.Shafer AB, Williams GR, Shutler D, Rogers RE, Stewart DT. Cophylogeny of Nosema (Microsporidia: Nosematidae) and bees (Hymenoptera: Apidae) suggests both cospeciation and a host-switch. J Parasitol. 2009;95:198–203. doi: 10.1645/GE-1724.1. [DOI] [PubMed] [Google Scholar]
- 107.Zhu F, Shen Z, Xu X, Tao H, Dong S, Tang X, Xu L. Phylogenetic analysis of complete rRNA gene sequence of Nosema philosamiae isolated from the lepidopteran Philosamia cynthia ricini. J Eukaryot Microbiol. 2010;57:294–6. doi: 10.1111/j.1550-7408.2010.00475.x. [DOI] [PubMed] [Google Scholar]
- 108.Baker MD, Vossbrinck CR, Becnel JJ, Andreadis TG. Phylogeny of amblyospora (Microsporida: amblyosporidae) and related genera based on small subunit ribosomal DNA data: A possible example of host parasite cospeciation. J Invertebr Pathol. 1998;71:199–206. doi: 10.1006/jipa.1997.4725. [DOI] [PubMed] [Google Scholar]
- 109.Andreadis TG, Simakova AV, Vossbrinck CR, Shepard JJ, Yurchenko YA. Ultrastructural characterization and comparative phylogenetic analysis of new microsporidia from Siberian mosquitoes: evidence for coevolution and host switching. J Invertebr Pathol. 2012;109:59–75. doi: 10.1016/j.jip.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 110.Bell AS, Aoki T, Yokoyama H. Phylogenetic relationships among microsporidia based on rDNA sequence data, with particular reference to fish-infecting Microsporidium balbiani 1884 species. J Eukaryot Microbiol. 2001;48:258–65. doi: 10.1111/j.1550-7408.2001.tb00313.x. [DOI] [PubMed] [Google Scholar]
- 111.Lom J. A catalogue of described genera and species of microsporidians parasitic in fish. Syst Parasitol. 2002;53:81–99. doi: 10.1023/a:1020422209539. [DOI] [PubMed] [Google Scholar]
- 112.Muller A, Trammer T, Chioralia G, Seitz HM, Diehl V, Franzen C. Ribosomal RNA of Nosema algerae and phylogenetic relationship to other microsporidia. Parasitol Res. 2000;86:18–23. doi: 10.1007/pl00008501. [DOI] [PubMed] [Google Scholar]
- 113.Chen Y, Evans JD, Zhou L, Boncristiani H, Kimura K, Xiao T, Litkowski AM, Pettis JS. Asymmetrical coexistence of Nosema ceranae and Nosema apis in honey bees. J Invertebr Pathol. 2009;101:204–9. doi: 10.1016/j.jip.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 114.Becnel JJ. Life cycles and host-parasite relationships of microsporidia in culicine mosquitoes. Folia Parasitol. 1994;41:91–96. [PubMed] [Google Scholar]
- 115.Refardt D, Canning EU, Mathis A, Cheney SA, Lafranchi-Tristem NJ, Ebert D. Small subunit ribosomal DNA phylogeny of microsporidia that infect Daphnia (Crustacea: Cladocera) Parasitology. 2002;124:381–9. doi: 10.1017/s0031182001001305. [DOI] [PubMed] [Google Scholar]
- 116.Ebert D. Host-parasite coevolution: Insights from the Daphnia-parasite model system. Curr Opin Microbiol. 2008;11:290–301. doi: 10.1016/j.mib.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 117.Keeling PJ, Luker MA, Palmer JD. Evidence from beta-tubulin phylogeny that microsporidia evolved from within the fungi. Molecular Biology and Evolution. 2000;17:23–31. doi: 10.1093/oxfordjournals.molbev.a026235. [DOI] [PubMed] [Google Scholar]
- 118.Edlind T, Katiyar S, Visvesvara G, Li J. Evolutionary origins of Microsporidia and basis for benzimidazole sensitivity: an update. J Eukaryot Microbiol. 1996;43:109S. doi: 10.1111/j.1550-7408.1996.tb05029.x. [DOI] [PubMed] [Google Scholar]
- 119.Hirt RP, Healy B, Vossbrinck CR, Canning EU, Embley TM. A mitochondrial Hsp70 orthologue in Vairimorpha necatrix: molecular evidence that microsporidia once contained mitochondria. Curr Biol. 1997;7:995–8. doi: 10.1016/s0960-9822(06)00420-9. [DOI] [PubMed] [Google Scholar]
