Abstract
γ-glutamylcysteine synthetase (Gcs) is a vital enzyme catalyzing the first and rate limiting step in the trypanothione biosynthesis pathway, the ATP-dependent ligation of L-Glutamate and L-Cysteine to form gamma-glutamylcysteine. The Trypanothione biosynthesis pathway is unique metabolic pathway essential for trypanosomatid survival rendering Gcs as a potential drug target. Here we report the cloning, expression, purification and characterization of L. donovani Gcs. Three other constructs of Gcs (GcsN, GcsC and GcsT) were designed on the basis of S. cerevisiae and E. coli Gcs crystal structures. The study shows Gcs possesses ATPase activity even in the absence of substrates L-glutamate and L-Cysteine. Divalent ions however plays an indispensable role in LdGcs ATPase activity. Isothermal titration calorimetry and fluorescence studies illustrates that L. donovani Gcs binds substrate in order ATP >L-glutamate>L-cysteine with Glu 92 and Arg 498 involved in ATP hydrolysis and Glu 92, Glu 55 and Arg 498 involved in glutamate binding. Homology modeling and molecular dynamic simulation studies provided the structural rationale of LdGcs catalytic activity and emphasized on the possibility of involvement of three Mg2+ ions along with Glutamates 52, 55, 92, 99, Met 322, Gln 328, Tyr 397, Lys 483, Arg 494 and Arg 498 in the catalytic function of L. donovani Gcs.
Keywords: Glutamyl cysteine synthetase, Polyamine biosynthesis pathway, L donovani, Divalent ion, Substrate binding
Highlights
-
•
L. donovani Gamma glutamylcysteine synthetase is a divalent dependent ATPase.
-
•
Substrate binds in order ATP>> L-Glutamate> L-cysteine.
-
•
Glu 92 and Arg 498 involved in ATP hydrolysis.
-
•
Glu 92, Glu 55 and Arg 498 involved in glutamate binding.
1. Introduction
Visceral leishmaniasis (VL) is the most devastating and fatal form of leishmaniasis caused primarily by the parasitic protozoan Leishmania donovani, leading to an estimated 300,000 new cases of disease annually with more than 20,000 deaths [1]. Co-infection with HIV results in a significant increase in VL pervasiveness [2]. The disease is prevalent throughout the tropical world and current drug treatment regimen have limitations due to toxicity, high cost, difficult dosage regime and development of resistance to antimonial drugs. These drawbacks have severely reduced the options for clinical treatment of VL and till date no effective drug or vaccine is available. The crucial step in the development of the new drug in a rationale-driven drug discovery approach is the identification, functional and structural characterization of a protein or a pathway in the pathogen that is either absent or sufficiently different from its host. One such pathway is the trypanothione biosynthesis pathway [3], [4], [5], [6]. Trypansomatids including Leishmania utilize a potent machinery employing the bis glutathione-spermidine conjugate trypanothione, as a major thiol along with universally present thiol glutathione. In the absence of the ubiquitously present enzymes catalase and glutathione peroxidase which are responsible for neutralizing reactive oxygen species (ROS) in other organisms, trypanothione plays a crucial role in fighting large amount of reactive oxidative species (ROS) generated by the macrophages during the initial phase of parasite–host interactions in trypanosomatids [7], [8], [9], [10].
Independently synthesised glutathione and spermidine conjugate to form trypanothione. Glutathione biosynthesis takes place in two steps : the first step, catalysed by γ-glutamylcysteine synthetase (Gcs EC 6.3.2.2) is the ATP-dependent ligation of L-Cysteine to L-Glutamate to form γ-glutamylcysteine [11]; the second step is the conjugation of L-glycine forming glutathione, catalysed by glutathione synthetase in an ATP dependent manner [12]. Spermidine is obtained from putrescine in the reactions catalyzed by ornithine decarboxylase and spermidine synthase. The next two steps resulting in trypanothione biosynthesis are unique to trypanosomatids wherein glutathione couples with spermidine resulting in the formation of glutathionyl spermidine [4], to which another molecule of glutathione is conjugated to produce trypanothione. The former reaction is catalysed by the enzyme glutathionyl spermidine synthetase (EC 6.3.1.8) while the latter is catalysed by trypanothione synthetase (EC 6.3.1.9) [13].
Among the enzymes involved in trypanothione biosynthesis pathway, Gcs catalyses the initial and rate-limiting step in glutathione synthesis and plays an indispensable role in the survival of the organism [14], [15]. Null mutants of Gcs in fungi, mammals, Trypanosoma brucei (T. brucei) and also in Leishmania infantum (L. infantum) could not survive unless rescued by exogenous glutathione [16], [17], [18], [19], [20], [21], [22]. The high resistance of human neuroblastoma cells against oxidative damage, has been correlated with the higher expression levels of Gcs at mRNA and protein (catalytic subunit) level [23]. L-buthionine-S, R-sulfoximine (BSO), a specific inhibitor of Gcs cures and prolongs survival of mice infected with T. brucei implicating Gcs as a potential drug target [24]. It was also observed that vaccination with L. donovani Gcs (LdGcs) fusion protein or DNA based vaccine provided protection against L. donovani infection in BALB/c mouse model which further validates the pharmaceutical importance of Gcs [25], [26].
Though an essential protein in all class of organisms, Gcs sequences show significant diversity and can be categorized into three distinct phylogenetic groups - the first group consists of Gcs from proteobacteria like E. coli, the second group contains non-plant eukaryotes such as Homo sapiens, Saccharomyces cerevisiae and trypanosomatids while the third group consists of alpha-proteobacteria and plants such as Pisum sativum and Glycine max [27]. Despite having insignificant pairwise sequence identities (<10%) between these groups, the structures share a core architecture comprised of 6 antiparallel β-strands surrounded by an α-helix [28], [29], [30].
LdGcs belongs to the second group sharing significant sequence identity with Saccharomyces cerevisiae Gcs (ScGcs), whose crystal structure had been elucidated. Further, residues essential for metal binding and catalysis have also been determined using structural information gained from glutamine synthetase in T. brucei ortholog (TbGcs) [31], [32], [33]. The structural and functional knowledge gained from ScGcs and TbGcs prompted us to study the active site, substrate binding and catalytic features of LdGcs in comparison with these organisms. Although recombinant full-length LdGcs has been purified from inclusion bodies under denaturising condition with 6 M urea, lack of a homogenous population has prevented this study [25]. In the present manuscript, we report the cloning and purification of L. donovani Gcs full length (Gcs), 45 residues N-terminal and 167 residue C-terminal truncated Gcs construct (GcsT) retaining all functionally important residues, in its properly folded and active conformation, and a C-terminal construct lacking first 291 residues (GcsC). The study provides first biochemical, biophysical and structural insights of LdGcs with the substrate binding aspect rationalized using computational studies.
2. Results and discussion
2.1. Sequence and phylogenetic analysis
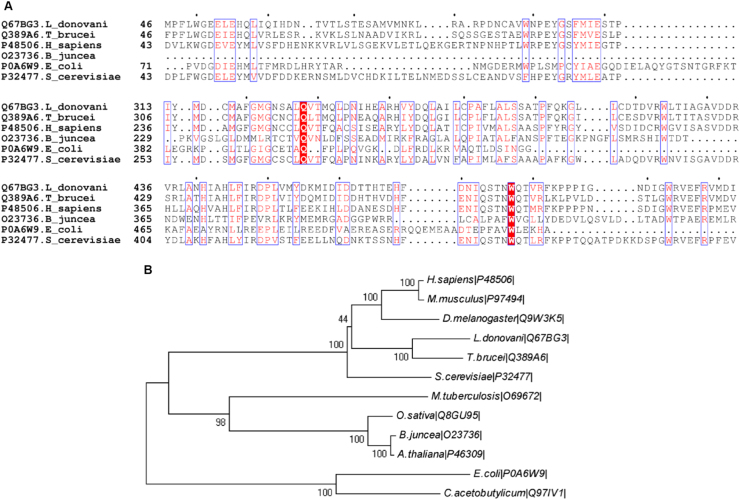
Leishmania donovani Gcs (UniProt Accession No Q67BG), is a member of the glutamylcysteine synthetase superfamily. PSI-BLAST against the Protein Data Bank recommend S. cereviseae Gcs (ScGcs) as the closest homologue with 31% sequence identity. Pairwise alignment of LdGcs with H. sapiens Gcs catalytic subunit and T. brucei Gcs shows 47% and 58% sequence identity respectively. Multiple Sequence alignment of Gcs from L. donovani, T. brucei, S. cereviseae, H. sapiens, B. juncea and E. coli shows that most of the functional residues are conserved among L. donovani, S. cerevisiae, T. brucei, while H. sapiens, while E. coli and B. juncea Gcs show differences, consistent with Gcs sequence classification studies (Fig. 1A). Interestingly, Cys 319, implicated in cystamine inactivation of T. brucei Gcs, is not conserved in L. donovani and is replaced by Asparagine. The other active site residues that differ in L. donovani are Glu 52, Asn 324 and Ala 326 which are replaced by Asp 49, Cys 264 and Cys 266 respectively (residue numbering as in S. cereviseae).