- 120.Hirt RP, Logsdon JM, Jr, Healy B, Dorey MW, Doolittle WF, Embley TM. Microsporidia are related to Fungi: evidence from the largest subunit of RNA polymerase II and other proteins. Proc Natl Acad Sci U S A. 1999;96:580–5. doi: 10.1073/pnas.96.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Keeling PJ, Doolittle WF. Alpha-tubulin from early-diverging eukaryotic lineages and the evolution of the tubulin family. Mol Biol Evol. 1996;13:1297–305. doi: 10.1093/oxfordjournals.molbev.a025576. [DOI] [PubMed] [Google Scholar]
- 122.Arisue N, Sanchez LB, Weiss LM, Muller M, Hashimoto T. Mitochondrial-type hsp70 genes of the amitochondriate protists, Giardia intestinalis, Entamoeba histolytica and two microsporidians. Parasitol Int. 2002;51:9–16. doi: 10.1016/s1383-5769(01)00093-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Germot A, Philippe H. Critical analysis of eukaryotic phylogeny: a case study based on the HSP70 family. J Eukaryot Microbiol. 1999;46:116–24. doi: 10.1111/j.1550-7408.1999.tb04594.x. [DOI] [PubMed] [Google Scholar]
- 124.Germot A, Philippe H, Le GH. Evidence for loss of mitochondria in microsporidia from a mitochondrial-type hsp70 in Nosema locustae. Molecular And Biochemical Parasitology. 1997;87:159–168. doi: 10.1016/s0166-6851(97)00064-9. [DOI] [PubMed] [Google Scholar]
- 125.Peyretaillade E, Broussolle V, Peyret P, Metenier G, Gouy M, Vivares CP. Microsporidia, amitochondrial protists, possess a 70-kDa heat shock protein gene of mitochondrial evolutionary origin. Mol Biol Evol. 1998;15:683–9. doi: 10.1093/oxfordjournals.molbev.a025971. [DOI] [PubMed] [Google Scholar]
- 126.Fast NM, Logsdon JM, Jr, Doolittle WF. Phylogenetic analysis of the TATA box binding protein (TBP) gene from Nosema locustae: evidence for a microsporidia-fungi relationship and spliceosomal intron loss. Mol Biol Evol. 1999;16:1415–9. doi: 10.1093/oxfordjournals.molbev.a026052. [DOI] [PubMed] [Google Scholar]
- 127.Vivares C, Biderre C, Duffieux F, Peyretaillade E, Peyret P, Metenier G, Pages M. Chromosomal localization of five genes in Encephalitozoon cuniculi (Microsporidia) J Eukaryot Microbiol. 1996;43:97S. doi: 10.1111/j.1550-7408.1996.tb05021.x. [DOI] [PubMed] [Google Scholar]
- 128.Edlind T. Phylogenetics of protozoan tubulin with reference to the amitochondriate eukaryotes. In: Coombs GH, Vickerman K, Sleigh MA, Warren A, editors. Evolutionary Relationships Among Protozoa. Chapman & Hall; London: 1998. pp. 91–108. [Google Scholar]
- 129.Lee SC, Weiss LM, Heitman J. Generation of genetic diversity in microsporidia via sexual reproduction and horizontal gene transfer. Commun Integr Biol. 2009;2:414–7. doi: 10.4161/cib.2.5.8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lee SC, Corradi N, Doan S, Dietrich FS, Keeling PJ, Heitman J. Evolution of the sex-related locus and genomic features shared in microsporidia and fungi. PLoS One. 2010;5:e10539. doi: 10.1371/journal.pone.0010539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Weiss LM, Vossbrinck CR. Molecular biology, molecular phylogeny, and molecular diagnostic approaches to the microsporidia. In: Wittner M, Weiss LM, editors. The Microsporidia and Microsporidia. ASM Press; Washington D.C: 1999. pp. 129–171. [Google Scholar]