Fig. 1.
Phylogeny and sequence analysis (A) Multiple sequence alignment of γ-glutamylcysteine synthetase of S. cerevisiae, H. sapiens, T. brucei,L. donovani, B. juncea and E. coli. Sequences were retrieved from Uniprot gene database with their Uniprot gene id given proceeding name in multiple sequence alignment. (B) Phylogenetic tree showing three different classes of Gcs.
Phylogenetic analysis shows three different clusters of Gcs enzyme (Fig. 1B), validating the multiple sequence alignment analysis. As can be seen from the analysis the second and third groups of Gcs share the same evolutionary lineage while first group Gcs evolving earlier. Also amongst members of group two, mammals, trypanosomatids, insects and yeast evolved distinctly over the course of time.
The pair-wise sequence alignment of LdGcs with ScGcs sequence suggests most of the functional residues are distributed within the central region (Supplementary Fig. 1) and therefore, a construct consistent with this (GcsT, Residues 45 – 520) was also designed in addition to the N- (Residues 1-291) and C- (Residues 292–697) terminal constructs named as GcsN and GcsC respectively (Supplementary Fig. 1).
2.2. Cloning and purification of GcsF, GcsT and GcsC
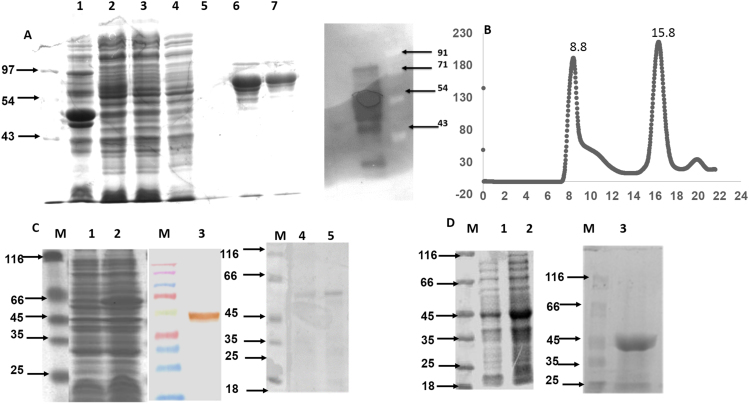
The L. donovani Gcs constructs were cloned, over-expressed and purified using immobilized metal affinity chromatography, and confirmed on a 10% SDS PAGE. While a band was observed at the expected full length size of 78 kDa, other bands (to ~ 45 kDa) are also present (Fig. 2A) suggesting the recombinant GcsF is not stable and degrades in ~24 h (Fig. 2A). Consequently, the experiments involving this construct were accomplished with freshly purified protein. Size exclusion chromatography of GcsF (Fig. 2B) shows that the recombinant protein is monomeric with some higher order oligomer population. The presence of higher order oligomers is probably observed for the first time in LdGcs, as other reported Gcs are monomers. The monomer population of LdGcs was used for enzymatic studies.
Fig. 2.
Purification of LdGcs. (A) 10% SDS PAGE of GcsF purification samples Lane 1–5 correspond to unstained protein marker; cell lysate loaded on NiNTA column; flow through of NiNTA column; wash sample conataining 10 mM imidazole; wash sample containing 50 mM imidazole; lane 6–7 elution sample containing NiNTA purified Gcs; right panel anti-His western blot of elution sample showing bands are degradation products of Gcs. (B) Size exclusion chromatography profile of Gcs with elution volume 15.8 ml corresponding to reference protein conalbumin (75 kDa) showing molecular weight corresponding to monomer. Peak at 8.8 corresponds to molecular weight above 440 corresponding to higher order oligomer. (C) 10% SDS PAGE showing M, unstained standards; lanes 1 and 2, over-expression of GcsT (DE3) pre-induction (lane 1) and post-induction (lane 2), with 1 mM IPTG. Lane 3, Western immunoblotting of over-expressed sample with anti-His antibody. Lane 4–5 purified GCST on 10% SDS PAGE (D) 10% SDS PAGE showing M unstained standards; lanes 1 and 2, over-expression of pET28a-GcsC (DE3) pre-induction and post-induction with 1 mM IPTG, M, prestained protein molecular weight marker; lane 3, Purified GcsC at expected size 45 kDa.
The recombinant GcsT and the GcsC constructs after IMAC purification were observed at their expected sizes i.e., 52 kDa and 43 kDa (Fig. 2C, D) respectively, without any degradation. Functional assays of the former two constructs were carried out after size exclusion chromatography. Prior to functional assays being carried out, GcsT and GcsC were analyzed for structural integrity using Circular Dichorism as their purification involved urea denaturation and both constructs showed properly folded structures (Supplementary Fig. 2A-D).
2.3. ATPase activity
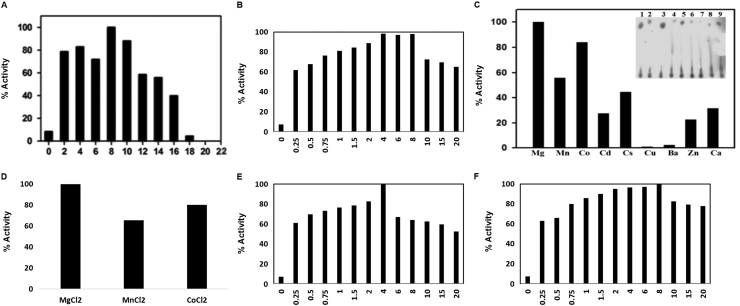
ATPase activity of GcsT and GcsC were studied by the protocol given in Ohno et al. [34] using 1μCi 32P ATP. The Optimum ATPase activity was displayed by 2.5 µM GcsT in buffer B in 40 min. Isotopic assays were carried out in the absence of the substrates L-Glutamate and L-Cysteine validating GcsT possess substrate independent ATPase activity. Isotopic assays in presence of Mg2+ displayed significant enhancement of activity at 2 mM with maximum activity at 8 mM concentration above which it has inhibitory effect (Fig. 3A-B). NADH coupled assays also substantiate this observation with comparable rise in ATPase activities in the presence of 0.25–8 mM Mg2+, with a significant fall on further increase in Mg2+ concentration.
Fig. 3.
Factors affecting ATPase activity Effect of MgCl2 on ATPase activity of GcsT (A) isotopic assay, (B) NADH coupled assay. The results were plotted as % relative activity versus MgCl2 concentration. Optimum activity was found at 8 mM MgCl2.for GcsT. (C-D) Effect of divalent cations on ATPase activity. Extent of hydrolysis was plotted as percentage activity as a function of individual divalent cations Mg2+, Mn2+, Co2+, Cd2+, Cs2+, Zn2+ and Ca2+. Mg2+ show maximum increase in ATPase activity. Inset shows radiographs having Gcs with the same divalent cations. ATPase activity in presence of different concentrations (0.25–20 mM) of (E) Mn2+ and (F) Co2+. The results were plotted as % relative activity versus divalent concentration. Three independent experiments were performed and results were averaged for relative activity calculation.
The presence of magnesium increases ATPase activity by ~8 fold. This encouraged us to determine the effect of other divalent ions such as Mn2+, Co2+, Cd2+, Cs2+, Zn2+ Ca2+ on Gcs ATPase activity. Isotopic assays recommend Mg2+ as most favorable divalent ion with Cobalt being second and manganese being third good substitute (Fig. 3C) and as before, verified with NADH coupled assays (Fig. 4D). The optimum concentrations of Mn2+ and Co2+ were also determined. Mn2+ was found to increase ATPase activity up to 4 mM concentration, after which it exhibits inhibitory effect, with the corresponding value for cobalt being 8 mM (Fig. 3E and F). This is in agreement with earlier studies of Abbott et al. [32] where too Mn2+ has stimulated activity at relatively lower concentrations than Mg2+ and has inhibitory effects at higher concentration [32]. However, the optimum concentration of Mn2+ was 0.75 mM in TbGcs unlike LdGcs where the optimum concentration in 2 mM. The plausible explanation behind this phenomenon lies in the fact that LdGcs possess three metal binding sites, instead of two detected in TbGcs, as observed in ScGcs crystal structure and corroborated with MD simulation studies. These three metal binding sites can separately bind metal ion alone or coordinated with substrates L-glutamate and ADP [29]. Magnesium in lower concentrations plays a crucial role in forming Mg-ATP complex and ATPase activity. However, at higher concentrations the complex is disrupted leading to inhibitory effect on ATPase reaction. This observation is consistent with earlier findings in other class of ATPase as well [35].
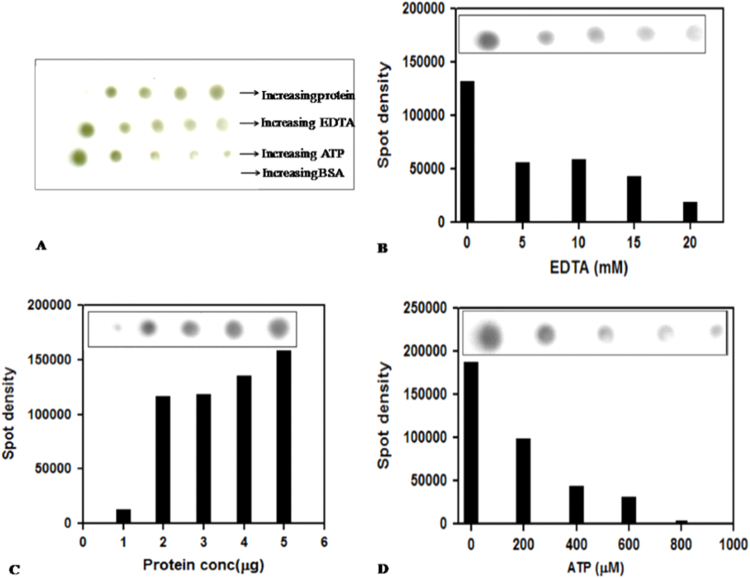
Fig. 4.
Dot blot assay for ATP binding. GcsT in buffer comprised of 50 mM Tris-HCl pH 8.5, 50 mM NaCl, 3 mM β-me and 4 mM MgCl2 was immobilized on a nitrocellulose membrane and incubated with labeled 50 μM of γ−32P ATP. (A) Nitrocellulose dot blot autoradiograph on X-ray film, showing effect of increasing concentration of protein, ATP and EDTA on ATP binding. (B) Intensity of spots representing bound radiolabelled ATP was plotted against increasing EDTA concentration. Bar diagram representing decrease in ATP binding with increasing EDTA concentration. (C) Bar diagram showing increase in ATP binding with increasing protein concentration. (D) Bar diagram showing displacement of radiolabeled ATP by unlabelled ATP as revealed by decrease in spot intensity.
2.4. Role of divalent ion in ATP binding
The role of divalent ion(s) in ATP binding of Gcs was further validated by a filter based nucleotide binding assay. GcsT was immobilized on a nitrocellulose membrane with increasing concentration of protein, EDTA, ATP and BSA (as control) and incubated with 50 μM γ−32P ATP. The autoradiographs in Fig. 4 A,C clearly show increase in spot density with increasing protein concentration, indicative of GcsT binding radiolabeled ATP. With increasing concentrations of EDTA, the spot density decreases, reaffirming the essential role of the divalent ion (Fig. 4B). Spot density decreases in the presence of excess unlabelled ATP as a result of competitive binding (Fig. 4D). As a control blotted BSA has no spots.
2.5. Kinetic parameters of ATPase activity
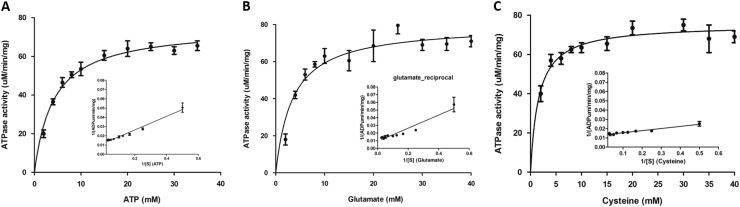
Kinetic parameters of L. donovani Gcs ATPase activity were analyzed by simultaneously measuring ATPase activity in varying concentrations of ATP, L-Glutamate and L-Cysteine with saturating concentration of other two substrates using a NADH-coupled assay as described in Materials and Methods section. The calculated specific activity of GcsF was 19 units/mg protein, which is comparable to the reported value for rat Gcs (16 units/mg) [36] and similar to the reported T. brucei Gcs (9.8 units/mg) [37] specific activity. The LdGcs and its constructs have basal ATPase activity even in the absence of L-glutamate and L-Cysteine. However, as mentioned earlier, the presence of Mg2+ was crucial for ATPase activity. Apparent Km and Vmax were determined to be in the milli molar range (Fig. 5; Table 1).
Fig. 5.
Kinetic parameters of ATPase activity. (A) ATP; (B) L-Glutamate; (C) L-Cysteine. Rate of ADP formed was plotted against substrate concentration. Km and Vmax values were obtained by the fit of Michaelis-Menten equation in Prism 5.0 software. Lineweaver Burk plot are shown in inset. All experiments were performed two times.
Table 1.
Km and Vmax values were determined by the Michaelis-Menten equation. The lineweaver burk plot was also considered for determining these paramerters.
| Kmapp(mM) | Vmaxapp(uM/min/mg) | |
|---|---|---|
| ATP | 6.25±.4 | 85.32 |
| L-Glutamate | 9.2±1.1 | 108.26 |
| L-Cysteine | 1.7±0.15 | 75.18 |
2.6. Substrate binding
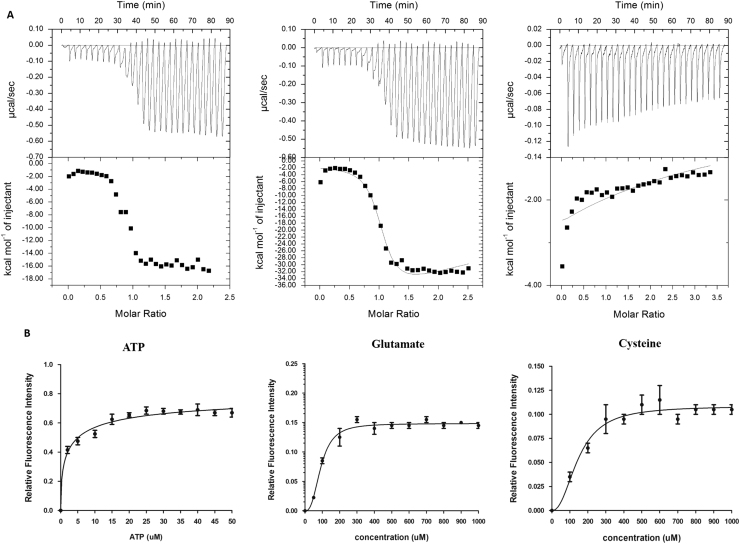
Gcs catalyzes the ATP-dependent ligation of L-glutamate and L-cysteine and has a large active site to accommodate all three substrates simultaneously. Despite the large active site, the binding and catalysis of substrates cannot be completely random. The available structural and biochemical knowledge from homologous Gcs [28], [29], [30], [31], [32], [33] have shown significant affinity with both L-glutamate and ATP and but the relative affinities for these are still not clear. Further, the binding affinity of Gcs to L-cysteine is also not known till date. To address these questions, a series of Isothermal titration calorimetry (ITC) and intrinsic tryptophan fluorescence quenching studies were carried out. Results of the ITC experiment exemplify that all interactions are thermodynamically favorable, with significant differences in relative affinities. The binding affinity of ATP is ~1000 fold higher then L-Glutamate which again has a ~1000 fold higher affinity than L-Cysteine (Fig. 6). This observation was verified by tryptophan fluorescence quenching studies as well which shows a similar trend in binding order i.e., ATP"-LGlutamate>L-Cysteine.
Fig. 6.
Substrate binding of LdGcs. (A) Thermogram of binding of LdGcs, left, middle and right panel corresponds to ATP, L-Glutamate and L-Cysteine. 15 μM recombinant protein in HEPES pH 7.5 was titrated against 200 μM of substrate. Kd values were obtained by fitting the data using Origin 7.0 software. (B) Tryptophan fluorescence saturation binding isotherms of Gcs.
In a subsequent set of experiments, Gcs was incubated with either ATP or L-glutamate and titrations carried out with the other ligand (L-glutamate or ATP) to determine if the binding site is shared between the ligands or a preferential order of binding, if any. ITC studies of ATP bound Gcs with L-Glutamate and L-Glutamate bound Gcs with ATP shows comparable Kd values in micro-molar range (Table 2). This suggests that binding of these substrates are independent of one another, validating the presence of distinct binding sites of ATP and L-Glutamate, later validated by computational studies. Further, binding affinity of glutamate with native Gcs is sufficiently lower than with Gcs-ATP complex favoring the possibility of ATP endorsing glutamate binding. This might also be the result of catalysis taking place due to the presence of two substrates. A similar experiment was carried out with the enzyme incubated with ATP and L-glutamate and titrated against the third substrate, Cysteine, but the results were inconclusive. GcsT and C-terminal constructs also show comparable binding with ATP, though these constructs have a lower affinity for L-Glutamate and L-Cysteine (Table 2). The apparent Kd values of all the substrates are summarized in Table 2.
Table 2.