- 132.Kamaishi T, Hashimoto T, Nakamura Y, Nakamura F, Murata S, Okada N, Okamoto K, Shimizu M, Hasegawa M. Protein phylogeny of translation elongation factor EF-1 alpha suggests microsporidians are extremely ancient eukaryotes. J Mol Evol. 1996;42:257–63. doi: 10.1007/BF02198852. [DOI] [PubMed] [Google Scholar]
- 133.Keeling PJ, Corradi N. Shrink it or lose it: balancing loss of function with shrinking genomes in the microsporidia. Virulence. 2011;2:67–70. doi: 10.4161/viru.2.1.14606. [DOI] [PubMed] [Google Scholar]
- 134.Desportes I. Ultrastructure de Stempellia mutabilis Leger et Hesse, microsporidie parasite de l’ephemere Ephemera vulgata L. Protistologica. 1976;12:121–150. [Google Scholar]
- 135.Flegel TW, Pasharawipas T. A proposal for typical eukaryotic meiosis in microsporidians. Can J Microbiol. 1995;41:1–11. [Google Scholar]
- 136.Xu Y, Takvorian PM, Cali A, Orr G, Weiss LM. Glycosylation of the major polar tube protein of Encephalitozoon hellem, a microsporidian parasite that infects humans. Infect Immun. 2004;72:6341–50. doi: 10.1128/IAI.72.11.6341-6350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Keeling PJ. Congruent evidence from alpha-tubulin and beta-tubulin gene phylogenies for a zygomycete origin of microsporidia. Fungal Genet Biol. 2003;38:298–309. doi: 10.1016/s1087-1845(02)00537-6. [DOI] [PubMed] [Google Scholar]
- 138.James TY, Pelin A, Bonen L, Ahrendt S, Sain D, Corradi N, Stajich JE. Shared signatures of parasitism and phylogenomics unite Cryptomycota and microsporidia. Curr Biol. 2013;23:1548–53. doi: 10.1016/j.cub.2013.06.057. [DOI] [PubMed] [Google Scholar]
- 139.Jones MD, Forn I, Gadelha C, Egan MJ, Bass D, Massana R, Richards TA. Discovery of novel intermediate forms redefines the fungal tree of life. Nature. 2011;474:200–3. doi: 10.1038/nature09984. [DOI] [PubMed] [Google Scholar]
- 140.Jones MD, Richards TA, Hawksworth DL, Bass D. Validation and justification of the phylum name Cryptomycota phyl. nov. IMA Fungus. 2011;2:173–5. doi: 10.5598/imafungus.2011.02.02.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Evans TN, Seviour RJ. Estimating biodiversity of fungi in activated sludge communities using culture-independent methods. Microb Ecol. 2012;63:773–86. doi: 10.1007/s00248-011-9984-7. [DOI] [PubMed] [Google Scholar]
- 142.Livermore JA, Mattes TE. Phylogenetic detection of novel Cryptomycota in an Iowa (United States) aquifer and from previously collected marine and freshwater targeted high-throughput sequencing sets. Environ Microbiol. 2013;15:2333–41. doi: 10.1111/1462-2920.12106. [DOI] [PubMed] [Google Scholar]
- 143.Manohar CS, Raghukumar C. Fungal diversity from various marine habitats deduced through culture-independent studies. FEMS Microbiol Lett. 2013;341:69–78. doi: 10.1111/1574-6968.12087. [DOI] [PubMed] [Google Scholar]
- 144.Haag KL, James TY, Pombert JF, Larsson R, Schaer TM, Refardt D, Ebert D. Evolution of a morphological novelty occurred before genome compaction in a lineage of extreme parasites. Proc Natl Acad Sci U S A. 2014;111:15480–5. doi: 10.1073/pnas.1410442111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Corsaro D, Walochnik J, Venditti D, Muller KD, Hauroder B, Michel R. Rediscovery of Nucleophaga amoebae, a novel member of the Rozellomycota. Parasitol Res. 2014;113:4491–8. doi: 10.1007/s00436-014-4138-8. [DOI] [PubMed] [Google Scholar]
- 146.Corsaro D, Walochnik J, Venditti D, Steinmann J, Muller KD, Michel R. Microsporidia-like parasites of amoebae belong to the early fungal lineage Rozellomycota. Parasitol Res. 2014;113:1909–18. doi: 10.1007/s00436-014-3838-4. [DOI] [PubMed] [Google Scholar]