Kd values of LdGcs obtained by ITC and tryptophan fluorescence quenching studies.
| LdGcs | KdμM (ITC) | KdμM (Tryptophan Fluorescence quenching) |
|---|---|---|
| ATP | 0.0005 | 2.7 |
| L-Glutamate | 0.25 | 93.7 |
| L-Cysteine | 204 | 177 |
| Gcs(ATP) + L-Glutamate | 0.046 | 53 |
| Gcs(L-Glutamate) + ATP | 0.032 | 22 |
2.7. Substrate binding studies of Gcs
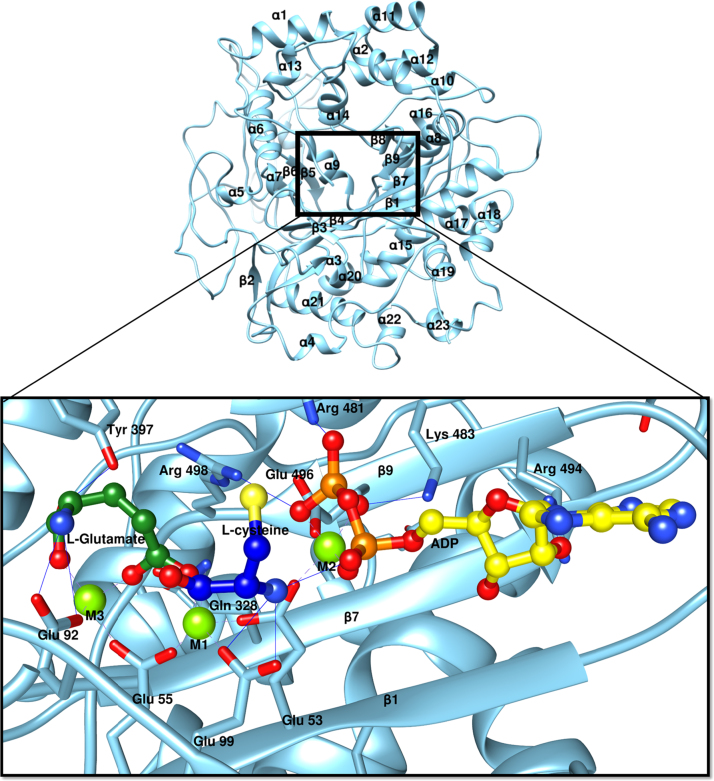
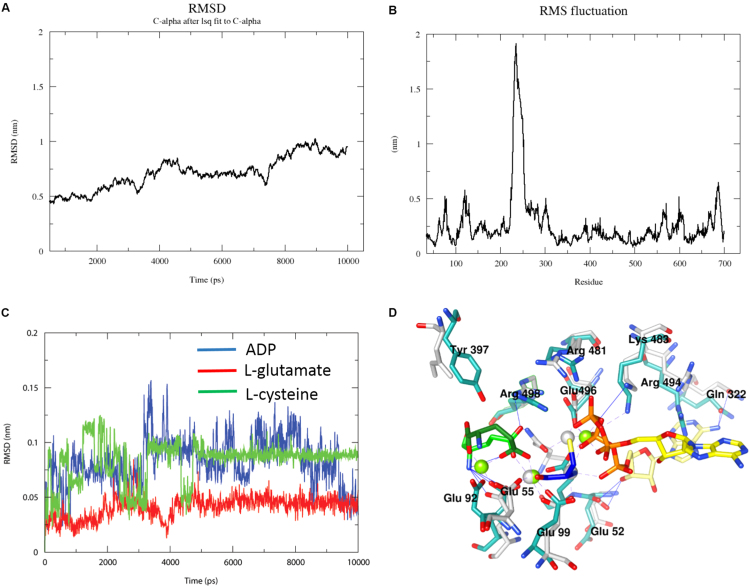
The residues essential for substrate binding and catalysis have been determined from several homologous Gcs [28], [29], [30], [31], [32], [33]. In the absence of three dimensional structure of Gcs, the active site determinants of nearest ortholog TbGcs has been determined earlier with the aid of homology model based on structurally similar protein glutamine synthase. The availability of ScGcs crystal structure reinvigorated the determination of substrate binding and catalysis determinants from LdGcs. Homology modeling was used to build the L. donovani Gcs structure, to provide a structural rationale for the ligand binding aspects, using the S. cereviseae Gcs as the template (details in Mat and Methods). The homology model has a core fold similar to other members of the ATP-grasp superfamily with central β-sheet comprised of six antiparallel β-strands surrounded by α-helices (Supplementary Fig. 3). The probable binding site of ligands were obtained from the homologous ScGcs–ligand co-crystal structures [28], [29] and 10 ns of molecular dynamics simulations were performed using Gromacs4.6 with CHARMM force field to obtain stable conformations for them. The ligands undergo a minor conformational change over the simulation that are summarized in Fig. 8. The root mean square (rms) deviation plot of Cα residues over the simulation period clearly shows that the complex is fairly stable with <1 nm deviation throughout the simulation (Fig. 7A). The rms fluctuation is still lower <0.5 nm if residues 225–270, corresponding to loop region are not included (Fig. 7B). Residues corresponding to this loop are not present in the ScGcs structure and are distant from active site pocket negating its involvement in catalytic mechanism of LdGcs. Although the binding site for all the ligands were confined to a common cavity, the simulation confirms that ATP and L-Glutamate bind to distinct pockets, sharing some key binding residues some of the residues involved in interaction were shared by ATP and L-Glutamate (Fig. 8A; Table 3).
Fig. 8.
Substrate binding site of Gcs. Homology model of LdGcs (sea green) with the ligand binding region enhanced. In this figure, the ligands, L-Glutamate (green), L-Cysteine (blue) and ADP (yellow) are shown in ball and stick , and residues that interact with these ligands are shown as sticks.
Fig. 7.
MD simulation. (A) RMSD plot of Cα during MD trajectories of protein substrate complex. (B) root-mean-square fluctuations during molecular dynamics trajectories for proposed protein-substrate complex. Large RMS fluctuation in residues 225–270 corresponds to loop region which is stabilized during simulation. The rmsd plot for substrate ADP (C), L-cysteine (D) and L-glutamate (E) during 10 ns long simulation. (F) Relative positions of ligands in ScGcs crystal structure and LdGcs after simulation. The ligands after simulation are shown as ADP (yellow), L-glutamate (green) and L-cysteine (Blue). The corresponding ScGcs conformations of ADP and L-glutamate are shown in pale yellow and light green respectively.
Table 3.
Substrate binding residues and bonds in LdGcs and ScGcs.
| LdGcs | ScGcs | ||||||
|---|---|---|---|---|---|---|---|
| ADP | Bond | Distance (Å) | Bond | Distance (Å) | |||
| Gln 332 OE1-H…N6 | 3.3 | ||||||
| Thr 101 OG1…. O2′ | 4.0 | ||||||
| Arg 481 NH2…. O2′ | 2.9 | Asp 49 O….. O3′ | 2.8 | ||||
| Lys 483 NZ…. O2B | 3.2 | Lys 451 NZ…. O1B | 3.9 | ||||
| Arg 498 NH2…. O3B | 2.7 | ||||||
| L-Glutamate | Tyr 397 OH…OXT | 2.6 | Tyr 362 OH…OXT | 3.0 | |||
| Glu 92 OE1…N | 2.7 | Glu 52 OE2…N | 3.7 | ||||
| Glu 92 OE2…N | 3.4 | Arg 472 NH2…OE2 | 3.1 | ||||
| 3.18 | Arg 472 NH1…OE2 | 3.9 | |||||
| L-Cysteine | Glu 99 OE2…N | 3.1 | Glu 96 OE2…N | 3.2 | |||
| Glu 96 OE1…N | 2.9 | ||||||
| Trp 445 NE1…O | 2.7 | ||||||
| Mg2+1 | Glu 53 OE1….. Mg | 1.84 | Glu 52 OE2….. Mg | 2.19 | |||
| Glu 55 OE1….. Mg | 1.72 | Glu 96 OE2….. Mg | 2.22 | ||||
| Glu 99 OE2…Mg | 1.79 | Glu 103 OE1…. Mg | 2.18 | ||||
| Glu OE2…. Mg | 1.75 | Glu OE2…Mg | 2.02 | ||||
| Mg2+2 | Glu 53 OE1…. Mg | 1.8 | Glu 50 OE1…. Mg | 2.16 | |||
| Glu 50 OE2…. Mg | 2.77 | ||||||
| Glu 496 OE2…Mg | 1.8 | Glu 470 OE2…Mg | 2.17 | ||||
| ADP O2B….. Mg | 1.85 | ADP O2B….. Mg | 2.17 | ||||
| ADP O2A….. Mg | 1.87 | ||||||
| Mg2+3 | Glu 55 OE2…. Mg | 1.86 | Glu 50 OE2…. Mg | 2.26 | |||
| Glu 92 OE2…Mg | 1.96 | Glu 103 OE2…Mg | 2.18 | ||||
| Glu O…. Mg | 1.83 | ADP O2A….. Mg | 2.65 | ||||
| Glu OE1…. Mg | 1.84 | ADP O3B….. Mg | 2.19 | ||||
2.8. Nucleotide binding site
The nucleotide binding site of LdGcs can be explained on the basis of ScGcs crystal structure with ADP (PDB ID 3IG8). 10 ns MD simulation of LdGcs complex has shown slight change in the conformation of adenine and sugar moiety of ADP while the phosphate moiety was stable throughout the simulation (Fig. 7C, D). The adenine ring is stabilized by hydrophobic interactions with Ile 491, Lys 487. The ribose moiety interacts with Arg 494 through one of the hydroxyl group. The phosphate moiety forms hydrogen bonds with Lys 483 and Arg 498 through its terminal beta phosphate group as shown in Fig. 7. In LdGcs single Mg2+ ion co-ordinate the alpha and beta phosphate moiety through conserved Glu 53and Glu 496 as shown in Fig. 8.