- 147.Vávra J, Ronny Larsson J. Structure of microsporidia. In: Weiss LM, Becnel J, editors. Microsporidia: Pathogens of Opportunity. Wiley Blackwell; Oxford: 2014. pp. 1–70. [Google Scholar]
- 148.Vavra JCJ. Fluorescence staining of microsporidian spores with the brightener “Calcofluor White M2k”. J Protozool. 1982;29:503. [Google Scholar]
- 149.Brosson D, Kuhn L, Prensier G, Vivares CP, Texier C. The putative chitin deacetylase of Encephalitozoon cuniculi: a surface protein implicated in microsporidian spore-wall formation. FEMS Microbiol Lett. 2005;247:81–90. doi: 10.1016/j.femsle.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 150.Han B, Zhou K, Li Z, Sun B, Ni Q, Meng X, Pan G, Li C, Long M, Li T, Zhou C, Li W, Zhou Z. Characterization of the first fungal glycosyl hydrolase family 19 chitinase (NbchiA) from Nosema bombycis (Nb) J Eukaryot Microbiol. 2015;63:37–45. doi: 10.1111/jeu.12246. [DOI] [PubMed] [Google Scholar]
- 151.Southern TR, Jolly CE, Lester ME, Hayman JR. EnP1, a microsporidian spore wall protein that enables spores to adhere to and infect host cells in vitro. Eukaryotic cell. 2007;6:1354–1362. doi: 10.1128/EC.00113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Undeen AH, Frixione E. The role of osmotic pressure in the germination of Nosema algerae spores. J Protozool. 1990;37:561–7. doi: 10.1111/j.1550-7408.1990.tb01265.x. [DOI] [PubMed] [Google Scholar]
- 153.Bohne W, Ferguson DJ, Kohler K, Gross U. Developmental expression of a tandemly repeated, glycine-and serine-rich spore wall protein in the microsporidian pathogen Encephalitozoon cuniculi. Infection and immunity. 2000;68:2268–2275. doi: 10.1128/iai.68.4.2268-2275.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hayman JR, Hayes SF, Amon J, Nash TE. Developmental expression of two spore wall proteins during maturation of the microsporidian Encephalitozoon intestinalis. Infection and immunity. 2001;69:7057–7066. doi: 10.1128/IAI.69.11.7057-7066.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Polonais V, Mazet M, Wawrzyniak I, Texier C, Blot N, El Alaoui H, Delbac F. The human microsporidian Encephalitozoon hellem synthesizes two spore wall polymorphic proteins useful for epidemiological studies. Infection and immunity. 2010;78:2221–2230. doi: 10.1128/IAI.01225-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Peuvel-Fanget I, Polonais V, Brosson D, Texier C, Kuhn L, Peyret P, Vivares C, Delbac F. EnP1 and EnP2, two proteins associated with the Encephalitozoon cuniculi endospore, the chitin-rich inner layer of the microsporidian spore wall. Int J Parasitol. 2006;36:309–18. doi: 10.1016/j.ijpara.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 157.Xu Y, Takvorian P, Cali A, Wang F, Zhang H, Orr G, Weiss LM. Identification of a new spore wall protein from Encephalitozoon cuniculi. Infect Immun. 2006;74:239–47. doi: 10.1128/IAI.74.1.239-247.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hayman JR, Southern TR, Nash TE. Role of sulfated glycans in adherence of the microsporidian Encephalitozoon intestinalis to host cells in vitro. Infection and immunity. 2005;73:841–848. doi: 10.1128/IAI.73.2.841-848.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ghosh K, Nieves E, Keeling P, Pombert J-F, Henrich PP, Cali A, Weiss LM. Branching network of proteinaceous filaments within the parasitophorous vacuole of Encephalitozoon cuniculi and Encephalitozoon hellem. Infection and immunity. 2011;79:1374–1385. doi: 10.1128/IAI.01152-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wu Z, Li Y, Pan G, Zhou Z, Xiang Z. SWP25, a novel protein associated with the Nosema bombycis endospore. J Eukaryot Microbiol. 2009;56:113–8. doi: 10.1111/j.1550-7408.2008.00375.x. [DOI] [PubMed] [Google Scholar]