2.9. L-Glutamate binding Site
Our studies have shown that Gcs has the highest affinity for ATP, and can hydrolyze ATP even in the absence of L-Glutamate. Also the affinity of L-glutamate increases to ATP-Gcs complex than Gcs alone, as evidenced from ITC experiments (Table 2). These results suggests L-Glutamate probably binds to the site after ATP binding and hydrolysis. MD simulation studies of all the three substrates within the active site of LdGcs validates that the L-glutamate binding pocket is comprised of Glu 53, Glu 55, Glu 92, Glu 99, Met 322, Asn 328, Arg 367, Tyr 397 and Arg 498. L-Glutamate is stabilized by hydrogen bond interaction with Glu 52 and Tyr 397. The terminal carboxyl oxygen forms the hydrogen bond with Tyr 397 while the amino group interacts directly with Glu 52. These interactions are consistent with ScGcs crystal structure with slight variation in side chains conformations of the interacting residues (Fig. 8, Table 3). Two of three Mg2+ ion in LdGcs complex were found to facilitate L-glutamate binding via coordination with Glu 53, Glu 55, Glu 99 and Glu 92. The conserved residues corresponding to Arg 373 and Arg 498 have been shown to influence L-glutamate binding in TbGcs [33]. We later performed simulation studies of LdGcs with phosphorylated glutamate and ADP. The 10 ns MD simulation shows both phosphoglutamate and ADP are stable within the LdGcs active site (Supplementary Fig. 4). The Arg 498 residues was found to interact with the phosphate moiety of phosphorylated glutamate validating its essential role in LdGcs activity (Supplementary Fig. 4). Detailed analysis of L-glutamate-Gcs interactions throughout the simulation corroborates the consistent involvement of Glu 92, Tyr 397, Arg 498, Met 322 and Gln 328 in the same order as observed in by the occupancy values of their interaction with L-glutamate. This validates the direct involvement of Glu 92, Met 322 and Gln 328 in addition to Tyr 397 and Arg 498 reported to determine L-glutamate binding in T. brucei.
2.10. L-Cysteine binding residues
L-Cysteine shows very weak or transient binding with Gcs. In the ScGCS structure also, the cysteine moiety is not present separately, but its relative orientation can be obtained from the crystal structures of ScGcs with glutathione and BSO (PDB ID 3LVV and 3LVW), as they possess L-Cysteine moiety in them. MD simulation studies shows Cysteine binding is weakly stabilized by Glu 99 in LdGcs by interacting with the amino group of cysteine moiety (Figs. 7–8; Table 3). This interaction is conserved throughout the MD simulation with an occupancy value of 87.23%.
2.11. Magnesium binding site
Divalent ion plays an essential role in the functional mechanism of LdGcs. The crystal structure of ScGcs clearly shows three Mg2+ ions within the active site [29], [30]. The presence of two Mg2+ binding site has been verified for T. brucei Gcs [32]. We considered the three Mg2+ ion along with ligands in our MD simulation studies. The final snapshot of protein after simulation confirms their presence with in LdGcs active site. The relative position of two Mg2+ ions (M1 and M2) are similar to that observed in ScGcs crystal structure while the third Mg2 ion (M3) adopts a different position >1 Å away from that observed in ScGcs (Fig. 8 and Table 3). In the ScGCS structure, both M2 and M3 are involved in coordinating ADP phosphate moiety while in LdGcs only M2 co-ordinates with alpha and beta phosphate moieties of ADP. Further in LdGcs in addition to M1, M2 ion also stabilizes L-glutamate (Figs. 7–8). These different positions might be dominant in different steps of catalysis.
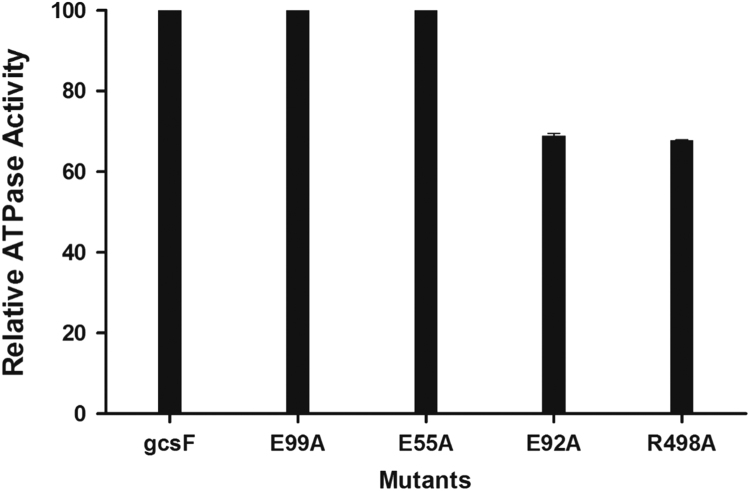
The computational analysis clearly shows the importance of Glutamates 52, 55, 92, 99, Tyr 397, Lys 483, Arg 494 and Arg 498 in the function of L. donovani Gcs. Single mutants of these residues in L. donovani GcsF and GcsT were designed using site directed mutagenesis to confirm their role in substrate binding. E55A, E92A, E99A and R498A mutants of GcsF and GcsT have been cloned and purified using a similar protocol as their native constructs. Substrate binding and ATPase activity of mutants were carried out as before and the results show that out of these four residues E92 and R498 play a significant role in ATP binding and hydrolysis, as the corresponding Ala mutants possess a 30% reduction in their relative ATPase activity with an enhanced Kd value (Fig. 9 and Table 3). However, E55A and E99A mutants do not show much variation in ATPase activity, although there is an ~8 fold increase in the apparent Kd (Table 4). The observations are similar in GcsT single mutants as well though the deletion mutants have significantly lower activity than GcsF (Data not shown). This is in agreement with our computational studies where Glu 55, and Arg 498 are involved in interaction with L-Glutamate while Glu 99 makes a Mg2+ dependent interaction.
Fig. 9.
Relative ATPase activity of LdGcs mutants obtained by NADH coupled assay. The experiments were performed three times. GcsF activity was considered 100% and relative activity of each construct was determined. Results shows 30% reduced activity for E92A and R498A mutant revealing their role in ATP binding or hydrolysis.
Table 4.
Apparent Kd values of ATP, L-Glutamate and L-Cysteine for different mutants of LdGcs. Kd values were calculated as mentioned in materials and methods.
| Constructs | Kd Value (μM) | ||
|---|---|---|---|
| ATP | L-Glutamate | L-Cysteine | |
| GcsF | 2.7± 0.3 | 93.7± 1.8 | 177± 2.6 |
| E55A | 2.5±1 | 793±.182 | 95±2.1 |
| E92A | 3235±.937 | 107±.16 | 115±.35 |
| R498A | 1550±238 | 890±.167 | 164±13 |
| E99A | 5.8±.3 | 666±.3 | 173±13 |
3. Conclusion
Gamma-glutamylcysteine synthetase is an essential enzyme of L. donovani metabolism. This manuscript reports the purification of L. donovani GcsF and its two constructs to homogeneity. Functional assays shows that all the constructs purified are active with specific activity GcsF 19 units/mg. Detailed nucleotide binding and hydrolysis studies shows that L. donovani possess substrate independent ATPase activity. Though, L-Glutamate and L-Cysteine are not crucial for ATPase activity their presence affects the rate of ATP hydrolysis. The Km value L-Glutamate and L-Cysteine were 9.2 and 1.7 mM respectively.
Substrate binding studies reveals significantly different Kd values of all three substrates. On the basis of Kd values, probable order of substrate binding might be ATP "L-Glutamate >L-Cysteine. This observation suggest that despite significantly large active site of Gcs binding of substrates is not random consistent with substrate binding proposed by Biterova et al. [29]. Divalent ions plays an exceptional role in ATP binding and thus ATPase activity of L. donovani Gcs with Mg2+ being the most appropriate divalent ion followed by Co2+ and Mn2+. The proposed LdGcs complex model was found stable during MD simulation studies, suggesting the stable relative orientation of substrates within LdGcs active site. The visual analysis of final snapshot after MD shows three Mg2+ ions similar to that observed in ScGcs crystal structure. While the M1 and M2 binding sites are similar to that observed in ScGcs crystal structure, M3 is involved in coordinating L-glutamate instead of ADP. This observation proposes that during different stages of catalysis the position of first two Mg2+ ions is constant while third Mg2+ while third Mg2+ might adopt different position, or might be absent as observed in TbGcs [32]. Computational studies were used to depict the active site architecture, in comparative manner with the substrate bound crystal structure of ScGcs. These studies show that most of the active site residues of LdGcs are conserved with ScGcs.
The role of Glu 55, Glu 92, Glu 99 and Arg 498 in ATP binding and hydrolysis was also studied. Mutation of Glu 92 and Arg 498 modestly decreased ATPase activity as well as binding. The Arg 498 has direct interaction with ADP while Glu 92 might be involved in stabilizing γ-phosphate moiety ATP via Mg2+ ion. The role of conserved Arg residue corresponding to Arg 498 has also been verified in T. brucei as well as ScGcs [29], [39]. L-Glutamate binding requires Glu 55 in addition to Glu 92 and Arg 498 as shown by ~8fold increase in apparent Kd values of the mutants. This in agreement with computational studies where Glu 55 and Glu 92 clearly shows Mg2+ mediated interaction with L-glutamate and Arg 498 lies within the hydrogen bonding distance.