- 161.Wang JY, Chambon C, Lu CD, Huang KW, Vivarès CP, Texier C. A proteomic based approach for the characterization of some major structural proteins involved in host–parasite relationships from the silkworm parasite Nosema bombycis (Microsporidia) Proteomics. 2007;7:1461–1472. doi: 10.1002/pmic.200600825. [DOI] [PubMed] [Google Scholar]
- 162.Yang D, Pan G, Dang X, Shi Y, Li C, Peng P, Luo B, Bian M, Song Y, Ma C. Interaction and assembly of two novel proteins in the spore wall of the microsporidian species Nosema bombycis and their roles in adherence to and infection of host cells. Infection and immunity. 2015;83:1715–1731. doi: 10.1128/IAI.03155-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chen J, Geng L, Long M, Li T, Li Z, Yang D, Ma C, Wu H, Ma Z, Li C. Identification of a novel chitin-binding spore wall protein (NbSWP12) with a BAR-2 domain from Nosema bombycis (microsporidia) Parasitology. 2013;140:1394–1402. doi: 10.1017/S0031182013000875. [DOI] [PubMed] [Google Scholar]
- 164.Li Z, Pan G, Li T, Huang W, Chen J, Geng L, Yang D, Wang L, Zhou Z. SWP5, a spore wall protein, interacts with polar tube proteins in the parasitic microsporidian Nosema bombycis. Eukaryot Cell. 2012;11:229–37. doi: 10.1128/EC.05127-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Thelohan P. Sur la presence d’une capsule a filament dans les spores des microsporidies. CR Acad Sci. 1894;118:1425–1427. [Google Scholar]
- 166.Thelohan P. Observations sur les Myxosporidies et essai de classification de ces organismes. Bull Soc Philom Paris. 1892;4:173–174. [Google Scholar]
- 167.Sanders JL, Watral V, Kent ML. Microsporidiosis in zebrafish research facilities. ILAR Journal. 2012;53:106–113. doi: 10.1093/ilar.53.2.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lom J, Vavra J. The mode of sporoplasm extrusion in microsporidian spores. Acta Protozool. 1963;1:81–89. [Google Scholar]
- 169.Weidner E. The microsporidian spore invasion tube. The ultrastructure, isolation, and characterization of the protein comprising the tube. J Cell Biol. 1976;71:23–34. doi: 10.1083/jcb.71.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ohshima K. On the function of the polar filament of Nosema bombycis. Parasitology. 1937;29:220–224. [Google Scholar]
- 171.Weidner E. The microsporidian spore invasion tube. III. Tube extrusion and assembly. The Journal of Cell Biology. 1982;93:976–979. doi: 10.1083/jcb.93.3.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Undeen AH, Epsky ND. In vitro and in vivo germination of Nosema locustae (Microspora: Nosematidae) spores. Journal of Invertebrate Pathology. 1990;56:371–379. [Google Scholar]
- 173.Keohane EM, Orr GA, Takvorian PM, Cali A, Tanowitz HB, Witter M, Weiss LM. Purification and characterization of a microsporidian polar tube protein. Molecular and Biochemical Parasitology. 1996;79:255–259. doi: 10.1016/0166-6851(96)02666-7. [DOI] [PubMed] [Google Scholar]
- 174.Keohane EM, Orr GA, Takvorian PM, Cali A, Tanowitz HB, Wittner M, Weiss LM. Purification and characterization of a microsporidian polar tube proteins. J Eukaryot Microbiol. 1996;43:100S. doi: 10.1111/j.1550-7408.1996.tb05023.x. [DOI] [PubMed] [Google Scholar]