The structural insights gained from the crystal structure of ScGcs [28], [29] and the results obtained in this study were used to explain the functional catalytic mechanism of L. donovani Gcs as most residues are conserved. The catalytic mechanism can be proposed to take place in three steps. The first step involves binding of ATP and its cleavage into ADP and subsequent activation of the γ-carboxyl group of L-Glutamate by released phosphate to form a γ-glutamylphosphate intermediate. The released phosphate moiety might be co-ordinated and transferred to L-Glutamate via Mg2+ ion making ATP hydrolysis possible even in absence of L-Glutamate. The second catalytic step involves nucleophilic attack of the amino group of L-Cysteine on the carbonyl group of phosphorylated L-Glutamate to generate a tetrahedral transition state [9], [10]. In the third step the transition state rearranges to form gamma glutamylcysteine as product and phosphate moiety is released. The detailed role of all the residues needs to be studied further by designing single and double mutants and this information can be used for designing novel specific inhibitors against L. donovani Gcs.
3.1. Experimental procedure
3.1.1. Computational analysis
The amino acid sequences of Gcs from L. donovani, H. sapiens, T. brucei, S. cerevisiae, B. juncea, and E. coli were retrieved from the UniProt data base (http://uniprot.org). The pairwise and multiple sequence alignment were performed using CLUSTALW version 1.8 [40]. The evolutionary traces of LdGcs and members of all the three classes of Gcs were studied by phylogenetic analysis with the aid of MEGA 5.0 (Molecular Evolutionary Genetics Analysis) using neighbor-joining methods with bootstrap test of phylogeny applying 500 bootstrap replications [41].
3.1.2. Cloning of L. donovani native GcsF and its deletion mutants
The coding regions of GcsF, GcsT, GcsC and GcsN 2 kb, 1.4 kb, 1.2 kb and 0.8 kb respectively were PCR amplified from the L. donovani genomic DNA using specific sense and anti-sense primers designed with sites for NheI and BamHI restriction enzymes respectively (Supplementary Table 1). The amplified PCR products were cloned in T/A vector pTZ57R/T (InsTA cloneTM PCR cloning kit, Fermentas International Inc.) and sub-cloned downstream pET28a vector (Novagen). Clones were confirmed by double digestion and nucleotide sequencing. E. coli BL21 (DE3) host cell was transformed with the recombinant plasmid pET28a and used for over-expression.
3.1.3. Purification of recombinant GcsF, GcsT and GcsC
E. coli BL21 (DE3) strain containing pET28a-Gcs constructs clones were grown at 37 °C in Luria-Bertani (LB) broth supplemented with 50 µg/ml kanamycin to an OD600 of 0.5–0.6, induced with 1 mM IPTG and grown further for 10–12h at 16 °C. Cells were harvested and resuspended in 50 mM Tris-HCl pH 8.5 buffer, 200 mM NaCl (Buffer A) with 8 mM MgCl2 and lysed by sonication and centrifuged at 12,000 rpm for 40 min and supernatant, in case of GcsF, was loaded to NiNTA column pre-equilibrated with buffer A. For GcsT and GcsC construct inclusion bodies collected in pellet were resuspended in Buffer A with 2 M urea and incubated for 3 h after which they were lysed, centrifuged and loaded on a Ni-NTA column as earlier. The column was incubated for an hour and subsequently washed twice with Buffer A containing 10 mM and 60 mM imidazole. Protein was then eluted in Buffer A with 300 mM imidazole. The eluted protein was dialyzed overnight into buffer B (50 mM Tris-HCl pH 8.5, 50 mM NaCl, 3 mM β-me and 8 mM MgCl2) concentrated using 10 kDa cutoff centricon (Amicon) and loaded on size exclusion chromatography for the second step purification and oligomerization analysis.
The GcsN construct was insoluble even under denaturing conditions and could not be used any further.
3.1.4. Size exclusion chromatography
Size-exclusion chromatography (SEC) experiments were performed at 25 °C using a Superdex 200 10/300 pre-packed column connected to an ÄKTA FPLC chromatograph (GE Healthcare). Calibration of the column was performed using the high molecular weight standard kit (GE Healthcare Cat No. 28-4038-42). 500 μl of 1 mg/ml protein was loaded in the column equilibrated with the buffer B. The isocratic elution at a flow rate of 0.3–0.4 ml/min was carried out and profiles were recorded by monitoring absorbance at 280 nm. The purified protein was concentrated as before and utilized for further structural and biochemical characterization.
3.1.5. Determination of protein concentration
The protein concentration was determined by using Bradford method [42]. The standard curve was plotted with bovine serum albumin in the range of 0–22 mg/ml.
3.1.6. Isothermal titration calorimetry
The thermodynamic parameters of interaction were determined by isothermal titration calorimetry (VP-ITC, Microcal, Northampton, MA, USA). GcsF (13 μM) dialyzed in the buffer containing HEPES pH 7.5, 300 mM NaCl and 8 mM MgCl2 was titrated against 200 μM of substrate. The thermogram was analyzed using Origin 7.0 software (Microcal, USA).
3.1.7. Fluorescence measurement
The intrinsic fluorescence emission spectra for the three protein constructs (2 μM) were recorded on a Cary Eclipse fluorescence spectrophotometer (Agilent Technologies) at 25 °C in 5 mm path length cuvettes, with excitation at 295 nm and the emission spectra recorded in the range 300–400 nm.
Steady-state fluorescence experiments were performed using identical protein concentration in buffer B and the change in tryptophan fluorescence spectra was observed at 339 nm with increasing concentration of ligands. Titrations with buffer alone were performed as control. The change in fluorescence was then related to binding of nucleotide by the following standard equation
Where, ∆F is the magnitude of the difference between the observed fluorescence intensity at a given concentration of substrate and the fluorescence intensity in the absence of substrate, ∆Fmax is the difference between the observed fluorescence intensities at zero and saturating substrate concentration], [Substrate] tot and Kd is the apparent dissociation constant. The Kd values were determined from non-linear least-squares regression analysis of titration data. Fluorescence spectra with all samples were corrected for the background fluorescence of the solution (buffer + substrate). Deconvolution of curves was performed using the Prism software (GraphPad Software, Inc., La Jolla, CA, USA).
3.1.8. Circular dichroism measurements
The far-UV CD measurements were made on a Jasco J810 spectropolarimeter and Chirascan™ CD spectropolarimeter (Applied Photophysics) calibrated with ammonium (+)−10-camphorsulfonate. The average of three spectra (200–260 nm, scan-speed 10 nm/min) from 2 µM protein samples, dissolved in buffer B was taken for far-UV CD spectra following standard protocols [43], [44].
3.1.9. Filter based nucleotide binding assay
GcsT (1–5 μg) in Buffer B was immobilized on a nitrocellulose membrane with increasing concentrations of BSA taken as negative control. The membrane was blocked with 2% BSA and then incubated with 50 μM [γ-32P] ATP diluted in 2% BSA in PBS at 25 °C for 25 min and then washed with PBS containing 0.5% Tween-20. Autoradiogram of the blot was taken, and densitometry of the spots was performed using alpha imager software (GE healthcare). ATP binding was further confirmed by incubating protein with increasing concentration of unlabelled ATP ranging from 0 to 800 μM. The role of magnesium ion was also analyzed by increasing concentration of EDTA (0–20 mM).
3.1.10. ATPase activity
The ATPase activity was carried out using radioactive and NADH coupled assays. Isotopic ATPase reaction mixture (10 μl) comprise 2.5 μM of protein in buffer B with 1 μCi of radiolabelled γ-32P ATP as substrate. Effect of MgCl2 was studied in ATPase reaction without MgCl2 at 25 °C. Reactions were stopped by adding 0.5 μl of 10% SDS. 1 μl of the reaction mixture was then spotted onto a PEI-cellulose TLC plate (10 cm×20 cm) and resolved in 0.5 M LiCl, 0.5 M formic acid and dried at 37 °C. The TLC plate was auto radiographed and the released γ-32P was quantified using the Image Master 1D Elite software (Amersham Biosciences). The percentage of ATP hydrolysis was calculated using the formula.
Percentage of ATP hydrolysis ={quantity of [γ-32P] P /(quantities of [γ-32P] P +[γ-32P] ATP} x 100.
Background values (without protein) were subtracted. Optimum protein concentration was identified by plotting released γ-32P as a function of increasing protein concentration (0–5 μM), the reaction was also setup with varying time at fixed concentration of GcsT to find out optimum time. The most suitable divalent was explored by using different divalent in 2 mM concentration. Optimum magnesium concentration for ATPase reaction was screened in the range of 0–20 mM.
Quantitative measurements of activity were done spectrophotometrically at 340 nm by determining the rate of ADP formation using a coupled assay with pyruvate kinase and lactate dehydrogenase [45]. For determination of steady state kinetics parameters, two substrates were fixed at saturation and the third substrate was varied in concentration. Km and Vmax values were determined using Michaelis-Menten kinetics incorporated in Prism 5.0 software (Graphpad Inc.).