- 175.Keohane EM, Orr GA, Takvorian PM, Cali A, Tanowitz HB, Wittner M, Weiss LM. Polar tube proteins of microsporidia of the family Encephalitozoonidae. J Eukaryot Microbiol. 1999;46:1–5. doi: 10.1111/j.1550-7408.1999.tb04569.x. [DOI] [PubMed] [Google Scholar]
- 176.Keohane EM, Orr GA, Takvorian PM, Cali A, Tanowitz HB, Wittner M, Weiss LM. Analysis of the major microsporidian polar tube proteins. J Eukaryot Microbiol. 1999;46:29S–30S. [PubMed] [Google Scholar]
- 177.Keohane EM, Orr GA, Zhang HS, Takvorian PM, Cali A, Tanowitz HB, Wittner M, Weiss LM. The molecular characterization of the major polar tube protein gene from Encephalitozoon hellem, a microsporidian parasite of humans. Mol Biochem Parasitol. 1998;94:227–36. doi: 10.1016/s0166-6851(98)00071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Keohane EM, Takvorian PM, Cali A, Tanowitz HB, Wittner M, Weiss LM. Identification of a microsporidian polar tube protein reactive monoclonal antibody. J Eukaryot Microbiol. 1996;43:26–31. doi: 10.1111/j.1550-7408.1996.tb02468.x. [DOI] [PubMed] [Google Scholar]
- 179.Keohane EM, Takvorian PM, Cali A, Tanowitz HB, Wittner M, Weiss LM. Monoclonal antibodies to cytoplasmic antigens of Nosema locustae (Microsporida: Nosematidae) J Invertebr Pathol. 2001;77:81–2. doi: 10.1006/jipa.2000.4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Delbac F, Peuvel I, Metenier G, Peyretaillade E, Vivares CP. Microsporidian invasion apparatus: identification of a novel polar tube protein and evidence for clustering of ptp1 and ptp2 genes in three Encephalitozoon species. Infect Immun. 2001;69:1016–24. doi: 10.1128/IAI.69.2.1016-1024.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Peuvel I, Delbac F, Metenier G, Peyret P, Vivares CP. Polymorphism of the gene encoding a major polar tube protein PTP1 in two microsporidia of the genus Encephalitozoon. Parasitology. 2000;121(Pt 6):581–7. doi: 10.1017/s0031182000006910. [DOI] [PubMed] [Google Scholar]
- 182.Peuvel I, Peyret P, Metenier G, Vivares CP, Delbac F. The microsporidian polar tube: evidence for a third polar tube protein (PTP3) in Encephalitozoon cuniculi. Mol Biochem Parasitol. 2002;122:69–80. doi: 10.1016/s0166-6851(02)00073-7. [DOI] [PubMed] [Google Scholar]
- 183.Xu Y, Takvorian P, Cali A, Weiss LM. Lectin binding of the major polar tube protein (PTP1) and its role in invasion. J Eukaryot Microbiol. 2003;50(Suppl):600–1. doi: 10.1111/j.1550-7408.2003.tb00644.x. [DOI] [PubMed] [Google Scholar]
- 184.Polonais V, Prensier G, Méténier G, Vivarès CP, Delbac F. Microsporidian polar tube proteins: highly divergent but closely linked genes encode PTP1 and PTP2 in members of the evolutionarily distant Antonospora and Encephalitozoon groups. Fungal Genetics and Biology. 2005;42:791–803. doi: 10.1016/j.fgb.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 185.Bouzahzah B, Nagajyothi F, Ghosh K, Takvorian PM, Cali A, Tanowitz HB, Weiss LM. Interactions of Encephalitozoon cuniculi polar tube proteins. Infect Immun. 2010;78:2745–53. doi: 10.1128/IAI.01205-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Bouzahzah B, Weiss LM. Glycosylation of the major polar tube protein of Encephalitozoon cuniculi. Parasitol Res. 2010;107:761–4. doi: 10.1007/s00436-010-1950-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hatjina F, Tsoktouridis G, Bouga M, Charistos L, Evangelou V, Avtzis D, Meeus I, Brunain M, Smagghe G, de Graaf DC. Polar tube protein gene diversity among Nosema ceranae strains derived from a Greek honey bee health study. Journal of Invertebrate Pathology. 2011;108:131–134. doi: 10.1016/j.jip.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 188.Delbac F, David D, Metenier G, Vivares C. First complete amino acid sequence of a polar tube protein in a microsporidian species, Encephalitozoon cuniculi. J Eukaryot Microbiol. 1997;44:77S. doi: 10.1111/j.1550-7408.1997.tb05791.x. [DOI] [PubMed] [Google Scholar]