3.1.11. Site directed mutagenesis
Site directed mutagenesis was carried out using standard PCR based methods [46]. Primers for single mutations were designed using Oligo software and are mentioned in supplementary table 2. PET28a-GcsF and pET28a-GcsT were used as template for PCR amplification using pfu DNA polymerase (Fermentas International Inc.). Amplified reaction product was digested with dpn1 (Fermentas International Inc.) and transformed into E. coli DH5α strain. Mutants were confirmed by double digestion and sequencing.
3.1.12. Homology modeling
Homology model was built using the crystal structure of Gcs from S. cerevisiae (ScGcs) (PDB ID 3IG8) as the template identified from Position specific iterative BLAST (Psi-BLAST) of LdGcs amino acid sequence against sequences in the Protein Data Bank S. cerevisiae Gcs (PDB ID: 3IG8) [29], [30] was chosen as template, based on maximum sequence identity and other statistical parameters. Homology models were built using Modeller 9.10 [47], based on spatial restraints method, using default parameters with pairwise sequence alignment file of the target and template as input. Five models were obtained as output for the full length Gcs as well as truncated construct. The top 5 models were considered for visual analysis and they possess acceptable stereochemistry, with the ~ 96% of the residues located in the generously allowed regions. The models were ranked on the basis of root mean square deviation (rmsd) and visual examination. Models with minimum rmsd were used for further validation using Molprobity and PROCHECK [48], [49].
3.1.13. Molecular dynamic simulations
Substrate binding sites were obtained by extracting ligands from ScGcs crystal structures (PDB ID- 3IG8, 3IG5 and 3LVW). Molecular dynamic (MD) simulations and analysis were performed using GROMACS 4.6.5 simulation package [50] adopting CHARMM force field parameters [51]. LdGcs homology model with ADP, L-glutamate, L-cysteine and 3 Mg2+ ions was taken as starting point for simulation. The topology of ligands ADP, L-glutamate and L-cysteine were created using swissparam server [52]. The protein ligand complex was solvated into a cubic box of TIP3P water model. The complex was neutralized by using 18 Na+ ion. Energy minimization was done using steepest descent method and convergent crieteria of 10 KJ/Mol followed by dynamics simulations of the whole system in the NVT and NPT ensemble at 293 K temperature with a time step of 2 ps. The electrostatic interactions were calculated using the Particle Mesh Ewald summation method [53] while constraints were applied on all bonds using the LINCS [54] algorithm. The equilibrated system was subjected to final MD production run of 10 ns. Root mean square deviation (RMSD) of Cα residues and ligands and root mean square fluctuation (RMSF) of complex were calculated using g_rms and g_rmsf commands respectively. Graphs were generated using Grace Program. The various frames generated were visually analyzed by Chimera 1.6.1 [55].
Acknowledgment
The financial support for this work was provided by Department of Science & Technology (DST) (GAP0030), Government of India. We acknowledge Dr. Karthikeyan Subramanian, Institute of Microbial Technology, Chandigarh and providing facility for FarUV-CD experiments. Dr R Ravishankar and Sonal Shree are duly acknowledged for ITC experiments. We acknowledge Nidhi Singh and Dr MI Siddiqi for their help in MD simulation studies. Pragati Agnihotri acknowledge fellowship Indian Council of Medical Research respectively. Saurabh Pratap Singh and Anil Kumar Shakya acknowledge fellowships from Council of Scientific and Industrial Research. This manuscript bears CDRI communication Number 9307.
Footnotes
Transparency data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.bbrep.2016.08.016.
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.bbrep.2016.08.016.
Appendix A. Transparency document
Supplementary material
.
Appendix B. Supplementary material
Supplementary material
.
Supplementary material
.
Supplementary material
.
References
- 1.Leishmaniasis Fact sheet N°375 2014 World Health Organization.
- 2.Alvar J., Velez I.D., Bern C., Herrero M., Desjeux P. Leishmaniasis worldwide and global estimates of its incidence. PLoS One. 2012;7(5):e35671. doi: 10.1371/journal.pone.0035671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fairlamb A.H., Blackburn P., Ulrich P., Chait B.T., Cerami A. Trypanothione: a novel bis(glutathionyl)spermidine cofactor for glutathione reductase in trypanosomatids. Science. 1985;227:1485–1487. doi: 10.1126/science.3883489. [DOI] [PubMed] [Google Scholar]
- 4.Nogoceke E., Gommel D.U., Kiess M., Kalisz H.M., Flohe L. A unique cascade of oxidoreductases catalyses trypanothione-mediated peroxide metabolism in Crithidia fasciculata. Biol. Chem. 1997;378(8):827–836. doi: 10.1515/bchm.1997.378.8.827. [DOI] [PubMed] [Google Scholar]
- 5.Levick M.P., Tetaud E., Fairlamb A.H., Blackwell J.M. Identification and characterisation of a functional peroxidoxin from Leishmania major. Mol. Biochem. Parasitol. 1998;96(1–2):125–137. doi: 10.1016/s0166-6851(98)00122-4. [DOI] [PubMed] [Google Scholar]
- 6.Ludemann H., Dormeyer M., Sticherling C., Stallmann D., Follmann H. Trypanosoma brucei tryparedoxin, a thioredoxin-like protein in African trypanosomes. FEBS Lett. 1998;431(3):381–385. doi: 10.1016/s0014-5793(98)00793-5. [DOI] [PubMed] [Google Scholar]
- 7.Flohe L., Hecht H.J., Steinert P. Glutathione and trypanothione in parasitic hydroperoxide metabolism. Free Radic. Biol. Med. 1999;27(9–10):966–984. doi: 10.1016/s0891-5849(99)00172-0. [DOI] [PubMed] [Google Scholar]
- 8.Schlecker T., Schmidt A., Dirdjaja N., Voncken F., Clayton C. Substrate specificity, localization, and essential role of the glutathione peroxidase-type tryparedoxin peroxidases in Trypanosoma brucei. J. Biol. Chem. 2005;280(15):14385–14394. doi: 10.1074/jbc.M413338200. [DOI] [PubMed] [Google Scholar]
- 9.Castro H., Tomas A.M. Peroxidases of trypanosomatids. Antioxid. Redox Signal. 2008;10(9):1593–1606. doi: 10.1089/ars.2008.2050. [DOI] [PubMed] [Google Scholar]
- 10.Miller-Flemming L., Olin-Sandoval V., Campbell K., Ralser M. Remaining mysteries of molecular biology: the role of polyamine in the Cell. J. Mol. Biol. 2015;427(21):3389–3406. doi: 10.1016/j.jmb.2015.06.020. [DOI] [PubMed] [Google Scholar]
- 11.Griffith O.W., Mulcahy R.T. The enzymes of glutathione synthesis: gamma-glutamylcysteine synthetase. Adv. Enzymol. Relat. Areas Mol. Biol. 1999;73:209–267. doi: 10.1002/9780470123195.ch7. [DOI] [PubMed] [Google Scholar]
- 12.Meister A. The gamma-glutamyl cycle. Diseases associated with specific enzyme deficiencies. Ann. Intern. Med. 1974;81(2):247–253. doi: 10.7326/0003-4819-81-2-247. [DOI] [PubMed] [Google Scholar]
- 13.Oza S.L., Shaw M.P., Wyllie S., Fairlamb A.H. Trypanothione biosynthesis in Leishmania major. Mol. Biochem. Parasitol. 2005;139(1):107–116. doi: 10.1016/j.molbiopara.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Orlowski M., Meister A. Partial reactions catalyzed by gamma glutamylcysteine synthetase and evidence for an activated glutamate intermediate. J. Biol. Chem. 1971;246(23):7095–7105. [PubMed] [Google Scholar]
- 15.Griffith O.W. Biologic and pharmacologic regulation of mammalian glutathione synthesis. Free Radic. Biol. Med. 1999;27(9–10):922–935. doi: 10.1016/s0891-5849(99)00176-8. [DOI] [PubMed] [Google Scholar]
- 16.Wu A.L., Moye-Rowley W.S. GSH1, which encodes gamma-glutamylcysteine synthetase, is a target gene for yAP-1 transcriptional regulation. Mol. Cell. Biol. 1994;14(9):5832–5839. doi: 10.1128/mcb.14.9.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaudhuri B., Ingavale S., Bachhawat A.K. Apd1+, a gene required for red pigment formation in ade6 mutants of Schizosaccharomyces pombe, encodes an enzyme required for glutathione biosynthesis: a role for glutathione and a glutathione-conjugate pump. Genetics. 1997;145(1):75–83. doi: 10.1093/genetics/145.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baek Y.U., Kim Y.R., Yim H.S., Kang S.O. Disruption of gamma-glutamylcysteine synthetase results in absolute glutathione auxotrophy and apoptosis in Candida albicans. FEBS Lett. 2004;556(1–3):47–52. doi: 10.1016/s0014-5793(03)01363-2. [DOI] [PubMed] [Google Scholar]