- 189.van Gool T, Vetter JC, Weinmayr B, Van Dam A, Derouin F, Dankert J. High seroprevalence of Encephalitozoon species in immunocompetent subjects. J Infect Dis. 1997;175:1020–4. doi: 10.1086/513963. [DOI] [PubMed] [Google Scholar]
- 190.Keohane EM, Weiss LM. Characterization and function of the microsporidian polar tube: a review. Folia Parasitol (Praha) 1998;45:117–27. [PubMed] [Google Scholar]
- 191.Peek R, Delbac F, Speijer D, Polonais V, Greve S, Wentink-Bonnema E, Ringrose J, van Gool T. Carbohydrate moieties of microsporidian polar tube proteins are targeted by immunoglobulin G in immunocompetent individuals. Infection and immunity. 2005;73:7906–7913. doi: 10.1128/IAI.73.12.7906-7913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Dolgikh VV, Semenov PB. The spore wall and polar tube proteins of the microsporidian Nosema grylli: the major spore wall protein is released before spore extrusion. Tsitologiia. 2003;45:324–9. [PubMed] [Google Scholar]
- 193.Peyretaillade E, Parisot N, Polonais V, Terrat S, Denonfoux J, Dugat-Bony E, Wawrzyniak I, Biderre-Petit C, Mahul A, Rimour S, Goncalves O, Bornes S, Delbac F, Chebance B, Duprat S, Samson G, Katinka M, Weissenbach J, Wincker P, Peyret P. Annotation of microsporidian genomes using transcriptional signals. Nature Communications. 2012;3:1137. doi: 10.1038/ncomms2156. [DOI] [PubMed] [Google Scholar]
- 194.Cornman RS, Chen YP, Schatz MC, Street C, Zhao Y, Desany B, Egholm M, Hutchison S, Pettis JS, Lipkin WI, Evans JD. Genomic analyses of the microsporidian Nosema ceranae, an emergent pathogen of honey bees. PLoS Pathog. 2009;5:e1000466. doi: 10.1371/journal.ppat.1000466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Pan GQ, Xu JS, Li T, Xia QY, Liu SL, Zhang GJ, Li SG, Li CF, Liu HD, Yang L, Liu T, Zhang X, Wu ZL, Fan W, Dang XQ, Xiang H, Tao ML, Li YH, Hu JH, Li Z, Lin LP, Luo J, Geng LN, Wang LL, Long MX, Wan YJ, He NJ, Zhang Z, Lu C, Keeling PJ, Wang J, Xiang ZH, Zhou ZY. Comparative genomics of parasitic silkworm microsporidia reveal an association between genome expansion and host adaptation. BMC Genomics. 2013;14:186. doi: 10.1186/1471-2164-14-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Polonais V, Belkorchia A, Roussel M, Peyretaillade E, Peyret P, Diogon M, Delbac F. Identification of two new polar tube proteins related to polar tube protein 2 in the microsporidian Antonospora locustae. FEMS Microbiol Lett. 2013;346:36–44. doi: 10.1111/1574-6968.12198. [DOI] [PubMed] [Google Scholar]
- 197.Slamovits CH, Fast NM, Law JS, Keeling PJ. Genome compaction and stability in microsporidian intracellular parasites. Curr Biol. 2004;14:891–6. doi: 10.1016/j.cub.2004.04.041. [DOI] [PubMed] [Google Scholar]
- 198.Brosson D, Kuhn L, Delbac F, Garin J, PVC, Texier C. Proteomic analysis of the eukaryotic parasite Encephalitozoon cuniculi (microsporidia): a reference map for proteins expressed in late sporogonial stages. Proteomics. 2006;6:3625–35. doi: 10.1002/pmic.200500796. [DOI] [PubMed] [Google Scholar]
- 199.Foucault C, Drancourt M. Actin mediates Encephalitozoon intestinalis entry into the human enterocyte-like cell line, Caco-2. Microb Pathog. 2000;28:51–8. doi: 10.1006/mpat.1999.0329. [DOI] [PubMed] [Google Scholar]