- 19.Shi Z.Z., Osei-Frimpong J., Kala G., Kala S.V., Barrios R.J. Glutathione synthesis is essential for mouse development but not for cell growth in culture. Proc. Natl. Acad. Sci. USA. 2000;97(10):5101–5106. doi: 10.1073/pnas.97.10.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huynh T.T., Huynh V.T., Harmon M.A., Phillips M.A. Gene knockdown of gamma-glutamylcysteine synthetase by RNAi in the parasitic protozoa Trypanosoma brucei demonstrates that it is an essential enzyme. J. Biol. Chem. 2003;278(41):39794–39800. doi: 10.1074/jbc.M306306200. [DOI] [PubMed] [Google Scholar]
- 21.Dalton T.P., Dieter M.Z., Yang Y., Shertzer H.G., Nebert D.W. Knockout of the mouse glutamate cysteine ligase catalytic subunit (Gclc) gene: embryonic lethal when homozygous, and proposed model for moderate glutathione deficiency when heterozygous. Biochem. Biophys. Res. Commun. 2000;279(2):324–329. doi: 10.1006/bbrc.2000.3930. [DOI] [PubMed] [Google Scholar]
- 22.Mukherjee A., Roy G., Guimond C., Ouellette M. The gamma-glutamylcysteine synthetase gene of Leishmania is essential and involved in response to oxidants. Mol. Microbiol. 2009;74(4):914–927. doi: 10.1111/j.1365-2958.2009.06907.x. [DOI] [PubMed] [Google Scholar]
- 23.Langston W., Circu M.L., Aw T.Y. Insulin stimulation of gamma-glutamylcysteine ligase catalytic subunit expression increases endothelial GSH during oxidative stress: influence of low glucose. Free Radic. Biol. Med. 2008;45(11):1591–1599. doi: 10.1016/j.freeradbiomed.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arrick B.A., Griffith O.W., Cerami A. Inhibition of glutathione synthesis as a chemotherapeutic strategy for trypanosomiasis. J. Exp. Med. 1981;153(3):720–725. doi: 10.1084/jem.153.3.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carter K.C., Henriquez F.L., Campbell S.A., Roberts C.W., Nok A. DNA vaccination against the parasite enzyme gamma-glutamylcysteine synthetase confers protection against Leishmania donovani infection. Vaccine. 2007;25(22):4502–4509. doi: 10.1016/j.vaccine.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 26.Henriquez F.L., Campbell S.A., Roberts C.W., Mullen A.B., Burchmore R. Vaccination with recombinant Leishmania donovani gamma-glutamylcysteine synthetase fusion protein protects against L. donovani infection. J. Parasitol. 2010;96(5):929–936. doi: 10.1645/GE-2360.1. [DOI] [PubMed] [Google Scholar]
- 27.Copley S.D., Dhillon J.K. Lateral gene transfer and parallel evolution in the history of glutathione biosynthesis genes. Genome Biol. 2002;3(5) doi: 10.1186/gb-2002-3-5-research0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hibi T., Nii H., Nakatsu T., Kimura A., Kato H. Crystal structure of gamma-glutamylcysteine synthetase: insights into the mechanism of catalysis by a key enzyme for glutathione homeostasis. Proc. Natl. Acad. Sci. USA. 2004;101(42):15052–15057. doi: 10.1073/pnas.0403277101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biterova E.I., Barycki J.J. Mechanistic details of glutathione biosynthesis revealed by crystal structures of Saccharomyces cerevisiae glutamate cysteine ligase. J. Biol. Chem. 2009;284(47):32700–32708. doi: 10.1074/jbc.M109.025114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biterova E.I., Barycki J.J. Structural basis for feedback and pharmacological inhibition of Saccharomyces cerevisiae glutamate cysteine ligase. J. Biol. Chem. 2010;285(19):14459–14466. doi: 10.1074/jbc.M110.104802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brekken D.L., Phillips M.A. Trypanosoma brucei γ-Glutamylcysteine Synthetase- Characterization of the kinetic mechanism and the role of cys-319 In cystamine inactivation. J. Biol. Chem. 1998;273(41):26317–26322. doi: 10.1074/jbc.273.41.26317. [DOI] [PubMed] [Google Scholar]
- 32.Abbott J.J., Pei J., Ford J.L., Qi Y., Grishin V.N. Structure prediction and active site analysis of the metal binding determinants in γ-glutamylcysteine synthetase. J. Biol. Chem. 2001;276(45):42009–42107. doi: 10.1074/jbc.M104672200. [DOI] [PubMed] [Google Scholar]
- 33.Abbott J.J., Ford J.L., Phillips M.A. Substrate binding determinants of Trypanosoma brucei ç-glutamylcysteine synthetase. Biochemistry. 2002;41:2741–2750. doi: 10.1021/bi0159128. [DOI] [PubMed] [Google Scholar]
- 34.Ohno T., Tsuchiya M., Osago H., Hara N., Jidoi J., Shimoyama M. Detection of arginine-ADP-ribosylated protein using recombinant ADP-ribosylarginine hydrolase. Anal. Biochem. 1995;231:115–122. doi: 10.1006/abio.1995.1510. [DOI] [PubMed] [Google Scholar]
- 35.Thom M., Komor E. Effect of Magnesium and ATP on ATPase of sugarcane vacuoles. Planta. 1984;161(4):361–365. doi: 10.1007/BF00398727. [DOI] [PubMed] [Google Scholar]
- 36.Chen Y., Shertzer H.G., Schneider S.N., Nebert D.W., Dalton T.P. Glutamate Cysteine Ligase Catalysis. Dependence on ATP and modifier subunit for regulation of tissue glutathione levels. J. Biol. Chem. 2005;280(40):33766–33774. doi: 10.1074/jbc.M504604200. [DOI] [PubMed] [Google Scholar]
- 37.Lueder D.V., Phillips M.A. Characterization of Trypanosoma brucei gamma-glutamylcysteine synthetase, an essential enzyme in the biosynthesis of trypanothione (diglutathionylspermidine) J. Biol. Chem. 1996;271(29):17485–17490. doi: 10.1074/jbc.271.29.17485. [DOI] [PubMed] [Google Scholar]
- 39.Fraser J.A., Saunders R.D., McLellan L.I. Drosophila melanogaster glutamate-cysteine ligase activity is regulated by a modifier subunit with a mechanism of action similar to that of the mammalian form. J. Biol. Chem. 2002;277(2):1158–1165. doi: 10.1074/jbc.M106683200. [DOI] [PubMed] [Google Scholar]
- 40.Thompson J.D., Gibson T.J., Higgins D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinforma. Ed. Board. 2002 doi: 10.1002/0471250953.bi0203s00. Andreas D Baxevanis (Chapter 2), Unit 2 3. [DOI] [PubMed] [Google Scholar]
- 41.Tamura K., Peterson D., Peterson N., Stecher G., Nei M. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2010;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 43.Greenfield N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006;1(6):2876–2890. doi: 10.1038/nprot.2006.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Louis-Jeune C., Andrade-Navarro M.A., Perez-Iratxeta C. Prediction of protein secondary structure from circular dichroism using theoretically derived spectra. Proteins. 2012;80(2):374–381. doi: 10.1002/prot.23188. [DOI] [PubMed] [Google Scholar]
- 45.Jez J.M., Cahoon R.E. Kinetic Mechanism of Glutatione synthetase from Arabidopsis thaliana. J. Biol. Chem. 2004;279(41):42726–42731. doi: 10.1074/jbc.M407961200. [DOI] [PubMed] [Google Scholar]
- 46.Tessier D.C., Thomas D.Y. PCR assisted mutagenesis for site-directed insertion/deletion of large DNA fragments. Methods Mol. Biol. 1996;57:229–237. doi: 10.1385/0-89603-332-5:229. [DOI] [PubMed] [Google Scholar]
- 47.N. Eswar, B. Webb, M.A. Marti-Renom, M.S. Madhusudhan D. Eramian. et al. Comparative protein structure modeling using Modeller. Current Protocols in Bioinformatics editoral board & Andreas D Baxevanis Chapter 5: Unit 5 6. [DOI] [PMC free article] [PubMed]
- 48.Chen V.B., Arendall W.B., Headd J.J., Keedy D.A., Immormino R.M. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lrmmdt J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 50.Hess B.K., van der Spoel D., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theo. Comp. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 51.Vanommeslaeghe K., Hatcher E., Acharya C., Kundu, Zhong S., Shim J., Darian E., Guvench O., Lopes P., Vorobyov I., MacKerell A.D., Jr CHARMM General Force Field (CGenFF): a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zoete V., Cuendet M.A., Grosdidier A., Michielin O. SwissParam, a fast force field generation tool for small organic molecules. J. Comput. Chem. 2011;32(11):2359–2368. doi: 10.1002/jcc.21816. [DOI] [PubMed] [Google Scholar]
- 53.Darden T., York D., Pedersen L. Particle Mesh Ewald - An N.Log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 54.Hess B., Bekker H., Berendsen H.J.C., Fraaije JGEM LINCS: A linear constraint solver for molecular simulations. J. Comp. Chem. 1997;18:1463–1472. [Google Scholar]
- 55.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material